Abstract
Intracellular levels of reactive oxygen species (ROS) increase as a consequence of oxidative stress and represent a major source of damage to biomolecules. Due to its high cellular abundance RNA is more frequently the target for oxidative damage than DNA. Nevertheless the functional consequences of damage on stable RNA are poorly understood. Using a genome-wide approach, based on 8-oxo-guanosine immunoprecipitation, we present evidence that the most abundant non-coding RNA in a cell, the ribosomal RNA (rRNA), is target for oxidative nucleobase damage by ROS. Subjecting ribosomes to oxidative stress, we demonstrate that oxidized 23S rRNA inhibits the ribosome during protein biosynthesis. Placing single oxidized nucleobases at specific position within the ribosome's catalytic center by atomic mutagenesis resulted in markedly different functional outcomes. While some active site nucleobases tolerated oxidative damage well, oxidation at others had detrimental effects on protein synthesis by inhibiting different sub-steps of the ribosomal elongation cycle. Our data provide molecular insight into the biological consequences of RNA oxidation in one of the most central cellular enzymes and reveal mechanistic insight on the role of individual active site nucleobases during translation.
INTRODUCTION
Oxidative damage through reactive oxidative species (ROS) is known to represent a serious threat to cell function and homeostasis. RNA oxidation is more prevalent than DNA oxidation and forms stable oxidized base lesions that persist in the nucleotide sequence (1,2). Beside nucleobase oxidations yielding 8-oxo-purines and 5-hydroxy-pyrimidines, ROS introduce in addition RNA backbone cleavage as well as abasic sites, respectively (Figure 1A). In vivo, total RNA is oxidized at a rate of 1 in 105 residues under unstressed conditions, but these lesions accumulate up to 10-fold during oxidative stress conditions (3). Since accumulating oxidative lesions disrupt normal RNA function, RNA oxidation is considered as a possible causal link to neurodegenerative conditions such as Alzheimer's disease (AD), Parkinson's disease (PD) and amyotrophic lateral sclerosis (ALS) (reviewed in (4,5)). While oxidized mRNA is detrimental to the speed and fidelity of protein biosynthesis (6), damaged molecules are rapidly identified and degraded by the translation-dependent no-go decay (7). However, it is unknown whether equivalent quality control mechanisms exist for stable non-protein-coding RNAs (ncRNAs), such as ribosomal RNA (rRNA) which represents an integral and functionally crucial part of the ribosome. Indeed rRNA from AD patients’ brain cells contains more 8-oxo-G, while also being less abundant compared to cells from healthy peers (6,8–9). The ribosomes extracted from AD-affected neuronal tissues are heavily oxidized and contain high levels of redox-cycling Fe(II) (6), which can generate ROS from within the ribosome via the Fenton reaction. Additionally, it was shown that AD-ribosomes translate significantly slower (10) and that oxidizing ribosomes reduces translation in vitro as well (6). Mounting evidence correlates ribosome oxidation with loss of neurons, specifically since RNA oxidation (11), ribosomal dysfunction (12), and altered protein synthesis rates (13) are all hallmarks for early events in AD. Bacterial rRNA is similarly damaged by oxidation, indicating that ribosomes in all species likely face the same challenges when it comes to the consequences of oxidative stress (14). Since the ribosome is the central hub for protein synthesis in all cells in all domains of life, any functional damage will have potentially severe consequences on protein homeostasis and cell survival. To our knowledge, there is no data available on the oxidation of specific rRNA residues inside the ribosome in cells and thus on the consequences on ribosomal functions can only be speculated (5).
Figure 1.
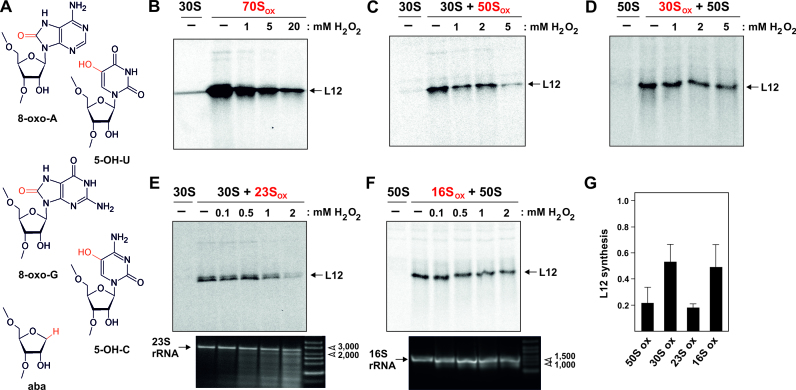
Oxidative stress to the ribosome disrupts translation and damages ribosomal RNA. (A) Structures of the most stable oxidation products found in RNA: 8-oxo-7,8-dihydroadenosine (8-oxo-A), 8-oxo-7,8-dihydroguanosine (8-oxo-G), 5-hydroxyuridine (5-OH-U), 5-hydroxycytidine (5-OH-C), and the abasic site (aba). The oxidations are highlighted in red. (B–D) In vitro translation activity of ribosomal components previously oxidized in vitro with increasing stress conditions (10 μM Fe(II)ascorbate and 0.1–20 mM H2O2). Translation of mRNA coding for r-protein L12 was tested by separating the [35S]-labeled products by SDS-PAGE and autoradiography. (B) In vitro translation with oxidized E. coli 70S ribosomes (70Sox). (C) Translation with oxidized T. aquaticus 50S subunits (50Sox) complemented with unstressed E. coli 30S subunits. (D) Translation with oxidized E. coli 30S subunits (30Sox) complemented with native E. coli 50S subunits. In (B), (C) and (D) complete translation reactions containing gradient-purified E. coli 30S or 50S ribosomal subunits, respectively, served as negative controls. The minor L12 product bands in these controls originate from minute amounts of contaminating 50S or 30S particles in the respective subunit preparation. (E) Translation with non-oxidized 30S subunits combined with reconstituted T. aquaticus 50S particles containing unstressed r-proteins and oxidized 23S rRNA (23Sox). The negative control (30S) contained unstressed E. coli 30S subunits and complete 50S reconstitution samples but lacking 23S rRNA. (F) Translation with non-oxidized 50S subunits combined with reconstituted E. coli 30S particles containing unstressed r-proteins and oxidized 16S rRNA (16Sox). The negative control (50S) contained unstressed E. coli 50S subunits and complete 30S reconstitution samples but lacking rRNA. The agarose gels at the bottom in (E) and (F) depict the integrity of the 23S and 16S rRNA, respectively, used for subunit reconstitution after increasing exposure to oxidative stress. (G) Quantification of relative translation activities with oxidized 50S or oxidized 30S subunits, and subunits reconstituted from oxidized 23S or 16S rRNA. Data shown represent the mean (n = 3) at the endpoints at the highest H2O2 levels in experiments shown in (C) through (F), normalized to translation with unstressed ribosomal subunits. Error bars indicate standard deviation.
The ribosome is a large molecular machine composed of two unequal subunits: the small subunit (30S and 40S in pro- and eukaryotes, respectively) is tasked with decoding the mRNA sequence information by pairing every codon with its cognate transfer RNA (tRNA) that carries a particular amino acid at its 3′-end. The large ribosomal subunit (50S and 60S in pro- and eukaryotes, respectively) then performs peptidyl transfer, which links together amino acids provided by the tRNA via peptide bonds in order to form a nascent polypeptide chain. While the subunits consist of both ribosomal proteins (r-proteins) and rRNA, it has been established by numerous biochemical, genetic and structural studies that most functions are directly performed or at least strongly promoted by rRNA in both subunits (15,16). Most impressively, peptide bonds are formed in an active site residing on the large subunit, called peptidyl transferase center (PTC), which is devoid of any r-protein side chains, thus establishing the ribosome as a ribozyme (17). The mechanism of catalysis and the residues involved are universally conserved in all domains of life. Therefore, whatever we learn about the PTC of prokaryotic ribosomes will most certainly also hold true for the larger and more complex ribosomes of multicellular organisms such, as humans.
For protein production ribosomes progress through the so-called elongation cycle, which is characterized by four well-orchestrated phases, namely initiation, elongation, termination and recycling (16). During elongation the peptide chain extension by one amino acid occurs immediately after aminoacyl-tRNA accommodation into the A-site. This results in a ribosomal complex carrying deacylated tRNA in the P-site and peptidyl-tRNA, elongated by one amino acid, at the A-site. In order to add a subsequent amino acid to the nascent peptide, the complex has to move one mRNA codon downstream. In bacteria, this is accomplished by the GTPase EF-G (eEF2 in eukarya), which is specifically targeted to and activated by the large subunit rRNA in the so-called pre-translocational complex (18,19). Once the ribosome has reached the end of the open reading frame and in response to an A-site located stop codon, class I release factors (RF1 or RF2 in bacteria) bind the A-site and trigger the hydrolysis of the P-site bound peptidyl-tRNA during the termination reaction.
