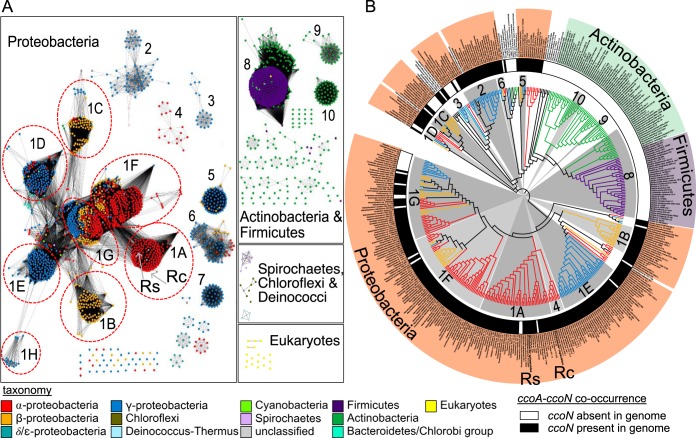
FIG 5 .
The CcoA-like family of putative transporters. (A) Protein similarity network of CcoA-like putative transporters identified in the Uniprot database. Each node (circle) represents a single protein sequence, and each edge (solid line) represents similarity between two proteins (threshold set at an alignment score of 80). Nodes are colored by taxonomy as shown at the bottom, and cluster designations (1 to 10) correspond to Table S4 and Fig. S1 PSN_CcoA.cys, which can be viewed in detail by using Cytoscape software. The locations of the nodes representing CcoA from R. sphaeroides (Rs) and R. capsulatus (Rc) are shown with gray arrows. (B) Evolutionary relationship between CcoA homologues identified in the SEED database. Branch lengths were ignored, and branch points with Shimodaira-Hasegawa scores of <0.5 were deleted. Branches are colored by taxonomy as shown at the bottom. The presence of ccoN (inner circle) in each genome is represented by black shading. Gray shading is used to color those clades that correspond to the numbered clusters (1A to 10) shown in panel A. Corresponding protein IDs are listed in Table S5. The positions of CcoA of R. sphaeroides (Rs) and R. capsulatus (Rc) in the tree are also indicated. Note that ccoA is present in Actinobacteria and Firmicutes (clusters 8 to 10) that are devoid of ccoN (i.e., cbb3-Cox).