Abstract
OBJECTIVE
The rs7903146 T allele in transcription factor 7 like 2 (TCF7L2) is strongly associated with type 2 diabetes (T2D), but the mechanisms for increased risk remain unclear. We evaluated the physiologic and hormonal effects of TCF7L2 genotype before and after interventions that influence glucose physiology.
RESEARCH DESIGN AND METHODS
We genotyped rs7903146 in 608 individuals without diabetes and recorded biochemical data before and after 1) one dose of glipizide (5 mg) on visit 1 and 2) a 75-g oral glucose tolerance test (OGTT) performed after administration of metformin 500 mg twice daily over 2 days. Incretin levels were measured in 150 of the 608 participants.
RESULTS
TT risk-allele homozygotes had 1.6 mg/dL higher baseline fasting glucose levels and 2.5 pg/mL lower glucagon levels per T allele than carriers of other genotypes at baseline. In a subset of participants, the T allele was associated with higher basal glucagon-like peptide 1 (GLP-1) levels at visit 1 (β = 1.52, P = 0.02 and β = 0.96, P = 0.002 for total and active GLP-1, respectively), and across all points of the OGTT after metformin administration. Regarding drug response, the T allele was associated with a shorter time (β = −7.00, P = 0.03) and a steeper slope (β = 0.23, P = 0.04) to trough glucose levels after glipizide administration, and lower visit 2 fasting glucose level adjusted for visit 1 fasting glucose level (β = −1.02, P = 0.04) and a greater decline in glucose level between visits (β = −1.61, P = 0.047) after metformin administration.
CONCLUSIONS
Our findings demonstrate that common variation at TCF7L2 influences acute responses to both glipizide and metformin in people without diabetes and highlight altered incretin signaling as a potential mechanism by which TCF7L2 variation increases T2D risk.
Introduction
Metformin and sulfonylureas are two of the most widely prescribed medications for the treatment of type 2 diabetes with millions of users worldwide. Metformin, the first-line drug for the treatment of type 2 diabetes, has a long-standing evidence base for efficacy and safety, is inexpensive, and may reduce the risk of cardiovascular events and death (1). Similarly, sulfonylureas are extensively used based on their glucose-lowering efficacy, universal local availability and low cost (2). Yet, there remains significant interindividual variation in the responses to these medications, and, despite our vision of precision medicine, clinicians continue to make therapeutic decisions based on the population average parameters of efficacy and side-effect profile of a given drug, rather than on individual level characteristics.
In recent years, the field of type 2 diabetes and related glycemic traits has experienced an explosion of genetic discovery; there are now ∼100 loci associated with type 2 diabetes or related traits through large-scale genome-wide association studies and high-throughput sequencing technology (3–8). Despite our knowledge of the associations of several genetic variants with type 2 diabetes and their involvement in physiological pathways involved in drug response, their impact on pharmacological interventions has not been systematically examined. Similarly, we know that genetic factors influence the glycemic response to metformin, with common genetic variants explaining between 21% and 34% of the variation in metformin response, depending on how glycemic response was measured (9). However, small pharmacokinetic studies of metformin response have not yielded significant results, and to date only two significant genome-wide efforts of metformin response have been published, yielding minimal results (10,11). There has also been limited success in pharmacogenetics studies of sulfonylurea response (12). Overall, there is a crucial need for strategies that integrate physiological and pharmacogenetic information with genetic discoveries to better understand biological pathways and drug response (13).
The T allele of the intronic single nucleotide polymorphism (SNP) rs7903146 located within the transcription factor 7 like 2 (TCF7L2) gene is strongly associated with type 2 diabetes (14,15). This finding has been consistently replicated across multiple populations including children, and TCF7L2 remains among the strongest common genetic risk factors for type 2 diabetes (16–19). However, our knowledge of the mechanisms for this increased risk still remains uncertain. TCF7L2 encodes a transcription factor associated with the Wnt/β-catenin signaling pathway and is widely expressed in various tissues (20). The key effector of the WNT signaling pathway is the transcription factor β-catenin/TCF, formed by heterodimerization of β-catenin and a member of the TCF family. Several potential mechanisms have been posited for the increased type 2 diabetes risk conferred by genetic variation in TCF7L2, related to mechanisms both within and beyond the β-cell. These include diminished β-cell mass, impaired proinsulin processing, reduced insulin secretion, and an altered incretin effect. To improve our understanding of the role of common variation in TCF7L2 in relation to medication response in human participants free of disease and naive to diabetes drugs prospectively, we leveraged our Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH). The primary objective of our study was to evaluate the physiologic and hormonal effects of TCF7L2 genotype before and after interventions that influence glucose physiology using two commonly prescribed medications for type 2 diabetes: the sulfonylurea glipizide and the biguanide metformin. A secondary objective of our study was to evaluate the incretin response in relation to the TCF7L2 genotype, the strongest common genetic risk factor for type 2 diabetes, as a potential mechanism that contributes to type 2 diabetes risk.