In this study, we demonstrate that the bacterial ribosome is damaged by ROS in vivo and that nucleobase oxidation of the large ribosomal subunit rRNA negatively affects protein biosynthesis. Focusing on the large ribosomal subunit we elucidated in a bottom-up approach the effects of nucleobase oxidation at the five inner shell residues of the catalytic center A2451, C2063, U2585, U2506 and A2602 (Escherichia coli nomenclature used throughout), as well as at the second tier residue G2447 located in immediate proximity to the PTC. By means of atomic mutagenesis, an experimental approach allowing the site-specific introduction of nucleoside modifications in the context of the whole ribosome (20,21), we have revealed differential effects of nucleobase oxidations on particular sub-steps of protein biosynthesis in a position-dependent manner.
MATERIALS AND METHODS
Oxidation of native ribosomes
Native 70S, 50S and 30S subunits were isolated from E. coli and Thermus aquaticus as described previously (20,22). For oxidation, 2 pmol native E. coli 70S or T. aquaticus 50S were incubated 1 h at 37°C in TMN buffer (50 mM Tris/HCl pH 7.6, 100 mM NH4Cl, 10 mM MgCl2 and 6 mM β-mercaptoethanol) containing 10 μM Fe(II)ascorbate and 0.1 to 20 mM H2O2. The ribosomal particles were precipitated in 9.5 vol EtOH, and while 70S were directly tested in the in vitro translation (see below), 50S were allowed to re-associate with 2 pmol native E. coli 30S in TMN buffer prior to translation. 23S or 16S rRNA was isolated from oxidized T. aquaticus 50S subunits or oxidized E. coli 30S subunits by double PCI extraction and the RNA quality was checked on 1% agarose gels. The oxidized 23S rRNA was subsequently used for in vitro reconstitution of 50S subunits with native 50S ribosomal proteins and non-oxidized 5S rRNA in vitro transcripts as described (22). Oxidized 16S rRNA was reconstituted in vitro with native 30S ribosomal proteins to form 30S subunits as described (23).
Purification of RNA from stressed E. coli cells
Escherichia coli K12 MG1655 cells were cultured at 37°C in LB. At an OD600 of ∼0.7 H2O2 was added to a final concentration of 2.5 mM. An equal volume of water was added to control. After 10 min incubation, 2 ml of culture were centrifuged (2 min at 13,000 rpm, 4°C). The pellet was cleaned with cold LB and resuspended in 50 μl lysis buffer (80 mM Tris/HCl pH 6.8, 15 mM EDTA pH 8.0, 1.6% SDS, 3.2% 2-mercaptoetanol). After 3 min incubation at 37°C, 1.5 ml Trizol was added. Samples were incubated 5 min at room temperature, and then 200 μl chloroform were added. Samples were shaken and incubated 3 min at room temperature and the aqueous phase was recovered after centrifugation (15 min, 12,000 rpm, 4°C). The supernatant was extracted again with 200 μl of chloroform and RNA was precipitated with 750 μl isopropanol (10 min incubation at room temperature and centrifugation of 20 min at 14,000 rpm, 4°C). RNA was washed twice with 75% ethanol at room temperature. After drying, the RNA was resuspended in 14 μl of TE (0.5 mM EDTA, 20 mM Tris/HCl pH 8.0).
Construction of a library of immuno-purified RNA
10 μl of total RNA (1.2 μg/μl, in TE) were denatured by incubation at 80°C for 3 min. Samples were rapidly cooled on ice and mixed with 12 μl of micrococcal nuclease mix containing 0.03 U/μl micrococcal nuclease (Thermo Fisher Scientific), 10 mM CaCl2, 0.2 mg/ml BSA and 20 mM Tris/HCl pH 8.0. The final concentration of 0.015 U/μl micrococcal nuclease is standardized to produce fragments in the range of 50–200 nt under these conditions. Additionally, 2 U of DNAse I (NEB) and 1 μl MgCl2 (24 mM) were added to eliminate possible DNA contamination. Samples were incubated for 40 min and fragmentation was stopped by adding 6.5 μl of EDTA (50 mM, pH 8.0). Fragmented RNA samples were heat denatured and rapidly cooled on ice. Two aliquot of 3.8 μl each were separated and stored at –80°C. One aliquot was used to confirm size distribution of fragmented RNAs and the others were used later for deep sequencing library construction encoding total fragmented RNA from control and oxidiatively stressed samples, respectively (referred to as fragment input samples). PCR reaction using primers for the glyW gene confirmed the absence of DNA in the fragment input samples after 35 cycles. 22.9 μl of the remaining input sample RNA (9 μg) were mixed with 15A3 antibody (100 μg/ml, 3 μg total, Santa Cruz Biotechnology) and 247.1 μl PBS pH 7.4 with 1 mM EDTA. Samples were incubated 1 hour at room temperature with constant gentle mixing. Then 30 μl of protein G agarose beads prewashed in PBS with 1 mM EDTA were added. Samples were incubated for additional 90 min, then 600 μl PBS with 1 mM EDTA were added. The supernatant was discarded after centrifugation of 2 min at 2,500 g at room temperature. Beads were subsequently washed four additional times using 600 μl of PBS with 1 mM EDTA and 0.004% NP40 (5 min incubations at 37°C, centrifugation at room temperature, centrifugation for 2 min 2,500 g). For elution, RNA beads were incubated 3 min at 37°C in 45 μl SDS 1% followed by addition of 450 μl Trizol, yielding immune-purified RNA fragments (referred to 8-oxo-G-enriched samples). Fragment input RNA samples were extracted using 30 μl of Trizol. All samples were incubated 5 min at room temperature after which chloroform was added (105 μl for 8-oxo-G-enriched samples and 7 μl for fragment input samples). Supernatants were collected after centrifugation (15 min, 12,000 rpm, 4°C) and precipitated adding 2 μl glycogen (20 mg/ml) and 600 or 40 μl isopropanol, respectively. All samples were incubated 10 min and then centrifuged (13,000 rpm, 30 min, 4°C). RNA was washed twice with 75% ethanol and stored at –80°C. Four cDNA libraries for deep sequencing were then prepared from one in vivo oxidation experiment using the Peregrine protocol (24) and sequenced in single end mode at the Biomedical Genomics Core, at Nationwide Children's Hospital (Columbus, OH, USA). Two to three million reads per sample were obtained.
Analysis of deep sequencing data
Primer sequences were eliminated and reads longer than 15 nt and with quality score Q bigger than 30 were selected using cutadapt (25). Reads were subsequently aligned using bowtie (26) to a fragment of the genome of E. coli K-12 (Accession number U00096.3) containing the operon A for rRNA genes (positions 4035331–4040836). Coverage at each position was calculated using BEDtools (27) and normalized by dividing the summation of coverage per position. 8-oxo-G-enrichment was expressed as the ratio between normalized coverage counts from the 8-oxo-G-enriched fragments and the input fragments. In order to reduce local fluctuations and to highlight larger-scale trends, the 8-oxo-G enrichment data was convoluted via a moving mean window of 31. Therefore, the enrichment at every position was expressed as the mean of its own value and the 15 positions before and after. Oxidation heat maps were created using RiboVision (28).
