ABSTRACT
The adaptations that protect pathogenic microorganisms against the cytotoxicity of nitric oxide (NO) engendered in the immune response are incompletely understood. We show here that salmonellae experiencing nitrosative stress suffer dramatic losses of the nucleoside triphosphates ATP, GTP, CTP, and UTP while simultaneously generating a massive burst of the alarmone nucleotide guanosine tetraphosphate. RelA proteins associated with ribosomes overwhelmingly synthesize guanosine tetraphosphate in response to NO as a feedback mechanism to transient branched-chain amino acid auxotrophies. Guanosine tetraphosphate activates the transcription of valine biosynthetic genes, thereby reestablishing branched-chain amino acid biosynthesis that enables the translation of the NO-consuming flavohemoglobin Hmp. Guanosine tetraphosphate synthesized by RelA protects salmonellae from the metabolic stress inflicted by reactive nitrogen species generated in the mammalian host response. This research illustrates the importance of nucleotide metabolism in the adaptation of salmonellae to the nutritional stress imposed by NO released in the innate host response.
KEYWORDS: Salmonella, adaptive resistance, animal models, cellular redox status, guanosine tetraphosphate, nitric oxide, nitrosative stress, nucleotide metabolism, stringent response
IMPORTANCE
Nitric oxide triggers dramatic drops in nucleoside triphosphates, the building blocks that power DNA replication; RNA transcription; translation; cell division; and the biosynthesis of fatty acids, lipopolysaccharide, and peptidoglycan. Concomitantly, this diatomic gas stimulates a burst of guanosine tetraphosphate. Global changes in nucleotide metabolism may contribute to the potent bacteriostatic activity of nitric oxide. In addition to inhibiting numerous growth-dependent processes, guanosine tetraphosphate positively regulates the transcription of branched-chain amino acid biosynthesis genes, thereby facilitating the translation of antinitrosative defenses that mediate recovery from nitrosative stress.
INTRODUCTION
Salmonella enterica serovar Typhimurium is a causative agent of zoonotic nontyphoidal salmonellosis. Although typically self-limiting in healthy individuals, this syndrome can be life threatening in immunocompromised patients with gamma interferon signaling defects or HIV or malaria comorbidities (1–4). Far from alleviating symptoms, antibiotic therapy can prolong the shedding of salmonellae in immunocompetent individuals (5) and often select for increasingly resistant Salmonella populations (6, 7). The World Health Organization has called for a greater focus on the development of novel treatments against fluoroquinolone-resistant salmonellae (8). Basic knowledge of bacterial pathogenesis provides new targets for the development of next-generation antibiotic therapies. Growth in macrophages is a key step in Salmonella pathogenesis. Interactions of salmonellae with professional phagocytes activate the translation of inducible nitric oxide synthase (iNOS), a flavohemoprotein that synthesizes nitric oxide (NO) from l-arginine, O2, and NADPH (9, 10). NO and its nitrosative and oxidative congeners exert bacteriostasis against salmonellae. The concerted actions of low-molecular-weight thiols, cytochrome bd, and the flavohemoglobin Hmp protect salmonellae against the nitrosative stress encountered in mammalian hosts (11–13). Despite these antinitrosative defenses, NO attacks redox-active moieties in dihydroxy-acid dehydratase and lipoamide-dependent lipoamide dehydrogenase, thus inhibiting the biosynthesis of branched-chain amino acids, methionine, and lysine (14, 15). The molecular mechanisms that help salmonellae resolve the resulting functional amino acid auxotrophies remain inadequately understood.
The alarmones guanosine tetraphosphate (ppGpp) and pentaphosphate (pppGpp) [collectively referred to as (p)ppGpp] are synthesized by bacteria in their adaptive response to nutritional starvation (16). The enzymes RelA and SpoT synthesize (p)ppGpp by transferring pyrophosphate from ATP to the 3′-OH group of ribose in GDP or GTP, yielding AMP and ppGpp or pppGpp, respectively (17). Typically, both ppGpp and pppGpp are produced at once in roughly equimolar concentrations (18), depending on the source of stress. RelA proteins respond to amino acid starvation by directly sensing unaminoacylated tRNAs erroneously placed in the A site of the ribosome (19, 20). Cytosolic SpoT proteins synthesize (p)ppGpp in response to drops in physiological concentrations of iron, fatty acids, phosphate, or nitrogen (16). SpoT is a bifunctional enzyme that can synthesize and hydrolyze (p)ppGpp (21–23). Transcription is the best-characterized target of (p)ppGpp, which binds to a hydrophobic pocket between the β′ and ω subunits of the RNA polymerase (RNAP) and at the interface of DksA and the rim of the β′ subunit (24, 25). Binding of (p)ppGpp to these sites destabilizes the RNAP holoenzyme-DNA open complex, repressing genes encoding rRNAs, tRNAs, and ribosomal proteins while activating amino acid biosynthesis operons (16). In addition, (p)ppGpp can inhibit translation, ribosome biogenesis, protein stability, and lipid and cell wall biosynthesis (16, 26–28). By doing so, (p)ppGpp influences major aspects of bacterial cell physiology.
Many bacterial pathogens have incorporated (p)ppGpp into regulatory circuits to control the expression of virulence determinants (29). Functional RelA and SpoT proteins are required for the contextual expression of Salmonella virulence programs associated with epithelial cell invasion and intracellular survival (30). Genetic evidence has also implicated (p)ppGpp in the antinitrosative defenses of salmonellae (31); however, the mechanisms by which these nucleotides enhance the resistance of salmonellae to NO are unknown. Our investigations described here indicate that (p)ppGpp aids with the recovery of salmonellae from nitrosative stress by activating the biosynthesis of branched-chain amino acids that fuel the translation of antinitrosative defenses.
RESULTS
Salmonellae experiencing nitrosative stress synthesize ppGpp.
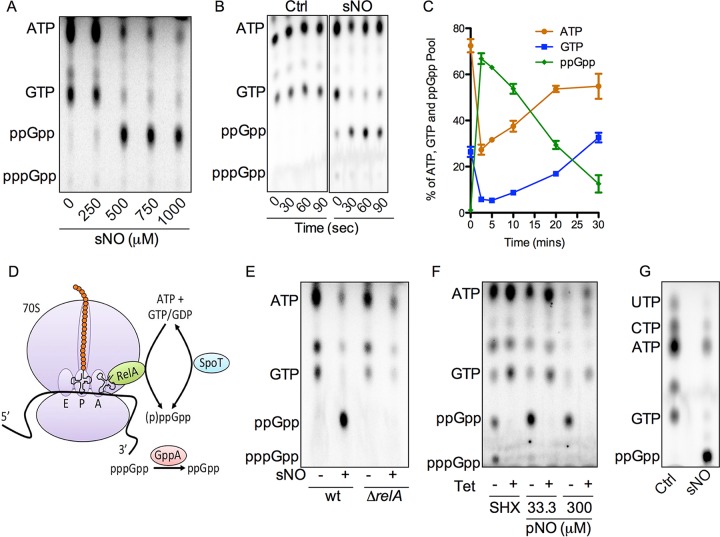
Salmonellae bearing mutations in relA and spoT are hypersusceptible to NO (31), suggesting that (p)ppGpp increases the fitness of salmonellae undergoing nitrosative stress. Accordingly, salmonellae treated for 5 min with a 500 μM concentration of the NO donor spermine NONOate abundantly accumulated ppGpp while experiencing marked depletions of ATP and GTP (Fig. 1A). NO treatment did not affect bacterial viability. The NO donor propylamine propylamine (PAPA) NONOate also stimulated the synthesis of ppGpp in salmonellae (see Fig. S1A in the supplemental material). Curiously, neither spermine NONOate nor PAPA NONOate triggered pppGpp synthesis. Kinetic analyses revealed that ppGpp was produced as early as 30 s after spermine NONOate treatment (Fig. 1B), lasting about 30 min thereafter (Fig. 1C; Fig. S1B).
FIG 1 .
NO dramatically affects the nucleotide pools of salmonellae. (A) TLC autoradiogram of 32P-labeled nucleotides extracted from log-phase salmonellae 5 min after exposure to increasing concentrations of spermine NONOate (sNO). (B) Nucleotides were examined 0 to 90 s after treatment with 750 μM sNO. (C) Relative abundance of ATP, GTP, and ppGpp in salmonellae treated with 750 μM sNO (see Fig. S1B for an example of the autoradiograms used in these analyses). n = 4. Data are shown as the mean value ± the standard error of the mean. (D) Model of (p)ppGpp metabolism. RelA synthesizes (p)ppGpp from ATP and GTP or GDP in response to unaminoacylated tRNAs erroneously loaded in the A site of the ribosome. SpoT can both synthesize and hydrolyze (p)ppGpp, whereas GppA hydrolyzes pppGpp to ppGpp. (E) Nucleotides from wild-type (wt) and ΔrelA mutant salmonellae treated for 5 min with 750 μM sNO. (F) Nucleotides in salmonellae treated with 70 μg/ml tetracycline for 3 min before the addition of 0.4 mg/ml serine hydroxamate (SHX) or 33.3 or 300 μM PAPA NONOate (pNO) for 2 min. All of the autoradiograms shown are representative of two to six independent experiments. (G) Nucleotides from salmonellae treated for 5 min with sNO were examined as described for panel A. Extracts in panels A, B, E, and F were separated with 1.25 M KH2PO4 buffer, pH 3.4. Specimens in panel G were separated with 0.9 M KH2PO4 buffer, pH 3.4. Ctrl, control.
