Abstract
Artificial intelligence (AI) aims to mimic human cognitive functions. It is bringing a paradigm shift to healthcare, powered by increasing availability of healthcare data and rapid progress of analytics techniques. We survey the current status of AI applications in healthcare and discuss its future. AI can be applied to various types of healthcare data (structured and unstructured). Popular AI techniques include machine learning methods for structured data, such as the classical support vector machine and neural network, and the modern deep learning, as well as natural language processing for unstructured data. Major disease areas that use AI tools include cancer, neurology and cardiology. We then review in more details the AI applications in stroke, in the three major areas of early detection and diagnosis, treatment, as well as outcome prediction and prognosis evaluation. We conclude with discussion about pioneer AI systems, such as IBM Watson, and hurdles for real-life deployment of AI.
Keywords: big data, deep learning, neural network, support vector machine, stroke
Overview of the medical artificial intelligence (AI) research
Recently AI techniques have sent vast waves across healthcare, even fuelling an active discussion of whether AI doctors will eventually replace human physicians in the future. We believe that human physicians will not be replaced by machines in the foreseeable future, but AI can definitely assist physicians to make better clinical decisions or even replace human judgement in certain functional areas of healthcare (eg, radiology). The increasing availability of healthcare data and rapid development of big data analytic methods has made possible the recent successful applications of AI in healthcare. Guided by relevant clinical questions, powerful AI techniques can unlock clinically relevant information hidden in the massive amount of data, which in turn can assist clinical decision making.1–3
In this article, we survey the current status of AI in healthcare, as well as discuss its future. We first briefly review four relevant aspects from medical investigators’ perspectives:
motivations of applying AI in healthcare
data types that have be analysed by AI systems
mechanisms that enable AI systems to generate clinical meaningful results
disease types that the AI communities are currently tackling.
Motivation
The advantages of AI have been extensively discussed in the medical literature.3–5 AI can use sophisticated algorithms to ‘learn’ features from a large volume of healthcare data, and then use the obtained insights to assist clinical practice. It can also be equipped with learning and self-correcting abilities to improve its accuracy based on feedback. An AI system can assist physicians by providing up-to-date medical information from journals, textbooks and clinical practices to inform proper patient care.6 In addition, an AI system can help to reduce diagnostic and therapeutic errors that are inevitable in the human clinical practice.3 4 6–10 Moreover, an AI system extracts useful information from a large patient population to assist making real-time inferences for health risk alert and health outcome prediction.11
Healthcare data
Before AI systems can be deployed in healthcare applications, they need to be ‘trained’ through data that are generated from clinical activities, such as screening, diagnosis, treatment assignment and so on, so that they can learn similar groups of subjects, associations between subject features and outcomes of interest. These clinical data often exist in but not limited to the form of demographics, medical notes, electronic recordings from medical devices, physical examinations and clinical laboratory and images.12
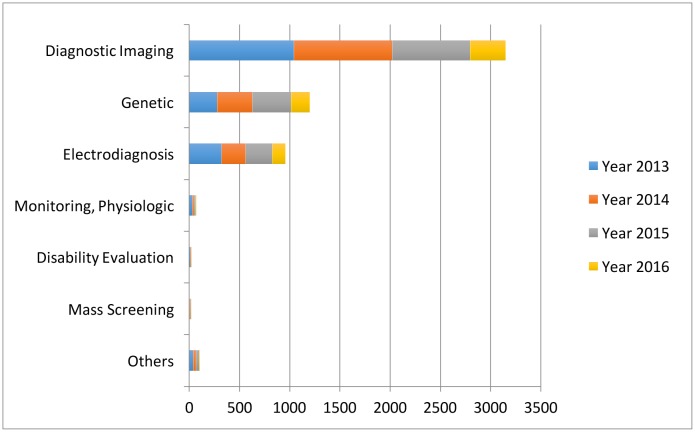
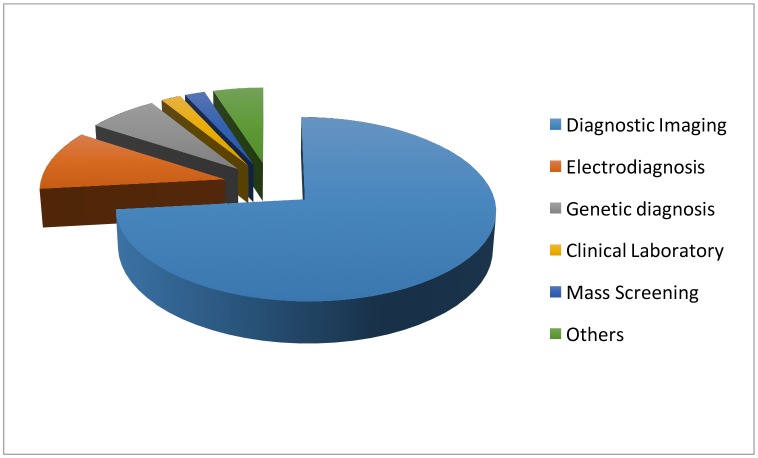
Specifically, in the diagnosis stage, a substantial proportion of the AI literature analyses data from diagnosis imaging, genetic testing and electrodiagnosis (figure 1). For example, Jha and Topol urged radiologists to adopt AI technologies when analysing diagnostic images that contain vast data information.13 Li et al studied the uses of abnormal genetic expression in long non-coding RNAs to diagnose gastric cancer.14 Shin et al developed an electrodiagnosis support system for localising neural injury.15
Figure 1.
The data types considered in the artificial intelligence artificial (AI) literature. The comparison is obtained through searching the diagnosis techniques in the AI literature on the PubMed database.
In addition, physical examination notes and clinical laboratory results are the other two major data sources (figure 1). We distinguish them with image, genetic and electrophysiological (EP) data because they contain large portions of unstructured narrative texts, such as clinical notes, that are not directly analysable. As a consequence, the corresponding AI applications focus on first converting the unstructured text to machine-understandable electronic medical record (EMR). For example, Karakülah et al used AI technologies to extract phenotypic features from case reports to enhance the diagnosis accuracy of the congenital anomalies.16
AI devices
The above discussion suggests that AI devices mainly fall into two major categories. The first category includes machine learning (ML) techniques that analyse structured data such as imaging, genetic and EP data. In the medical applications, the ML procedures attempt to cluster patients’ traits, or infer the probability of the disease outcomes.17 The second category includes natural language processing (NLP) methods that extract information from unstructured data such as clinical notes/medical journals to supplement and enrich structured medical data. The NLP procedures target at turning texts to machine-readable structured data, which can then be analysed by ML techniques.18
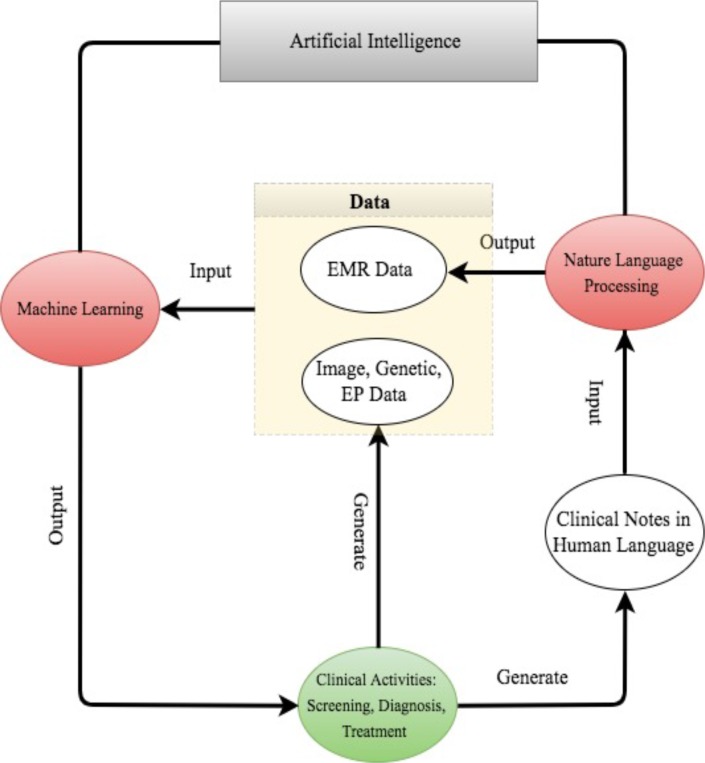
For better presentation, the flow chart in figure 2 describes the road map from clinical data generation, through NLP data enrichment and ML data analysis, to clinical decision making. We comment that the road map starts and ends with clinical activities. As powerful as AI techniques can be, they have to be motivated by clinical problems and be applied to assist clinical practice in the end.