Synthetic RNA oligonucleotides
Non-modified synthetic RNA oligoribonucleotides were purchased from Microsynth (Balgach, Switzerland). Synthetic RNA oligoribonucleotides containing oxidized RNA residues were synthesized by solid phase synthesis using the known 2′-O-TBDMS protected phosphoramidite precursors for the base-oxidized ribonucleosides 8-oxo-A (29), 8-oxo-G (30) and 5-OH-C (31). For 5-OH-U, a 5-(2-nitrobenzyl) protected building block was used, which was prepared in analogy to 5-OH-C. Unmodified 2′-O-TBDMS protected RNA phosphoramidites were purchased from GlenResearch (Sterling, VA, USA) and CPG solid supports were purchased from Link Technologies Ltd. (Bellshill, UK). Syntheses were performed on a Pharmacia Gene Assembler Plus DNA synthesizer on a 1.3 or 10 μmol scale, following standard phosphoramidite protocols with 5-(benzylthio)-1H-tetrazole (0.25 M in acetonitrile) as the activator and coupling times of 6 min. Modified oligoribonucleotides were deprotected at room temperature for 17 h using NH4OH/EtOH 3:1. The sequence containing the 8-oxo-G modification was deprotected at 55°C for 17 h using 0.25 M ethanethiol in NH4/EtOH 3:1. Solid supports were filtered off and washed with water and the resulting solutions were evaporated to dryness at room temperature in a Speedvac evaporator. The resulting pellets were further dried by addition and evaporation of dry ethanol. The obtained product of a 1.3 μmol synthesis (amounts are 6-fold for a 10 μmol synthesis) was dissolved in anhydrous DMSO (100 μl) and neat triethylamine trihydrofluoride (125 μl) was added. After shaking at room temperature for 24 h, an aqueous solution of NaOAc (3 M, pH 5.2, 25 μl) and n-butanol (1 ml) were added and the mixture was chilled on dry ice for 45 min. After centrifugation for 20 min in an Eppendorf centrifuge, the supernatant was discarded, the pellet washed twice with cold ethanol (80%, 1 ml) and dried under high vacuum at room temperature, yielding the crude oligonucleotides. The modified oligoribonucleotides were purified on preparative 15–20% denaturing polyacrylamide gels, product bands were identified by UV shadowing, cut out and extracted by passive elution into buffer (0.3 M NaOAc, 1 mM EDTA) and EtOH precipitated. Deprotection of the 2-nitrobenzyl groups in 5-OH-C and 5-OH-U containing oligoribonucleotides was achieved by irradiating aliquots (300 μl, 50 mM) in water for 1 h using a 25 W tungsten lamp (31). Wavelengths below 300 nm were filtered off by a glass filter. Fully deprotected oligoribonucleotides were again purified by PAGE as described above and precipitated from EtOH. For the 2506_5-OH-U oligo, just a 22-mer containing the 5-OH-U was synthesized and subsequently ligated to an unmodified 5′ 17-mer fragment in order to obtain the full-length 39-mer required for atomic mutagenesis. The 22-mer was chemically phosphorylated following the RNA synthesis with phosphorylating reagent from GlenResearch. 15 nmol of each RNA fragment were combined in ligase buffer with an equal amount of 20-mer DNA splint (Supplementary Table S2). The reaction was heated to 75°C for 2 min, cooled to 30°C for 20 min and incubated for at 30°C for 16 h with 60 U T4 DNA Ligase (Thermo Scientific). Ligations and corresponding unmodified control RNA were gel-purified using a 12% denaturing (7 M urea) polyacrylamide gel, extracted and EtOH precipitated. Supplementary Table S1 gives an overview of all synthetic RNA oligoribonucleosides used in this project.
Reconstitution of chemically engineered ribosomes
The method of atomic mutagenesis allows for a single modified nucleoside to be site-specifically incorporated into 23S rRNA in the context of the entire 50S ribosome of T. aquaticus, as described previously (20). We utilized five different circularly permuted (cp) 23S rRNA constructs harboring a short sequence gap encompassing the 23S rRNA nucleoside under investigation (see Supplementary Figure S1). Production and reconstitution of the cp-23S rRNAs to form 50S subunits was performed as described previously (20,21) (see Supplementary Table S1 for synthetic RNA strands used and Supplementary Table S3 for the sequences of the PCR primers employed to generate cp23S rRNA templates). The reconstituted 50S were associated with either E. coli 30S (for in vitro translation and release assays) or with T. aquaticus 30S (for the puromycin reaction, the translocation, EF-G GTPase activation and tRNA binding assays) to form 70S ribosomes. For all functional assays, the negative control was a reconstitution reaction that contained no complementing synthetic RNA oligo, but was otherwise treated the same and associated with 30S.
Poly(Phe) synthesis
Assessment of the in vitro translation products in the poly(U)-directed poly(Phe) synthesis approach was performed as described (19,20). After 3 h incubation at 42°C, the produced poly(Phe) peptides were precipitated with 5% TCA at 95°C for 15 min, filtrated through glass-fiber filters, and detected by scintillation counting (32).
In vitro translation of genuine mRNA
In vitro oxidized 70S ribosomes or 50S subunits or in vitro restituted ribosomes were tested for their ability to translate mRNA coding for the ribosomal protein L12 of Methanococcus thermolithotrophicus. This assay was performed as previously described (20). The [35S]-Met-labeled translation products were separated on 18% SDS poly-acrylamide gel and visualized by phorphorimaging.
Peptide bond formation
Peptide bond formation was measured by the puromycin reaction as described previously (21), using 1 pmol Ac-[3H]Phe-tRNAPhe (15,000 cpm/pmol) as P-site substrate and 2 mM puromycin as the acceptor substrate.
EF-G GTP hydrolysis
We investigated the ribosome-dependent multiple-turnover EF-G GTP hydrolysis as described previously (19). 0.15 μM reconstituted ribosomes with pre-bound 0.37 μM deacylated tRNAPhe to the P-site and 25 μg poly(U) were added to 50 μM [γ-32P]GTP and 1.13 μM E. coli EF-G at 37°C. The reaction was terminated by the addition of 20% (v/v) formic acid and detection by TLC was performed as described.
Translocation assessed by toeprinting
In order to detect the translocation movement of the ribosome along an mRNA analog (coding for MFKSIRYV; (32)), we employed the toeprinting assay as described previously (33). See Supplementary Table S2 for the used DNA reverse transcription primer and the mRNA sequence. The reverse transcription products were separated by denaturing 7.5% PAGE and visualized by phosphorimaging. Comparing the relative toeprint intensity at position +19 (if position +1 is defined as the first nucleotide of the AUG start codon) on the used mRNA analog (indicating POST-state ribosomal complexes) versus position +16/+17 (PRE-state ribosomes) indicates the translocated fraction of ribosomes. The time-resolved translocation experiments were based on a protocol described in (34). A toeprinting/translocation master mix was assembled as in Koch et al. (33), aliquots were removed in 15 s intervals and added immediately to the reverse transcription mix containing 0.2 mM neomycin, which instantaneously stops translocation. In the time-course samples, concentration of the dNTPs used for the reverse transcription was increased to a final concentration of 5 mM.
pept-tRNA hydrolysis (release assay)
The release reaction with reconstituted ribosomes was assessed as described previously (35), by binding 0.5 pmol formyl-[3H]Met-tRNAfMet (45,000 cpm/pmol) to the P-site of reconstituted ribosomes programmed with 250 pmol synthetic mRNA UUCAUGUAA (Dharmacon Research, Inc.) before adding 15 pmol of E. coli RF-1 or RF-2. N-terminally His-tagged RF-1 and RF-2 were overexpressed in BL21 DE3 cells and purified over Ni-NTA-columns. Purified release factors were dialyzed against RF storage buffer (20 mM HEPES/KOH pH 7.6, 6 mM MgAc2, 150 mM NH4Cl, 4 mM β-mercaptoethanol, 20% glycerol) and stored at –80°C.
Filter binding assays
In order to assess tRNA binding to the P-site, Ac-[3H]Phe-tRNAPhe (15,000 cpm/pmol) was added in a 1:1 ratio to reconstituted ribosomes in binding buffer containing 20 mM HEPES/KOH (pH 7.6), 6 mM MgAc2, 150 mM NH4Cl, 4 mM β-mercaptoethanol, 2 mM spermidine and 50 μM spermine and the same mRNA analog as used for the toeprinting assay (see above). To measure tRNA binding to the A-site, the P-site was first filled by adding deacetlyated tRNAfMet in an 1.5 fold excess over the reconstituted and mRNA-programmed ribosomes. Subsequently, Ac-[3H]Phe-tRNAPhe (15,000 cpm/pmol) added in a 1:1 ratio was forced to bind to the A-site. In both cases, the resulting ribosomal complexes were diluted in cold binding buffer, passed through nitrocellulose filters (Millipore 0.45 μm), and washed. The radioactivity retained on the filter was determined by scintillation counting.
SHAPE probing
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) (36) was used to probe the reactivity of the backbone ribose 2′-OH of the synthetic RNA oligos used for in vitro reconstitutions of 50S subunits. To this end, reconstituted 50S subunits containing either A or 8-oxo-A in position 2451 of the PTC were subjected to 50 mM N-methylisatoic anhydride (NMIA, Sigma-Aldrich) for 45 min at 37°C. Subsequently, fully reconstituted 50S subunits (from a 40 pmol reconstitution sample) were separated from possible non-incorporated synthetic oligos by a 10–40% sucrose gradient and centrifugation for 15 h at 115,600 g in an SW-41 rotor. RNA extracted and PCI-purified from the isolated subunits was then analyzed by extension of primer Taq2451_rev (see Supplementary Table S2) with AMV RT (Promega) in the presence of 1.25 mM of each dNTP, 25 mM DTT and 12.5% DMSO. The SHAPE experiments using 3.6 pmol non-incorporated free oligoribonucleosides were performed in an analogous manner in binding buffer (see above; filter binding assays). The radiolabeled products were analyzed as described for toeprinting.