Effect of NO on the nucleotide pools of salmonellae. (A) TLC autoradiogram of 32P-labeled nucleotides extracted from salmonellae treated for 30 s with increasing concentrations of PAPA NONOate (pNO). (B) Nucleotides were visualized as described for panel A at the times of treatment with 750 μM spermine NONOate (sNO) indicated. (C) Quantification of (ppGpp + pppGpp)/(GTP + ppGpp + pppGpp) ratios from nucleotide autoradiograms from salmonellae treated with either 0.4 mg/ml serine hydroxamate (SHX) or pNO for 90 s, followed by the addition of 70 μg/ml tetracycline (Tet). Samples were collected from 0 to 10 min after the addition of Tet. Ctrl, control. (D) Nucleotides from wild-type (wt) and ΔgppA salmonellae were visualized as described for panel A 5 min after treatment with either 0.4 mg/ml SHX or 750 μM sNO. All autoradiograms are representative of two or three independent experiments. (E) Firefly luciferase quantification of ATP pools in salmonellae left untreated (Untr.) or treated for 5 min with 750 μM sNO. n = 8 or 9. Data are shown as the mean value ± the standard error of the mean. ***, P < 0.001 (determined by Student t test). Download FIG S1, PDF file, 3.9 MB (4MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Gammaproteobacteria such as salmonellae synthesize (p)ppGpp through the enzymatic activity of RelA or SpoT protein (Fig. 1D) (32). ΔrelA mutant salmonellae failed to accumulate any measureable ppGpp after a NO challenge (Fig. 1E), implying that RelA may be solely responsible for the burst of ppGpp recorded in salmonellae experiencing nitrosative stress. RelA is classically activated during amino acid starvation in response to its direct contact with unaminoacylated tRNAs erroneously loaded into the A site of the ribosome (19). NO modifies cysteines and [4Fe-4S] clusters in lipoamide-dependent lipoamide dehydrogenase and dihydroxy-acid dehydratase, respectively, inhibiting methionine, lysine, and branched-chain amino acid biosynthesis (14, 15). The consequent accumulation of unaminoacylated tRNAs could activate RelA. To test this model, salmonellae were pretreated with tetracycline, an antibiotic that blocks the entry of tRNAs into the A site of ribosomes and thereby prevents classical RelA activation (18). Tetracycline completely prevented the synthesis of (p)ppGpp in response to serine hydroxamate (Fig. 1F), a classical RelA activator that irreversibly inhibits seryl-tRNA synthetases (26). Tetracycline also completely inhibited ppGpp synthesis in response to the NO donor PAPA NONOate. These data suggest that NO activates ppGpp production from RelA by increasing the amount of unaminoacylated tRNAs.
To examine the rates of SpoT-dependent (p)ppGpp decay, salmonellae were pretreated with either serine hydroxamate or PAPA NONOate before tetracycline was added to the cultures. The burst of (p)ppGpp decayed quickly (5 min) and with similar kinetics after the addition of serine hydroxamate or 33.3 μM PAPA NONOate (Fig. S1C) (estimated half-lives of 1.3 and 0.7 min, respectively). In contrast, the ppGpp pool in salmonellae challenged with 300 μM PAPA NONOate diminished at a substantially lower rate (i.e., a half-life of 8.5 min), suggesting that high concentrations of NO inhibit the ability of SpoT to hydrolyze ppGpp.
In the canonical (p)ppGpp cycle, pppGpp is the primary nucleotide synthesized by RelA, but the 5′-monophosphatase activity of GppA converts pppGpp to ppGpp (Fig. 1D) (18, 33). According to this model, serine hydroxamate-treated ΔgppA mutant salmonellae synthesized nearly four times as much pppGpp as ppGpp, whereas serine hydroxamate stimulated nearly equimolar concentrations of pppGpp and ppGpp in wild-type salmonellae (Fig. S1D). As described for wild-type controls, NO stimulated the exclusive production of ppGpp in ΔgppA mutant salmonellae. These data demonstrate that NO selectively induces ppGpp synthesis independently of the enzymatic activity of GppA.
Because nitrosative stress diminishes ATP and GTP synthesis (see above), the thin-layer chromatography (TLC) solvent system was modified to determine the global effects of NO on nucleoside triphosphates (NTPs). NO-treated salmonellae suffered massive drops in ATP, UTP, CTP, and GTP (Fig. 1G), perhaps reflecting the fact that NTP synthesis requires ATP. Consistent with the patterns seen by TLC, firefly luciferase analyses revealed that NO reduces the ATP pool by roughly 60% in salmonellae (Fig. S1E). Using ATP as an internal standard, we estimate that the intracellular concentration of ppGpp reaches nearly 2 mM after NO treatment. Collectively, these data indicate that salmonellae dramatically reprogram nucleotide metabolism in response to NO, with overall decreases in NTPs and a parallel accumulation of ppGpp. The overall changes in nucleotides could have substantial consequences for bacterial physiology, NO toxicity, and the antinitrosative defenses of salmonellae.
Branched-chain amino acids reestablish the antinitrosative defenses of salmonellae.
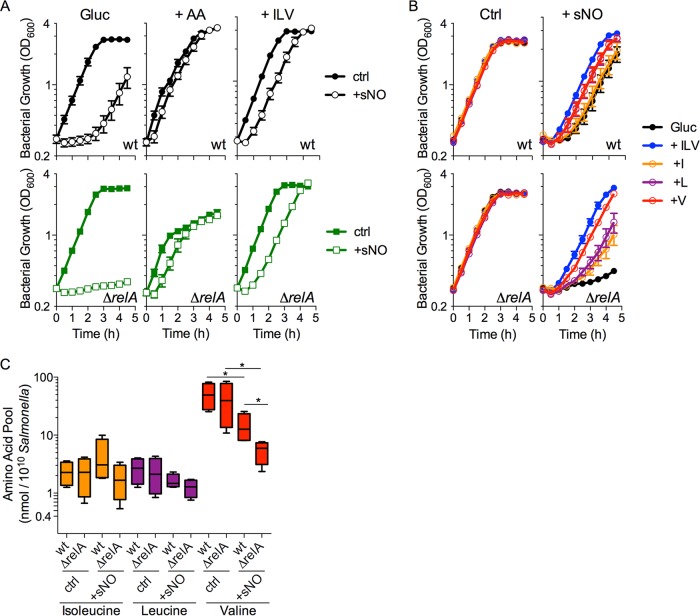
We next examined whether ppGpp synthesized in response to NO contributes to the antinitrosative defenses of salmonellae. Wild-type salmonellae grown to log phase in glucose minimal medium resumed growth at about 2.5 h after treatment with 750 μM spermine NONOate (Fig. 2A). On the other hand, ΔrelA mutant salmonellae never recovered from the bacteriostasis induced by NO in the 5-h time frame examined. The hypersusceptibility of ΔrelA mutant salmonellae to NO could be rescued by genetic complementation with wild-type relA but not by complementation with the catalytically deficient D275G or E335Q (22) relA allele (Fig. S2A). Addition of all amino acids to glucose minimal medium significantly increased the resistance of salmonellae to the antimicrobial activity of NO (Fig. 2A). These observations are consistent with previous studies (15, 31). The addition of amino acids to glucose minimal medium also stimulated the growth of ΔrelA mutant salmonellae undergoing nitrosative stress, suggesting that NO induces transient, functional amino acid auxotrophies that are resolved by RelA-derived ppGpp.
FIG 2 .
RelA and branched-chain amino acids accelerate the recovery of NO-treated salmonellae. (A) Growth of wild-type (wt) and ΔrelA mutant salmonellae in MOPS glucose minimal medium (Gluc) in the presence or absence of 750 μM spermine NONOate (+sNO). Where indicated, all 20 amino acids (+AA) or branched-chain amino acids (+ILV) were added to the medium. n = 4 to 6. Data are shown as the mean value ± the standard error of the mean. (B) Growth of wild-type and ΔrelA mutant salmonellae challenged with 750 μM sNO in MOPS glucose minimal medium supplemented with branched-chain amino acids (+ILV), isoleucine (+I), leucine (+L), or valine (+V). n = 4. Data are shown as the mean value ± the standard error of the mean. (C) Intracellular concentrations of isoleucine, leucine, and valine were determined by LC-MS in wild-type and ΔrelA mutant salmonellae grown in M9 glucose minimal medium. Where indicated, 750 μM sNO was added to the cultures. n = 4. The data are represented as box-and-whiskers plots. *; P < 0.05 (determined by Mann-Whitney analysis). Ctrl, control.
Effect of amino acids on the recovery of salmonellae from a NO challenge. (A) Growth of wild-type (wt) salmonellae and ΔrelA and ΔrelA mutant salmonellae complemented with the wild-type, D275G, or E335Q relA allele in MOPS minimal medium supplemented with glucose. Where indicated, bacterial cultures were treated with 750 μM spermine NONOate (+sNO). n = 3. Data are shown as the mean value ± the standard error of the mean. (B) The effects of sNO on the growth of salmonellae were also tested in MOPS glucose minimal medium (Gluc) supplemented with the aromatic amino acids (FWY), all amino acids except the aromatic amino acids (AA − FWY), or all 20 amino acids (AA). (C) Growth of salmonellae left untreated (Ctrl.) or challenged with 750 μM sNO in MOPS minimal medium supplemented with glucose or glucose and methionine and lysine (MK), all amino acids except methionine and lysine (AA − MK), or all 20 amino acids (AA). (D) Stationary-phase salmonellae were challenged with 750 μM sNO in MOPS glucose minimal medium supplemented with the branched-chain amino acids (ILV), methionine and lysine (MK), all amino acids except the branched-chain amino acids (AA − ILV), all amino acids except methionine or lysine (AA − MK), or all 20 amino acids (AA). (E, F) The time at which the initial OD600 doubled was calculated from data presented in Fig. 2A or B. *, P < 0.05; ***, P < 0.001 (determined by one-way ANOVA). Download FIG S2, PDF file, 3 MB (3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
NO and its oxidative and nitrosative congeners inhibit branched-chain amino acid (leucine, isoleucine, and valine), aromatic amino acid (phenylalanine, tyrosine, and tryptophan), methionine, and lysine biosynthetic pathways (14, 15). To gain insights into the amino acid biosynthetic pathways that might be targeted by NO, salmonellae were challenged with spermine NONOate in glucose minimal medium supplemented with different combinations of amino acids. The addition of branched-chain amino acids rendered both wild-type and ΔrelA mutant salmonellae resistant to nitrosative stress (Fig. 2A; Fig. S2E). The addition of aromatic amino acids (Fig. S2B) or methionine and lysine (Fig. S2C) did not greatly affect the recovery of NO-treated salmonellae. Moreover, amino acid mixtures lacking aromatic amino acids or lysine and methionine still rendered log-phase salmonellae highly resistant to NO (Fig. S2B and C). These findings indicate that under the conditions examined here, NO exerts considerable pressure on the biosynthesis of branched-chain amino acids but not aromatic amino acids or lysine and methionine. Given previous findings (15), the apparent dispensability of lysine and methionine in the recovery of salmonellae from NO might seem surprising. The apparent inconsistencies between these studies could be due to the growth phase at which salmonellae were challenged with NO, as suggested by the fact that methionine and lysine, not branched-chain amino acids, aided the recovery of stationary-phase salmonellae from NO toxicity (Fig. S2D). Cumulatively, salmonellae seem to develop different transient, functional amino acid auxotrophies, depending on the growth phase in which they encounter NO.
