Abstract
Non-coding RNAs play a vital role in diverse cellular processes. Pseudogenes, which are non-coding homologs of protein-coding genes, were once considered non-functional evolutional relics. However, recent studies have shown that pseudogene transcripts can regulate their parental transcripts by sequestering shared microRNAs (miRNAs), thus acting as competing endogenous RNAs (ceRNAs). In this study, we utilize an unbiased screen to identify the ferritin heavy chain 1 (FTH1) transcript and multiple FTH1 pseudogenes as targets of several oncogenic miRNAs in prostate cancer (PCa). We characterize the critical role of this FTH1 gene:pseudogene:miRNA network in regulating tumorigenesis in PCa, whereby oncogenic miRNAs downregulate the expression of FTH1 and its pseudogenes to drive oncogenesis. We further show that impairing miRNA binding and subsequent ceRNA crosstalk completely rescues the slow growth phenotype in vitro and in vivo. Our results also demonstrate the reciprocal regulation between the pseudogenes and intracellular iron levels, which are crucial for multiple physiological and pathophysiological processes. In summary, we describe an extensive gene:pseudogene network comprising multiple miRNAs and multiple pseudogenes derived from a single parental gene. The network could be regulated through multiple mechanisms to modulate iron storage in various signaling pathways, the deregulation of which results in PCa development and progression.
INTRODUCTION
Of the 70–90% of the transcribed mammalian genome, only a minute 1–2% code for proteins (1,2). The remainder of the transcriptome comprises non-coding RNAs (ncRNAs), consisting of diverse classes of RNA species, such as long ncRNAs (lncRNAs), pseudogenes, circular RNAs (circRNAs) and short ncRNAs, such as microRNAs (miRNAs), snoRNAs, tRNAs, rRNAs and other emerging variants. To date, many studies have demonstrated the coding-independent functions of ncRNAs in various important biological processes, such as the regulation of development, differentiation and proliferation (3–7). As such, the dysregulation of the non-coding transcriptome is frequently observed in disease development, especially cancer.
MiRNAs represent one of the most widely studied ncRNA species. They post-transcriptionally regulate gene expression by binding to specific recognition sites known as miRNA response elements (MREs) on target transcripts (8). Studies have shown that the functions of miRNAs are often context specific (9,10). MiR-638 is exemplary as it has been shown to exhibit both tumor-suppressive and oncogenic properties that are cancer type- or subtype-dependent. Reports have highlighted its contrasting roles in triple-negative breast cancer (TNBC) compared to non-TNBC (11–13). Furthermore, it could play dual roles in colorectal carcinoma, by targeting different genes, such as tumor suppressors PTEN and p53 or oncogenes SOX2 and TSPAN1 (14–16), which could be differentially expressed in a subtype- or cell line-specific manner.
Recent studies have described an additional layer of post-transcriptional regulation whereby multiple RNA transcripts that contain MREs for common miRNAs could co-regulate one another (17). These transcripts act as competing endogenous RNAs (ceRNAs) as they compete for and sequester shared miRNAs. Apart from mRNAs, a few ncRNA classes, including lncRNAs, pseudogenes and circRNAs, have also been shown to participate in ceRNA regulation (18).
Pseudogenes are a class of ncRNAs previously presumed to be ‘junk’ DNA due to their multiple mutations and the lack of coding potential and function (19). However, human transcriptomic and proteomic studies have evidenced the presence of transcripts and proteins derived from pseudogenes (2,20). More importantly, pseudogene transcripts have recently been shown to sponge miRNAs from their ancestral genes and thus possess ceRNA activity. Poliseno et al. first uncovered the non-coding functions of the pseudogene of the tumor suppressor PTEN, PTENP1 (21). The authors demonstrated the ability of PTENP1 to sponge PTEN-targeting miRNAs and regulate cellular PTEN expression to exert a tumor-suppressive effect. Similarly, the pseudogene of oncogenic BRAF, BRAFP1, has been shown to act as a ceRNA that regulates BRAF expression, thus affecting MAPK signaling and proliferation, and driving tumorigenesis in vivo (22). To date, only a handful of studies have identified other pseudogenes with miRNA-sponging potential, such as TUSC2P, INST6P1, OCT4-pg4, OCT4-pg5 and CYP4Z2P, all of which were selected for studies solely based on their parental gene (23–28).
In this study, we used a miRNA pulldown to identify novel and context-specific targets for miR-638 in prostate cancer (PCa). Intriguingly, the top targets include FTH1 and several of its pseudogenes, which form a gene:pseudogene network. FTH1, a key subunit of the ferritin ‘nanocage’ that stores iron in its non-toxic ferric form, plays a critical role in the maintenance of iron homeostasis in cells to prevent harmful effects caused by iron overload (29,30). Apart from miR-638, we further validated other oncogenic miRNAs that target and downregulate FTH1 and its pseudogenes. In contrast, overexpression of the pseudogenes simultaneously inhibits growth and downregulates cellular iron. Furthermore, we perform mutagenesis experiments to demonstrate that impairing miRNA binding to the pseudogenes results in rescue in growth phenotype in vitro and in vivo. To summarize, we establish and characterize an extensive FTH1 gene:pseudogene:miRNA network, encompassing many miRNAs and multiple pseudogenes derived from a single parental gene, which is a critical regulator of iron homeostasis and tumorigenesis in PCa.
MATERIALS AND METHODS
Reagents
Reagents are as follows: anti-HSP90 antibody (Santa Cruz); anti-GAPDH antibody (Santa Cruz); anti-FTH1 antibody (Cell Signaling); miRIDIAN miRNA mimics for non-targeting control (NC) and miRNAs (638, 19b-3p, 19b-1-5p, 181a-5p, 210–3p, 362–5p and 616–3p), miRIDIAN miRNA inhibitors for NC (NC-AS) and miR-638 (638-AS), biotinylated miRNA reagents (NC1, NC2 and 638), siGENOME siRNA reagents for NC (siNC) and FTH1 (siFTH), Dharmafect 1 (Dharmacon); phosphorothioate modified antisense oligonucleotides (ASO) (IDT); Trizol LS, Lipofectamine 3000, Dulbecco’s modified Eagle medium (DMEM), Iscove’s modified Dulbecco’s medium (IMDM), OptiMem reduced serum media, fetal bovine serum (FBS), Dynabeads M-280 Streptavidin (Life Technologies); psiCHECK-2 vector (Promega); pcDNA4-Puro vector (Addgene); normal and PCa tissue RNA samples (Origene).
Plasmids and mutagenesis
Oligonucleotides for specific MREs were annealed by heating at 95°C for 5 min and cooled gradually to room temperature. MREs were cloned into psiCHECK-2 vector using the XhoI and NotI restriction sites (Supplementary Tables S1 and 2). Full length FTH1 and its pseudogenes were amplified by polymerase chain reaction (PCR) and cloned by restriction digest and ligation via the NotI and XhoI restriction sites into psiCHECK-2, and the HindIII and BamHI sites into pcDNA4-Puro vector. Specific MREs on the full-length constructs from above were mutated using the Quickchange Lightning Multi Site-Directed Mutagenesis Kit (Agilent) following the manufacturer's instructions (Supplementary Table S3).
Cell culture and transfection
DU145 and PC3 cells were grown in DMEM supplemented with 10% FBS, penicillin/streptomycin and glutamine. Hap1 FTH1 knockout cells were grown in IMDM supplemented with 10% FBS and penicillin/streptomycin. All cells were grown at 37°C in a humidified atmosphere with 5% CO2. For the transfection of miRNA mimics, inhibitors and siRNAs, cells were transfected with 50 nM of each reagent per well using Dharmafect 1 in 12-well dishes as per the manufacturer's instructions. A seeding density of 150 000 per well was used for DU145 and 200 000 cells for PC3. For plasmid and ASO transfections, DU145 and PC3 cells were seeded in 12-well dishes at a density of 100 000 and 140 000 cells per well respectively 24 h before transfection. Lipofectamine 3000 was used to transfect 500 ng of plasmids or 100 nM of ASO per well following the manufacturer’s protocol.