Research Design and Methods
Study Design and Description of Participants
The study design and baseline participant characteristics of SUGAR-MGH (Clinical trial reg. no. NCT01762046, clinicaltrials.gov) have been described in detail previously (21). Briefly, SUGAR-MGH is a National Institutes of Health (NIH)-funded clinical research study and novel resource of genetic and biochemical data in 1,000 adults who were enrolled at three Boston academic medical centers from 2008 to 2015. Participants were preferentially enrolled if they had risk factors for type 2 diabetes or lifestyle-controlled type 2 diabetes, but had never received antidiabetic therapy. During the study protocol, they were sequentially administered glipizide and metformin, two medications commonly used to treat type 2 diabetes and chosen to perturb two different limbs of the glucose homeostatic system (insulin secretion and insulin action) under basal and hyperglycemic conditions. Plasma glucose and insulin were drawn at regular intervals before and after 1) a single dose of 5 mg glipizide (visit 1) and 2) a 75-g oral glucose tolerance test (OGTT) after 2 days of treatment with 500 mg of metformin twice a day. Glucagon-like peptide 1 (GLP-1), glucose-dependent insulinotropic polypeptide (GIP), proinsulin, and glucagon were measured in a subset of these samples. The SUGAR-MGH protocol was approved by the Partners Human Research Committee, and written informed consent was obtained from all participants. The SUGAR-MGH protocol required two visits to the Clinical Research Center (CRC) and 1 week of active treatment per subject. No participant experienced a hypoglycemia-related serious adverse event, and no participant required intravenous glucose or glucagon to treat hypoglycemia. In this study, we describe findings from the first 608 subjects that were genotyped consecutively of 1,000 subjects who were recruited.
Genotyping
DNA was extracted and genotyping performed using the iPLEX-GOLD Assay from Sequenom by allele-specific primer extension of amplified products with detection by mass spectroscopy. Genotype data for SNP rs7903146 (TCF7L2) was available for the first 608 participants who completed both visits and took all four doses of metformin. A subset of these participants (150) had GLP-1 and GIP assay measurements.
Measurements
A medical history was obtained by the consenting practitioner from all participants; and weight, height, blood pressure, and heart rate were measured at each visit to the CRC. Plasma glucose was measured by a hexokinase assay (Roche, Indianapolis, IN). Insulin level was determined using a radioimmunoassay (Beckman Coulter, Fullerton, CA). The intra-assay coefficient of variability (CV) for the insulin assay was 2.2–4.4% and the interassay CV was 2.9–6.0%. C-peptide was measured by radioimmunoassay (KPED1; Siemens, Erlangen, Germany); glucagon was measured by radioimmunoassay (LINCOplex Kit, HENDO-65K-Rev; Linco Research, St. Charles, MO). The CV of the glucagon assay was 10.9–13.3%. Incretin hormones (GLP-1 and GIP) were measured from blood samples collected in prechilled EDTA tubes containing DPP-IV inhibitor (Millipore, Billerica, MA). Active GLP-1 (7–36,7–37) was measured using the GLP-1 (Active) ELISA Kit (Millipore); total GLP-1 was measured using the GLP-1 (7–36,9–36) ELISA kit from Alpco Immunoassays (Salem, NH); and GIP was measured by using the Human GIP Total ELISA Kit from Millipore.
Statistical Analyses
The area under the curve (AUC) for insulin and glucagon during the glipizide challenge and for glucose, insulin, GLP-1, and GIP during the OGTT challenge was calculated by the trapezoidal method and adjusted for baseline values. The area over the curve for decreases in glucose during the glipizide challenge was calculated by subtracting AUC by the trapezoidal method from the baseline glucose value at visit 1 × total time for the glipizide challenge. Mean (± SD) or median (interquartile range) is provided for continuous normally or non-normally distributed traits, respectively, unless otherwise specified. Missing data were not imputed. Multilevel mixed-effects linear regression models, with adjustments for age, sex, self-reported race/ethnicity, and BMI were used to test the association of genotype on selected end points. The threshold for statistical significance in all analyses was set at a P value of 0.05. Confounding was assessed in these linear regression models by age, sex, race, and BMI, and consistent results were found. Statistical analyses were performed by using STATA (version 13). Figures were constructed using GraphPad Prism (version 6).
Results
Demographic Characteristics of the Participants and Baseline Associations
The baseline demographic characteristics of the 608 SUGAR-MGH participants included in the study by genotype at TCF7L2 are summarized in Table 1. Nine percent of the participants included in this study were homozygous for the diabetes risk–conferring T allele, 40% were heterozygous, and 51% were homozygous for the protective C allele. Over 35% of the participants came from ethnic minority populations, participants were nearly evenly distributed between men and women, and their average BMI fell within the obese category based on the definition of the Centers for Disease Control and Prevention (22). Allele frequencies were similar to what has been previously described in the literature (23) for this variant (Supplementary Table 1). In support of previous findings, we detected a significant association between fasting glucose level and TCF7L2 genotype, with T–risk allele homozygotes having ∼6 mg/dL higher fasting glucose than heterozygotes or C-allele homozygotes (Supplementary Fig. 1). The T–risk allele homozygotes had a 1.61 mg/dL higher baseline fasting glucose level per T allele than carriers of other genotypes (Table 2). Fasting insulin, proinsulin, and C-peptide concentrations were not different across the genotypes at baseline (Table 1).
Table 1.