To identify which one of the branched-chain amino acids contributes to the antinitrosative defenses of salmonellae, wild-type and ΔrelA mutant salmonellae were challenged with NO in morpholinepropanesulfonic acid (MOPS) glucose minimal medium supplemented with valine, leucine, or isoleucine. Compared to isoleucine and leucine, valine had the greatest impact on the recovery of wild-type and ΔrelA mutant salmonellae from the bacteriostatic activity of NO (Fig. 2B; Fig. S2F). Consistent with these observations, the intracellular valine pools were substantially reduced in both wild-type and ΔrelA mutant salmonellae experiencing nitrosative stress (Fig. 2C). It should be noted that the diminution of valine was more pronounced in NO-treated ΔrelA mutant salmonellae than in similarly treated wild-type controls. In contrast to valine, other amino acid pools were largely unaffected by the ΔrelA mutation, a NO challenge, or both (Fig. S3). The only exceptions were aspartate, which was reduced after a NO challenge in wild-type salmonellae; arginine, which was lower in ΔrelA mutant salmonellae; and threonine, which was reduced in NO-treated ΔrelA mutant salmonellae (Fig. S3). NO-mediated damage of IlvB would curtail the synthesis of all three branched-chain amino acids, yet it only seems to affect valine pools. This might be due to the high levels of valine, which is about 20-fold more abundant than isoleucine and leucine, maintained by salmonellae (Fig. 2C). Altogether, these data demonstrate that NO depletes the intracellular pools of valine in exponentially growing salmonellae and that ppGpp synthesized by RelA mitigates the NO-induced valine starvation, thereby helping resolve the auxotrophy of this branched-chain amino acid.
Intracellular concentrations of amino acids in salmonellae under nitrosative stress. Amino acids were determined by LC-MS (n = 4, box-and-whisker plot) from wild-type (wt) and ΔrelA mutant salmonellae grown to log phase in M9 glucose minimal medium. Where indicated, the bacterial cultures were treated with 750 μM spermine NONOate (+sNO) for 5 min. Isoleucine, leucine, and valine are represented in Fig. 2C. Asparagine, cysteine, glutamine, phenylalanine, and tryptophan were not detected by this method. *, P < 0.05 (determined by Mann-Whitney analysis). Untr., untreated. Download FIG S3, PDF file, 2.3 MB (2.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Branched-chain amino acid biosynthetic genes are reexpressed in a relA-dependent manner following a NO challenge.
In the first step of valine biosynthesis, acetohydroxy acid synthase I decarboxylates pyruvate and transfers the active aldehyde to a second molecule of pyruvate (Fig. S4A) (34). However, the addition of pyruvate to glucose minimal medium did not alter the toxicity of NO against wild-type or ΔrelA mutant salmonellae (Fig. S5), suggesting that pyruvate is not limiting for the recovery of salmonellae from nitrosative stress. We therefore focused on the expression of valine biosynthetic enzymes. Valine is synthesized by the sequential enzymatic activities of acetohydroxy acid synthase I, ketol-acid reductoisomerase, dihydroxy-acid dehydratase, and transaminase B (encoded by ilvBN, ilvC, ilvD, and ilvE, respectively) (Fig. S4B). Valine transamination is a reversible enzymatic reaction, and thus valine can be a substrate for leucine biosynthesis. The transcription of ilvB and ilvD was repressed in wild-type salmonellae after 30 min of NO treatment (Fig. 3A and C); that of ilvD was also repressed (P < 0.001) in NO-treaed ΔrelA mutant salmonellae (Fig. 3C). Wild-type salmonellae recovered the transcription of ilvB, ilvC, ilvD, and leuA after 1 h of NO treatment. One hour after NO treatment, the transcription of these genes was significantly lower in ΔrelA mutant controls than in wild-type salmonellae. Expression of IlvB-3×FLAG (Fig. 3E) and IlvD-3×FLAG (Fig. 3G) protein levels matched mRNA expression patterns. IlvC-3×FLAG (Fig. 3F) and LeuD-3×FLAG (Fig. 3H) did not appreciably change in NO-treated salmonellae. The NO-mediated repression of valine biosynthetic genes may explain why the addition of valine enhances the resistance of salmonellae to nitrosative stress. To study further the effects of ppGpp on the expression of branched-chain amino acid genes, we measured the in vitro transcription of ivbL and ilvD (Fig. 3I), whose promoters drive ilvB and ilvD expression, respectively (Fig. S4B). Although ppGpp by itself did not activate ivbL or ilvD in vitro transcription, this alarmone synergized with the RNAP-binding protein DksA (Fig. 3I). The synergism was most evident at low concentrations (i.e., 10 to 100 μM) of ppGpp.
FIG 3 .
RelA regulates the expression of branched-chain amino acid biosynthetic loci in salmonellae undergoing nitrosative stress. (A to D) Transcription of ilvB, ilvC, ilvD, and leuA mRNAs in wild-type (wt) and ΔrelA mutant salmonellae grown in MOPS glucose minimal medium was quantified by qRT-PCR. Bacterial cultures were treated with 750 μM spermine NONOate for the times indicated. The results were normalized to internal levels of the ampD housekeeping gene. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (determined by two-way ANOVA). n = 4. Data are shown as the mean value ± the standard error of the mean. (E to H) Western blot analysis of IlvB-3×FLAG, IlvC-3×FLAG, IlvD-3×FLAG, and LeuD-3×FLAG from the Salmonella strains indicated grown in MOPS glucose minimal medium after the addition of 750 μM sNO. Blots are representative of two or three independent experiments. (I) Transcription of ivbL (top) and ilvD (bottom) promoters by RNAP in reaction mixtures containing the concentrations of ppGpp indicated and/or 5 μM DksA. The results are representative of three independent experiments.
Organization of the Salmonella branched-chain amino acid biosynthetic pathway and operons. (A) Model of the branched-chain amino acid biosynthetic pathway. Valine is synthesized from pyruvate by the sequential enzymatic actions of acetohydroxy acid synthase I (encoded by ilvBN), ketol-acid reductoisomerase (encoded by ilvC), dihydroxy-acid dehydratase (encoded by ilvD), and transaminase B (encoded by ilvE). Leucine is synthesized from pyruvate by the enzymatic actions of IlvBN, IlvC, IlvD, 2-isopropylmalate synthase (encoded by leuA), 3-isopropylmalate dehydratase (encoded by leuCD), and 3-isopropylmalate dehydrogenase (encoded by leuB). Isoleucine is synthesized from threonine by the enzymatic actions of threonine dehydratase (encoded by ilvA), acetohydroxy acid synthase II (or AHAS II, encoded by ilvGM), IlvC, IlvD, and IlvE. (B) Genetic organization of branched-chain amino acid biosynthesis genes. Genes are depicted as block arrows; small arrows represent promoters. ilvX, ivbL, and leuL encode leader peptides. Download FIG S4, PDF file, 2.9 MB (2.9MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of pyruvate supplementation on the recovery of salmonellae from nitrosative stress. Growth of wild-type (wt) and ΔrelA mutant salmonellae in MOPS glucose minimal medium supplemented with 100 μg/ml pyruvate (Pyr). Selected samples were challenged with 750 μM spermine NONOate (sNO). ctrl, control. Download FIG S5, PDF file, 0.5 MB (575.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The translation of antinitrosative defenses is dependent on ppGpp and branched-chain amino acids.
The flavohemoglobin Hmp is the most potent antinitrosative defense yet described in salmonellae (12, 35). We examined whether RelA-derived ppGpp and branched-chain amino acids affect the expression of hmpA. A cocktail of all 20 amino acids or branched-chain amino acids, in this order of importance, enhanced the amount of Hmp translated in NO-treated salmonellae (Fig. 4A). NO-treated ΔrelA mutant salmonellae expressed lower Hmp concentrations than did wild-type controls, perhaps explaining why ΔrelA mutant salmonellae still recover more slowly than wild-type controls in the presence of all 20 amino acids or branched-chain amino acids (Fig. 2A; Fig. S2E). Wild-type salmonellae grown in glucose required longer times and expressed smaller amounts of Hmp than controls grown in all 20 amino acids or branched-chain amino acids. In contrast to wild-type salmonellae, ΔrelA mutant bacteria grown in glucose minimal medium barely expressed Hmp 2 h after NO treatment (Fig. 4A). The inability of ΔrelA mutant salmonellae to express Hmp cannot be explained by defects in hmpA transcription (Fig. 4B).
FIG 4 .