Figure 2.
The road map from clinical data generation to natural language processing data enrichment, to machine learning data analysis, to clinical decision making. EMR, electronic medical record; EP, electrophysiological.
Disease focus
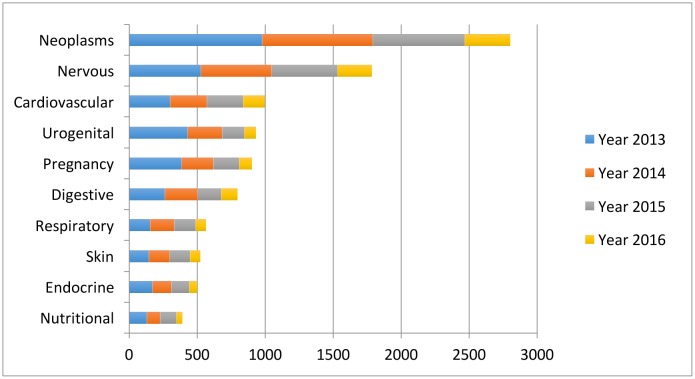
Despite the increasingly rich AI literature in healthcare, the research mainly concentrates around a few disease types: cancer, nervous system disease and cardiovascular disease (figure 3). We discuss several examples below.
Figure 3.
The leading 10 disease types considered in the artificial intelligence (AI) literature. The first vocabularies in the disease names are displayed. The comparison is obtained through searching the disease types in the AI literature on PubMed.
Cancer: Somashekhar et al demonstrated that the IBM Watson for oncology would be a reliable AI system for assisting the diagnosis of cancer through a double-blinded validation study.19 Esteva et al analysed clinical images to identify skin cancer subtypes.20
Neurology: Bouton et al developed an AI system to restore the control of movement in patients with quadriplegia.21 Farina et al tested the power of an offline man/machine interface that uses the discharge timings of spinal motor neurons to control upper-limb prostheses.22
Cardiology: Dilsizian and Siegel discussed the potential application of the AI system to diagnose the heart disease through cardiac image.3 Arterys recently received clearance from the US Food and Drug Administration (FDA) to market its Arterys Cardio DL application, which uses AI to provide automated, editable ventricle segmentations based on conventional cardiac MRI images.23
The concentration around these three diseases is not completely unexpected. All three diseases are leading causes of death; therefore, early diagnoses are crucial to prevent the deterioration of patients’ health status. Furthermore, early diagnoses can be potentially achieved through improving the analysis procedures on imaging, genetic, EP or EMR, which is the strength of the AI system.
Besides the three major diseases, AI has been applied in other diseases as well. Two very recent examples were Long et al, who analysed the ocular image data to diagnose congenital cataract disease,24 and Gulshan et al, who detected referable diabetic retinopathy through the retinal fundus photographs.25
The rest of the paper is organised as follows. In section 2, we describe popular AI devices in ML and NLP; the ML techniques are further grouped into classical techniques and the more recent deep learning. Section 3 focuses on discussing AI applications in neurology, from the three aspects of early disease prediction and diagnosis, treatment, outcome prediction and prognosis evaluation. We then conclude in section 4 with some discussion about the future of AI in healthcare.
The AI devices: ML and NLP
In this section, we review the AI devices (or techniques) that have been found useful in the medial applications. We categorise them into three groups: the classical machine learning techniques,26 the more recent deep learning techniques27 and the NLP methods.28
Classical ML
ML constructs data analytical algorithms to extract features from data. Inputs to ML algorithms include patient ‘traits’ and sometimes medical outcomes of interest. A patient’s traits commonly include baseline data, such as age, gender, disease history and so on, and disease-specific data, such as diagnostic imaging, gene expressions, EP test, physical examination results, clinical symptoms, medication and so on. Besides the traits, patients’ medical outcomes are often collected in clinical research. These include disease indicators, patient’s survival times and quantitative disease levels, for example, tumour sizes. To fix ideas, we denote the jth trait of the ith patient by Xij, and the outcome of interest by Yi.
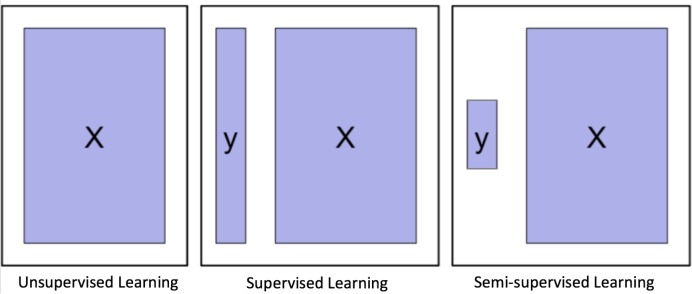
Depending on whether to incorporate the outcomes, ML algorithms can be divided into two major categories: unsupervised learning and supervised learning. Unsupervised learning is well known for feature extraction, while supervised learning is suitable for predictive modelling via building some relationships between the patient traits (as input) and the outcome of interest (as output). More recently, semisupervised learning has been proposed as a hybrid between unsupervised learning and supervised learning, which is suitable for scenarios where the outcome is missing for certain subjects. These three types of learning are illustrated in figure 4.
Figure 4.
Graphical illustration of unsupervised learning, supervised learning and semisupervised learning.
Clustering and principal component analysis (PCA) are two major unsupervised learning methods. Clustering groups subjects with similar traits together into clusters, without using the outcome information. Clustering algorithms output the cluster labels for the patients through maximising and minimising the similarity of the patients within and between the clusters. Popular clustering algorithms include k-means clustering, hierarchical clustering and Gaussian mixture clustering. PCA is mainly for dimension reduction, especially when the trait is recorded in a large number of dimensions, such as the number of genes in a genome-wide association study. PCA projects the data onto a few principal component (PC) directions, without losing too much information about the subjects. Sometimes, one can first use PCA to reduce the dimension of the data, and then use clustering to group the subjects.
On the other hand, supervised learning considers the subjects’ outcomes together with their traits, and goes through a certain training process to determine the best outputs associated with the inputs that are closest to the outcomes on average. Usually, the output formulations vary with the outcomes of interest. For example, the outcome can be the probability of getting a particular clinical event, the expected value of a disease level or the expected survival time.