Baseline demographics of the study participants
| Total | Genotype |
P value | |||
|---|---|---|---|---|---|
| TT | CT | CC | |||
| SUGAR-MGH, n | 608 | 55 | 242 | 311 | |
| Male sex, n (%) | 25 (45.6) | 125 (51.7) | 142 (45.7) | 0.347 | |
| Age (years) | 48.7 ± 16 | 50 ± 16 | 50 ± 16 | 47 ± 16.3 | 0.111 |
| BMI (kg/m2) | 30.6 ± 7 | 29.6 ± 7 | 30.4 ± 6 | 30.9 ± 7 | 0.339 |
| Fasting glucose (mg/dL) | 93.2 ± 14 | 98.3 ± 20 | 93 ± 13 | 92 ± 12 | 0.012 |
| Fasting insulin (mU/L) | 7.2 ± 8 | 7.6 ± 6 | 7.2 ± 6 | 7.5 ± 6 | 0.867 |
| C-peptide (pmol/L) | 810 ± 391 | 820 ± 368 | 830 ± 390 | 793 ± 396 | 0.559 |
| Lifestyle-controlled T2D, n (%) | 15 (2.5) | 5 (9.1) | 6 (2.5) | 4 (1.3) | 0.003 |
Data are mean ± SD unless otherwise noted. T2D, type 2 diabetes.
Table 2.
Magnitude of change per T allele after glipizide and metformin administration
| β-coefficient | 95% CI | P value | |
|---|---|---|---|
| Fasting glucose (mg/dL) | 1.61 | 0.036, 3.18 | 0.04 |
| Glucose AOC (mg/dL) | 160.2 | −166.6, 487.1 | 0.34 |
| Fasting glucagon (pg/mL) | −2.5 | −4.97, −0.013 | 0.049 |
| Fasting glucose V2 (mg/dL) | −1.02 | −1.96, −0.06 | 0.04 |
| Fasting glucose V2–V1 (mg/dL) | −1.61 | −3.22, 0.02 | 0.04 |
| GLP-1 total V1 (pmol/L) | 1.52 | 0.22, 2.81 | 0.02 |
| GLP-1 active V1 (pmol/L) | 0.96 | 0.35, 1.57 | 0.002 |
| Time to trough glucose at V1 adjusted for baseline glucose (min) | −7.00 | −13.21, −0.79 | 0.027 |
| (Glu0 to Glutrough)/time (mg/dL/time) | 0.023 | 0.002, 0.045 | 0.035 |
| (Glu240 to Glutrough)/time (mg/dL/time) | 0.024 | 0.004, 0.044 | 0.017 |
| Peak insulin at V1 (mU/L) | 0.121 | −3.146, 3.387 | 0.942 |
| Time to peak insulin at V1 (min) | −2.139 | −9.998, 5.719 | 0.593 |
| Glucose trough at V1 (mg/dL) | −0.072 | −1.477, 1.334 | 0.920 |
| Glucose trough adjusted for baseline glucose (mg/dL) | −0.727 | −1.984, 0.529 | 0.256 |
| HOMA-β V1 | −3.35 | −21.72, 15.01 | 0.720 |
| HOMA-β V2 | −4.34 | −19.198, 10.50 | 0.566 |
| HOMA-IR V1 | 0.068 | −0.16, 0.29 | 0.563 |
| HOMA-IR V2 | −0.42 | −0.26, 0.18 | 0.706 |
| Stumvoll index V2 | −0.0014 | −0.004, 0.001 | 0.248 |
AOC, area over the curve; Glu0, baseline fasting glucose level; Glu240, glucose level at 240 min of OGTT; Glutrough, trough glucose level; V, visit.
Biochemical Response to Glipizide Differs by TCF7L2 Genotype
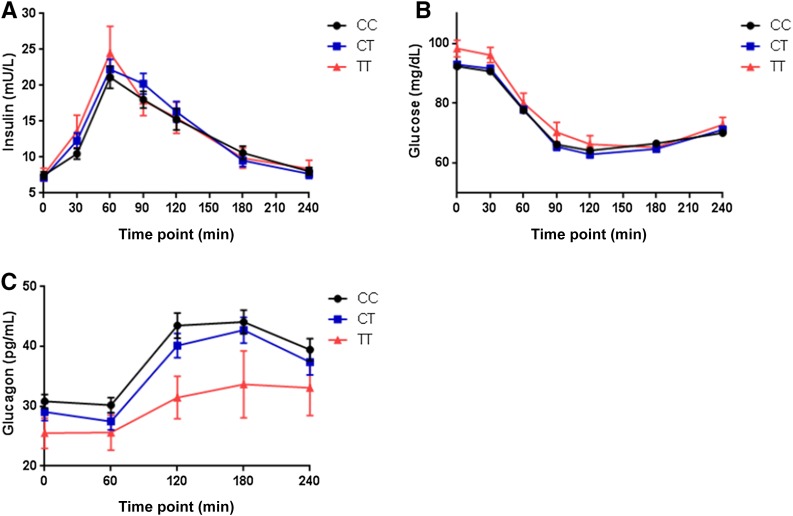
Glipizide raised serum insulin levels and lowered blood glucose levels in all participants as expected, with the insulin peak achieved at 60 min (Fig. 1A) and blood glucose level reaching a nadir at 120 min (Fig. 1B). Despite their higher baseline glucose levels, T–risk allele homozygotes had a similar magnitude of glucose decline after a single dose of glipizide compared with heterozygotes or C-allele homozygotes (Table 2). Likewise, there were no differences in insulin levels across genotype during the challenge. Glucagon levels during the glipizide challenge peaked at 180 min, at a time point after the mean blood glucose reached its lowest value. The T allele was associated with lower glucagon levels at baseline (Table 2), and the fasting glucagon level adjusted for baseline glucose showed a linear relationship across genotypes (Fig. 1C). The AUC for glucagon did not achieve statistically significant differences across genotype groups.