Cell proliferation
At 24 h post-transfection DU145 and PC3 cells were trypsinized, resuspended and seeded equally in five separate 12-well plates. Six hours after seeding (Day 0), one plate was washed once in phosphate-buffered saline (PBS), fixed in 10% formalin solution for 10 min at room temperature, washed again with PBS and kept in PBS at 4°C. Subsequently one plate per day was washed, fixed and stored as described above. On the last day, the wells were stained with crystal violet, solubilized with 10% acetic acid and optical density measured at 595 nm.
Soft agar assay
A 0.6% agarose base was prepared in 6-well dishes. At 24 h post-transfection, DU145 and PC3 cells were trypsinized, resuspended and counted. Seeding densities of 25 000 cells and 20 000 cells per well were used for DU145 and PC3 respectively. The cells were mixed with growth medium and agarose to a final agarose concentration of 0.3%, which was added above the base. The cells were grown at 37°C in a humidified atmosphere with 5% CO2 and were fed with growth medium every two days. After 10–14 days, the colonies were imaged under 4× magnification and quantified using ImageJ.
Luciferase reporter assay
DU145 and PC3 cells were seeded at a density of 30 000 and 50 000 cells per well, respectively, in 24-well dishes. After 24 h, Lipofectamine 3000 was used to co-transfect 50 nM miRNA mimic and 10 ng psiCHECK-2 plasmids per well per the manufacturer’s instructions. At 48 h post-transfection, cells were washed in PBS and lysed, and luminescence was measured following instructions of the dual luciferase reporter assay kit (Promega).
Iron assay and treatments
DU145 and PC3 cells transfected with pcDNA4-Puro overexpression plasmids were harvested 48 h post-transfection. The Iron Assay Kit (Metallogenics) utilizing the Ferrozine chromogenic method was used to measure cellular iron content according to the manufacturer’s protocol. For treatments, iron chloride and chelator, 1,10-Phenanthroline (Sigma) were prepared as described previously (31). Cells were seeded and treated in 96-well plates and assayed 24 h post-treatment using CellTiter-Glo Luminescent Cell Viability Assay (Promega) per the manufacturer’s instructions.
Biotinylated miRNA pulldown
Pulldown experiments in DU145 and PC3 cells with control miRNAs NC1 and NC2, and miR-638 mimic (Dharmacon), were conducted as described previously (32). RNA extracted from streptavidin beads using Trizol LS was further purified using phenol: chloroform:isoamyl (25:24:1) and chloroform:isoamyl (24:1) (Sigma).
RNA extraction, real-time quantitative PCR and microarray
For real-time PCR (RT-PCR) analyses, total RNA was extracted using Trizol reagent followed by column purification using the PureLink RNA Mini Kit (Ambion). cDNA was generated using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). MiRNA reverse transcription was performed using the TaqMan miRNA Reverse Transcription Kit following the manufacturer’s instructions (Applied Biosystems). RT-PCR was subsequently performed using the 7900HT Fast or QuantStudio 5 RT-PCR System (Applied Biosystems). For microarray, the TargetAmp-NanoLabeling Kit (Epicentre) was used to generate cRNA from purified total RNA. cRNAs were hybridized to Human HT-12 v4 Expression BeadChips (Illumina) per the manufacturer’s protocols for microarray analyses. Raw expression data were obtained from the GenomeStudio software with the subtraction of the background. Prior to the identification of differentially expressed genes, raw data were normalized based on the cross-correlation method (33). Significantly changed genes were identified based on the average fold change cutoff of 1.5 and the cutoff of the FDR-corrected P-value across all replicates at 0.05. Genes selected as negative controls were based on the average fold change cutoff range of 0.85–1.15.
Protein extraction and western blot analysis
Cells were harvested by trypsinization. Cell pellets were washed in chilled PBS and lysed on ice for 15 min with Protein Lysis Buffer [10 mM Hepes pH 7, 0.1 M KCl, 5 mM MgCl2, 25 mM EDTA pH 8, 0.5% (v/v) NP-40, 20 μM DTT, supplemented with Proteinase Inhibitor (Roche)]. Lysates were precleared by centrifugation at 16 000 × g for 15 min at 4°C and protein concentrations were measured using Bradford Protein Assay (Bio-Rad). For western blot analysis, sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to size fractionate 10–15 μg of proteins in 4–12% Bis-Tris acrylamide NuPAGE gels in MOPS SDS running buffer (Invitrogen) and transferred to PVDF membranes (Thermo Scientific) in transfer buffer [25 mM Tris, 192 mM glycine, 20% (v/v) methanol (pH 8.3)]. The membranes were probed with specific primary antibodies.
Xenograft
DU145 and PC3 cells transfected with miRNA mimics, siRNA or expression plasmids were harvested 48 h post-transfection. The cells were trypsinized, washed in PBS and counted. The appropriate number of cells were pelleted, resuspended in culture medium and mixed with Matrigel matrix (Corning) in a 1:1 ratio. The cell suspension was subcutaneously injected into nude mice and tumor sizes were measured every three days. The mice were sacrificed after 15–21 days.
MRE prediction and sequence alignment
MRE predictions were performed using RNA22 which is available at https://cm.jefferson.edu/rna22 (34). Multiple sequence alignments of FTH1 and its pseudogenes were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Low- and high-homology regions between each pseudogene and FTH1 were determined arbitrarily, whereby regions with greater than 90% sequence identity were defined as ‘high homology’ and lower than 20% ‘low homology’.
Statistical analysis
Unpaired Student’s t-test was used to analyze all in vitro data derived from three or more independent experiments. Values calculated from multiple independent experiments are presented as mean ± SEM, whilst data shown from a representative experiment are presented as mean ± SD. Values of P < 0.05 were considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001.
RESULTS
MiR-638 possesses oncogenic properties in prostate cancer
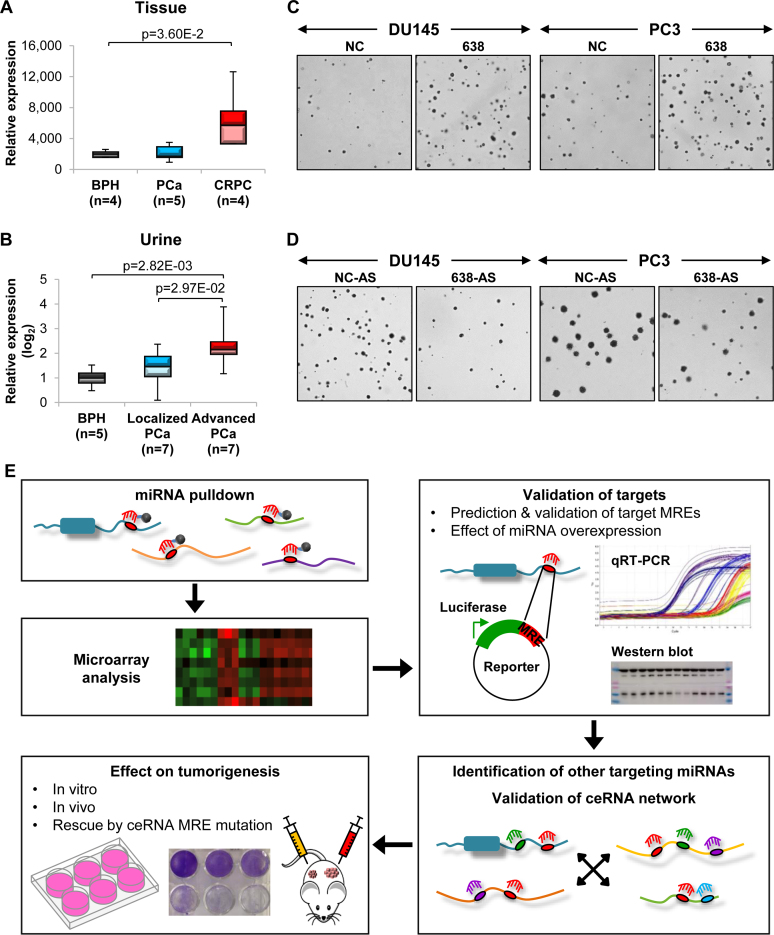
We first examined the expression profile of miR-638 in PCa. Using publicly available datasets from the ArrayExpress and Gene Expression Omnibus (GEO) databases, we found that miR-638 is upregulated in PCa compared to normal tissues (Supplementary Figure S1A), and even more significantly so in castration-resistant prostate cancer (CRPC) (Figure 1A). Interestingly, miR-638 expression is also increased in urine samples of patients with advanced PCa (Figure 1B), suggesting that miR-638 could be a potential biomarker and non-invasive diagnostic tool for more aggressive and later stage PCa.