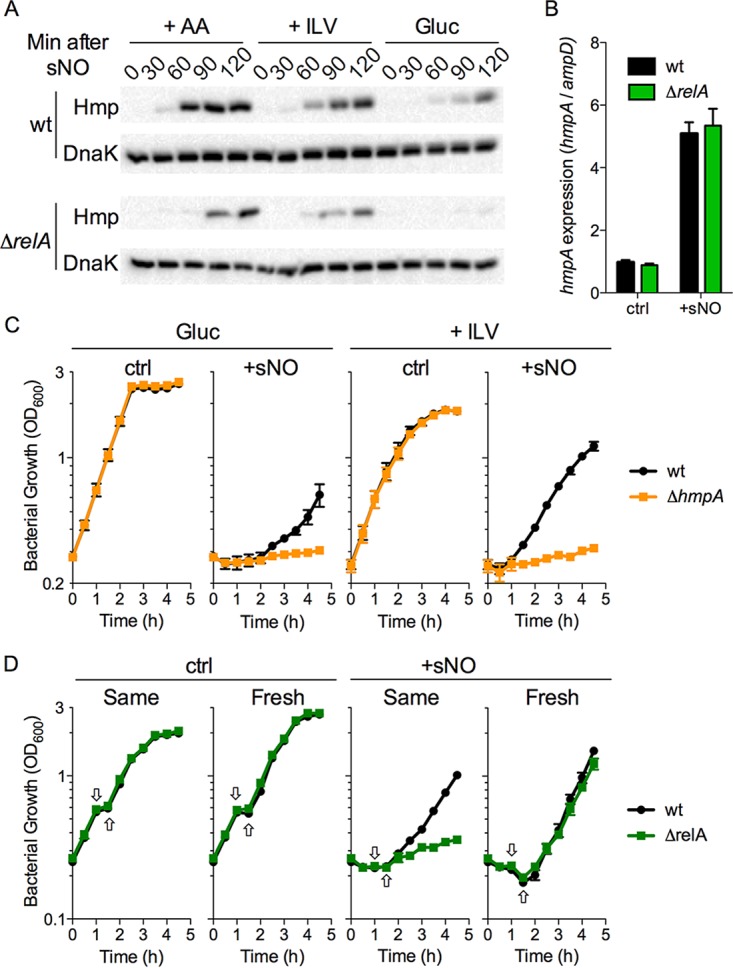
RelA aids the translation of the antinitrosative defense Hmp in salmonellae experiencing nitrosative stress. (A) Western blot analysis of Hmp-3×FLAG in wild-type (wt) or ΔrelA mutant salmonellae grown in MOPS glucose minimal medium (Gluc) after the addition of 750 μM spermine NONOate (sNO). Where indicated, the medium was supplemented with all 20 amino acids (+AA) or branched-chain amino acids (+ILV). Blots are representative of five independent experiments. (B) Transcription of hmpA mRNA in salmonellae grown in MOPS glucose minimal medium 60 min after treatment with 750 μM sNO. (C) Growth of wild-type and ΔhmpA mutant salmonellae in M9 glucose minimal medium. Selected samples were treated with 750 μM spermine NONOate (sNO). Where noted, the medium was supplemented with branched-chain amino acids (+ILV). (D) Growth of wild-type and ΔrelA mutant salmonellae in M9 glucose minimal medium. One hour after the addition of 750 μM sNO, bacteria were collected by centrifugation (down arrow) and the pellets were resuspended in either the same medium (up arrow) or fresh M9 medium lacking sNO. n = 4 to 6. Data are shown as the mean value ± the standard error of the mean. ctrl, control.
As expected (12), ΔhmpA mutant salmonellae were hypersusceptible to the bacteriostatic activity of NO (Fig. 4C). However, in contrast to ΔrelA mutant salmonellae (Fig. 2A), the extreme susceptibility of ΔhmpA mutant salmonellae to nitrosative stress was not ameliorated by branched-chain amino acids (Fig. 4C). Cumulatively, these investigations suggest that defective expression of NO-consuming Hmp could contribute to the hypersusceptibility of ΔrelA mutant salmonellae to NO. Consistent with this idea, ΔrelA mutant salmonellae recovered from NO toxicity as well as wild-type controls upon switching to fresh glucose minimal medium (Fig. 4D).
Branched-chain amino acid biosynthesis and RelA-derived ppGpp support Salmonella pathogenesis.
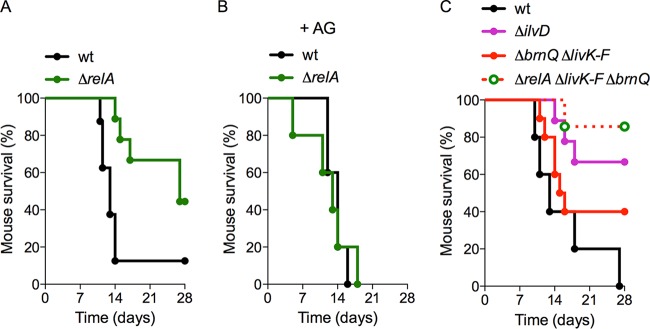
Our data here demonstrate that ppGpp enhances the recovery of salmonellae from NO-associated cytotoxicity, in part by reestablishing branched-chain amino acid biosynthesis. We found that ΔrelA mutant salmonellae are attenuated in this murine model of infection (P < 0.01) (Fig. 5A). To determine if the attenuation of ΔrelA mutant salmonellae is due to NO-dependent immunity, the drinking water of infected mice was supplemented with the iNOS inhibitor aminoguanidine. ΔrelA mutant salmonellae became as virulent (P = 0.937) as wild-type controls in aminoguanidine-treated C3H/HeN mice (Fig. 5B).
FIG 5 .
Importance of relA and ilvD in the pathogenesis of salmonellae. (A to C) C3H/HeN mice were infected p.o. with 5 × 106 CFU of the Salmonella strains indicated. Murine survival was monitored for 28 days. The drinking water in panel B was treated with a 2.5% solution of the iNOS inhibitor aminoguanidine (AG). n = 5 to 10 mice. Log rank analysis results: wild type versus ΔrelA mutant, P < 0.01; wild type versus ΔilvD mutant, P < 0.01; wild type versus ΔlivD-F ΔbrnQ mutant, P = 0.141; wild type versus ΔrelA ΔlivD-F ΔbrnQ mutant, P < 0.001. No statistically significant difference between the wild type and the ΔrelA mutant in AG-treated C3H/HeN mice was found (P = 0.937).
Salmonella is equipped with two specialized branched-chain amino acid uptake systems, LIV-I and LIV-II, encoded by livKHMGF (and livJ) and brnQ, respectively (Fig. S6) (36, 37). To determine if either branched-chain amino acid biosynthesis or import is important for Salmonella pathogenesis, C3H/HeN mice were infected orally (p.o.) with wild-type or ΔilvD or ΔlivKHMGF ΔbrnQ mutant salmonellae (Fig. 5C). Wild-type and ΔlivKHMGF ΔbrnQ mutant salmonellae killed C3H/HeN mice with similar kinetics (P = 0.141) (Fig. 5C). In contrast, ΔilvD mutant salmonellae were attenuated (P < 0.01) (Fig. 5C). These data point to an important role for branched-chain amino acid biosynthesis, not import, in Salmonella pathogenesis. Nonetheless, a combination of ΔrelA and ΔlivKHMGF ΔbrnQ mutations greatly attenuated salmonellae (P < 0.001) (Fig. 5C), suggesting that the LIV-I and LIV-II systems can partially complement the defects of ΔrelA mutant salmonellae to maintain valine biosynthesis.
Model of branched-chain amino acid specialized transport systems. (A) The LIV-I system is an ABC-type, high-affinity, ATP-dependent transporter composed of the proteins LivJKHGMF. The LIV-II system, which is formed by the single protein BrnQ, has 12 transmembrane domains and a lower affinity for branched-chain amino acids than the LIV-I system and is energized by the proton motive force (PMF). OM, outer membrane; Peri, periplasm; IM, inner membrane. (B) The LIV-I system is encoded by the livKHGMF operon and the livJ gene, whereas the LIV-II system is encoded by brnQ. Download FIG S6, PDF file, 1.7 MB (1.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
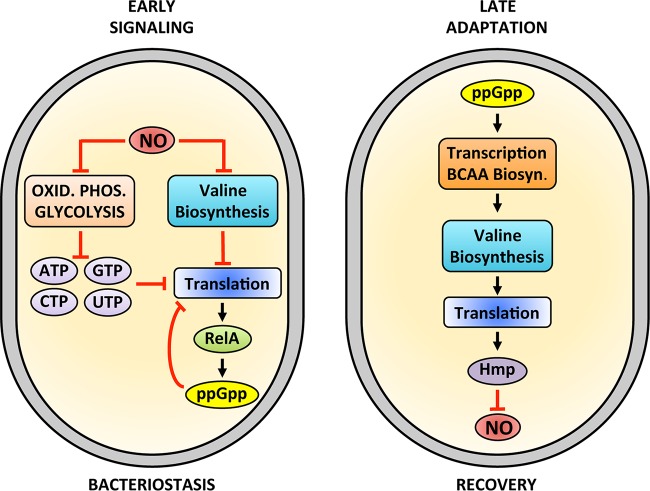
The NTPs ATP, GTP, CTP, and UTP power cellular energetics, serve as building blocks for DNA replication and RNA transcription, regulate key steps in translation and cell division, and are substrates in lipid and peptidoglycan biosynthesis. The nucleotide alarmone ppGpp inhibits many of these processes (16, 27). Thus, the massive depletion of NTPs, along with a simultaneous burst of ppGpp, likely contributes to the bacteriostasis recorded in NO-treated salmonellae (Fig. 6). Global reprogramming of nucleotide metabolism may allow salmonellae to avoid deleterious damage of DNA, lipids, and proteins by reactive nitrogen species. In addition to blocking numerous cellular processes, ppGpp offers a feedback mechanism to reestablish Salmonella homeostasis, as this alarmone activates branched-chain amino acid biosynthesis, which is a prerequisite for the translation of antinitrosative defenses.
FIG 6 .
Integrated model of the role of nucleotide metabolism and branched-chain amino acids in the adaptation of salmonellae to nitrosative stress. Early signaling events in response to nitrosative stress. NO inhibits respiration and glycolysis. The depletion of ATP that follows the NO-mediated inhibition of substrate level and oxidative phosphorylation leads to drops in GTP, CTP, and UTP. NO also inhibits valine biosynthesis. In addition, drops in valine stimulate the accumulation of unaminoacylated tRNAVal, stimulating the synthesis of ppGpp by RelA associated with ribosomes. Depletion of NTPs and valine and synthesis of ppGpp inhibit translation and bacterial growth. Late adaptive responses by which ppGpp aids in the recovery of salmonellae from nitrosative stress are shown on the right. The ppGpp produced by RelA activates the transcription of branched-chain amino acid (BCAA) biosynthetic genes, restoring valine levels that are needed for the translation of the NO-consuming flavohemoprotein Hmp.