Figure 1.
A: Change in plasma insulin across genotypes at visit 1 after glipizide administration. B: Change in plasma glucose across genotypes at visit 1 after glipizide administration. C: Change in plasma glucagon across genotypes at visit 1 after glipizide administration. Data depict mean ± SEM, n = 608 (CC = 311, CT = 242, TT = 55).
Ninety-four (30%) CC homozygotes, 75 (31%) CT heterozygotes, and 18 (33%) TT homozygotes terminated the glipizide challenge early due to hypoglycemia or hypoglycemia-related symptoms, without statistically significant differences between the groups (P = 0.935). However, overall, participants who completed the entire glipizide challenge had higher fasting glucose values prior to receiving glipizide and higher trough glucose during the glipizide challenge than did participants who terminated the glipizide challenge because of hypoglycemia or hypoglycemia-related symptoms.
We evaluated predefined phenotypes that capture the acute pharmacological and physiological responses to the medications used in the SUGAR-MGH and characterized clinically relevant and pharmacodynamic parameters. There were no significant differences in glucose trough adjusted for baseline glucose level by genotype at TCF7L2 (Table 2). There were significant genotype-driven differences in the time to glucose trough after glipizide administration, which was shorter by 7 min per T allele after adjustment for baseline glucose level (Table 2).The T allele was also associated with a steeper slope in glucose drop between the start of the visit (0 min) and the trough level (Table 2). There were also significant differences across genotypes in the upward slope of glucose recovery between the glucose trough and the end of the study visit (240 min) with the T allele associated with a steeper slope (Table 2). There were no genotype-driven differences in any of the insulin-based predefined end points (Table 2).
Biochemical Response to Metformin Differs by TCF7L2 Genotype
Although there were no differences across genotypes in fasting glucose levels at visit 2 after four doses of metformin, the T allele was associated with lower fasting glucose levels at visit 2 after adjusting for the fasting glucose level at visit 1 (Table 2). The T allele was also associated with a greater drop in glucose between visits 1 and 2 (Fig. 1C and Table 2). There were no differences in fasting insulin levels or in surrogate measures of β-cell function and insulin resistance (IR) by homeostasis model assessment (HOMA-β or HOMA-IR) (24) between visits 1 and 2 and in the modified Stumvoll index (25) by genotype at TCF7L2 (Table 2).
Alterations in Incretin Levels During the OGTT
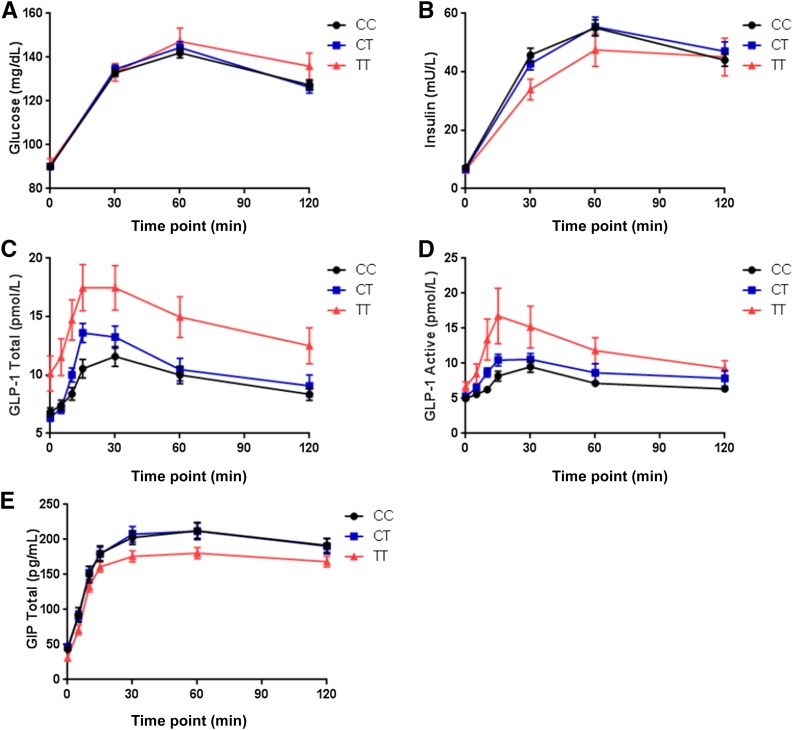
There were no genotype-driven differences in glucose or insulin levels during the OGTT (Fig. 2A and B). Incretin levels were measured in a subset of participants (50 from each genotype group at TCF7L2 rs7903146). The baseline characteristics of this subset of participants were similar to those observed in the overall cohort, suggesting that they were drawn from the same underlying distribution. The T allele was associated with higher baseline total and active GLP-1 at visit 1, even after adjusting for age, sex, race, BMI, and fasting glucose level (Table 2). At visit 2, the T allele was again associated with higher fasting total and active GLP-1 levels, and these differences remained across all time points of the OGTT, with a higher AUC for both total and active GLP-1 levels (P < 0.005 for both). These differences were mainly driven by TT homozygotes (Fig. 2C and D). Although there were no differences observed in GIP levels at baseline in visit 1, during the OGTT in visit 2 the T allele was associated with lower GIP levels (P = 0.02) (Fig. 2E).