Figure 1.
miR-638 possesses oncogenic properties in PCa. (A) ArrayExpress (E-MEXP-2319) data analysis of miR-638 expression in CRPC and PCa compared to benign prostate hyperplasia (BPH). (B) GEO data (GSE54010) analysis of miR-638 expression in patient urine samples of localized and advanced PCa compared to BPH. (C and D) Effect of miR-638 overexpression (C) and inhibition (D) on anchorage-independent growth in vitro. (See Supplementary Figure S1C and F for quantification). (E) Schematic outlining the integrated computational analysis and experimental validation strategies to establish the ceRNA network involved in PCa development. NC and NC-AS denote NC mimic and NC antisense mimic respectively. (C and E) Mean ± SD; n ≥ 3. *P < 0.05; **P < 0.01; ***P < 0.001.
In agreement with the expression data that point toward an oncogenic role for miR-638 in PCa, overexpression of miR-638 enhanced cell proliferation (Supplementary Figure S1B) and anchorage-independent growth in soft agar (Figure 1C and Supplementary Figure S1C) in two PCa cell lines, DU145 and PC3. Similar effects were observed in xenograft tumors overexpressing miR-638 (Supplementary Figure S1D). In contrast, inhibition of miR-638 significantly reduced cell growth (Figure 1D; Supplementary Figure S1E and F). These observations support the oncogenic potential of miR-638 in PCa. Critically, PC3 cells are null for PTEN and p53, two key tumor-suppressor genes previously shown to be miR-638 targets (14,35,36). This suggests that miR-638 could exert its oncogenic effects through the regulation of other target transcripts. With this in mind, we designed a multidimensional scheme to identify and characterize a post-transcriptional regulatory network, centered around miR-638 and other potentially oncogenic miRNAs, that could be involved in PCa development (Figure 1E).
The FTH1 gene:pseudogene network is a potential miR-638 target
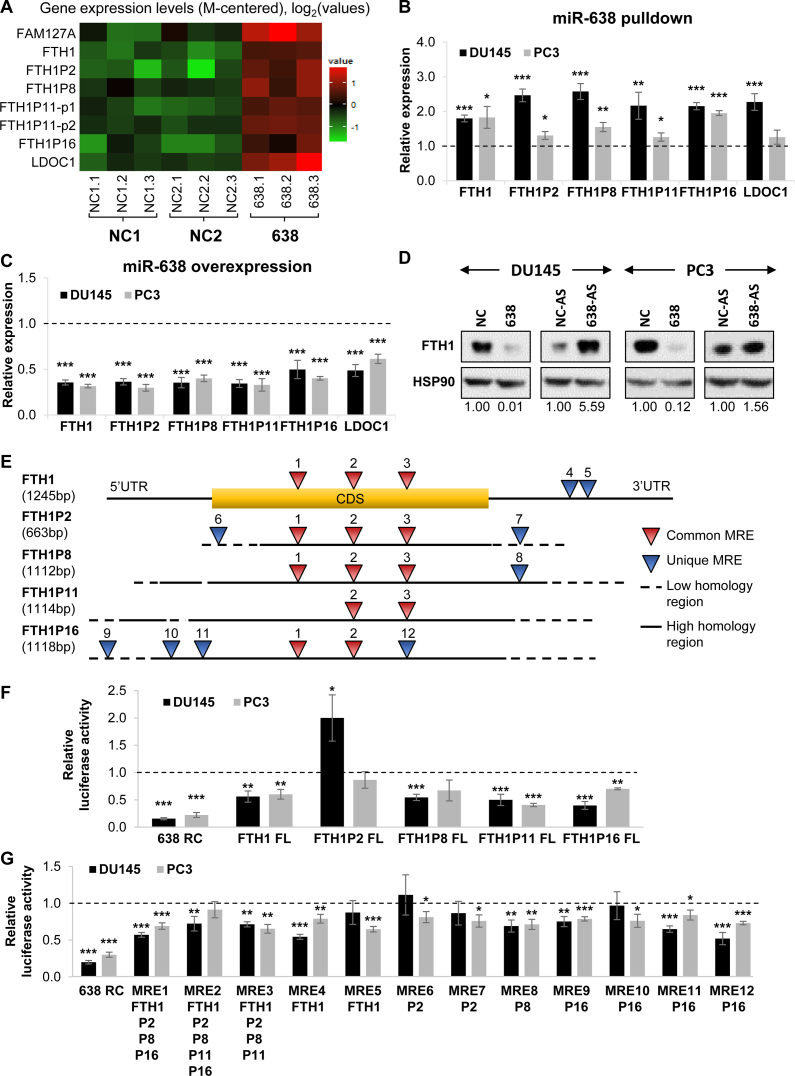
Next, we sought to identify targets of miR-638 which may be involved in PCa tumorigenesis. Whilst algorithm-derived predictions of new miRNA targets are robust, they do not account for cell line- and tissue-dependent differential gene expression. To identify context-specific targets, we performed biotinylated miRNA pulldown coupled with microarray analysis (32). Transcripts that were significantly enriched in the pulldown and downregulated upon miR-638 overexpression were selected as potential candidates for further experimental validation.
The shortlisted candidates were FAM127A, FTH1, FTH1P2, FTH1P8, FTH1P11, FTH1P16 and LDOC1 (Figure 2A). Intriguingly, the candidates from the unbiased pulldown and microarray analysis consist mainly of FTH1 and several of its pseudogenes (FTH1P2, FTH1P8, FTH1P11 and FTH1P16). There are currently 21 annotated FTH1 pseudogenes, however, only four were significantly enriched against both controls in the pulldown. These targets showed similar enrichment in a further validation using qRT-PCR in DU145 and PC3 cells (Figure 2B). We also validated the downregulation of the candidate genes at the transcript level when miR-638 was overexpressed (Figure 2C). Additionally, miR-638 overexpression reduced FTH1 protein expression, whereas miR-638 inhibition had the opposite effect (Figure 2D). To verify the specificity of these targets, we tested and confirmed genes which were not differentially expressed in the miR-638 pulldown and overexpression samples as negative controls (Supplementary Figure S2A and B).
Figure 2.
Identification of the FTH1 gene:pseudogene network as a potential miR-638 target. (A) Microarray analysis of the expression of candidate miR-638 targets in biotinylated miR-638 pulldown compared to NCs (NC1 and NC2) in DU145 cells. (B and C) qRT-PCR validation of the expression of miR-638 target candidates in biotinylated miR-638 pulldown (B) and miR-638 overexpression samples (C). (D) Effect of miR-638 overexpression and inhibition on FTH1 protein expression. (E) Schematic of the predicted miR-638 MREs on FTH1 and its pseudogenes. (F and G) Effect of miR-638 overexpression on the luciferase reporter activity of full-length (FL) constructs (F) and the predicted MREs of FTH1 and its pseudogenes (G). RC denotes reverse complement and acts as a positive control; P2, P8, P11 and P16 denote FTH1P2, FTH1P8, FTH1P11 and FTH1P16, respectively. (B, C, F and G) Mean ± SEM; n ≥ 3. *P < 0.05; **P < 0.01; ***P < 0.001.
As pseudogenes are evolutionary products of their parental genes, their sequences are highly homologous and they could function as competitive molecular sponges for miRNAs targeting the parental gene as illustrated by recent studies on the pseudogenes of cancer-related genes (21,22). To test this hypothesis, we performed miR-638 MRE predictions for FTH1 and its pseudogenes (Supplementary Table S1). We found that a number of these MREs are highly conserved and shared across the parental gene and pseudogenes (Figure 2E). We next generated luciferase reporter constructs for FTH1, its four pseudogenes and the predicted MREs. Overexpression of miR-638 resulted in a consistent and significant decrease in the signal of FTH1, FTH1P11 and FTH1P16 reporters (Figure 2F) and selected MRE reporters, validating at least seven of the 12 predicted MREs (Figure 2G). This also confirmed FTH1 and its selected pseudogenes as miR-638 targets.