Global transcriptional analyses had already indicated that NO elicits the stringent response (38). The RNAP-binding protein DksA is a bona fide sensor of reactive nitrogen species that coordinates the transcription of several Salmonella antinitrosative defenses (39–41). The phenotypes of relA spoT mutants had also suggested roles for (p)ppGpp in the adaptation of salmonellae to nitrosative stress (31). Here, we demonstrate fast accumulation of high concentrations of ppGpp in salmonellae responding to NO. This nucleotide activates the expression of valine biosynthesis as part of the adaptation that enables the translation of NO detoxification systems (Fig. 6). The indirect role ppGpp plays in the translation of Hmp protein acts in concert with the transcriptional derepression of hmpA initiated by nitrosylated NsrR (42).
As GTP is the most abundant and preferred substrate of RelA (18), amino acid starvation mostly induces pppGpp synthesis. The phosphatase activity of GppA is responsible for the nearly equimolar amounts of ppGpp and pppGpp synthesized in amino acid-starving bacteria (this study and reference 33). In contrast to classical nutritional starvation, RelA appears to favor ppGpp synthesis in NO-treated salmonellae. The depletion of the GTP pool could make GDP the more abundant substrate for RelA in NO-treated salmonellae. However, salmonellae exposed to low concentrations of NO still overwhelmingly produced ppGpp, despite abundant GTP pools in the cell. Investigations to identify the mechanism by which NO selectively induces ppGpp are beyond the scope of the present work. Regardless of the mechanism, the predominant production of ppGpp could have major implications for Salmonella physiology, as this nucleotide is more potent than its pppGpp analog (17).
GTPases are major targets of (p)ppGpp (27). These nucleotides can inhibit GTPases by indirectly depleting GTP or through direct competitive binding with GTP. Ribosomal GTPases such as EF-Tu, EF-G, EF-Ts, RF3, IF2, and CgtA/Obg can bind (p)ppGpp (43–46). The accumulation of 2 mM ppGpp coupled with a steep depletion of GTP (from ~1 to 0.22 mM) could drastically reduce, if not temporarily eliminate, translation. Rapid and thorough inhibition of translation by ppGpp could preserve small amounts of aminoacylation in tRNA pools in amino acid-starved bacteria. As seen in ΔrelA mutant Escherichia coli for a variety of amino acids (47), ΔrelA mutant salmonellae suffered a greater depletion of valine after NO treatment than did wild-type controls. Moreover, (p)ppGpp activates the hydrolysis of ribosomal proteins and antitoxin modules by Lon protease (48). Amino acids released in this process could counteract NO-mediated valine starvation. Together, these independent mechanisms may explain why NO-treated ΔrelA mutant salmonellae do not translate Hmp as quickly or to the same levels as wild-type controls, even when all amino acids are readily available.
Hydrolysis of ppGpp is greatly diminished in salmonellae experiencing high fluxes of nitrosative stress. Hydrolysis of (p)ppGpp by SpoT relies on energy provided by the proton motive force (49), which is inhibited upon the nitrosylation of terminal cytochromes in the electron transport chain (50). Thus, repression of respiration by NO could prevent the hydrolysis of ppGpp by SpoT. Inhibition of ATP synthesis may also explain the overall depletion of NTPs in NO-treated salmonellae (Fig. 6).
Stationary-phase salmonellae undergoing nitrosative stress require methionine and lysine for recovery (this study and reference 15), while exponentially growing cells require branched-chain amino acids. We find it curious that salmonellae display specific amino acid auxotrophies, depending on the growth phase at which they are exposed to NO. Methionine and lysine are derived from the tricarboxylic acid (TCA) cycle intermediate succinyl coenzyme A. The TCA cycle is poorly expressed in stationary-phase salmonellae (51), a situation that can be further aggravated by the NO-dependent modification of aconitase [4Fe-4S] cluster and lipoamide-dependent lipoamide dehydrogenase catalytic cysteine residues (15, 52). NO also prevents the expression of frdABCD and sdhCDAB in the reductive branch of the TCA cycle in stationary-phase, but not log-phase, salmonellae (15, 51). These mechanisms might explain why the addition of lysine and methionine is inconsequential to exponentially growing salmonellae but helps stationary-phase bacteria recover from nitrosative stress. Nonetheless, NO seems to put pressure on lysine, methionine, and branched-chain amino acid biosynthetic pathways during Salmonella infection in mice. MetD, LIV-I, and LIV-II transport systems help salmonellae overcome shortages of methionine and branched-chain amino acids during periods of nitrosative stress in vertebrate hosts, especially if the generation of these amino acids is hampered by mutations in biosynthetic genes or relA (this study and reference 15). Taken together, these investigations suggest that Salmonella must resist the antimicrobial activity of NO in periods of both active and inactive growth.
Branched-chain amino acids enable immunity (53). Dietary restrictions of branched-chain amino acids increase the susceptibility of mice to salmonellae (54). Human mononuclear phagocytes use branched-chain aminotransferase 1 to generate TCA cycle intermediates to support iNOS expression in response to lipopolysaccharide (55). Given the reliance of the immune system on branched-chain amino acids and the fact that host-derived NO induces branched-chain amino acid auxotrophies in salmonellae, we were surprised to find that the LIV-I and LIV-II transport systems appear to be dispensable for Salmonella pathogenesis. Nonetheless, LIV-I and LIV-II contribute to the pathogenesis of ΔrelA mutant salmonellae. It is possible that in the absence of the ppGpp-mediated upregulation in the biosynthesis of branched-chain amino acids, scavenging of branched-chain amino acids becomes an important mechanism by which salmonellae resist the antimicrobial activity of iNOS.
Dinitrosyl-iron complexes in the [4Fe-4S] cluster of dihydroxy-acid dehydratase induce functional auxotrophies for branched-chain amino acids (14). Repair of the [Fe-4S] cluster of dihydroxy-acid dehydratase is a major aspect of the regeneration of branched-chain amino acid biosynthesis. According to this model, we noted that nitrosative stress diminishes the valine pools of salmonellae. Our investigations also add a new perspective to this model and suggest that ppGpp-mediated activation of the transcription of ilvB-encoded acetohydroxy acid synthase I is a previously unknown aspect that facilitates the recovery of salmonellae from nitrosative stress. Our data suggest that ppGpp acts in concert with DksA for activation of the ivbL and ilvD promoters, an observation that is consistent with the auxotrophies of dksA and relA spoT mutants for leucine and valine (56). Salmonellae experiencing nitrosative stress translate ilvB faster than hmpA, suggesting that this enteropathogen prioritizes amino acid biosynthesis over the enzymatic detoxification of NO. Amino acid biosynthesis genes are enriched for starvation-resistant codons, whose cognate tRNAs maintain significant levels of aminoacylation during amino acid starvation (57). However, valine codons are not notable for resistance to starvation, and neither the ilvB nor the hmpA gene is enriched in starvation-resistant codons (Table S1). Additional regulation by small RNAs and the RNA-binding protein Hfq could contribute to the delayed translation of hmpA (58, 59).
ilvB and hmpA codon frequencies. Download TABLE S1, PDF file, 0.1 MB (67.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Salmonellae employ (p)ppGpp in the regulation of numerous virulence programs. Poor transcription of genes encoding SPI1 and SPI2 type III secretion systems likely contributes to the attenuation of ΔrelA ΔspoT mutant salmonellae and their inability to invade epithelial cells or grow intracellularly (30). As is the case in other pathogenic bacteria (29), ΔrelA single mutant salmonellae have minor phenotypes, as illustrated by the fact that ΔrelA mutant salmonellae can invade HeLa cells and replicate intracellularly in J774 macrophages (not shown). This has led to the idea that (p)ppGpp synthesized by either RelA or SpoT is sufficient for most aspects of bacterial pathogenesis. The investigations presented here have identified a unique role for RelA in the antinitrosative defenses of salmonellae that cannot be compensated by SpoT. Our work sheds light on the specialized roles (p)ppGpp and (p)ppGpp synthetases can play in bacterial pathogenesis.
In summary, salmonellae undergoing nitrosative stress suffer radical changes in their nucleotide pools. Because nucleotides serve as energy sources, substrates for macromolecule synthesis, and signaling molecules, changes in nucleotide metabolism will have a major impact on the response of salmonellae to NO. As part of the nucleotide metabolic reprogramming, ppGpp generated by RelA enhances the expression of antinitrosative defenses in salmonellae by activating valine biosynthesis, which fuels the translation of antinitrosative defenses.
MATERIALS AND METHODS
Bacterial strains.
S. enterica serovar Typhimurium 14028s and its derivatives were maintained on LB (Lennox broth). Genetic manipulations of salmonellae were made with the λ Red recombinase system as previously described (60, 61). Mutations were transduced between strains with P22 phage (62), and antibiotic resistance genes were excised by FLP-mediated recombination (63). Wild-type and relA point mutant alleles were moved into the relA locus of ΔrelA mutant Salmonella strain AV0663 with the λ Red recombinase system. Tables S2 to S4 list the primers, plasmids, and strains used in the course of our investigations.
Primers used in this study. Download TABLE S2, PDF file, 0.04 MB (47.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Plasmids used in this study. Download TABLE S3, PDF file, 0.03 MB (34.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Strains used in this study. Download TABLE S4, PDF file, 0.1 MB (70.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Minimal media.
Salmonella strains were grown either in MOPS minimal medium [40 mM MOPS buffer (pH 7.2), 4 mM Tricine, 2 mM K2HPO4, 10 μM FeSO4 ⋅ 7H2O, 9.5 mM NH4Cl, 276 μM K2SO4, 500 nM CaCl2, 50 mM NaCl, 525 pM MgCl2, 2.9 nM (NH4)6Mo7O24 ⋅ 4H2O, 400 nM H3BO3, 30 nM CoCl2, 9.6 nM CuSO4, 80.8 nM MnCl2, 9.74 nM ZnSO4] (64) supplemented with 0.4% glucose (22.2 mM) or in M9 minimal medium (49.31 mM Na2HPO4, 22.04 mM KH2PO4, 8.56 mM NaCl, 0.997 mM MgSO4, 135.16 μM CaCl2) supplemented with 11.1 mM glucose and 10 μM FeSO4 ⋅ 7H2O. Where indicated, defined amino acid mixtures (each amino acid at 40 μg/ml) were added to the minimal media.