Figure 2.
A and B: n = 608 (CC = 311, CT = 242, TT = 55). A: Change in plasma glucose across the various time points of the OGTT at visit 2 after four doses of metformin. B: Change in plasma insulin across the various time points of the OGTT at visit 2 after four doses of metformin. C–E: n = 150 (CC = 50, CT = 50, TT = 50). C: Change in plasma GLP-1 (total) across the various time points of the OGTT at visit 2 after four doses of metformin. D: Change in plasma GLP-1 (active) across the various time points of the OGTT at visit 2 after four doses of metformin. E: Change in plasma GIP across the various time points of the OGTT at visit 2 after four doses of metformin. Data depict mean ± SEM.
Conclusions
Using a novel prospective human pharmacogenetic resource, which is free of the uncontrolled nature of retrospective clinical data sets, we have shown that the type 2 diabetes–associated variant rs7903146 in TCF7L2 influences the acute response to both glipizide and metformin in persons free of overt diabetes. In parallel, through pharmacological perturbations and hormonal measurements we have been able to generate additional evidence in support of altered incretin signaling as a potential mechanism by which TCF7L2 increases type 2 diabetes risk. We have demonstrated that SUGAR-MGH is adequately powered to detect differences, because participants who carried the high-risk T allele at rs7903146 in the TCF7L2 gene had a higher fasting glucose level at baseline before the glycemic perturbations, a finding that is consistent with previous work (26). Despite this higher baseline, TT homozygotes achieved a trough glucose level faster than CC homozygotes after glipizide administration and also demonstrated a significantly steeper slope to glucose trough. The T allele was associated with lower glucagon levels at baseline even after adjusting for baseline glucose level. Because this measure excludes participants who received rescue carbohydrate for hypoglycemia, it likely reflects the endogenous response to hypoglycemia driven by counter-regulatory hormones. In terms of metformin response, high-risk T-allele carriers had a lower fasting glucose level after having received a short course of metformin, as well a greater decrease in fasting glucose levels from visit 1 to visit 2. These findings suggest that they had a better response to metformin than subjects who did not carry the high-risk allele, and the persistence of a significant difference after adjustment for baseline glucose at visit 1 suggests that the drug response is driven by genotype and not by the higher baseline glucose. Although the clinical implications of the individual end points may be subtle, it is noteworthy that we are able to demonstrate significant differences based on genotype just after a single dose of 5 mg of glipizide and after four doses of 500 mg of metformin.
With regard to the mechanism at this locus, high-risk T-allele carriers also had lower glucagon levels at baseline, ruling out a primary effect on higher glucagon levels as the process by which TCF7L2 increases type 2 diabetes risk and suggesting that lower glucagon levels in T-allele carriers are a compensatory response for a primary effect on fasting glucose. This may also explain why rates of hypoglycemia were not significantly higher in TT carriers. Lower glucagon levels can also be explained by reduced expression of proglucagon in α-cells or altered post-translational processing to glucagon. Our study adds to a mounting body of evidence that suggests that the α-cell may play a significant role in TCF7L2-mediated diabetes risk (27,28). In a study of 120 individuals evenly distributed between TT and CC homozygotes at this locus, TT carriers exhibited impaired the suppression of glucagon after an oral challenge, suggesting that genetic variation in TCF7L2 is associated with impaired postprandial suppression of glucagon (29).
In a small subset of our participants, we observed that the TCF7L2 high-risk genotype is associated with higher total and active GLP-1 levels at baseline and across all points of the OGTT; together with the lack of a detectable change in glucose or insulin levels across genotypes, this observation is suggestive of GLP-1 resistance in these individuals. These findings expand on the existing knowledge base (30–32) and continue to implicate altered incretin signaling as a potential contributory mechanism for increased TCF7L2-associated type 2 diabetes risk.
