Abstract
Adenosine monophosphate (AMP)–activated protein kinase (AMPK) acts as a master regulator of cellular energy homeostasis by directly phosphorylating metabolic enzymes and nutrient transporters and by indirectly promoting the transactivation of nuclear genes involved in mitochondrial biogenesis and function. We explored the mechanism of AMPK-mediated induction of gene expression. We identified AMPK consensus phosphorylation sequences in three proteins involved in nucleosome remodeling: DNA methyltransferase 1 (DNMT1), retinoblastoma binding protein 7 (RBBP7), and histone acetyltransferase 1 (HAT1). DNMT1 mediates DNA methylation that limits transcription factor access to promoters and is inhibited by RBBP7. Acetylation of histones by HAT1 creates a more relaxed chromatin-DNA structure that favors transcription. AMPK-mediated phosphorylation resulted in the activation of HAT1 and inhibition of DNMT1. For DNMT1, this inhibition was both a direct effect of phosphorylation and the result of increased interaction with RBBP7. In human umbilical vein cells, pharmacological AMPK activation or pulsatile shear stress triggered nucleosome remodeling and decreased cytosine methylation, leading to increased expression of nuclear genes encoding factors involved in mitochondrial biogenesis and function, such as peroxisome proliferator–activated receptor gamma coactivator–1α (PGC-1α), transcription factor A (Tfam), and uncoupling proteins 2 and 3 (UCP2 and UCP3). Similar effects were seen in the aortas of mice given pharmacological AMPK activators, and these effects required AMPK2α. These results enhance our understanding of AMPK-mediated mitochondrial gene expression through nucleosome remodeling.
INTRODUCTION
Mitochondria mediate various functions, including β-oxidation and oxidative phosphorylation, contributing to cellular energy homeostasis (1). Dysregulation of mitochondrial biogenesis and function results in impaired adenosine triphosphate (ATP) production, increased reactive oxygen species (ROS), and altered Ca2+ signaling, all of which play a role in metabolic and cardiovascular disorders (1). These effects are mitigated by a complex signaling network, partly controlled by adenosine monophosphate (AMP)–activated protein kinase (AMPK) (2).
AMPK participates in various processes involved in energy homeostasis. Kinases such as liver kinase B1 (LKB1) and calcium/calmodulin-dependent protein kinase kinase β (CaMKKβ) phosphorylate and activate AMPK (3, 4). AICAR (5-aminoimidazole-4-carboxamide ribonucleotide), metformin, and pulsatile shear stress activate AMPK (5–9). Active AMPK phosphorylates components of signaling pathways that enhance mitochondrial biogenesis and energy bioavailability such as peroxisome proliferator–activated receptor gamma coactivator–1α (PGC-1α). PGC-1α acts as a master transcription factor for mitochondrial biogenesis by transactivating nuclear respiratory factors 1 and 2 (NRF1 and NRF2), which increase the expression of the gene encoding transcription factor A (Tfam), a key activator of mitochondrial gene transcription (10, 11).
Nucleosome remodeling is required for promoter activation and involves multiple enzymatic reactions including those catalyzed by kinases, methyltransferases, acetyltransferases, and poly[adenosine diphosphate (ADP)–ribose] polymerases (PARPs). The discovery of AMPK as a kinase that phosphorylates histone 2B (H2B) suggests that AMPK regulates nucleosome remodeling (12). To identify AMPK targets that modulate mitochondrial function at the nucleosomal level, we searched for AMPK phosphorylation sequences in epigenetic factors (13, 14). We found phosphorylation sequences in DNA methyltransferase 1 (DNMT1), retinoblastoma binding protein 7 (RBBP7), and histone acetyltransferase 1 (HAT1). We report here that AMPK phosphorylated this epigenetic regulatory network, contributing to nucleosome remodeling and reducing methylation of the promoters for the genes encoding PGC-1α, NRF1, NRF2, Tfam, and uncoupling proteins 2 and 3 (UCP2 and UCP3), thereby increasing mitochondrial biogenesis and function.