FTH1 pseudogenes sequester multiple miRNAs
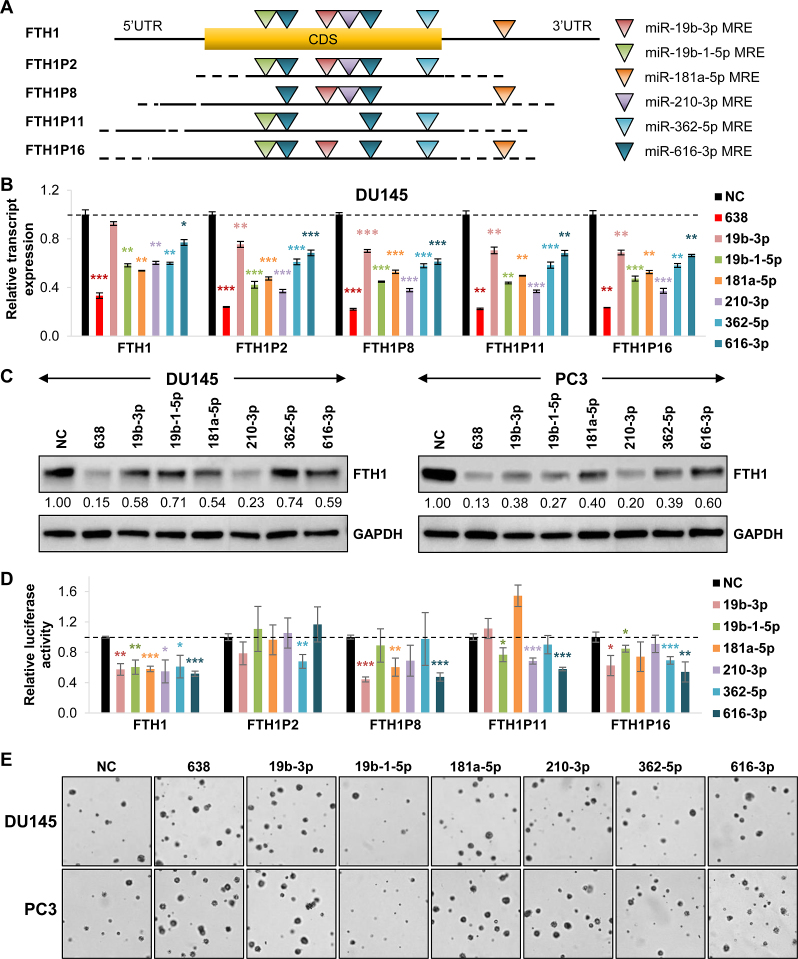
To establish a wider gene:pseudogene network for FTH1 and miR-638, we performed predictions for miRNAs that bind to each of the four shortlisted FTH1 pseudogenes. From the predictions, we selected miRNAs that are upregulated in PCa (Supplementary Figure S3A), hence potentially oncogenic, and targeted two or more of the FTH1 pseudogenes for further studies: miR-19b-3p, 19b-1-5p, 181a-5p, 210-3p, 362-5p and 616-3p (Figure 3A and Supplementary Table S2).
Figure 3.
FTH1 pseudogenes sequester multiple miRNAs. (A) Schematic of the predicted MREs for the selected miRNAs targeting FTH1 and its pseudogenes. CDS denotes coding sequence. (B) Effect of the overexpression of selected miRNAs on the transcript expression of FTH1 and its pseudogenes in DU145 cells. (C) Effect of selected miRNA overexpression on FTH1 protein expression. (See Supplementary Figure S3C for quantification). (D) Effect of selected miRNA overexpression on the luciferase reporter activity of full-length constructs of FTH1 and its pseudogenes in DU145 cells. (E) Effect of the overexpression of selected miRNAs on anchorage-independent growth in vitro. (See Supplementary Figure S3E for quantification). (B and D) Mean ± SEM; n ≥ 3. *P < 0.05; **P < 0.01; ***P < 0.001.
Overexpression of the shortlisted miRNAs downregulated transcript expression of FTH1 and the four pseudogenes (Figure 3B and Supplementary Figure S3B), as well as FTH1 protein expression (Figure 3C and Supplementary Figure S3C). Furthermore, they reduced the luciferase reporter activity of FTH1 and its selected pseudogenes, with FTH1P8, FTH1P11 and FTH1P16 being the most targeted ones (Figure 3D). Notably, all six miRNA candidates had a significant effect on FTH1; whereas, of the five miRNAs predicted to target FTH1P2, only miR-362 significantly reduced its luciferase reporter activity. Although the overexpression of these miRNAs did not exhibit a strong phenotype in anchorage-dependent growth (Supplementary Figure S3D), five of the miRNAs (miR-19b-3p, 181a-5p, 210-3p, 362-5p and 616-3p) consistently increased anchorage-independent growth in soft agar, which is more representative of physiological tumor growth (Figure 3E and Supplementary Figure S3E). These observations validated the oncogenic capacity of the selected miRNAs and their targeting of FTH1 and its pseudogenes, therefore, implicating them in the FTH1 regulatory network and PCa tumorigenesis.
The FTH1 gene:pseudogene network possesses tumor-suppressive properties in prostate cancer
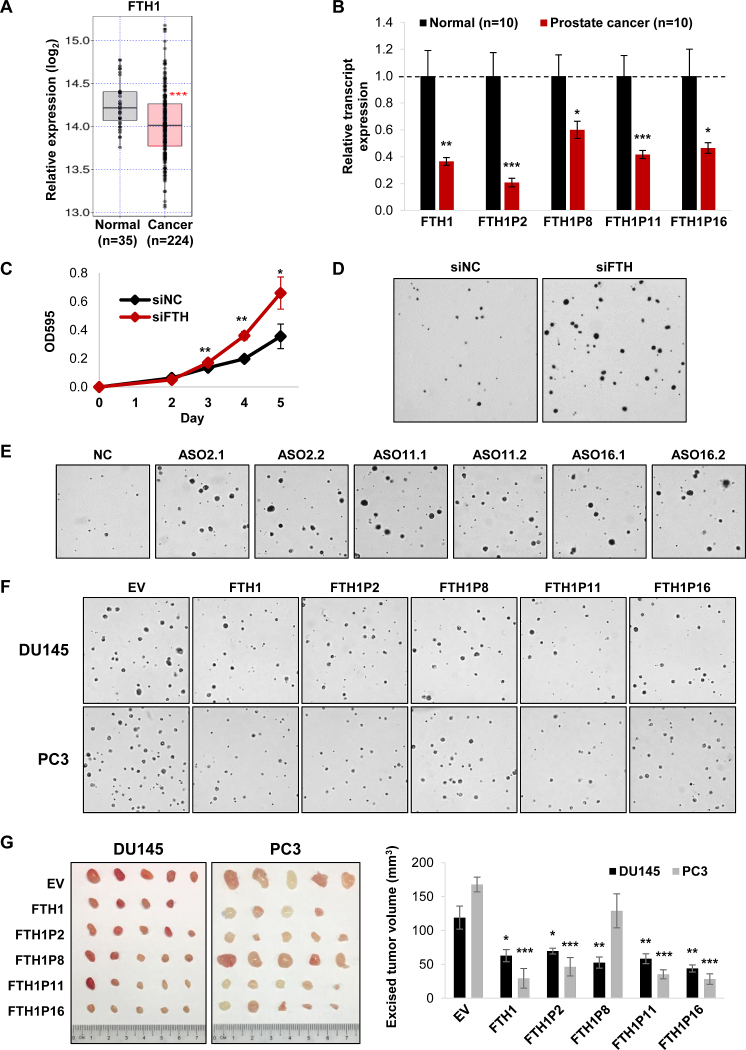
Downregulation of FTH1 has been linked to tumorigenesis in breast cancer and colorectal carcinoma, suggesting a tumor-suppressive role for this critical ferritin subunit that maintains iron balance (37,38). In PCa, FTH1 is significantly downregulated based on TCGA data analysis (Figure 4A). In accordance with this and its potential tumor-suppressive function, we find that FTH1 expression is decreased in PCa tissue samples (Figure 4B). Moreover, its expression level is linked to PCa patient survival, whereby patients with high FTH1 expression have a higher survival rate (Supplementary Figure S4A). Intriguingly, the transcript expression of the FTH1 pseudogenes is also downregulated in PCa tissue samples, suggesting a possible tumor-suppressive role for these pseudogenes (Figure 4B).