With the exception of some monogenic forms of diabetes (33), few pharmacogenetic studies of metformin and sulfonylurea response have been published. Small-scale studies have shown differences in sulfonylurea metabolism, but studies on actual measured drug response are conflicting (12). Similarly, despite the status of metformin as the most commonly prescribed medication for type 2 diabetes, few pharmacogenetic studies of its response have been published. Two genome-wide association studies have implicated common variants in the genes ATM (10) and SLC2A2 (11) in metformin response, but the physiological or pharmacological implications of these findings still remain unclear. Several studies have focused on the rs7903146 variant in TCF7L2 as a determinant of drug response, given its relatively strong impact on type 2 diabetes risk among common variants. In a large GoDARTS retrospective study of subjects with type 2 diabetes, of 901 users of sulfonylureas, homozygotes for the type 2 diabetes T risk allele at TCF7L2 rs7903146 were less likely to respond to sulfonylureas, with an odds ratio of failure of 1.73 (95% CI 1.11, 2.70; P = 0.015), which is in the opposite direction to our findings in the SUGAR-MGH. Furthermore, no effect of the TCF7L2 genotype was observed in 945 individuals evaluated for metformin response (34), which again is in contrast to our present results. Several reasons may explain these discrepancies. Although the GoDARTS authors were careful to account for potential confounders, the limitations inherent to an observational study make it susceptible to prescriber bias, which may influence treatment and dosage decisions and therefore response. Furthermore, it is possible that the genotype has a differential effect in subjects in whom diabetes has not yet developed compared with subjects with established type 2 diabetes, in whom some degree of β-cell failure may have already occurred. This concept was demonstrated in a study of 1,576 subjects with varying degrees of glycemia, in whom the level of glycemia determined the effect of TCF7L2 variation on surrogate measures of insulin secretion (35). Under this paradigm, if variation at TCF7L2 impairs β-cell function and predisposes β-cells to failure, sulfonylureas could be more effective in risk allele carriers in whom β-cell mass is preserved early in the type 2 diabetes disease course, but become less effective as the disease progresses. Similarly, metformin might be more effective in this at-risk group before pathological changes have had the opportunity to occur, although the DPP results suggest that this effect may not be sustained.
Numerous investigations have been conducted to understand the molecular mechanisms by which TCF7L2 exerts its functions in pancreatic and intestinal endocrine cells (36–38). The TCF7L2 transcription factor binds to the promoter of the proglucagon gene, which in turn encodes glucagon, GLP-1, and GLP-2 (39). The total form of GLP-1, GLP-1 (7–36) NH2, is rapidly metabolized to the active form GLP-1 (9–36) NH2, which is the predominant form of GLP-1 in postprandial plasma because of its relatively slower clearance (40). The WNT signaling pathway is required for the normal development of pancreatic islets and also regulates GLP-1 expression and secretion in the intestinal L cell. Therefore, one early hypothesis postulated that polymorphisms in TCF7L2 could reduce incretin-stimulated insulin secretion and thereby increase type 2 diabetes risk. However, rather than showing a deficiency in GLP-1 production, previous studies have demonstrated evidence of impaired incretin-mediated insulin response in individuals who carry the high-risk allele at TCF7L2 (30,32,38,41). Consistent with our results, Schäfer et al. (30) reported normal GLP-1 secretion but reduced effect of GLP-1 upon stimulation of insulin secretion in rs7903146 T-allele carriers, indicating a state of relative incretin resistance. Our results expand on current knowledge by demonstrating an impaired incretin effect (higher glucose levels despite higher GLP-1 levels and reduced GIP levels) in a clinical research study of a multiethnic cohort of individuals who have risk factors for type 2 diabetes but are naive to antidiabetic therapy.
A limitation of our study design is the lack of an OGTT prior to the administration of metformin. Although this was a consideration when designing the study, logistical considerations that would enable recruitment of a large cohort with excellent retention rates, and short duration did not make this feasible. Therefore, although we are able to effectively demonstrate genotype-driven differences in metformin response, we are unable to provide a more refined explanation based on a dynamic glucose challenge. We emphasize that SUGAR-MGH, although providing a unique pharmacogenetic resource to probe the influence of genetic variation on physiology and the response to metformin and glipizide, was not designed to be a clinical trial seeking to modify or expand current treatment indications for metformin or sulfonylureas. Rather, it allows for the ongoing systematic evaluation of how known genetic variants modify acute antidiabetic drug responses in humans. Our study provides clinically relevant results that genetic variation can alter the response to therapy in type 2 diabetes. Our findings build on a growing body of evidence to suggest that raising incretin levels pharmacologically in carriers of TCF7L2 variants who have a pathological background of incretin resistance may be a less desirable therapeutic choice. For instance, testing of DPP-IV inhibitors, which block the degradation of incretins, could be of clinical relevance in this population (42). In sum, a unique data set like SUGAR-MGH enables the study of genetic determinants of drug response, as a crucial test of whether genetically based precision medicine might predict a therapeutic benefit in the clinical management of patients. Further studies are needed to better understand the physiological and clinical impacts of specific genetic variants to help guide the design of prospective pharmacogenetic clinical trials.
Supplementary Material
Article Information
Acknowledgments. The authors thank the Massachusetts General Hospital (MGH) CRC and staff nurses at MGH, the Brigham and Women’s Hospital Center for Clinical Investigation (CCI) and CCI nurses and staff, and the Joslin Diabetes Center CRC and CRC nurses and staff for help and support. The authors also thank Mary Lukowski (Cardiology Division, MGH) for assistance with the MGH Cardiology and Metabolic Patient cohort database (CAMP MGH), the Partners Human Research Committee, and Dr. Enrico Cagliero (Diabetes Unit, MGH) for his role as a safety officer. In addition, the authors thank all the members of the Florez laboratory at MGH and at the Broad Institute for helpful discussion and feedback.