32P-labeled nucleotide pools.
Nucleotides were examined as originally described by Cashel, with minor modifications (65). Briefly, salmonellae grown overnight in MOPS glucose minimal medium supplemented with all amino acids and 2 mM K2HPO4 were diluted 1:100 in fresh medium supplemented with 0.4 mM K2HPO4. The cultures were grown to early exponential phase (optical density at 600 nm [OD600] of ~0.2). One-milliliter culture aliquots were labeled with 10 μCi of 32P-labeled orthophosphate for approximately 2.5 doubling times (roughly 1 h to an OD600 of 0.5). Salmonellae were treated with NaOH (vehicle control for the NO donors), spermine NONOate, PAPA NONOate, serine hydroxamate, or tetracycline before 0.4 ml of ice-cold 50% formic acid was added to the cultures. Extracts were incubated on ice for at least 20 min before the specimens were centrifuged at 16,000 × g for 5 min. A 5- or 10-μl volume of ice-cold extracts was spotted along the bottom of polyethyleneimine-cellulose TLC plates (20 by 20 cm; EDM Millipore, Billerica, MA). The spots were air dried, and the TLC plates were put into a chamber containing 1.25 M K2HPO4, pH 3.4. To resolve all NTPs, 0.9 M K2HPO4, pH 3.4, was used as the solvent. 32P-labeled nucleotides in the TLC plates were visualized with phosphor screens and a phosphorimager (Bio-Rad, Hercules, CA), and relative nucleotide levels were quantified with the ImageJ software (NIH, Rockville, MD).
Luciferase ATP assay.
Intracellular pools of ATP were quantified with the luciferase-based ATP determination kit (Molecular Probes, Eugene, OR). Briefly, wild-type salmonellae grown to early exponential phase (OD600 of 0.2 to 0.4) were challenged for 5 min with 750 μM spermine NONOate in MOPS glucose minimal medium supplemented with each amino acid at 40 μg/ml. Aliquots were set aside for serial 10-fold plate dilutions, and 0.5-ml samples of cultures were thoroughly mixed with 0.6 ml of ice-cold, freshly prepared 380 mM formic acid containing 17 mM EDTA. Samples stored at −80°C were centrifuged for 1 min at 16,000 × g. Supernatants were diluted 25-fold into 100 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) buffer, pH 7.4. Ten-microliter volumes of samples or ATP standards were mixed with 90 μl of reaction master mix (8.9 ml of water, 500 μl of 20× buffer, 500 μl of 10 mM d-luciferin, 100 μl of 100 mM dithiothreitol [DTT], 2.5 μl of 5-mg/ml firefly luciferase). Luminescence was recorded in an LMax 1.1L luminometer, and the data were analyzed with SoftMax Pro software (Molecular Devices, Sunnyvale, CA). ATP concentrations in the lysates were calculated by linear regression using known standards. The intracellular concentration of ATP was calculated by taking into account the ATP concentrations in the lysates, the number of CFU per milliliter, and an estimated bacterial cell volume of 1 fl.
NO recovery assays.
Salmonellae grown overnight in either MOPS or M9 glucose minimal medium were subcultured 1:100 into the same medium and grown until they reached early exponential phase. The cultures were diluted to an OD600 of 0.2, and selected specimens were challenged with 750 μM spermine NONOate. The OD600 was recorded in a 96-well microtiter plate every 30 min for 4.5 h. The time required for each culture to double the initial OD600 was calculated by exponential regression. Where indicated, stationary-phase salmonellae grown overnight in LB broth were diluted in MOPS glucose minimal medium and challenged with spermine NONOate.
Liquid chromatography (LC)-mass spectrometry (MS) of amino acid pools.
Amino acids were extracted from 1010 Salmonella cells/ml of ice-cold lysis buffer (5:3:2 ratio of methanol-acetonitrile-water [Fisher Scientific, Pittsburgh, PA] containing a 3 μM concentration of a mixture of amino acid standards [Cambridge Isotope Laboratories, Inc., Tewksbury, MA]). Samples were vortexed for 30 min at 4°C in the presence of 1-mm glass beads. Insoluble proteins and lipids were pelleted by centrifugation at 12,000 × g for 10 min at 4°C. The supernatants were collected and dried with a SpeedVac concentrator. The pellets resuspended in 0.1% formic acid were analyzed in a Thermo Vanquish ultrahigh-performance liquid chromatography (UHPLC) device coupled online to a Thermo Q Exactive mass spectrometer. The UHPLC-MS methods and data analysis approaches used were described previously (66).
Total RNA isolation, cDNA synthesis, and quantitative real-time (qRT)-PCR.
Salmonellae were grown to early exponential phase in MOPS glucose minimal medium. Bacterial cultures were challenged with 750 μM spermine NONOate for 0, 30, 60, or 120 min, and 5-ml aliquots were incubated on ice for at least 10 min after being mixed with 1 ml of ice-cold RNA stabilization solution (95% ethanol, 5% aqueous phenol). The specimens were centrifuged at 16,000 × g for 10 min at 4°C, and the pellets were saved at −80°C until extraction. Total RNA was extracted and purified with the High Pure RNA Isolation kit (Roche, Basel, Switzerland). RNA samples were stored at −80°C until cDNA synthesis. Briefly, 1 μg of RNA was mixed with 1.3 μl of 10 mM random DNA hexamers in a final volume of 16.5 μl. The RNA solution was heated to 70°C for 5 min and then cooled for 5 min at 4°C. An 8.5-μl volume of master mix (5 μl of 5× Moloney murine leukemia virus [MMLV] buffer, 2.5 μl of 4 mM each deoxynucleoside triphosphate, 0.5 μl of RNasin, 0.5 μl of MMLV reverse transcriptase) was added to each sample, and cDNA was synthesized at 42°C for 1 h. The amounts of cDNA in the samples were quantified by real-time PCR. Briefly, 25-μl reaction mixtures were prepared in 96-well Hard-Shell plates in a CFX Connect real-time system with Bio-Rad CFX Manager 3.1 software (Bio-Rad, Hercules, CA). Each reaction mixture contained 5 μl of water, 12.5 μl of 2× SYBR Select master mix (Applied Biosystems, Thermo Fisher, Waltham, MA), 1 μl of each gene-specific primer at 10 μM, 0.5 μl of gene-specific Probe (IDT, Coralville, IA) at 10 μM, and 5 μl of cDNA or gene-specific PCR product. The quantification cycle was calculated by exponential regression by using PCR product standards. Gene expression was normalized to the internal housekeeping gene ampD (67).
In vitro transcription.
pivbL and pivlD PCR templates were purified with the GeneJET gel extraction kit (Thermo Scientific). RNA synthesized in in vitro transcription reactions was labeled with [α-32P]UTP after incubation of 1 nM template DNA, 5 nM E. coli RNA polymerase (holoenzyme; NEB), and 5 μM DksA in reaction buffer (40 mM HEPES [pH 7.4]; 2 mM MgCl2; 60 mM potassium glutamate; 0.1% NP-40; 200 μM each ATP, GTP, and UTP; 8 U of RiboLock RNase inhibitor [Thermo Scientific]; 1 mM DTT) in the presence or absence of ppGpp (40). DksA was purified as previously described (41). The specimens were incubated at 37°C for 10 min, and reactions were terminated by heating at 70°C for 10 min. Products of in vitro transcription reactions were separated on 7 M urea–6% polyacrylamide gels and visualized by phosphorimaging.
Western blotting.
Salmonella strains expressing 3×FLAG epitope-tagged alleles were grown in MOPS glucose or M9 minimal medium to an OD600 of 0.2 to 0.5. Bacterial cells were challenged with 750 μM spermine NONOate. At regular time intervals after the addition of spermine NONOate, 1-ml samples were mixed with 111 μl of ice-cold 100% trichloroacetic acid and the specimens were placed on ice for at least 10 min. Trichloroacetic acid-treated samples were centrifuged at 16,000 × g for 10 min, and pellets were stored at −80°C until analysis. Pellets were resuspended with either 10 mM Tris buffer (pH 7.4) or phosphate-buffered saline containing 50 mM NaOH. Samples were mixed with 6× Laemmli loading buffer (416 mM sodium dodecyl sulfate [SDS], 0.9 mM bromophenol blue, 47% [vol/vol] glycerol, 60 mM Tris [pH 6.8]) containing 5% β-mercaptoethanol, boiled for 10 min, and loaded onto reducing 10% polyacrylamide gels. Proteins were separated by SDS-PAGE (85 V for 25 min, followed by 110 V for 50 min) in SDS running buffer. Proteins were transferred onto nitrocellulose membranes with transfer buffer (50 mM Tris base, 40 mM glycine, 1 mM SDS, 20% [vol/vol] methanol) at 25 V for 30 min on a semidry transfer apparatus (Bio-Rad, Hercules, CA). Membranes were blocked overnight at 4°C in 5% skim milk in TBST buffer (25 mM Tris [pH 7.6], 150 mM NaCl, 0.1% [vol/vol] Tween 20). Membranes were then probed with 1:500 anti-FLAG or 1:2,942 anti-DnaK mouse antibody diluted in 5% skim milk in TBST buffer while gently rocking for 2 h at room temperature. After washing with TBST buffer, membranes were probed for 1 h at room temperature with 1:4,000 goat anti-mouse horseradish peroxidase-labeled secondary antibody prepared in 5% skim milk in TBST buffer. Chemiluminescence signals were visualized after the addition of horseradish peroxidase detection solution (GE Healthcare Life Sciences, Pittsburgh, PA) on a Gel Doc (Bio-Rad, Hercules, CA).
Murine infections.
C3H/HeN mice were housed and bred at the University of Colorado Denver Animal Care and Use Committee guidelines. Six- to 8-week-old mice were infected by oral gavage with approximately 5 × 106 CFU of either wild-type or mutant salmonellae. Murine survival was monitored daily for 28 days. Sick mice were humanely euthanized by CO2-mediated asphyxiation, followed by cervical dislocation. For selected studies, the drinking water was treated daily with a 2.5% solution of the iNOS inhibitor aminoguanidine.
