Abstract
Hepatic miR-122 can serve as a pro-apoptotic factor to suppress tumorigenesis. The underlying mechanism, however, remains incompletely understood. Here we present the first evidence that miR-122 promotes hepatocellular carcinoma cell apoptosis through directly silencing the biogenesis of cell survival oncomiR miR-21 at posttranscriptional level. We find that miR-122 is strongly expressed in primary liver cell nucleus but its nuclear localization is markedly decreased in transformed cells particularly in chemoresistant tumor cells. MiRNA profiling and RT-qPCR confirm an inverse correlation between miR-122 and miR-21 in hepatocellular carcinoma tissues/cells, and increasing or decreasing nuclear level of miR-122 respectively reduces or increases miR-21 expression. Mechanistically, nuclear miR-122 suppresses miR-21 maturation via binding to a 19-nt UG-containing recognition element in the basal region of pri-miR-21 and preventing the Drosha-DGCR8 microprocessor's conversion of pri-miR-21 into pre-miR-21. Furthermore, both in vitro and in vivo studies demonstrate that nuclear miR-122 participates in the regulation of HCC cell apoptosis through modulating the miR-21-targeted programmed cell death 4 (PDCD4) signal pathway.
INTRODUCTION
MicroRNAs (miRNAs), a class of noncoding RNAs of ∼22nt in length, play an essential role in gene regulation in animals and plants (1,2). In the canonical pathway of miRNA biogenesis, a long primary transcript (pri-miRNA) is initially cleaved by RNase III DROSHA and its cofactor, DGCR8 to release a relative short hairpin intermediate, pre-miRNAs (3,4). The pre-miRNAs are then exported by exportin-5 to cytoplasm (5,6) and then cleaved by Dicer, another RNase III type protein to generate a miRNA duplexes. One strand of the duplexes becomes a mature miRNA and is preferentially assembled into the effector complex called miRNA-induced silencing complex (RISC). In the RISC, the mature miRNA acts as a guide by base pairing with its cognate mRNAs and induces translational repression or mRNA destabilization in cytoplasm (7–9), while the other strand of the duplexes is degraded immediately. Although the prevailing view is that miRNAs execute their function in the cytoplasm, accumulating evidence has shown that miRNAs together with functional proteins such as Argonaute 2 (Ago2) can localize in nucleus (10–19), suggesting that nuclear miRNAs may also regulate protein expression at the level of DNA as well as after transcription (10,13,14,20–22). Using superquencher molecular beacon probes, Foldes-Papp et al. (12) first showed that the cytoplasm-assembled mature miR-122 could re-enter into the nucleus in human liver cells. Subsequently, the distribution of miRNAs in both nucleus and cytoplasm has been widely shown by many investigators using systematic and microarray profiling approaches (15–19), suggesting that the presence of mature miRNAs in the nucleus is a general phenomenon in mammalian cells. Interestingly, Hwang et al. showed that miR-29b was predominantly present in the nuclei of HeLa and 3T3 cells, whereas the relevant miR-29a was mainly localized in the cytoplasm (11), implying that a unique sequence may serve as signal to guide specific miRNA entering the nucleus.
It has been also reported that the level of miRNAs in the nucleus was decreased following the cell's conversion to a differentiated state (18), suggesting that nuclear miRNAs might play a role in maintaining the undifferentiated state and cortical development. Offering further evidence that mature miRNA can influence the maturation of primary miRNA (pri-miRNA), we demonstrated that mouse miR-709 acted as a posttranscriptional regulator of the miR-15a/16–1 transcript expression by directly binding to a recognition element on the pri-miR-15a/16–1 in the nucleus (23). In C. elegans, Zisoulis et al. (24) showed that mature let-7 miRNA could bind to a specific site at the 3′ end of its own primary transcripts and promote the maturation of primary let-7. Although these two studies revealed a novel picture of miRNA transcripts as the targets by other miRNAs, various functions of nuclear miRNAs especially the underlying mechanisms governing the gene regulation mediated by nuclear miRNAs remain largely unknown.
Previous studies showed that miR-122, the most abundant miRNA in the liver, could serve as a pro-apoptotic factor in suppressing hepatocellular carcinoma cell migration and invasion (25–28). During hepatocyte tumorigenesis, miR-122 was strongly repressed (26,29). Although the underlying mechanism remains unclear, Bai et al. (30) have reported that miR-122 sensitizes hepatocellular carcinoma (HCC) cells to sorafenib. In line with this, Xu et al. (31) found reduction of miR-122 in sorafenib-resistant cells, and their study further showed that miR-122 overexpression induced cell apoptosis and re-sensitized drug-resistant tumor cells to sorafenib treatment. Programmed cell death 4 (PDCD4), a tumor suppressor protein targeted by miR-21, has been shown to suppresses tumor cell drug-resistance and chemo-resistance (32,33). However, it remains unknown whether and how PDCD4 is involved in the suppressive effect of miR-122 on HCC drug-resistance and chemo-resistance.
In the present study, we demonstrated that miR-122 promotes liver cancer cell apoptosis through blocking the maturation of cell survival oncomiR miR-21 (34,35). Using miRNA tracing, in site hybridization and RT-qPCR studies, we found a considerable amount of miR-122 re-entering into liver cell nucleus. Microarray profiling and RT-qPCR assays showed an inverse relationship between miR-122 and miR-21 was validated in HCC tissues and cells, and that increasing or decreasing nuclear miR-122 level in liver cancer cells significantly reduced or increased miR-21 expression, respectively. Mechanistic studies further revealed that nuclear miR-122 bound to a 19-nt UG-containing recognition element in the basal region of pri-miR-21, preventing the processing of pri-miR-21 into pre-miR-21 by Drosha-DGCR8 microprocessor. Finally, functional assays showed that nuclear miR-122 decreased liver cancer cell drug-resistance and suppressed HCC growth via downregulating miR-21 but increasing cellular events downstream of PDCD4 activity.
MATERIALS AND METHODS
HCC specimens and animals
All protocols concerning the use of patient samples in this study were approved by the Medical Ethics Committee of the Affiliated Gulou Hospital of Nanjing University (Nanjing, China). A signed consent form was obtained from each donor. Sixteen HCC patients who underwent primary surgical resection were enrolled in this study. Paired HCC and ANCT tissues were obtained from each patient. Tissue sections were immediately frozen in liquid nitrogen at the time of surgery and stored at -80°C. Clinical and pathological data, including age, gender and pathological grading are listed in Supplementary Table S1. All protocols involving animal experiments were approved by the Ethics Committee of Nanjing University. The 6–8 weeks old male C57BL/6J mice and SCID (severe combined immune deficiency) nude mice were purchased form Model Animal Research Center of Nanjing University (Nanjing, China) and fed under specific pathogen-free conditions at Nanjing University.
Cell culture, reagents and antibodies
Hepatic carcinoma Huh-7 cells, Hepa1–6 cells and human embryonic kidney 293T (HEK293T) cells were purchased from the cell bank of Chinese Academy of Sciences (Shanghai, China). The cells were maintained at 37°C in a humidified 5% CO2 incubator in Dulbecco's Modified Eagle's Medium (Gibco, CA, USA) containing 10% fetal bovine serum (Gibco), and 100 units/ml penicillin and 100 μg/ml streptomycin. Drug-resistant cells were treated with sorafenib at the concentration of 10 μM for three rounds, each lasting 72 h. Viable cells remaining attached to the dish at the end of the third round of drug treatment were considered to be sorafenib-resistant and were collected for experiments. Resistant cell lines were maintained in the continuous presence of 10 μM sorafenib, supplemented every 72 h. Stable transfection with constructed lentivirus Lenti-LV3-NC, Lenti-LV3-pri-21-WT and Lenti-LV3-pri-21-MUT in Huh-7 cells (5 × 108 TU/ml) was selected through multiple rounds against puromycin (2 μg/ml) treatment until the GFP-positive cells reached >90% of total cells. These antibodies were purchased from indicated sources: anti-PDCD4, anti-H2A, anti-Histone H3, anti-ERp72, anti-Drosha, anti-DGCR8 antibodies (Cell Signaling, Danvers, MA, USA); anti-GAPDH antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-cytochrome C antibody (Abcam, Cambridge, MA, USA). Secondary antibodies against mouse or rabbit IgGs were purchased from Santa Cruz Biotechnology. Sorafenib Tosylate was purchased from Selleckchem (Houston, TX, USA). Actinomycin D and puromycin were purchased from Sigma Aldrich (St Louis, MO, USA).
Preparation of nuclear extracts
The nuclear fraction of cells was extracted using a PARISTM kit (Ambion, AM1921). Cells were rinsed three times with PBS on ice followed by centrifugation at 500 × g for 5 min. Cell pellets were re-suspended in cell fraction buffer from the PARISTM kit, incubated on ice for 10 min, and then centrifuged at 500 × g for 5 min at 4°C. The supernatant, containing the cytoplasmic fraction, was used for RNA or protein purification. Nuclear pellets underwent another wash with cell fraction buffer and final homogenized with the cell disruption buffer from the PARISTM kit.
Isolation and culture of primary mouse hepatocytes
Isolation and culture of adult mouse hepatocytes was described previously (36). Briefly, the 6–8 weeks old C57BL/6J male mice were sacrificed and the viable hepatocytes were isolated by two step collagenase perfusion. The isolated cells were inoculated and cultured into 10 cm culture dishes with culture media. Cultured cells were then placed in an incubator with 5% CO2 at 37°C to continue culturing until required. Culture media contained the following: Williams E medium (Sigma), 1% (v/v) penicillin/streptomycin (10 000 U/μg/ml; Sigma), 0.5% (v/v) gentamycin (10 mg/ml; Gibco), 0.04% (v/v) fungizone (amphotericin B, 250 μg/ml; Gibco), 1% (v/v) 200 mM l-glutamine (Sigma), 1% (v/v) nonessential amino acids (Gibco).
Transfection of cells with miRNA mimics, ASO and scrambled control RNAs
Synthetic RNA molecules, including the miR-122 mimic, miR-122-MUT mimic, miR-122 agomir, miR-122 ASO, single strand miR-122, miR-21 mimic, miR-21 ASO and corresponding scrambled control RNAs were purchased from Ruibo Company (Guangzhou, China). Synthetic 5′-Cy3-labeled miR-122, miR-29a and miR-29b oligonucleotide duplexes were purchased from Invitrogen (Shanghai, China). Cells were seeded in six-well plates or 10 cm dishes, and then were transfected the following day (∼60–80% confluence) using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. For the transfection, 20 pmol RNA or 2.5 μg plasmid DNA per 105 cells was used. Cells were harvested 24 h after transfection for real-time PCR analysis or western blotting.
MicroRNA microarray
MiRNA microarray analyzed as described previously (23). Total and nuclear RNA was purified using the mirVANA miRNA isolation kit (Ambion). Purified RNA was labeled with fluorescein, and hybridization was performed on a miRNA microarray chip (miRNA microarray V4.0, CapitalBio Corp., Beijing, China). Finally, hybridization signals were detected and scanner images were quantified.
RNA isolation and miRNA quantification by RT-qPCR and low density array
Total RNA was extracted from cells or tissues using miRNeasy Mini Kit (Qiagen). The RT-qPCR was carried out using TaqMan miRNA probes (Applied Biosystems) according to the manufacturer's instructions for mature miRNAs or SYBR Green (Takara, Mountain View, CA, USA) for mRNA. Briefly, total RNA was reverse-transcribed to cDNA using AMV reverse transcriptase (Takara) and a stem-loop RT primer or reverse primer (Applied Biosystems). Real-time PCR was performed on an Applied Biosystems 7900 HT Fast system (Applied Biosystems). All of the reactions, including the no-template controls, were run in triplicate. The used RNA sample was equal for every reaction and the miRNA expression in the cells or nuclei was normalized to U6 snRNA, and mRNA expression in the cells was normalized to GAPDH. The fold changes of miRNA were calculated using the equation 2−ΔΔCt. The expression profiling of miRNA in huh-7 cells was scanned using TaqMan Low Density Array on a 7900 HT Fast Real-Time PCR System (Applied Biosystems) as described preciously (37). The RT-qPCR of pri-miR-21 and pre-miR-21 was described previously (38). Sequences of the primers used were as follows: human pri-miR-21: 5′-TTTTGTTTTGCTTGGGAGGA-3′ and 5′-AGCAGACAGTCAGGCAGGAT-3′. human pre-miR-21: 5′-TGTCGGGTAGCTTATCAGAC-3′ and 5′-TGTCAGACAGCCCATCGACT-3′. GFP: 5′-ATGGTGAGCAAGGGCGAGGA-3′ and 5′-CTTGTACAGCTCGTCCATGCCGAGA-3′. β-actin: 5′-CTGGAACGGTGAAGGTGACA-3′ and 5′-AAGGGACTTCCTGTAACAATGCA-3′.
Pri-miR-21 pull-down assay
A DNA probe complementary to human pri-miR-21 was synthesized from Invitrogen (Shanghai, China), labeled with biotin at both 5′ and 3′ terminals and dissolved in 500 μl of wash/binding buffer (0.5 M NaCl, 20 mM Tris–HCl, 1 mM EDTA, pH 7.4) at a final concentration of 8 pmol/μl. The probe was then incubated with streptavidin-coated magnetic beads (New England BioLabs, S1420S) at 25°C for 1 h to generate probe-coated magnetic beads. Purified nuclear lysate was pretreated with DNase I (Takara) and then incubated with probe-coated beads at 37°C for 3 h. After washing four times with the wash/binding buffer, a magnet was applied to cause the beads/RNA complex to bind to the side of the tube. RNA was eluted from the streptavidin beads with 10 mM EDTA (pH 8.2, 90°C, 10 min), and the RNA was analyzed by RT-qPCR. Following primer sequences were used: anti-pri-miR-21 pull-down probe, ATGACATAATGCTCATAATGCTCCA; and random pull-down probe, ATCATGTCTAGCGCTTGGGCTTTGA.
Luciferase reporter assay
A whole sequence of human PDCD4 3′-UTR, the miR-122 binding site (CTACCATCGTGACATCTCCAT), the miR-122 binding mutant site (CGTGGCACCAGTGTTCTCCAT) and mouse pri-miR-21 sequences were synthesized and inserted into the pMIR-report plasmid (Ambion) by GenScript Corporation (Nanjing, China). Efficient insertion was confirmed by sequencing. For the luciferase reporter assays, cells were cultured in 24-well plates, and each well was transfected with 0.5 μg firefly luciferase reporter plasmid, 0.5 μg β-galactosidase expression vector (Ambion), and equal amount of scrambled ncRNA or miR-122 mimic or miR-21 mimic using Lipofectamine 3000 (Invitrogen). The β-galactosidase vector was used as a transfection control. Cells were assayed using the luciferase assay kits (Promega, Madison, WI, USA) 24 h after transfection. The reported data represent three independent experiments.
Immunoprecipitation and immunoblotting
Cells were lysed with lysis buffer (10 mM Tris, 140 mM NaCl, 1.5 mM MgCl2, 0.5% Nonidet P-40, 0.5 mM DTT, 1 mM NaF, 1 mM PMSF and 1% protease inhibitor cocktail from Sigma, pH 7.4) for 30 min on ice. The lysates were cleared by centrifugation for 10 min (1000 × g) at 4°C. Nuclear pellets were sonicated in nuclear lysis buffer (cell lysis buffer containing 1% Tween-40 and 0.5% deoxycholic acid) and then immunoprecipitated with anti-Drosha (Cell Signaling, D28B1) rabbit mAb, or normal IgG (Sigma-Aldrich) followed by SureBeads Protein G Magnetic Beads (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions. After the elution, the protein products were further used for in vitro processing assay or detected by western blot assay by using anti-Drosha and anti-DGCR8 (Cell Signaling, D78E4) antibodies.
Western blot analysis
Tissues and cultured cells were lysed in a buffer (50 mM Tris–HCl, 150 mM NaCl, 1% NP-40, 0.1% SDS, pH 7.4), sonicated and centrifuged at 12 000 × g for 10 min at 4°C. The supernatant fraction was removed, and the protein concentration was determined by a BCA assay (Pierce, Rockford). Aliquots of proteins (60–100 μg) were separated on 10% sodium dodecyl sulfate-polyacrylamide gels (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Roche). The membranes were blocked for 1 h at room temperature (RT) with 5% non-fat milk in Tris-buffered saline (TBS) plus 0.1% Tween-20 (TBST) followed by an overnight incubation at 4°C with antibodies diluted in blocking buffer. After three 10 min washes with TBST, blots were incubated at RT for 1 h with the appropriate secondary antibody conjugated to horseradish peroxidase, and protein expression was detected with an enhanced chemiluminescent reagent (Cell Signaling). The autoradiographic intensity of each band was scanned and quantified through densitometric analysis with Image J software.
Fluorescence in situ hybridization (FISH)
FISH was used to identify the subcellular localization of miR-122 in Huh-7 cells by using Dig labeled microRNA Detection Probes (Exiqon) (39,40). Cells were cultured on four-well chamber slides. At the time of harvest, cells were fixed with 4% paraformaldehyde for 10 min and washed three times for 5 min with PBST at RT. Then slides were immersed in acetylation solution (5 ml DEPC-treated water, 80 μl triethanolamine, 10.5 μl HCl (37%), 15 μl acetic anhydride) and stirred gently for 10 min. The slides were pre-incubated in hybridization solution (10 ml formamide, 5 ml 20 × SSC, 200 μl 20 mg/ml yeast RNA, 1000 μl 10 mg/ml salmon sperm DNA, 0.4 g Roche blocking reagents, 500 μl of 10% CHAPS, 20 μl of 20% Tween) for 3 h at 45°C. For each slide, the probe was diluted with hybridization buffer. The probes were denatured by heating them up to 80°C for 5 min after which the probes were quickly placed on ice. The probes were added carefully onto the slides and hybridized at 53°C overnight followed by extensive wash with 0.1 × SSC (300 mM NaCl, 300 mM sodium citrate, pH 7.0). Slides were incubated in block solution (10% FBS in 100 mM Tris, 150 mM NaCl, pH 7.4) for 1 h, and then in color reaction solution buffer diluted with anti-digoxigenin-rhodamine, Fab fragments (Roche) at 4°C overnight. After three washes, slides were equilibrated for 10 min in solution (100 mM Tris, 100 mM NaCl, 50 mM MgCl2, pH 9.5). All samples were treated with DAPI dye for nuclear staining. For confocal microscopy, the Nikon C2 Plus confocal microscope (Nikon Corp, Japan) was used.
GFP and miR-21 expression plasmid construction
To construct GFP expression plasmid, the miR-122 binding site (CTACCATCGTGACATCTCCAT) and mutant binding site (CGTCCGCCCAGTGTTGTGGTT) were cloned and inserted into the pcDNA6.2- GW/EmGFP-miR plasmid (Invitrogen). Construction of miR-21 expression plasmid was described previously (23). Briefly, to construct the pcDNA-miR-21*-WT, the pri-miR-21 substrate (the mature miR-21 sequence in the vector was mutated by replacing three nucleotides) consisted of about 1 kb of human pri-miR-21 was synthesized and inserted into pcDNA3.1 plasmid by GenScript Corporation (Nanjing, China). The pcDNA-miR-21*-MUT was substituted for mutant binding site as described above.
In vitro analysis of processing pri-miRNAs
In vitro pri-miRNA processing assay and construction of pri-miRNA substrates were performed as described previously (4,41). Firstly, the pri-miR-21/pri-mut-miR-21 and pri-miR-150 substrates consisted of 320nt were synthesized and inserted into the pCMV-MIR plasmid by GenScript Corporation (Nanjing, China). The pri-miRNA substrates were then amplified by pri-miR-21 and pri-miR-150 primers using Q5 High-Fidelity 2X Master Mix (New England BioLabs, M0492S) according to the manufacturer's protocol. Secondly the PCR products were separated by 1% agarose gel electrophoresis and extracted from the gel using GenElute Gel Extraction Kit (Sigma-Aldrich). Each purified PCR product was confirmed by sequencing. The templates were then used in vitro transcription reaction by RiboMAX Large-Scale RNA Production System-T7 (Promega, P1300). The in vitro transcribed pri-miRNA substrates were then incubated with single strand miR-122 oligonucleotide (10 μM) in Annealing Buffer for RNA Oligos (Beyotime, Shanghai, China), at 90°C for 1 min and then underwent programmed cooling to 25°C for 1 h. Then the mixtures were incubated with ribonuclease inhibitor (1.5 U/μl, Promega), MgCl2 (6.4 mM) and the immunoprecipitated microprocessor which was acquired as described above or nuclear extracts for 90 min at 37°C. Reaction mixtures were subjected to RNA extraction, followed by northern blot analysis. Pri-miR-21 primer sequences: 5′-TAATACGACTCACTATAGGGTTCGATCTTAACAGGC-3′ and 5′-GACTCTAAGTGCCACCAGACA-3′; Pri-miR-150 primer sequences: 5′-TAATACGACTCACTATAGGGCCCTCTTTGATGCGG-3′ and 5′-CCCGAAGGGGAGAGACGCATA-3′.
Northern blotting analysis
A sensitive non-radioactive northern blot method to detect miRNAs was performed as above (42). The northern blot analysis was carried out using microRNA Detection Probes with DIG-labeling (Exiqon) and a chemiluminescent reaction by enzyme-immunoassay. Firstly, equal amount of in vitro processing RNA substrate was dissolved in TBE-Urea Sample Buffer (Invitrogen), and heated at 95°C for 5 min then rapidly cooled on ice. The RNA was then loaded onto a denaturing 15% polyacrylmide–7.5 M urea gel and transferred electrophoretically to Hybond Nmembranes (Amersham Pharmacia Biotech). The membrane was dried at 80°C for 10 min and then pre-hybridized in 10 ml of pre-heated DIG Easy Hyb (Roche) at 45°C for 1 h. The membrane was hybridized overnight with the hsa-miR-21 DIG labeled LNA-DNA probe (Exiqon) at the concentration of 5 pmol/ml in hybridization oven at 53°C. After hybridization and washing, the membrane was detected with DIG Luminescent Detection Kit (Roche) according to the manufacturer's instructions. Lastly, the membrane was exposed to Amersham Hyperfilm ECL (GE Healthcare Life Sciences, Piscataway, NJ). The bar graphs corresponding to the northern blots were generated through densitometric analysis with ImageJ software.
Flow cytometric assays
Cells were treated for the respective time and harvested by centrifugation at 300 × g for 5 min at 4 °C. The Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences) was used to stain the cells for the evaluation of apoptosis according to the manufacturer's protocol by flow cytometry (BD Biosciences).
In vivo assays
To overexpress or deplete miR-122 in C57BL/6J mouse liver, mice were injected with lentivirus expressing miR-122 mimic or sponge (5 × 108 TU/ml lentivirus in 100 μl per mouse, five mice in each group) via tail vein once every 3 days for one month. Mice were then sacrificed and RNAs were extracted from the livers. The stable infected Huh-7 cells were injected subcutaneously into the nude mice (5 × 106 cells per mouse, five mice per group) for tumor implantation. After 2 weeks, miR-122 agomir (10 nmol in 100 μl saline per mouse) was injected intratumorally every 3 days for 3 weeks. After 5 weeks, mice were sacrificed and tumors were excised for further analysis.
Statistical analysis
All of the images of western blot and northern blot are representative of at least three independent experiments. RT-qPCR, the luciferase reporter assays, and apoptosis assays were performed in triplicate, and each experiment was repeated several times. The data shown are presented as the mean ± SD of at least three independent experiments. The differences were considered statistically significant at P < 0.05 using Student's t-test.
RESULTS
Detection of human mature miR-122 in the nucleus of liver cells
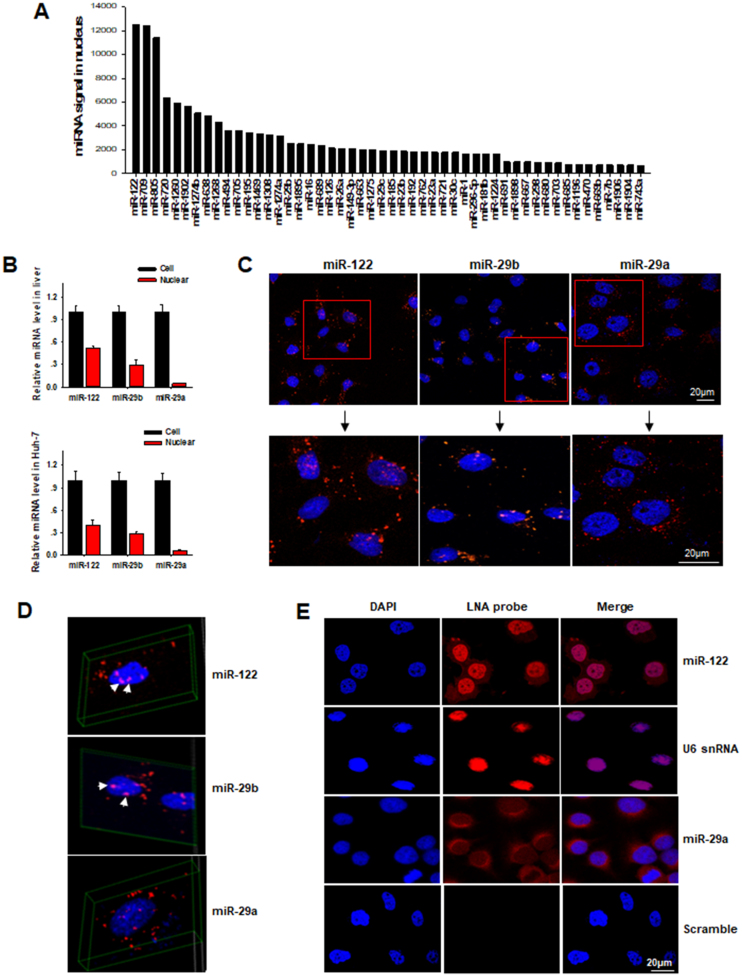
To detect the nuclear distribution of miRNAs, we purified the nuclear and cytoplasmic compartments from human liver cell line Huh-7 cells and C57BL/6J mouse livers using Ambion PARIS™ kit (23,43). The high purity of isolated nuclear fraction was confirmed by using nuclear/cytoplasmic RNA and protein markers including U6 small nuclear RNA (U6), H2A histone protein (H2A), β-actin and GAPDH (Supplementary Figure S1A and B). As shown, the isolated nuclear fraction displayed high level of nuclear markers (U6 and H2A) but nearly no cytoplasmic markers (β-actin and GAPDH). We also detected ERp72 and cytochrome C in isolated nuclear and cytoplasmic fractions. As shown in supplementary Figure S1C, no signal was detected by antibody against ERp72 or cytochrome c in the extracted nuclear fractions, indicating no ER and mitochondria contamination in purified nuclear fractions. A survey of mouse miRNA expression profile in the nuclei was then performed using a microarray assay. As shown in Figure 1A, miR-122 and miR-709 had the highest signal in the nucleus. The localization of miR-122 in the mouse liver cells was validated by a TaqMan probe-based quantitative real time PCR (RT-qPCR). In the assay, miR-29a, a typical cytoplasmic miRNA and miR-29b, a miRNA that can re-enter into the nuclear compartment (11), were used as negative and positive control, respectively. The RT-qPCR results indicated that miR-122, as well as miR-29b, were distributed to both nucleus and cytoplasm, whereas miR-29a was only detected in the cytoplasm (Figure 1B, upper). A similar distribution pattern of miR-122 was observed in human Huh-7 cells (Figure 1B, lower). Localization of fluorescently-labeled miR-122 mimics further showed the re-entry of mature miR-122 into the nuclei in Huh-7 cells (Figure 1C). In this experiment, Huh-7 cells were transfected with synthetic Cy3-labled miR-122, miR-29a and miR-29b mimics. After 24 h transfection, fluorescent miR-122 and miR-29b were located in the nuclei of Huh-7 cells, whereas the majority of fluorescent miR-29a was located in the cytoplasm. Re-composed figure (top view) of a series of Z-stacks images of Cy3-labeled miR-122, miR-29b and miR-29a mimics in Huh-7 cells (Supplementary Figure S2) confirmed the distribution of Cy3-labeled miR-122, miR-29b but not miR-29a in the nucleus, indicated by the merged pink colored spots (Figure 1D, white arrowheads). To further validate the nuclear localization of miR-122, we performed fluorescence in situ hybridization (FISH) using Digoxigenin (Dig)-labeled miRCURY LNA™ microRNA Detection Probes to detect the subcellular distribution of miR-122 in Huh-7 cells. As shown in Figure 1E, significant miR-122 probe signal was detected in the nucleus which was heavily stained by U6 probe while miR-29a was mostly detected in the cytoplasm.
Figure 1.
MiR-122 localizes in the nucleus of liver cells. (A) Nuclear miRNA expression profile in mouse liver cells via detecting by miRNA microarray. The most 50 enriched nuclear miRNAs were plotted in the figure. (B) MiR-122, miR-29b and miR-29a expression levels in mouse hepatocytes and nuclei (upper panel), human Huh-7 cells and nuclei (lower panel) by using TaqMan probe-based RT-qPCR assay. (C) Nuclear localization of synthetic Cy3-labeled miR-122, miR-29b and miR-29a mimics in Huh-7 cells. (D) 3-D re-constituted images of Z-stacks of Cy3-labeled miR-122, miR-29b and miR-29a mimics in Huh-7 cells. White arrowheads indicated the merged pink colored spots. (E) Distribution of miR-122 in Huh-7 cells by FISH using Dig-labeled miRCURY LNA™ microRNA Detection Probes. All images were obtained with the confocal microscope. The nuclear expression of U6 was visualized using the miRCURY LNA™ U6 probe as a positive control. DAPI was dyed with blue and probes were labeled with red by anti-Dig-rhodamine. Bars, 20 μm.
Nuclear miR-122 level in HCC tissues and drug-resistant liver cancer cells are markedly decreased
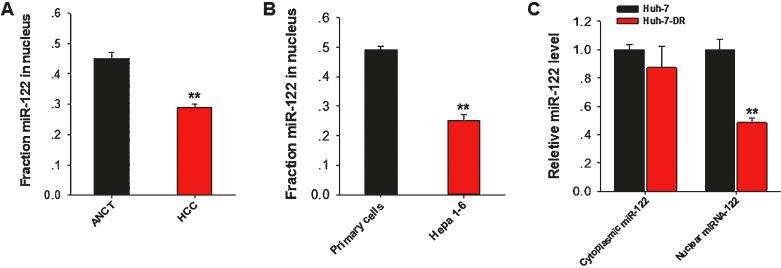
It has been widely shown that miR-122 suppresses hepatocarcinogenesis process (27,28,30,44–46). To explore whether nuclear distribution of miR-122 is involved in the tumor-suppressive function of miR-122, we next assessed the nuclear miR-122 level in various liver tissues and cells. In this experiment, paired tumor tissues and adjacent noncancerous tissue (ANCT) specimens were collected from 16 HCC patients (Gulou Hospital, Nanjing). Tissues were digested and then the nuclear fractions were isolated for further assay of miR-122. As shown in Figure 2A, ratio of nuclear/cellular miR-122 was significantly decreased in HCC tissues compared to that in ANCT, suggesting that nuclear miR-122 may suppress tumorigenesis. In line with this, we also found that the ratio of nuclear miR-122 vs cellular miR-122 in mouse cancer Hepa1–6 cells was significantly lower than that in mouse primary liver cells (Figure 2B). Moreover, to our surprise, when compare the miR-122 expression pattern in Huh-7 cells with that in sorafenib-resistant Huh-7 cells (Huh-7-DR) generated as previously described (31), we found that, although two cell types had similar cytoplasmic miR-122 expression, the nuclear miR-122 level in Huh-7-DR cells was significantly lower than that in Huh-7 cells (Figure 2C). Taken together, these results suggest that decrease of nuclear miR-122 level may be associated with hepatocarcinogenesis and development of tumor cell sorafenib-resistance.
Figure 2.
Nuclear miR-122 level in HCC tissues, HCC cells and anti-drug HCC cells are decreased. (A) Relative nuclear miR-122 percentage in HCC tissues and corresponding adjacent noncancerous tissue (ANCT) specimens. (B) Relative nuclear miR-122 percentage in mouse primary hepatocytes and mouse Hepa 1–6 cells. (C) Sorafenib resistance Huh-7 cells (Huh-7-DR) was established by treating Huh-7 cells with sorafenib (10 μM). The cytoplasmic and nuclear fractions were extracted and detected the expression levels of miR-122 and miR-21 in Huh-7 and Huh-7-DR cells respectively. The results are presented as the mean ± SD (N = 3) of three independent experiments. **P < 0.01.
Nuclear miR-122 suppresses miR-21 biogenesis via blocking the processing of pri-miR-21 to pre-miR-21
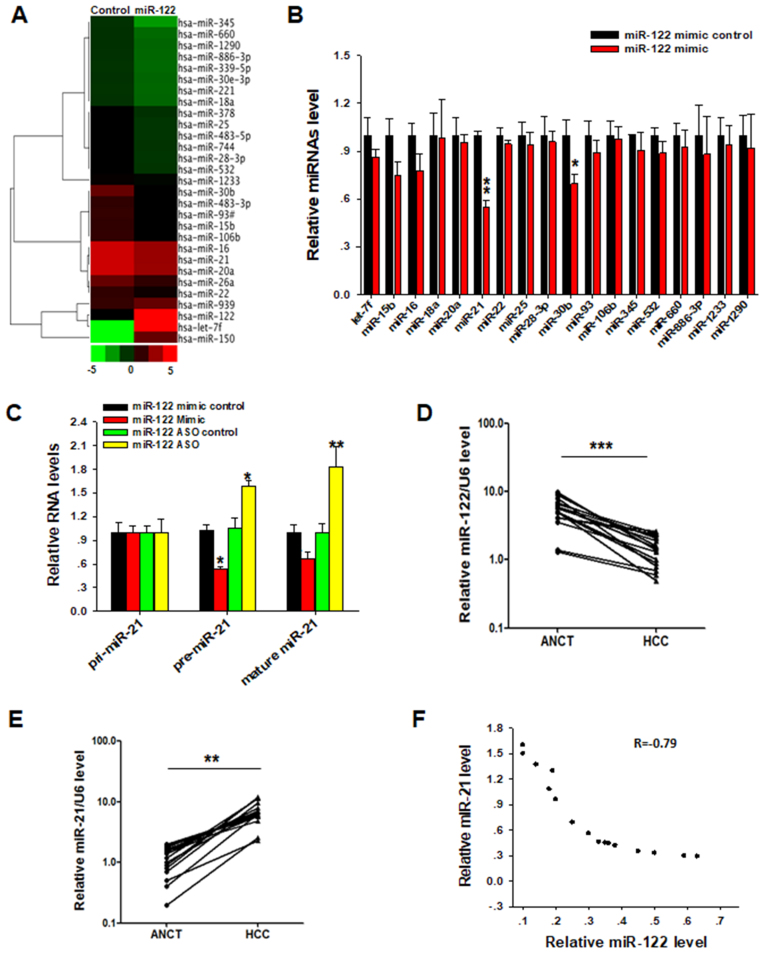
Given that a miRNA with high nuclear distribution can affect the biogenesis of other miRNAs (23), we postulated that nuclear miR-122 might execute its tumor suppressive function through inhibiting the biogenesis of certain cell survival oncomiRs. To test this hypothesis, a TaqMan Low Density Array was applied to screen for target miRNAs of miR-122 in Huh-7 cells following overexpressing miR-122 or control oligonucleotides. As shown in Supplementary Figure S3A, transfection of miR-122 mimic upregulated the miR-122 level in both nuclei and total cells, while the miR-122 antisense oligonucleotide (ASO) depleted the miR-122 level in total cells or cell nuclei. The differential expression of miRNAs by TaqMan low density array was shown in the cluster figure (Figure 3A). We next validated the miRNA candidates that were downregulated by miR-122 using RT-qPCR assay and confirmed miR-21 as the most significantly downregulated miRNA after miR-122 overexpression (Figure 3B). The decrement of miR-21 level affected by miR-122 was also observed in mouse liver cells. As shown in Supplementary Figure S3B, miR-21 in primary mouse hepatocytes was downregulated by miR-122 mimic transfection but upregulated by miR-122 ASO, which depletes cellular miR-122 level. To test whether miR-122 suppresses miR-21 biogenesis in vivo, we intravenously injected lentivirus that expresses miR-122 mimic or a miR-122 sponge into mice via tail vein and then evaluated the miR-21 level in mouse liver. As shown in Supplementary Figure S3C, miR-21 in mouse livers was downregulated by miR-122-expressing lentivirus but upregulated by miR-122 sponge-expressing lentivirus. Together, both in vitro and in vivo assays reveal that miR-122 negatively regulates the biogenesis of miR-21 in liver cells.
Figure 3.
Nuclear miR-122 inhibits miR-21 biogenesis. (A) TaqMan Low Density Array screening for target miRNAs of miR-122. Huh-7 cells were transfected with miR-122 mimic or control oligonucleotide and then harvested 24 h after transfection. The miRNA expression profile was sorted using a hierarchical clustering method (Cluster 3.0 and Java TreeView). Twenty eight most significantly changed miRNAs were shown in the cluster. (B) RT-qPCR validation of decreased miRNAs screened by Low Density Array following ectopic expression of miR-122 mimic or control. (C) Relative pri-miR-21, pre-miR-21 and miR-21 expression levels in Huh-7 cells after miR-122 overexpression or depletion. (D) RT-qPCR analysis of expression levels of miR-122 in 16 paired HCC and ANCT tissue samples. (E) RT-qPCR analysis of miR-21 level in 16 paired HCC and ANCT samples. (F) Pearson's correlation scatter plot of the levels of miR-122 and miR-21 in 16 paired HCC tissues. The results are presented as the mean ± SD (N = 3) of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001.
In mammalian cells, miRNA biosynthesis generally follows two steps: first, primary miRNAs (pri-miRNAs) are transcribed and cleaved by DROSHA complexes to produce precursor miRNAs (pre-miRNAs) in nucleus; second, pre-miRNAs are transported from nucleus to cytoplasm where they are further cleaved by Dicer, generating mature miRNAs (2). We next determined which step along miR-21 biogenesis was affected by nuclear miR-122. For this purpose, we overexpressed or depleted nuclear miR-122 by transfecting Huh-7 cells with miR-122 mimic, miR-122 ASO or control oligonucleotides, and then measured the relative expression levels of various miR-21 gene transcripts (pri-miR-21, pre-miR-21 or mature miR-21) by RT-qPCR (47). As shown in Figure 3C, upregulation of nuclear miR-122 in Huh-7 cells by transfection of miR-122 mimic resulted in a significant decrease of pre-miR-21 level whereas downregulation of nuclear miR-122 by anti-miR-122 ASO increased pre-miR-21 level. In contrast, no significant change in the expression of pri-miR-21 was observed under either condition. Together, these results suggest that repression of miR-21 biogenesis by miR-122 may occur during the processing of pri-miR-21 to pre-miR-21. This conclusion was confirmed by the same results obtained in primary mouse liver cells (Supplementary Figure S4A and B). To exclude the possibility that alteration of pre-miR-21 level is due to differential transcription of pri-miR-21, we treated the Huh-7 cells with 5 μg/ml Actinomycin D (ActD) to block the transcription process (48), and then performed the same experiments. After 24 h treatment, pri-miR-21, pre-miR-21 and mature miR-21 were detected by RT-qPCR. Although transcription of pri-miR-21 was stopped due to transcriptional shutoff, miR-122-mediated reduction of pre-miR-21 was not affected by ActD treatment (Supplementary Figure S4C). The mature miR-21 level was significantly reduced or increased in ActD-treated Huh-7 cells after overexpressing or depleting miR-122, respectively.
In supporting the notion that nuclear miR-122 suppresses miR-21 biogenesis, an inverse relationship between miR-21 and miR-122 level (R = –0.79) was revealed in HCC and ANCT tissues using Pearson's correlation scatter plot. As shown in Figure 3D–F, miR-122 level in HCC tissues was significantly decreased compared to ANCT (Figure 3D), while the miR-21 in HCC was markedly higher relative to the counterpart tissues (Figure 3E). The strong inverse relationship between miR-21 level and miR-122 level in HCC tissues was further illustrated using Pearson's correlation scatter plot (Figure 3F). One may argue that the impact of miR-122 on miR-21 biogenesis is due to a general competition for the miRNA processing apparatus by overexpressing miR-122. To exclude this possibility, we transfected Huh-7 cells with miR-21 mimic or miR-21 ASO and then examined cellular miR-122 levels. As shown in Supplementary Figure S4D, cellular miR-122 levels were not affected by transfection with miR-21 mimics, miR-21 ASO and control oligonucleotides.
Nuclear miR-122 directly binds to a 19-nt cognate element in the basal region of pri-miR-21 transcript
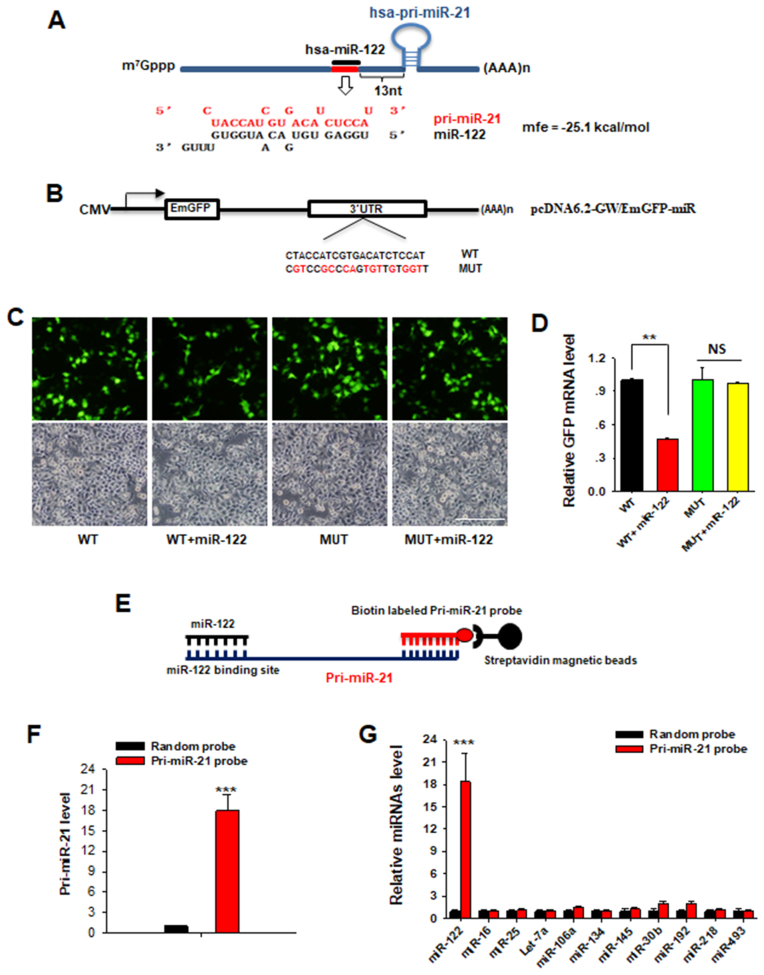
To explore the mechanism underneath the blockade of miR-21 biogenesis by nuclear miR-122, we used RNAhybrid (49) to analyze the potential cognate element on pri-miR-21 for miR-122, and identified a putative binding site for miR-122 on human pri-miR-21. As shown in Figure 4A, a near perfect complementary site for miR-122 is located 13 bp upstream of the pre-miR-21 gene loci. The minimum free energy (ΔG) predicted for hybridization of miR-122 with pri-miR-21 at this site is –25.1 kcal/mol. Moreover, the seed region (the sequence that encompasses the first 2–8 bases of miR-122) displayed a perfect base-pairing with cognate target transcript. Interestingly, a similar putative binding site was also found on mouse pri-miR-21 (Supplementary Figure S5B). The location of the binding element (∼25 bp upstream of the mmu-miR-21 gene loci) resembles the hsa-miR-122 binding site. To further investigate whether the recognition binding element on human pri-miR-21 was responsible for miR-122 binding and blockade of miR-21 biogenesis, we modified GFP expression plasmid by inserting the recognition binding site (Binding-WT) and the mutated binding site (Binding-MUT) into the 3′-UTR of GFP (Figure 4B). HEK-293T cells were then co-transfected with the modified vector and miR-122 mimic. As shown in Figure 4C, co-transfection of miR-122 mimic and the modified pcDNA-GFP-Binding-WT vector into HEK-293T cells significantly reduced the GFP expression (∼30%) compared to co-transfection of miR-122 mimic with the pcDNA-GFP-Binding-MUT vector. We also examined the GFP mRNA expression level in each assay and found that the decreased GFP signal also correlated to the reduced mRNA level (Figure 4D). This result demonstrated that human miR-122 can bind to the predicted binding site on pri-miR-21. To further test the binding of miR-122 with pri-miR-21 in human, we constructed luciferase reporter plasmids containing the predicted miR-122-binding sequence of human pri-miR-21 (WT) or its mutated form (MUT) (Supplementary Figure S6A), and then transfected HEK-293T cells with these plasmids to obtain WT cells and MUT cells. As expected, the WT luciferase reporter activity was significantly reduced following miR-122 transfection, whereas the MUT luciferase reporter activity was unaffected (Supplementary Figure S6B), suggesting that miR-122 can directly bind to the cognate site on pri-miR-21. Similarly, luciferase reporter assay also verified that murine miR-122 could specifically bind to the recognition site on mouse pri-miR-21 (Supplementary Figure S6C).
Figure 4.
Nuclear miR-122 directly binds to a cognate element on pri-miR-21 transcript. (A) Putative binding site for miR-122 on human pri-miR-21 as predicted using RNAhybrid. As shown in the schematic diagram, a near perfect complementary site (red rectangle) for miR-122 (black rectangle) was located 13 bp upstream of the pre-miR-21 gene loci. mfe: minimum free energy. (B) Schematic representations of modified GFP expression plasmid. MiR-122 binding sequences (Binding-WT) and mutant sequences (Binding-MUT) were inserted into the 3′-UTR of GFP in GFP expression plasmid. (C) HEK-293T cells were co-transfected with the modified vector and miR-122 mimic. After 48 h, fluorescence microscopy was used to detect GFP expression. Scale bar: 1.0 mm. (D) The GFP mRNA expression level was detected by RT-qPCR. NS: no significance. (E) Schematic illustration of pri-miR-21 pull-down strategy. (F and G) Levels of pri-miR-21 (F) or miR-122 and other candidate miRNAs (G) in pull-down products which were co-precipitated by anti-pri-miR-21 probe or random probe. Data are presented as mean ± SD (N = 3) of three independent experiments. **P < 0.01; ***P < 0.001.
To corroborate the above finding, we further carried out a biotin–streptavidin pulldown experiment to assess the direct binding of miR-122 to pri-miR-21. Briefly, a biotin-labeled specific anti-pri-miR-21 probe was designed and incubated with the nuclear lysate from Huh-7 cells. Pri-miR-21 and miR-122 were co-precipitated via streptavidin-conjugated magnetic beads, and the levels of pri-miR-21 and miR-122 in the pulldown products were analyzed by RT-qPCR (Figure 4E). As expected, pri-miR-21 was significantly enriched (∼18-fold higher) in the pri-miR-21 pull-down product as compared to the random probe product (Figure 4F), indicating that pri-miR-21 transcript could be immunoprecipitated by pri-miR-21 probe. Intriguingly, miR-122 was also immunoprecipitated by pri-miR-21 probe and displayed a similar enrichment as pri-miR-21 in the pulldown product (Figure 4G). We also tested other miRNAs, including miR-16, miR-25, let-7a, etc., in pulldown products and found that these miRNAs were not immunoprecipitated by pri-miR-21 probe, suggesting the specific binding manner between pri-miR-21 and miR-122.
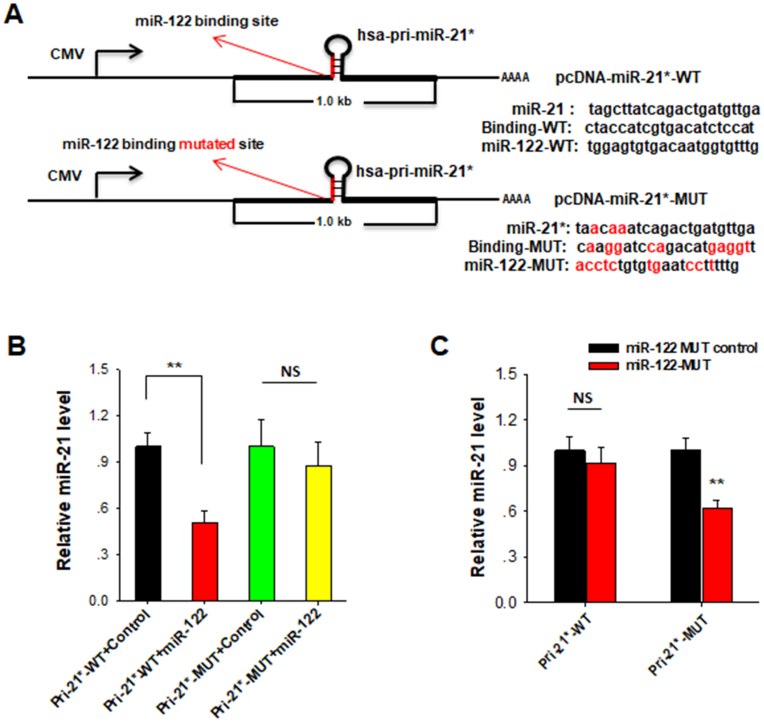
To evaluate the impact of nuclear miR-122 on the maturation of miR-21 in a cellular environment, we constructed the human pri-miR-21 expression plasmid by cloning 1 kb of miR-21 gene (510 bp flanking sequences on its 5′ end and 490 bp flanking sequences on its 3′ end) into a modified vector. The mature miR-21 sequence in the vector was mutated by replacing three nucleotides (miR-21*) so our construct could be distinguished from the endogenous miR-21. In this experiment, the pri-miR-21 plasmids with or without mutation of miR-122 binding site (pcDNA-miR-21*-WT or pcDNA-miR-21*-MUT) were constructed (Figure 5A), and co-transfected with miR-122 mimic into HEK-293T cells, respectively. The mature miR-21* was then detected using the customized probe after harvesting cells 48 h post-infection. As shown in Figure 5B, miR-122 mimic significantly reduced the miR-21* level in cells co-transfected with pcDNA-miR-21*-WT, but did not affected the miR-21* level in the cells co-transfected with pcDNA-miR-21*-MUT. Furthermore, we mutated miR-122 mimic to miR-122-MUT, which is complementary to the mutated miR-122 binding site in the pcDNA-miR-21*-MUT construct (Figure 5A). In a similar fashion, cells were co-transfected with miR-122-MUT or control oligonucleotide and pcDNA-miR-21*-WT or pcDNA-miR-21*-MUT plasmids, respectively. Compared with control group, miR-122-MUT inhibited the biogenesis of miR-21 in cells transfected with pcDNA-miR-21*-MUT but not with pcDNA-miR-21*-WT (Figure 5C). These results suggest that miR-122 can regulate the miR-21 biogenesis by directly binding to the recognition element on pri-miR-21 in a cell.
Figure 5.
MiR-122 reduces the level of miR-21* which was generated from artificial miR-21* expression vector. (A) Modification of human pri-miR-21 expression vector. The pri-miR-21 substrate consisted of about 1 kb of human pri-miR-21, with 510 nt flanking pre-miR-21 on its 5′ side and 490 nt flanking sequences on its 3′ side. The mature-miR-21 sequences were mutated with three nucleotides and marked miR-21* so can be distinguished from the endogenous miR-21. The pri-miR-21 plasmids with (red) or without mutation of miR-122 binding site (pcDNA-miR-21*-WT or pcDNA-miR-21*-MUT) were constructed. (B) pcDNA-miR-21*-WT or pcDNA-miR-21*-MUT plasmids were co-transfected with miR-122 mimic into HEK-293T cells, respectively. The mature miR-21* was then detected using the customized probe after harvesting cells 48 h post-infection. (C) miR-122-MUT mimic was co-transfected with pcDNA-miR-21*-WT or pcDNA-miR-21*-MUT plasmids into HEK-293T cells, respectively. The mature miR-21* level was then detected 48 h post-transfection. NS: No Significance. The results are presented as the mean ± SD (N = 3) of three independent experiments. **P < 0.01.
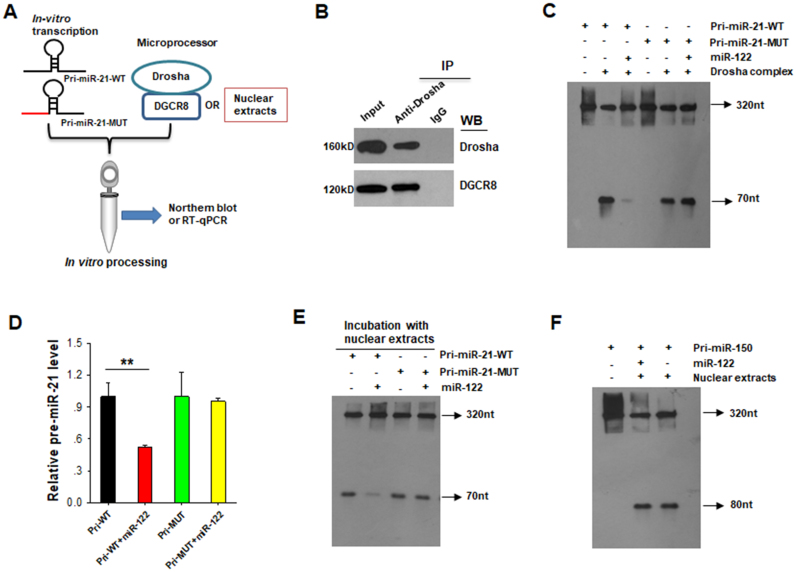
In vitro miRNA processing assay has recently been shown as a powerful tool to study miRNA biogenesis and regulatory function (4). To obtain more visible and robust evidence, we applied an in vitro miRNA processing system to evaluate the inhibiting effect of miR-122 on the maturation of miR-21. The schematic diagram of in vitro miRNA processing assay strategy is presented in Figure 6A. A pri-miR-21 plasmid of ∼320 bp (pri-miR-21-WT) containing the pre-miR-21 hairpin was synthesized (41). Using the same method, we constructed a pri-miR-21 plasmid with the miR-122-binding site being mutated (pri-miR-21-MUT). We first amplified the pri-miR-21 products from both pri-miR-21-WT and pri-miR-21-MUT plasmids and transcribed them in vitro by T7 RNA polymerase to obtain the pri-miR-21 or pri-mut-miR-21 transcripts. We then purified Drosha-based microprocessor from nuclear lysate via immunoprecipitation using anti-Drosha antibody (4). As shown in Figure 6B, the protein Drosha, as well as DGCR8 which acts as a cofactor protein, was co-immunoprecipitated by anti-Drosha antibody. Finally, we incubated pri-miR-21 or pri-mut-miR-21 transcripts with miR-122 in the presence or absence of isolated Drosha-based microprocessor. After in vitro processing, the cleaved processing products were separated on a denaturing polyacrylamide urea gel and detected by northern blot analysis. As shown in Figure 6C, the pri-miR-21 band at the 320 nt was present in all lanes, suggesting that the pri-miR-21 and pri-mut-miR-21 were successfully transcribed. Additionally, a band at 70 nt, the location of pre-miR-21, was present for both the pri-miR-21 and pri-mut-miR-21 products in the absence of miR-122. However, miR-122 displayed a differential effect on the processing of pri-miR-21 and pri-mut-miR-21 by microprocessor. As shown, miR-122 significantly repressed the cleavage of pri-miR-21 into pre-miR-21 by microprocessor, but had no effect on processing of pri-mut-miR-21 into pre-miR-21 under the same condition. We also analyzed the levels of pre-miR-21 in these in vitro processing assays by RT-qPCR and the results confirmed the northern blot observation that miR-122 blocks the cleavage of pri-miR-21 but not pri-mut-miR-21 by Drosha-based microprocessor (Figure 6D). In a parallel fashion, we performed the same assay using the nuclear extracts as the catalyzed system. In this experiment, in addition to pri-mut-miR-21, another unrelated miRNA primary transcript, pri-miR-150, was used as a control. As shown in Figure 6E and F, miR-122 significantly repressed the cleavage of pri-miR-21 into pre-miR-21, but had no effect on processing of pri-mut-miR-21 and pri-miR-150 into pre-miR-21 and pre-miR-150, respectively. These data strongly support that nuclear miR-122 can prevent Drosha's processing of pri-miR-21 to pre-miR-21 via binding to the recognition element on pri-miR-21.
Figure 6.
MiR-122 blocks the primary miR-21 processing by in vitro pri-miRNA processing assay. (A) Schematic illustration of pri-miR-21 in vitro processing assay strategy. (B) For immunoprecipitation (IP) assays, Huh-7 cell lysates were incubated with anti-Drosha antibody and IgG control antibody, The IP-product was then detected by western blotting (WB) using anti-Drosha and anti-DGCR8 antibody. (C) Northern blotting analysis of the miR-122 blocking in vitro processing of the pri-miR-21-WT and pri-miR-21-MUT. The pri-miR-21-WT and pri-miR-21-MUT transcripts were incubation of synthetic mature single strand miR-122, respectively, and then cleaved by Drosha-complex in vitro. The in vitro processing products were analyzed by northern blot. (D) RT-qPCR validation of pre-miR-21 level in processing products. (E) The pri-miR-21-WT and pri-miR-21-MUT transcripts were incubation of synthetic mature single strand miR-122, respectively, and then cleaved by nuclear extracts. The in vitro processing products were analyzed by northern blot. (F) An unrelated pri-miR-150 was incubation of synthetic mature single strand miR-122 and then cleaved by nuclear extracts. The in vitro processing products were analyzed by northern blot. The results are presented as the mean ± SD (N = 3) of three independent experiments. **P < 0.01.
Nuclear miR-122 promotes liver cell apoptosis via downregulating miR-21 but increasing PDCD4 activity
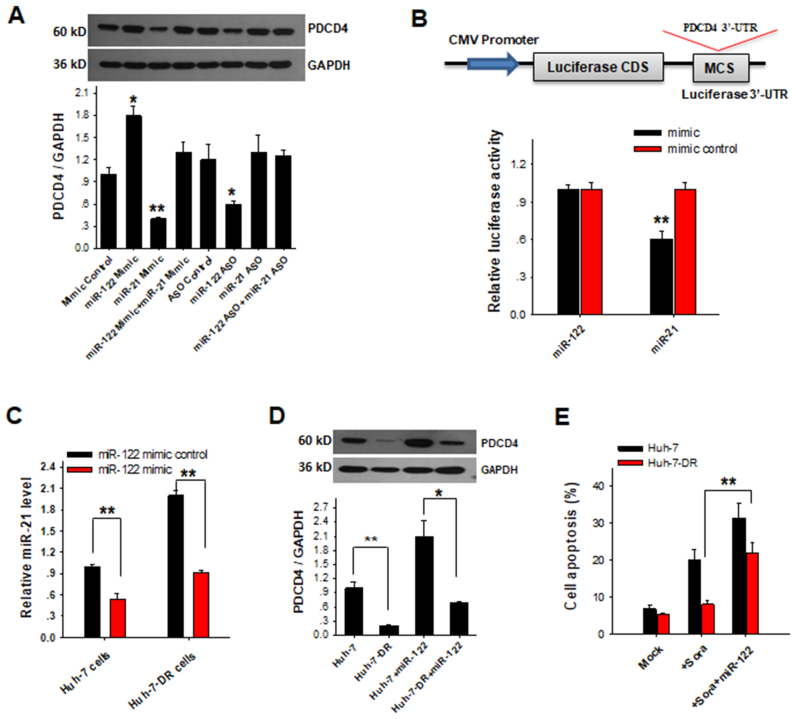
As a cell survival oncomiR, miR-21 is usually upregulated in most tumors (34,35,50–54) and can target and suppress the tumor suppressor protein PDCD4 thereby promoting tumor cell survival (33,35,55,56). We next tested if alteration of miR-21 by nuclear miR-122 had a biological role in HCC cells. As shown in Figure 7A, ectopic expression of miR-122 induced the upregulation of PDCD4 in Huh-7 cells and this effect of miR-122 was abolished by co-expression with miR-21, suggesting that miR-122 overexpression may affect the miR-21/PDCD signal pathway. In agreement with the notion that nuclear miR-122 enhances PDCD4 pathway via reducing cellular miR-21 level, depleting miR-122 resulted in an elevation of cellular miR-21 level but downregulation of PDCD4. To exclude the possibility of miR-122 directly binding to the PDCD4 gene 3′-UTR and thus repressing its expression, we constructed the whole PDCD4 3′-UTR sequences into the firefly luciferase reporter plasmid and used luciferase reporter assay to detect the binding activity of the PDCD4 gene 3′-UTR with miR-122 or miR-21 (Figure 7B, upper panel). The plasmid was then introduced into 293T cells along with a transfection control plasmid expressing β-galactosidase and miR-122 mimic, miR-21 mimic or the mimic control. As shown in Figure 7B, lower panel, miR-122 overexpression failed to result in significant luciferase activity change compared to control oligonucleotides. In contrast, transfection with miR-21 mimic showed a significant decrease in the luciferase reporter activity.
Figure 7.
Nuclear miR-122 regulates cell apoptosis via modulating miR-21/PDCD4 signal pathway. (A) PDCD4 protein level in Huh-7 cells was assessed at 24 h post-transfection with various oligonucleotides including mimic control, miR-122 mimic, miR-21 mimic, miR-122 mimic plus miR-21 mimic, ASO control, miR-122 ASO, miR-21 ASO or miR-122 ASO plus miR-21 ASO. Upper panels, representative western blot. Bottom panels representative quantitative analysis of western blot. (B) Luciferase reporter assay testing the possible binding of miR-122 on PDCD4 3′-UTR. The whole PDCD4 3′-UTR sequences were cloned into the luciferase plasmid. The plasmid was co-transfected into HEK-293T cells along with miR-122 mimic or mimic control RNA. As a positive control, miR-21 mimic or scrambled RNA was also co-transfected with luciferase plasmid. At 24 h post-transfection, the cells were assayed using a luciferase assay kit. Firefly luciferase values were normalized to β-galactosidase activity and luciferase activity in the scrambled RNA-transfected cells was set as 1. (C) Relative miR-21 level in Huh-7 cells and Huh-7-DR cells by ectopic expression of miR-122 mimic or mimic control respectively. (D) PDCD4 protein levels in Huh-7 cells and Huh-7-DR cells were assessed at 24 h post-transfection with miR-122 mimic: representative western blot (upper panel) and quantitative analysis (bottom panel). (E) Huh-7 and Huh-7-DR cells were treated with sorafenib (Sora) for 12 h and then transfected with miR-122 mimic or control respectively. Apoptosis was detected by PI/Annexin V staining at 24 h post-transfection. The results are presented as the mean ± SD (N = 3) of three independent experiments. *P < 0.05; **P < 0.01.
Given that PDCD4 plays a critical role in suppressing tumor cell chemo-resistance (32,33), we next tested whether nuclear miR-122 could reduce tumor cell chemo-resistance through enhancing PDCD4 expression. For this purpose, Huh-7 cells resistant to sorafenib (Huh-7-DR) were generated according to strategy previously described (31). As shown in Supplementary Figure S7A, miR-122 expression was markedly reduced in Huh-7-DR cells compared with that in Huh-7 cells. In contrast, miR-21 level was significantly increased in Huh-7-DR cells, which was in agreement with previous finding of elevation of miR-21 level in sorafenib-resistant HCC cells (31). We further determined the effect of miR-122 on tumor cell drug-resistance by overexpressing miR-122 in both Huh-7-DR and Huh-7 cells. The result showed that miR-122 overexpression significantly reduced miR-21 level in Huh-7-DR cells after 24 h post-transfection (Figure 7C). In line with this, the level of PDCD4, a tumor suppressor targeted by miR-21, was dramatically increased in the Huh-7-DR cells after ectopic expression of miR-122 (Figure 7D). Comparing the levels of pri-miR-21 and pre-miR-21 in Huh-7-DR cells after ectopic expression of miR-122, we found that pre-miR-21 was significantly downregulated compared to control group, though pri-miR-21 levels were almost the same in two groups (Supplementary Figure S7B).
We next tested whether the alteration of miR-122 plays a role in cell apoptosis through the miR-21-targeting PDCD4 pathway. In this experiment, sorafenib-induced cell apoptosis was assessed by flow cytometry analysis. As shown in Figure 7E, soranfenib treatment (5 μM, 12 h) in Huh-7 cells caused a higher apoptosis (27.4%) than in Huh-7-DR cells (6.8%), confirming the drug-resistance of Huh-7-DR cells. However, transfection with miR-122 mimic significantly increased sorafenib-induced apoptosis rate in Huh-7-DR cells (from 6.8 to 23%) compared to that in Huh-7 cells (from 27.4 to 33%). Taken together, these results suggest that the pro-apoptotic function of nuclear miR-122 on liver tumor cells is likely through modulating miR-21-PDCD4 pathway.
MiR-122 inhibits HCC growth in mice through reducing the expression of cell survival miR-21
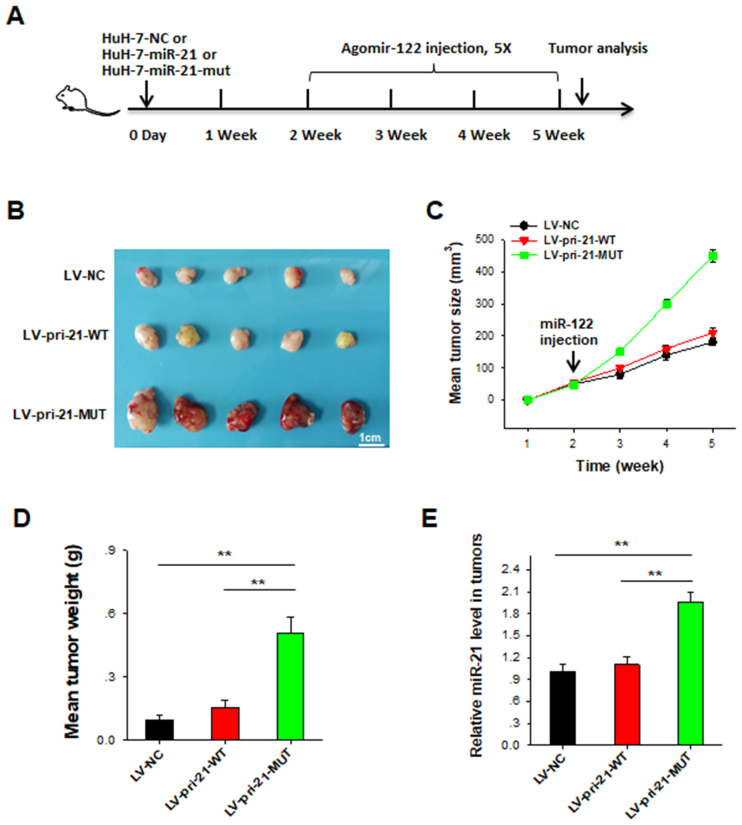
In order to investigate the antitumor effect of miR-122 in vivo, we established a nude mice hepatoma xenograft model using Huh-7 stable infected cell lines. Firstly, Huh-7 cells stably expressing miR-21 or miR-21-MUT were generated by infecting cells with lentivirus-packaged pri-miR-21-WT (LV-pri-21-WT) or lentivirus pri-miR-21-MUT (LV-pri-21-MUT) followed by several rounds of selection under pressure of puromycin (2 μg/ml) (57). Lentivirus expressing control oligonucleotide (LV-NC) served as the control in the experiment. As designed in Figure 8A, 2 weeks after subcutaneously implanting the stable transfected Huh-7 cells in nude mice, the mice were intratumorally injected with cholesterol-conjugated miR-122 agomir (10 nmol in 0.1 ml saline per mouse) once every 3 days for 3 weeks. After 5 weeks, mice were sacrificed and tumors were excised for further analysis. The results showed that treatment with miR-122 significantly inhibited the tumor growth (Figure 8B–D) in mice implanted with LV-NC or LV-pri-21-WT cells, but displayed no effect on tumor growth in mice implanted with LV-pri-21-MUT cells. In line with this, RT-qPCR analysis of RNA samples extracted from excised tumor showed that miR-21 level in tumors from LV-pri-21-MUT group was significantly higher than those from LV-pri-21-WT and LV-NC groups (Figure 8E). These results suggest that miR-122 might suppress HCC growth through decreasing the production of cell survival miR-21 in tumor cells.
Figure 8.
MiR-122 inhibits HCC growth in mice through suppressing the expression of cell survival oncomiR miR-21. (A) The schematic diagram illustrating the experimental design. Huh-7 cells were stable transfected with LV-pri-21-WT, LV-pri-21-MUT and LV-NC. A nude mice hepatoma xenograft model was established by using the stable infected cell lines. Two weeks after subcutaneously implanting the stable transfected Huh-7 cells into nude mice, the mice were then intratumorally injected with miR-122 agomir once every 3 days for 3 weeks. (B) Representative images of tumors after 5 weeks. (C) The time course of tumor size. (D) The quantitative analysis of tumor weight. (E) RT-qPCR analysis of miR-21 level from excised tumor RNA samples. The results are presented as the mean ± SD (N = 3) of three independent experiments. **P < 0.01.
DISCUSSION
MiR-122 is the most abundant miRNA in healthy hepatocytes. However, in cancerous hepatocytes the level of miR-122 is strongly reduced (26,29). Our current study articulates the mechanisms, involving the interactions between miR-122 and miR-21, which result in alteration of PDCD4-mediated cancer cells re-sensitization to chemotherapy. In identifying these mechanisms, we have revealed yet another example of a miRNA performing a function in the nucleus of the cell.
Consistent with the findings by others, we showed that the level of miR-122 particularly the level of nuclear miR-122 was strongly suppressed in cancerous hepatocytes in comparison to healthy hepatocytes. Although the mechanism of transportation remains unclear, mature miR-122 can enter into liver cell nucleus (12). We were able to identify a specific function that miR-122 performs in the nucleus of the cell. Previous studies by us (23) and others (24) have shown nuclear miRNAs can regulate the biogenesis of other miRNAs. These prior findings prompted the hypothesis that nuclear miR-122 may execute a critical tumor-suppressive role through blocking the biogenesis of certain tumor cell survival factors, particularly oncomiRs. TaqMan Low Density Array and RT-qPCR assays selected miR-21 as one such oncomiR regulated by nuclear miR-122. Our study generated several findings confirming that nuclear miR-122 inhibits the biosynthesis of miR-21: First, increase or decrease of nuclear miR-122 respectively resulted in downregulation or upregulation of miR-21. An inverse relationship between miR-122 and miR-21 was observed in liver tissues and cells. Second, we identified a 19-nt recognition element for miR-122 in the basal region of pri-miR-21 and we confirmed the binding of miR-122 to this recognition element, through the mutated version and by luciferase reporter assay and immunoprecipitation. In line with this, our data indicated that nuclear miR-122 only reduced pre-miR-21 level but not pri-miR-21 level, suggesting that nuclear miR-122 affect the biogenesis of miR-21 at posttranscriptional level, specifically, at the stage of processing of pri-miR-21 into pre-miR-21. Finally, we used an in vitro pri-miRNA processing system to show that miR-122 can block the processing of pri-miR-21 into pre-miR-21 by Drosha-based microprocessor through binding to the recognition element on pri-miR-21. Previous study suggests that regulation of mature miRNA redistribution from cytoplasm to nucleus is critical for its functions (58). Although the molecular basis modulating miR-122 redistribution in transformed vs primary cells is not clear, our recent report implies that RNA import and export factors such as Importin 8 may be involved in guiding mature miRNA re-entering into the nucleus (43). The nucleo-cytoplasmic trafficking of mature miRNAs mediated by RNA import/export factors have been identified by other groups (21,59).
We went on to investigate the mechanisms through which the lack of miR-122 influences oncomiR miR-21 and confers resistance to chemotherapy induced apoptosis in tumor cells. MiR-21 is a well-studied cell survival factor and is found to be upregulated in various cancer cells (35,51,53,60,61). As a cell survival oncomiR, miR-21 can target and suppress the tumor suppressor protein PDCD4 thereby promoting tumor cell survival (33,35,55,56). We confirmed that when miR-122 is overexpressed then the levels of PDCD4 are increased. Conversely, when miR-122 is absent, levels of PDCD4 decline. We further confirmed that miR-122 does not directly affect the processing of PDCD4 mRNA, although miR-21 does. Moreover, we showed that miR-122 overexpression restores vulnerability to sorafenib-induced-apoptosis in previously drug-resistant tumor cells. In our study, we only focused on the impact of miR-122 on vulnerability to sorafenib induced apoptosis. Future work may confirm the impact of nuclear miR-122 on other cancer related events such as cell transformation, epithelial to mesenchymal transition, and metastasis.
Our study contributes to the knowledge base regarding the function of miR-122. In the cytosol, miR-122 controls the translation of proteins involved in fatty acid metabolism including CD320, AldoA, BCKDK and the genes involved in IFN signaling. We showed that miR-122 performs a function in the nucleus as well, viz., inhibiting the maturation of miR-21 so that miR-21 cannot inhibit PDCD4. Thus, miR-122, indirectly through inhibiting miR-21 levels, can act as a pro-apoptotic factor to promote tumor cell apoptosis and control tumor invasion. Given that miR-122 is a predominant miRNA in liver cell and normally is abundantly located in nucleus, it may control the biogenesis of other miRNAs as well. Supporting this hypothesis, we found that decrease of nuclear miR-122 level resulted in rapid increase of miR-21 and other miRNAs including miR-30b and miR-16. Sequence analysis also predicted the miR-122-binding sites on the primary transcripts of these miRNAs (Supplementary Figure S5A).
Previously, the work of others focused on factors controlling the posttranscriptional maturation of miR-21 (41,47,62). For example, Trabucchi et al. showed that KSRP protein can bind to the terminal loops of a subset of miRNA precursor, including pri-miR-21, to promote their cleavage by Drosha (62). In a similar manner, SMADA protein was reported to promote miR-21 biogenesis via promoting the processing of pri-miR-21 into pre-miR-21 by the Drosha complex (47). In contrast, Diaz et al. found a small molecule that can specifically inhibit the cleavage of pri-miR-21 by the microprocessor complex of Drosha and DGCR8 (41). Our study further contributes to the knowledge base regarding the control of miR-21 maturation. We showed that nuclear miR-122 inhibits miR-21 biogenesis through directly binding to the flanking sequence of pri-miR-21, blocking the processing of pri-miR-21 by the microprocessor complex of Drosha and DGCR8.
Our study also contributes to the knowledge base regarding the configurations through which mature miRNAs can control the maturation of other primary miRNAs. The sequences flanking the stem loop have been shown to be critical for efficient processing of miRNA primary transcripts (63). Auyeung et al. recently showed that certain sequence elements in the basal region (the UG motif (–13) and the CNNC (+17) motif) and terminal loop (the UGUG motif) of human pri-miRNAs play an important role in facilitating pri-miRNA processing, and they found that at least one of these three motifs is present in 79% of human miRNAs (64). Different from the relative remote location of miR-709 binding sequence on pri-miR-15a/16-1 (23), miR-122 binding site on pri-miR-21 is very close to the stem loop of miR-122 (13 bp upstream of the pre-miR-21 gene loci). Interestingly, the UG motif was also found in the miR-122-binding sequence element in the basal region of pri-miR-21. It is likely that miR-122 binds to the UG motif (13 bp) of pri-miR-21 to block the processing of pri-miR-21 by the microprocessor complex of Drosha and DGCR8.
In summary, our study demonstrates that miR-122 can enter into cell nucleus where it directly targets the primary transcript of miR-21 and thus controls miR-21 biogenesis through blocking the processing of pri-miR-21 to pre-miR-21 by the microprocessor complex of Drosha and DGCR8. The consequences of blocking the maturation of pri-miR-21 to mature miR-21 are profound for hepatic tumor cells. When levels of miR-122 are restored in tumor cells, then maturation of miR-21 is prevented and levels of PDCD4 increase and vulnerability to chemotherapy induced apoptosis is restored. Thus, the level of nuclear miR-122 level may be a key factor in tumor cell chemoresistance.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Jill Leslie Littrell (Georgia State University, Atlanta, GA) for critical reading and constructive discussion of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
FUNDING
National Natural Science Foundation of China [31670917, 91640103, 31770981]; Natural Science Foundation of Jiangsu Province [BK20170076]; Six talent peaks project of Jiangsu Province; Fundamental Research Funds for the Central Universities [020814380039, 020814380082]. Funding for open access charge: Natural Science Foundation of Jiangsu Province [BK20170076].
Conflict of interest statement. None declared.
REFERENCES
- 1. Ambros V. The functions of animal microRNAs. Nature. 2004; 431:350–355. [DOI] [PubMed] [Google Scholar]
- 2. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 3. Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003; 425:415–419. [DOI] [PubMed] [Google Scholar]
- 4. Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N.. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002; 21:4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lund E., Guttinger S., Calado A., Dahlberg J.E., Kutay U.. Nuclear export of microRNA precursors. Science. 2004; 303:95–98. [DOI] [PubMed] [Google Scholar]
- 6. Kim V.N. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol. 2004; 14:156–159. [DOI] [PubMed] [Google Scholar]
- 7. Zeng Y., Yi R., Cullen B.R.. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Filipowicz W., Jaskiewicz L., Kolb F.A., Pillai R.S.. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005; 15:331–341. [DOI] [PubMed] [Google Scholar]
- 9. Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T.. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004; 15:185–197. [DOI] [PubMed] [Google Scholar]
- 11. Hwang H.W., Wentzel E.A., Mendell J.T.. A hexanucleotide element directs microRNA nuclear import. Science. 2007; 315:97–100. [DOI] [PubMed] [Google Scholar]
- 12. Foldes-Papp Z., Konig K., Studier H., Buckle R., Breunig H.G., Uchugonova A., Kostner G.M.. Trafficking of mature miRNA-122 into the nucleus of live liver cells. Curr. Pharmaceut. Biotechnol. 2009; 10:569–578. [DOI] [PubMed] [Google Scholar]
- 13. Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R.. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014; 6:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Robb G.B., Brown K.M., Khurana J., Rana T.M.. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005; 12:133–137. [DOI] [PubMed] [Google Scholar]
- 15. Politz J.C., Hogan E.M., Pederson T.. MicroRNAs with a nucleolar location. RNA. 2009; 15:1705–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liao J.Y., Ma L.M., Guo Y.H., Zhang Y.C., Zhou H., Shao P., Chen Y.Q., Qu L.H.. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010; 5:e10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeffries C.D., Fried H.M., Perkins D.O.. Nuclear and cytoplasmic localization of neural stem cell microRNAs. RNA. 2011; 17:675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khudayberdiev S.A., Zampa F., Rajman M., Schratt G.. A comprehensive characterization of the nuclear microRNA repertoire of post-mitotic neurons. Front. Mol. Neurosci. 2013; 6:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park C.W., Zeng Y., Zhang X., Subramanian S., Steer C.J.. Mature microRNAs identified in highly purified nuclei from HCT116 colon cancer cells. RNA Biol. 2010; 7:606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu Y., Yue X., Younger S.T., Janowski B.A., Corey D.R.. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010; 38:7736–7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ohrt T., Mutze J., Staroske W., Weinmann L., Hock J., Crell K., Meister G., Schwille P.. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008; 36:6439–6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nishi K., Nishi A., Nagasawa T., Ui-Tei K.. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. R,NA. 2013; 19:17–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang R., Li L., Zhu D., Hou D., Cao T., Gu H., Zhang J., Chen J., Zhang C.Y., Zen K.. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: evidence for a microRNA hierarchy system. Cell Research. 2012; 22:504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zisoulis D.G., Kai Z.S., Chang R.K., Pasquinelli A.E.. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature. 2012; 486:541–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R. et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004; 1:106–113. [DOI] [PubMed] [Google Scholar]
- 26. Lin C.J., Gong H.Y., Tseng H.C., Wang W.L., Wu J.L.. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2008; 375:315–320. [DOI] [PubMed] [Google Scholar]
- 27. Coulouarn C., Factor V.M., Andersen J.B., Durkin M.E., Thorgeirsson S.S.. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009; 28:3526–3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tsai W.C., Hsu P.W., Lai T.C., Chau G.Y., Lin C.W., Chen C.M., Lin C.D., Liao Y.L., Wang J.L., Chau Y.P. et al. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009; 49:1571–1582. [DOI] [PubMed] [Google Scholar]
- 29. Kutay H., Bai S., Datta J., Motiwala T., Pogribny I., Frankel W., Jacob S.T., Ghoshal K.. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell. Biochem. 2006; 99:671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bai S., Nasser M.W., Wang B., Hsu S.H., Datta J., Kutay H., Yadav A., Nuovo G., Kumar P., Ghoshal K.. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J. Biol. Chem. 2009; 284:32015–32027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu Y., Huang J., Ma L., Shan J., Shen J., Yang Z., Liu L., Luo Y., Yao C., Qian C.. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Cancer Lett. 2016; 371:171–181. [DOI] [PubMed] [Google Scholar]
- 32. Jansen A.P., Camalier C.E., Stark C., Colburn N.H.. Characterization of programmed cell death 4 in multiple human cancers reveals a novel enhancer of drug sensitivity. Mol. Cancer Therapeut. 2004; 3:103–110. [PubMed] [Google Scholar]
- 33. Tomimaru Y., Eguchi H., Nagano H., Wada H., Tomokuni A., Kobayashi S., Marubashi S., Takeda Y., Tanemura M., Umeshita K. et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br. J. Cancer. 2010; 103:1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Medina P.P., Nolde M., Slack F.J.. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010; 467:86–90. [DOI] [PubMed] [Google Scholar]
- 35. Liu C., Yu J., Yu S., Lavker R.M., Cai L., Liu W., Yang K., He X., Chen S.. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J. Hepatol. 2010; 53:98–107. [DOI] [PubMed] [Google Scholar]
- 36. Li W.C., Ralphs K.L., Tosh D.. Isolation and culture of adult mouse hepatocytes. Methods Mol. Biol. 2010; 633:185–196. [DOI] [PubMed] [Google Scholar]
- 37. Luo Y., Wang C., Chen X., Zhong T., Cai X., Chen S., Shi Y., Hu J., Guan X., Xia Z. et al. Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin. Chem. 2013; 59:658–666. [DOI] [PubMed] [Google Scholar]
- 38. Schmittgen T.D., Lee E.J., Jiang J., Sarkar A., Yang L., Elton T.S., Chen C.. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008; 44:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dixon-McIver A., East P., Mein C.A., Cazier J.B., Molloy G., Chaplin T., Andrew Lister T., Young B.D., Debernardi S.. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS One. 2008; 3:e2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sempere L.F., Preis M., Yezefski T., Ouyang H., Suriawinata A.A., Silahtaroglu A., Conejo-Garcia J.R., Kauppinen S., Wells W., Korc M.. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin. Cancer Res. 2010; 16:4246–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diaz J.P., Chirayil R., Chirayil S., Tom M., Head K.J., Luebke K.J.. Association of a peptoid ligand with the apical loop of pri-miR-21 inhibits cleavage by Drosha. RNA. 2014; 20:528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim S.W., Li Z., Moore P.S., Monaghan A.P., Chang Y., Nichols M., John B.. A sensitive non-radioactive northern blot method to detect small RNAs. Nucleic Acids Res. 2010; 38:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wei Y., Li L., Wang D., Zhang C.Y., Zen K.. Importin 8 regulates the transport of mature microRNAs into the cell nucleus. J. Biol. Chem. 2014; 289:10270–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang H.W., Wang N., Wang Y., Wang F., Fu Z., Yan X., Zhu H., Diao W., Ding Y., Chen X. et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J. Hepatol. 2016; 64:278–291. [DOI] [PubMed] [Google Scholar]
- 45. Zeisel M.B., Pfeffer S., Baumert T.F.. miR-122 acts as a tumor suppressor in hepatocarcinogenesis in vivo. J. Hepatol. 2013; 58:821–823. [DOI] [PubMed] [Google Scholar]
- 46. Wen J., Friedman J.R.. miR-122 regulates hepatic lipid metabolism and tumor suppression. J. Clin. Invest. 2012; 122:2773–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Davis B.N., Hilyard A.C., Lagna G., Hata A.. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008; 454:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bail S., Swerdel M., Liu H., Jiao X., Goff L.A., Hart R.P., Kiledjian M.. Differential regulation of microRNA stability. RNA. 2010; 16:1032–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R.. Fast and effective prediction of microRNA/target duplexes. RNA. 2004; 10:1507–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Buscaglia L.E., Li Y.. Apoptosis and the target genes of microRNA-21. Chin. J. Cancer. 2011; 30:371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chan J.A., Krichevsky A.M., Kosik K.S.. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005; 65:6029–6033. [DOI] [PubMed] [Google Scholar]
- 52. Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T.. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007; 133:647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Si M.L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.Y.. miR-21-mediated tumor growth. Oncogene. 2007; 26:2799–2803. [DOI] [PubMed] [Google Scholar]
- 54. Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y.Y.. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008; 18:350–359. [DOI] [PubMed] [Google Scholar]
- 55. Asangani I.A., Rasheed S.A., Nikolova D.A., Leupold J.H., Colburn N.H., Post S., Allgayer H.. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008; 27:2128–2136. [DOI] [PubMed] [Google Scholar]
- 56. Frankel L.B., Christoffersen N.R., Jacobsen A., Lindow M., Krogh A., Lund A.H.. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 2008; 283:1026–1033. [DOI] [PubMed] [Google Scholar]
- 57. Gao J., Zhao N., Knutson M.D., Enns C.A.. The hereditary hemochromatosis protein, HFE, inhibits iron uptake via down-regulation of Zip14 in HepG2 cells. J. Biol. Chem. 2008; 283:21462–21468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pitchiaya S., Heinicke L.A., Park J.I., Cameron E.L., Walter N.G.. Resolving Subcellular miRNA Trafficking and Turnover at Single-Molecule Resolution. Cell Rep. 2017; 19:630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Castanotto D., Lingeman R., Riggs A.D., Rossi J.J.. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:21655–21659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Iorio M.V., Ferracin M., Liu C.G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M. et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005; 65:7065–7070. [DOI] [PubMed] [Google Scholar]
- 61. Schetter A.J., Leung S.Y., Sohn J.J., Zanetti K.A., Bowman E.D., Yanaihara N., Yuen S.T., Chan T.L., Kwong D.L., Au G.K. et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008; 299:425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Trabucchi M., Briata P., Garcia-Mayoral M., Haase A.D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M.G.. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009; 459:1010–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N.. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006; 125:887–901. [DOI] [PubMed] [Google Scholar]
- 64. Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P.. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013; 152:844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.