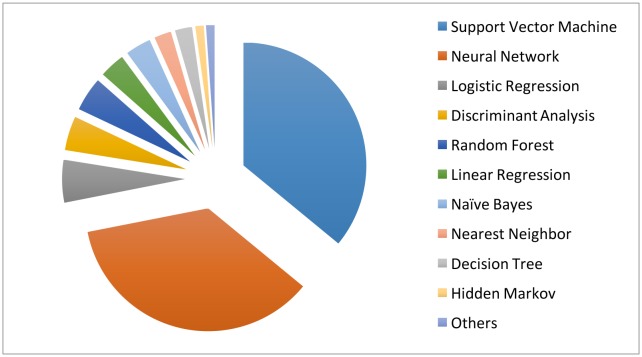
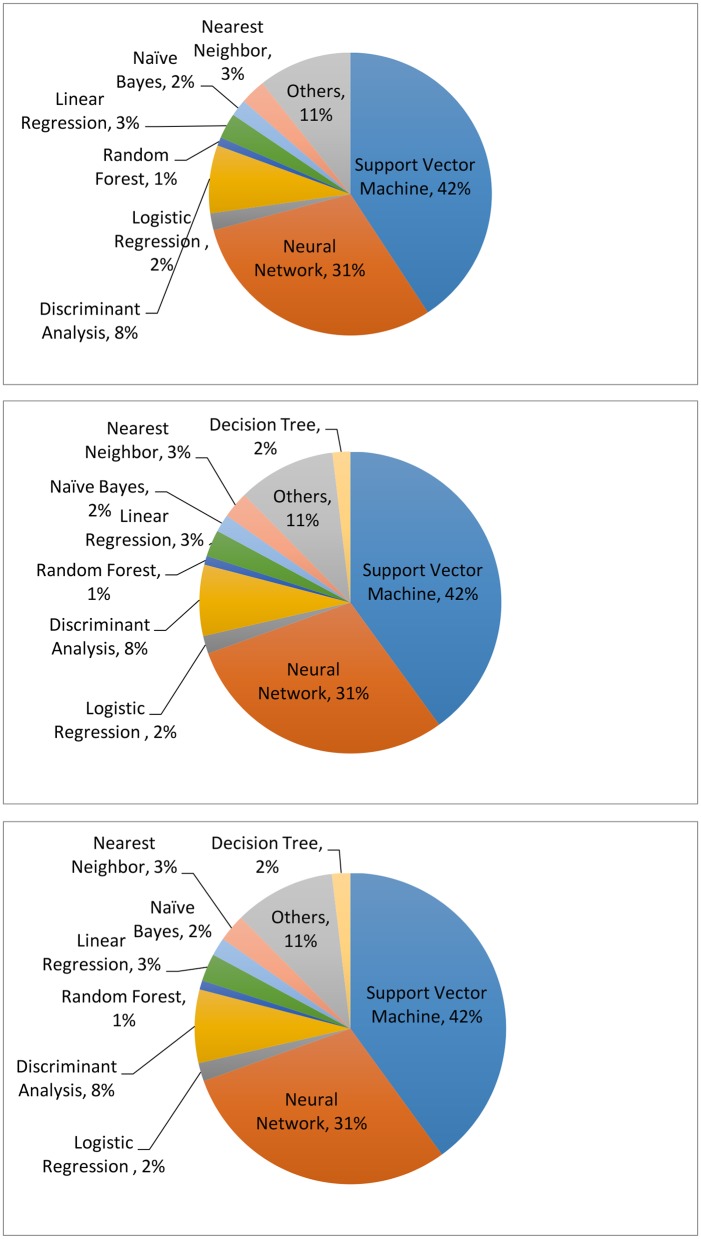
Clearly, compared with unsupervised learning, supervised learning provides more clinically relevant results; hence AI applications in healthcare most often use supervised learning. (Note that unsupervised learning can be used as part of the preprocessing step to reduce dimensionality or identify subgroups, which in turn makes the follow-up supervised learning step more efficient.) Relevant techniques include linear regression, logistic regression, naïve Bayes, decision tree, nearest neighbour, random forest, discriminant analysis, support vector machine (SVM) and neural network.27 Figure 5 displays the popularity of the various supervised learning techniques in medical applications, which clearly shows that SVM and neural network are the most popular ones. This remains the case when restricting to the three major data types (image, genetic and EP), as shown in figure 6.
Figure 5.
The machine learning algorithms used in the medical literature. The data are generated through searching the machine learning algorithms within healthcare on PubMed.
Figure 6.
The machine learning algorithms used for imaging (upper), genetic (middle) and electrophysiological (bottom) data. The data are generated through searching the machine learning algorithms for each data type on PubMed.
Below we will provide more details about the mechanisms of SVM and neural networks, along with application examples in the cancer, neurological and cardiovascular disease areas.
Support vector machine
SVM is mainly used for classifying the subjects into two groups, where the outcome Yi is a classifier: Yi = −1 or 1 represents whether the ith patient is in group 1 or 2, respectively. (The method can be extended for scenarios with more than two groups.) The basic assumption is that the subjects can be separated into two groups through a decision boundary defined on the traits Xij, which can be written as:
where wj is the weight putting on the jth trait to manifest its relative importance on affecting the outcome among the others. The decision rule then follows that if ai >0, the ith patient is classified to group 1, that is, labelling Yi = −1; if ai <0, the patient is classified to group 2, that is, labelling Yi=1. The class memberships are indeterminate for the points with ai=0. See figure 7 for an illustration with , , a1=1, and a2=−1.
Figure 7.
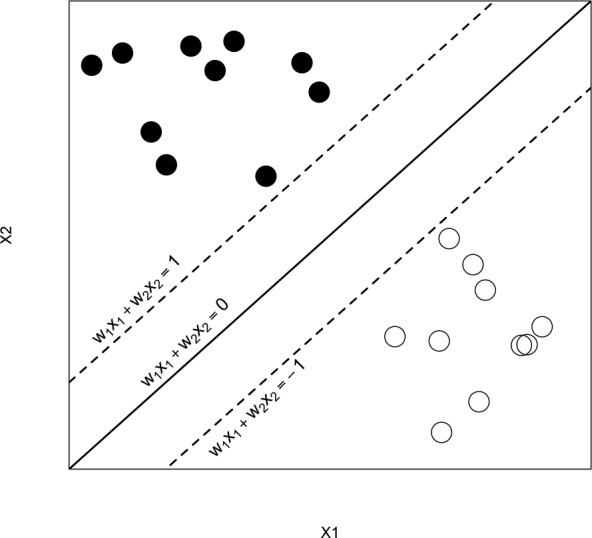
An illustration of the support vector machine.
The training goal is to find the optimal wjs so that the resulting classifications agree with the outcomes as much as possible, that is, with the smallest misclassification error, the error of classifying a patient into the wrong group. Intuitively, the best weights must allow (1) the sign of ai to be the same as Yi so the classification is correct; and (2) |ai| to be far away from 0 so the ambiguity of the classification is minimised. These can be achieved by selecting wjs that minimise a quadratic loss function.29 Furthermore, assuming that the new patients come from the same population, the resulting wjs can be applied to classify these new patients based on their traits.