Statistical analysis.
The Student t test, the Mann-Whitney t test, one-way analysis of variance (ANOVA), two-way ANOVA, and log rank analyses were performed with GraphPad Prism version 5.0b. Differences were considered significant when the P value was <0.05.
ACKNOWLEDGMENTS
We appreciate the technical comments and thoughtful discussions kindly given by Michael Cashel. We thank the University of Colorado Denver Mass Spectrometry Core for analysis of amino acids.
These studies were funded by VA merit grant BX0002073, NIH R01 AI54959, T32 AI052066, F31 AI118223, and the Burroughs Wellcome Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Fitzsimmons LF, Liu L, Kim J-S, Jones-Carson J, Vázquez-Torres A. 2018. Salmonella reprograms nucleotide metabolism in its adaptation to nitrosative stress. mBio 9:e00211-18. https://doi.org/10.1128/mBio.00211-18.
REFERENCES
- 1.Schreiber F, Lynn DJ, Houston A, Peters J, Mwafulirwa G, Finlay BB, Brinkman FS, Hancock RE, Heyderman RS, Dougan G, Gordon MA. 2011. The human transcriptome during nontyphoid Salmonella and HIV coinfection reveals attenuated NFκB-mediated inflammation and persistent cell cycle disruption. J Infect Dis 204:1237–1245. doi: 10.1093/infdis/jir512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takem EN, Roca A, Cunnington A. 2014. The association between malaria and non-typhoid Salmonella bacteraemia in children in sub-Saharan Africa: a literature review. Malar J 13:400. doi: 10.1186/1475-2875-13-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorman SE, Picard C, Lammas D, Heyne K, van Dissel JT, Baretto R, Rosenzweig SD, Newport M, Levin M, Roesler J, Kumararatne D, Casanova JL, Holland SM. 2004. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet 364:2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- 4.de Jong R, Altare F, Haagen IA, Elferink DG, Boer T, van Breda Vriesman PJ, Kabel PJ, Draaisma JM, van Dissel JT, Kroon FP, Casanova JL, Ottenhoff TH. 1998. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science 280:1435–1438. doi: 10.1126/science.280.5368.1435. [DOI] [PubMed] [Google Scholar]
- 5.Wen SC, Best E, Nourse C. 2017. Non-typhoidal Salmonella infections in children: review of literature and recommendations for management. J Paediatr Child Health 53:936–941. doi: 10.1111/jpc.13585. [DOI] [PubMed] [Google Scholar]
- 6.Crump JA, Medalla FM, Joyce KW, Krueger AL, Hoekstra RM, Whichard JM, Barzilay EJ, Emerging Infections Program NWG . 2011. Antimicrobial resistance among invasive nontyphoidal Salmonella enterica isolates in the United States: National Antimicrobial Resistance Monitoring System, 1996 to 2007. Antimicrob Agents Chemother 55:1148–1154. doi: 10.1128/AAC.01333-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ercis S, Erdem B, Hasçelik G, Gür D. 2006. Nalidixic acid resistance in Salmonella strains with decreased susceptibility to ciprofloxacin isolated from humans in Turkey. Jpn J Infect Dis 59:117–119. [PubMed] [Google Scholar]
- 8.Tacconelli E. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics, p 1–7. World Health Organization, Geneva, Switzerland: http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/. [Google Scholar]
- 9.Vazquez-Torres A, Jones-Carson J, Mastroeni P, Ischiropoulos H, Fang FC. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J Exp Med 192:227–236. doi: 10.1084/jem.192.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacMicking J, Xie QW, Nathan C. 1997. Nitric oxide and macrophage function. Annu Rev Immunol 15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 11.Song M, Husain M, Jones-Carson J, Liu L, Henard CA, Vázquez-Torres A. 2013. Low-molecular-weight thiol-dependent antioxidant and antinitrosative defences in Salmonella pathogenesis. Mol Microbiol 87:609–622. doi: 10.1111/mmi.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bang IS, Liu L, Vazquez-Torres A, Crouch ML, Stamler JS, Fang FC. 2006. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J Biol Chem 281:28039–28047. doi: 10.1074/jbc.M605174200. [DOI] [PubMed] [Google Scholar]
- 13.Henard CA, Vázquez-Torres A. 2011. Nitric oxide and Salmonella pathogenesis. Front Microbiol 2:84. doi: 10.3389/fmicb.2011.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hyduke DR, Jarboe LR, Tran LM, Chou KJ, Liao JC. 2007. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc Natl Acad Sci U S A 104:8484–8489. doi: 10.1073/pnas.0610888104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC. 2011. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar Typhimurium. Cell Host Microbe 10:33–43. doi: 10.1016/j.chom.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 17.Mechold U, Potrykus K, Murphy H, Murakami KS, Cashel M. 2013. Differential regulation by ppGpp versus pppGpp in Escherichia coli. Nucleic Acids Res 41:6175–6189. doi: 10.1093/nar/gkt302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sy J. 1974. Reversibility of the pyrophosphoryl transfer from ATP to GTP by Escherichia coli stringent factor. Proc Natl Acad Sci U S A 71:3470–3473. doi: 10.1073/pnas.71.9.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haseltine WA, Block R. 1973. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci U S A 70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agirrezabala X, Fernández IS, Kelley AC, Cartón DG, Ramakrishnan V, Valle M. 2013. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep 14:811–816. doi: 10.1038/embor.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem 266:5980–5990. [PubMed] [Google Scholar]
- 22.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. 2004. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117:57–68. doi: 10.1016/S0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 23.Mechold U, Murphy H, Brown L, Cashel M. 2002. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J Bacteriol 184:2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. 2013. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross W, Sanchez-Vazquez P, Chen AY, Lee JH, Burgos HL, Gourse RL. 2016. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell 62:811–823. doi: 10.1016/j.molcel.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pizer LI, Merlie JP. 1973. Effect of serine hydroxamate on phospholipid synthesis in Escherichia coli. J Bacteriol 114:980–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 28.Kanjee U, Ogata K, Houry WA. 2012. Direct binding targets of the stringent response alarmone (p)ppGpp. Mol Microbiol 85:1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x. [DOI] [PubMed] [Google Scholar]
- 29.Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. 2010. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev 74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pizarro-Cerdá J, Tedin K. 2004. The bacterial signal molecule, ppGpp, regulates Salmonella virulence gene expression. Mol Microbiol 52:1827–1844. doi: 10.1111/j.1365-2958.2004.04122.x. [DOI] [PubMed] [Google Scholar]
- 31.Henard CA, Vázquez-Torres A. 2012. DksA-dependent resistance of Salmonella enterica serovar Typhimurium against the antimicrobial activity of inducible nitric oxide synthase. Infect Immun 80:1373–1380. doi: 10.1128/IAI.06316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Atkinson GC, Tenson T, Hauryliuk V. 2011. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keasling JD, Bertsch L, Kornberg A. 1993. Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc Natl Acad Sci U S A 90:7029–7033. doi: 10.1073/pnas.90.15.7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Epelbaum S, LaRossa RA, VanDyk TK, Elkayam T, Chipman DM, Barak Z. 1998. Branched-chain amino acid biosynthesis in Salmonella typhimurium: a quantitative analysis. J Bacteriol 180:4056–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones-Carson J, Husain M, Liu L, Orlicky DJ, Vázquez-Torres A. 2016. Cytochrome bd-dependent bioenergetics and antinitrosative defenses in Salmonella pathogenesis. mBio 7:e02052-16. doi: 10.1128/mBio.02052-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahmanian M, Claus DR, Oxender DL. 1973. Multiplicity of leucine transport systems in Escherichia coli K-12. J Bacteriol 116:1258–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guardiola J, De Felice M, Klopotowski T, Iaccarino M. 1974. Multiplicity of isoleucine, leucine, and valine transport systems in Escherichia coli K-12. J Bacteriol 117:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bourret TJ, Porwollik S, McClelland M, Zhao R, Greco T, Ischiropoulos H, Vázquez-Torres A. 2008. Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS One 3:e1833. doi: 10.1371/journal.pone.0001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crawford MA, Henard CA, Tapscott T, Porwollik S, McClelland M, Vázquez-Torres A. 2016. DksA-dependent transcriptional regulation in Salmonella experiencing nitrosative stress. Front Microbiol 7:444. doi: 10.3389/fmicb.2016.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawford MA, Tapscott T, Fitzsimmons LF, Liu L, Reyes AM, Libby SJ, Trujillo M, Fang FC, Radi R, Vázquez-Torres A. 2016. Redox-active sensing by bacterial DksA transcription factors is determined by cysteine and zinc content. mBio 7:e02161-15. doi: 10.1128/mBio.02161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henard CA, Tapscott T, Crawford MA, Husain M, Doulias PT, Porwollik S, Liu L, McClelland M, Ischiropoulos H, Vázquez-Torres A. 2014. The 4-cysteine zinc finger motif of the RNA polymerase regulator DksA serves as a thiol switch for sensing oxidative and nitrosative stress. Mol Microbiol 91:790–804. doi: 10.1111/mmi.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodenmiller DM, Spiro S. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J Bacteriol 188:874–881. doi: 10.1128/JB.188.3.874-881.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rojas AM, Ehrenberg M, Andersson SG, Kurland CG. 1984. ppGpp inhibition of elongation factors Tu, G and Ts during polypeptide synthesis. Mol Gen Genet 197:36–45. doi: 10.1007/BF00327920. [DOI] [PubMed] [Google Scholar]
- 44.Kihira K, Shimizu Y, Shomura Y, Shibata N, Kitamura M, Nakagawa A, Ueda T, Ochi K, Higuchi Y. 2012. Crystal structure analysis of the translation factor RF3 (release factor 3). FEBS Lett 586:3705–3709. doi: 10.1016/j.febslet.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 45.Buglino J, Shen V, Hakimian P, Lima CD. 2002. Structural and biochemical analysis of the Obg GTP binding protein. Structure 10:1581–1592. doi: 10.1016/S0969-2126(02)00882-1. [DOI] [PubMed] [Google Scholar]