RESULTS
Oxidative stress damages rRNA and disrupts translation in vitro
To globally investigate the effects of ROS on ribosomal function, we extracted native ribosomes from the bacterial species E. coli and T. aquaticus and oxidized them in vitro. For oxidation, we employed the Fenton reaction driven by an iron-ascorbate system and hydrogen peroxide, which is known to produce hydroxyl radicals. Oxidatively stressed 70S ribosomes lost their in vitro translation activities in a hydrogen peroxide-dependent manner (Figure 1B). This effect was retained even when only the large 50S ribosomal subunit was oxidized and re-associated with unstressed small 30S subunits (Figure 1C), indicating that at least some of the responsible oxidative damages reside on the large ribosomal subunit which also harbors the peptidyl transferase center. Oxidizing the small 30S subunits only, and combining it with unstressed 50S particles resulted in a less severe reduction of translational activity (Figure 1D), indicating that the small ribosomal subunit is less susceptible to oxidative damage. To gain further insight into the molecular species responsible for these effects, rRNA or r-proteins, we isolated the 23S rRNA from oxidized 50S and reconstituted the large subunit using r-proteins from unstressed 50S particles. These reconstituted 50S subunits were associated with unstressed 30S particles to form 70S ribosomes and used in translation. The fact that in vitro translation activity was still impaired demonstrates that oxidized 23S rRNA is, at least in part, responsible for the loss of ribosomal function (Figure 1E). When we conducted the reciprocal experiment with the small ribosomal subunit by isolating and reconstituting 16S rRNA from oxidized 30S subunits, translation was also impaired, albeit to a lesser extent compared to oxidized 23S (Figure 1F). This indicates that 16S rRNA carries either fewer oxidized lesions under these conditions or its functions are less impaired by oxidative damage (Figure 1G).
Oxidative stress damages rRNA in vivo
Previous reports have shown that rRNA is a target for oxidation in E. coli cells (14). Nevertheless, there is no information regarding specific rRNA oxidation regions, nor whether specifically 23S rRNA is a target of oxidative modification in vivo. To gain more detailed insight we adapted previous methods used to immuno-purify oxidized RNA (37) to be compatible with current deep sequencing technologies. We purified total RNA from E. coli cells cultured in either control (in LB media) or oxidative stress conditions (in the presence of H2O2). Total RNA was fragmented and a fraction was immuno-purified using an antibody against 8-oxo-G, one of the common products of RNA oxidation (14). The 8-oxo-G antibody is a widespread tool for detecting RNA oxidation and its application is considered also as proxy for the relative levels of other oxidative lesions as well. Subsequent to deep sequencing analysis, the 23S, 16S and 5S rRNA reads were aligned to the operon A of the rRNA genes. We expected enrichment in read numbers for sequences around oxidized guanosines. When mapping the rRNA sequence reads enriched in 8-oxo-G back to the known primary and secondary structure of rRNA, we found that the 23S rRNA is most frequently oxidized, while 16S rRNA has relatively few oxidative lesions, and 5S rRNA seems to be not oxidized at all (Supplementary Figure S2). The 23S rRNA exhibits oxidations throughout its sequence, mostly in domains II, III, V (which also encompasses the PTC) and VI. Some of these oxidation hotspots show a clear ∼4–5-fold increase in sequence reads, indicative of 8-oxo-G enrichment under oxidative stress conditions. Unexpectedly, we found that many oxidation hotspots showed increased read numbers under both control and oxidative stress conditions (Supplementary Figure S2 A). These in vivo findings generally coincide with our results from in vitro stress and subsequent translation, namely that 23S rRNA oxidation affects protein synthesis more severely than 16S rRNA oxidation (Figure 1). Interestingly, some of these possible 23S rRNA oxidation hotspots are in the inner core of the ribosome, which also harbors the PTC, the catalytically active site of the ribosome.
Atomic mutagenesis of the oxidized peptidyl transferase center
In order to specifically characterize the effects of 23S rRNA oxidation on ribosome function, we investigated single nucleobase oxidations within the ribosome's catalytic heart, the PTC. Using the atomic mutagenesis approach (20), we reconstituted 50S subunits containing a single oxidized base in one of the five central PTC positions A2451, C2063, U2585, U2506 or A2602, and the PTC-adjacent position G2447 (Figure 1A). The basic principle of the atomic mutagenesis approach is the use of circularly permuted 23S rRNA (cp-23S rRNA) that places its new ends in such a way that a short sequence gap is introduced encompassing the rRNA nucleoside under investigation (Supplementary Figure S1). During in vitro assembly of the T. aquaticus 50S subunits the missing rRNA fragment is provided as synthetic RNA carrying the desired nucleobase oxidation (Supplementary Table S1) and is combined with the total r-protein fraction of the large subunit. Subsequently, the chemically engineered 50S were associated with native E. coli 30S subunits and tested in different in vitro assays. This experimental design allows assessing the effects of individual nucleobase oxidations on specific sup-steps of the ribosomal elongation cycle.
8-oxo-A2451 slows peptide bond formation
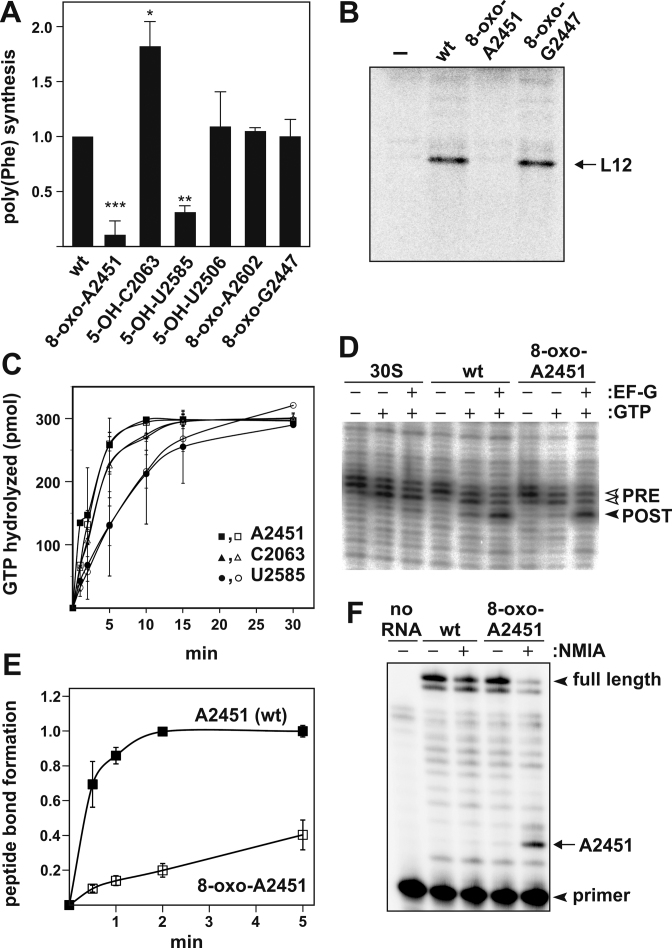
In order to assess the overall consequence of oxidized nucleobases in the PTC on translation we used the poly(U)-directed poly(Phe) synthesis assay (20,32,38). This is a comprehensive functional test for ribosomes since it involves essentially all sub-steps of protein biosynthesis, with the exception of translation termination. Ribosomes containing 8-oxo-A2451 in the PTC showed significantly reduced total peptide synthesis in the poly(Phe) assay compared to their reconstituted wild-type (wt) counterparts containing an intact adenine (Figure 2A and Table 1). When providing the physiologically more relevant heteropolymeric mRNA coding for r-protein L12, no protein synthesis product was detectable with ribosomes harboring 8-oxo-A2451 in the PTC (Figure 2B), indicating that at least one sub-step of protein biosynthesis is severely compromised by this single oxidation. By testing 8-oxo-A2451 ribosomes in reactions assessing the EF-G GTPase activation or the EF-G-mediated tRNA translocation step (via the toeprinting approach) it became evident that this oxidation does not affect the translocation phase of the elongation cycle or tRNA binding per se (Figure 2C, D and Table 1). This leaves the actual transpeptidation step as the most likely affected reaction left that could explain the reduced in vitro translation activities. Indeed peptide bond formation itself was significantly inhibited as evidenced by a 12-times reduced rate in the puromycin reaction (Figure 2E and Table 1). This reduced efficiency of transpeptidation greatly affects the processive amino acid polymerization during protein synthesis and therefore explains the very low translation activities observed with 8-oxo-A2451 ribosomes.
Figure 2.