Figure 4.
The FTH1 gene:pseudogene network possesses tumor-suppressive properties in PCa. (A) TCGA data analysis of FTH1 expression in PCa compared to normal prostate. (B) qRT-PCR analysis of the expression of FTH1 and its pseudogenes in clinical samples of PCa compared to normal prostate. (C and D) Effect of siRNA-mediated knockdown of FTH1 and its pseudogenes on proliferation (C) and anchorage-independent growth (D) in DU145 cells. (See Supplementary Figure S4E for quantification). (E) Effect of ASO-mediated knockdown of the FTH1 pseudogenes on anchorage-independent growth in DU145 cells. (See Supplementary Figure S5E for quantification). (F and G) Effect of overexpressing FTH1 and its pseudogenes on anchorage-independent growth in vitro (F) and tumor xenograft growth of DU145 and PC3 cells subcutaneously injected into nude mice (G). (See Supplementary Figure S6C for quantification of anchorage-independent growth). EV denotes empty vector control. (B and G) Mean ± SEM; n ≥ 3. (C) Mean ± SD; n ≥ 3. *P < 0.05; **P < 0.01; ***P < 0.001.
To investigate the role of FTH1 and the gene:pseudogene network in PCa, we first knocked down its expression using a siRNA pool (siFTH) (Supplementary Figure S4B), which not only silences FTH1 but also its pseudogenes due to their high sequence homology. A combined knockdown of FTH1 and its pseudogenes resulted in an increase in cell proliferation (Figure 4C and Supplementary Figure S4C), anchorage-independent growth in soft agar (Figure 4D; Supplementary Figure S4D and E) and xenograft tumor growth (Supplementary Figure S4F).
FTH1 pseudogenes possess tumor-suppressive properties in prostate cancer
As the siRNA pool did not selectively target FTH1 or its individual pseudogenes, we next employed ASOs for more specific gene knockdown. To investigate the possibility of non-specificity, we performed pairwise sequence alignments between each ASO and the various FTH transcripts to determine their similarities. These yielded a substantial number of nucleotide mismatches for most pairings (Supplementary Figure S5A). We next validated these ASOs and observed a decrease in the transcript expression of all pseudogenes except FTH1P8 (Supplementary Figure S5B). Although FTH1P8 and ASO16.1 had the smallest number of mismatches, the latter had no effect on FTH1P8 expression, indicating that the phenomenon observed is unlikely to be purely due to off-target effects, instead could be partially driven by ceRNA regulation.
The ASO-mediated downregulation of the pseudogenes also resulted in an increase in proliferation (Supplementary Figure S5C) and anchorage-independent growth in soft agar (Figure 4E; Supplementary Figure S5D and E). To reinforce these findings and the underlying ceRNA regulation, we demonstrated that each ASO pool targeting the same pseudogene significantly reduced the FTH1 protein expression (Supplementary Figure S5F). We also combined the ASOs targeting the two pseudogenes, FTH1P11 and FTH1P16, which have so far shown the most consistent phenotype, and observed similar effects. Therefore, we hypothesized that the FTH1 pseudogenes could play a role in regulating FTH1 expression via a gene:pseudogene network by competing with FTH1 for the same pool of miRNAs.
Since we cannot completely eliminate off-target effects of the ASOs, we next overexpressed each pseudogene separately as a complementary approach to assess their individual capacity as tumor suppressors (Supplementary Figure S6A). Overexpression of FTH1P2, FTH1P11 and FTH1P16 reduced cell proliferation (Supplementary Figure S6B), anchorage-independent growth in soft agar (Figure 4F and Supplementary Figure S6C) and xenograft tumor growth (Figure 4G). FTH1P8 had no effect on anchorage-dependent cell proliferation (Supplementary Figure S6B). However, it significantly reduced growth in soft agar and xenograft tumors of DU145 cells; whereas its effect in PC3 cells was negligible (Figure 4F and Supplementary Figure S6C). Collectively, these findings illustrate the tumor-suppressive properties of the FTH1 pseudogenes which could be mediated in part through ceRNA regulation involving FTH1.
The FTH1 gene:pseudogene network disrupts iron regulation in prostate cancer
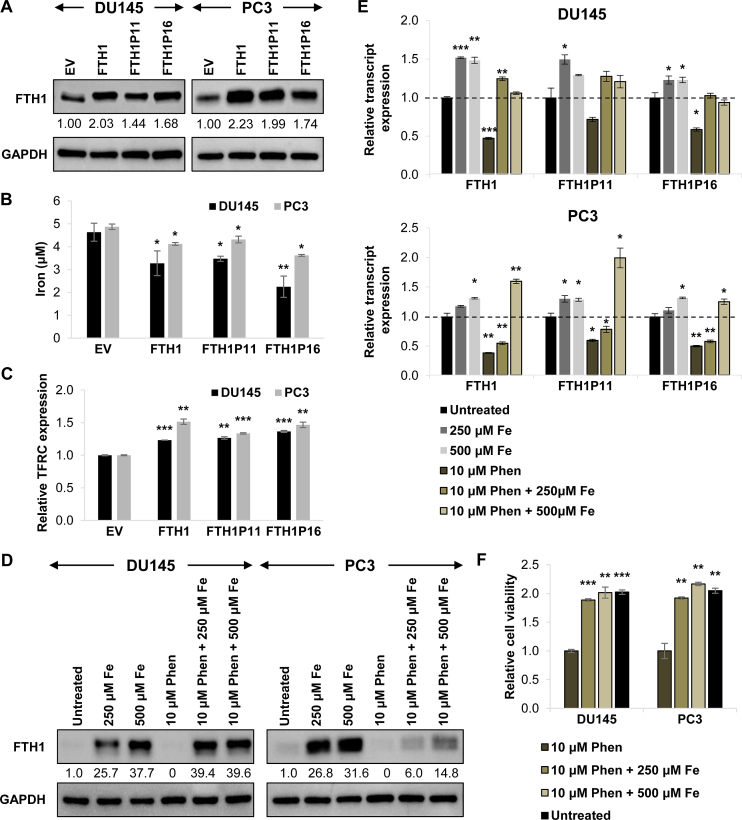
Due to the role of FTH1 in iron storage, we hypothesized that its regulation by the pseudogenes could affect iron balance. After establishing the tumor-suppressive functions of the FTH1 gene:pseudogene network, we next explored the possible regulatory mechanisms involved. We selected two FTH1 pseudogenes, FTH1P11 and FTH1P16, which have shown the most consistent phenotype as tumor suppressors and are similar in abundance to FTH1, for more in-depth studies (Figures 2F, 3D, 4E–G; Supplementary Figures S5C–F, S6B and C). To verify that FTH1P11 and FTH1P16 function primarily through their non-coding properties, we overexpressed the pseudogenes in a FTH1 knockout cell line and confirmed that these two pseudogenes do not express functional FTH1 protein despite containing predicted open reading frames within their sequences (Supplementary Figure S7A and B).
We first overexpressed FTH1, FTH1P11 and FTH1P16 separately, and found that the overexpression of each of these transcripts could upregulate FTH1 protein expression (Figure 5A), thus we hypothesized that the FTH1 pseudogenes could possess tumor-suppressive properties in part by acting as decoys for FTH1-targeting miRNAs to enhance FTH1 expression and its downstream effects. Previous studies have emphasized the importance of near-equimolar ratio of ceRNA molecules for an optimal ceRNA network (39,40). To analyze the stoichiometry of FTH1 and its pseudogenes in DU145 and PC3 cells, we quantified the absolute copy number of their transcripts by qRT-PCR using gene-specific primers calibrated by standard curves. The expression of FTH1P11 and FTH1P16 is within the same magnitude as FTH1 (Supplementary Figure S7C), which suggests effective ceRNA crosstalk may occur within this gene:pseudogene network.
Figure 5.
The FTH1 gene:pseudogene network disrupts iron regulation in PCa. (A) Effect of overexpressing FTH1 and its pseudogenes individually on FTH1 protein expression. (B and C) Effect of FTH1, FTH1P11 and FTH1P16 overexpression on intracellular iron content (B) and TFRC transcript expression (C) in DU145 and PC3 cells. (D–F) Effect of 24-h iron chloride (Fe) and iron chelator 1,10-Phenanthroline (Phen) treatment on FTH1 protein expression (D), FTH1, FTH1P11 and FTH1P16 transcript expression (E) and cell viability (F) in DU145 and PC3 cells. (B and F) Mean ± SD; n ≥ 3. (C and E) Mean ± SEM; n ≥ 3. *P < 0.05; **P < 0.01; ***P < 0.001.
As FTH1 is a major player in iron storage, we postulated that its inhibition via miRNA targeting could affect cellular iron levels. Direct measurements of the cellular iron concentration showed that iron levels were reduced upon FTH1, FTH1P11 and FTH1P16 overexpression in both DU145 and PC3 cells (Figure 5B). We also used the transcript expression of transferrin receptor (TFRC) as an indicator for intracellular iron content. Previous studies have shown that during iron depletion, iron-regulatory protein (IRP) binds to iron-responsive elements within the TFRC 3’UTR and stabilizes its transcript (41). We overexpressed FTH1, FTH1P11 and FTH1P16, and found that these upregulated TFRC transcript expression, conceivably due to the decrease in cellular iron caused by increased storage in ferritin (Figure 5C).
To reinforce the regulatory effects between FTH1 gene:pseudogene network and iron, we treated cells with extracellular iron and iron chelator, 1,10-Phenanthroline. Consistent with previous studies (42) and our findings above, excess iron increased FTH1 protein expression in a dose-dependent manner (Figure 5D). Remarkably, iron treatment upregulated the transcript expression of not only FTH1, but also that of FTH1P11 and FTH1P16 (Figure 5E). Conversely, iron depletion through a chelator downregulated FTH1 and the pseudogenes, which could be rescued by restoring iron availability (Figure 5D and E).
We further tested the phenotypic output of these treatments and found that chelating iron significantly reduced cell viability, which was rescued by iron reconstitution (Figure 5F). This highlights the importance of iron balance in sustaining PCa cell proliferation. Our data also indicate an interlinked role between ceRNA and iron regulation in eliciting the tumor-suppressive effects of the FTH1 gene:pseudogene network.
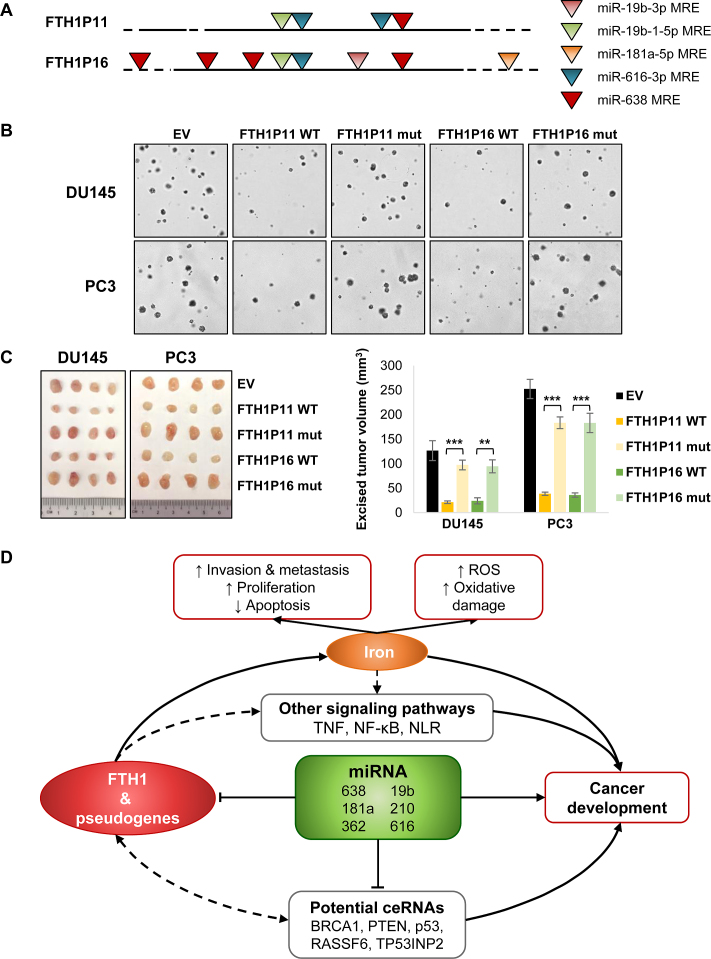
Mutation of MREs on FTH1 pseudogenes results in phenotype rescue
To further investigate whether the functions of FTH1P11 and FTH1P16 were mediated by ceRNA interactions, we mutated the MREs of miR-638 and other selected miRNAs on the pseudogenes to disrupt miRNA binding (Figure 6A and Supplementary Table S3). We only mutated nucleotides in regions corresponding to the miRNA seed sequences whilst retaining the purine/pyrimidine composition to preserve the secondary structures of the RNA transcripts.
Figure 6.
MRE mutations on FTH1 pseudogenes rescue growth phenotype. (A) Schematic of the MREs selected for mutation on FTH1P11 and FTH1P16. (B and C) Effect of overexpressing wild-type (WT) FTH1P11 and FTH1P16 and their mutants (mut) on anchorage-independent growth in soft agar (See Supplementary Figure S7G for quantification) (B) and tumor xenograft growth of DU145 and PC3 cells subcutaneously injected into nude mice (C). EV denotes empty vector control. Mean ± SEM; n ≥ 3. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Schematic of the FTH1 gene:pseudogene:miRNA network in iron regulation and cancer.
We confirmed that the miRNAs were not able to downregulate the luciferase reporters which contain MRE mutations, therefore the sequences mutated were essential in facilitating miRNA binding (Supplementary Figure S7D). Moreover, contrary to the wild-type pseudogenes, the overexpression of the FTH1P11 and FTH1P16 MRE mutants also had no effect on the FTH1 protein expression (Supplementary Figure S7E). Additionally, we observed phenotypic rescue in cell proliferation (Supplementary Figure S7F) and anchorage-independent growth in soft agar (Figure 6B and Supplementary Figure S7G) in both DU145 and PC3 cells. Likewise, the xenograft tumors overexpressing the FTH1P11 and FTH1P16 mutants grew at a similar rate to the empty vector control (Figure 6C). These findings supported our hypothesis that the tumor-suppressive properties of FTH1P11 and FTH1P16 are, at least in part, mediated by their ceRNA crosstalk with FTH1 in a gene:pseudogene network involved in iron regulation.
DISCUSSION
In this study, we characterize the tumor-suppressive FTH1 gene:pseudogene network. This network is targeted by miR-638 and other oncogenic miRNAs, including miR-19b, 181a, 210, 362 and 616, to inhibit its tumor-suppressive functions and promote PCa cell growth by disrupting iron regulation (Figure 6D). The major findings are (i) the expansive gene:pseudogene network encompassing FTH1 and its multiple pseudogenes, (ii) the tumor-suppressive properties of the FTH1 gene:pseudogene network facilitated by ceRNA crosstalk, (iii) phenotypic rescue by pseudogenes with impaired miRNA interactions and (iv) the potential regulation of iron by the FTH1 gene:pseudogene:miRNA network.
Our data suggest that the FTH1 gene:pseudogene network elicits its tumor-suppressive properties partially through the regulation of iron storage. It is well established that iron is an essential dietary element required for many important cellular functions, including DNA synthesis, adenosine triphosphate generation and proliferation (42). In recent years, the implication of dysregulated iron metabolism in tumorigenesis has been gaining widespread interest. Studies have demonstrated regulatory links between ferritin and genes that are frequently deregulated in cancer, including MYC proto-oncogene and tumor suppressor p53 (43,44). The latter has previously been shown to induce ferritin expression by inactivating IRPs, which could serve as an additional layer of gene regulation that could feedback to the FTH1 gene:pseudogene network through ceRNA crosstalk.