- 46.Mitkevich VA, Ermakov A, Kulikova AA, Tankov S, Shyp V, Soosaar A, Tenson T, Makarov AA, Ehrenberg M, Hauryliuk V. 2010. Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J Mol Biol 402:838–846. doi: 10.1016/j.jmb.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 47.Sørensen MA. 2001. Charging levels of four tRNA species in Escherichia coli Rel+ and Rel− strains during amino acid starvation: a simple model for the effect of ppGpp on translational accuracy. J Mol Biol 307:785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- 48.Christensen SK, Mikkelsen M, Pedersen K, Gerdes K. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci U S A 98:14328–14333. doi: 10.1073/pnas.251327898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tétu C, Dassa E, Boquet PL. 1980. The energy-dependent degradation of guanosine 5′-diphosphate 3′-diphosphate in Escherichia coli. Lack of correlation with ATP levels in vivo and role of the transmembrane proton gradient. Eur J Biochem 103:117–124. doi: 10.1111/j.1432-1033.1980.tb04295.x. [DOI] [PubMed] [Google Scholar]
- 50.Jones-Carson J, Zweifel AE, Tapscott T, Austin C, Brown JM, Jones KL, Voskuil MI, Vázquez-Torres A. 2014. Nitric oxide from IFNγ-primed macrophages modulates the antimicrobial activity of β-lactams against the intracellular pathogens Burkholderia pseudomallei and nontyphoidal Salmonella. PLoS Negl Trop Dis 8:e3079. doi: 10.1371/journal.pntd.0003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Gardner PR, Costantino G, Szabó C, Salzman AL. 1997. Nitric oxide sensitivity of the aconitases. J Biol Chem 272:25071–25076. doi: 10.1074/jbc.272.40.25071. [DOI] [PubMed] [Google Scholar]
- 53.Calder PC. 2006. Branched-chain amino acids and immunity. J Nutr 136:288S–293S. doi: 10.1093/jn/136.1.288S. [DOI] [PubMed] [Google Scholar]
- 54.Petro TM, Bhattacharjee JK. 1981. Effect of dietary essential amino acid limitations upon the susceptibility to Salmonella typhimurium and the effect upon humoral and cellular immune responses in mice. Infect Immun 32:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papathanassiu AE, Ko JH, Imprialou M, Bagnati M, Srivastava PK, Vu HA, Cucchi D, McAdoo SP, Ananieva EA, Mauro C, Behmoaras J. 2017. BCAT1 controls metabolic reprogramming in activated human macrophages and is associated with inflammatory diseases. Nat Commun 8:16040. doi: 10.1038/ncomms16040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown L, Gentry D, Elliott T, Cashel M. 2002. DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol 184:4455–4465. doi: 10.1128/JB.184.16.4455-4465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elf J, Nilsson D, Tenson T, Ehrenberg M. 2003. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 58.Troxell B, Fink RC, Porwollik S, McClelland M, Hassan HM. 2011. The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol 11:236. doi: 10.1186/1471-2180-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hao Y, Updegrove TB, Livingston NN, Storz G. 2016. Protection against deleterious nitrogen compounds: role of σS-dependent small RNAs encoded adjacent to sdiA. Nucleic Acids Res 44:6935–6948. doi: 10.1093/nar/gkw404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc Natl Acad Sci U S A 98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCollister BD, Bourret TJ, Gill R, Jones-Carson J, Vázquez-Torres A. 2005. Repression of SPI2 transcription by nitric oxide-producing, IFNγ-activated macrophages promotes maturation of Salmonella phagosomes. J Exp Med 202:625–635. doi: 10.1084/jem.20050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 64.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cashel M. 1974. Preparation of guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) from Escherichia coli ribosomes. Anal Biochem 57:100–107. doi: 10.1016/0003-2697(74)90056-6. [DOI] [PubMed] [Google Scholar]
- 66.Nemkov T, D’Alessandro A, Hansen KC. 2015. Three-minute method for amino acid analysis by UHPLC and high-resolution quadrupole Orbitrap mass spectrometry. Amino Acids 47:2345–2357. doi: 10.1007/s00726-015-2019-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mühlig A, Behr J, Scherer S, Müller-Herbst S. 2014. Stress response of Salmonella enterica serovar Typhimurium to acidified nitrite. Appl Environ Microbiol 80:6373–6382. doi: 10.1128/AEM.01696-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Effect of NO on the nucleotide pools of salmonellae. (A) TLC autoradiogram of 32P-labeled nucleotides extracted from salmonellae treated for 30 s with increasing concentrations of PAPA NONOate (pNO). (B) Nucleotides were visualized as described for panel A at the times of treatment with 750 μM spermine NONOate (sNO) indicated. (C) Quantification of (ppGpp + pppGpp)/(GTP + ppGpp + pppGpp) ratios from nucleotide autoradiograms from salmonellae treated with either 0.4 mg/ml serine hydroxamate (SHX) or pNO for 90 s, followed by the addition of 70 μg/ml tetracycline (Tet). Samples were collected from 0 to 10 min after the addition of Tet. Ctrl, control. (D) Nucleotides from wild-type (wt) and ΔgppA salmonellae were visualized as described for panel A 5 min after treatment with either 0.4 mg/ml SHX or 750 μM sNO. All autoradiograms are representative of two or three independent experiments. (E) Firefly luciferase quantification of ATP pools in salmonellae left untreated (Untr.) or treated for 5 min with 750 μM sNO. n = 8 or 9. Data are shown as the mean value ± the standard error of the mean. ***, P < 0.001 (determined by Student t test). Download FIG S1, PDF file, 3.9 MB (4MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of amino acids on the recovery of salmonellae from a NO challenge. (A) Growth of wild-type (wt) salmonellae and ΔrelA and ΔrelA mutant salmonellae complemented with the wild-type, D275G, or E335Q relA allele in MOPS minimal medium supplemented with glucose. Where indicated, bacterial cultures were treated with 750 μM spermine NONOate (+sNO). n = 3. Data are shown as the mean value ± the standard error of the mean. (B) The effects of sNO on the growth of salmonellae were also tested in MOPS glucose minimal medium (Gluc) supplemented with the aromatic amino acids (FWY), all amino acids except the aromatic amino acids (AA − FWY), or all 20 amino acids (AA). (C) Growth of salmonellae left untreated (Ctrl.) or challenged with 750 μM sNO in MOPS minimal medium supplemented with glucose or glucose and methionine and lysine (MK), all amino acids except methionine and lysine (AA − MK), or all 20 amino acids (AA). (D) Stationary-phase salmonellae were challenged with 750 μM sNO in MOPS glucose minimal medium supplemented with the branched-chain amino acids (ILV), methionine and lysine (MK), all amino acids except the branched-chain amino acids (AA − ILV), all amino acids except methionine or lysine (AA − MK), or all 20 amino acids (AA). (E, F) The time at which the initial OD600 doubled was calculated from data presented in Fig. 2A or B. *, P < 0.05; ***, P < 0.001 (determined by one-way ANOVA). Download FIG S2, PDF file, 3 MB (3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Intracellular concentrations of amino acids in salmonellae under nitrosative stress. Amino acids were determined by LC-MS (n = 4, box-and-whisker plot) from wild-type (wt) and ΔrelA mutant salmonellae grown to log phase in M9 glucose minimal medium. Where indicated, the bacterial cultures were treated with 750 μM spermine NONOate (+sNO) for 5 min. Isoleucine, leucine, and valine are represented in Fig. 2C. Asparagine, cysteine, glutamine, phenylalanine, and tryptophan were not detected by this method. *, P < 0.05 (determined by Mann-Whitney analysis). Untr., untreated. Download FIG S3, PDF file, 2.3 MB (2.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Organization of the Salmonella branched-chain amino acid biosynthetic pathway and operons. (A) Model of the branched-chain amino acid biosynthetic pathway. Valine is synthesized from pyruvate by the sequential enzymatic actions of acetohydroxy acid synthase I (encoded by ilvBN), ketol-acid reductoisomerase (encoded by ilvC), dihydroxy-acid dehydratase (encoded by ilvD), and transaminase B (encoded by ilvE). Leucine is synthesized from pyruvate by the enzymatic actions of IlvBN, IlvC, IlvD, 2-isopropylmalate synthase (encoded by leuA), 3-isopropylmalate dehydratase (encoded by leuCD), and 3-isopropylmalate dehydrogenase (encoded by leuB). Isoleucine is synthesized from threonine by the enzymatic actions of threonine dehydratase (encoded by ilvA), acetohydroxy acid synthase II (or AHAS II, encoded by ilvGM), IlvC, IlvD, and IlvE. (B) Genetic organization of branched-chain amino acid biosynthesis genes. Genes are depicted as block arrows; small arrows represent promoters. ilvX, ivbL, and leuL encode leader peptides. Download FIG S4, PDF file, 2.9 MB (2.9MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of pyruvate supplementation on the recovery of salmonellae from nitrosative stress. Growth of wild-type (wt) and ΔrelA mutant salmonellae in MOPS glucose minimal medium supplemented with 100 μg/ml pyruvate (Pyr). Selected samples were challenged with 750 μM spermine NONOate (sNO). ctrl, control. Download FIG S5, PDF file, 0.5 MB (575.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Model of branched-chain amino acid specialized transport systems. (A) The LIV-I system is an ABC-type, high-affinity, ATP-dependent transporter composed of the proteins LivJKHGMF. The LIV-II system, which is formed by the single protein BrnQ, has 12 transmembrane domains and a lower affinity for branched-chain amino acids than the LIV-I system and is energized by the proton motive force (PMF). OM, outer membrane; Peri, periplasm; IM, inner membrane. (B) The LIV-I system is encoded by the livKHGMF operon and the livJ gene, whereas the LIV-II system is encoded by brnQ. Download FIG S6, PDF file, 1.7 MB (1.8MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ilvB and hmpA codon frequencies. Download TABLE S1, PDF file, 0.1 MB (67.9KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Primers used in this study. Download TABLE S2, PDF file, 0.04 MB (47.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Plasmids used in this study. Download TABLE S3, PDF file, 0.03 MB (34.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Strains used in this study. Download TABLE S4, PDF file, 0.1 MB (70.7KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.