Oxidation in the ribosome's active site differentially affects protein synthesis. (A) Endpoint measurements in poly(Phe) synthesis employing in vitro reconstituted ribosomes containing either synthetic RNA oligonucleotides with the wildtype sequence (wt) in the active site, or oligos harboring specific nucleobase oxidations in the PTC or nearby regions. In all reconstituted cp-23S rRNA constructs the minute translational activities of ribosomes containing no oligo was subtracted as background, and the activity of ribosomes containing the corresponding wt oligonucleotide was taken as 1.00. Values represent the mean of at least three independent experiments, with SD shown as error bars. Significance according to paired Student's t-test: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. (B) Representative autoradiogram of an SDS-PAGE showing in vitro translation of L12 r-protein mRNA by reconstituted ribosomes carrying 8-oxo-adenosine at 23S rRNA position A2451 or 8-oxo-guanosine G2447. Reactions with ribosomes reconstituted without a complementing synthetic RNA oligo (–) served as negative control. (C) EF-G GTPase activation as measured by the rate of GTP hydrolysis using reconstituted ribosomes with the wt sequence (filled symbols) or oxidized PTC bases (open symbols). Values represent the mean of at least two independent time course experiments, with error bars indicating SD. (D) Translocation of ribosomes oxidized at position A2451 along a heteropolymeric mRNA as determined by toeprinting. Toeprint bands indicate the positions of mRNA-bound ribosome complexes before (PRE) and after (POST) addition of EF-G and GTP. Ribosomal complexes carried deacylated tRNA and peptidyl-tRNA in the P- and A-site in the PRE state, and in the E- and P-site in the POST complexes, respectively. Reactions in the presence of T. aquaticus 30S subunits alone served as negative controls. (E) Time course of peptide bond formation in ribosomes containing either the wt sequence or 8-oxo-A2451. Values represent the mean of 3 independent time course experiments. The activities of in vitro assembled ribosomes without a synthetic oligo were subtracted as background. The endpoint value of ribosomes containing the wt sequence was taken as 1.00. (F) SHAPE probing of the 8-oxo-A2451 oligo inside the reconstituted large ribosomal subunit. Reconstituted 50S were treated with NMIA to determine ribose 2′-OH reactivity, which was detected by primer extension analysis. The location of the radiolabeled primer, the full length reverse transcription product and the position of A2451 in the denaturing polyacrylamide gel are indicated by arrow heads and an arrow, respectively. Note that the NMIA reaction product halts reverse transcription one position 3′ of the reacted site. Primer extension reaction in the absence of any RNA was used as negative control (no RNA).
Table 1. In vitro functional activities of ribosomes carrying RNA oxidative lesions in the PTC. Final values observed at end points or rates in the reaction's linear range relative to corresponding reconstituted wt ribosomes are shown.
| Site | Modification | poly(Phe) synthesis1 | mRNA translation1 | Peptide bond formation2 | EF-G GTPase2 | Translocation | pept-tRNA hydrolysis | A-site tRNA binding1 |
|---|---|---|---|---|---|---|---|---|
| A2451 | 8-oxo-A | 0.11 (±0.13) | 0.00 (±0.02) | 0.08 (±0.01) | 1.05 (±0.06) | 1.16 (±0.12)1 | 0.97 (±0.18)1 | n.d. |
| abasic | 0.72 d | + e | 0.53 a | n.d. | n.d. | 0.56 (±0.25)2 c | n.d. | |
| C2063 | 5-OH-C | 1.82 (±0.23) | 1.61 (±0.25) | 1.15 (±0.21) | 0.99 (±0.12) | 0.93 (±0.08)1 | n.d. | n.d. |
| 0.88 (±0.14)2 | ||||||||
| abasic | 0.22 d | n.d. | 0.98 c | n.d. | n.d. | 0.20 (±0.08)2 c | n.d. | |
| U2585 | 5-OH-U | 0.31 (±0.07) | 0.18 (±0.30) | 0.93 (±0.12) | 1.07 (±0.12) | 0.98 (±0.08)1 | 1.04 (±0.05)1 | 0.52 (±0.16) |
| abasic | 1.56 d | ++ f | 1.49 b | n.d. | n.d. | 2.57 (±0.41)2 c | n.d. | |
| U2506 | 5-OH-U | 1.09 (±0.32) | 1.07 (±0.39) | n.d. | n.d. | n.d. | n.d. | n.d. |
| abasic | 1.80 d | ++ f | 2.21 b | n.d. | n.d. | 1.14 (±0.18)2 c | n.d. | |
| A2602 | 8-oxo-A | 1.05 (±0.04) | 1.00 (±0.08) | n.d. | n.d. | n.d. | 1.07 (±0.03)1 | n.d. |
| abasic | n.d. | n.d. | 1.09 c | n.d. | n.d. | 1.52 (±0.44)2 c | n.d. | |
| G2447 | 8-oxo-G | 1.01 (±0.16) | 0.85 (±0.11) | n.d. | n.d. | n.d. | n.d. | n.d. |
1Value at end point.
2Initial rate constant (±Stdv).
n.d.: not determined.
Qualitative assessments: ++ slightly reduced compared to wt (approx. 0.5); + significantly reduced compared to wt (approx. 0.1).
aData from (21).
bData from (41).
cData from (35).
dData from (40).
eData from (20).
fData from (54).
The central PTC residue A2451, specifically its ribose 2′-OH group, is essential for catalyzing peptide bond formation (39). However, this catalytic effect seems to be independent of the nucleobase, since 2451 abasic ribosomes were active in transpeptidation and in vitro translation as long as the ribose 2′-OH remained in place (20–21,40) (Table 1). 8-oxo-A2451 is in fact the first nucleobase modification identified thus far at this PTC residue that has a significant impact on transpeptidation (21,39,41). In the latest catalytic model of peptide bond formation the 2′-OH of A2451 is part of a proton wire that shuttles a proton away from the α-amine of aminoacyl-tRNA during the formation of the peptide bond and relays a proton back to the leaving group as the reaction intermediate breaks down (42). Therefore the polarity and electronegative balance of the functional groups involved in this proton wire (such as the A2451 ribose 2′-OH) are crucial. To gain insight into the reactivity of the ribose 2′-OH as a function of the C8 oxidation at A2451, we performed SHAPE experiments. This RNA probing approach relies on the protonation status and flexibility of ribose 2′-OH groups (36). Ribosomes either carrying the wt sequence or the 8-oxo-A2451 in the PTC were treated with N-methylisatoic anhydride (NMIA) and the extent of 2′-OH alkylation was monitored by primer extension analysis. Within the context of the ribosomal PTC, as well as within the unincorporated oligoribonucleotide strand free in solution, nucleobase oxidation at A2451 resulted in a clearly increased SHAPE reactivity, indicating a slight pKa downshift of its ribose 2′-OH (Figure 2F and Supplementary Figure S3).
5-OH-C2063 facilitates translation
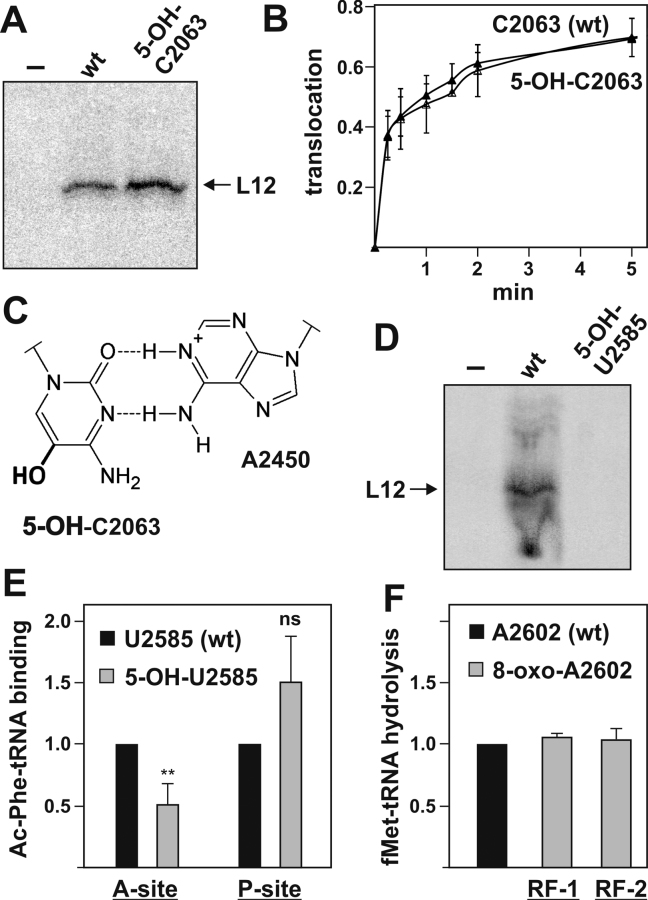
The second inner core PTC residue investigated was C2063. Previous studies have shown that C2063 forms a highly conserved non-Watson–Crick base pair with A2450 (40,43–44) and is important for establishing the active site architecture. We observed that ribosomes harboring 5-OH-C2063 produced increased amounts of poly(Phe) peptides in the poly(U) translation assay (Figure 2A and Table 1). To validate these unexpected findings and to repeat in vitro translation under physiologically more relevant conditions, we used ribosomes with an oxidized nucleobase at C2063 in a translation reaction utilizing a natural heteropolymeric mRNA coding for r-protein L12. Also under these conditions higher amounts of synthesized L12 was detectable compared to the wt control (Figure 3A). The rates of peptide bond formation and EF-G-triggered GTP hydrolysis were not affected by 5-OH-C2063 (Table 1). Also the extent of the translocation reaction along the mRNA was unaffected by this oxidation as assessed by the toe-printing assay (Table 1). To gain insight into possible kinetic differences in translocation we repeated the toeprinting reaction in a time-resolved manner using the antibiotic neomycin to halt translocation at defined time points (see Materials and Methods). Nevertheless, also under these conditions, no differences between 5-OH-C2063 and the wt control were apparent (Figure 3B) that could explain the previously observed increase in peptide and protein production (Figures 2A and 3A).