An important property of SVM is that the determination of the model parameters is a convex optimisation problem so the solution is always global optimum. Furthermore, many existing convex optimisation tools are readily applicable for the SVM implementation. As such, SVM has been extensively used in medical research. For instance, Orrù et al applied SVM to identify imaging biomarkers of neurological and psychiatric disease.30 Sweilam et al reviewed the use of SVM in the diagnosis of cancer.31 Khedher et al used the combination of SVM and other statistical tools to achieve early detection of Alzheimer’s disease.32 Farina et al used SVM to test the power of an offline man/machine interface that controls upper-limb prostheses.22
Neural network
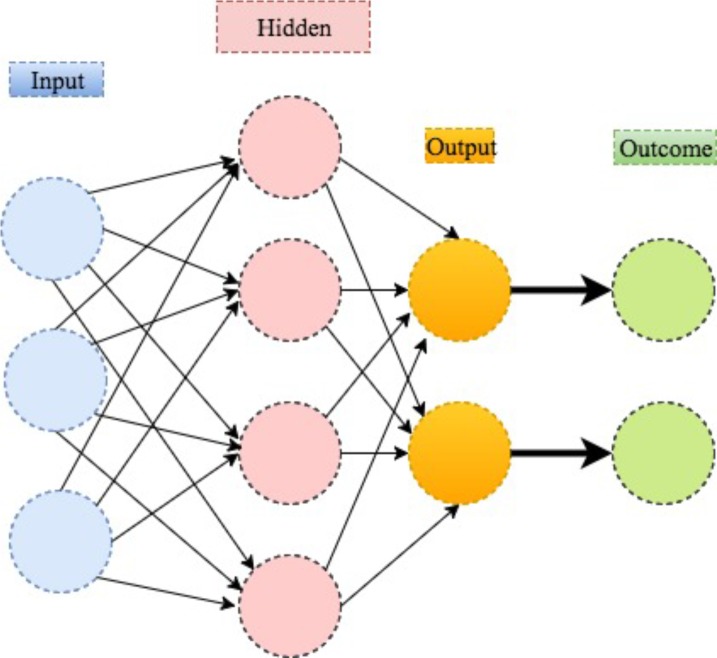
One can think about neural network as an extension of linear regression to capture complex non-linear relationships between input variables and an outcome. In neural network, the associations between the outcome and the input variables are depicted through multiple hidden layer combinations of prespecified functionals. The goal is to estimate the weights through input and outcome data so that the average error between the outcome and their predictions is minimised. We describe the method in the following example.
Mirtskhulava et al used neural network in stroke diagnosis.33 In their analysis, the input variables Xi1, …, Xip are p=16 stroke-related symptoms, including paraesthesia of the arm or leg, acute confusion, vision, problems with mobility and so on. The outcome Yi is binary: Yi=1/0 indicates the ith patient has/does not have stroke. The output parameter of interest is the probability of stroke, ai, which carries the form of
In the above equation, the w10 and w20≠0 guarantee the above form to be valid even when all Xij, fk are 0; the w1l and 2ls are the weights to characterise the relative importance of the corresponding multiplicands on affecting the outcome; the fks and are prespecified functionals to manifest how the weighted combinations influence the disease risk as a whole. A stylised illustration is provided in figure 8.
Figure 8.
An illustration of neural network.
The training goal is to find the weights wij, which minimise the prediction error 2. The minimisation can be performed through standard optimisation algorithms, such as local quadratic approximation or gradient descent optimisation, that are included in both MATLAB and R. If the new data come from the same population, the resulting wij can be used to predict the outcomes based on their specific traits.29
Similar techniques have been used to diagnose cancer by Khan et al, where the inputs are the PCs estimated from 6567 genes and the outcomes are the tumour categories.34 Dheeba et al used neural network to predict breast cancer, with the inputs being the texture information from mammographic images and the outcomes being tumour indicators.35 Hirschauer et al used a more sophisticated neural network model to diagnose Parkinson’s disease based on the inputs of motor, non-motor symptoms and neuroimages.36
Deep learning: a new era of ML
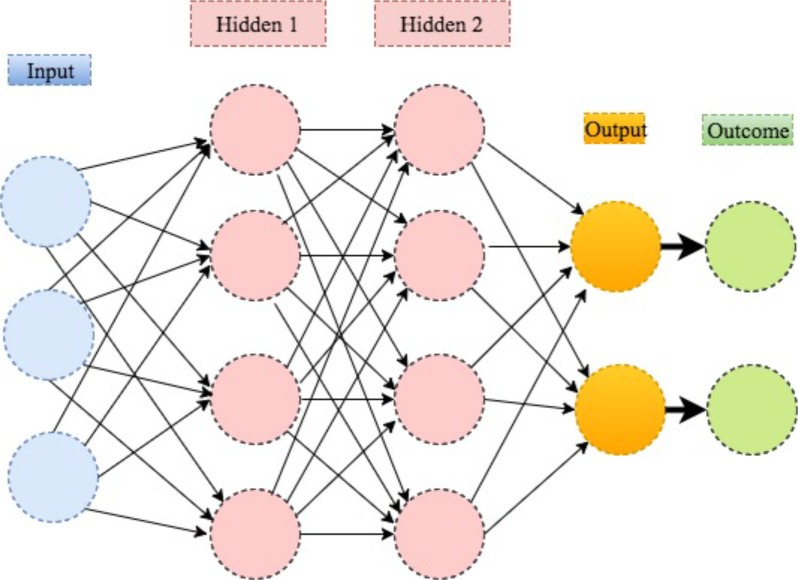
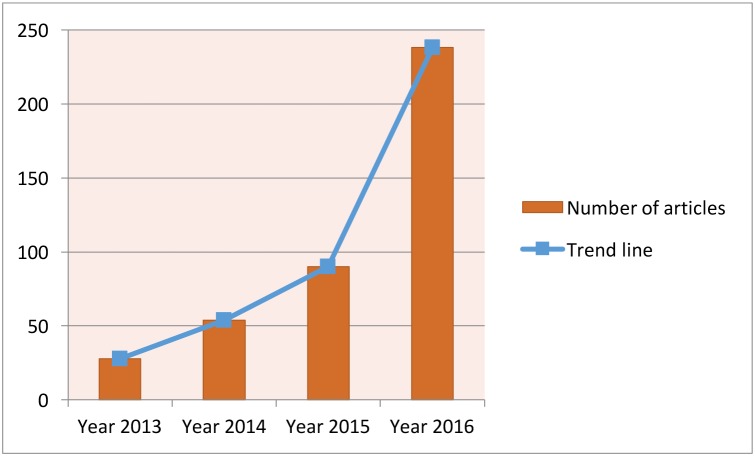
Deep learning is a modern extension of the classical neural network technique. One can view deep learning as a neural network with many layers (as in figure 9). Rapid development of modern computing enables deep learning to build up neural networks with a large number of layers, which is infeasible for classical neural networks. As such, deep learning can explore more complex non-linear patterns in the data. Another reason for the recent popularity of deep learning is due to the increase of the volume and complexity of data.37 Figure 10 shows that the application of deep learning in the field of medical research nearly doubled in 2016. In addition, figure 11 shows that a clear majority of deep learning is used in imaging analysis, which makes sense given that images are naturally complex and high volume.
Figure 9.
An illustration of deep learning with two hidden layers.
Figure 10.
Current trend for deep learning. The data are generated through searching the deep learning in healthcare and disease category on PubMed.
Figure 11.
The data sources for deep learning. The data are generated through searching deep learning in combination with the diagnosis techniques on PubMed.
