Abstract
Modified uridine containing taurine, 5-taurinomethyluridine (τm5U), is found at the anticodon first position of mitochondrial (mt-)transfer RNAs (tRNAs). Previously, we reported that τm5U is absent in mt-tRNAs with pathogenic mutations associated with mitochondrial diseases. However, biogenesis and physiological role of τm5U remained elusive. Here, we elucidated τm5U biogenesis by confirming that 5,10-methylene-tetrahydrofolate and taurine are metabolic substrates for τm5U formation catalyzed by MTO1 and GTPBP3. GTPBP3-knockout cells exhibited respiratory defects and reduced mitochondrial translation. Very little τm5U34 was detected in patient’s cells with the GTPBP3 mutation, demonstrating that lack of τm5U results in pathological consequences. Taurine starvation resulted in downregulation of τm5U frequency in cultured cells and animal tissues (cat liver and flatfish). Strikingly, 5-carboxymethylaminomethyluridine (cmnm5U), in which the taurine moiety of τm5U is replaced with glycine, was detected in mt-tRNAs from taurine-depleted cells. These results indicate that tRNA modifications are dynamically regulated via sensing of intracellular metabolites under physiological condition.
INTRODUCTION
RNA molecules contain a wide variety of chemical modifications that are introduced post-transcriptionally. RNA modifications confer chemical diversity on simple RNA molecules, endowing them with a wider range of biological functions. To date, >130 species of RNA modifications have been reported in various RNA molecules from all domains of life (1–4). Many of these modifications were discovered in transfer RNAs (tRNAs), and tRNA modifications are involved in various molecular and physiological functions (5,6). In particular, tRNA modifications at the first (wobble) position of the anticodon (position 34) play critical roles in regulating the codon–anticodon interaction at the A-site of ribosome during the decoding process in translation (7).
The mitochondrion is a eukaryotic intracellular organelle that produces adenosine triphosphate (ATP) by oxidative phosphorylation (OXPHOS). Mitochondria contain their own genome, called mitochondrial DNA (mtDNA), which encodes 13 essential subunits of the respiratory chain complexes, two mt-rRNAs and 22 mt-tRNAs. The mammalian mitochondrial genetic code deviates from the universal code by using four dialectal codons: AUA for Met, UGA for Trp and AGA and AGG (AGR; R = A or G) for the stop signal. The 22 species of mt-tRNAs participate in translation of 13 proteins encoded in the mtDNA. RNA modifications in mt-tRNAs play critical roles in accurate and efficient translation in mitochondria (8). Previously, we mapped all modifications in 22 species of mammalian mt-tRNAs, and found 15 species of modified nucleosides at 118 positions (9). Three of the modifications, 5-formylcytidine (f5C), 5-taurinomethyluridine (τm5U) and 5-taurinomethyl-2-thiouridine (τm5s2U) are specific to mt-tRNAs.
f5C is present at the first position of the anticodon in mt-tRNAMet (10), and f5C34 plays an essential role in translation of the dialectal AUA codon as Met (8,11,12). We and other groups, recently demonstrated that f5C34 is synthesized via consecutive reactions, initiated by 5-methylcytidine (m5C) formation catalyzed by NSUN3 (13,14), followed by hydroxylation and oxidation of m5C to generate f5C, mediated by ALKBH1 (15,16). Both enzymes are required for efficient mitochondrial translation and respiratory activity. We also identified two pathogenic point mutations in mt-tRNAMet that impair f5C34 formation (13). In addition, loss-of-function mutations in NSUN3 were detected in a patient with symptoms of mitochondrial disease (14). Together, these observations indicate that loss of f5C34 results in pathological consequences.
Two taurine-containing uridines are present at the wobble positions of five mt-tRNAs: τm5U in the mt-tRNAs for Leu(UUR) and Trp, and τm5s2U in the mt-tRNAs for Glu, Lys and Gln (9,17) (Figure 1A). These modifications promote accurate decoding of purine-ending codons (NNR; N = U, C, A or G) and prevent misreading of pyrimidine-ending codons (NNY; Y = U or C) (8). During decoding, τm5U34 stabilizes U–G wobble pairing by assuming Watson–Crick geometry, thus engaging in a favorable stacking interaction with the 3′ adjacent base (A35) (18). We found that τm5(s2)U modifications do not occur in mutant tRNAs isolated from cells of patients with mitochondrial encephalomyopathies (8,19). The mt-tRNALeu(UUR) harboring one of five pathogenic mutations associated with MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) lacks τm5U34 (20,21). Biochemical studies revealed that the hypomodified mt-tRNALeu(UUR) lacking τm5U34 decodes the UUA codon normally, but failed to efficiently decode the UUG codon efficiently (22), strongly suggesting that inefficient decoding by the hypomodified tRNA results in defective translation of protein subunits of respiratory chain complexes. In addition, τm5s2U34 is absent from mt-tRNALys with the A8344G point mutation associated with MERRF (myoclonus epilepsy associated with ragged red fibers) (23,24). The hypomodified mt-tRNALys also lacks the ability to decode its cognate codons, leading to defective mitochondrial translation and respiratory activity (25). The pathogenic point mutations associated with MELAS and MERRF are thought to prevent recognition by the tRNA-modifying enzymes responsible for τm5U formation. These findings represent the first instance of human disease caused by aberrant RNA modifications (19). Hence, we propose ‘RNA modopathy’ as a distinct category of human disease.
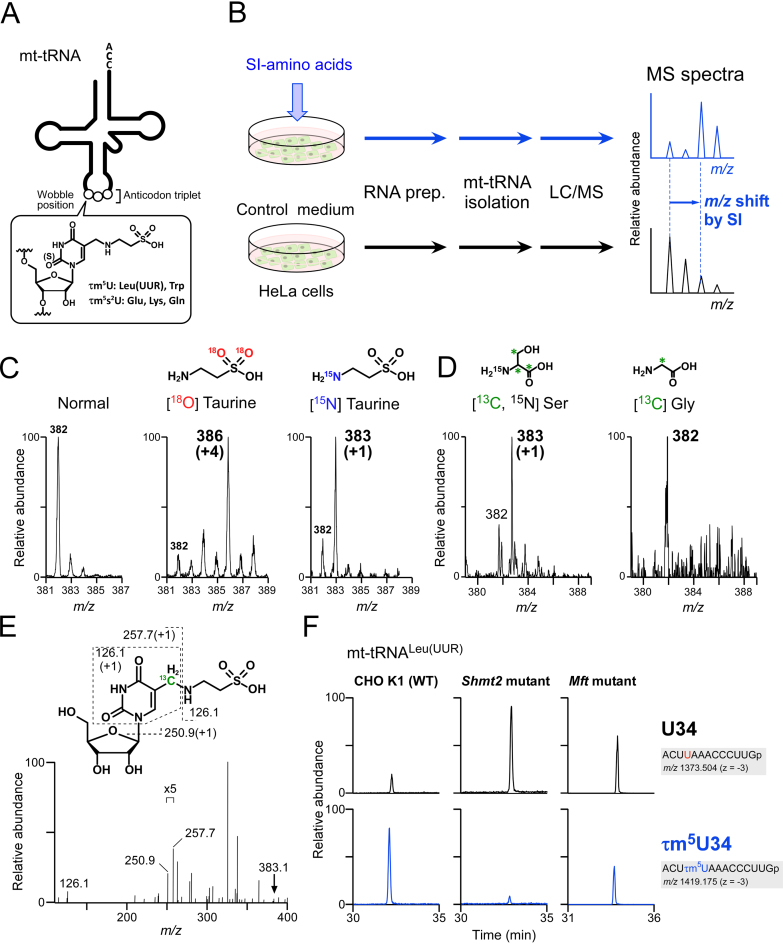
Figure 1.
Metabolic labeling of mt-tRNAs to determine metabolic sources of τm5U. (A) Chemical structure of τm5(s2)U which is present at the wobble (first) position of anticodon of five mt-tRNAs. (B) Scheme for metabolic labeling experiments. HeLa cells were cultured with stable isotope (SI)-labeled or non-labeled amino acids. Total RNA was extracted from each culture, and individual mt-tRNAs were isolated and digested into nucleosides, followed by liquid chromatography–mass spectrometry (LC/MS) analysis to detect SI-labeled τm5(s2)U. (C) Mass spectra of τm5U in mt-tRNATrp isolated from HeLa cells cultured in normal medium (first from the left), [18O] taurine–supplemented medium (second) and [15N] taurine–supplemented medium (third). Labeled atoms are colored as indicated. Increased m/z values of τm5U due to the presence of SI-atoms are shown in parentheses. (D) Mass spectra of τm5U in mt-tRNAs for Trp and Leu(UUR) isolated from HeLa cells cultured in [15N, 13C3] Ser–supplemented medium (left) and [13C] Gly–supplemented medium (right). 13C atoms in their chemical structures are indicated by asterisks. (E) Collision-induced dissociation (CID) spectrum of [13C]-labeled τm5U nucleoside. The precursor ion (m/z 383.1) for CID is marked by an arrow. Product ions in the CID spectrum are assigned in the chemical structure of τm5U. (F) Mitochondrial 1C metabolism involved in τm5U biosynthesis (see Figure 7). Extracted mass chromatograms (XICs) of anticodon-containing RNase T1-digested fragments bearing U34 (upper panels) or τm5U34 (lower panels) from mt tRNALeu(UUR) isolated from Chinese hamster ovary (CHO) wild-type (WT) (left panels), Shmt2 mutant (center panels) and Mft mutant (right panels) cells. Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right.
A metabolic labeling experiment revealed that extracellular taurine supplied in the medium is directly incorporated to 5-taurinomethyl groups in mt-tRNAs (17). However, a comprehensive understanding of τm5U biogenesis remains elusive. Based on a genetic study in yeast (26), we previously predicted the tRNA modifying enzymes responsible for τm5s2U34. MTO1 and GTPBP3 are likely to be involved in the formation of the 5-taurinomethyl group, while MTU1 is responsible for the 2-thio group of τm5s2U34 in mt-tRNAs. Genetic mutations in MTU1 were identified in patients of reversible infantile liver failure (RILF) (27–29). Liver-specific knockout of Mtu1 mice exhibited severe impairment of mitochondrial translation and respiratory activities in the hepatocytes, demonstrating that mitochondrial dysfunction due to loss of 2-thiolation of τm5s2U34 is a primary cause of RILF (30). Regarding 5-taurinomethyl modification, exome sequencing identified pathogenic mutations in both MTO1 (31–33) and GTPBP3 (34,35) associated with hypertrophic cardiomyopathy and lactic acidosis, indicating that lack of taurine modification results in mitochondrial dysfunction via a mechanism similar to those of MELAS and MERRF. However, it has not been directly demonstrated that the frequency of τm5(s2)U34 is reduced in patients’ cells or tissues.
In regard to the biogenesis of τm5U34, we demonstrate here that the β-carbon of L-serine is a source of the methylene group of τm5(s2)U34 via 5,10-methylene-tetrahydrofolate (THF) in one-carbon (1C) metabolism. This finding is supported by the observation that the level of τm5(s2)U34 was reduced in cells harboring mutations in serine hydroxymethyltransferase 2 (SHMT2) and mitochondrial folate transporter (MFT). In addition, we found that the τm5U34 frequency in mt-tRNAs was downregulated by taurine starvation. Intriguingly, we detected 5-carboxymethylaminomethyluridine (cmnm5U), in which the taurine moiety of τm5U is replaced with glycine, in mt-tRNAs of taurine-starved cells, indicating that tRNA modification is dynamically regulated by sensing of intracellular metabolites under physiological condition. In addition, in the cells of domestic cat (Felis catus) and Japanese flounder (Paralichthys olivaceus), neither of which have the biosynthetic pathway for taurine, we observed that the τm5U34 frequency in mt-tRNAs decreased upon taurine starvation during feeding, indicating that dietary taurine deficiency decreases the abundance of the taurine modification in mt-tRNAs, leading to pathological consequences including cardiomyopathy and other manifestation of mitochondrial diseases. The physiological importance of the taurine modifications was demonstrated by the finding that GTPBP3-knockout cells exhibited respiratory defects and reduced mitochondrial translation. We here clearly demonstrated that τm5(s2)U34 was absent from mt-tRNAs isolated from GTPBP3 KO cells, Mto1 KO cells, as well as fibroblasts of a patient with pathogenic mutations in GTPBP3. Finally, we successfully reconstituted τm5U34 formation in mt-tRNA in vitro in a system consisting of GTPBP3 and MTO1 in the presence of taurine, the methylene-THF, guanosine triphosphate (GTP) and other co-factors.
