Abstract
The DNA-dependent protein kinase (DNA-PK), consisting of the DNA binding Ku70/80 heterodimer and the catalytic subunit DNA-PKcs, has been well characterized in the non-homologous end-joining mechanism for DNA double strand break (DSB) repair and radiation resistance. Besides playing a role in DSB repair, DNA-PKcs is required for the cellular response to replication stress and participates in the ATR-Chk1 signaling pathway. However, the mechanism through which DNA-PKcs is recruited to stalled replication forks is still unclear. Here, we report that the apoptosis mediator p53-induced protein with a death domain (PIDD) is required to promote DNA-PKcs activity in response to replication stress. PIDD is known to interact with PCNA upon UV-induced replication stress. Our results demonstrate that PIDD is required to recruit DNA-PKcs to stalled replication forks through direct binding to DNA-PKcs at the N’ terminal region. Disruption of the interaction between DNA-PKcs and PIDD not only compromises the ATR association and regulation of DNA-PKcs, but also the ATR signaling pathway, intra-S-phase checkpoint and cellular resistance to replication stress. Taken together, our results indicate that PIDD, but not the Ku heterodimer, mediates the DNA-PKcs activity at stalled replication forks and facilitates the ATR signaling pathway in the cellular response to replication stress.
INTRODUCTION
The catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs) is a key component of the non-homologous end joining (NHEJ) pathway in DNA double strand break (DSB) repair (1). DNA-PKcs and the closely related members of the phosphatidylinositol-3 kinase-like kinase (PIKK) family, including ataxia-telangiectasia mutated (ATM) and ATM and Rad3-related (ATR), are key regulators in the cellular response to various types of DNA damage (2). Together they are critical for the maintenance of genomic integrity to prevent genetic disorders, aging and carcinogenesis.
The primary role of DNA-PKcs in DSB repair has been well characterized (1). DNA-PKcs is recruited to DSBs by the DNA-binding Ku70/80 heterodimer. Together they form the kinase active DNA-PK holoenzyme and promote NHEJ-mediated DSB repair. The regulation of DNA-PKcs activity also relies on its own phosphorylation status particularly at the Thr2609 cluster region, which is crucial for DSB repair, cellular resistance to radiation and tissue stem maintenance (3–5). DNA-PKcs phosphorylation at the Thr2609 cluster may contribute to its conformational changes and modulates the association of DNA-PKcs with the Ku70/80 heterodimer or other DNA repair molecules to impact DSB repair (1). In addition, we have demonstrated that DNA-PKcs is rapidly phosphorylated by ATR at the Thr2609 cluster upon replication stress (6), and also maintains the stability of the Chk1-Claspin complex for optimal intra-S-phase checkpoint (7). Cells lacking a functional Thr2609 cluster confer sensitivity to ultraviolet (UV) and other replication stress-inducing agents, suggesting that ATR-dependent DNA-PKcs phosphorylation is necessary for the cellular response to replication stress (5,6). This is consistent with the finding that DNA-PKcs plays a role in regulating RPA2 phosphorylation (8–11). However, the mechanism through which DNA-PKcs is recruited to stalled replication forks and whether the Ku70/80 heterodimer is involved in the ATR pathway and regulation is still unknown.
To further delineate the mechanism of DNA-PKcs in the replication stress response, we have discovered that the p53-induced protein with a death domain (PIDD) participates in the recruitment of DNA-PKcs to stalled replication forks. PIDD is known to play multiple roles in the response to DNA damage, possibly leading to either cell death or survival (12). For example, PIDD is known as a mediator of apoptosis through assembly of the PIDDosome complex containing Caspase-2 for the initiation of apoptosis upon genotoxic stress (13). In contrast, PIDD also promotes the NF-κB pathway activation for cellular survival upon DNA damage (14). PIDD also participates in DNA damage repair and stimulates PCNA monoubiquitination during translesion DNA synthesis in response to UV irradiation (15). Here, we showed that PIDD is required for DNA-PKcs association and stimulation of the ATR pathway. Disruption of the interaction between DNA-PKcs and PIDD compromises the intra-S-phase checkpoint and cell survival against UV irradiation. Our results suggest that DNA-PKcs and PIDD are integral components of the cellular response to replication stress.
MATERIALS AND METHODS
Cell culture, siRNA transfection and plasmid
Human cervical cancer HeLa cells, human embryonic kidney 293 cells, human colon cancer HCT116 Ku86flox/− cells (16), Chinese hamster ovary cells deficient in DNA-PKcs (V3) and V3-complemented cells were maintained in α–minimum essential medium (HyClone) supplemented with 10% fetal bovine serum in a humidified incubator at 37°C with 5% CO2. Small interfering RNA (siRNA) oligonucleotide duplexes designed against PIDD (siPIDD#1: CAGACUGUUCCUGACCUCAGA, siPIDD#2: GCAGCCCUCAUUCCAGAAA, siPIDD#3: UCCUUGUCCUGCACAGCAA, Invitrogen) were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Full-length or truncated PIDD were amplified and cloned into pIRES2-DsRed2 (Clontech) or pcDNA3.1+/N-HA (Invitrogen) for mammalian expression. DNA-PKcs cDNA fragments were cloned into pGEX-6P1 (GE Healthcare Life Sciences) for Glutathione S-transferase (GST) fusion protein, and PIDD cDNA fragments were cloned into pET28a (Novagen) for His-tagged protein.
Clonogenic survival assay and growth curve
Exponentially growing V3 and human DNA-PKcs-complemented V3 cells were exposed to γ-rays with a J. L. Shepherd and Associates irradiator that emitted 137Cs γ-rays at a dose rate of 3.44 Gy/min to the cells’ positions. Cells were immediately plated at the appropriate number and incubated for 7–9 days to yield 30–60 surviving colonies for each 60-mm dish (Greiner). For UV irradiation, cells growing exponentially on culture dishes were washed with phosphate-buffered saline (PBS) and exposed to UV-C (254 nm) at a rate of 0.8–1 J/m2/s to achieve the desired cumulative dose. Fresh culture medium was added to the culture dishes immediately after irradiation. The colonies were fixed with 100% ethanol and stained with 0.1% crystal violet. A colony with more than 50 cells was counted as a survivor. To generate a growth curve, cells were plated with 5 × 104 cells for each 60-mm dish on day 0 and counted daily with a Z2 Coulter Counter (Beckman) according to the manufacturer’s instructions.
Western blot, immunoprecipitation and immunofluorescence staining
Whole cell lysates were prepared with lysis buffer (50 mM Tris pH 7.5, 0.2 M NaCl, 1% Tween-20, 1% NP40, 1 mM sodium orthovanadate, 2 mM β–glycerophosphate and protease inhibitors) and then used for western blotting (WB). For immunoprecipitation (IP), antibodies or the normal IgG control (Santa Cruz Biotechnology) were first incubated with protein A Sepharose beads CL-4B (GE Healthcare Life Sciences) for 1 h. Cells were lysed in IP buffer (20 mM Tris pH 7.5, 120 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.2% NP-40, 10% glycerol and protease inhibitors) and incubated overnight with pulled antibody-protein A beads. The beads were washed with IP buffer and resuspended in Laemmli buffer for WB. To avoid any interference from the heavy chains and the light chains of IgG used for IP, either EasyBlot HRP-conjugated anti-mouse IgG or EasyBlot HRP-conjugated anti-rabbit IgG was used as the secondary antibody in WB. For immunofluorescence (IF) staining, cells were fixed with 4% paraformaldehyde (PFA) in PBS for 15 min, treated with 0.5% Triton X-100 in PBS for 5 min, and blocked with 5% normal goat serum in PBS for 1 h at room temperature. Cells were incubated with primary antibodies for 2 h, washed three times with PBS, and incubated with Alexa Fluor 488-anti-rabbit and Texas Red-conjugated anti-mouse secondary antibodies for 1 h (Invitrogen). Cells were washed three times with PBS and mounted in Vectashield mounting medium with 4,6-diamidino-2-phenylindole (Vector Laboratories). For EdU (5-ethynyl-2′-deoxyuridine) labeling, cells were labeled with 50 μM EdU for 1 h and then exposed to UV. Cells were fixed with 4% PFA, stained with a Click-iT EdU imaging kit (Invitrogen) according to the manufacturer’s instructions and treated for IF staining as described above. For RPA2 IF staining, cells were pre-extracted with cytoskeleton buffer (10 mM HEPES, pH 7.4, 300 mM sucrose, 100 mM NaCl, 3 mM MgCl2, 0.1% Triton X-100) for 5 min and then fixed with 4% PFA. Cells were incubated with anti-RPA2 antibody (EMD Millipore, NA19L, 1:500) overnight and proceeded with the staining procedure as described above. For single-strand DNA (ssDNA) staining, cells were cultured in regular medium containing 10 μM BrdU for 30 h. Following hydroxyurea (HU) (2 mM) treatment, cells were fixed with 4% PFA and proceeded with the staining procedure using anti-BrdU antibody (BD Bioscience). Images were acquired using a Zeiss AxioImager M2 microscope system equipped with a Plan-Apochromat 63X /NA 1.40 objective, an AxioCam MRm CCD camera and AxioVision software (Carl Zeiss). Anti-pS2056, anti-pT2609 and anti-pT2647 phospho-specific rabbit polyclonal antibodies have been used and described previously (3,4,17). DNA-PKcs mouse monoclonal antibody clones 18–2 and 25–4 (Thermo Fisher Scientific) for IP, anti-ATR N19 (Santa Cruz Biotechnology), anti-ATR pT1989 (GeneTex), anti-SMC1 (Bethyl), anti-phosphor-S966 SMC1 (Bethyl), anti-Chk1 (Cell Signaling), anti-phospho-S317 Chk1 (Cell Signaling), anti-PIDD Anto-1 (Enzo Life Sciences), anti-phospho-S139 H2AX (EMD Millipore), anti-FLAG M2 (Sigma), anti-Ku80 (Santa Cruz Biotechnology), anti-Orc2 (BD Biosciences) and anti-Actin (Sigma) were commercially available from the indicated vendors.
Protein purification and GST pull down assay
Recombinant GST-fused DNA-PKcs fragments and His-tagged PIDD were expressed in BL21 Escherichia coli. GST-DNA-PKcs fusion pellets were lysed and sonicated in STE buffer (150 mM NaCl, 25 mM Tris pH 8, 1 mM EDTA, 100 μg/ml lysozyme, 1.5% N-Lauroylsarcosine, 2% Triton X-100), and cleared by centrifugation (13000 rpm, 20 min at 4°C). Recombinant His-tagged PIDD-DD proteins were purified by the His-affinity (Qiagen) column. For the GST pull down assay, GST lysates were affinity purified with Glutathione-Sepharose beads (GE Healthcare Life Sciences) and incubated with His-tagged PIDD-DD in the presence of EtBr (40 ng/μl). Proteins bound to Glutathione-Sepharose beads were analyzed by WB.
Proximity ligation assay
Exponentially growing cells were exposed to UV radiation and harvested after 30 min of incubation. Cells were trypsinized, counted, and deposited onto microscope slides (1 × 105/slide) by a Cytospin 4 cytocentrifuge (Thermo Fisher Scientific) at 500 rpm for 5 min. Proximity ligation assay (PLA) was performed using Duolink PLA reagents (Sigma-Aldrich) according to the manufacturer’s instructions. Briefly, deposited cells were fixed with 4% PFA in PBS for 20 min, treated with 0.5% Triton X-100 in PBS for 10 min and blocked with Duolink in situ blocking solution for 1 h at room temperature. Cells were incubated with primary antibodies in Duolink in situ antibody diluent for 2 h, washed with PBS twice, and left in PBS overnight at room temperature. Cells were incubated with oligonucleotides-conjugated secondary antibodies (PLA probe anti-rabbit PLUS and anti-mouse MINUS) followed by ligation and rolling circle amplification with a fluorophore labeled oligonucleotide probe (λex = 495 nm and λem = 527 nm). Cells were mounted in Vectashield mounting medium with 4,6-diamidino-2-phenylindole (Vector Laboratories) and visualized using a Zeiss AxioImager M2 microscope equipped with a EC Plan-Neofluar 40X/0.75 objective. PLA spot counting was performed using the ImageJ (NIH, USA) software. Mouse anti-DNA-PKcs (Thermo Fisher Scientific, 1:100 dilution), rabbit anti-ATR (Bethyl, 1:100), mouse anti-RPA2 (EMD Millipore, 1:50) and rabbit anti-FLAG (Sigma, 1:100) antibodies were commercially available at the indicated vendors.
Fractionation
Soluble nuclear and chromatin-bound fractions were prepared according to a published fractionation protocol (18). Briefly, cell pellets were resuspended (2 × 107 cells/ml) in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM Dithiothreitol (DTT), protease inhibitors and phosphatase inhibitors) containing 0.1% Triton X-100 and incubated for 5 min on ice. Nuclei (P1) was collected by low-speed centrifugation (1300 × g, 4 min, 4°C) and washed once with buffer A. Washed nuclei were lysed with buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease inhibitors and phosphatase inhibitors) and incubated for 30 min on ice. The soluble nuclear fraction (S3) was separated from insoluble chromatin by centrifugation (1700 × g, 4 min, 4°C). The chromatin pellet was washed once with buffer B and centrifuged again. The final pellet (P3) was resuspended with Laemmli buffer and sonicated for 30 s.
DNA fiber analysis
Cells were labeled sequentially with 100 μM iododeoxyuridine (IdU) for 10 min and chlorodeoxyuridine (CldU) for 20 min in a mock experiment. To monitor the replication stress response, cells were first labeled with IdU followed by UV exposure and then labeled with CldU. DNA fibers were spread as described (19), and stained with primary antibodies (mouse anti-BrdU/IdU, BD Bioscience; rat anti-BrdU/CldU, Accurate Chemical) and fluorescence-conjugated secondary antibodies (Alexa Fluor 488 anti-rat and Texas Red anti-mouse, Invitrogen). Fibers were imaged using the Zeiss AxioImager M2 and measured using the AxioVision software (x64 version 4.9.1).
Statistical analysis
Statistical analyses were performed using a two-way analysis of variance (ANOVA) followed by the Bonferroni’s post-hoc test. All statistical analyses were performed using the Prism GraphPad (version 6.02) software. Statistical significance was defined as P < 0.05, and values of P < 0.0001 were also shown to indicate the level of confidence. The length or CldU to IdU ratios of the DNA fiber tracks were diagnosed with the equal variance assumption by F-test, and analyzed by the significance by unpaired t-test.
RESULTS
A PIP-like motif of DNA-PKcs is required for cellular resistance to replication stress
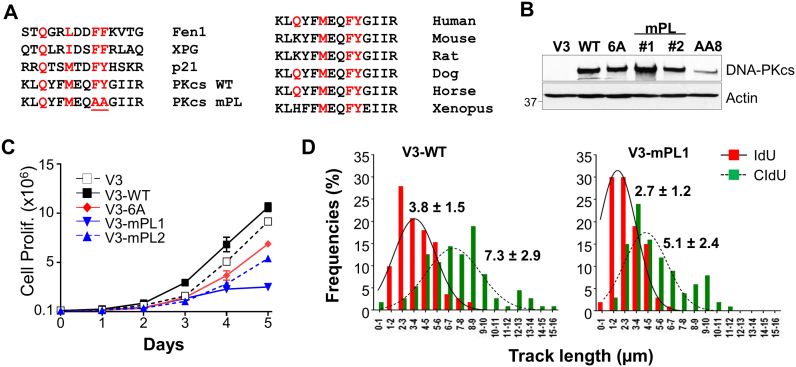
We have identified a motif resembling the consensus PCNA-interacting protein (PIP) motif (20) at the N’-terminal of DNA-PKcs (Figure 1A). To determine whether this PIP-like (abbreviates as PL) motif is functional, DNA-PKcs-deficient CHO V3 cells were complemented with a DNA-PKcs mutant harboring alanine substitutions at F345/Y346 within the PL motif (V3-mPL #1 and #2), and were analyzed together with other V3 derivative cell lines (Figure 1B). In cell proliferation assays, we observed that V3 cells complemented with wild-type DNA-PKcs (V3-WT) showed a slightly improved growth rate than V3 cells complemented with the empty vector (V3) or a mutant DNA-PKcs carrying alanine substitutions at the Thr2609 phosphorylation cluster (V3–6A). On the contrary, both V3-mPL1 and V3-mPL2 cells exhibited slower growth rates than other V3 cells (Figure 1C). Furthermore, when subjected to sequential pulse-labeling with iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU) in the DNA fiber assay (19,21), we observed that the progression of on-going DNA replication tracks (measured by the length of the DNA fibers) were significantly attenuated in V3-mPL1 cells than in V3-WT cells (Figure 1D). IdU-labeled tracks were 2.7 ± 1.2 μm in V3-mPL1 cells compared to 3.8 ± 1.5 μm in V3-WT cells (P < 0.0001), whereas CldU-labeled tracks were 5.1 ± 2.4 μm in V3-mPL1 cells compared to 7.3 ± 2.9 μm in V3-WT cells (P < 0.0001). These results indicate that impairment of the DNA-PKcs PL motif compromises normal DNA replication and cell proliferation, although flow cytometry analysis revealed similar cell cycle profiles in V3-mPL cells and other V3 derivative cells (Supplementary Figure S1A).
Figure 1.
The PL motif of DNA-PKcs is required for robust DNA replication. (A) Sequence comparison of human DNA-PKcs amino acid 337–350 with the PIP box motif of other PIPs (left panel) and homology with other vertebrates (right panel). Consensus sequence of the PIP box, QxxΨxxϑϑ. Ψ = M, L, I; ϑ = F, Y. The DNA-PKcs PL mutant (mPL) carries alanine substitutions at F345 and Y346. (B) Expression of DNA-PKcs in wild-type CHO AA8 cells, DNA-PKcs deficient V3 cells and V3 cells complemented with empty vector (V3), wild-type DNA-PKcs (WT) and mutants carrying alanine substitutions at the Thr2609 cluster (V3–6A) or the PL motif (V3-mPL). (C) Growth curve of V3 and derived cell lines. Cell numbers were analyzed by a cell counter (Beckman Coulter Z2). Data are presented as mean ± s.d., N = 2. (D) V3-WT and V3-mPL cells were pulse-labeled with iododeoxyuridine (IdU, 10 min) and chlorodeoxyuridine (CldU, 20 min) sequentially, and were analyzed by the DNA fiber assay. The lengths of IdU (red) and CldU (green) tracks were analyzed from >100 ongoing DNA replication tracks.
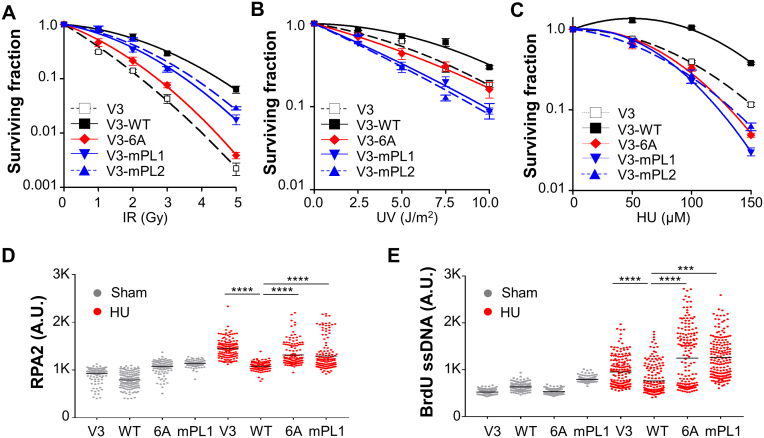
To further delineate the role of the PL motif on DNA-PKcs activity and regulation, V3-mPL cells were analyzed for clonogenic survival against ionizing radiation (IR), UV and HU. V3-mPL1 and V3-mPL2 cells exhibited an intermediate sensitivity toward IR compared with the extreme radiosensitive V3 and V3–6A cells (4,6) (Figure 2A). However, V3-mPL cells were highly sensitive to UV irradiation (Figure 2B) and HU exposure (Figure 2C), suggesting that V3-mPL cells are defective in cellular response to replication stress or stalled replication forks. This notion was further supported by the stronger induction of ssDNA in response to HU treatment in V3-mPL cells as well as in V3 and V3–6A cells than V3-WT cells (Figure 2E). Similarly, we also observed significant increases of chromatin-bound RPA2 in V3-mPL, V3 and V3–6A cells compared to V3-WT cells (Figure 2D and Supplementary Figure S1B). These results suggest that the PL motif or phosphorylation at the Thr2609 cluster region are critical for the role of DNA-PKcs in cellular response to replication stress.
Figure 2.
The PL motif of DNA-PKcs is required for DNA replication stress response. (A–C) V3 derivative cell lines were analyzed for their colony forming ability against IR (A), UV (B) and HU (C). Data are presented as mean ± s.e.m., N = 3. (D) V3 derived cells were pulse-labeled with 50 μM EdU, treated with HU (2 mM) for 2 h, pre-extracted with 0.1% TX-100, and stained against anti-RPA2 antibody and EdU. Data are presented as RPA2 densities in EdU+ nuclei (N > 150). ****P < 0.0001. (E) V3 derived cells were labeled with BrdU (10 μM) for 30 h and then treated with HU (2 mM) for 2 h. BrdU-labeled single-stranded DNA was stained with anti-BrdU antibody under non-denaturing condition. Data are presented as BrdU densities in nuclei (N > 150). ****P < 0.0001; ***P < 0.001.
DNA-PKcs PL motif participates in the ATR signaling pathway and intra-S-phase checkpoint
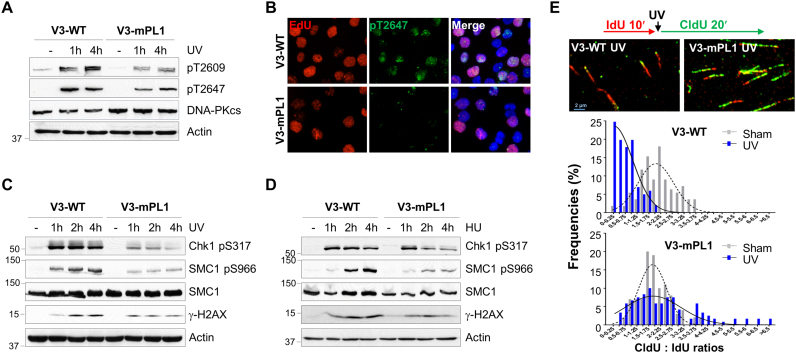
Since both the PL motif and Thr2609 cluster contribute to the cellular resistance to replication stress, we investigated whether the mutated PL motif affects DNA-PKcs phosphorylation at the Thr2609 cluster. In V3-WT cells, UV irradiation induced robust DNA-PKcs phosphorylation at both Thr2609 and Thr2647; however, both DNA-PKcs phosphorylations were attenuated in V3-mPL1 cells (Figure 3A). A similar reduction in DNA-PKcs Thr2647 phosphorylation was observed in V3-mPL1 cells after IF staining; importantly, UV-induced Thr2647 phosphorylation in V3-WT cells occurred primarily in the EdU labeled S-phase population (Figure 3B), indicating that the PL motif is required for ATR dependent DNA-PKcs phosphorylation upon UV-induced replication stress (6).
Figure 3.
DNA-PKcs PL motif participates in the ATR signaling pathway and intra-S checkpoint. (A) V3-WT and V3-mPL1 cells were exposed to UV (20 J/m2) and harvested at the indicated time points for WB using regular and phospho-specific antibodies against DNA-PKcs. (B) V3-WT and V3-mPL1 cells were pulse-labeled with 50 μM EdU, exposed to UV and then stained against EdU (red) and anti-pT2647 antibody (green). (C and D) Cells were treated with UV or HU and harvested at the indicated time points. Whole cell lysates were analyzed with indicated antibodies. (E) V3-WT and V3-mPL1 cells were sequentially labeled with IdU (100 μM, 10 min) and CldU (100 μM, 20 min) with or without UV exposure in between labels, and then analyzed by DNA fiber assay. The length of DNA tracks labeled with IdU (red) and CldU (green) were measured. The ratios of CldU to IdU in length were calculated from ongoing replication tracks (red-green, N ≥ 100).
We next asked whether this phenomenon is specific to ATR-dependent DNA-PKcs phosphorylation or if it generally affects the ATR signaling pathway. ATR downstream signaling events including Chk1 phosphorylation at S317 (Chk1 pS317) and SMC1 phosphorylation at S966 (SMC1 pS966) were rapidly induced in both V3-WT and V3-mPL1 cells upon UV treatment (Figure 3C). However, the intensities of UV-induced Chk1 and SMC1 phosphorylations were significantly decreased in V3-mPL1 cells. In contrast, we observed similar levels of H2AX phosphorylation (γH2AX) in both V3-WT and V3-mPL1 cells in response to UV. In response to HU treatment, reduction in SMC1 phosphorylation was obvious in V3-mPL cells whereas reduction in Chk1 phosphorylation was subtle initially and became obvious at 2–4 h (Figure 3D).
Attenuation in the ATR signaling pathway would lead to a compromised intra-S-phase checkpoint. For validation, the DNA fiber assay was performed on V3-WT and V3-mPL1 cells under pulse-labeling with IdU for 10 min, with or without UV treatment and followed by secondary pulse-labeling with CIdU for 20 min (Figure 3E). The intra-S-phase checkpoint response can be determined by comparing the length of the IdU tracks (before UV irradiation) versus the CIdU tracks (after UV irradiation). Our analysis revealed that normal DNA replication was similar in V3-WT and V3-mPL1 cells in the absence of UV radiation. The average ratios of the CldU tracks against the IdU tracks were 2.06 ± 0.77 in V3-WT cells and 1.96 ± 0.67 in V3-mPL1 cells (P = 0.3658). Upon UV exposure, the progression of DNA replication in V3-WT cells was attenuated due to the ATR-Chk1-dependent intra-S-phase checkpoint, whereas this checkpoint response was compromised in V3-mPL1 cells. The average of the CldU to IdU ratios was 0.64 ± 0.46 in V3-WT cells and 2.38 ± 1.64 in V3-mPL1 cells (P < 0.0001).
The PL motif of DNA-PKcs is required for PIDD interaction
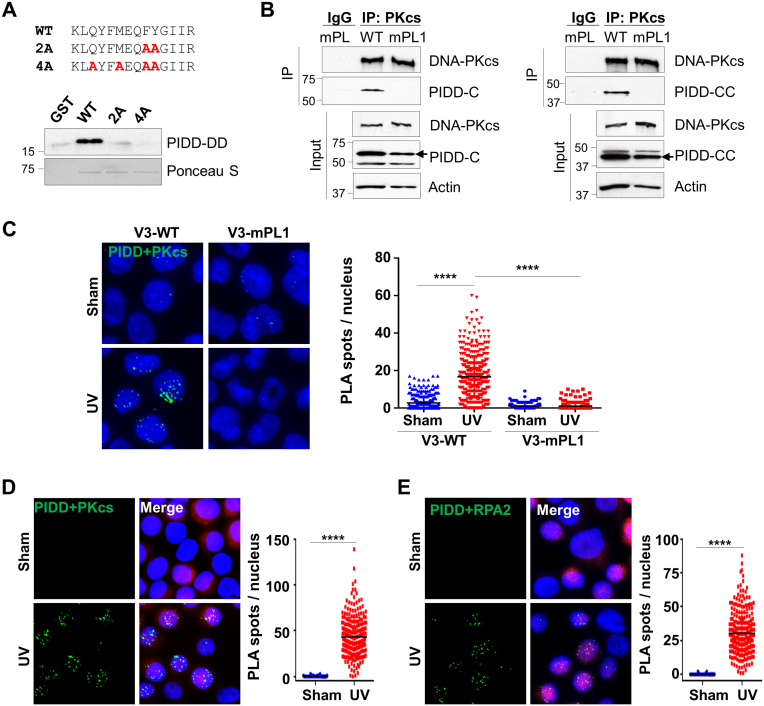
We initially hypothesized that the PL motif of DNA-PKcs could bridge the interaction between DNA-PKcs and PCNA in the DNA damage response. Using the GST pulldown assay, we showed that endogenously purified DNA-PKcs from HeLa cells could bind to GST-PCNA fusion proteins in vitro (Supplementary Figure S2A). However, in a reciprocal GST pulldown with the N’-terminal DNA-PKcs fragment containing the PL motif, recombinant PCNA proteins displayed only a weak affinity to the GST-PKcs fragment as compared with GST-p21 fusion protein (Supplementary Figure S2B). Alanine substitutions at the DNA-PKcs PL motif also did not alter the binding of PCNA, suggesting that the PL motif of DNA-PKcs interacts with other molecules but not PCNA. In collaboration with C. Du, we have identified that DNA-PKcs could form a protein complex with the PIDD (data not shown). Our finding was supported by an independent study that DNA-PKcs was identified among PIDD immunoprecipitated protein complexes (15).
PIDD normally undergoes auto-proteolysis to produce the truncated PIDD-C and PIDD-CC fragments (22–24). Using co-IP assays, we observed that DNA-PKcs interacts with not only the full length PIDD but also both PIDD-C and PIDD-CC fragments (Supplementary Figure S3B), suggesting that DNA-PKcs associates with PIDD at the C’ terminal region containing the death domain (DD). Conversely, domain mapping analysis revealed that PIDD binds to DNA-PKcs directly at the N’ terminal region (Supplementary Figure S3C). To determine whether the DNA-PKcs PL motif mediates the direct protein–protein interaction between DNA-PKcs and PIDD, recombinant PIDD-DD fragments were subjected to GST pulldown with the wild-type DNA-PKcs N-terminal fragments or PL mutant fragments. The result showed that alanine substitutions at either F345/Y346 (2A) or Q339/M342/F345/Y346 (4A) residues completely abolished the binding of PIDD to DNA-PKcs (Figure 4A). Furthermore, when transfecting FLAG-tagged PIDD-C or PIDD-CC fragments into V3-WT and V3-mPL1 cells, PIDD was co-precipitated with the wild-type DNA-PKcs but not the mPL mutant (Figure 4B). This verifies that the DNA-PKcs PL motif is required for PIDD interaction under the full-length DNA-PKcs context in vivo. This notion is supported by in situ PLA that UV exposure stimulates the association between DNA-PKcs and PIDD in V3-WT cells but not in V3-mPL1 cells (Figure 4C). Similar results generated in HeLa cells showed that UV exposure stimulates the association of PIDD with DNA-PKcs (Figure 4D) and RPA2 (Figure 4E). These analyses further suggest that UV irradiation induces DNA-PKcs and PIDD association at stalled replication forks.
Figure 4.
The DNA-PKcs PL motif mediates the DNA-PKcs and PIDD association in vitro and in vivo. (A) Recombinant His-tagged PIDD death domain (a.a. 778–873) was pulled down by the GST fusion protein carrying the wild-type DNA-PKcs N’ terminal fragment (a.a. 1–403) but not the PL mutant fragments (2A or 4A), as indicated. The loading of the GST fusions was demonstrated by Ponceau S staining. (B) V3-WT or V3-mPL1 cells were transfected with FLAG-tagged PIDD-C (a.a. 446–910) or PIDD-CC (a.a. 588–910) constructs. Whole cell lysates were subjected to co-IP with anti-DNA-PKcs antibody and western blotted against anti-DNA-PKcs or anti-FLAG antibodies. (C) V3-WT and V3-mPL1 cells transfected with the FLAG-PIDD construct were UV (20 J/m2) irradiated and harvested at 30 min for PLA (green) using anti-DNA-PKcs and anti-FLAG antibodies. The right panel shows the quantification of PLA spots per nucleus. N > 200. ****P < 0.0001. (D and E) HeLa cells transfected with an IRES-DsRed/FLAG-PIDD plasmid were exposed to UV followed by PLA analysis using anti-FLAG coupled with (D) anti-DNA-PKcs or (E) anti-RPA2 antibodies. Right, quantification. N > 200. ****P < 0.0001.
PIDD facilitates the association of DNA-PKcs with ATR in response to UV radiation
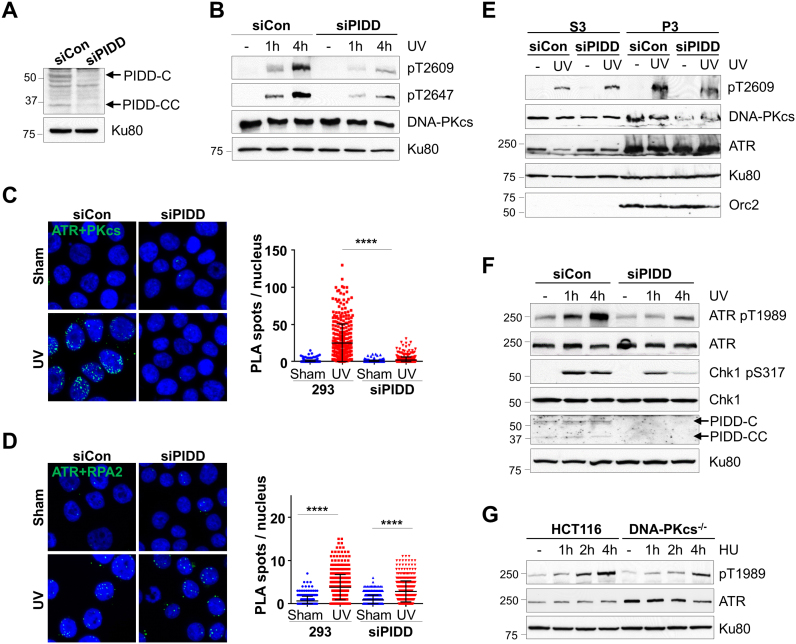
Upon UV irradiation, PIDD is recruited to stalled replication forks through direct binding to PCNA (15). We hypothesize that the PIDD-dependent recruitment of DNA-PKcs facilitates its association with ATR. To test this possible scenario, 293FT cells were transfected with control and siRNA against PIDD (siPIDD) (Figure 5A and Supplementary Figure S4), and examined for ATR-dependent DNA-PKcs phosphorylation upon UV irradiation. Our analyses revealed that DNA-PKcs phosphorylations at Thr2609 and Thr2647 were attenuated in PIDD knockdown cells (Figure 5B and Supplementary Figure S5A). Using in situ PLA analyses, we observed that the association between DNA-PKcs and ATR was significantly induced upon UV irradiation in control 293FT cells, and that this stimulation was abolished in siPIDD-transfected 293FT cells (Figure 5C). On the contrary, the PIDD knockdown did not affect the direct association between ATR and RPA2 in response to UV irradiation (Figure 5D). Similar results were generated in HeLa cells (Supplementary Figure S5C). In agreement with the PLA analysis, we also observed that chromatin-bound DNA-PKcs and its phosphorylation were compromised in the PIDD knockdown cells, whereas chromatin-bound ATR remained the same regardless of the PIDD status (Figure 5E).
Figure 5.
PIDD bridges DNA-PKcs and ATR upon replication stress. (A) 293FT cells were transfected with control (siCon) and siRNA against PIDD (siPIDD), and analyzed for PIDD protein expression. (B) Control and siPIDD-transfected 293FT cells were exposed to UV (20 J/m2) and harvested at the indicated time points. Whole cell lysates were subjected to western blot. (C and D) Control and siPIDD-transfected 293FT cells were exposed to UV and harvested at 30 min for PLA (green) using anti-ATR in combination with (C) anti-DNA-PKcs or (D) anti-RPA2 antibodies. The right panel shows the quantification of the PLA spots per nucleus. N > 300. ****P < 0.0001. (E) HeLa cells transfected with the control and siPIDD were exposed to UV. Soluble nuclear protein fraction (S3) and chromatin nuclear matrix fraction (P3) were prepared for WB as indicated. (F) 293FT-transfected siCon and siPIDD were analyzed for the ATR signaling pathway upon UV. (G) HCT116 and derivative DNA-PKcs−/− cells were treated with HU and analyzed for ATR autophosphorylation.
The PIDD knockdown not only affected DNA-PKcs phosphorylation and its association with ATR, but also attenuated the ATR signaling pathway. We observed that ATR pS1989 autophosphorylation (25,26) and Chk1 pS317 phosphorylation were attenuated in PIDD-depleted 293FT cells (Figure 5F and Supplementary Figure S5A). Additionally, UV-induced PRA2 hyperphosphorylation was also attenuated in PIDD-depleted 293FT cells (Supplementary Figure S5B). PIDD-dependent DNA-PKcs recruitment to stalled replication forks likely facilitates DNA-PKcs association with ATR and the ATR signaling pathway. This notion was supported by the finding that ATR autophosphorylation at S1989 upon HU was attenuated in DNA-PKcs−/− cells as compared with that in the parental HCT116 cells (Figure 5G). The steady state protein levels of ATR were elevated in the DNA-PKcs−/− cells, suggesting a feedback mechanism that compensates for the loss of DNA-PKcs or an increase in spontaneous replication stress.
Ku80 is dispensable for the association between DNA-PKcs and ATR in response to UV irradiation
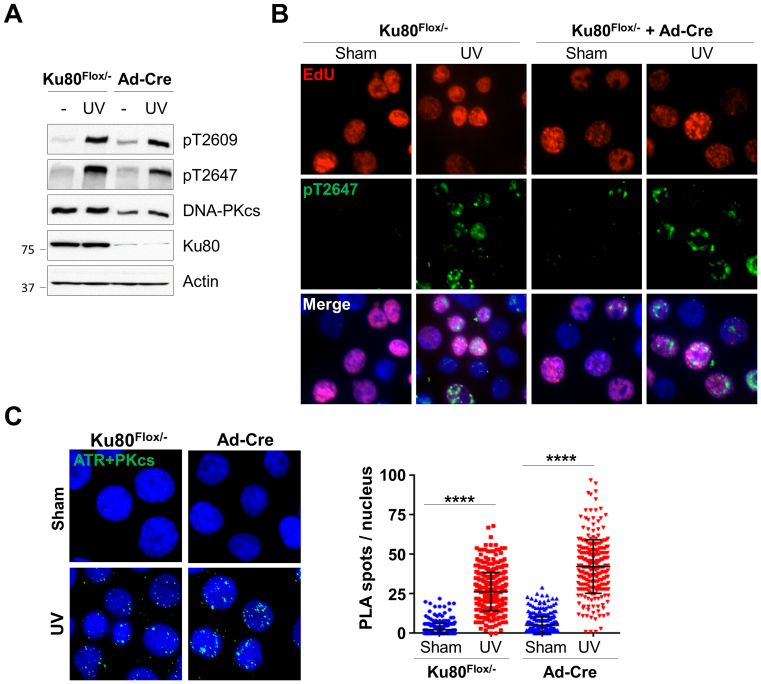
Our results consistently indicate that PIDD is required to mediate DNA-PKcs activity in response to replication stress, although the Ku70/Ku80 heterodimer, the DSB-binding partner of DNA-PKcs, may play a role in DNA-PKcs recruitment to stalled replication forks. For further clarification, HCT116-derived conditional null Ku80flox/− cells (16) were infected with an adenoviral vector carrying Cre recombinase (Ad-Cre) to knockout Ku80 gene expression (Supplementary Figure S6A). Our analysis revealed that UV-induced DNA-PKcs phosphorylation at the Thr2609 cluster was not affected in Ku80 knockout cells, although the protein levels of DNA-PKcs were decreased without Ku80 (Figure 6A). UV-induced DNA-PKcs phosphorylation occurred in EdU labeled S-phase cell population in both control Ku80flox/− cells and Ad-Cre infected Ku80flox/− cells (Figure 6B), confirming that Ku80 is not required for ATR-dependent DNA-PKcs phosphorylation upon replication stress. Indeed, PLA analysis revealed that the knockout of Ku80 did not affect UV stimulation on the association between DNA-PKcs and ATR (Figure 6C). A similar PLA result was generated using mouse embryonic fibroblasts derived from wild-type and Ku80 knockout mice (Supplementary Figure S6B). Taken together, we conclude that Ku80 is dispensable for DNA-PKcs recruitment and its association with ATR at stalled replication forks.
Figure 6.
Ku80 is dispensable for DNA-PKcs and ATR association in response to UV irradiation. HCT116 Ku86Flox/− cells were sham treated or incubated with Ad-Cre adenovirus expressing Cre recombinase for 4 days. (A) Cells were exposed to UV (20 J/m2) and analyzed for DNA-PKcs phosphorylation by western blot. (B) Cells were pulse-labeled with EdU, exposed to UV, followed by immunostaining against EdU (red) and anti-pT2647 (green) antibodies. (C) Cells were exposed to UV and analyzed by PLA using anti-ATR and anti-DNA-PKcs antibodies. Right, quantification. Ku86Flox/− sham, N > 300. ****P < 0.0001.
PIDD facilitates the intra-S-phase checkpoint in response to UV irradiation
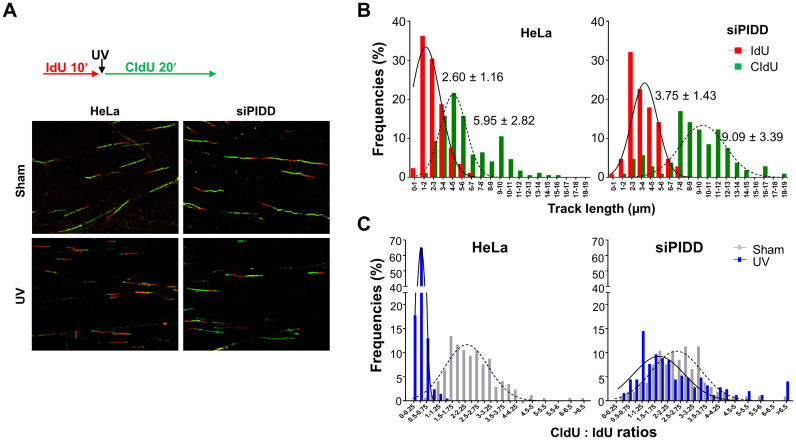
Our results predict that the PIDD deficiency will compromise the intra-S-phase checkpoint response since PIDD is required to promote the association between DNA-PKcs and ATR for optimal activation of the ATR signaling pathway. For validation, HeLa cells were transfected with siPIDD and subjected to DNA fiber assay with or without UV exposure. Our analysis revealed that the speed of the ongoing DNA replication forks increased in PIDD-depleted cells without UV treatment (Figure 7B). The averages of the IdU tracks (10-min labeling) were 2.60 ± 1.16 μm in HeLa cells and 3.75 ± 1.43 μm in siPIDD cells (P < 0.0001), whereas the averages of the CIdU tracks (20-min labeling) were 5.95 ± 2.82 μm in HeLa cells and 9.09 ± 3.39 μm in siPIDD cells (P < 0.0001). Nonetheless, the CldU to IdU ratios were similar in HeLa (average 2.44 ± 1.01) and siPIDD cells (average 2.60 ± 1.05) (Figure 7C). In response to UV irradiation, the progression of the replication forks was mostly attenuated or completely stalled in HeLa cells. In contrast, DNA tracks were still observed in siPIDD cells even after UV exposure (Figure 7A). The average CldU to IdU ratios were 0.39 ± 0.19 in HeLa cells compared to 2.45 ± 1.77 in siPIDD cells (Figure 7C). These results clearly demonstrated that PIDD is required for the intra-S-phase checkpoint in response to UV exposure.
Figure 7.
PIDD is required for intra-S-phase checkpoint response. (A) HeLa cells were sham-treated or transfected with siPIDD followed by DNA fiber analysis. DNA tracks labeled with IdU (red) and CldU (green) were detected using monoclonal mouse and rat anti-BrdU antibodies, respectively. (B) The lengths of IdU and CldU tracks were analyzed from ongoing replication tracks (red-green, N > 100). (C) The ratios of CldU to IdU in length were calculated from ongoing replication tracks. N > 100.
DISCUSSION
In the current study, we showed that the PIDD is required to recruit DNA-PKcs and its association with ATR at stalled replication forks. Our results demonstrate that DNA-PKcs interacts with PIDD dead domain through a PL motif at the N’ terminal region of DNA-PKcs (Supplementary Figure S3C). Although PIDD-DD might also bind to DNA-PKcs at the FAT (FRAP, ATM, TRRAP) region, alanine substitutions at the PL motif completely abolish the interaction between DNA-PKcs and PIDD in vitro and in vivo (Figure 3A–C), indicating that DNA-PKcs PL motif is essential for PIDD association. PIDD was reported to interact directly with PCNA upon UV irradiation and promote the recovery of stalled replication through the translesion synthesis mechanism (15). Our studies revealed that PIDD is also necessary to bridge the interaction between DNA-PKcs and ATR upon replication stress (Figure 5C), and may facilitate the ATR signaling pathway (Figure 5F). Disrupting the binding of DNA-PKcs to PIDD or depleting the expression of PIDD would result in attenuation of ATR-dependent DNA-PKcs phosphorylation at the Thr2609 cluster region (6) (Figures 3A, 5B and E), Chk1 phosphorylation (Figures 3C and D, 5F) and the intra-S-phase checkpoint regulation (Figures 3E and 7C). In contrast, the depletion of Ku80 did not affect the interaction between DNA-PKcs and ATR upon UV irradiation or ATR-dependent DNA-PKcs phosphorylation (Figure 5), suggesting that Ku is dispensable for the initial recruitment and regulation of DNA-PKcs upon stalled replication forks. Our study thus provides new insight that DNA-PKcs is recruited by PIDD to stalled replication forks and participates directly in the ATR signaling pathway.
The ATR pathway, the major signaling mechanism in the cellular response to replication stress, elicits the intra-S-phase checkpoint and promotes the recovery of stalled replication forks (27,28). Upon replication stress, the uncoupling of the MCM helicase complex and replication machinery leads to the formation of long stretch single-stranded DNA (ssDNA) (29), which provides the sensing mechanism as the loading of replication protein A (RPA) to ssDNA, recruits the ATR/ATRIP complex, and results in the initial activation of the ATR kinase (30). ATR targets Chk1 phosphorylation at S317 and S345, stimulating the Chk1 kinase and releasing it from chromatin to carry out intra-S-phase checkpoint activities (31,32). In addition to the initial recruitment and activation of ATR through RPA-coated ssDNA, the full-blown activation of ATR kinase requires the addition of cofactors, including TopBP1 and the recently identified ETAA1 (33–35). TopBP1 and ETAA1 possess a similar ATR-activation domain for binding and directly stimulating the ATR kinase activity, despite exhibiting distinctive regulation on the ATR signaling pathway. While TopBP1 is recruited to stalled replication forks through the Rad9/Hus1/Rad1 (9-1-1) complex and is required for Chk1 phosphorylation (33), ETAA1 is recruited through direct binding to the RPA complex and is dispensable for Chk1 phosphorylation (34,35). Our results demonstrated that Chk1 phosphorylation was attenuated in cells expressing the mPL DNA-PKcs mutant or depleted with PIDD preferentially at later time points (Figures 3C and D, 5F; Supplementary Figure S5A), suggesting that DNA-PKcs association with PIDD could facilitate the second phase of ATR activation through TopBP1. This is in agreement with our analysis in that DNA-PKcs is able to interact with the 9–1-1 complex (YFL and BPC unpublished result).
We observed that UV-induced RPA2 hyperphosphorylation was significantly attenuated in PIDD depleted cells (Supplementary Figure S5B). RPA2 hyperphosphorylation at its N’ terminal region is known to play a critical role in replication stress response (36–38), and is involved with multiple kinases including ATR, DNA-PKcs, ATM and CDK ((9) and references therein). Evidence suggests that, upon genotoxic stress, phosphorylation of Ser33 by ATR facilitates subsequent phosphorylations at Thr21, Ser12, Ser4 and Ser8 by DNA-PKcs and ATM (9). Although PIDD depletion or DNA-PKcs PL mutation did not alter the association between ATR and RPA in PLA analysis (Figure 5D), our results demonstrated that ATR downstream signaling as well as DNA-PKcs recruitment to stalled replication forks were attenuated under these circumstances. Consequently, RPA2 hyperphosphorylation (slower migrating species) and ATR-dependent phosphorylation at Ser33 were attenuated in PIDD depleted cells (Supplementary Figure S5B). It is likely that the phenotypic defects associated with PIDD depletion or DNA-PKcs PL mutation are in part due to the diminished RPA2 phosphorylation.
We demonstrated that the PL motif at the N’ terminal of DNA-PKcs is required for the protein–protein interaction between DNA-PKcs and PIDD, since the mPL mutant DNA-PKcs harboring alanine substitutions at the PL motif was unable to interact with PIDD (Figure 4B). We also observed a significant reduction of DNA-PKcs phosphorylation within the Thr2609 cluster in V3-mPL cells and PIDD-depleted cells, although the signaling was still detectable. DNA-PKcs may still be recruited to stalled replication forks without PIDD association. For example, DNA-PKcs could interact directly with the RPA complex (10) and is required for RPA2 phosphorylation (8–11). In parallel with the siPIDD approach, we generated PIDD knockout clones in 293FT cells using the CRISPR-Cas9 strategy (39,40) (Supplementary Figure S7A). We observed that UV stimulation of Chk1 phosphorylation was attenuated in PIDD knockout 293FT cells (Supplementary Figure S7B). However, we observed differential regulation of DNA-PKcs phosphorylation within the Thr2609 cluster. In PIDD knockout 293FT cells, phosphorylation at Thr2647 but not at Thr2609 was significantly attenuated (Supplementary Figure S7C), although this inhibitory effect decreased gradually in later passages of PIDD knockout cells (data not shown). Without PIDD, a complementary mechanism may be responsible for restoring ATR-dependent DNA-PKcs phosphorylation upon replication stress. This also implies that the association between DNA-PKcs and the ATR pathway is critical for the cellular response against DNA replication errors during normal cell proliferation and survival against replication stress.
We previously proposed a two-step mechanism regarding the role of DNA-PKcs during the initial replication stress response before fork collapse, and its subsequent role when DSB forms (41). The current study further supports the model that PIDD recruits DNA-PKcs upon initial stalled replication forks and mediates ATR-dependent DNA-PKcs phosphorylation. The presence of the DNA-PKcs protein and/or its phosphorylation will facilitate the ATR-Chk1 signaling pathway to elicit the intra-S-phase checkpoint and promote recovery of stalled replication (27,28). This notion is supported by our studies where the intra-S-phase checkpoint was compromised in cells expressing the mPL mutant DNA-PKcs or depleted with PIDD expression (Figures 3E and 7C). Additionally, our previous studies using the CHO V3 cell model (6) and the DNA-PKcs3A/3A mouse model (5) showed that DNA-PKcs phosphorylation at the Thr2609 cluster is required for cellular resistance against UV irradiation and other replication stress agents. Our analyses also revealed that the Ku heterodimer is dispensable for the recruitment and phosphorylation of DNA-PKcs at stalled replication forks (Figure 6). This is consistent with a previous report showing that the levels of the Ku heterodimer on nascent DNA were unchanged for 2 h after HU addition (42). When stalled replication forks cannot be resolved, a delay of DSB formation occurs through structure-selective endonucleases such as MUS81-EME2 (43) or EEPD1 (44) to facilitate replication fork restart. Although Ku may be dispensable during the initial response to stalled replication forks, upon replication associated DSBs, Ku-dependent DNA-PK kinase activation and subsequent Ku phosphorylation could modulate the affinity of Ku to the DSB ends to dictate the DSB repair pathway choice at stalled replication forks (45).
PIDD has been characterized in both pro-apoptosis and pro-survival regulations in the cellular response to genotoxic stress (12). PIDD is known to mediate the apoptotic response through assembling the PIDDosome complex containing Caspase-2 and RAIDD (receptor-interacting protein-associated ICE-1/CED-3 homologous protein with a death domain upon DNA damage (13). On the contrary, PIDD could promote the pro-survival NF-κB signaling pathway through the association with NEMO (NF-κB essential modulator) and RIP1 (receptor-interacting protein kinase 1) (14). The switch role of PIDD may depend on ATM-mediated PIDD phosphorylation at its death domain, which dictates the association of PIDD with Caspase-2/RAIDD for cell death or with NEMO/RIP1 for cell survival (46). PIDD also participates directly in DNA damage repair. PIDD binds to PCNA upon UV irradiation through its ZU-5 (ZO-1 and Unc5-like) domains and stimulates PCNA monoubiquitination during translesion DNA synthesis (15). Since DNA-PKcs could also interact directly with PCNA (Supplementary Figure S3A), it is likely that multiple protein–protein interactions among PCNA, PIDD and DNA-PKcs will stabilize DNA-PKcs recruitment at stalled replication forks to facilitate the ATR signaling pathway for the intra-S checkpoint and fork recovery.
In summary, we report here that PIDD, but not the Ku heterodimer, is required to recruit DNA-PKcs and bridge its interaction with ATR at stalled replication forks. Consequently, PIDD facilitates ATR-dependent DNA-PKcs phosphorylation and the ATR signaling pathway to promote the intra-S checkpoint. Taken together, our results demonstrate that the interaction between DNA-PKcs and PIDD is an integral part of the ATR signaling network and is required for cellular resistance to replication stress.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Eric Hendrickson for kindly providing the HCT116 Ku86Flox/− cells, and Dr Damiana Chiavolini and Emily Miller for editing the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [CA166677 to B.P.C., CA158323 to C.D.]; Cancer Prevention Research Institute of Texas [RP160268 to B.P.C.]; Ministry of Science and Technology Fellowship [104–2911-I-002-578 to C.T.K.]; China Scholarship Council, P.R. China Fellowship [201603170218 to J.G.]. Funding for open access charge: National Institutes of Health [CA166677].
Conflict of interest statement. None declared.
REFERENCES
- 1. Davis A.J., Chen B.P., Chen D.J.. DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair. 2014; 17:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lovejoy C.A., Cortez D.. Common mechanisms of PIKK regulation. DNA Repair (Amst). 2009; 8:1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan D.W., Chen B.P., Prithivirajsingh S., Kurimasa A., Story M.D., Qin J., Chen D.J.. Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 2002; 16:2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen B.P., Uematsu N., Kobayashi J., Lerenthal Y., Krempler A., Yajima H., Lobrich M., Shiloh Y., Chen D.J.. Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 2007; 282:6582–6587. [DOI] [PubMed] [Google Scholar]
- 5. Zhang S., Yajima H., Huynh H., Zheng J., Callen E., Chen H.T., Wong N., Bunting S., Lin Y.F., Li M. et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J. Cell Biol. 2011; 193:295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yajima H., Lee K.J., Chen B.P.. ATR-dependent phosphorylation of DNA-dependent protein kinase catalytic subunit in response to UV-induced replication stress. Mol. Cell. Biol. 2006; 26:7520–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin Y.F., Shih H.Y., Shang Z., Matsunaga S., Chen B.P.. DNA-PKcs is required to maintain stability of Chk1 and Claspin for optimal replication stress response. Nucleic Acids Res. 2014; 42:4463–4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cruet-Hennequart S., Coyne S., Glynn M.T., Oakley G.G., Carty M.P.. UV-induced RPA phosphorylation is increased in the absence of DNA polymerase eta and requires DNA-PK. DNA Repair (Amst). 2006; 5:491–504. [DOI] [PubMed] [Google Scholar]
- 9. Liu S., Opiyo S.O., Manthey K., Glanzer J.G., Ashley A.K., Amerin C., Troksa K., Shrivastav M., Nickoloff J.A., Oakley G.G.. Distinct roles for DNA-PK, ATM and ATR in RPA phosphorylation and checkpoint activation in response to replication stress. Nucleic Acids Res. 2012; 40:10780–10794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shao R.G., Cao C.X., Zhang H., Kohn K.W., Wold M.S., Pommier Y.. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 1999; 18:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liaw H., Lee D., Myung K.. DNA-PK-dependent RPA2 hyperphosphorylation facilitates DNA repair and suppresses sister chromatid exchange. PLoS One. 2011; 6:e21424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janssens S., Tinel A.. The PIDDosome, DNA-damage-induced apoptosis and beyond. Cell Death Differ. 2012; 19:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tinel A., Tschopp J.. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004; 304:843–846. [DOI] [PubMed] [Google Scholar]
- 14. Janssens S., Tinel A., Lippens S., Tschopp J.. PIDD mediates NF-kappaB activation in response to DNA damage. Cell. 2005; 123:1079–1092. [DOI] [PubMed] [Google Scholar]
- 15. Logette E., Schuepbach-Mallepell S., Eckert M.J., Leo X.H., Jaccard B., Manzl C., Tardivel A., Villunger A., Quadroni M., Gaide O. et al. PIDD orchestrates translesion DNA synthesis in response to UV irradiation. Cell Death Differ. 2011; 18:1036–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y., Ghosh G., Hendrickson E.A.. Ku86 represses lethal telomere deletion events in human somatic cells. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:12430–12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen B.P., Chan D.W., Kobayashi J., Burma S., Asaithamby A., Morotomi-Yano K., Botvinick E., Qin J., Chen D.J.. Cell cycle dependence of DNA-dependent protein kinase phosphorylation in response to DNA double strand breaks. J. Biol. Chem. 2005; 280:14709–14715. [DOI] [PubMed] [Google Scholar]
- 18. Mendez J., Stillman B.. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000; 20:8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Merrick C.J., Jackson D., Diffley J.F.. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 2004; 279:20067–20075. [DOI] [PubMed] [Google Scholar]
- 20. Boehm E.M., Washington M.T.. R.I.P. to the PIP: PCNA-binding motif no longer considered specific: PIP motifs and other related sequences are not distinct entities and can bind multiple proteins involved in genome maintenance. Bioessays. 2016; 38:1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seiler J.A., Conti C., Syed A., Aladjem M.I., Pommier Y.. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell. Biol. 2007; 27:5806–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pick R., Badura S., Bosser S., Zornig M.. Upon intracellular processing, the C-terminal death domain-containing fragment of the p53-inducible PIDD/LRDD protein translocates to the nucleoli and interacts with nucleolin. Biochem. Biophys. Res. Commun. 2006; 349:1329–1338. [DOI] [PubMed] [Google Scholar]
- 23. Telliez J.B., Bean K.M., Lin L.L.. LRDD, a novel leucine rich repeat and death domain containing protein. Biochim. Biophys. Acta. 2000; 1478:280–288. [DOI] [PubMed] [Google Scholar]
- 24. Tinel A., Janssens S., Lippens S., Cuenin S., Logette E., Jaccard B., Quadroni M., Tschopp J.. Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-kappaB pathway. EMBO J. 2007; 26:197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu S., Shiotani B., Lahiri M., Marechal A., Tse A., Leung C.C., Glover J.N., Yang X.H., Zou L.. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol. Cell. 2011; 43:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nam E.A., Zhao R., Glick G.G., Bansbach C.E., Friedman D.B., Cortez D.. Thr-1989 phosphorylation is a marker of active ataxia telangiectasia-mutated and Rad3-related (ATR) kinase. J. Biol. Chem. 2011; 286:28707–28714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cortez D. Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst). 2015; 32:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Byun T.S., Pacek M., Yee M.C., Walter J.C., Cimprich K.A.. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005; 19:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 31. Smits V.A., Reaper P.M., Jackson S.P.. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr. Biol. 2006; 16:150–159. [DOI] [PubMed] [Google Scholar]
- 32. Zhao H., Piwnica-Worms H.. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 2001; 21:4129–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Delacroix S., Wagner J.M., Kobayashi M., Yamamoto K., Karnitz L.M.. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007; 21:1472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. torBass T.E., Luzwick J.W., Kavanaugh G., Carroll C., Dungrawala H., Glick G.G., Feldkamp M.D., Putney R., Chazin W.J., Cortez D.. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 2016; 18:1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haahr P., Hoffmann S., Tollenaere M.A., Ho T., Toledo L.I., Mann M., Bekker-Jensen S., Raschle M., Mailand N.. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 2016; 18:1196–1207. [DOI] [PubMed] [Google Scholar]
- 36. Lindsey-Boltz L.A., Reardon J.T., Wold M.S., Sancar A.. In vitro analysis of the role of replication protein A (RPA) and RPA phosphorylation in ATR-mediated checkpoint signaling. J. Biol. Chem. 2012; 287:36123–36131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Murphy A.K., Fitzgerald M., Ro T., Kim J.H., Rabinowitsch A.I., Chowdhury D., Schildkraut C.L., Borowiec J.A.. Phosphorylated RPA recruits PALB2 to stalled DNA replication forks to facilitate fork recovery. J. Cell Biol. 2014; 206:493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oakley G.G., Tillison K., Opiyo S.A., Glanzer J.G., Horn J.M., Patrick S.M.. Physical interaction between replication protein A (RPA) and MRN: involvement of RPA2 phosphorylation and the N-terminus of RPA1. Biochemistry. 2009; 48:7473–7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R.. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013; 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang H., Wang H., Jaenisch R.. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 2014; 9:1956–1968. [DOI] [PubMed] [Google Scholar]
- 41. Yajima H., Lee K.J., Zhang S., Kobayashi J., Chen B.P.. DNA double-strand break formation upon UV-induced replication stress activates ATM and DNA-PKcs kinases. J. Mol. Biol. 2009; 385:800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sirbu B.M., Couch F.B., Feigerle J.T., Bhaskara S., Hiebert S.W., Cortez D.. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011; 25:1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pepe A., West S.C.. MUS81-EME2 promotes replication fork restart. Cell Rep. 2014; 7:1048–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu Y., Lee S.H., Williamson E.A., Reinert B.L., Cho J.H., Xia F., Jaiswal A.S., Srinivasan G., Patel B., Brantley A. et al. EEPD1 rescues stressed replication forks and maintains genome stability by promoting end resection and homologous recombination repair. PLoS Genet. 2015; 11:e1005675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee K.J., Saha J., Sun J., Fattah K.R., Wang S.C., Jakob B., Chi L., Wang S.Y., Taucher-Scholz G., Davis A.J. et al. Phosphorylation of Ku dictates DNA double-strand break (DSB) repair pathway choice in S phase. Nucleic Acids Res. 2016; 44:1732–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ando K., Kernan J.L., Liu P.H., Sanda T., Logette E., Tschopp J., Look A.T., Wang J., Bouchier-Hayes L., Sidi S.. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol. Cell. 2012; 47:681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.