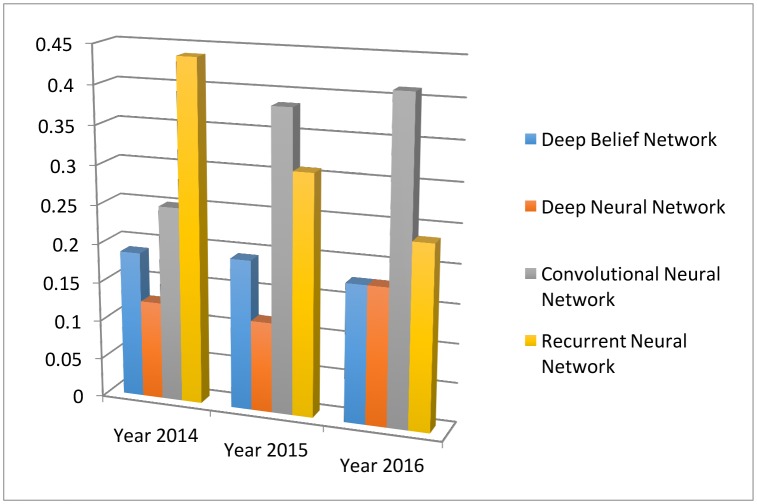
Different from the classical neural network, deep learning uses more hidden layers so that the algorithms can handle complex data with various structures.27 In the medical applications, the commonly used deep learning algorithms include convolution neural network (CNN), recurrent neural network, deep belief network and deep neural network. Figure 12 depicts their trends and relative popularities from 2013 to 2016. One can see that the CNN is the most popular one in 2016.
Figure 12.
The four main deep learning algorithm and their popularities. The data are generated through searching algorithm names in healthcare and disease category on PubMed.
The CNN is developed in viewing of the incompetence of the classical ML algorithms when handling high dimensional data, that is, data with a large number of traits. Traditionally, the ML algorithms are designed to analyse data when the number of traits is small. However, the image data are naturally high-dimensional because each image normally contains thousands of pixels as traits. One solution is to perform dimension reduction: first preselect a subset of pixels as features, and then perform the ML algorithms on the resulting lower dimensional features. However, heuristic feature selection procedures may lose information in the images. Unsupervised learning techniques such as PCA or clustering can be used for data-driven dimension reduction.
The CNN was first proposed and advocated for the high-dimensional image analysis by Lecun et al.38 The inputs for CNN are the properly normalised pixel values on the images. The CNN then transfers the pixel values in the image through weighting in the convolution layers and sampling in the subsampling layers alternatively. The final output is a recursive function of the weighted input values. The weights are trained to minimise the average error between the outcomes and the predictions. The implementation of CNN has been included in popular software packages such as Caffe from Berkeley AI Research,39 CNTK from Microsoft40 and TensorFlow from Google.41
Recently, the CNN has been successfully implemented in the medical area to assist disease diagnosis. Long et al used it to diagnose congenital cataract disease through learning the ocular images.24 The CNN yields over 90% accuracy on diagnosis and treatment suggestion. Esteva et al performed the CNN to identify skin cancer from clinical images.20 The proportions of correctly predicted malignant lesions (ie, sensitivity) and benign lesions (ie, specificity) are both over 90%, which indicates the superior performance of the CNN. Gulshan et al applied the CNN to detect referable diabetic retinopathy through the retinal fundus photographs.25 The sensitivity and specificity of the algorithm are both over 90%, which demonstrates the effectiveness of using the technique on the diagnosis of diabetes. It is worth mentioning that in all these applications, the performance of the CNN is competitive against experienced physicians in the accuracy for classifying both normal and disease cases.
Natural language processing
The image, EP and genetic data are machine-understandable so that the ML algorithms can be directly performed after proper preprocessing or quality control processes. However, large proportions of clinical information are in the form of narrative text, such as physical examination, clinical laboratory reports, operative notes and discharge summaries, which are unstructured and incomprehensible for the computer program. Under this context, NLP targets at extracting useful information from the narrative text to assist clinical decision making.28
An NLP pipeline comprises two main components: (1) text processing and (2) classification. Through text processing, the NLP identifies a series of disease-relevant keywords in the clinical notes based on the historical databases.42 Then a subset of the keywords are selected through examining their effects on the classification of the normal and abnormal cases. The validated keywords then enter and enrich the structured data to support clinical decision making.
The NLP pipelines have been developed to assist clinical decision making on alerting treatment arrangements, monitoring adverse effects and so on. For example, Fiszman et al showed that introducing NLP for reading the chest X-ray reports would assist the antibiotic assistant system to alert physicians for the possible need for anti-infective therapy.43 Miller et al used NLP to automatically monitor the laboratory-based adverse effects.44 Furthermore, the NLP pipelines can help with disease diagnosis. For instance, Castro et al identified 14 cerebral aneurysms disease-associated variables through implementing NLP on the clinical notes.45 The resulting variables are successfully used for classifying the normal patients and the patients with cerebral, with 95% and 86% accuracy rates on the training and validation samples, respectively. Afzal et al implemented the NLP to extract the peripheral arterial disease-related keywords from narrative clinical notes. The keywords are then used to classify the normal and the patients with peripheral arterial disease, which achieves over 90% accuracy.42
AI applications in stroke
Stroke is a common and frequently occurring disease that affects more than 500 million people worldwide. It is the leading cause of death in China and the fifth in North America. Stroke had cost about US$689 billion in medical expenses across the world, causing heavy burden to countries and families.46 47 Therefore, research on prevention and treatment for stroke has great significance. In recent years, AI techniques have been used in more and more stroke-related studies. Below we summarise some of the relevant AI techniques in the three main areas of stroke care: early disease prediction and diagnosis, treatment, as well as outcome prediction and prognosis evaluation.
Early detection and diagnosis
Stroke, for 85% of the time, is caused by thrombus in the vessel called cerebral infarction. However, for lack of judgement of early stroke symptom, only a few patients could receive timely treatment. Villar et al developed a movement-detecting device for early stroke prediction.48 Two ML algorithms — genetic fuzzy finite state machine and PCA — were implemented into the device for the model building solution. The detection process included a human activity recognition stage and a stroke-onset detection stage. Once the movement of the patient is significantly different from the normal pattern, an alert of stroke would be activated and evaluated for treatment as soon as possible. Similarly, Maninini et al proposed a wearable device for collecting data about normal/pathological gaits for stroke prediction.49 The data would be extracted and modelled by hidden Markov models and SVM, and the algorithm could correctly classify 90.5% of the subjects to the right group.
For diagnosis of stroke, neuroimaging techniques, including MRI and CT, are important for disease evaluation. Some studies have tried to apply ML methods to neuroimaging data to assist with stroke diagnosis. Rehme et al used SVM in resting-state functional MRI data, by which endophenotypes of motor disability after stroke were identified and classified.50 SVM can correctly classify patients with stroke with 87.6% accuracy. Griffis et al tried naïve Bayes classification to identify stroke lesion in T1-weighted MRI.51 The result is comparable with human expert manual lesion delineation. Kamnitsas et al tried three-dimensional CNN (3D CNN) for lesion segmentation in multimodel brain MRI.52 They also used fully connected conditional random field model for final postprocessing of the CNN’s soft segmentation maps. Rondina et al analysed stroke anatomical MRI images using Gaussian process regression, and found that the patterns of voxels performed better than lesion load per region as the predicting features.53
ML methods have also been applied to analyse CT scans from patients with stroke. Free-floating intraluminal thrombus may be formed as lesion after stroke, which is difficult to be distinguished with carotid plaque on the CT imaging. Thornhill et al used three ML algorithms to classify these two types by quantitative shape analysis, including linear discriminant analysis, artificial neural network and SVM.54 The accuracy for each method varies between 65.2% and 76.4%.