Figure 3.
Effects of oxidation at positions C2063, U2585 and A2602. (A) In vitro translation of r-protein L12 mRNA with chemically engineered ribosomes oxidized at position C2063. Reaction with ribosomes reconstituted without a complementing synthetic RNA oligo (–) served as negative control. (B) Time course of translocation of ribosomes oxidized at position C2063 along a heteroplymeric mRNA as determined by toeprinting. The translocation reaction was initiated by the addition of EF-G•GTP and stopped by the antibiotic neomycin at the indicated time points. The mean and standard deviation of three independent time course experiments are shown. (C) The non-Watson-Crick base pair formed in the 50S subunit between C2063 and A2450 is shown here with 5-OH-C (bold) in position 2063. The integrity of this interaction was shown previously to be vital for effective translation (40). (D) In vitro translation of r-protein L12 mRNA with chemically engineered ribosomes oxidized at position U2585. Reaction with ribosomes reconstituted without a complementing synthetic RNA oligo (–) served as negative control. (E) Ac-[3H]Phe-tRNAPhe binding affinity to the A- or P-site, respectively, in chemically engineered ribosomes oxidized at position U2585. Binding activities were normalized to ribosomes harboring the synthetic wt oligo, whereas unspecific tRNA binding to ribosomes containing no synthetic oligo was always subtracted as background. The mean and standard deviation of 5–7 independent binding experiments are shown. Paired Student's t-test: **P ≤ 0.01; ns, statistically not significant. (F) Release reaction as measured by P-site-bound f[3H]Met-tRNAfMet hydrolysis after RF-1 (n = 2) or RF-2 (n = 4) addition to chemically engineered ribosomes carrying an 8-oxo-adenosine at position A2602. f[3H]Met-tRNAfMet hydrolysis was normalized to wt activities and f[3H]Met release in ribosomes containing no oligo was subtracted as background.
5-OH-U2585 interferes with A-site tRNA accommodation
The oxidation of base U2585 inhibited in vitro translation in the poly(Phe) assay (Figure 2A and Table 1) and completely abolished the in vitro translation of a native mRNA (Figure 3D). However, both the rate of peptide bond formation and EF-G activation were unaffected by the oxidation (Table 1 and Figure 2C). Based on biochemical (45) and structural (46,47) investigations the nucleobase of U2585 has been proposed to be involved P- and A-site tRNA binding. The hydroxyl group of 5-OH-U2585 would face the ribosomal A-site of the PTC and thus could repel accommodation of the A-site tRNA to the 50S subunit. To test this possibility we established a tRNA binding assays which allows monitoring Ac-[3H]Phe-tRNAPhe binding to either the A- or the P-site, respectively. These experiments revealed that 5-OH-U2585 hindered the peptidyl-tRNA analog from fully entering the ribosomal A-site, whereas the P-site affinity was slightly increased, the latter effect though was statistically not significant (Figure 3E). The reduced affinity of the PTC A-site for peptidyl-tRNA therefore can explain the significantly hampered translation observed in 5-OH-U2585 ribosomes.
5-OH-U2506, 8-oxo-A2602 and 8-oxo-G2447 do not affect ribosome functions
The last two inner-core PTC residues investigated are U2506 and A2602. Additionally we also included 8-oxo-G2447 as a member of the second tier PTC residues since it has previously been reported to play a role in shaping the architecture of inner core PTC residues (17). In our functional assays nucleobase oxidation of neither U2506 and A2602 nor G2447 significantly affected the ribosome's performance (Table 1, and Figure 2A, B). In previous research the highly flexible A2602 has been shown to be crucial for the hydrolysis of peptidyl-tRNA and peptide release during the termination phase of protein biosynthesis (35,48–49). The poly(Phe) synthesis (Figure 2A) does not depend on canonical peptide release, as the poly-uridine acting as mRNA analog does not contain stop codons. We therefore additionally tested ribosomes carrying 8-oxo-A2602 in the PTC in a release factor-dependent termination assay. The amounts of hydrolyzed formyl[3H]-Met-tRNAfMet in the presence of bacterial release factor 1 or 2 were however comparable to the wt control. It therefore appears that the crucial function of A2602 during peptide release is unaffected by its nucleobase oxidation (Figure 3F).
DISCUSSION
Reactive oxygen species (ROS) represent a threat to all biological components of a cell. As oxidative stress arises from many unavoidable sources, cells have evolved strategies to disable ROS by both enzymes and small molecules (50) or to eliminate oxidized macromolecules (51). While DNA oxidation has been studied with increasing molecular insight over the past 40 years (52), research on RNA oxidative damage has only gained attention more recently (reviewed in (2)). Especially oxidative damage by ROS on non-coding transcripts with longer half-lives, such as tRNAs and rRNAs (t1/2 in the range of several hours to several days), represent a hazard for cell metabolism. Indeed, accumulating evidence links oxidized RNA to a variety of diseases, particularly age-related degeneration (4), thus shifting oxidized RNA and its consequences on biological systems into the focus of both basic as well as biomedical research.
In contrast to early theories, it was shown that the folded structure and protein interactions do not protect the rRNA from ROS in the ribosome (14). Our findings fully corroborate these conclusions and provide unprecedented molecular insight of rRNA nucleobase oxidation for ribosome functioning. We demonstrate that rRNA nucleobase oxidations interfere with efficient protein biosynthesis (Figure 1) and that bacterial ribosomes are targeted by ROS in vivo (Supplementary Figure S2). We furthermore observed that the 50S and its 23S rRNA, which harbors the ribosome's catalytic site, are more susceptible to oxidative stress than the small subunit, which contains the decoding center (Figure 1C–G). While our in vivo oxidation map cannot provide nucleotide resolution data due to inherent limitations of the immuno-precipitation method employed, we nevertheless observed clear rRNA oxidation hotspots, with some of them located in known functional regions (Supplementary Figure S2). These apparent oxidation hotspots lack an obvious correlation with the absence of bound r-proteins, the location of proximal Mg2+ binding sites, or less compact secondary rRNA structure. Thus the oxidation susceptibility of rRNA positions within the ribosome in vivo seems to be governed by a more complex combination of criteria. We also observed an unexpected amount of possible rRNA oxidation under non-stress conditions (Supplementary Figure S2A). Even when no external oxidative stress is applied, ROS arise from multiple cellular processes such as the respiratory chain. The fact that these rRNA regions are apparently oxidized even under normal conditions could indicate that they are especially sensitive to oxidation, yet do not pose a functional burden for the ribosome in vivo. In any case, further investigation of this phenomenon is required.
By utilizing the power of nucleoside analog interference, named atomic mutagenesis of the ribosome (20,21), we deciphered the functional consequences of specific nucleobase oxidations of 23S rRNA residues in the PTC on translation (Figures 2, 3 and Table 1). The outcomes varied from no effects (A2602, U2506, G2447), to strongly inhibitory (A2451, U2585), to even slightly stimulating effects (C2063). These findings also highlight the varied roles of the central PTC residues and add to our molecular understanding of their fine-tuned contributions to the mechanism of translation (see below). Beside nucleobase oxidations ROS are also known to trigger breakage of the glycosidic bond, yielding abasic sites in RNA (53). In our previous work we demonstrated that removal of the entire nucleobase at inner core PTC residues had surprisingly little effects on peptide bond formation and in vitro translation (Table 1). For the catalytic heart of the ribosome, ablation of a nucleobase is obviously less of a hurdle than carrying a hydroxyl or carbonyl oxygen at certain PTC nucleobases (e.g. A2451, U2585). This conclusion however cannot be generalized as abasic sites at other functional regions of the 50S subunit (G2421 in the E-site (33), or A2062 and U2586 in the nascent peptide exit tunnel (54)) do affect ribosomal functions.
In previous studies on A2451, even drastic nucleobase modifications such as methylation, amination, N to C replacements of heteroatoms, or the complete removal of the purine nucleobase had little or no effect on transpeptidation (21,39,41). 8-oxo-A2451 is the first nucleobase modification at this central PTC residue that significantly affected its catalytic role in peptide bond formation. This oxidation might sufficiently shift the electron density in the nucleoside to affect the pivotal role of its ribose 2′-OH. In the latest catalytic mechanism for peptide bond synthesis the A2451 2′-OH is central for establishing a proton wire (42). In the proposed model the A2451 2′-OH is part of an array of H-donors/acceptors that subtracts a proton from the nucleophilic α-amine, and thereby facilitating the nucleophilic attack of the α-amine of the A-site aminoacyl-tRNA onto the ester carbonyl carbon of the P-site peptidyl-tRNA. As the intermediate breaks down to form a new peptide bond, the proton wire then relays a proton back to saturate the 3′-O− of the P-site tRNA leaving group. Evidently, the positioning of all substrates, transition states, and ribosomal residues contributing to the concerted redistribution of charges must be tightly controlled to achieve efficient transpeptidation compatible with the observed in vivo rates of amino acid polymerization of about 20 s–1 (55). While oxidation at C8 of A2451 is expected to primarily impact the electron distribution within the purine base, it could also affect the pKa of its ribose 2′-OH group. By applying SHAPE experiments we were able to demonstrate a markedly enhanced reactivity of the A2451 ribose 2′-OH upon nucleobase oxidation (Figure 2F), indicating a pKa downshift within the PTC. Although this change might be very subtle, it is conceivable that even a small difference in the pKa of such a central member of the proton wire may significantly impact the overall efficiency of proton shuttling and thus peptide bond synthesis. Alternatively, the presence of the C8 carbonyl group could also lead to a shift from the anti to the syn conformation in the oxidized nucleoside. Since we do observe the same SHAPE reactivity pattern at 8-oxo-A2451 in the context of the free oligonucleotide in solution and the fact that an anti to syn rotation inside the PTC seems challenging from an architectural point of view, we favor electron re-distribution as cause for the acidified pKa at the ribose 2′-OH as described above. Indeed it has been observed that the presence of a 2′-fuorine group can affect electron distribution of the nucleobase π-electrons (56), thus an electron re-distribution force in the other direction (from the base to the ribose 2′-OH) is conceivable.
PTC residue C2063 forms a non-canonical, yet highly conserved base pair in the active site (Figure 3C) (43). This base pair appears to be crucial for efficient translation, since modifications that disrupt pairing inhibit the tRNA translocation step (40). Our new data suggest that the reverse is also true: strengthening of the A2450–C2063 base pair by the introduction of 5-OH-C2063, facilitates translation yields (Figures 2A and 3A). The oxidation of C5 of C2063 alters the pKa of the pyrimidine base, including the functional groups forming the H-bonds in the non-Watson-Crick base pair (Figure 3C). While the pKa’s of A2450 can be considered to remain constant, the pKa of C2063 N3 is expected to be lowered markedly upon oxidation of C5. Since the strength of an individual H-bond depends on the pKa of the donor and acceptor pair, a more acidic N3 would result in a stronger bond between the N6 amino of A2450 and the N3 of 5-OH-C2063 (Figure 3C). Introduction of 5-OH-C2063 into the PTC resulted in increased peptide and protein yields during in vitro translation (Figures 2A and 3A), yet leaving the rates of peptide bond formation, EF-G GTPase and tRNA translocation unaffected (Table 1 and Figures 2C, 3B). Therefore, the oxidized C2063 is expected to stabilize the functionally competent PTC architecture by its improved interaction with A2450. This active site stabilization might be especially beneficial for in vitro reconstituted ribosomes that are known for their low assembly efficiencies (41). However, even seemingly positive effects on protein synthesis as observed with oxidized C2063, might have detrimental effects in vivo: translation speed is specifically regulated by codon usage or tRNA abundance and represents an important factor for mRNA stability and correct protein folding (57,58).
The effects of nucleobase oxidations on 23S rRNA residues observed in this study demonstrate that oxidation can impair different steps in the ribosomal elongation cycle. As in any model, there is no guarantee that oxidized ribosomes behave exactly the same in vivo, but our experience in the past has revealed atomic mutagenesis as a reliable tool for predicting and understanding ribosomal functions (20,59). However, it is still an open question as to whether the central PTC residues investigated here are targeted by ROS in multicellular organisms and whether they are actually oxidized in neurodegenerative diseases like Alzheimer's. The answer to these questions would require a sequence-specific survey of all rRNA nucleobase oxidations in patient's samples, an undertaking that is clearly beyond of the scope of this study.
AVAILABILITY
BAM files were deposited at SRA database (bioproject ID: PRJNA401389).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Miriam Koch for valuable input and discussions and for critical reading of the manuscript. We kindly acknowledge Matthias Erlacher and Thomas Hoernes for providing plasmid clones of E. coli RF-1 and RF-2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss National Science Foundation [31003A_166527 to N.P.]; NCCR ‘RNA & Disease’ funded by the Swiss National Science Foundation (to N.P.); Fondo Nacional de Desarrollo Científico y Tecnológico, Chile [11140222]; Comisión Nacional de Investigación Científica y Tecnológica of Chile [79130044 to A.K.]. Funding for open access charge: Swiss National Science Foundation [31003A_166527 to N.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hofer T., Seo A.Y., Prudencio M., Leeuwenburgh C.. A method to determine RNA and DNA oxidation simultaneously by HPLC-ECD: greater RNA than DNA oxidation in rat liver after doxorubicin administration. Biol. Chem. 2006; 387:103–111. [DOI] [PubMed] [Google Scholar]
- 2. Küpfer P., Leumann C.. Erdmann VA. Chemical Biology of Nucleic Acids. 2014; 5:Berlin, Heidelberg: Springer-Verlag; 75–94. [Google Scholar]
- 3. Hofer T., Badouard C., Bajak E., Ravanat J.L., Mattsson A., Cotgreave I.A.. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 2005; 386:333–337. [DOI] [PubMed] [Google Scholar]
- 4. Poulsen H.E., Specht E., Broedbaek K., Henriksen T., Ellervik C., Mandrup-Poulsen T., Tonnesen M., Nielsen P.E., Andersen H.U., Weimann A.. RNA modifications by oxidation: a novel disease mechanism?. Free Radic. Biol. Med. 2012; 52:1353–1361. [DOI] [PubMed] [Google Scholar]
- 5. Simms C.L., Zaher H.S.. Quality control of chemically damaged RNA. Cell. Mol. Life Sci. 2016; 73:3639–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Honda K., Smith M.A., Zhu X., Baus D., Merrick W.C., Tartakoff A.M., Hattier T., Harris P.L., Siedlak S.L., Fujioka H. et al. Ribosomal RNA in Alzheimer disease is oxidized by bound redox-active iron. J. Biol. Chem. 2005; 280:20978–20986. [DOI] [PubMed] [Google Scholar]
- 7. Simms C.L., Hudson B.H., Mosior J.W., Rangwala A.S., Zaher H.S.. An active role for the ribosome in determining the fate of oxidized mRNA. Cell Rep. 2014; 9:1256–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ding Q., Markesbery W.R., Cecarini V., Keller J.N.. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer's disease. Neurochem. Res. 2006; 31:705–710. [DOI] [PubMed] [Google Scholar]
- 9. Ding Q., Zhu H., Zhang B., Soriano A., Burns R., Markesbery W.R.. Increased 5S rRNA oxidation in Alzheimer's disease. J. Alzheimer's Dis.: JAD. 2012; 29:201–209. [DOI] [PubMed] [Google Scholar]
- 10. Langstrom N.S., Anderson J.P., Lindroos H.G., Winblad B., Wallace W.C.. Alzheimer's disease-associated reduction of polysomal mRNA translation. Brain Res. Mol. Brain Res. 1989; 5:259–269. [DOI] [PubMed] [Google Scholar]
- 11. Nunomura A., Perry G., Aliev G., Hirai K., Takeda A., Balraj E.K., Jones P.K., Ghanbari H., Wataya T., Shimohama S. et al. Oxidative damage is the earliest event in Alzheimer disease. J. Neuropathol. Exp. Neurol. 2001; 60:759–767. [DOI] [PubMed] [Google Scholar]
- 12. Ding Q., Markesbery W.R., Chen Q., Li F., Keller J.N.. Ribosome dysfunction is an early event in Alzheimer's disease. J. Neurosci. 2005; 25:9171–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hernandez-Ortega K., Garcia-Esparcia P., Gil L., Lucas J.J., Ferrer I.. Altered machinery of protein synthesis in Alzheimer's: from the nucleolus to the ribosome. Brain Pathol. 2016; 26:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu M., Gong X., Alluri R.K., Wu J., Sablo T., Li Z.. Characterization of RNA damage under oxidative stress in Escherichia coli. Biol. Chem. 2012; 393:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Polacek N., Mankin A.S.. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit. Rev. Biochem. Mol. 2005; 40:285–311. [DOI] [PubMed] [Google Scholar]