MATERIALS AND METHODS
Cell culture and stable-isotope labeling
HeLa and HEK293T cells were cultured at 37°C in 5% CO2 in Dulbecco’s modified eagle’s medium (DMEM) (high-glucose) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. GTPBP3 KO cells were maintained at 37°C in 5% CO2 in DMEM (high-glucose) supplemented with 0.11 g/l sodium pyruvate, 0.05 g/l uridine, 10% FBS and 1% penicillin–streptomycin.
For stable isotope (SI) labeling with taurine, HeLa cells were cultured with taurine-18O2 or taurine-15N (Aldrich) as described (17). To determine the 1C carbon source, HeLa cells were cultured at 37°C in 5% CO2 for 48 h with a synthetic medium containing 1× minimal essential media (GIBCO), 10% dialyzed FBS (GIBCO), 20 mM HEPES-KOH (pH 7.5), 100 units/ml penicillin, 0.1 mg/ml streptomycin, 0.5 mM sodium pyruvate and 0.1 mM each of amino acids (L-alanine, L-asparagine, L-aspartic acid, L-glutamic acid, L-methionine and L-proline), supplemented with 0.5 mM Gly-2–13C (Taiyo Nippon Sanso) or Ser-15N, 13C3 (Taiyo Nippon Sanso).
For taurine starvation medium, 10% dialyzed FBS was used instead of 10% FBS. To restore taurine, 40 mM taurine (final concentration) was added to the taurine starvation medium.
Chinese hamster ovary (CHO) K1 cells were obtained from RIKEN BioResource Center (RCB2869). The glycine auxotrophic CHO glyA (Shmt2) and glyB (Mft) mutant strains (36) were kindly provided by Dr Lawrence Chasin (Columbia University). CHO strains were cultured at 37°C in 5% CO2 with DMEM (high-glucose) containing 10% FBS and 1% penicillin–streptomycin.
Mto1 heterozygous mice were generated by conventional homologous recombination to replace exons 1 and 2 of Mto1 with the neomycin-resistance cassette (37). Mouse ES cell clones were prepared by in vitro fertilization of male and female Mto1 heterozygous mice. The Animal Ethics Committee of Kumamoto University reviewed and approved all animal procedures (Approval ID: A27−037 R1). ES cells were cultured under feeder-free conditions in DMEM supplemented with 1% FBS (Hyclone), 15% knockout serum replacement (Invitrogen), CHIR99021 (WAKO Pure Chemical Industries) and PD184352 (WAKO Pure Chemical Industries).
Neonatal human dermal fibroblasts (NHDFs, Takara Bio) and patient fibroblasts (Pt751) were cultured as described (35). RNA analyses using these fibroblasts were approved by the ethics committee of the Saitama Medical University.
Construction of GTPBP3 KO cells
GTPBP3 KO cells were generated using the CRISPR/Cas9 system as described previously (13,38). In short, sense and antisense oligonucleotides encoding a single guide RNA (sgRNA) (Supplementary Table S1) were cloned into pX330 vector (Addgene plasmid 42230) (39). HEK293T cells seeded in 24-well plates were transfected using FuGENE HD (Promega) with 300 ng of the pX330 containing the designed sgRNA sequence, 100 ng of pEGFP-N1 (Clontech) and 100 ng of a modified pLL3.7 containing the puromycin-resistance gene. One day after transfection, cells were seeded at low density, and transfectants were selected with 1 μg/ml puromycin. Transfection efficiency was monitored by enhanced green fluorescent protein (EGFP) fluorescence. Eight days after transfection, several colonies were picked and further grown for several days. The target region of the genome in each clone was polymerase chain reaction (PCR)-amplified using primers listed in Supplementary Table S1 and sequenced.
Animal models
Animal experiments using domestic cats were approved by the Ethics Committee of Tokyo University of Agriculture and Technology (protocol 29–18). Domestic cats were obtained from Saitama Experimental Animals Supply Co., Ltd, Saitama, Japan. A custom-ordered taurine deficient cat diet was purchased from Bio-Serv, Inc. An individual cat fed the taurine-deficient diet was maintained for 6 months. Blood taurine concentration decreased from 169.7 to 42.9 nmol/ml in the first month, as described (40). At the end of the 6 months period, the animal was sacrificed and its organs were harvested. A healthy cat liver was obtained from a cat incidentally sacrificed due to a traffic accident.
For the flatfish experiment, the formulation of the taurine-deficient diet is described in Supplementary Table S2. All ingredients were mixed well, moistened by addition of tap water, formed into pellets using a mincer, freeze-dried and stored at −20°C until use. Japanese flounder P. olivaceus juveniles were purchased from a private hatchery (Nisshin Marinetech, Japan) and fed at the Nansei Laboratory, National Research Institute of Aquaculture. Initially, they were fed a commercial fishmeal-based flounder feed (Higashimaru, Japan) in a 500 l round tank with sand-filtered sea water. Then, 45 flatfish (mean weight, 16.3 g) were fed the taurine-deficient diet to apparent satiation four times a day. The water temperature was 21.7 ± 1.1°C (mean ± SD). Sixty days after the feeding, ten flatfish were anesthetized with 2-phenoxyethanol (200 ppm) and weighed (mean weight, 49.2 g). Control flatfish fed the commercial feed were taken from the 500 l tank. Individual muscles were dissected out, frozen in liquid nitrogen and stored at −80°C until analysis. Total RNA from isolated liver or muscle was prepared as described below.
RNA preparation and tRNA isolation
Total RNA from each cell or tissue was prepared using the TriPure Isolation Reagent (Roche Life Science). Individual mt-tRNAs were isolated by the reciprocal circulating chromatography method, as described (41). DNA probes for the target mt-tRNAs are listed in Supplementary Table S1. For nucleoside analysis, 450 μg of total RNAs extracted from HeLa cells cultured under taurine-depleted or taurine-restored conditions were subjected to an anion exchange resin (DEAE Sepharose Fast Flow, GE Healthcare) with low buffer [10 mM Hepes-KOH (pH 7.5), 250 mM NaCl and 2 mM dithiothreitol (DTT)] and small RNA fractions were eluted with high buffer [10 mM Hepes-KOH (pH 7.5), 1 M NaCl and 2 mM DTT].
Mass spectrometry
The isolated tRNAs were digested with RNase T1 (Ambion) and analyzed by capillary liquid chromatography (LC)/nano-electron spray ionization mass spectrometry (ESI-MS), as described (9,42,43). SI-labeled nucleoside analysis by LC/MS was performed as described (44,45). For nucleoside analysis of HeLa cells cultured under taurine-depleted condition, 5 μg of tRNA fraction was digested as follows: 10 μl reaction mixture consisting of 20 mM ammonium acetate (pH 5.3) and 0.01 units/μl nuclease P1 (Wako Pure Chemical Industries) was incubated at 37°C for 30 min, followed by adding 0.5 μl of 1 M ammonium bicarbonate (pH 8.0) and 0.25 units of phosphodiesterase I (Worthington Biochemical Corporation) and incubated at 37°C for 30 min, then 0.1 units of alkaline Phosphatase (Escherichia coli C75) (Takara Bio) was added and further incubated at 37°C for 30 min. The digests were subjected to hydrophilic interaction liquid chromatography (HILIC)/MS analysis as described (46). For detection of THF derivatives bound to Thermotoga maritima MnmE, 175 μg of recombinant protein was directly injected into the LC/MS under the same conditions used for the nucleoside analysis.
Northern blotting
Northern blotting analysis of mt-tRNAs was conducted essentially as described (43). The DNA probes used in this study are listed in Supplementary Table S1.
Oxygen consumption rate
An XF24 extracellular flux analyzer (Seahorse Bioscience) was used to measure oxygen consumption rate (OCR). Wild-type (WT) HEK293T and GTPBP3 KO cells were seeded in three wells each (3 × 105 cells per well) of an XF24 cell culture miniplate pre-coated with collagen, and then cultured for 24 h. The medium was then replaced with a solution consisting of 25 mM glucose and 1.25 mM pyruvic acid (adjusted to pH 7.4 with NaOH). The OCR of each well was measured over the course of programmed injections of oligomycin (f.c. 2.5 μg/ml), carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP, f.c. 1 μM) and rotenone/antimycin A (f.c. 2 μM/4 μM).
Respiratory complex activities
For isolation of mitochondrial fraction, about 1 mg of WT or GTPBP3 KO cells was homogenized in hypotonic buffer [80 mM sucrose and 10 mM 3-morpholinopropanesulfonic acid (MOPS) (pH 7.2)], followed by addition of an equal volume of hypertonic buffer [250 mM sucrose and 10 mM MOPS (pH 7.2)]. After centrifugation at 600 × g for 5 min, the supernatant was collected, homogenized again and centrifuged at 10 000 × g for 20 min. The precipitate was suspended in 100 μl of hypertonic buffer. Respiratory complex activities were measured as described (47). Absorbance at the indicated wave length was monitored on a UV–visible spectrophotometer V-630 equipped with a temperature control unit PAC-743R (JASCO).
Western blotting
The mitochondrial fraction was isolated from WT and GTPBP3 KO cells (1 × 107 cells) using the Mitochondria Isolation Kit (Miltenyi Biotec). Subunit proteins of mitochondrial respiratory chain complexes were detected by immunoblotting using Total OXPHOS Rodent WB Antibody Cocktail (1:250, ab110413, Abcam), anti-ND2 antibody (1:200, 19704–1-AP, Proteintech) and anti-mt-VDAC1 antibody (1:500, 10886–1-AP, Proteintech) as a control. HRP-conjugated anti-rabbit IgG (1:20000, 715–035-152, Jackson ImmunoResearch) was used as the secondary antibody.
Pulse-labeling of mitochondrial protein synthesis
The pulse-labeling experiment was performed essentially as described basically performed as described (13). WT and GTPBP3 KO cells (2.0 × 106 cells) were cultured at 37°C for 15 min in methionine/cysteine-free medium [D0422 (Sigma) with 10 mM HEPES-KOH (pH 7.5), 2 mM L-glutamine, 133 μM L-cysteine and 10% FBS] containing 50 μg/ml emetine to inhibit cytoplasmic protein synthesis, followed by addition of 7.4 MBq (0.2 mCi) of [35S]-methionine/[35S]-cysteine (EXPRE35S35S Protein Labeling Mix, [35S]-, PerkinElmer), and then cultured for 1 h to specifically label mitochondrial translation products. Cell lysates (100 μg of total proteins) were resolved by Tricine-sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (16.5%), and the gel was Coomassie brilliant blue (CBB)-stained and dried on a gel drier (AE-3750 RapiDry, ATTO). The radiolabeled mitochondrial protein products were visualized on an imaging plate (BAS-MS2040, Fujifilm) by a laser scanner imaging system (FLA-7000, Fujifilm).
Expression and isolation of the GTPBP3–MTO1 complex
The cDNAs of human GTPBP3 and MTO1 were cloned into pDEST12.2 (Invitrogen) and transferred to vector pEFh SBP vector (kindly provided by Dr Yamashita of Yokohama City University) (48) to construct expression vectors pEFh-GTPBP3 (without tag) and pEFh-MTO1-FLAG (C-terminal FLAG tag) using primers listed in Supplementary Table S1. The In-Fusion HD Cloning Kit (Clontech) was used for vector conversion.
For transient expression of GTPBP3 and MTO1, HEK293T cells (2.0 × 106) were transfected with 6 μg each of pEFh-GTPBP3 and pEFh-MTO1-FLAG using FuGENE HD (Promega), and then cultured for 48 h. The cells were harvested and suspended with 1 ml of lysis buffer [100 mM KCl, 10 mM Tris–HCl (pH8.0), 2.5 mM MgCl2, 1 mM DTT, 0.05% Triton X-100, 10% glycerol and 1× Complete EDTA-free protease inhibitor cocktail (Roche Life Science)] and lysed on ice by 20 passages through a 25G syringe needle. The lysate was centrifuged twice at 20 000 × g for 20 min to remove cell debris. The supernatant was pre-cleared by mixing with 30 μl of Mouse IgG-Agarose (Sigma), and immunoprecipitated with 30 μl of anti-FLAG beads (ANTI-FLAG M2 affinity gel, Sigma). The beads were washed four times with 1 ml of lysis buffer, and the GTPBP3–MTO1 complex was eluted with FLAG peptide in elution buffer [100 mM KCl, 10 mM Tris–HCl (pH 8.0), 2.5 mM MgCl2, 1 mM DTT, 0.05% Triton X-100, 10% glycerol and 250 μg/ml FLAG peptide]. To remove the FLAG peptide, the eluent was gel-filtered using Centri Sep spin columns (Princeton Separations) and concentrated using an Amicon Ultra-0.5 ml device [nominal molecular weight limit: 30 kDa]. The GTPBP3–MTO1 complex was quantified based on SDS-PAGE band intensity after staining with SYPRO Ruby (Thermo Fisher Scientific), using a BSA standard.
In vitro reconstitution of cmnm5U
MnmG (E. coli) and MnmE (E. coli and T. maritima) were cloned into pET28b (Novagen) to construct expression vectors for N-terminally His-tagged proteins. Expression and purification of the recombinant proteins were performed basically as described (49). Purified recombinant proteins were dialyzed and stored in 20 mM Tris–HCl (pH 8.0), 100 mM NaCl, 7 mM β-mercaptoethanol (for MnmG) or 10 mM Tris–HCl (pH 8.0), 50 mM NaCl, 1 mM DTT for (E. coli MnmE) or 10 mM Tris–HCl (pH 8.0), 50 mM NaCl, 5 mM MgCl2 and 1 mM DTT (for T. maritima MnmE).
The DNA template for in vitro transcription of tRNA was prepared using a set of synthetic DNAs shown in Supplementary Table S1. Escherichia coli tRNAGlyUCC was transcribed by T7 RNA polymerase essentially as described (50). The transcripts were gel-purified by 10% denaturing PAGE.
For in vitro reconstitution of cmnm5U, a reaction mixture (20 μl) consisting of 50 mM Tris–HCl (pH 8.0), 10 mM MgCl2, 5 mM DTT, 100 mM KCl, 2.5 mM FAD, 2.5 mM GTP, 2.5 mM NADH, 2 mM 5,10-CH2-THF (or 5-formyl-THF), 2.5 mM glycine (or taurine), 0.2 μM E. coli tRNAGlyUCC transcript and 10 μM each recombinant E. coli MnmG and MnmE was incubated at 37°C for 1.5 h. As controls, the same reaction was performed in the presence of THF or 5-formyl-THF instead of 5,10-CH2-THF, or without the enzymes. After the reaction, the tRNA was recovered using the QIAquick nucleotide removal kit (QIAGEN) and dialyzed for 1 h on a 0.025 μm VSWP filter (Millipore) floating on Milli-Q water. Modification frequency was measured by mass spectrometric analysis as described above.
In vitro reconstitution of τm5U
The reaction mixture (20 μl) consisting of 50 mM Tris–HCl (pH8.0), 10 mM MgCl2, 5 mM DTT, 100 mM KCl, 2.5 mM FAD, 1 mM ATP, 2.5 mM NADH, 2.5 mM NADPH, 2 mM 5,10-CH2-THF and 0.19 μM hypomodified mt-tRNALeu(UUR) (isolated from GTPBP3 KO cells), 2.5 mM GTP, 2.5 mM taurine and 2 μM GTPBP3–MTO1 complex was incubated at 37°C for 1 h. As negative controls, the same reaction was performed without GTP and taurine, or without enzyme complex. After the reaction, the mt-tRNALeu(UUR) was extracted from the mixture by phenol–chloroform treatment, cleaned up by with Q Sepharose High Performance (GE Healthcare), and recovered by two rounds of ethanol precipitation. Modification frequency was measured by mass spectrometric analysis as described above.
RESULTS
Identification of the metabolic sources of τm5U
To identify metabolic substrates for the 5-taurinomethyl group of τm5U in five mt-tRNAs (Figure 1A), we carried out metabolic labeling experiments in human cell culture. As illustrated in Figure 1B, HeLa cells were cultured in a defined medium with or without SI-labeled metabolite to label τm5U in mt-tRNAs. Total RNA was extracted from the cells, and mt-tRNAs were isolated and digested to nucleosides, which were then analyzed by LC/MS to detect the labeled τm5U with increased molecular mass derived from the SI-labeled metabolite.
As reported previously (17), when cells were cultured with [18O]-labeled taurine, we observed a proton adduct of τm5U nucleoside in mt-tRNATrp with molecular mass (m/z 386) 4-Da larger than that of the natural nucleoside (m/z 382), confirming that the sulfonic acid moiety of the labeled taurine was incorporated in τm5U (Figure 1C). To demonstrate that the entire molecule of taurine was incorporated into τm5U, the cells were cultured with [15N] taurine. We detected τm5U with molecular mass (m/z 383) 1 Da larger than the natural nucleoside (Figure 1C), demonstrating that free taurine is a direct metabolic substrate for τm5U biogenesis.
We still needed to determine the carbon source of the methylene group of τm5U. In bacteria, yeast and nematode mitochondria, cmnm5U (see Figure 6F) is present at the wobble position in tRNAs. cmnm5U is a modified uridine in which the taurine moiety of τm5U is replaced with Gly. Genetic and biochemical studies revealed that MnmE and MnmG employ glycine and one-carbon (1C) folate derivatives as substrates, and catalyze cmnm5U formation in the presence of essential co-factors including GTP, K+ and FAD (51–53). Given that GTPBP3 and MTO1 are mammalian homologs of bacterial MnmE and MnmG, respectively, we speculated that the methylene group of τm5U comes from a folate derivative in 1C metabolism. To determine which folate derivative is actually involved in cmnm5U formation, we conducted LC/MS analysis of THF derivatives bound to T. maritima MnmE recombinantly expressed in E. coli. We detected 5,10-methylene(CH2)-THF, along with THF, but no formyl-THF, indicating that 5,10-CH2-THF is a bona fide substrate for cmnm5U formation (Supplementary Figure S1A and B). We then reconstituted cmnm5U on E. coli tRNA with recombinant MnmE and MnmG in the presence of a series of folate derivatives and Gly, and found that 5,10-CH2-THF served as a substrate for cmnm5U formation (Supplementary Figure S1C).
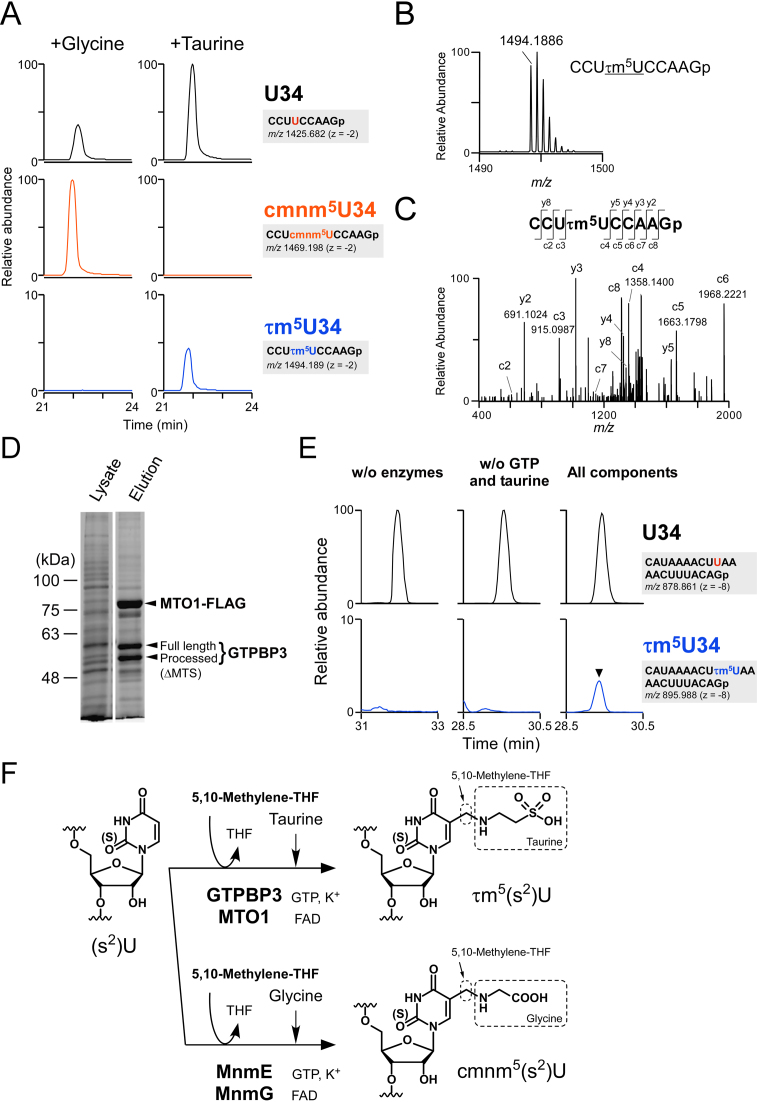
Figure 6.
In vitro reconstitution of τm5U34. (A) In vitro reconstitution of cmnm5U34 and τm5U34 on E. coli tRNAGly(UCC) in the presence of Gly (left panels) or taurine (right panels) along with other substrates and co-factors. XICs of anticodon-containing RNase T1-digested fragments with U34 (top panels), cmnm5U34 (middle panels) and τm5U34 (bottom panels) of E. coli tRNAGly(UCC) after the reaction are shown. Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right. (B) Mass spectrum of the anticodon-containing RNase T1-digested fragment of E. coli tRNAGly(UCC) containing τm5U34, reconstituted in vitro. The exact mass of the mono-isotopic ion is indicated. (C) CID spectrum of the above fragment with τm5U34. Assigned c- and y-product ions are indicated. (D) Co-immunoprecipitation of GTPBP3 and MTO1-FLAG complex. Both proteins were transiently co-expressed in HEK293T cells, pulled down with anti-FLAG antibody–agarose and eluted with FLAG peptide. Whole lysate and eluate were resolved by SDS-PAGE and stained with a fluorescent dye. MTS stands for mitochondrial targeting sequence. (E) In vitro reconstitution of τm5U on mt-tRNALeu(UUR) without enzymes (left panels), without GTP and taurine (middle panels), or with all components including substrates and co-factors (right panels). XICs of anticodon-containing RNase T1-digested fragments with U34 (top panels) and τm5U34 (bottom panels) of mt-tRNALeu(UUR) after the reaction. Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right. (F) Scheme of τm5U and cmnm5U biosynthesis. τm5U is synthesized by GTPBP3/MTO1 complex using 5,10-CH2-THF and taurine as substrates while glycine is used in place of taurine for cmnm5U by MnmE–MnmG complex. Co-factors including GTP, FAD and potassium ion are essential to cmnm5U formation, suggesting that they are also required for τm5U.
To identify the carbon source for the methylene group in τm5U biogenesis, we cultured HeLa cells in a medium supplemented with SI-labeled Gly or Ser, because the α-carbon of Gly or β-carbon of Ser is a source of 1C folate derivative (Figure 1B). Then, mt-tRNATrp and mt-tRNALeu(UUR) were isolated from each culture and subjected to nucleoside analysis by LC/MS (Figure 1D). When the cells were cultured with the SI-labeled Ser, we clearly detected a 1 Da larger species (m/z 383) of τm5U, which we then further probed by collision-induced dissociation (CID). Based on assignment of the product ions generated by CID (Figure 1E), the 13C atom derived from SI-labeled Ser resided in the product ion composed of uracil base and C5-carbon. Because uridines and pseudouridines contained no 13C derived from the SI-labeled Ser (data not shown), the 13C atom was attributed to the methylene group of τm5U. On the other hand, when the cells were cultured with SI-labeled Gly, no carbon derived from this amino acid could be detected in τm5U (Figure 1D). This result demonstrated that the β-carbon of Ser is a metabolic source of the methylene group of τm5U, via a 1C folate derivative that is presumably 5,10-CH2-THF.
Requirement of 1C metabolism in τm5U biosynthesis
In 1C metabolism in mammalian mitochondria, 5,10-CH2-THF is generated via two independent pathways mediated by SHMT2 and the glycine cleavage system (GCS) (see Figure 7). SHMT2 catabolizes Ser to Gly and transfers the β-carbon of Ser as a 1C unit to 5,10-CH2-THF, whereas GCS digests Gly into CO2 and NH3 and provides the α-carbon of Gly as a 1C unit to 5,10-CH2-THF. Judging from our metabolic labeling experiment, SHMT2-mediated 5,10-CH2-THF formation is a major mitochondrial pathway in HeLa cells, as in other cancer cells (54). To confirm whether 1C metabolism is actually involved in τm5U formation, we analyzed τm5U status in CHO glyA and glyB mutant cell lines, which exhibit glycine auxotrophy due to reduced Shmt2 and Mft activities, respectively (36,55,56). The mt-tRNALeu(UUR) was isolated from WT and mutant CHO cells, digested with RNase T1 and subjected to RNA–MS to analyze τm5U status (Figure 1F). In the WT cells, about 80% of mt-tRNALeu(UUR) contained τm5U, whereas in Shmt2 mutant cells, the τm5U frequency dropped sharply to 9%, demonstrating that loss of 5,10-CH2-THF due to impairment of SHMT2 activity results in τm5U deficiency. In addition, we observed hypomodification of τm5U (48%) in the Mft mutant strain, indicating that folate transport to mitochondria is required for efficient τm5U formation. Taken together, these observations indicate that τm5U biogenesis in mammalian cells requires 5,10-CH2-THF generated by SHMT2 in mitochondrial 1C metabolism.
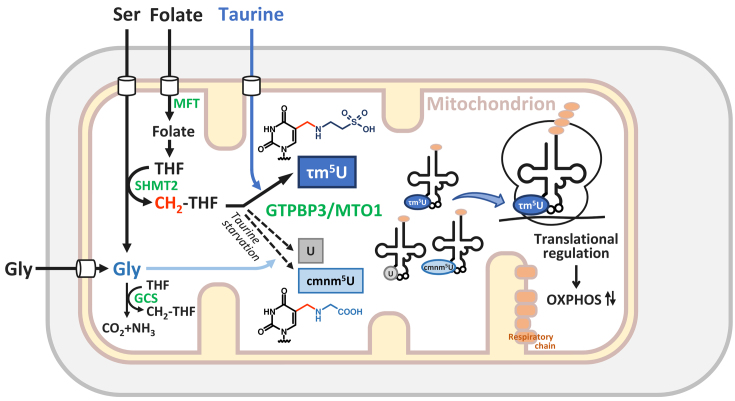
Figure 7.
Metabolic regulation and chemical switching of tRNA modification in mitochondria. Schematic depiction of 1C metabolism, τm5U biogenesis and potential translational regulation in mitochondria. τm5U is biosynthesized from CH2-THF and taurine catalyzed by GTPBP3–MTO1 complex. τm5U in mt-tRNAs is required for efficient mitochondrial translation to synthesize the respiratory chain complexes essential for OXPHOS on the mitochondrial inner membrane. Taurine starvation decreases τm5U frequency and increases unmodified U. In this condition, cmnm5U, in which the taurine moiety of τm5U is replaced with glycine, is also introduced in mt-tRNAs. The tRNA modification dynamically regulated in response to metabolic status implies translational regulation in a codon-specific manner under physiological conditions. Abbreviations are as follows: THF, tetrahydrofolate; SHMT2, serine hydroxymethyltransferase 2 (mitochondrial SHMT); MFT, mitochondrial folate transporter; GCS, glycine cleavage system.
Taurine starvation downregulates τm5(s2)U in HeLa cells and animal tissues
Taurine is an essential nutrient for carnivores, including cat and fox, which have little endogenous taurine biogenesis activity. In these animals, deficiency of dietary taurine causes developmental defects, central retinal degradation, hepatic lipidosis and dilated cardiomyopathy (57,58), which are also major manifestations of human mitochondrial encephalomyopathies (59). In humans and possibly primates, infants and young children have low capacity for taurine biosynthesis, so dietary taurine is essential for normal development (60).
These facts prompted us to investigate whether τm5U frequency could be affected by extracellular taurine concentration. In this experiment, HeLa cells were cultured in a medium containing dialyzed FBS, which does not contain taurine and other small compounds, and mt-tRNAs were isolated for analysis of τm5U status by RNA–MS. In HeLa cells cultured in a normal medium, the frequency of τm5s2U34 (and τm5U34) in mt-tRNALys was 45% (Figure 2A and Supplementary Table S3). When the cells were cultured in a medium with dialyzed FBS, the τm5U frequency in mt-tRNALys dropped to 17%. When 40 mM taurine was added back to medium containing dialyzed FBS, the τm5U frequency in mt-tRNALys was restored to 56%. This result was reproducible (Supplementary Figure S2 and Supplementary Table S3). We also analyzed the other four mt-tRNAs isolated from cells cultured with dialyzed FBS, and found that the τm5U frequency was significantly reduced in each of them (Supplementary Figure S3 and Supplementary Table S3). These results indicate that the τm5U frequency in mt-tRNAs is dynamically regulated by extracellular taurine concentration.
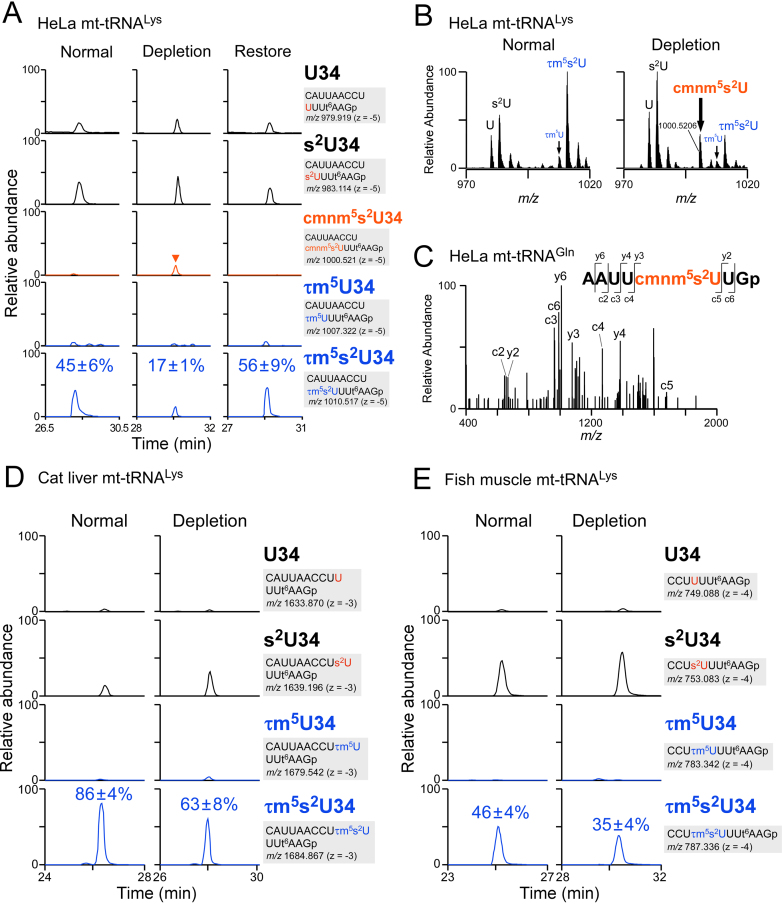
Figure 2.
Downregulation of τm5(s2)U in culture cells and animal tissues upon taurine starvation. (A) XICs of anticodon-containing RNase T1-digested fragments with different wobble modifications of mt-tRNALys isolated from HeLa cells cultured in normal (left panels), taurine-depleted (center panels) and taurine-restoring (right panels) conditions. Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right. Relative τm5(s2)U frequencies indicated in the bottom chromatograms are averaged values with standard deviations, calculated from the peak height ratio of the multiply charged negative ions (−3 to −7) for RNA fragments with different modifications. s2U, 2-thiouridine; t6A, N6-threonylcarbamoyladenosine. (B) Averaged mass spectra of the anticodon-containing fragments with different wobble modifications of mt-tRNALys isolated from HeLa cells cultured in normal (left panel) and taurine-depleted (right panel) conditions. (C) CID spectrum of the anticodon-containing RNase T1-digested fragment bearing cmnm5s2U of mt-tRNAGln isolated from HeLa cells cultured under taurine-depleted conditions. The c- and y-series of the product ions are assigned on the sequence. (D) XICs of anticodon-containing RNase T1-digested fragments with different wobble modifications of mt-tRNALys, isolated from livers of cats fed a normal diet (left panels) or a taurine-depleted diet (right panels). Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right. Relative τm5(s2)U frequencies indicated in the bottom chromatograms are averaged values with standard deviations, calculated from the peak height ratio of the multiply charged negative ions (−3 to −7) for RNA fragments with different modifications. (E) XICs of anticodon-containing RNase T1-digested fragments with different wobble modifications of mt-tRNALys, isolated from muscles of flatfish fed a normal diet (left panels) or a taurine-depleted diet (right panels). Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right. Relative τm5(s2)U frequencies indicated in the bottom chromatograms are averaged values with standard deviations, calculated from the peak height ratio of the multiply charged negative ions (−2 to −4) for RNA fragments with different modifications.
Upon taurine depletion, we happened to detect 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U), in which the taurine moiety of τm5s2U is replaced with Gly in mt-tRNALys (Figure 2A and B; Supplementary Figure S2). We also detected cmnm5U in mt-tRNAs for Leu(UUR) and Trp (Supplementary Figures S3A,B and S4A,B), and cmnm5s2U in mt-tRNAs for Gln and Glu (Supplementary Figures S3A,B and S4A,B), only in cells cultured under taurine-depleted conditions. We confirmed presence of cmnm5U (m/z 332) and cmnm5s2U (m/z 348) by conducting LC/MS nucleoside analyses of total RNAs extracted from HeLa cells cultured under taurine-depleted condition (Supplementary Figure S5). When taurine was restored to the medium, cmnm5(s2)U completely disappeared, while τm5(s2)U increased clearly. As a negative control, no signal of cmnm5(s2)U as well as τm5(s2)U was detected in GTPBP3 knockout cells (see below), indicating that cmnm5(s2)U are synthesized by the same pathway of τm5(s2)U biogenesis. Sequence analysis of the anticodon-containing fragment by CID revealed that cmnm5s2U was present at position 34 (Figure 2C). Judging from the mass-chromatographic intensities, 5–23% of mt-tRNAs contained cmnm5U or cmnm5s2U (Figure 2A; Supplementary Figures S2 and S3). This result suggests that cmnm5(s2)U34 is synthesized by utilizing Gly instead of taurine under taurine-depleted conditions.
Given that cats do not have the biosynthetic pathway for taurine, we asked whether the τm5U frequency in cat mt-tRNAs could be regulated by taurine starvation. For this purpose, we raised two domestic cats (Felis catus), fed either a control or taurine-deficient diet. mt-tRNALys was isolated from livers of those cats and subjected to RNA–MS for analysis of τm5U status, which revealed that the frequencies of τm5s2U34 (and τm5U34) in mt-tRNAsLys were 86% and 63% in the the control and taurine-deprived animals, respectively (Figure 2D and Supplementary Table S3). Thus, the τm5U frequency in cat liver was slightly but significantly decreased upon taurine depletion.
The Japanese flounder P. olivaceus has little taurine biogenesis activity. Accordingly, in aquaculture production, taurine promotes growth of the juvenile flatfish (61,62). To examine the effect of dietary taurine level on τm5U frequency, we cultivated P. olivaceus for 2 months on a control or taurine-deficient diet. mt-tRNALys was isolated from the flatfish muscle tissues and analyzed by RNA-MS. The τm5s2U frequencies of mt-tRNAsLys isolated from the flatfish cultivated on the control and taurine-deficient diets were 46% and 35%, respectively (Figure 2E and Supplementary Table S3). Thus, dietary taurine regulates τm5U frequency in flatfish mt-tRNAs.
GTPBP3 and MTO1 are essential for τm5U biogenesis
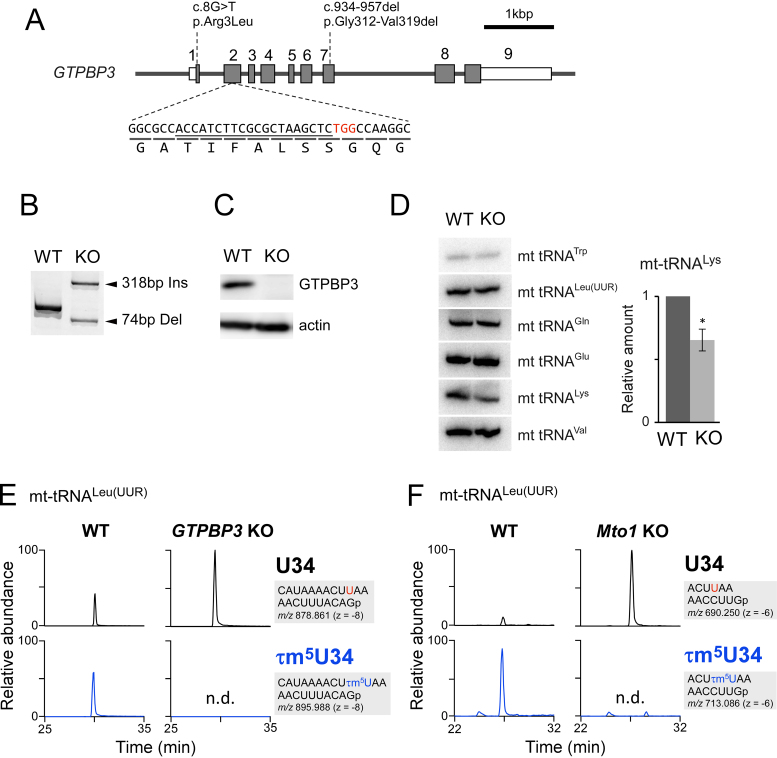
Based on our previous finding that MSS1 and MTO1 are responsible for cmnm5U formation of mt-tRNAs in yeast mitochondria (26), we investigated the involvement of their human homologs, GTPBP3 and MTO1, in τm5U biosynthesis. First, we knocked out the GTPBP3 gene in HEK293T cells using the CRISPR–Cas9 system (Figure 3A), and obtained a knockout cell line in which both alleles harbor indel mutations (Figure 3B). Endogenous GTPBP3 was not detectable by western blotting in GTPBP3 KO cells (Figure 3C). We then performed northern blotting to compare the steady-state levels of six mt-tRNAs, including five τm5U34-containing tRNAs, between WT HEK293T and GTPBP3 KO cells (Figure 3D). The results revealed that the level of mt-tRNALys decreased moderately, whereas the levels of the other five tRNAs were unchanged. We then isolated five τm5U-containing mt-tRNAs from WT and KO cells, digested them with RNase T1 and analyzed the RNA fragments by RNA-MS. The τm5(s2)U34-containing fragments detected in mt-tRNAs isolated from the WT cells completely disappeared, and were converted to the U34 or s2U34-containing fragments, in GTPBP3 KO cells (Figure 3E and Supplementary Figure S6).
Figure 3.
GTPBP3 and MTO1 are essential for τm5U biogenesis. (A) A diagram of the human GTPBP3 gene and the sites of mutations introduced by the CRISPR–Cas9 system. Shaded boxes indicate coding regions, open boxes indicate untranslated regions of exons and lines indicate introns. Heterozygous mutations found in the patient with mitochondrial disease are indicated above the diagram (34,35). The sgRNA sequence is underlined; the PAM sequence is in red. Reading frame and translated amino acids are shown below the sequence. (B) Genomic PCR products covering the sgRNA target site from WT HEK293T and GTPBP3 KO cells. One allele of the KO cell has a 318 bp insertion, and the other has a 74 bp deletion. (C) Western blotting of GTPBP3 and actin proteins in WT and KO cells. (D) Steady-state levels of six mt-tRNAs from WT and KO cells, evaluated by northern blotting. The bar graph shows relative amounts of mt-tRNALys from WT and KO cells, quantified in three independent replicates. Error bars indicate standard deviation. *p = 0.02, Student’s paired t-test. (E) XICs of anticodon-containing RNase T1-digested fragments with U34 (upper panels) and τm5U34 (lower panels) of mt tRNALeu(UUR), isolated from WT (left panels) and GTPBP3 KO cells (right panels). Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right. (F) XICs of anticodon-containing RNase T1-digested fragments with U34 (upper panels) and τm5U34 (lower panels) of mt tRNALeu(UUR), isolated from WT (left panels) and Mto1 KO mES cells (right panels). Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right.
Next, to examine the role of MTO1, we obtained Mto1 KO ES cells and isolated five mt-tRNAs from the cells. RNA–MS analysis revealed that all five mt-tRNAs isolated from the Mto1 KO ES cells lacked τm5(s2)U34 and remained unmodified at this position (Figure 3F and Supplementary Figure S7). Together, these results clearly demonstrated that GTPBP3 and MTO1 are responsible for τm5U34 formation of five mt-tRNAs in mammalian cells.
Mitochondrial dysfunction in GTPBP3 KO cells
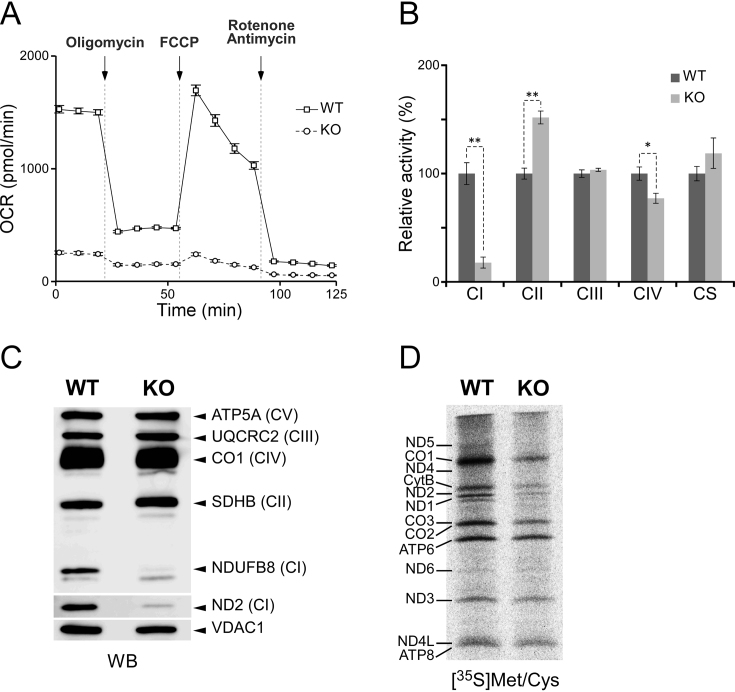
We then assessed mitochondrial function in GTPBP3 KO cells. The OCR of the KO cells, measured using a flux analyzer, was significantly lower than that of WT cells (Figure 4A). Adding of an uncoupler, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone, barely increased OCR in the KO cells, suggesting that knockout of GTPBP3 results in loss of electron transfer activity. We then compared the biochemical activities of respiratory complexes between GTPBP3 KO and WT cells (Figure 4B). In GTPBP3 KO cells, we observed severe reduction in complex I, mild reduction in complex IV and no significant change in complex III. On the other hand, the activity of complex II, all of whose components are encoded in the nuclear genome, was elevated in GTPBP3 KO cells, probably to compensate for the reduction in complex I activity. We then analyzed steady-state levels of protein components in the respiratory complexes by western blotting (Figure 4C), and found severe reductions in the levels of ND2 and NDUFB8, both of which are subunit proteins in complex I, indicating defective assembly of complex I in GTPBP3 KO cells.
Figure 4.
Mitochondrial dysfunction in GTPBP3 KO cells. (A) Oxygen consumption rate in HEK293T WT (square) and GTPBP3 KO (circle) cells, measured using a flux analyzer. Each plot represents the mean value of three independent samples. Error bars indicate standard deviations. (B) Relative activities of respiratory chain complexes I–IV and citrate synthase of WT (black bar) and GTPBP3 KO (gray bar) cells. Error bars indicate standard deviation. *p < 0.05, **p< 0.01, Student’s t-test. (C) Western blotting of subunit proteins (indicated on the right) of respiratory chain complexes in isolated mitochondria from WT and GTPBP3 KO cells. The CBB-stained gel images are shown in Supplementary Figure S8A. (D) Pulse-labeling of mitochondrial protein synthesis. WT and GTPBP3 KO cells were labeled with [35S] Met and [35S] Cys after cytoplasmic protein synthesis was halted by emetine. Assignment of mitochondrial proteins is indicated on the left. The CBB-stained gel images are shown in Supplementary Figure S8B.
To evaluate mitochondrial protein synthesis, we conducted a pulse-labeling experiment (Figure 4D). Cytoplasmic translation was inhibited by addition of emetine, and mitochondrial translation products were labeled with [35S] Met and Cys. Mitochondrial protein synthesis was drastically lower in GTPBP3 KO cells than in WT cells; in particular, the levels of CO1 and ND2 were markedly reduced in GTPBP3 KO cells. Given that τm5(s2)U34 in mt-tRNAs plays a critical role in the decoding process of mitochondrial translation, these phenotypes can be explained by the hypomodification of mt-tRNAs in GTPBP3 KO cells. Reduced activity of complex I in GTPBP3 KO cells explains complex I deficiency as a clinical symptom of patients harboring pathogenic mutations of the GTPBP3 gene (34,35) and mtDNA mutations associated with MELAS (63).
Hypomodification of the taurine modification in a GTPBP3 patient
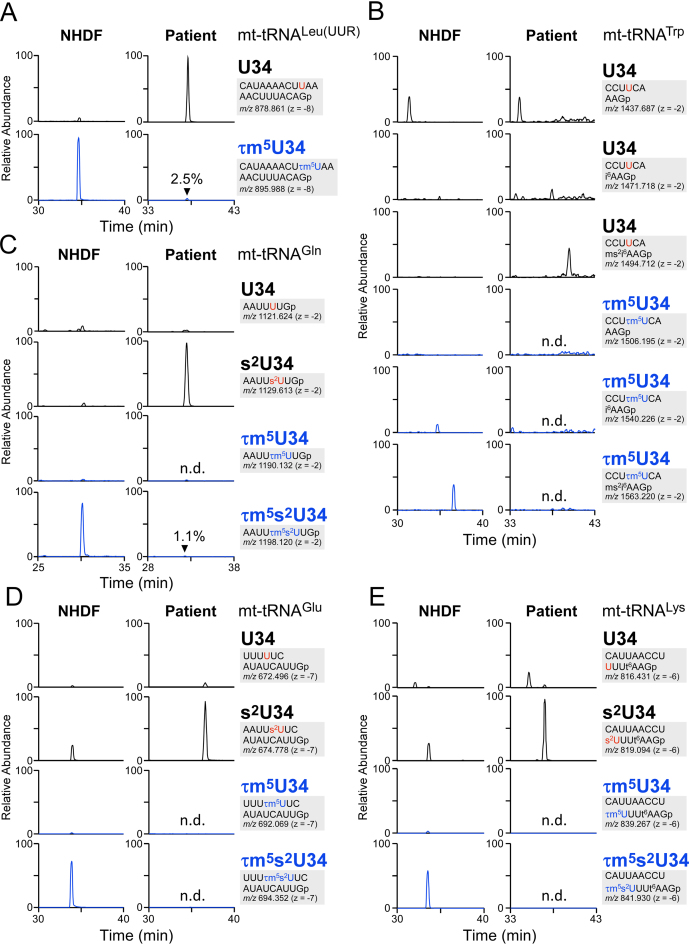
To determine whether pathogenic mutations in the GTPBP3 gene actually impair τm5(s2)U34 in mt-tRNAs, we isolated five mt-tRNAs from the fibroblasts of a 2-year-old female patient harboring pathogenic mutations in GTPBP3 genes (Figure 3A) (35), and analyzed τm5(s2)U status by RNA–MS (Figure 5). No τm5(s2)U34 was detected in three mt-tRNAs (for Glu, Trp and Lys), whereas the frequencies of τm5(s2)U in mt-tRNALeu(UUR) and mt-tRNAGln were 2.5% and 1.1%, respectively, indicating that most GTPBP3 enzymatic activity was lost due to the pathogenic mutations. This phenomenon could represent a primary cause of disease.
Figure 5.
Hypomodification of the taurine modification in a patient with pathogenic mutations in GTPBP3. XICs of anticodon-containing RNase T1-digested fragments with different modifications of mt-tRNAs for Leu(UUR) (A), Trp (B), Gln (C), Glu (D) and Lys (E), isolated from neonatal human dermal fibroblast (NHDF) (left panels) and patient fibroblasts (right panels). Sequence of the detected fragment, with its m/z value and charge state, is indicated on the right. Relative frequencies of τm5(s2)U in the patient sample are indicated in the bottom chromatograms. n.d., not detected. ms2i6A, 2-methylthio-N6-isopentenyladenosine; i6A, N6-isopentenyladenosine.
In vitro reconstitution of τm5U
Although pathogenic point mutations in mt-tRNAs associated with MELAS and MERRF might act as negative determinants that prevent τm5(s2)U34 formation mediated by GTPBP3 and MTO1, no direct evidence confirming this prediction has been provided to date. In vitro reconstitution of τm5U34 formation would help us to directly examine the effect of these pathogenic mutations on τm5U34 formation.
Given that cmnm5U is a structural homolog of τm5U in which the taurine moiety is replaced with glycine, we first tried to reconstitute τm5U on E. coli tRNA using recombinant E. coli MnmE and MnmG in the presence of 5,10-CH2-THF and taurine as substrates. The products were digested with RNase T1 and analyzed by RNA–MS. In a positive control reaction, cmnm5U was efficiently reconstituted in the presence of glycine (Figure 6A and Supplementary Figure S1C). When taurine was added instead of glycine to the reaction mixture, a small amount of τm5U was introduced into the tRNA (Figure 6A). The exact mass of the fragment containing τm5U was consistent with the calculated m/z value (m/z 1494.1889, z = −2), with an error rate of 0.2 ppm (Figure 6B). This fragment was further probed by CID to determine the position of τm5U reconstituted in vitro. The data confirmed that τm5U resided at the wobble position, as determined by assignment of the product ions of CID (Figure 6C). This result demonstrated that E. coli MnmE and MnmG have a limited ability to synthesize τm5U on E. coli tRNA.
Next, we tried to reconstitute τm5U using human GTPBP3 and MTO1. Because yeast Mss1p and Mto1p formed a stable complex in mitochondria (64), we first examined the direct interaction between GTPBP3 and MTO1 in the cell. For this purpose, C-terminally FLAG-tagged MTO1 (MTO1-FLAG) and GTPBP3 without tag were transiently expressed in HEK293T cells. MTO1-FLAG was immunoprecipitated with anti-FLAG antibody, and the precipitants were resolved by SDS-PAGE. As shown in Figure 6D, GTPBP3 was co-precipitated with MTO1. The processed form of GTPBP3 lacking the N-terminal mitochondrial targeting sequence (MTS) was detected, indicating that the MTS is cleaved off by a mitochondrial processing protease after import into the mitochondria. Both proteins are predominantly localized to mitochondria (65,66), and they form a stable complex involved in τm5U34 formation. Because tRNA modifications other than τm5U34 might be required for efficient τm5U34 formation, as a substrate, we prepared mt-tRNALeu(UUR) lacking τm5U34 isolated from GTPBP3 KO cells. Based on our finding that 5,10-CH2-THF is employed as a substrate employed by MnmE (Supplementary Figure S1C), we conducted in vitro reconstitution of τm5U34 on mt-tRNALeu(UUR) with the GTPBP3–MTO1-FLAG complex in the presence of taurine, 5,10-CH2-THF and co-factors including GTP, FAD and potassium ion. In negative control reactions, no product was formed without the enzyme complex or in the absence of GTP and taurine (Figure 6E). In the presence of all components, the τm5U34-containing fragment was clearly detected (Figure 6E). Judging from the ratio of modified (τm5U34) versus unmodified (U34) fragments, 3.3% of mt-tRNALeu(UUR) was modified. The observed m/z value of the τm5U-containing fragment (m/z 895.9884, z = −8) was nearly identical to the calculated one (m/z 895.9881, z = −8), with a small error of 0.33 ppm, suggested that τm5U was successfully reconstituted in this reaction (Figure 6F). This is the first demonstration that GTPBP3–MTO1 complex can form τm5U34 on mt-tRNA.
DISCUSSION
In this study, a metabolic labeling experiment revealed that the taurinomethyl group of τm5U34 is synthesized by taurine and the β-carbon of Ser which is a carbon source for 1C metabolism mediated by the folate pathway (Figure 1D). τm5U was down-regulated in glycine auxotroph CHO cells bearing mutations in Shmt2 and Mft (Figure 1F), indicating that 5,10-CH2-THF and mitochondrial folate metabolism are involved in τm5U formation. We clearly showed that cmnm5U on tRNA could be efficiently reconstituted by bacterial MnmE and MnmG utilizing 5,10-CH2-THF and Gly as substrates (Supplementary Figure S1C and Figure 6A). Moreover, we directly detected 5,10-CH2-THF bound to T. maritima MnmE (Supplementary Figure S1A). Based on these results, we conclude that 5,10-CH2-THF is a bona fide substrate for τm5U formation. In fact, we successfully reconstituted a small amount of τm5U on mt-tRNALeu(UUR)in vitro using the GTPBP3/MTO1 complex in the presence of taurine, 5,10-CH2-THF and co-factors (Figure 6E).
In mitochondria, SHMT2 digests Ser into Gly, and transfers the β-carbon of Ser to THF to synthesize 5,10-CH2-THF (Figure 7). SHMT2 is involved in cellular growth regulation by producing Gly for de novo nucleotide synthesis (54). The 1C carbon of 5,10-CH2-THF is redundantly derived from the β-carbon of Ser mediated by SHMT2 or the α-carbon of Gly mediated by the GCS pathway. Both pathways are active in stem cells, whereas little GCS pathway activity can be detected in most cancer cells, indicating that 5,10-CH2-THF is mainly produced via the SHMT2 pathway in cancer cells (54). Indeed, SHMT2 expression is elevated in various cancer cells, and high expression defines poor-prognosis subtypes (67). Becasue downregulation of SHMT2 suppresses tumorigenesis in human hepatocellular carcinoma (68), SHMT2 is regarded to be a potential therapeutic target of cancers. Moreover, SHMT2 plays a critical role in activation and survival of T cells by stimulating mitochondrial 1C metabolism (69). Intriguingly, the mitochondrial respiratory defect in elderly humans can be partly explained by down regulation of SHMT2 (70). Consistent with this, the OCR was decreased by SHMT2 knockdown. Considering the fact that hypomodification of τm5U was observed in Shmt2 mutant CHO cells in this study (Figure 1F), it is likely that age-associated downregulation of SHMT2 decreases the frequency of τm5U, leading to mitochondrial dysfunction and respiratory defects.
Taurine is one of the most abundant amino acids, and ubiquitously distributed in various organs and tissues in mammals. Taurine is involved in a wide variety of cellular functions, including bile salt synthesis and cholesterol metabolism, antioxidation, osmoregulation and modulation of neuronal excitability (71,72). Mammals not only take up taurine from the diet but also synthesize it de novo from cysteine. Carnivores like cat and fox do not synthesize taurine de novo due to low activity of cysteine sulfinate decarboxylase, and therefore must acquire taurine through the diet. Taurine deficiency in cats leads to dilated cardiomyopathy and blindness due to retina failure (57,73). Because human infants have low de novo taurine synthesis activity, dietary taurine is essential for normal human development (60). Juvenile flatfish do not synthesize taurine de novo, and therefore require it as a nutrient for rapid growth in aquaculture production (61,74). We clearly showed that extracellular taurine is incorporated to the cells and used as a direct metabolic substrate for τm5U in mt-tRNAs (17). The lack of τm5U34 results in a decoding deficiency of mt-tRNAs and decreases the rate of mitochondrial translation (75). Thus, if τm5U frequency is modulated by intracellular taurine concentration, taurine depletion could downregulate the corresponding mt-tRNA modification, resulting in impaired mitochondrial translation and respiratory activity. In this study, we observed a marked reduction in the τm5(s2)U frequency in mt-tRNAs isolated from HeLa cells cultured under taurine-depleted conditions (Figure 2A; Supplementary Figures S2 and S3). When taurine was added back to the medium, the τm5(s2)U frequency was restored (Figure 2A and Supplementary Figure S2). These results demonstrated that τm5(s2)U frequency can be dynamically regulated by cellular taurine concentration, indicating that taurine supplied from the extracellular environment is necessary for maintaining a high frequency of τm5(s2)U, even though human cells can biosynthesize taurine from cysteine de novo. Downregulation of τm5(s2)U was also observed in animal models under taurine-depleted conditions. Specifically, we observed a reduction in the τm5(s2)U frequency in mt-tRNAs isolated from the livers of cats fed a taurine-deficient diet (Figure 2D). This result revealed the physiological importance of taurine, which is key to maintaining a high τm5(s2)U frequency in cat. Moreover, we observed that the level of τm5(s2)U decreased in mt-tRNAs isolated from flatfish cultivated on taurine-deficient diets for 2 months (Figure 2E). Although the reduction rate of τm5(s2)U was not large in either animal model, mitochondrial translation could nonetheless be affected by alteration of the τm5(s2)U frequency following taurine depletion. These findings suggest that tRNA modification is dynamically regulated by sensing of cellular metabolic status, challenging accepted notions that tRNA modifications are stable, static and rarely regulated.
Taurine plays a critical role in mitochondrial translation and respiratory activity by maintaining τm5(s2)U in mt-tRNAs, and at least one other physiological connection between taurine and mitochondria has been reported. Taurine-upregulated gene 1 (Tug1), a long non-coding RNA induced by taurine (76), physically interacts with PGC1α, a master transcription factor involved in mitochondrial biogenesis and facilitates the binding of PGC1α to its own promoter, leading to activation of mitochondrial biogenesis in mouse kidney (77). Hence, taurine synchronizes mitochondrial protein synthesis by maintaining τm5(s2)U34 and mitochondrial biogenesis via PGC1α activation.
cmnm5U, a structural homolog of τm5U in which taurine is replaced with Gly, is used as the wobble modification in bacterial tRNAs, as well as in yeast and nematode mt-tRNAs (26,78), suggesting that taurine took over for Gly as s structural component of the wobble modification over the course of animal mitochondrial evolution. Intriguingly, we happened to detect cmnm5(s2)U modification of mt-tRNAs in HeLa cells cultured under taurine-depleted conditions (Figure 2A, B and C; Supplementary Figures S2, S3 and S4): this is the first observation of cmnm5(s2)U detected in mammalian cells. This finding suggests that the GTPBP3–MTO1 complex retains an intrinsic specificity for Gly, which is a substrate of the bacterial MnmE–MnmG complex. Curiously, bacterial MnmE–MnmG complex had a limited ability to use taurine to synthesize τm5U in vitro (Figure 6A), implying that the GTPBP3–MTO1 complex gradually acquired taurine specificity over the course of mitochondrial evolution. τm5(s2)U34 is primarily synthesized when the taurine concentration is high enough in mitochondria, whereas cmnm5(s2)U34 is synthesized by utilizing Gly instead of taurine under taurine-starved conditions, indicating that cmnm5(s2)U could be used as a molecular marker for taurine starvation in mammals. In a sense, cmnm5(s2)U34 formation can be regarded as a backup system for mitochondrial translation under taurine-depleted conditions. However, as the decoding efficiency of cmnm5(s2)U34 in mammalian mitochondria has not been characterized, functional and physiological impact of this modification remains unclear. According to a chemical characterization of these nucleosides (79), the electron density of the uracil rings and the acidity of the N3H protons in these compounds are modulated by different C5-substituents, suggesting that the decoding efficiency and fidelity of NNG codons is affected by mt-tRNAs bearing cmnm5(s2)U34. High Gly concentration in blood is a characteristic feature of nonketotic hyperglycinemia, a type of mitochondrial disease caused by pathogenic mutations in the mitochondrial GCS pathway (80). The levels of cmnm5(s2)U34 might be elevated in patients with this disease, and the modification status of mt-tRNAs might be involved in its molecular pathogenesis.
Several groups studied the physiological impact of GTPBP3 and MTO1 using genetic approaches, and observed mitochondrial dysfunction when either protein was depleted (81–83). However, none of these studies provided direct evidence that these genes are actually involved in τm5U34 formation. In this study, we clearly showed that τm5U34 was absent from five mt-tRNAs in GTPBP3 KO cells (Figure 3E and Supplementary Figure S6), as well as in Mto1 KO cells (Figure 3F and Supplementary Figure S7). GTPBP3 KO cells exhibited marked reductions in OCR and mitochondrial translation. We also observed reduced levels of complex I subunit proteins and deficient complex I activity (Figure 4A and B). Taken together, our findings demonstrate that τm5U34 is essential for efficient mitochondrial translation and respiratory activity. Because τm5(s2)U34 is present in five mt-tRNAs whose codons are widely distributed throughout all genes encoded in mtDNA, lack of the taurine modification is predicted to result in global reduction of mitochondrial protein synthesis. To modulate respiratory complex assembly, mitochondrial protein synthesis is coordinated with inner membrane insertion on the matrix side (84,85). Thus, repression of mitochondrial translation due to lack of τm5U34 in GTPBP3 KO might disrupt maturation and stability of respiratory chain complexes, leading to defective assembly of complex I. We also observed defective mitochondrial translation and respiratory activity in NSUN3 KO and ALKBH1 KO, in which formation of f5C34 in mt-tRNAMet is impaired (13,15). However, the pulse-labeling pattern of mitochondrial translation and respiratory activities in these KO cells are quite different from those observed in GTPBP3 KO cells, indicating that a codon-specific decoding defect mediated by different tRNA species arises in each system. In particular, because f5C34 is essential for AUA decoding (11), translational defects caused by lack of f5C34 could be related to the AUA codon distribution. Likewise, the distribution of codons decoded by five mt-tRNAs bearing τm5(s2)U should be associated with the characteristic phenotype and translational deficiency observed in GTPBP3 KO cells. To obtain mechanistic insights into the decoding disorder caused by deficiency in wobble modification, mitoribosome profiling of each type of KO cell could provide a snapshot of the decoding status at single-codon resolution (86).
Several approaches to restoring mitochondrial dysfunction in MELAS patients have been explored at both the basic research and clinical levels (87–92). Considering that lack of τm5(s2)U in mutant mt-tRNA is a primary cause of MELAS and MERRF, restoration of the taurine modification represents a promising therapeutic approach for treatment of these diseases. The pathogenic point mutations found in MELAS and MERRF are thought to prevent mutant tRNA recognition by GTPBP3–MTO1 complex, hampering τm5(s2)U formation. Although we showed here that GTPBP3–MTO1 complex is responsible for τm5U formation in all five mt-tRNA species, these tRNAs contain no consensus sequence or characteristic structural features, indicating that tRNA recognition by the GTPBP3–MTO1 complex has broad specificity. Nevertheless, it is puzzling that only one point mutation in mt-tRNAs associated with MELAS and MERRF impaired τm5(s2)U formation. To obtain mechanistic insights into τm5(s2)U formation and the tRNA specificity of GTPBP3–MTO1 complex, it is necessary to recapitulate τm5U formation. In vitro τm5U34 formation would be a powerful tool for directly examining the impact of pathogenic point mutations associated with MELAS and MERRF. In this study, we successfully introduced τm5U in mt-tRNA in vitro using the GTPBP3–MTO1 complex expressed in HEK293T cells in the presence of taurine, 5,10-CH2-THF, GTP and co-factors (Figure 6E), although the efficiency was quite low in comparison with that of in vitro cmnm5U34 formation by bacterial MnmE–MnmG complex (Figure 6A). To advance these studies, it will be necessary to optimize the reaction conditions to improve the efficiency as much as possible, such as adequate salt concentration, optimal pH, divalent cations, coenzymes and so on. Otherwise, some accessory proteins and/or unidentified co-factors might be required for efficient τm5U formation. In this study, we used monoglutamate form of CH2-THF for in vitro reconstitution. But, polyglutamate form is a major folate derivative in mitochondria (93). It's worth examining polyglutamate form of CH2-THF for further study. Using in vitro τm5U34 formation, it might be possible to investigate other pathogenic mutations in mt-tRNAs that impair τm5U formation. We could also analyze enzymatic activities of GTPBP3 and MTO1 with the reported pathogenic mutations associated with respiratory deficiency. In the future, it could be possible to engineer a GTPBP3–MTO1 complex that is capable of recognizing mt-tRNAs with pathogenic mutations and introducing τm5U into these molecules.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Suzuki laboratory, in particular, K. Miyauchi and A. Nagao for many insightful discussions. We are grateful to A. Yamashita (Yokohama City University) for providing vectors, L. Chasin (Columbia University) for providing CHO mutant cell lines and Merck Ltd. Japan for providing tetrahydrofolate derivatives.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-Aid for Scientific Research from Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT); Japan Society for the Promotion of Science (JSPS) (to Ta.S., Ts.S.). Funding for open access charge: JSPS; Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1. Machnicka M.A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K.M. et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 2013; 41:D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen K., Zhao B.S., He C.. Nucleic acid modifications in regulation of gene expression. Cell Chem. Biol. 2016; 23:74–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gilbert W.V., Bell T.A., Schaening C.. Messenger RNA modifications: form, distribution and function. Science. 2016; 352:1408–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frye M., Jaffrey S.R., Pan T., Rechavi G., Suzuki T.. RNA modifications: what have we learned and where are we headed. Nat. Rev. Genetics. 2016; 17:365–372. [DOI] [PubMed] [Google Scholar]
- 5. Duechler M., Leszczynska G., Sochacka E., Nawrot B.. Nucleoside modifications in the regulation of gene expression: focus on tRNA. Cell Mol. Life Sci. 2016; 73:3075–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bjork G. Soll D, Rajbhandary UL. Biosynthesis and function of modified nucleosides. tRNA: structure, biosynthesisand function. 1995; Washington, D.C.: American Society for Microbiology; 165–205. [Google Scholar]
- 7. Suzuki T. Grosjean H. Biosynthesis and function of tRNA wobble modifications. Fine-tuning of RNA functions by modification and editing. 2005; NY: Springer; 24–69. [Google Scholar]
- 8. Suzuki T., Nagao A., Suzuki T.. Human mitochondrial tRNAs: biogenesis, function, structural aspects and diseases. Annu. Rev. Genet. 2011; 45:299–329. [DOI] [PubMed] [Google Scholar]
- 9. Suzuki T., Suzuki T.. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014; 42:7346–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moriya J., Yokogawa T., Wakita K., Ueda T., Nishikawa K., Crain P.F., Hashizume T., Pomerantz S.C., McCloskey J.A., Kawai G. et al. A novel modified nucleoside found at the first position of the anticodon of methionine tRNA from bovine liver mitochondria. Biochemistry. 1994; 33:2234–2239. [DOI] [PubMed] [Google Scholar]
- 11. Takemoto C., Spremulli L.L., Benkowski L.A., Ueda T., Yokogawa T., Watanabe K.. Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system. Nucleic Acids Res. 2009; 37:1616–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sloan K.E., Hobartner C., Bohnsack M.T.. How RNA modification allows non-conventional decoding in mitochondria. Cell Cycle. 2017; 16:145–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakano S., Suzuki T., Kawarada L., Iwata H., Asano K., Suzuki T.. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNA(Met). Nat. Chem. Biol. 2016; 12:546–551. [DOI] [PubMed] [Google Scholar]
- 14. Van Haute L., Dietmann S., Kremer L., Hussain S., Pearce S.F., Powell C.A., Rorbach J., Lantaff R., Blanco S., Sauer S. et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016; 7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kawarada L., Suzuki T., Ohira T., Hirata S., Miyauchi K., Suzuki T.. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017; 45:7401–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haag S., Sloan K.E., Ranjan N., Warda A.S., Kretschmer J., Blessing C., Hubner B., Seikowski J., Dennerlein S., Rehling P. et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J. 2016; 35:2104–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Suzuki T., Suzuki T., Wada T., Saigo K., Watanabe K.. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 2002; 21:6581–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kurata S., Weixlbaumer A., Ohtsuki T., Shimazaki T., Wada T., Kirino Y., Takai K., Watanabe K., Ramakrishnan V., Suzuki T.. Modified uridines with C5-methylene substituents at the first position of the tRNA anticodon stabilize U.G wobble pairing during decoding. J. Biol. Chem. 2008; 283:18801–18811. [DOI] [PubMed] [Google Scholar]
- 19. Suzuki T., Nagao A., Suzuki T.. Human mitochondrial diseases caused by lack of taurine modification in mitochondrial tRNAs. Wiley Interdiscip. Rev. RNA. 2011; 2:376–386. [DOI] [PubMed] [Google Scholar]
- 20. Yasukawa T., Suzuki T., Suzuki T., Ueda T., Ohta S., Watanabe K.. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 2000; 275:4251–4257. [DOI] [PubMed] [Google Scholar]
- 21. Kirino Y., Goto Y., Campos Y., Arenas J., Suzuki T.. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:7127–7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirino Y., Yasukawa T., Ohta S., Akira S., Ishihara K., Watanabe K., Suzuki T.. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15070–15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yasukawa T., Suzuki T., Ishii N., Ueda T., Ohta S., Watanabe K.. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 2000; 467:175–178. [DOI] [PubMed] [Google Scholar]
- 24. Yasukawa T., Kirino Y., Ishii N., Holt I.J., Jacobs H.T., Makifuchi T., Fukuhara N., Ohta S., Suzuki T., Watanabe K.. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett. 2005; 579:2948–2952. [DOI] [PubMed] [Google Scholar]
- 25. Yasukawa T., Suzuki T., Ishii N., Ohta S., Watanabe K.. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 2001; 20:4794–4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Umeda N., Suzuki T., Yukawa M., Ohya Y., Shindo H., Watanabe K., Suzuki T.. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005; 280:1613–1624. [DOI] [PubMed] [Google Scholar]
- 27. Zeharia A., Shaag A., Pappo O., Mager-Heckel A.M., Saada A., Beinat M., Karicheva O., Mandel H., Ofek N., Segel R. et al. Acute infantile liver failure due to mutations in the TRMU gene. Am. J. Hum. Genet. 2009; 85:401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schara U., von Kleist-Retzow J.C., Lainka E., Gerner P., Pyle A., Smith P.M., Lochmuller H., Czermin B., Abicht A., Holinski-Feder E. et al. Acute liver failure with subsequent cirrhosis as the primary manifestation of TRMU mutations. J. Inherit. Metab. Dis. 2011; 34:197–201. [DOI] [PubMed] [Google Scholar]
- 29. Gaignard P., Gonzales E., Ackermann O., Labrune P., Correia I., Therond P., Jacquemin E., Slama A.. Mitochondrial infantile liver disease due to TRMU gene mutations: three new cases. JIMD Rep. 2013; 11:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu Y., Wei F.Y., Kawarada L., Suzuki T., Araki K., Komohara Y., Fujimura A., Kaitsuka T., Takeya M., Oike Y. et al. Mtu1-mediated thiouridine formation of mitochondrial tRNAs is required for mitochondrial translation and is involved in reversible infantile liver injury. PLoS Genet. 2016; 12:e1006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taylor R.W., Pyle A., Griffin H., Blakely E.L., Duff J., He L., Smertenko T., Alston C.L., Neeve V.C., Best A. et al. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. Jama. 2014; 312:68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghezzi D., Baruffini E., Haack T.B., Invernizzi F., Melchionda L., Dallabona C., Strom T.M., Parini R., Burlina A.B., Meitinger T. et al. Mutations of the mitochondrial-tRNA modifier MTO1 cause hypertrophic cardiomyopathy and lactic acidosis. Am. J. Hum. Genet. 2012; 90:1079–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baruffini E., Dallabona C., Invernizzi F., Yarham J.W., Melchionda L., Blakely E.L., Lamantea E., Donnini C., Santra S., Vijayaraghavan S. et al. MTO1 mutations are associated with hypertrophic cardiomyopathy and lactic acidosis and cause respiratory chain deficiency in humans and yeast. Hum. Mutat. 2013; 34:1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kopajtich R., Nicholls T.J., Rorbach J., Metodiev M.D., Freisinger P., Mandel H., Vanlander A., Ghezzi D., Carrozzo R., Taylor R.W. et al. Mutations in GTPBP3 cause a mitochondrial translation defect associated with hypertrophic cardiomyopathy, lactic acidosis, and encephalopathy. Am. J. Hum. Genet. 2014; 95:708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kohda M., Tokuzawa Y., Kishita Y., Nyuzuki H., Moriyama Y., Mizuno Y., Hirata T., Yatsuka Y., Yamashita-Sugahara Y., Nakachi Y. et al. A comprehensive genomic analysis reveals the genetic landscape of mitochondrial respiratory chain complex deficiencies. PLoS Genet. 2016; 12:e1005679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kao F., Chasin L., Puck T.T.. Genetics of somatic mammalian cells. X. complementation analysis of glycine-requiring mutants. Proc. Natl. Acad. Sci. U.S.A. 1969; 64:1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thomas K.R., Capecchi M.R.. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987; 51:503–512. [DOI] [PubMed] [Google Scholar]
- 38. Pyzocha N.K., Ran F.A., Hsu P.D., Zhang F.. RNA-guided genome editing of mammalian cells. Methods Mol. Biol. 2014; 1114:269–277. [DOI] [PubMed] [Google Scholar]
- 39. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pacioretty L., Hickman M.A., Morris J.G., Rogers Q.R.. Kinetics of taurine depletion and repletion in plasma, serum, whole blood and skeletal muscle in cats. Amino Acids. 2001; 21:417–427. [DOI] [PubMed] [Google Scholar]
- 41. Miyauchi K., Ohara T., Suzuki T.. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007; 35:e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miyauchi K., Kimura S., Suzuki T.. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013; 9:105–111. [DOI] [PubMed] [Google Scholar]
- 43. Ohira T., Suzuki T.. Precursors of tRNAs are stabilized by methylguanosine cap structures. Nat. Chem. Biol. 2016; 12:648–655. [DOI] [PubMed] [Google Scholar]
- 44. Suzuki T., Ikeuchi Y., Noma A., Suzuki T., Sakaguchi Y.. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007; 425:211–229. [DOI] [PubMed] [Google Scholar]
- 45. Ikeuchi Y., Kimura S., Numata T., Nakamura D., Yokogawa T., Ogata T., Wada T., Suzuki T.. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 2010; 6:277–282. [DOI] [PubMed] [Google Scholar]
- 46. Sakaguchi Y., Miyauchi K., Kang B.I., Suzuki T.. Nucleoside analysis by hydrophilic interaction liquid chromatography coupled with mass spectrometry. Methods Enzymol. 2015; 560:19–28. [DOI] [PubMed] [Google Scholar]
- 47. Trounce I.A., Kim Y.L., Jun A.S., Wallace D.C.. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996; 264:484–509. [DOI] [PubMed] [Google Scholar]
- 48. Melero R., Uchiyama A., Castano R., Kataoka N., Kurosawa H., Ohno S., Yamashita A., Llorca O.. Structures of SMG1-UPFs complexes: SMG1 contributes to regulate UPF2-dependent activation of UPF1 in NMD. Structure. 2014; 22:1105–1119. [DOI] [PubMed] [Google Scholar]
- 49. Osawa T., Ito K., Inanaga H., Nureki O., Tomita K., Numata T.. Conserved cysteine residues of GidA are essential for biogenesis of 5-carboxymethylaminomethyluridine at tRNA anticodon. Structure. 2009; 17:713–724. [DOI] [PubMed] [Google Scholar]
- 50. Sampson J.R., Uhlenbeck O.C.. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Suzuki T., Suzuki T.. 22nd tRNA Workshop. 2007; Sweden: Uppsala. [Google Scholar]
- 52. Meyer S., Wittinghofer A., Versees W.. G-domain dimerization orchestrates the tRNA wobble modification reaction in the MnmE/GidA complex. J. Mol. Biol. 2009; 392:910–922. [DOI] [PubMed] [Google Scholar]
- 53. Moukadiri I., Prado S., Piera J., Velazquez-Campoy A., Bjork G.R., Armengod M.E.. Evolutionarily conserved proteins MnmE and GidA catalyze the formation of two methyluridine derivatives at tRNA wobble positions. Nucleic Acids Res. 2009; 37:7177–7193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ducker G.S., Rabinowitz J.D.. One-carbon metabolism in health and disease. Cell Metab. 2017; 25:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. McCarthy E.A., Titus S.A., Taylor S.M., Jackson-Cook C., Moran R.G.. A mutation inactivating the mitochondrial inner membrane folate transporter creates a glycine requirement for survival of chinese hamster cells. J. Biol. Chem. 2004; 279:33829–33836. [DOI] [PubMed] [Google Scholar]
- 56. Narkewicz M.R., Sauls S.D., Tjoa S.S., Teng C., Fennessey P.V.. Evidence for intracellular partitioning of serine and glycine metabolism in Chinese hamster ovary cells. Biochem. J. 1996; 313:991–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Morris J.G. Idiosyncratic nutrient requirements of cats appear to be diet-induced evolutionary adaptations. Nutr. Res. Rev. 2002; 15:153–168. [DOI] [PubMed] [Google Scholar]
- 58. Verbrugghe A., Bakovic M.. Peculiarities of one-carbon metabolism in the strict carnivorous cat and the role in feline hepatic lipidosis. Nutrients. 2013; 5:2811–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wallace D.C. Mitochondrial defects in cardiomyopathy and neuromuscular disease. Am. Heart. J. 2000; 139:S70–S85. [DOI] [PubMed] [Google Scholar]
- 60. Sturman J.A. Taurine in development. Physiol. Rev. 1993; 73:119–147. [DOI] [PubMed] [Google Scholar]
- 61. Kim S.K., Takeuchi T., Yokoyama M., Murata Y.. Effect of dietary supplementation with taurine, β-alanine and GABA on the growth of juvenile and fingerling Japanese flounder Paralichthys olivaceus. Fish. Sci. 2003; 69:242–248. [Google Scholar]
- 62. Seikai T., Takeuchi T., Park G.S.. Comparison of growth, feed efficiency, and chemical composition of juvenile flounder fed live mysids and formula feed under laboratory conditions. Fish. Sci. 1997; 63:520–526. [Google Scholar]
- 63. Goto Y., Horai S., Matsuoka T., Koga Y., Nihei K., Kobayashi M., Nonaka I.. Mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS): a correlative study of the clinical features and mitochondrial DNA mutation. Neurology. 1992; 42:545–550. [DOI] [PubMed] [Google Scholar]
- 64. Colby G., Wu M., Tzagoloff A.. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 1998; 273:27945–27952. [DOI] [PubMed] [Google Scholar]
- 65. Li X., Guan M.X.. A human mitochondrial GTP binding protein related to tRNA modification may modulate phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol. Cell. Biol. 2002; 22:7701–7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li X., Li R., Lin X., Guan M.X.. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J. Biol. Chem. 2002; 277:27256–27264. [DOI] [PubMed] [Google Scholar]
- 67. Ye J., Fan J., Venneti S., Wan Y.W., Pawel B.R., Zhang J., Finley L.W., Lu C., Lindsten T., Cross J.R. et al. Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov. 2014; 4:1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Woo C.C., Chen W.C., Teo X.Q., Radda G.K., Lee P.T.. Downregulating serine hydroxymethyltransferase 2 (SHMT2) suppresses tumorigenesis in human hepatocellular carcinoma. Oncotarget. 2016; 7:53005–53017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ron-Harel N., Santos D., Ghergurovich J.M., Sage P.T., Reddy A., Lovitch S.B., Dephoure N., Satterstrom F.K., Sheffer M., Spinelli J.B. et al. Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 2016; 24:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hashizume O., Ohnishi S., Mito T., Shimizu A., Ishikawa K., Nakada K., Soda M., Mano H., Togayachi S., Miyoshi H. et al. Epigenetic regulation of the nuclear-coded GCAT and SHMT2 genes confers human age-associated mitochondrial respiration defects. Sci. Rep. 2015; 5:10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Huxtable R.J. Physiological actions of taurine. Physiol. Rev. 1992; 72:101–163. [DOI] [PubMed] [Google Scholar]
- 72. Bouckenooghe T., Remacle C., Reusens B.. Is taurine a functional nutrient. Curr. Opin. Clin. Nutr. Metab. Care. 2006; 9:728–733. [DOI] [PubMed] [Google Scholar]
- 73. Pion P.D., Kittleson M.D., Rogers Q.R., Morris J.G.. Myocardial failure in cats associated with low plasma taurine: a reversible cardiomyopathy. Science. 1987; 237:764–768. [DOI] [PubMed] [Google Scholar]
- 74. Kim S.K., Matsunari H., Takeuchi T., Yokoyama M., Furuita H., Murata Y., Goto T.. Comparison of taurine biosynthesis ability between juveniles of Japanese flounder and common carp. Amino Acids. 2008; 35:161–168. [DOI] [PubMed] [Google Scholar]
- 75. Suzuki T., Miyauchi K., Suzuki T., Yokobori S., Shigi N., Kondow A., Takeuchi N., Yamagishi A., Watanabe K.. Taurine-containing uridine modifications in tRNA anticodons are required to decipher non-universal genetic codes in ascidian mitochondria. J. Biol. Chem. 2011; 286:35494–35498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Young T.L., Matsuda T., Cepko C.L.. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr. Biol. 2005; 15:501–512. [DOI] [PubMed] [Google Scholar]
- 77. Long J., Badal S.S., Ye Z., Wang Y., Ayanga B.A., Galvan D.L., Green N.H., Chang B.H., Overbeek P.A., Danesh F.R.. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J. Clin. Invest. 2016; 126:4205–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sakurai M., Ohtsuki T., Suzuki T., Watanabe K.. Unusual usage of wobble modifications in mitochondrial tRNAs of the nematode Ascaris suum. FEBS Lett. 2005; 579:2767–2772. [DOI] [PubMed] [Google Scholar]
- 79. Sochacka E., Lodyga-Chruscinska E., Pawlak J., Cypryk M., Bartos P., Ebenryter-Olbinska K., Leszczynska G., Nawrot B.. C5-substituents of uridines and 2-thiouridines present at the wobble position of tRNA determine the formation of their keto-enol or zwitterionic forms - a factor important for accuracy of reading of guanosine at the 3-end of the mRNA codons. Nucleic Acids Res. 2017; 45:4825–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kikuchi G., Motokawa Y., Yoshida T., Hiraga K.. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2008; 84:246–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen D., Li F., Yang Q., Tian M., Zhang Z., Zhang Q., Chen Y., Guan M.X.. The defective expression of gtpbp3 related to tRNA modification alters the mitochondrial function and development of zebrafish. Int. J. Biochem. Cell Biol. 2016; 77:1–9. [DOI] [PubMed] [Google Scholar]
- 82. Martinez-Zamora A., Meseguer S., Esteve J.M., Villarroya M., Aguado C., Enriquez J.A., Knecht E., Armengod M.E.. Defective expression of the mitochondrial-trna modifying enzyme GTPBP3 triggers AMPK-mediated adaptive responses involving complex I assembly factors, uncoupling protein 2, and the mitochondrial pyruvate carrier. PLoS One. 2015; 10:e0144273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Becker L., Kling E., Schiller E., Zeh R., Schrewe A., Holter S.M., Mossbrugger I., Calzada-Wack J., Strecker V., Wittig I. et al. MTO1-deficient mouse model mirrors the human phenotype showing complex I defect and cardiomyopathy. PLoS One. 2014; 9:e114918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ott M., Herrmann J.M.. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim. Biophys. Acta. 2010; 1803:767–775. [DOI] [PubMed] [Google Scholar]
- 85. Herrmann J.M., Woellhaf M.W., Bonnefoy N.. Control of protein synthesis in yeast mitochondria: the concept of translational activators. Biochim. Biophys. Acta. 2013; 1833:286–294. [DOI] [PubMed] [Google Scholar]
- 86. Rooijers K., Loayza-Puch F., Nijtmans L.G., Agami R.. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 2013; 4:2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Karicheva O.Z., Kolesnikova O.A., Schirtz T., Vysokikh M.Y., Mager-Heckel A.M., Lombes A., Boucheham A., Krasheninnikov I.A., Martin R.P., Entelis N. et al. Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria. Nucleic Acids Res. 2011; 39:8173–8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hornig-Do H.T., Montanari A., Rozanska A., Tuppen H.A., Almalki A.A., Abg-Kamaludin D.P., Frontali L., Francisci S., Lightowlers R.N., Chrzanowska-Lightowlers Z.M.. Human mitochondrial leucyl tRNA synthetase can suppress non cognate pathogenic mt-tRNA mutations. EMBO Mol. Med. 2014; 6:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Perli E., Fiorillo A., Giordano C., Pisano A., Montanari A., Grazioli P., Campese A.F., Di Micco P., Tuppen H.A., Genovese I. et al. Short peptides from leucyl-tRNA synthetase rescue disease-causing mitochondrial tRNA point mutations. Hum. Mol. Genet. 2016; 25:903–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Perli E., Giordano C., Pisano A., Montanari A., Campese A.F., Reyes A., Ghezzi D., Nasca A., Tuppen H.A., Orlandi M. et al. The isolated carboxy-terminal domain of human mitochondrial leucyl-tRNA synthetase rescues the pathological phenotype of mitochondrial tRNA mutations in human cells. EMBO Mol. Med. 2014; 6:169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rikimaru M., Ohsawa Y., Wolf A.M., Nishimaki K., Ichimiya H., Kamimura N., Nishimatsu S., Ohta S., Sunada Y.. Taurine ameliorates impaired the mitochondrial function and prevents stroke-like episodes in patients with MELAS. Intern. Med. 2012; 51:3351–3357. [DOI] [PubMed] [Google Scholar]
- 92. Koga Y., Akita Y., Nishioka J., Yatsuga S., Povalko N., Katayama K., Matsuishi T.. MELAS and L-arginine therapy. Mitochondrion. 2007; 7:133–139. [DOI] [PubMed] [Google Scholar]
- 93. McBurney M.W., Whitmore G.F.. Isolation and biochemical characterization of folate deficient mutants of Chinese hamster cells. Cell. 1974; 2:173–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.