Treatment
ML has also been applied for predicting and analysing the performance of stroke treatment. As a critical step of emergency measure, the outcome of intravenous thrombolysis (tPA) has strong relationship with the prognosis and survival rate. Bentley et al used SVM to predict whether patients with tPA treatment would develop symptomatic intracranial haemorrhage by CT scan.55 They used whole-brain images as the input into the SVM, which performed better than conventional radiology-based methods. To improve the clinical decision-making process of tPA treatment, Love et al proposed a stroke treatment model by analysing practice guidelines, meta-analyses and clinical trials using Bayesian belief network.56 The model consisted of 56 different variables and three decisions for analysing the procedure of diagnosis, treatment and outcome prediction. Ye et al used interaction trees and subgroup analysis to explore appropriate tPA dosage based on patient characteristics, taking into account both the risk of bleeding and the treatment efficacy.57
Outcome prediction and prognosis evaluation
Many factors can affect stroke prognosis and disease mortality. Compared with conventional methods, ML methods have advantages in improving prediction performance. To better support clinical decision-making process, Zhang et al proposed a model for predicting 3-month treatment outcome by analysing physiological parameters during 48 hours after stroke using logistic regression.58 Asadi et al compiled a database of clinical information of 107 patients with acute anterior or posterior circulation stroke who underwent intra-arterial therapy.59 The authors analysed the data via artificial neural network and SVM, and obtained prediction accuracy above 70%. They also used ML techniques to identify factors influencing outcome in brain arteriovenous malformation treated with endovascular embolisation.60 While standard regression analysis model could only achieve a 43% accuracy rate, their methods worked much better with 97.5% accuracy.
Birkner et al used an optimal algorithm to predict 30-day mortality and obtained more accurate prediction than existing methods.61 Similarly, King et al used SVM to predict stroke mortality at discharge.62 In addition, they proposed the use of the synthetic minority oversampling technique to reduce the stroke outcome prediction bias caused by between-class imbalance among multiple data sets.
Brain images have been analysed to predict the outcome of stroke treatment. Chen et al analysed CT scan data via ML for evaluating the cerebral oedema following hemispheric infarction.63 They built random forest to automatically identify cerebrospinal fluid and analyse the shifts on CT scan, which is more efficient and accurate than conventional methods. Siegel et al extracted functional connectivity from MRI and functional MRI data, and used ridge regression and multitask learning for cognitive deficiency prediction after stroke.64 Hope et al studied the relationship between lesions extracted from MRI images and the treatment outcome via Gaussian process regression model.65 They used the model to predict the severity of cognitive impairments after stroke and the course of recovery over time.
Conclusion and discussion
We reviewed the motivation of using AI in healthcare, presented the various healthcare data that AI has analysed and surveyed the major disease types that AI has been deployed. We then discussed in details the two major categories of AI devices: ML and NLP. For ML, we focused on the two most popular classical techniques: SVM and neural network, as well as the modern deep learning technique. We then surveyed the three major categories of AI applications in stroke care.
A successful AI system must possess the ML component for handling structured data (images, EP data, genetic data) and the NLP component for mining unstructured texts. The sophisticated algorithms then need to be trained through healthcare data before the system can assist physicians with disease diagnosis and treatment suggestions.
The IBM Watson system is a pioneer in this field. The system includes both ML and NLP modules, and has made promising progress in oncology. For example, in a cancer research, 99% of the treatment recommendations from Watson are coherent with the physician decisions.66 Furthermore, Watson collaborated with Quest Diagnostics to offer the AI Genetic Diagnostic Analysis.66 In addition, the system started to make impact on actual clinical practices. For example, through analysing genetic data, Watson successfully identified the rare secondary leukaemia caused by myelodysplastic syndromes in Japan.67
The cloud-based CC-Cruiser in24 can be one prototype to connect an AI system with the front-end data input and the back-end clinical actions. More specifically, when patients come, with their permission, their demographic information and clinical data (images, EP results, genetic results, blood pressure, medical notes and so on) are collected into the AI system. The AI system then uses the patients’ data to come up with clinical suggestions. These suggestions are sent to physicians to assist with their clinical decision making. Feedback about the suggestions (correct or wrong) will also be collected and fed back into the AI system so that it can keep improving accuracy.
Stroke is a chronic disease with acute events. Stroke management is a rather complicated process with a series of clinical decision points. Traditionally clinical research solely focused on a single or very limited clinical questions, while ignoring the continuous nature of stroke management. Taking the advantage of large amount of data with rich information, AI is expected to help with studying much more complicated yet much closer to real-life clinical questions, which then leads to better decision making in stroke management. Recently, researchers have started endeavours along this direction and obtained promising initial results.57
Although the AI technologies are attracting substantial attentions in medical research, the real-life implementation is still facing obstacles. The first hurdle comes from the regulations. Current regulations lack of standards to assess the safety and efficacy of AI systems. To overcome the difficulty, the US FDA made the first attempt to provide guidance for assessing AI systems.68 The first guidance classifies AI systems to be the ‘general wellness products’, which are loosely regulated as long as the devices intend for only general wellness and present low risk to users. The second guidance justifies the use of real-world evidence to access the performance of AI systems. Lastly, the guidance clarifies the rules for the adaptive design in clinical trials, which would be widely used in assessing the operating characteristics of AI systems. Not long after the disclosure of these guidances, Arterys’ medical imaging platform became the first FDA-approved deep learning clinical platform that can help cardiologists to diagnose cardiac diseases.23
The second hurdle is data exchange. In order to work well, AI systems need to be trained (continuously) by data from clinical studies. However, once an AI system gets deployed after initial training with historical data, continuation of the data supply becomes a crucial issue for further development and improvement of the system. Current healthcare environment does not provide incentives for sharing data on the system. Nevertheless, a healthcare revolution is under way to stimulate data sharing in the USA.69 The reform starts with changing the health service payment scheme. Many payers, mostly insurance companies, have shifted from rewarding the physicians by shifting the treatment volume to the treatment outcome. Furthermore, the payers also reimburse for a medication or a treatment procedure by its efficiency. Under this new environment, all the parties in the healthcare system, the physicians, the pharmaceutical companies and the patients, have greater incentives to compile and exchange information. Similar approaches are being explored in China.
Footnotes
Competing interests: None declared.
Provenance and peer review: Commissioned; internally peer reviewed.
Data sharing statement: No additional data are available.
Correction notice: This paper has been corrected since it was published Online First. Owing to a scripting error, some of the publisher names in the references were replaced with ’BMJ Publishing Group'. This only affected the full text version, not the PDF. We have since corrected these errors and the correct publishers have been inserted into the references. Figures 6-9 have also been corrected.
References
- 1. Murdoch TB, Detsky AS. The inevitable application of big data to health care. JAMA 2013;309:1351–2. doi:10.1001/jama.2013.393 [DOI] [PubMed] [Google Scholar]
- 2. Kolker E, Özdemir V, Kolker E. How Healthcare can refocus on its Super-Customers (Patients, n =1) and Customers (Doctors and Nurses) by Leveraging Lessons from Amazon, Uber, and Watson. OMICS 2016;20:329–33. doi:10.1089/omi.2016.0077 [DOI] [PubMed] [Google Scholar]
- 3. Dilsizian SE, Siegel EL. Artificial intelligence in medicine and cardiac imaging: harnessing big data and advanced computing to provide personalized medical diagnosis and treatment. Curr Cardiol Rep 2014;16:441 doi:10.1007/s11886-013-0441-8 [DOI] [PubMed] [Google Scholar]
- 4. Patel VL, Shortliffe EH, Stefanelli M, et al. The coming of age of artificial intelligence in medicine. Artif Intell Med 2009;46:5–17. doi:10.1016/j.artmed.2008.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jha S, Topol EJ. Adapting to Artificial Intelligence: radiologists and pathologists as information specialists. JAMA 2016;316:2353–4. doi:10.1001/jama.2016.17438 [DOI] [PubMed] [Google Scholar]
- 6. Pearson T. How to replicate Watson hardware and systems design for your own use in your basement. 2011. https://www.ibm.com/developerworks/community/blogs/InsideSystemStorage/entry/ibm_watson_how_to_build_your_own_watson_jr_in_your_basement7?lang=en (accessed 1 Jun 2017).