RESULTS
AMPK phosphorylates a network of epigenetic regulators
Promoter activation depends upon the extent and pattern of nucleosome remodeling. Using an in-house generated R script, we searched mouse, rat, and human proteomes for putative AMPK substrates involved in epigenetic regulation (table S1). This database was integrated into Gaggle software to identify central nodes of epigenetic regulation (fig. S1) (15), revealing AMPK phosphorylation sequences in both DNMT1 and RBBP7 (Fig. 1A and fig. S1). Promoter methylation, which is partially regulated by DNMT1, determines DNA accessibility by transcription factors (16). Our nodal analysis predicted that association with RBBP7 could inhibit DNMT1 (Fig. 1B). Although this interaction may be facilitated by AMPK-mediated phosphorylation of RBBP7, DNMT1 may be independently phosphorylated and inhibited by AMPK (Fig. 1B). AMPK phosphorylation of DNMT1 and RBBP7 may synergistically decrease promoter methylation, thereby facilitating AMPK-regulated gene transcription. Epigenetic regulation also occurs through histone modifications. Histone acetyltransferases, including HAT1, acetylate histone N-terminal tails relaxing histone-DNA interactions, remodeling the nucleosome to a euchromatic state. RBBP7 is a coactivator of HAT1 (17). Our bioinformatics analysis also revealed an AMPK phosphorylation sequence in HAT1 (Fig. 1A and fig. S1). Consequently, we explored whether AMPK regulates nucleosome remodeling and promoter activation through the phosphorylation of DNMT1, RBBP7, and HAT1.
Fig. 1. AMPK regulated a nucleosome remodeling network by phosphorylating DNMT1, RBBP7, and HAT1.
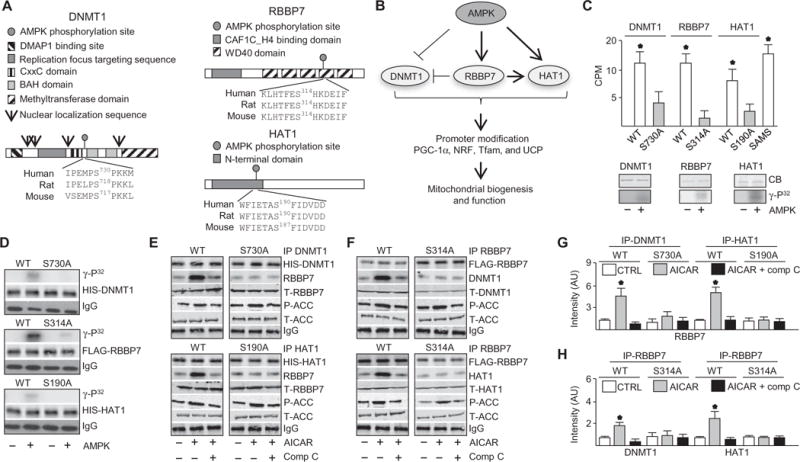
(A) Protein domains and putative AMPK-mediated phosphorylation sequence of DNMT1, RBBP7, and HAT1. DMAP1, DNA methyltransferase 1–associated protein 1 binding domain; BAH, bromo-adjacent homology domain. (B) Illustrated hypothesis of AMPK-mediated phosphorylation of DNMT1, RBBP7, and HAT1 and effects on mitochondrial biogenesis and function. (C) Kinase assays using recombinant target protein in the presence or absence of activated recombinant AMPKα2β1γ1. Top: Kinase assays conducted with peptides. Bottom: Kinase assays with full-length proteins. n = 4 independent experiments. CPM, counts per minute. (D) Kinase assays using immunoprecipitated wild-type (WT) and mutated DNMT1, RBBP7, and HAT1 proteins from HUVECs. