Abstract
The formation of 3′ single-stranded DNA overhangs is a first and essential step during homology-directed repair of double-stranded breaks (DSB) of DNA, a task that in Escherichia coli is performed by RecBCD. While this protein complex has been well characterized through in vitro single-molecule studies, it has remained elusive how end resection proceeds in the crowded and complex environment in live cells. Here, we develop a two-color fluorescent reporter to directly observe the resection of individual inducible DSB sites within live E. coli cells. Real-time imaging shows that RecBCD during end resection degrades DNA with remarkably high speed (∼1.6 kb/s) and high processivity (>∼100 kb). The results show a pronounced asymmetry in the processing of the two DNA ends of a DSB, where much longer stretches of DNA are degraded in the direction of terminus. The microscopy observations are confirmed using quantitative polymerase chain reaction measurements of the DNA degradation. Deletion of the recD gene drastically decreased the length of resection, allowing for recombination with short ectopic plasmid homologies and significantly increasing the efficiency of horizontal gene transfer between strains. We thus visualized and quantified DNA end resection by the RecBCD complex in live cells, recorded DNA-degradation linked to end resection and uncovered a general relationship between the length of end resection and the choice of the homologous recombination template.
INTRODUCTION
Double-stranded breaks (DSB) are exceptionally toxic DNA damage events because of the simultaneous loss of integrity of both strands of the DNA duplex. If left unrepaired or when repaired incorrectly, these breaks can have dramatic consequences such as cell death, deleterious mutations, chromosomal translocations, or genomic rearrangements (1). DSBs can be repaired with a template-independent non-homologous end joining (NHEJ) reaction, or by the homologous recombination (HR), where the genetic information is recovered from an intact, homologous template. While NHEJ is present only in a eukaryotes, archaea and some phyla of bacteria (2), HR seems to be common in all domains of life (3). The most fundamental steps in HR share a great resemblance in their mechanism across different organisms. Accordingly, findings established in simple model organism, such as budding yeast, or bacteria, have been successfully extrapolated and helped to understand processes found in mammalian cells (4).
In Escherichia coli, DSB repair is initiated by binding of the complex molecular machine RecBCD, a nuclease/helicase that recognizes and binds to the ends of a broken double-stranded DNA (dsDNA). Both RecB and RecD subunits are helicases, but with different polarities, as RecB translocates in the 3′ to 5′ direction, whereas RecD moves from 5′ to 3′ and is the dominant motor initially (5). The activity of RecBCD is regulated by an asymmetric octamer DNA sequence called χ (Chi = crossover hotspot instigator = 5′-GCTGGTGG-3′, Figure 1A) (6). In the initial step of DSB processing, RecBCD digests the dsDNA while traversing away from the break site, but after successful recognition of a χ site, RecD helicase activity is repressed, RecB becomes the dominant motor and the complex starts to produce a 3′ ssDNA overhang onto which RecA protein is loaded (7). In vitro, the functionality of RecBCD was found to depend on the ratio between the concentration of Mg2+ and adenosine triphosphate (ATP). When Mg2+ is present in excess over ATP, RecBCD initially degrades both strands of the DNA duplex. The recognition of χ attenuates degradation of the 3′ end, but does not stop the degradation of 5′ end, which leads to the formation of a 3′ tail. Alternatively, when ATP is in excess over Mg2+, RecBCD merely acts as a simple helicase and recognition of χ only produces a single nick in dsDNA, which subsequentlythen, starting from χ, produces a 3′ ssDNA coated with RecA (8). In both situations, it is postulated that a ssDNA–RecA filament forms after recognition of χ, but the question whether ‘degradation’ or ‘nick-at-χ’ occurs in vivo has remained unanswered (9). In the next steps of the HR process, the ssDNA/RecA nucleofilament created by RecBCD undergoes a search for a suitable homologous template and the repair process proceeds (10). A major function of HR in E. coli seems to be the reactivation of collapsed replication forks, where the sister chromosome is used as a template (11,12), which may explain the fact that the majority of chromosomal χ sites are oriented to activate RecBCD molecules translocating toward the direction of oriC (Figure 1A). Interestingly, live E. coli cell studies showed that the search for template homology during DSB repair is physically directed toward the replicated sister chromosome (13).
Figure 1.
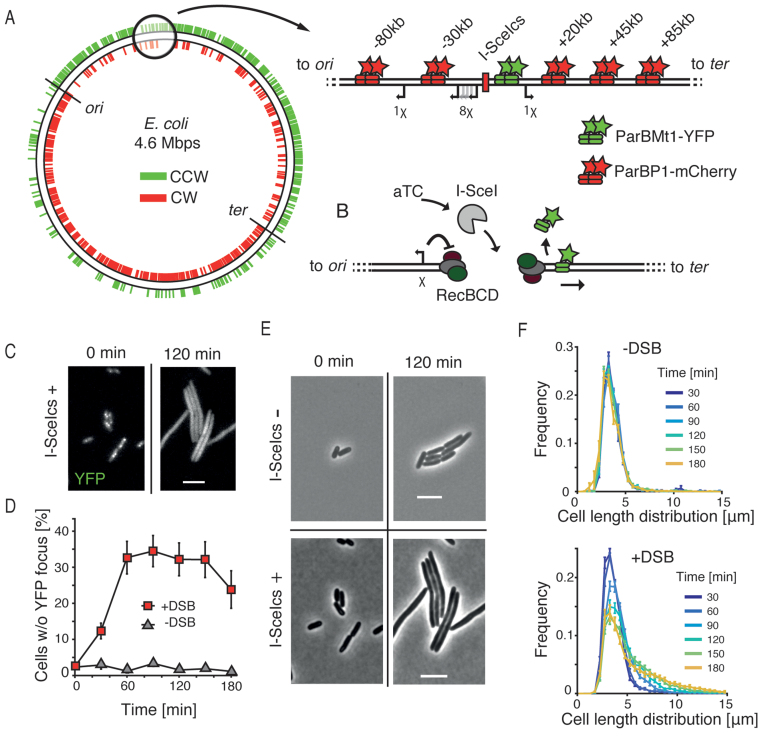
Double-stranded DNA break processing in Escherichia coli. (A) Schematic showing the asymmetric distribution of χ sites plotted on a circular representation of the E. coli chromosome. Sites oriented to activate RecBCD translocating in counterclockwise direction are shown in green; and those in clockwise direction shown in red. Positions of oriC and the terminus region are indicated. Zoom shows the region into which the I-SceI cut site (I-SceIcs) was integrated. Green stars indicate a position of parSPMT1 to which ParBMT1-YFP binds, red stars show the position of parSP1 to which ParBP1-mCherry binds. Distances from the I-SceIcs are shown on top; directions toward the terminus are denoted with a ‘+’ sign; directions toward the origin of replication are denoted with a ‘−’ sign. Arrows show positions of chromosomal χ sites that are oriented to activate RecBCD during translocation from the I-SceIcs position. (B) Schematic representing the experimental procedure. aTC is inducing the expression of I-SceI (gray) from the aTC-inducible promoter. I-SceI creates the site-specific DSB that is recognized and processed by RecBCD complexes. RecBCD end resection is controlled by a correctly oriented χ site (shown as an arrow on left). Translocation of RecBCD and degradation of parS sites results in displacement of the fluorescent ParB proteins. (C) Bacterial cells carrying the chromosomal I-SceIcs and expressing the I-SceI enzyme shown in the YFP channel. Scale bar is 5 μm. (D) Percentage of cells in the population that lost the YFP focus after induction of DSBs (red squares, mean ± SEM, n = 15), and results for the control, a non-induced culture (gray triangles, mean ± SEM, n = 4). (E) Effects of I-SceI expression on cell length in bacteria without the I-SceIcs (top) or with the I-SceIcs (top) sequence on the chromosome. Scale bar is 5 μm. (F) Distribution of cells sizes in time in the population in which DSBs were not induced (top, n = 4) or induced (bottom, n = 15).
While great efforts were made to elucidate the mechanism of RecBCD activity through a series of elegant in vitro experiments with controlled conditions, the direct observation of the end resection process in vivo has remained lacking. It is however of particular interest to measure the dynamics, speed, lengths and processing steps of the resection process in the actual complexity of live cells. In vitro, the RecBCD complex is extraordinarily fast and processive, but it is unknown whether this also holds true in its natural environment of the cell which features steric constraints due to e.g. crowding and roadblocks on the DNA. New features may also appear in vivo. For example, as the majority of chromosomal χ sites are oriented in the direction that is recognized by a RecBCD translocating toward the oriC, one might possibly expect an asymmetric processing of two of the ends of a DSB (Figure 1A).
Here we report end resection as it happens in live cells, as studied with both a fluorescent microscopy assay and with quantitative polymerase chain reaction (qPCR) probes. Our study shows that in vivo the resection proceeds surprisingly fast (∼1.6 kb/s), is temperature dependent, is highly processive (>∼100 kb) and features a preference for the terminus-directed end of chromosomal DSBs. In mutants where the RecD motor is removed, we observe that the length of the end resection is drastically reduced and we show that this shortened end resection has important implications for the choice of template homology during the repair. We find that inactivation of the long end resection significantly increases the chances of recombination with linear E. coli chromosome fragments, which may direct horizontal gene transfer (HGT) in cells carrying mutations in the RecBCD pathway. Screening a library of E. coli genomes, we find that mutations in recBCD genes are common in nature, illustrating the importance of understanding of early steps in recombination in bacteria.
MATERIALS AND METHODS
Strains
Cut site, fluorescent markers and repair templates were integrated into E. coli K12 TB28 (14) (MG1655, ΔlacIZYA) cells. Cloning reactions were transformed into Top10 cells (Thermo Fisher Scientific). To construct the fluorescent reporter of end resection, we first integrated the construct containing the I-SceI recognition cut site, flanked by 2 χ sites on each side and a parS-MT1 sequence (15) (amplified from pBlueDSBarms-parSMT1-BglII-frtCmR) using the lambda-red method (16) into TB28 cells. Distal parS-P1 (15) sequences were integrated into TB28 cells (amplified from pBlueHTarms-parSMT1-BglII-frtCmR) and then combined with the cut site strain using P1 phage transduction. MG1655 deletion of recD was done with a lambda-red protocol in a MG1655 background, followed by P1 transduction. Resistances were removed using the pCP20 plasmid (16). Due to the length of lambda-red PCR products required to integrate HR reporter (RS) from one template plasmid two fragments from pLS6 were amplified using with Phusion polymerase (Thermo Scientific) with primer pairs Jw246 and Jw358, and Jw245 and Jw359. The resulting fragments shared 20 bp of homology and the full length fragment was assembled using Phusion polymerase. The full-length fragment was integrated into genome of TB28 cells using a lambda-red protocol.
Plasmids
All plasmids were constructed using the Gibson assembly protocol (NEB) with the exception of pBlueDSBarms-parSMT1-BglII-frtCmR and pBlueHTarms-parSMT1-BglII-frtCmR, which were constructed using restriction digestion and ligation as follows: first pBlueHTarms and pBlueDSBarms (both were gifts from Hugo Snippert) were linearized using BglIL restriction enzyme (New England Biolabs). Next, fragments containing parS-P1 and parS-Mt1 followed by chloramphenicol resistance cassettes were amplified with primer Jw024 and JW025 and cut with BglII from plasmid pGBKD3-ParSP1 and pGBKD3-ParSPMT1 (17), fragments were ligated with linearized backbone.
Plasmid p15aSceI_deg was cloned using Gibson assembly by fusing fragment amplified from the plasmid pdCas9degRNA3 (18) with Jw201 and Jw202, then a fragment from a plasmid pDL2655 (19) amplified with primers Jw205 and Jw206 to create p15aSceI_deg_kan. The resulting plasmid was kanamycin resistant, and to change the resistance to chloramphenicol, we performed an additional round of Gibson assembly. A fragment amplified from p15aSceI_deg_kan with primers Jw210 and Jw211 was fused with a fragment amplified from pKD13 (16) with primers Jw212 and Jw213 to create a plasmid p15aSceI, which was chloramphenicol resistant.
Cloning of pSC101SceI_deg was similar to the previous plasmid, but in the first step we amplified a backbone from pHippACMR (gift from Helena Shomar Monges) using primers Jw203 and Jw204 and fused it with a fragment amplified from pDL2655 with primers Jw205 and Jw207 to create pSC101SceI_deg_kan. Kanamycin resistance gene was exchanged with the chloramphenicol resistance gene by fusing a fragment amplified from pSC101SceI_deg_cm with primers Jw208 and Jw209 with a fragment amplified from pKD13 with primers Jw212 and Jw213.
Plasmids pLS2χ+, and pLS2χ− were created using Gibson assembly, and their sequences can be found in supplementary material.
Mix & Go E. coli transformation kit (Zymo Research) was used to transform plasmids into the cells. A list of plasmids and primers used in this study can be found in the Supplementary Table S1.
Growth conditions and media
Bacteria used for microscopy experiments were grown in M9 minimal medium supplemented with 0.2% glucose at 37°C. qPCR experiments were carried out on bacteria grown in Lysogeny Broth (LB) at 37°C with the addition of antibiotics. The overnight LB culture was refreshed on the following day in a 1–500 dilution and incubated until reached OD ∼0.1, then I-SceI expression was induced with addition of aTC to the media (2 ng/ml) for 1 h. For microscopy experiments with snapshots, the overnight culture was grown in M9-glucose media in 37°C and refreshed on the following day in a 1–100 dilution. When the culture reached OD ∼0.1, the I-SceI expression was induced by the addition of aTC (2 ng/ml) for 15 min. Then the culture was centrifuged and cells were resuspended in a fresh M9-glucose media without aTC and grown at 37°C. For timelapse experiments, cells were grown as in snapshot the experiments, but the imaging started after an initial 15 min induction without the washing step. Ampicillin (100 μg/ml), chloramphenicol (34 μg/ml) and kanamycin (50 μg/ml) were added to cultures when required.
Microscopy and image analysis
Microscopy experiments were performed using a Nikon Ti-E microscope with a 100x oil-immersion phase-contrast objective (CFI Plan Apochromoat λ DM 100x). Fluorescence was excited using a Lumencor SpectraX LED light source and images were acquired using an Andor Zyla 4.2 CMOS camera. Fluorescent image exposure was selected to provide a good signal-to-noise ratio maintaining a minimal exposure time (for snapshot experiments exposure times were 400 ms, and for 30 s time-lapse experiment 200 ms). Intensity of the LED light source was fixed at 25% of the maximum intensity. The 30 and 10 s time-lapse experiments were imaged in a custom-made environmental chamber held at 37°C, 30°C or 23°C. Cells were imaged on M9 media pads containing 1% agarose and aTC (2 ng/ml). The 2.5 μl of the culture was transferred on the pad and once cells were absorbed onto the M9-agarose the pad was placed on the microscope slide and imaged.
Bacterial cells in phase-contrast images were segmented using MATLAB and Oufti package (20). Foci were detected using a Crocker and Grier routine (adapted for MATLAB by Blair and Dufresne, http://site.physics.georgetown.edu/matlab/) embedded in a custom automated image-processing pipeline. For each detected cell, a region of interest was cropped from fluorescent images. In the next step, each cell was validated based on fluorescent signal and only cells which mean fluorescent intensity above global background level were selected. After initial foci detection, only foci which intensity exceed the cellular background noise level were selected. Degradation events in fast time-lapse experiments were selected manually in FIJI software (21).
Quantitative PCR
Induction of the DSB for qPCR measurements of degradation was done in LB media with induction with aTC for 1 h in 37°C. Genomic DNA was isolated with the Wizard Genomic DNA purification kit (Promega). DNA concentration was measured using Quant-iT™ (ThermoFisher) and for each qPCR reaction equal amounts of template DNA were used. For end resection experiments, the Ct value of each marker was compared to the Ct value of the oriC primer pair, used as an integral frequency normalization marker. Normalized values were then compared to corresponding values obtained for the un-induced strain. We used dye-based GoTaq® qPCR Master Mix (Promega) and Eco Real-Time PCR system (Illumina) for all degradation qPCR reactions. Multiple primer pairs were tested for each chromosomal marker were tested and only the pairs which resulted in an efficiency close to 100% were used for final experiments.
For the recombination-efficiency experiment, cells were induced as for the qPCR degradation experiments, but after the addition of aTC (2 ng/ml) the culture was incubated for 4 h, and then plasmid DNA was isolated using Wizard® Plus SV Miniprep (Promega). Plasmid DNA was further purified with ethanol precipitation: 30 μl of plasmid DNA was mixed with 300 μl of 100% ethanol and 30 μl of 3 M sodium acetate and incubated in −20°C for 1 h and next, centrifuged at 4°C for 45 min at maximum speed (Eppendorf 5418R). Supernatant was discarded and the DNA pellet was suspended in 300 μl of 70% ethanol solution, upon which the centrifugation step was repeated. Supernatant was discarded, the DNA pellet was dried and re-suspended in 100 μl of milliQ water. DNA concentration was measured using Quant-iT™ (ThermoFisher) and for each qPCR reaction, equal amounts of template DNA were used. To calculate the recombination efficiency, we first calculated Ct values for primer pair specific to the recombination product (with primers HR product f. and HR product r.) and compared these values to the Ct values for primer pair specific to the p15a_SceIdeg plasmid (with primers p15a f., and p15a r.). We assumed that the concentration of p15a_SceIdeg was not affected by the recombination, and therefore we could use it as a reference to measure the concentration of the product of recombination. Finally, normalized Ct values were compared to the normalized values obtained with uninduced culture of wild-type cell with χ+ plasmid.
Purification of genomic DNA for HGT experiments
Genomic DNA of a strain carrying the CmR gene was prepared using Promega Wizard® Genomic DNA Kit following the manufacturers protocol. Shortly, 1 ml of overnight culture of Jx097 and Jx098 cells was used to isolate the DNA. After the isolation, the DNA sample was further purified using ethanol precipitation protocol. Electrocompetent cells were prepared as follows: cells were grown in LB in 37°C until the culture reached an OD ∼0.6. Then 1 ml of culture was centrifuged (10 000 RPM, 4°C, Eppendorf 5418R) and washed three times in 1 ml of milliQ water. After a final wash, cell pellet was resuspended in 50 μl of milliQ water. All steps were done in 4°C. A total of 100 ng of purified genomic DNA was used for electroporation into electrocompetent cells.
Probability of χ site recognition
The probability of degradation of a DNA marker after n χ sites can be written as pd = 1 – (1 – pχ)n, where (1 – pχ) is the probability of not recognizing a χ site. From the measured value of pd = 0.586 obtained in our experiments for 3 χ sites. This yields (1 – pχ) = 0.745, and therefore a probability of recognizing single χ site pχ = 0.255.
Bioinformatic analysis
Mutation rates in recABCD genes were calculated using a collection of genomes downloaded from the RefSeq release 82 database. First, genomes were screened for annotated sequences coding recABCD genes using custom MATLAB scripts, resulting in 285 genomes where all genes were found (see Supplementary Table S2 for list of genomes). Next, genes were translated into amino-acid sequence and aligned with the canonical amino-acid sequence of the protein (found in K12 MG1655 genome) using the BLASTP algorithm. Alignments were further processed by custom MATLAB scripts to identify unique non-synonymous substitutions in the aligned sequences. To calculate the mutation rates, we counted the mean number of unique non-synonymous mutations in the amino-acid sequence from each genome and divided it by the total number of amino-acids coding a given protein.
RESULTS
Fluorescent reporters of end resection in living cells
To study the dynamics of end resection after DSB formation in live E. coli cells, we developed an in vivo fluorescent reporter system consisting of three components: a site-specific I-SceI inducible cut site (22) integrated at the codA locus, a parSMt1/ParBMt1-YFP (15) marker placed close (+1.5 kb) to the I-SceI recognition site (I-SceIcs) and a parSP1/ParBP1-mCherry (23) marker placed at variable distances from the break (Figure 1A, −80, −30, +20, +45, or +85 kb from the DSB site). The I-SceI cut site was flanked on each end by 2 χ sites to induce RecBCD recombination activity (Supplementary Figure S1) (5). The I-SceI coding gene (24), fused to a LAA degradation tag (25), was placed on a low-copy pSC101-origin plasmid under the control of an anhydrotetracycline (aTC) inducible promoter. Genetic integrations and expression of ParB proteins did not affect chromosome organization and segregation, as the fluorescent microscopy analysis showed native positioning of the integrated sites comparable to previously published studies (Supplementary Figure S2) (26). Induction of the DSB with I-SceI creates free dsDNA ends, which in E. coli are recognized and processed by the RecBCD complex (5). In vitro studies have shown that RecBCD can move over very large distances on the DNA during end resection and can displace DNA-bound proteins during translocation (27–29). We hypothesized that the degradation of a DNA fragment containing parS sites would cause the release of localized ParB proteins and consequently a loss of a focus (Figure 1B). Such loss of focus could then be used as an in vivo reporter of the dynamics of end resection.
To test if the induction of I-SceI in our engineered cells would lead to the formation and processing of DSBs, we used time-lapse microscopy with cells growing in the presence of aTC. Cells with integrated I-SceIcs that were expressing I-SceI enzyme showed an elongated phenotype and lost the ParB foci (Figure 1C and Supplementary Figure S3), confirming that DSBs were formed, recognized and processed. To show that DSBs observed in those cells are site specific, we used a strain without an integrated chromosomal cut-site, that showed no effects of DNA damage in presence of I-SceI (Figure 1E and Supplementary Figure S3). Importantly, we did not observe recovery of fluorescent foci during 4 h experiments, what strongly suggested that the DSB ends, together with parS sequences were degraded and not just unwound during the end resection.
To study the synchronized progression of DSB repair, we used a short, 15-min pulse of aTC induction, followed by a wash with fresh media. Analysis of fluorescent microscopy images on the pulse-induced culture showed a growing proportion of cells that lost all yellow fluorescent protein (YFP) foci due to end resection. The proportion of induced cells increased rapidly after the induction of DSBs and remained stable after 60 min at about 30% (red squares, Figure 1D). In the non-induced culture, by contrast, almost all cells contained at least one YFP focus throughout the time of an experiment, showing no traces of DSB damage. The short duration of the induction pulse allowed to trigger the formation of DSBs in a limited time-window, enabling the observation of the advancement of synchronized end resection in bacterial culture.
I-SceI expression in E. coli creates stable DSBs
Formation of a DSB induces an SOS response in bacteria, which stalls the progress toward cell division and leads to an increase in cell length (30). Analysis of the cell sizes in an unperturbed E. coli culture showed a time-stable distribution, with a mean cell length distribution of 3.7 ± 1.7 μm (mean ± SD, pooled data from all time points, n = 12 575 cells, Figure 1F), characteristic to cells grown in M9 supplemented medium (18). In the pulse-induced culture, a clear elongation of cells was visible, which accompanied the aforementioned disappearance of the YFP foci. The cell-size lengthening in induced cells was rapid in the first 90 min after the induction of DSB and slowly increased after that (Figure 1F). Fitting log-normal distributions to the pooled cell length data for each of the time points showed that the induced cultures consisted of two populations with mean lengths characteristic of DSB-induced and uninduced cells (Supplementary Figure S4). We did not observe a recovery of induced cultures to their initial state that would indicate the repair of DSBs during the course of experiments. Such a lack of repair can be explained by the efficient cleaving of all chromosomal copies in induced cells, i.e. not merely one. Because the presence of an intact sister homology is necessary for the repair by HR, the completion of the repair would be rendered impossible as in our assay the I-SceI enzyme can target all available sites. Lack of recovery of induced cells to the initial uninduced phenotype supports this explanation (Figure 1C, D and F). Lowering the concentration and period of aTC expression in our experiments had an influence only on the fraction of induced cells, in which all I-SceI cut sites were cleaved nevertheless. This is to be expected since freely diffusing protein can scan the entire volume of bacterial cell in a matter of seconds (31), and hence, even small bursts of I-SceI expression are likely sufficient to cut all copies of sister chromosomes in a single cell. This all-or-nothing phenotype is less fit to study the completion of HR, but provided us with a robust framework to look into the early steps of the repair, where here we focus on end resection by RecBCD.
Single-DSB analysis shows χ site recognition within live cells
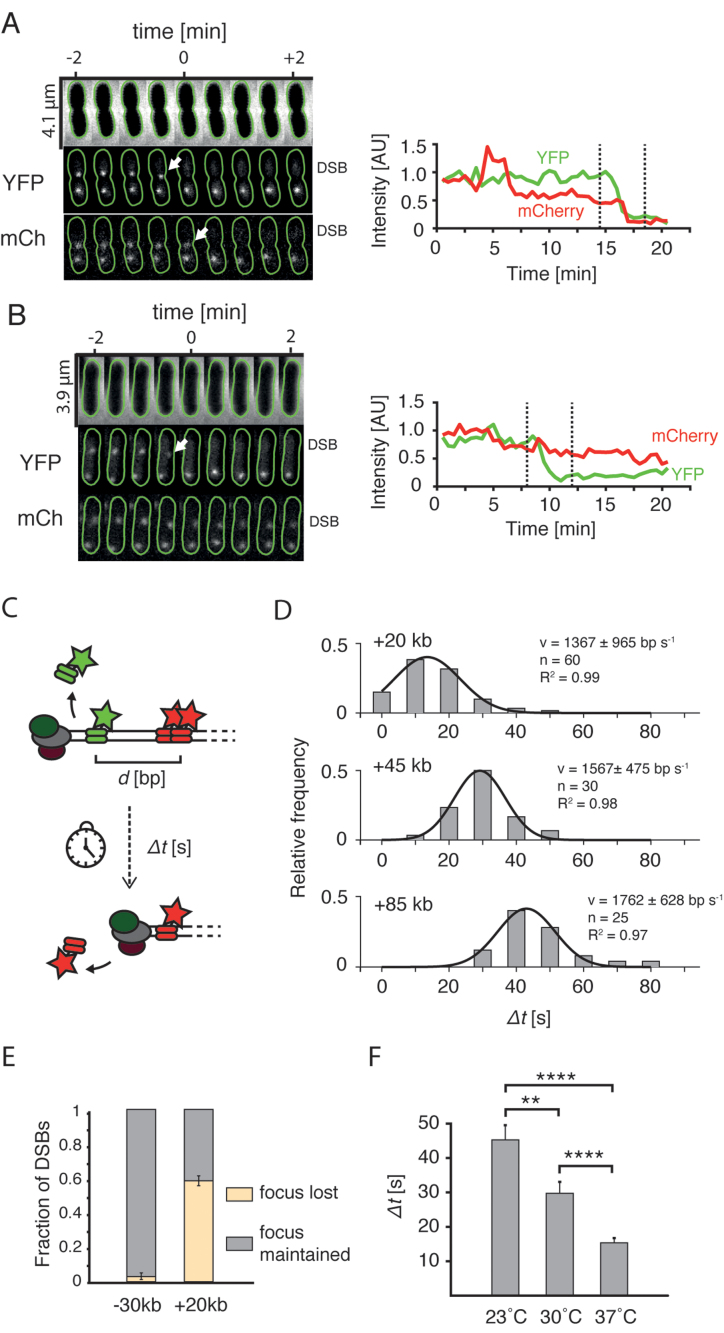
Fast (10–30 s/frame) time lapse imaging of cells undergoing end resection allowed to study the fate of single DSBs in the native environment of a cell. We could distinguish YFP and mCherry foci related to the same DSB site even in cells with multiple genome copies, as these markers positioned closely on the chromosome are separated by a short physical distance in an E. coli cell (32). In some cells, we observed that the loss of a YFP focus was followed by a rapid disappearance of the mCherry spot, as could be expected for end resection by RecBCD (Figure 2A). In cells that lost the YFP focus, the +20 kb mCherry marker was lost in 59 ± 2% (mean ± SD, n = 46 DSBs) of cases, while in the remaining cells that lost YFP spot, a localized mCherry signal was maintained through the duration of 30-min experiment (Figure 2B). The cells with a −30 kb marker showed a significantly different behaviour than the +20 kb cells, despite the similar genetic distance between the DSB and mCherry marker. Here, the mCherry focus rarely disappeared after the loss of YFP focus (3 ± 3% mean ± SD, of cells, n = 61 DSBs, Figure 2E), even though we used exactly the same inducing conditions for both strains. The contrast in marker disappearance is likely explained by the different number of χ sites between the I-SceI cut-site and the mCherry marker (10 χ sites toward the −30 kb marker site versus 3 χ sites toward the +20 kb site). Given that the RecBCD complex encounters 3 χ sites in the direction of +20 kb marker, the calculated efficiency of χ site recognition equals 26% (see supplementary information for details), which is in good agreement with previously reported values of 20–40% (33).
Figure 2.
End resection in live cells. (A) Cell in which loss of the YFP focus is linked to the disappearance of mCherry focus. Top: phase contrast channel; middle: +1.5 kb marker in the YFP channel; bottom: +20 kb marker in the mCherry channel. Green lines show cell outlines obtained using Oufti software (20). Arrows point at the events of disappearance of foci. Plots on the right represent integrated intensity of the labeled DSB. Dashed lines mark the time-interval shown on microscopy montage on right. (B) Example cell in which the loss of the YFP focus was not linked to the loss of the mCherry focus. (C) Experimental procedure to measure the speed of resection with the two-color assay. The time between the events of loss of two foci (Δt) divided by the genetic distance (d) yields an average speed of resection. (D) Histograms of processing times (Δt) measured for cells with +20, +45 and +85 kb markers. Black lines represent Gaussian fits to the experimental data. Goodness of fit, number of data points, and calculated mean (±SD) are shown. (E) Fraction (mean ± SD; −30 kb: n = 61; +20 kb: n = 46) of DSBs in which the mCherry foci were lost, or maintained after the YFP focus was displaced. (F) Mean processing times in cells with +20 kb measured at different temperatures (mean ± SEM, 23°C: n = 40; 30°C: n = 40; 37°C: n = 60).
Events of foci loss in cells with the +20 kb marker were very abrupt and happened between two consecutive time frames suggesting that cellular end resection by RecBCD is very fast. To accurately measure the speed of end resection, we used 10 s time-lapse imaging on strains with +20, +45 and +85 kb integrations, in which YFP and mCherry foci occasionally disappear sequentially. After the induction of DSBs, we measured the time between events of disappearance of foci, from which we can extract the average speed of RecBCD as we know the genomic distance between the foci (Figure 2C). These times were distributed normally and, as expected, were larger for strains with integrations located further away from the DSB (Figure 2D). The analysis yielded a mean speed of end resection of 1565 ± 198 bp/s (mean ± SD, for mean speed in individual measurements see Figure 2D). Rather surprisingly, we found that the resection of even the distant +85 kb marker happened with similar speed as the closer markers. In vitro studies estimated the RecBCD processivity at 30 kb on average (33), but our data indicate a processivity of the end resection in a cell that can easily exceed this value. This may be explained by either a much larger in vivo processivity of a single RecBCD, or by a fast replacement of a RecBCD complex that detaches from the DNA strand by another cellular RecBCD.
We were curious how the temperature affects the dynamics of end resection and thus repeated similar 10 s time-lapse measurements in the +20 kb strain at lower temperatures. We find that the mean processing time increased with lowering temperature from 37°C to 30°C to 23°C (Figure 2F). Furthermore, the distributions became broader, suggesting that end resection may be more stochastic at lower temperatures (Supplementary Figure S5). While one might expect to observe a decreased speed of the RecBCD enzymatic reaction at lower temperatures, the broadening of speed distribution is nontrivial which potentially reflects that collisions with DNA-associated proteins, that binds more stably in low temperatures, may have a more disruptive effect.
We thus used our microscopy assay to directly observe the process of end resection in live cells and measure its speed and response to χ sites. A very high speed of 1.6 ± 0.2 kb/s was measured. Such a high rate is rarely observed in biology, but similar numbers were reported in biochemical in vitro experiments on RecBCD (27,29). The fact that we measure a similar speed in vivo is remarkable, given the much less favorable conditions of the interior of the cell, where RecBCD must compete with other DNA-binding factors, yet does not seem to slow down even at the distance of 85 kb away from the DSB site. Because of the direct and straight-forward nature of our two-color assay, we expect that it is possible to adapt it to characterize processivity and rates of end resection also in other organisms.
Asymmetric large-scale end resection of DSB ends
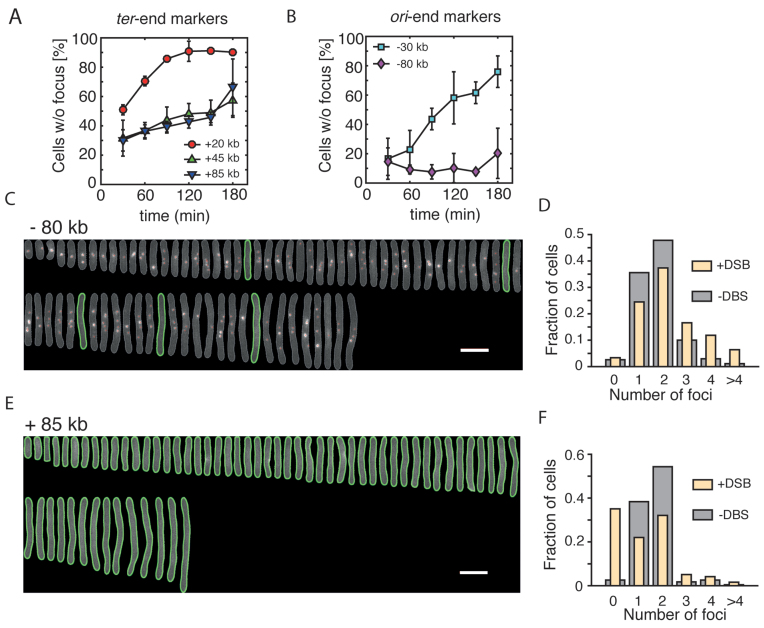
Having showed that we can accurately trace the fate of DSB sites in live E. coli cells, we set out to characterize a possible asymmetry of end resection, using mCherry markers integrated at various positions. We used pulse aTC induction and selected only cells which lost the YFP foci for analysis, that is, cells where a DSB was induced and resection was initiated. We observed substantial differences in the processing of the ori-oriented and ter-oriented ends of the DSB. The +20 kb ter-oriented marker was lost in 51 ± 3% (mean ± SD) of cells at 30 min after the induction of a DSB, consistent with the time-lapse measurements (Figures 2E and 3A). More distant ter-oriented markers, +45 and +85 kb showed a somewhat slower resection, that was progressing monotonously at a similar rates over the course of the experiment (Figure 3A). By contrast, the resection of the closest ori-end marker, −30 kb, was very limited. For example, at 30 min after induction, only 16 ± 14% (mean ± SD) of cells that lost YFP focus also did not contain mCherry marker, again consistent with the time-lapse experiments (Figures 2E and 3B). Because of an extended duration of this snap shot experiment, it is very likely that, in contrast to fast time-lapse experiments, the DSBs that are observed here may have undergone multiple rounds of end resection and recombination, thus extending the distance of DNA degradation in later time points. Still, surprisingly, the most distant ori-end marker, −80 kb, was not processed at all, even in the latest time-point (Figure 3B and C), but instead its copy number increased. Indeed, 3 h after induction, we found a large fraction of elongated cells with an unusually high number of −80 kb foci, reflecting active initiation of replication in DSB-positive cells (Figure 3C and D). On the contrary, the +85 kb marker placed on the ter-end of the DSB was mostly resected and elongated cells contained generally no mCherry spots (Figure 3E and F).
Figure 3.
Asymmetric processing of DSBs. (A) Percentage (mean ± SD, n = 3) of induced cells that lost the mCherry at different time points after the induction of DSBs. Three markers positioned on the ter-end of the DSB are shown. (B) Percentage (mean ± SD, n = 3) of induced cells that lost the mCherry at different time points after the induction of DSBs. Two markers positioned on the ori-end of the DSB are shown. (C) Aligned representative cells from a population of DSB-induced cells showing multiple −80 kb markers. Images were taken 3 h after induction of the DSB. The mCherry channel is shown. Green lines represent outlines of cells that did not contain mCherry foci, detected using Oufti software. White scale bar is 5 μm. (D) mCherry foci number distribution for the −80 kb strain 3 h after induction of DSBs (yellow; n = 271 cells) and in a non-induced culture (gray; n = 1327 cells). (E) Same as in (C) showing the +85 kb strain. (F) mCherry foci number distribution in the +85 kb strain 3 h after induction of DSBs (yellow; n = 232 cells) and in a non-induced culture (gray; n = 2034 cells).
These results show a pronounced difference in the RecBCD processing of the two ends of DSB, where the ori-oriented DNA is well protected from resection while the ter end is highly degraded. This difference can simply be explained by the different number of properly oriented χ sites within the DNA, rendering the ori-end much more resistant to RecBCD processing. Hence, the bias of end resection reflects the bias of chromosomal χ site orientations. An unexpected effect of the asymmetric end resection is the over-replication of the −80 kb marker observed in our experiments. Replication forks established at oriC will progress to the DSB and then will drop off from the free DNA end, hence increasing the number of free DNA ends in a cell with each round of replication.
qPCR assay shows DNA degradation during end resection
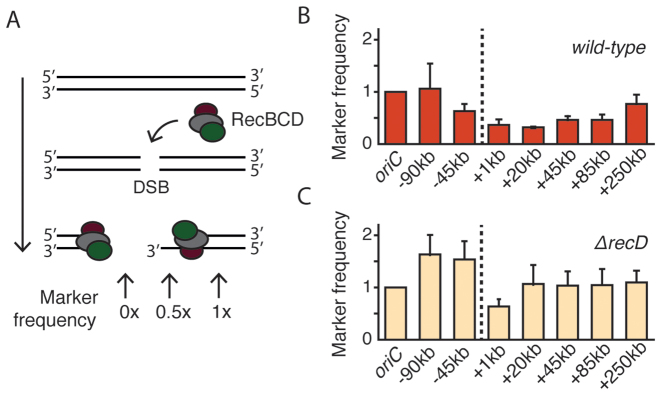
In order to verify that the large extent and the asymmetry of the end resection were not caused or distorted by the imaging conditions or by the presence of fluorescent ParB proteins, we developed a qPCR-based method to independently monitor the progression of RecBCD in a population of synchronously resecting E. coli cells. A similar qPCR-based methodology was used before to study resection in yeast cells (34) and recently in Caulobacter cresentus (35). Using qPCR probes, we measured the concentrations of chromosomal markers in the proximity of the DSB as they were processed by RecBCD (Figure 4A). We induced the formation of DSBs by addition of aTC to the LB medium and compared the frequency of chromosomal markers to a non-induced culture. Similar to the microscopy assay, we observed long-distance and asymmetric end resection of DNA flanking the DSB, with a shape nearly identical to the microscopy assay (Figure 4B and Supplementary Figure S6). Again, the ter-oriented end of DSB was resected to a much greater extent than the ori-oriented end, and we observed that end resection reached markers as far as +250 kb within an hour after the induction. In the qPCR assay, the signal originating from a chromosomal marker can only be reduced when the dsDNA in the region of interest is actually degraded. If the RecBCD complex was instead only acting as a helicase that unzips the DNA without the degradation, as has been postulated in a ‘nick-at-χ’ scenario (8), we would not expect to detect the resection with the qPCR assay. Our data instead support the model where RecBCD is a helicase/nuclease that degrades significant amounts of DNA. Moreover, because the end resection profile obtained with the qPCR assay matches very well with the microscopy results, we conclude that the fluorescent ParB fusions had no significant influence on the DSB processing.
Figure 4.
End resection profile studied with qPCR. (A) Degradation of DNA during end resection removes one or two strands of DNA duplex. In the schematic illustration, the RecBCD complex is loaded onto the dsDNA ends that are created as a result of DSB formation. DNA degradation during end resection removes both DNA strands (as shown on left) or just one strand of the DNA helix (right). Degradation of DNA removes the primers binding site, thus reducing qPCR signal. (B) Profile of the wild-type end resection measured with qPCR 1 h after the induction of DSBs (mean ± SD, n = 3). Marker frequencies are relative to those in the non-induced culture. Dashed line shows the position of the DSB. (C) Profile of the end resection in ΔrecD cells measured with qPCR 1 h after the induction of DSBs (mean ± SD, n = 3).
Deletion of recD strongly reduces end resection
Next, we characterized end resection in cells where the RecBCD machinery is perturbed. Escherichia coli strains lacking the recD gene are recombination proficient and can integrate homologous linear DNA into chromosome (36). In vitro, the RecBC complex lacks activity for DNA degradation and can load RecA onto ssDNA in a χ-independent manner (37). We asked whether end resection in ΔrecD cells is significantly different from that in wild-type cells. To test this, we deleted the recD gene from the bacterial chromosome and used this strain to assay the end resection with the same qPCR assay as for wild-type bacteria. Indeed, the end resection process in the mutant cells was very strongly reduced and showed ∼50% resection only for the closest +1 kb marker, while the resection of any farther-out markers was not detectable at all (Figure 4C). The RecBC complex has the potential to unwind the DNA and initiate the recombination directly at the end of DSB, and accordingly the ∼50% signal may be attributed to the presence of only the 3′ end. The results directly show that in vivo the end resection in ΔrecD mutants is dramatically reduced, and lacks the pronounced degradation characteristics of RecBCD.
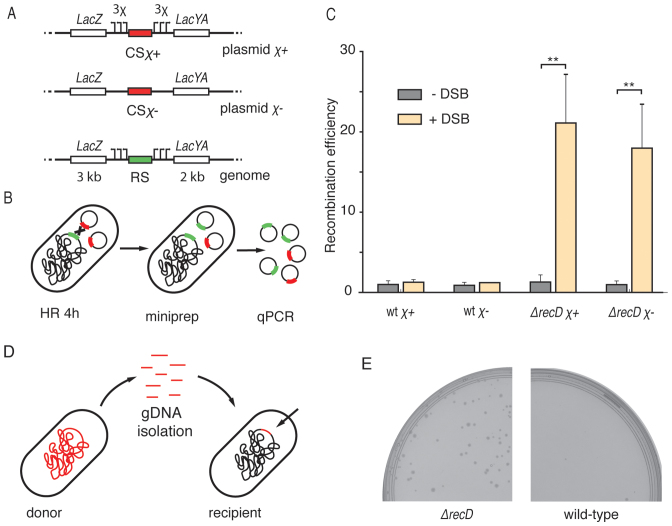
What is the importance of this reduced resection length at the DSB site? For example, how does it influence the selection of repair templates in E. coli? We developed a reporter system consisting of a low-copy-number plasmid carrying an inducible I-SceI cut site flanked by homologous arms, together with a chromosomal repair template with the corresponding homologous arms. We constructed two versions of the inducible DSB site on the plasmids, one flanked by 3 χ sites at each end (named the χ+ plasmid, pLS2χ+,), and one lacking any χ sites (χ− plasmid pLS2χ−) (Figure 5A). DSBs formed on the plasmids could be repaired either by another, intact copy of the plasmid, or by the recombination with an ectopic chromosomal site. Gene conversion with the chromosome would however alter the sequence of the plasmid, allowing detection with a repair-specific primer pair on plasmids isolated from cells. After induction of DSBs and purification of the plasmids, we used qPCR to quantify the ratio of recombinant sequence to a reference plasmid coding I-SceI (Figure 5B). After 4h induction, the difference was very pronounced for the case of ΔrecD cells (∼20-fold increase, t-test P ≤ 0.01, Figure 5C). Interestingly, recombination with the chromosome was not detected in the wild-type cells, even for the χ+ construct. Very likely, the probability of χ recognition by RecBCD complex is too low to initiate the recombination of a cut plasmid at the regions of homologous arms due to the short length of used homologies (∼5 kb). The repair of a χ− DSB plasmid was monitored in a strain that carried a chromosomal repair template with χ sites, thereby introducing a mismatched region between χ− plasmid and chromosome, and interestingly, the efficiency of recombination was observed to be slightly lower for the χ− plasmid than for the χ+ plasmid (Figure 5C). Mismatches between templates were recently shown to influence the efficiency of recombination in yeast (38), and possibly this also occurred in our assay.
Figure 5.
Length of end resection influences HR. (A) Genetic maps of components of the HR reporter. The I-SceI cut site (red) was placed on the low copy number plasmid. The repair template was integrated into the genome. Arrows represent χ sites. Lengths of the homologies flanking I-SceI cut site are shown on the bottom of genomic cassette. (B) Recombination was induced by the formation of chromosomal DSBs. After 4 h, reporter plasmids were isolated from cells and the concentration of recombinant plasmids was measured with qPCR. (C) The efficiency of recombination (mean ± SD; n = 3) in wild-type and in ΔrecD backgrounds. P-values were obtained using the two-tailed t-test (**P ≤ 0.01). (D) For the HGT assay, chromosomal DNA was isolated from a donor strain as a library of short linear fragments. After delivery of fragments into the recipient strain, the antibiotic-resistance marker was integrated into the genome. (E) Growth of recombinant colonies in the presence of chloramphenicol.
We thus have developed and used a plasmid reporter system to study the efficiency of HR and found out that recombination with short linear sequences is strikingly more efficient in ΔrecD than in wild-type cells. Our data suggest that the wild-type RecBCD recombination is fastidious and ignores short ectopic homologies, such as the plasmids used in this study, even if those were marked by χ sequences. On the other hand, when end resection is shortened and independent of χ, recombination is efficient with short homologous sequences, which was demonstrated before with linear DNA (36) and here with plasmids.
Deletion of ΔrecD increases efficiency of horizontal gene transfer
Can a shortened end resection influence the process of HGT as much as it influences recombination with chromosomal homologies? In the natural environment of bacteria, uptake of linear DNA, next to transduction and conjugation, is one of the mechanism enabling HGT (39). To test how the ΔrecD mutation influences HGT between different E. coli cultures, we designed an experimental approach where we first isolated a library of linear DNA fragments from a donor carrying a chromosomal chloramphenicol (Cm) resistance marker, then electroporated it into a recipient Cm-sensitive strain and finally screened for Cm-resistant colonies (Figure 5D). We indeed found colonies when ΔrecD cells were used as the recipients of the donor DNA, but not when wild-type cells were used (Figure 5E). PCR genotyping showed that the Cm-resistant ΔrecD colonies contained the Cm-resistance marker as well as a ΔrecD specific genomic watermark, confirming integration into the genome (Supplementary Figure S7). End resection of linear DNA ends by RecBCD was previously shown to degrade some of the incoming DNA during conjugation in E. coli (40). If long and χ-rich conjugating DNA fragments were degraded by RecBCD, then complete degradation of short linear fragments should be expected in wild-type cells. Here we showed that HGT from a collection of short, linear chromosomal fragments was possible when the natural end resection was inactivated by the ΔrecD mutation.
In the light of this result, it is of interest to examine how commonly mutations of recBCD genes occur within the E. coli genomes. When we screened the E. coli genomes deposited in the NCBI Reference Sequence Database (Supplementary Table S2), we found that majority of genomes (n = 285 sequences) carried at least one non-synonymous substitution in the recBCD genes (73%, n = 285, 74%, n = 283 and 67%, n = 285, for recB, recC and recD respectively). To compare, an equivalent analysis of the recA gene showed variability in only 5% of the screened sequences (Supplementary Figure S8). Although it is uncertain whether or how these mutations modify the mechanism of RecBCD, the high rates may suggest that, in naturally growing E. coli, the end resection machinery is more likely to be targeted by a mutation than the core strand-exchange protein RecA.
DISCUSSION
End resection in live cells
In this study, we directly observed and quantified the process of end resection during DSB repair within individual E. coli cells. With fluorescence microscopy, we demonstrated that the end resection in bacteria is surprisingly fast and extensive, processing tens of kilobases in just a matter of seconds. We provided a detailed characterization of the speed of the end resection by RecBCD and estimated it at 1.6 ± 0.2 kb/s. This high value for the resection speed in live cells is close to the fastest speeds achieved by RecBCD complexes in in vitro assays based on DNA curtains (29), or flow stretched DNA (27). Although helicases translocating along cellular DNA will necessarily encounter numerous roadblocks such as RNA polymerases or proteins organizing the genome, RecBCD seems not to be hindered by these. Indeed, RecBCD was shown to be able to knock off multiple substrates from the DNA in vitro (41). Speeds of eukaryotic and prokaryotic end resection were estimated before from indirect measurements such as southern blots, DAPI signal decay or quantitative modeling to fit degradation profiles obtained with high-throughput sequencing. Those speed values were reported to be 1 bp/s for vegetative yeast cells (42), 10 bp/s for yeast mitotic resection (43), 190 bp/s for E. coli (13) and 400 bp/s for C. cresentus (35). Direct measurements of the dynamics of end resection in different species would be of future interest, to explore why some species invest in a very fast end-processing, whereas some others seem to be satisfied with much lower speeds.
We observed very long extents of end resection in live E. coli cells, up to 85 kb of DNA created within a minute (or even 250 kb in our qPCR experiments), which suggests that the processivity of RecBCD is larger than anticipated previously. Degradation of such large portions of DNA is surprising if motivated only by the DSB repair process. One explanation of the extensive end resection may lay in the involvement of RecBCD in the protection of E. coli from bacteriophages with linear genomes, as any unprotected linear DNA molecule will quickly be degraded by RecBCD complexes. DNA degradation by RecBCD is also involved in the production of short DNA fragments that are used as CRISPR memory (44). Another explanation of the high processivity, especially in the direction of ter, resides in the fact that the most common role of HR in E. coli involves the restart of collapsed replication forks (45). Such event creates only a single-ended DSB from the direction of oriC, and therefore the evolutionary pressure to protect the ter region from end resection is less pronounced. We have limited the study of end resection to one position of the E. coli genome. Further studies may elucidate how the local genomic context influences the speed and processivity of resection by monitoring the reaction at different positions.
End resection involves DNA degradation
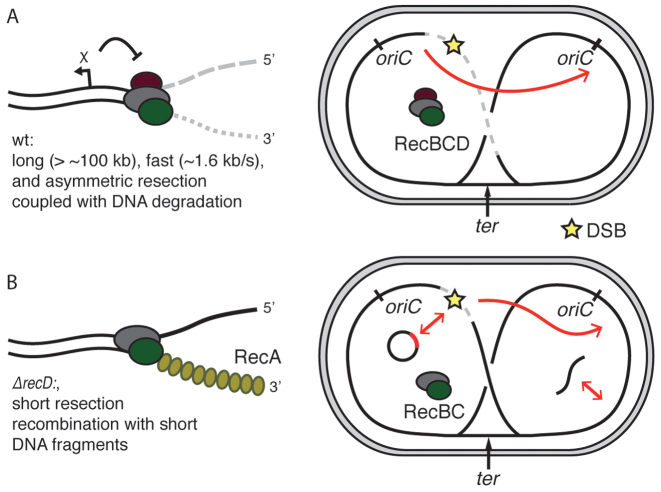
Our experiments showed that end resection is linked to the permanent disappearance of ParB signals localized on parS sequences, and a decrease of the qPCR signal. Both of those effects are explained by the DNA degradation linked to end resection, consistent with the nuclease/helicase model and characteristic of excess Mg2+/ATP conditions. Alternatively, a ‘nick-at-χ’ reaction, would lead to regeneration of foci once RecBCD passed over parS sequences, and would not lead to differences in marker frequencies in qPCR assay. It has been a subject of scientific debate which reaction was the more biologically relevant process. Based on our data, we suggest that DNA degradation is an intrinsic step in the RecBCD end resection in live cells (Figure 6A). Degradation during end resection is also more fit to support the observations of CRISPR-spacers acquisition during DNA degradation by RecBCD as well as the protection against phage infections.
Figure 6.
Models of end resection in Escherichia coli. (A) HR in wild-type cells. Initially, during resection, RecBCD degrades both strand of DNA; recombination is activated by recognition of a χ site. After formation of a DSB, large fragments of the chromosome are rapidly degraded, preferentially in the direction of ter. Such end resection drives the recombination to use the intact sister chromosome as a repair template. (B) Deletion of the recD gene removes the influence of χ sites and greatly shortens the resection and allows for RecA-loading and recombination close to the ends of DSBs. As a consequence of the limited resection, short homologies such as plasmids or short linear fragments can now be used as templates for recombination.
Consequences of shortened end resection in ΔrecD cells
Our data highlight the importance of the length of end resection for the understanding of bacterial HR. In vivo deletion of the RecD subunit resulted in a very short length of end resection, likely leading to repair starting right at the ends of the DSB. On the other hand, the extended wild-type resection initiates recombination only after successful recognition of properly oriented χ-site. In experiments that we performed using a plasmid system, we detected successful recombination of linear fragments facilitated by short end resection in ΔrecD mutants, but not in wild-type cells (Figure 6B). Short end resection was shown to result in abnormalities during HR in eukaryotic cells, mostly due to recombination with short homologous repeats that are common in eukaryotic genomes (46), but E. coli lacks such repeats. We showed that E. coli genome fragments could be integrated by HGT in ΔrecD, but not in wild-type cells. While E. coli cells are generally deficient in natural transformation, its genome encodes machinery for the uptake of DNA from the environment (47). We noted that mutations in recBCD genes are much more common than mutations of recA, suggesting that the variability in the end resection machinery is relevant for bacterial recombination.