Funding. This work was conducted with support from NIH/National Institute of Diabetes and Digestive and Kidney Diseases awards R01-DK-088214, R03-DK-077675, and P30-DK-036836; the Joslin Clinical Research Center and its philanthropic donors; and the Harvard Catalyst: The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences; NIH Awards M01-RR-01066, 1UL1-RR-025758-04, and 8UL1-TR-000170-05; and financial contributions from Harvard University and its affiliated academic health care centers).
Duality of Interest. S.S. is a current employee of the University of California, San Francisco, but was a clinical and research fellow at the Massachusetts General Hospital when this study was conducted. B.C. is a consultant for VeroScience, LLC. M.K.T. is a current employee of Eli Lilly and Company but was an employee at the Massachusetts General Hospital when the SUGAR-MGH was designed. A.G. is a current employee of Novartis but was an employee at the Joslin Diabetes Center when this study was conducted. J.C.F. has received consulting honoraria from Merck, Boehringer Ingelheim, and Intarcia Therapeutics. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. S.S. assisted in analysis of the results, interpretation of the data, wrote the first draft of the manuscript, and edited and approved the manuscript. V.K. assisted in the collection and interpretation of the data, analyzed the results, generated the tables and figures, and edited and approved the manuscript. B.C. assisted in the collection, analysis, and interpretation of data and edited and approved the manuscript. K.R.L. assisted in the collection and analysis of the data. L.C. assisted in the collection and interpretation of the data and statistical analysis and edited and approved the manuscript. A.K.M. assisted in the statistical analysis and edited and approved the manuscript. J.M. assisted in the analysis of the results and edited and approved the manuscript. M.K.T. assisted in the design of the study and the interpretation of the results. M.H. assisted in the recruitment of study subjects and interpretation of the data and edited and approved the manuscript. A.G. assisted in the recruitment of study subjects and the interpretation of data and edited and approved the manuscript. J.C.F. designed the study, supervised the collection and interpretation of the data and the analysis of the results, and extensively edited and approved the manuscript. J.C.F. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 62nd Annual Meeting of the American Society of Human Genetics, San Francisco, CA, 6–10 November 2012.
Footnotes
Clinical trial reg. no. NCT01762046, clinicaltrials.gov.
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc17-1386/-/DC1.
S.S. is currently affiliated with the Division of Pediatric Endocrinology and Diabetes, University of California, San Francisco, San Francisco, CA.
References
- 1.Holman RR, Paul SK, Bethel MA, Matthews DR, Neil HA. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med 2008;359:1577–1589 [DOI] [PubMed] [Google Scholar]
- 2.Bressler R, Johnson DG. Pharmacological regulation of blood glucose levels in non-insulin-dependent diabetes mellitus. Arch Intern Med 1997;157:836–848 [PubMed] [Google Scholar]
- 3.Mahajan A, Go MJ, Zhang W, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium; Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; Mexican American Type 2 Diabetes (MAT2D) Consortium; Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium . Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet 2014;46:234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho YS, Chen CH, Hu C, et al.; DIAGRAM Consortium; MuTHER Consortium . Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet 2011;44:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma RC, Hu C, Tam CH, et al.; DIAGRAM Consortium; MuTHER Consortium . Genome-wide association study in a Chinese population identifies a susceptibility locus for type 2 diabetes at 7q32 near PAX4. Diabetologia 2013;56:1291–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voight BF, Scott LJ, Steinthorsdottir V, et al.; MAGIC investigators; GIANT Consortium . Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 2010;42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris AP, Voight BF, Teslovich TM, et al.; Wellcome Trust Case Control Consortium; Meta-Analyses of Glucose and Insulin-related traits Consortium (MAGIC) Investigators; Genetic Investigation of ANthropometric Traits (GIANT) Consortium; Asian Genetic Epidemiology Network–Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinthorsdottir V, Thorleifsson G, Sulem P, et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet 2014;46:294–298 [DOI] [PubMed] [Google Scholar]
- 9.Zhou K, Donnelly L, Yang J, et al.; Wellcome Trust Case Control Consortium 2 . Heritability of variation in glycaemic response to metformin: a genome-wide complex trait analysis. Lancet Diabetes Endocrinol 2014;2:481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou K, Bellenguez C, Spencer CC, et al.; GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group; Wellcome Trust Case Control Consortium 2; MAGIC investigators . Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 2011;43:117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou K, Yee SW, Seiser EL, et al.; MetGen Investigators; DPP Investigators; ACCORD Investigators . Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet 2016;48:1055–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becker ML, Visser LE, Trienekens PH, Hofman A, van Schaik RH, Stricker BH. Cytochrome P450 2C9 *2 and *3 polymorphisms and the dose and effect of sulfonylurea in type II diabetes mellitus. Clin Pharmacol Ther 2008;83:288–292 [DOI] [PubMed] [Google Scholar]