- 7. Weingart SN, Wilson RM, Gibberd RW, et al. Epidemiology of medical error. BMJ 2000;320:774–7. doi:10.1136/bmj.320.7237.774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Graber ML, Franklin N, Gordon R. Diagnostic error in internal medicine. Arch Intern Med 2005;165:1493–9. doi:10.1001/archinte.165.13.1493 [DOI] [PubMed] [Google Scholar]
- 9. Winters B, Custer J, Galvagno SM, et al. Diagnostic errors in the intensive care unit: a systematic review of autopsy studies. BMJ Qual Saf 2012;21:894–902. doi:10.1136/bmjqs-2012-000803 [DOI] [PubMed] [Google Scholar]
- 10. Lee CS, Nagy PG, Weaver SJ, et al. Cognitive and system factors contributing to diagnostic errors in radiology. AJR Am J Roentgenol 2013;201:611–7. doi:10.2214/AJR.12.10375 [DOI] [PubMed] [Google Scholar]
- 11. Neill DB. Using artificial intelligence to improve hospital inpatient care. IEEE Intell Syst 2013;28:92–5. doi:10.1109/MIS.2013.51 [Google Scholar]
- 12. Administration UFaD. Guidance for industry: electronic source data in clinical investigations. 2013. https://www.fda.gov/downloads/drugs/guidances/ucm328691.pdf (accessed 1 Jun 2017).
- 13. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563–77. doi:10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li CY, Liang GY, Yao WZ, et al. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric Cancer. Int J Oncol 2016;48:1965–76. doi:10.3892/ijo.2016.3407 [DOI] [PubMed] [Google Scholar]
- 15. Shin H, Kim KH, Song C, et al. Electrodiagnosis support system for localizing neural injury in an upper limb. J Am Med Inform Assoc 2010;17:345–7. doi:10.1136/jamia.2009.001594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karakülah G, Dicle O, Koşaner O, et al. Computer based extraction of phenoptypic features of human congenital anomalies from the digital literature with natural language processing techniques. Stud Health Technol Inform 2014;205:570–4. [PubMed] [Google Scholar]
- 17. Darcy AM, Louie AK, Roberts LW. Machine Learning and the Profession of Medicine. JAMA 2016;315:551–2. doi:10.1001/jama.2015.18421 [DOI] [PubMed] [Google Scholar]
- 18. Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA 2011;306:848–55. doi:10.1001/jama.2011.1204 [DOI] [PubMed] [Google Scholar]
- 19. Somashekhar SP, Kumarc R, Rauthan A, et al. Abstract S6-07: double blinded validation study to assess performance of IBM artificial intelligence platform, Watson for oncology in comparison with manipal multidisciplinary tumour board ? first study of 638 breast Cancer cases. Cancer Res 2017;77(4 Suppl):S6-07 doi:10.1158/1538-7445.SABCS16-S6-07 [Google Scholar]
- 20. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin Cancer with deep neural networks. Nature 2017;542:115–8. doi:10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bouton CE, Shaikhouni A, Annetta NV, et al. Restoring cortical control of functional movement in a human with quadriplegia. Nature 2016;533:247–50. doi:10.1038/nature17435 [DOI] [PubMed] [Google Scholar]
- 22. Farina D, Vujaklija I, Sartori M, et al. Man/machine interface based on the discharge timings of spinal motor neurons after targeted muscle reinnervation. Nat Biomed Eng 2017;1:0025 doi:10.1038/s41551-016-0025 [Google Scholar]
- 23. Marr B. First FDA approval for clinical Cloud-Based Deep Learning in Healthcare. 2017. https://www.forbes.com/sites/bernardmarr/2017/01/20/first-fda-approval-for-clinical-cloud-based-deep-learning-in-healthcare/#7a0ed8dc161c (accessed 1 Jun 2017).
- 24. Long E, Lin H, Liu Z, et al. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts, 2017. [Google Scholar]
- 25. Gulshan V, Peng L, Coram M, et al. Development and Validation of a Deep Learning Algorithm for detection of Diabetic Retinopathy in retinal fundus photographs. JAMA 2016;316:2402–10. doi:10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 26. James G, Witten D, Hastie T, et al. An introduction to Statistical Learning with applications in R. First Edition: Springer, 2013. [Google Scholar]
- 27. Goodfellow I, Bengio Y, Courville A. Deep Learning. First Edition: The MIT Press, 2016. [Google Scholar]
- 28. Kantor P. Foundations of statistical natural language processing: MIT Press, 1999:91–2. [Google Scholar]
- 29. Bishop CM, ed Pattern recognition and machine Learning (Information Science and Statistics, 2007. [Google Scholar]
- 30. Orrù G, Pettersson-Yeo W, Marquand AF, et al. Using support Vector Machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neurosci Biobehav Rev 2012;36:1140–52. doi:10.1016/j.neubiorev.2012.01.004 [DOI] [PubMed] [Google Scholar]
- 31. Sweilam NH, Tharwat AA, Abdel Moniem NK, Moniem NKA. Support vector machine for diagnosis Cancer disease: a comparative study. Egyptian Informatics Journal 2010;11:81–92. doi:10.1016/j.eij.2010.10.005 [Google Scholar]
- 32. Khedher L, Ram?rez J, G?rriz JM, et al. Early diagnosis of Alzheimer?s disease based on partial least squares, principal component analysis and support vector machine using segmented MRI images. Neurocomputing 2015;151:139–50. doi:10.1016/j.neucom.2014.09.072 [Google Scholar]
- 33. : Mirtskhulava L, Wong J, Al-Majeed S, Pearce G, et al. ; Artificial Neural Network Model in Stroke diagnosis. modelling and simulation (UKSim), 2015 17th UKSim-AMSS International Conference on: IEEE, 2015. [Google Scholar]
- 34. Khan J, Wei JS, Ringnér M, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med 2001;7:673–9. doi:10.1038/89044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dheeba J, Albert Singh N, Tamil Selvi S. Computer-aided detection of breast Cancer on mammograms: a swarm intelligence optimized wavelet neural network approach. J Biomed Inform 2014;49:45–52. doi:10.1016/j.jbi.2014.01.010 [DOI] [PubMed] [Google Scholar]
- 36. Hirschauer TJ, Adeli H, Buford JA. Computer-Aided diagnosis of Parkinson’s Disease Using Enhanced Probabilistic Neural Network. J Med Syst 2015;39:179 doi:10.1007/s10916-015-0353-9 [DOI] [PubMed] [Google Scholar]
- 37. Ravi D, Wong C, Deligianni F, et al. Deep Learning for Health Informatics. IEEE J Biomed Health Inform 2017;21:4–21. doi:10.1109/JBHI.2016.2636665 [DOI] [PubMed] [Google Scholar]
- 38. Lecun Y, Bottou L, Bengio Y, et al. Gradient-based learning applied to document recognition. Proc IEEE Inst Electr Electron Eng 1998;86:2278–324. doi:10.1109/5.726791 [Google Scholar]
- 39. Research BA. Caffe. 2017. http://caffe.berkeleyvision.org/ (accessed 1 Jun 2017).