CONCLUSION
This work captures bacterial end resection in action inside living cells. We find a high-speed (1.6 kb/s) and extensive (>∼100 kb) asymmetric end resection. We conclude that cellular resection involves the full degradation of DNA ends near a DSB, and not merely a nick at χ sites. Furthermore, we showed how a shortened end resection has a very pronounced effect on recombination and on HGT. We expect that the results and methodology presented in this work can adapted to other biological systems.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Félicia Tjien-Fooh and Amelie Erben for valuable experimental contributions; theo van Laar and Jaco van der Torre for the experimental advices; Hugo Snippert for the gift of the I-SceI cut site plasmids; Helena Shomar for the gift of the pSC101 origin plasmid; Jacob Kerssemakers for Matlab discussions; and Christian Lesterlin for I-SceI, TB28 and DSB constructs, discussions, encouragement, and help with initial experiments. We thank Jorine Eeftens and Hyun Youk for a critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council (ERC) [NanoforBio No. 247072 to C.S., SynDiv 16 669598 to C.D.]; Wellcome Trust [SIA099204/Z/12Z to D.J.S.]; Leverhulme Trust [RP2013-K-017 to D.J.S.]; European Molecular Biology Organization [ASTF 393–2013 to J.W.]. Funding for open access charge: ERC SynDiv 16 669598.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wyman C., Kanaar R.. DNA double-strand break repair: all's well that ends well. Annu. Rev. Genet. 2006; 40:363–383. [DOI] [PubMed] [Google Scholar]
- 2. Shuman S., Glickman M.S.. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007; 5:852–861. [DOI] [PubMed] [Google Scholar]
- 3. Sung P., Klein H.. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006; 7:739–750. [DOI] [PubMed] [Google Scholar]
- 4. Lee J.Y., Terakawa T., Qi Z., Steinfeld J.B., Redding S., Kwon Y., Gaines W.A., Zhao W., Sung P., Greene E.C.. Base triplet stepping by the Rad51/RecA family of recombinases. Science. 2015; 349:977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dillingham M.S., Kowalczykowski S.C.. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008; 72:642–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lam S.T., Stahl M.M., McMilin K.D., Stahl F.W.. Rec mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974; 77:425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Singleton M.R., Dillingham M.S., Gaudier M., Kowalczykowski S.C., Wigley D.B.. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004; 432:187–193. [DOI] [PubMed] [Google Scholar]
- 8. Ponticelli A.S., Schultz D.W., Taylor A.F., Smith G.R.. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985; 41:145–151. [DOI] [PubMed] [Google Scholar]
- 9. Smith G.R. How RecBCD enzyme and chi promote DNA Break repair and recombination: a molecular biologist's view. Microbiol. Mol. Biol. Rev. 2012; 76:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Forget A.L., Kowalczykowski S.C.. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature. 2012; 482:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J.. The importance of repairing stalled replication forks. Nature. 2000; 404:37–41. [DOI] [PubMed] [Google Scholar]
- 12. Kuzminov A. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 1995; 16:373–384. [DOI] [PubMed] [Google Scholar]
- 13. Lesterlin C., Ball G., Schermelleh L., Sherratt D.J.. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014; 506:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernhardt T.G., De Boer P.A.. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 2003; 48:1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nielsen H.J., Ottesen J.R., Youngren B., Austin S.J., Hansen F.G.. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol. Microbiol. 2006; 62:331–338. [DOI] [PubMed] [Google Scholar]
- 16. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Espeli O., Mercier R., Boccard F.. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol. Microbiol. 2008; 68:1418–1427. [DOI] [PubMed] [Google Scholar]
- 18. Wiktor J., Lesterlin C., Sherratt D.J., Dekker C.. CRISPR-mediated control of the bacterial initiation of replication. Nucleic Acids Res. 2016; 44:3801–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. White M.A., Eykelenboom J.K., Lopez-Vernaza M.A., Wilson E., Leach D.R.F.. Non-random segregation of sister chromosomes in Escherichia coli. Nature. 2008; 455:1248–1250. [DOI] [PubMed] [Google Scholar]
- 20. Paintdakhi A., Parry B., Campos M., Irnov I., Elf J., Surovtsev I., Jacobs-Wagner C.. Oufti: An integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol. Microbiol. 2016; 99:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. . Fiji: an open source platform for biological image analysis. Nat. Methods. 2012; 9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bellaiche Y., Mogila V., Perrimon N.. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics. 1999; 152:1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tal A., Arbel-Goren R., Costantino N., Court D.L., Stavans J.. Location of the unique integration site on an Escherichia coli chromosome by bacteriophage lambda DNA in vivo. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:7308–7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monteilhet C., Perrin a, Thierry a, Colleaux L., Dujon B.. Purification and characterization of the in vitro activity of I-Sce I, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 1990; 18:1407–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keiler K.C., Waller P.R.H., Sauer R.T.. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996; 16:990–993. [DOI] [PubMed] [Google Scholar]
- 26. Wang X., Liu X., Possoz C., Sherratt J.D.. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006; 20:1727–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu B., Baskin R.J., Kowalczykowski S.C.. DNA unwinding heterogeneity by RecBCD results from static molecules able to equilibrate. Nature. 2013; 500:482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bianco P.R., Brewer L.R., Corzett M., Balhorn R., Yeh Y., Kowalczykowski S.C., Baskin R.J.. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001; 409:374–378. [DOI] [PubMed] [Google Scholar]
- 29. Finkelstein I.J., Visnapuu M.-L., Greene E.C.. Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature. 2010; 468:983–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schoemaker J.M., Gayda R.C., Markovitz A.. Regulation of cell division in Escherichia coli: SOS induction and cellular location of the SulA protein, a key to lon-associated filamentation and death. J. Bacteriol. 1984; 158:551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uphoff S., Reyes-Lamothe R., Leon F.G., de Sherratt D.J., Kapanidis A.N.. Single-molecule DNA repair in live bacteria. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8063–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marbouty M., Le Gall A., Cattoni D.I., Cournac A., Koh A., Fiche J.-B., Mozziconacci J., Murray H., Koszul R., Nollmann M.. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell. 2015; 59:588–602. [DOI] [PubMed] [Google Scholar]
- 33. Spies M., Bianco P.R., Dillingham M.S., Handa N., Baskin R.J., Kowalczykowski S.C.. A molecular throttle: The recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003; 114:647–654. [DOI] [PubMed] [Google Scholar]
- 34. Zierhut C., Diffley J.F.X.. Break dosage, cell cycle stage and DNA replication influence DNA double strand break response. EMBO J. 2008; 27:1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Badrinarayanan A., Le T.B.K., Spille J.-H., Cisse I.I., Laub M.T.. Global analysis of double-strand break processing reveals in vivo properties of the helicase-nuclease complex AddAB. PLOS Genet. 2017; 13:e1006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Russell C.B., Thaler D.S., Dahlquist F.W.. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J. Bacteriol. 1989; 171:2609–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Churchill J.J., Anderson D.G., Kowalczykowski S.C.. The RecBC enzyme loads recA protein onto ssDNA asymmetrically and independently of Chi, resulting in constitutive recombination activation. Genes Dev. 1999; 13:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anand R., Beach A., Li K., Haber J.. Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature. 2017; 544:377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ochman H., Lawrence J.G., Groisman E.A.. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000; 405:299–304. [DOI] [PubMed] [Google Scholar]
- 40. Babić A., Lindner A.B., Vulić M., Stewart E.J., Radman M.. Direct visualization of horizontal gene transfer. Science. 2008; 319:1533–1536. [DOI] [PubMed] [Google Scholar]
- 41. Terakawa T., Redding S., Silverstein T.D., Greene E.C.. Sequential eviction of crowded nucleoprotein complexes by the exonuclease RecBCD molecular motor. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E6322–E6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu Z., Chung W.H., Shim E.Y., Lee S.E., Ira G.. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008; 134:981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mimitou E.P., Yamada S., Keeney S.. A global view of meiotic double-strand break end resection. Science. 2017; 355:40–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Levy A., Goren M.G., Yosef I., Auster O., Manor M., Amitai G., Edgar R., Qimron U., Sorek R.. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015; 520:505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kowalczykowski S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000; 25:156–165. [DOI] [PubMed] [Google Scholar]
- 46. Chung W.H., Zhu Z., Papusha A., Malkova A., Ira G.. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010; 6:e1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sinha S., Redfield R.J.. Natural DNA Uptake by Escherichia coli. PLoS One. 2012; 7:e35620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.