Critically, we show, for the first time, that MRE mutations that impair miRNA binding to individual pseudogenes were capable of complete phenotypic rescue in both in vitro experiments and mouse xenograft models. Therefore, a ceRNA network could comprise of multiple pseudogenes from the same parental gene. Furthermore, we showed that the knockdown of FTH1 and its pseudogenes resulted in a concomitant decrease in the protein expression of p53, a major tumor-suppressor and known miR-638 target (Supplementary Figure S8A). Together, these data indicate possible reciprocal regulation between the FTH1 gene:pseudogene network and p53 through multiple mechanisms, thus implicating a wider ceRNA network in the modulation of the tumor-suppressive effects of FTH1.
Recent studies on ceRNA regulation have proposed that a significant ceRNA effect is dependent on a network of interactions, an equilibrium in the ceRNA molecules involved and the abundance of target sites that could act cooperatively (40,45,46). Upon meeting these criteria, even highly expressed miRNAs can be sponged (47). In line with these studies, FTH1, FTH1P11 and FTH1P16 are similar in abundance, thus conducive for optimal ceRNA crosstalk, which is evident in the tumor-suppressive effects for these pseudogenes. Additionally, FTH1 and its pseudogenes carry multiple and closely spaced MREs for miR-638 and other oncogenic miRNAs, thus could potentially sequester common miRNAs in a cooperative manner that drives their tumor-suppressive functions. This supports previous reports of the functionality of MREs located outside the 3’UTR region, as well as the coding-independent function of protein-coding genes (18,48). It also suggests that the presence of multiple similar pseudogenes from the same parental gene is of functional relevance. Moreover, other known or yet to be characterized targets of miR-638 and other oncogenic miRNAs could add to this synergy to facilitate the functions of this network (12,14,49,50).
Interestingly, microarray analysis of FTH1, FTH1P11 and FTH1P16 overexpression revealed a common list of differentially expressed genes enriched in KEGG pathways, including NF-κB, NOD-like receptor (NLR) and tumor necrosis factor (TNF) signaling pathways (Supplementary Figure S8B and C). Studies have demonstrated regulatory links among these signaling pathway which share a distinguished feature in their association with iron regulation. Both NF-κB and TNF pathways are involved in the regulation of reactive oxygen species (ROS)-mediated apoptosis, in which FTH1 plays a vital role as an antioxidant (51–53). Notably, iron homeostasis is also central in inflammatory responses, whereby NF-κB, TNF and NLR are known to be important regulators and have been implicated in ROS crosstalk and cancer-related inflammation (52–55). As the FTH1 pseudogenes could regulate FTH1 expression, the gene:pseudogene network is thus implicated in these regulatory pathways and its deregulation could promote tumorigenesis.
Previous studies that employed a candidate gene approach have so far yielded one pseudogene for each gene of interest, whereas the robust FTH1 gene:pseudogene network in this study was identified using an unbiased screen for the biochemical associations of miR-638. This demonstrates the efficiency of the pulldown approach, which could be developed into a high throughput platform for miRNA target identification in a more physiologically relevant setting. As with the FTH1 gene:pseudogene network explored here, this application could be utilized to detect and characterize potential ceRNA partners in different cancers as well as other diseases involving aberrant miRNA expression. Not only could this facilitate the identification of new druggable targets, it could also provide the groundwork for miRNA-based therapeutics in various diseases.
The FTH1 gene:pseudogene:miRNA network represents the characterization of the first ceRNA network comprising many miRNAs and multiple pseudogenes derived from a single parental gene which could form an extensive network involving many miRNAs. Through their multiple closely spaced MREs, they could synergistically propagate their tumor-suppressive effects and regulate key biological processes. Given that FTH1 is a major iron storage protein and a key player in important signaling pathways triggered by disrupted iron and ROS levels, it is critical to maintain the balance within the gene:pseudogene network to prevent detrimental outcomes. More importantly, as miRNAs are the bridges connecting the players in this tightly regulated network, their aberrant expression could tip the balance and lead to disease development, making miRNAs attractive diagnostic tools. More specifically, in addition to miRNAs that are upregulated in PCa, urinary miRNAs could be used as a group of non-invasive biomarkers for PCa diagnosis. In a more clinical point of view, new combinatorial therapies could be developed to include both conventional drugs as well as those targeting selected miRNAs or the iron network for more effective PCa treatment.
DATA AVAILABILITY
The microarray datasets generated and analyzed during the current study have been deposited with the Gene Expression Omnibus repository under the accession number GSE101837 (https://www.ncbi.nlm.nih.gov/geo/).
Supplementary Material
ACKNOWLEDGEMENTS
We apologize to all colleagues whose work could not be cited due to space constraints. We thank Gina M. DeNicola for her insightful suggestions. We also thank Tay lab members for critical reading of the manuscript, Ng Desi, Angel Madero, Jeffrey Adijanto and Chun You Lim for their technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore National Research Foundation Fellowship (to Y.T.); National University of Singapore President’s Assistant Professorship (to Y.T.); RNA Biology Center at CSI Singapore (to Y.T.); Singapore Ministry of Education’s Tier 3 [MOE2014-T3-1-006 to Y.T.]; National Medical Research Council Grant [NMRC/CBRG/0077/2014 to C.L.]. Funding for open access charge: Singapore National Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. et al. The transcriptional landscape of the mammalian genome. Science. 2005; 309:1559–1563. [DOI] [PubMed] [Google Scholar]
- 2. Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. et al. Landscape of transcription in human cells. Nature. 2012; 489:101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee R.C., Feinbaum R.L., Ambros V.. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993; 75:843–854. [DOI] [PubMed] [Google Scholar]
- 4. Wightman B., Ha I., Ruvkun G.. Post-transcriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993; 75:855–862. [DOI] [PubMed] [Google Scholar]
- 5. Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 6. Guttman M., Rinn J.L.. Modular regulatory principles of large non-coding RNAs. Nature. 2012; 482:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cech T.R., Steitz J.A.. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014; 157:77–94. [DOI] [PubMed] [Google Scholar]
- 8. Bartel D.P., Chen C.Z.. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004; 5:396–400. [DOI] [PubMed] [Google Scholar]
- 9. Bracken C.P., Scott H.S., Goodall G.J.. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016; 17:719–732. [DOI] [PubMed] [Google Scholar]
- 10. Svoronos A.A., Engelman D.M., Slack F.J.. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016; 76:3666–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tan X., Peng J., Fu Y., An S., Rezaei K., Tabbara S., Teal C.B., Man Y.G., Brem R.F., Fu S.W.. miR-638 mediated regulation of BRCA1 affects DNA repair and sensitivity to UV and cisplatin in triple-negative breast cancer. Breast Cancer Res. 2014; 16:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicoloso M.S., Sun H., Spizzo R., Kim H., Wickramasinghe P., Shimizu M., Wojcik S.E., Ferdin J., Kunej T., Xiao L. et al. Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility. Cancer Res. 2010; 70:2789–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ren Y., Chen Y., Liang X., Lu Y., Pan W., Yang M.. MiRNA-638 promotes autophagy and malignant phenotypes of cancer cells via directly suppressing DACT3. Cancer Lett. 2017; 390:126–136. [DOI] [PubMed] [Google Scholar]