- 40. Seide F, Agarwal A, eds CNTK: microsoft’s Open-Source Deep-Learning Toolkit. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016. [Google Scholar]
- 41. Abadi M, Agarwal A, Barham P, et al. ; TensorFlow: large-scale Machine Learning on heterogeneous distributed Systems, 2016. [Google Scholar]
- 42. Afzal N, Sohn S, Abram S, et al. Mining peripheral arterial disease cases from narrative clinical notes using natural language processing. J Vasc Surg 2017;65:1753–61. doi:10.1016/j.jvs.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fiszman M, Chapman WW, Aronsky D, et al. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc 2000;7:593–604. doi:10.1136/jamia.2000.0070593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miller TP, Li Y, Getz KD, et al. Using electronic medical record data to report laboratory adverse events. Br J Haematol 2017;177:283–6. doi:10.1111/bjh.14538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Castro VM, Dligach D, Finan S, et al. Large-scale identification of patients with cerebral aneurysms using natural language processing. Neurology 2017;88:164–8. doi:10.1212/WNL.0000000000003490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saenger AK, Christenson RH. Stroke biomarkers: progress and challenges for diagnosis, prognosis, differentiation, and treatment. Clin Chem 2010;56:21–33. doi:10.1373/clinchem.2009.133801 [DOI] [PubMed] [Google Scholar]
- 47. Heeley E, Anderson CS, Huang Y, et al. Role of health insurance in averting economic hardship in families after acute stroke in China. Stroke 2009;40:2149–56. doi:10.1161/STROKEAHA.108.540054 [DOI] [PubMed] [Google Scholar]
- 48. Villar JR, González S, Sedano J, et al. Improving human activity recognition and its application in early stroke diagnosis. Int J Neural Syst 2015;25:1450036 doi:10.1142/S0129065714500361 [DOI] [PubMed] [Google Scholar]
- 49. Mannini A, Trojaniello D, Cereatti A, et al. A machine Learning Framework for Gait classification using inertial sensors: application to Elderly, Post-Stroke and Huntington’s Disease Patients. Sensors 2016;16:134 doi:10.3390/s16010134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rehme AK, Volz LJ, Feis DL, et al. Identifying neuroimaging markers of Motor Disability in acute stroke by machine Learning Techniques. Cereb Cortex 2015;25:3046–56. doi:10.1093/cercor/bhu100 [DOI] [PubMed] [Google Scholar]
- 51. Griffis JC, Allendorfer JB, Szaflarski JP. Voxel-based gaussian naïve Bayes classification of ischemic stroke lesions in individual T1-weighted MRI scans. J Neurosci Methods 2016;257:97–108. doi:10.1016/j.jneumeth.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kamnitsas K, Ledig C, Newcombe VF, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61–78. doi:10.1016/j.media.2016.10.004 [DOI] [PubMed] [Google Scholar]
- 53. Rondina JM, Filippone M, Girolami M, et al. Decoding post-stroke motor function from structural brain imaging. Neuroimage Clin 2016;12:372–80. doi:10.1016/j.nicl.2016.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thornhill RE, Lum C, Jaberi A, et al. Can shape analysis differentiate free-floating internal carotid artery Thrombus from atherosclerotic plaque in patients evaluated with CTA?for stroke or transient ischemic attack? Acad Radiol 2014;21:345–54. doi:10.1016/j.acra.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 55. Bentley P, Ganesalingam J, Carlton Jones AL, et al. Prediction of stroke thrombolysis outcome using CT brain machine learning. Neuroimage Clin 2014;4:635–40. doi:10.1016/j.nicl.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Love A, Arnold CW, El-Saden S, et al. Unifying acute stroke treatment guidelines for a bayesian belief network. Stud Health Technol Inform 2013;192:1012. [PubMed] [Google Scholar]
- 57. Ye H, Shen H, Dong Y, et al. Using Evidence-Based medicine through Advanced Data Analytics to work toward a National Standard for Hospital-based acute ischemic Stroke treatment. Mainland China, 2017. [Google Scholar]
- 58. Zhang Q, Xie Y, Ye P, et al. Acute ischaemic stroke prediction from physiological time series patterns. Australas Med J 2013;6:280–6. doi:10.4066/AMJ.2013.1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Asadi H, Dowling R, Yan B, et al. Machine learning for outcome prediction of acute ischemic stroke post intra-arterial therapy. PLoS One 2014;9:e88225 doi:10.1371/journal.pone.0088225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Asadi H, Kok HK, Looby S, et al. Outcomes and complications after endovascular treatment of Brain Arteriovenous Malformations: a Prognostication Attempt using artificial Intelligence. World Neurosurg 2016;96:562–9. doi:10.1016/j.wneu.2016.09.086 [DOI] [PubMed] [Google Scholar]
- 61. Birkner MD, Kalantri S, Solao V, et al. Creating diagnostic scores using data-adaptive regression: an application to prediction of 30-day mortality among stroke victims in a rural hospital in India. Ther Clin Risk Manag 2007;3:475–84. [PMC free article] [PubMed] [Google Scholar]
- 62. Ho KC, Speier W, El-Saden S, et al. Predicting discharge mortality after acute ischemic stroke using balanced data. AMIA Annu Symp Proc 2014;2014:1787–96. [PMC free article] [PubMed] [Google Scholar]
- 63. Chen Y, Dhar R, Heitsch L, et al. Automated quantification of cerebral edema following hemispheric infarction: application of a machine-learning algorithm to evaluate CSF shifts on serial head CTs. Neuroimage Clin 2016;12:673–80. doi:10.1016/j.nicl.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Siegel JS, Ramsey LE, Snyder AZ, et al. Disruptions of network connectivity predict impairment in multiple behavioral domains after stroke. Proc Natl Acad Sci USA 2016;113:E4367–E4376. doi:10.1073/pnas.1521083113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hope TM, Seghier ML, Leff AP, et al. Predicting outcome and recovery after stroke with lesions extracted from MRI images. Neuroimage Clin 2013;2:424–33. doi:10.1016/j.nicl.2013.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. LOHR S. IBM is counting on its bet on Watson, and Paying Big Money for It. 2016. https://www.nytimes.com/2016/10/17/technology/ibm-is-counting-on-its-bet-on-watson-and-paying-big-money-for-it.html (accessed 1 Jun 2017).
- 67. Otake T. IBM Big Data used for rapid diagnosis of rare leukemia case in Japan. 2016. http://www.japantimes.co.jp/news/2016/08/11/national/science-health/ibm-big-data-used-for-rapid-diagnosis-of-rare-leukemia-case-in-japan (accessed 1 Jun 2017).
- 68. Graham J. Artificial Intelligence, Machine Learning, and the FDA. 2016. https://www.forbes.com/sites/theapothecary/2016/08/19/artificial-intelligence-machine-learning-and-the-fda/#4aca26121aa1 (accessed 1 Jun 2017).
- 69. Kayyali B, Knott D, Kuiken SV. The big-data revolution in US health care: accelerating value and innovation. 2013. http://www.mckinsey.com/industries/healthcare-systems-and-services/our-insights/the-big-data-revolution-in-us-health-care (accessed 1 Jun 2017).