- 16. Wilson D.N., Blaha G., Connell S.R., Ivanov P.V., Jenke H., Stelzl U., Teraoka Y., Nierhaus K.H.. Protein synthesis at atomic resolution: mechanistics of translation in the light of highly resolved structures for the ribosome. Curr. Protein Peptide Sci. 2002; 3:1–53. [DOI] [PubMed] [Google Scholar]
- 17. Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A.. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000; 289:920–930. [DOI] [PubMed] [Google Scholar]
- 18. Belardinelli R., Sharma H., Peske F., Wintermeyer W., Rodnina M.V.. Translocation as continuous movement through the ribosome. RNA Biol. 2016; 13:1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clementi N., Chirkova A., Puffer B., Micura R., Polacek N.. Atomic mutagenesis reveals A2660 of 23S ribosomal RNA as key to EF-G GTPase activation. Nat. Chem. Biol. 2010; 6:344–351. [DOI] [PubMed] [Google Scholar]
- 20. Erlacher M.D., Chirkova A., Voegele P., Polacek N.. Generation of chemically engineered ribosomes for atomic mutagenesis studies on protein biosynthesis. Nat. Prot. 2011; 6:580–592. [DOI] [PubMed] [Google Scholar]
- 21. Erlacher M.D., Lang K., Shankaran N., Wotzel B., Huttenhofer A., Micura R., Mankin A.S., Polacek N.. Chemical engineering of the peptidyl transferase center reveals an important role of the 2′-hydroxyl group of A2451. Nucleic Acids Res. 2005; 33:1618–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khaitovich P., Tenson T., Kloss P., Mankin A.S.. Reconstitution of functionally active Thermus aquaticus large ribosomal subunits with in vitro-transcribed rRNA. Biochemistry. 1999; 38:1780–1788. [DOI] [PubMed] [Google Scholar]
- 23. Schrode P., Huter P., Clementi N., Erlacher M.. Atomic mutagenesis at the ribosomal decoding site. RNA Biol. 2017; 14:104–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langevin S.A., Bent Z.W., Solberg O.D., Curtis D.J., Lane P.D., Williams K.P., Schoeniger J.S., Sinha A., Lane T.W., Branda S.S.. Peregrine: A rapid and unbiased method to produce strand-specific RNA-Seq libraries from small quantities of starting material. RNA Biol. 2013; 10:502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011; 17:10–12. [Google Scholar]
- 26. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bernier C.R., Petrov A.S., Waterbury C.C., Jett J., Li F., Freil L.E., Xiong X., Wang L., Migliozzi B.L., Hershkovits E. et al. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss. 2014; 169:195–207. [DOI] [PubMed] [Google Scholar]
- 29. Kim S.K., Kim J.Y., Baek A.K., Moon B.J.. Base pairing properties of 8-oxo-7,8-dihydroadenosine in cDNA synthesis by reverse transcriptases. Bioorg. Med. Chem. Lett. 2002; 12:1977–1980. [DOI] [PubMed] [Google Scholar]
- 30. Calabretta A., Kupfer P.A., Leumann C.J.. The effect of RNA base lesions on mRNA translation. Nucleic Acids Res. 2015; 43:4713–4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kupfer P.A., Leumann C.J.. Synthesis, base pairing properties and trans-lesion synthesis by reverse transcriptases of oligoribonucleotides containing the oxidatively damaged base 5-hydroxycytidine. Nucleic Acids Res. 2011; 39:9422–9432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Szaflarski W., Vesper O., Teraoka Y., Plitta B., Wilson D.N., Nierhaus K.H.. New features of the ribosome and ribosomal inhibitors: non-enzymatic recycling, misreading and back-translocation. J. Mol. Biol. 2008; 380:193–205. [DOI] [PubMed] [Google Scholar]
- 33. Koch M., Clementi N., Rusca N., Vogele P., Erlacher M., Polacek N.. The integrity of the G2421-C2395 base pair in the ribosomal E-site is crucial for protein synthesis. RNA Biol. 2015; 12:70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fredrick K., Noller H.F.. Catalysis of ribosomal translocation by sparsomycin. Science. 2003; 300:1159–1162. [DOI] [PubMed] [Google Scholar]
- 35. Amort M., Wotzel B., Bakowska-Zywicka K., Erlacher M.D., Micura R., Polacek N.. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res. 2007; 35:5130–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merino E.J., Wilkinson K.A., Coughlan J.L., Weeks K.M.. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005; 127:4223–4231. [DOI] [PubMed] [Google Scholar]
- 37. Shan X., Tashiro H., Lin C.L.. The identification and characterization of oxidized RNAs in Alzheimer's disease. J. Neurosci. 2003; 23:4913–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jelenc P.C., Kurland C.G.. Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl. Acad. Sci. U.S.A. 1979; 76:3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lang K., Erlacher M., Wilson D.N., Micura R., Polacek N.. The role of 23S ribosomal RNA residue A2451 in peptide bond synthesis revealed by atomic mutagenesis. Chem. Biol. 2008; 15:485–492. [DOI] [PubMed] [Google Scholar]
- 40. Chirkova A., Erlacher M.D., Clementi N., Zywicki M., Aigner M., Polacek N.. The role of the universally conserved A2450-C2063 base pair in the ribosomal peptidyl transferase center. Nucleic Acids Res. 2010; 38:4844–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Erlacher M.D., Lang K., Wotzel B., Rieder R., Micura R., Polacek N.. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2′-OH at A2451 of 23S rRNA. J. Am. Chem. Soc. 2006; 128:4453–4459. [DOI] [PubMed] [Google Scholar]
- 42. Polikanov Y.S., Steitz T.A., Innis C.A.. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat. Struct. Mol. Biol. 2014; 21:787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bayfield M.A., Thompson J., Dahlberg A.E.. The A2453-C2499 wobble base pair in Escherichia coli 23S ribosomal RNA is responsible for pH sensitivity of the peptidyltransferase active site conformation. Nucleic Acids Res. 2004; 32:5512–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hesslein A.E., Katunin V.I., Beringer M., Kosek A.B., Rodnina M.V., Strobel S.A.. Exploration of the conserved A+C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes. Nucleic Acids Res. 2004; 32:3760–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bocchetta M., Xiong L., Mankin A.S.. 23S rRNA positions essential for tRNA binding in ribosomal functional sites. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:3525–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Arenz S., Ramu H., Gupta P., Berninghausen O., Beckmann R., Vazquez-Laslop N., Mankin A.S., Wilson D.N.. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat. Commun. 2014; 5:3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schmeing T.M., Huang K.S., Strobel S.A., Steitz T.A.. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005; 438:520–524. [DOI] [PubMed] [Google Scholar]
- 48. Polacek N., Gomez M.G., Ito K., Nakamura Y., Mankin A.S.. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol.Cell. 2003; 11:103–112. [DOI] [PubMed] [Google Scholar]
- 49. Youngman E.M., Brunelle J.L., Kochaniak A.B., Green R.. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004; 117:589–599. [DOI] [PubMed] [Google Scholar]
- 50. Sies H. Oxidative stress: oxidants and antioxidants. Exp. Physiol. 1997; 82:291–295. [DOI] [PubMed] [Google Scholar]
- 51. Davies K.J. Oxidative stress, antioxidant defenses, and damage removal, repair, and replacement systems. IUBMB life. 2000; 50:279–289. [DOI] [PubMed] [Google Scholar]
- 52. Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J.. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003; 17:1195–1214. [DOI] [PubMed] [Google Scholar]
- 53. Tanaka M., Han S., Kupfer P.A., Leumann C.J., Sonntag W.E.. An assay for RNA oxidation induced abasic sites using the aldehyde reactive probe. Free Rad. Res. 2011; 45:237–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Koch M., Willi J., Pradere U., Hall J., Polacek N.. Critical 23S rRNA interactions for macrolide-dependent ribosome stalling on the ErmCL nascent peptide chain. Nucleic Acids Res. 2017; 45:6717–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bremer H., Dennis P.P.. Modulation of chemical composition and other parameters of the cell by growth rate Escherichia coli and Salmonella. Cell. Mol. Biol. 1996; 2:1553–1569. [Google Scholar]
- 56. Pallan P.S., Greene E.M., Jicman P.A., Pandey R.K., Manoharan M., Rozners E., Egli M.. Unexpected origins of the enhanced pairing affinity of 2′-fluoro-modified RNA. Nucleic Acids Res. 2011; 39:3482–3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nedialkova D.D., Leidel S.A.. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu C.H., Dang Y., Zhou Z., Wu C., Zhao F., Sachs M.S., Liu Y.. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015; 59:744–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Erlacher M.D., Polacek N.. Probing functions of the ribosomal peptidyl transferase center by nucleotide analog interference. Methods Mol. Biol. 2012; 848:215–226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.