- 14. Tay Y., Tan S.M., Karreth F.A., Lieberman J., Pandolfi P.P.. Characterization of dual PTEN and p53-targeting microRNAs identifies microRNA-638/Dnm2 as a two-hit oncogenic locus. Cell Rep. 2014; 8:714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma K., Pan X., Fan P., He Y., Gu J., Wang W., Zhang T., Li Z., Luo X.. Loss of miR-638 in vitro promotes cell invasion and a mesenchymal-like transition by influencing SOX2 expression in colorectal carcinoma cells. Mol. Cancer. 2014; 13:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang J., Fei B., Wang Q., Song M., Yin Y., Zhang B., Ni S., Guo W., Bian Z., Quan C. et al. MicroRNA-638 inhibits cell proliferation, invasion and regulates cell cycle by targeting tetraspanin 1 in human colorectal carcinoma. Oncotarget. 2014; 5:12083–12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P.. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011; 146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tay Y., Rinn J., Pandolfi P.P.. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014; 505:344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Z., Harrison P.M., Liu Y., Gerstein M.. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003; 13:2541–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim M.S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. et al. A draft map of the human proteome. Nature. 2014; 509:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P.. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010; 465:1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Karreth F.A., Reschke M., Ruocco A., Ng C., Chapuy B., Leopold V., Sjoberg M., Keane T.M., Verma A., Ala U. et al. The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. 2015; 161:319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rutnam Z.J., Du W.W., Yang W., Yang X., Yang B.B.. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat. Commun. 2014; 5:2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng H., Ishida M., Li L., Saito A., Kamiya A., Hamilton J.P., Fu R., Olaru A.V., An F., Popescu I. et al. Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget. 2015; 6:5666–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang L., Guo Z.Y., Zhang R., Xin B., Chen R., Zhao J., Wang T., Wen W.H., Jia L.T., Yao L.B. et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis. 2013; 34:1773–1781. [DOI] [PubMed] [Google Scholar]
- 26. Bai M., Yuan M., Liao H., Chen J., Xie B., Yan D., Xi X., Xu X., Zhang Z., Feng Y.. OCT4 pseudogene 5 upregulates OCT4 expression to promote proliferation by competing with miR-145 in endometrial carcinoma. Oncol. Rep. 2015; 33:1745–1752. [DOI] [PubMed] [Google Scholar]
- 27. Zheng L., Li X., Gu Y., Lv X., Xi T.. The 3′UTR of the pseudogene CYP4Z2P promotes tumor angiogenesis in breast cancer by acting as a ceRNA for CYP4Z1. Breast Cancer Res. Treat. 2015; 150:105–118. [DOI] [PubMed] [Google Scholar]
- 28. Zheng L., Li X., Meng X., Chou J., Hu J., Zhang F., Zhang Z., Xing Y., Liu Y., Xi T.. Competing endogenous RNA networks of CYP4Z1 and pseudogene CYP4Z2P confer tamoxifen resistance in breast cancer. Mol. Cell. Endocrinol. 2016; 427:133–142. [DOI] [PubMed] [Google Scholar]
- 29. Hentze M.W., Keim S., Papadopoulos P., O’Brien S., Modi W., Drysdale J., Leonard W.J., Harford J.B., Klausner R.D.. Cloning, characterization, expression, and chromosomal localization of a human ferritin heavy-chain gene. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:7226–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Torti F.M., Torti S.V.. Regulation of ferritin genes and protein. Blood. 2002; 99:3505–3516. [DOI] [PubMed] [Google Scholar]
- 31. Tai Y.K., Chew K.C., Tan B.W., Lim K.L., Soong T.W.. Iron mitigates DMT1-mediated manganese cytotoxicity via the ASK1-JNK signaling axis: Implications of iron supplementation for manganese toxicity. Sci. Rep. 2016; 6:21113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lal A., Thomas M.P., Altschuler G., Navarro F., O’Day E., Li X.L., Concepcion C., Han Y.C., Thiery J., Rajani D.K. et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011; 7:e1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chua S.W., Vijayakumar P., Nissom P.M., Yam C.Y., Wong V.V., Yang H.. A novel normalization method for effective removal of systematic variation in microarray data. Nucleic Acids Res. 2006; 34:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., Thomson A.M., Lim B., Rigoutsos I.. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006; 126:1203–1217. [DOI] [PubMed] [Google Scholar]
- 35. Carroll A.G., Voeller H.J., Sugars L., Gelmann E.P.. p53 oncogene mutations in three human prostate cancer cell lines. Prostate. 1993; 23:123–134. [DOI] [PubMed] [Google Scholar]
- 36. Vlietstra R.J., van Alewijk D.C., Hermans K.G., van Steenbrugge G.J., Trapman J.. Frequent inactivation of PTEN in prostate cancer cell lines and xenografts. Cancer Res. 1998; 58:2720–2723. [PubMed] [Google Scholar]
- 37. Liu N.Q., De Marchi T., Timmermans A.M., Beekhof R., Trapman-Jansen A.M., Foekens R., Look M.P., van Deurzen C.H., Span P.N., Sweep F.C. et al. Ferritin heavy chain in triple negative breast cancer: a favorable prognostic marker that relates to a cluster of differentiation 8 positive (CD8+) effector T-cell response. Mol. Cell. Proteomics. 2014; 13:1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Funauchi Y., Tanikawa C., Yi Lo P.H., Mori J., Daigo Y., Takano A., Miyagi Y., Okawa A., Nakamura Y., Matsuda K.. Regulation of iron homeostasis by the p53-ISCU pathway. Sci. Rep. 2015; 5:16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ala U., Karreth F.A., Bosia C., Pagnani A., Taulli R., Leopold V., Tay Y., Provero P., Zecchina R., Pandolfi P.P.. Integrated transcriptional and competitive endogenous RNA networks are cross-regulated in permissive molecular environments. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7154–7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bosson A.D., Zamudio J.R., Sharp P.A.. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014; 56:347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Muckenthaler M., Gray N.K., Hentze M.W.. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell. 1998; 2:383–388. [DOI] [PubMed] [Google Scholar]
- 42. Torti S.V., Torti F.M.. Iron and cancer: more ore to be mined. Nat. Rev. Cancer. 2013; 13:342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu K.J., Polack A., Dalla-Favera R.. Coordinated regulation of iron-controlling genes, H-ferritin and IRP2, by c-MYC. Science. 1999; 283:676–679. [DOI] [PubMed] [Google Scholar]
- 44. Zhang F., Wang W., Tsuji Y., Torti S.V., Torti F.M.. Post-transcriptional modulation of iron homeostasis during p53-dependent growth arrest. J. Biol. Chem. 2008; 283:33911–33918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M.. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014; 54:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M.. Impact of microRNA levels, target-site complementarity, and cooperativity on competing endogenous RNA-regulated gene expression. Mol. Cell. 2016; 64:565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Powers J.T., Tsanov K.M., Pearson D.S., Roels F., Spina C.S., Ebright R., Seligson M., de Soysa Y., Cahan P., Theissen J. et al. Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature. 2016; 535:246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Steinkraus B.R., Toegel M., Fulga T.A.. Tiny giants of gene regulation: experimental strategies for microRNA functional studies. Wiley Interdiscip. Rev. Dev. Biol. 2016; 5:311–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mi Y., Zhang D., Jiang W., Weng J., Zhou C., Huang K., Tang H., Yu Y., Liu X., Cui W. et al. miR-181a-5p promotes the progression of gastric cancer via RASSF6-mediated MAPK signalling activation. Cancer Lett. 2017; 389:11–22. [DOI] [PubMed] [Google Scholar]
- 50. Bhattacharya A., Schmitz U., Raatz Y., Schonherr M., Kottek T., Schauer M., Franz S., Saalbach A., Anderegg U., Wolkenhauer O. et al. miR-638 promotes melanoma metastasis and protects melanoma cells from apoptosis and autophagy. Oncotarget. 2015; 6:2966–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pham C.G., Bubici C., Zazzeroni F., Papa S., Jones J., Alvarez K., Jayawardena S., De Smaele E., Cong R., Beaumont C. et al. Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell. 2004; 119:529–542. [DOI] [PubMed] [Google Scholar]
- 52. Morgan M.J., Liu Z.G.. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011; 21:103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Brenner D., Blaser H., Mak T.W.. Regulation of tumour necrosis factor signalling: live or let die. Nat. Rev. Immunol. 2015; 15:362–374. [DOI] [PubMed] [Google Scholar]
- 54. Saxena M., Yeretssian G.. NOD-Like Receptors: Master Regulators of Inflammation and Cancer. Front. Immunol. 2014; 5:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blaser H., Dostert C., Mak T.W., Brenner D.. TNF and ROS Crosstalk in Inflammation. Trends Cell Biol. 2016; 26:249–261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray datasets generated and analyzed during the current study have been deposited with the Gene Expression Omnibus repository under the accession number GSE101837 (https://www.ncbi.nlm.nih.gov/geo/).