- 13.Florez JC. Pharmacogenetic perturbations in humans as a tool to generate mechanistic insight. Diabetes 2013;62:3019–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grant SF, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 2006;38:320–323 [DOI] [PubMed] [Google Scholar]
- 15.Helgason A, Pálsson S, Thorleifsson G, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 2007;39:218–225 [DOI] [PubMed] [Google Scholar]
- 16.Florez JC, Jablonski KA, Bayley N, et al.; Diabetes Prevention Program Research Group . TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 2006;355:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saxena R, Gianniny L, Burtt NP, et al. Common single nucleotide polymorphisms in TCF7L2 are reproducibly associated with type 2 diabetes and reduce the insulin response to glucose in nondiabetic individuals. Diabetes 2006;55:2890–2895 [DOI] [PubMed] [Google Scholar]
- 18.Scott LJ, Bonnycastle LL, Willer CJ, et al. Association of transcription factor 7-like 2 (TCF7L2) variants with type 2 diabetes in a Finnish sample. Diabetes 2006;55:2649–2653 [DOI] [PubMed] [Google Scholar]
- 19.Dabelea D, Dolan LM, D’Agostino R Jr, et al. Association testing of TCF7L2 polymorphisms with type 2 diabetes in multi-ethnic youth. Diabetologia 2011;54:535–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin T, Liu L. The Wnt signaling pathway effector TCF7L2 and type 2 diabetes mellitus. Mol Endocrinol 2008;22:2383–2392 [DOI] [PubMed] [Google Scholar]
- 21.Walford GA, Colomo N, Todd JN, et al. The Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH): design of a pharmacogenetic resource for type 2 diabetes. PLoS One 2015;10:e0121553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention [Internet], 2017. About adult BMI. Available from https://www.cdc.gov/healthyweight/assessing/bmi/adult_bmi/index.html. Accessed 1 February 2017
- 23.Accelerating Medicines Partnership (AMP). Type 2 diabetes knowledge portal [Internet], 2017. Available from http://www.type2diabetesgenetics.org/gene/geneInfo/TCF7L2. Accessed 1 February 2017
- 24.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985;28:412–419 [DOI] [PubMed] [Google Scholar]
- 25.Stumvoll M, Van Haeften T, Fritsche A, Gerich J. Oral glucose tolerance test indexes for insulin sensitivity and secretion based on various availabilities of sampling times. Diabetes Care 2001;24:796–797 [DOI] [PubMed] [Google Scholar]
- 26.Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey KA, Savic D, Zielinski M, et al. Evidence of non-pancreatic beta cell-dependent roles of Tcf7l2 in the regulation of glucose metabolism in mice. Hum Mol Genet 2015;24:1646–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boj SF, van Es JH, Huch M, et al. Diabetes risk gene and Wnt effector Tcf7l2/TCF4 controls hepatic response to perinatal and adult metabolic demand. Cell 2012;151:1595–1607 [DOI] [PubMed] [Google Scholar]
- 29.Shah M, Varghese RT, Miles JM, et al. TCF7L2 genotype and α-cell function in humans without diabetes. Diabetes 2016;65:371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schäfer SA, Tschritter O, Machicao F, et al. Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia 2007;50:2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villareal DT, Robertson H, Bell GI, et al. TCF7L2 variant rs7903146 affects the risk of type 2 diabetes by modulating incretin action. Diabetes 2010;59:479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pilgaard K, Jensen CB, Schou JH, et al. The T allele of rs7903146 TCF7L2 is associated with impaired insulinotropic action of incretin hormones, reduced 24 h profiles of plasma insulin and glucagon, and increased hepatic glucose production in young healthy men. Diabetologia 2009;52:1298–1307 [DOI] [PubMed] [Google Scholar]
- 33.Pearson ER, Flechtner I, Njølstad PR, et al.; Neonatal Diabetes International Collaborative Group . Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med 2006;355:467–477 [DOI] [PubMed] [Google Scholar]
- 34.Pearson ER, Donnelly LA, Kimber C, et al. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes 2007;56:2178–2182 [DOI] [PubMed] [Google Scholar]
- 35.Heni M, Ketterer C, Thamer C, et al. Glycemia determines the effect of type 2 diabetes risk genes on insulin secretion. Diabetes 2010;59:3247–3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z, Habener JF. Glucagon-like peptide-1 activation of TCF7L2-dependent Wnt signaling enhances pancreatic beta cell proliferation. J Biol Chem 2008;283:8723–8735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu Y, Brown C, Castro RA, et al. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin Pharmacol Ther 2008;83:273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyssenko V, Lupi R, Marchetti P, et al. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J Clin Invest 2007;117:2155–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yi F, Brubaker PL, Jin T. TCF-4 mediates cell type-specific regulation of proglucagon gene expression by beta-catenin and glycogen synthase kinase-3beta. J Biol Chem 2005;280:1457–1464 [DOI] [PubMed] [Google Scholar]
- 40.Kieffer TJ, Habener JF. The glucagon-like peptides. Endocr Rev 1999;20:876–913 [DOI] [PubMed] [Google Scholar]
- 41.Færch K, Pilgaard K, Knop FK, et al. Incretin and pancreatic hormone secretion in Caucasian non-diabetic carriers of the TCF7L2 rs7903146 risk T allele. Diabetes Obes Metab 2013;15:91–95 [DOI] [PubMed] [Google Scholar]
- 42.Zimdahl H, Ittrich C, Graefe-Mody U, et al. Influence of TCF7L2 gene variants on the therapeutic response to the dipeptidylpeptidase-4 inhibitor linagliptin. Diabetologia 2014;57:1869–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.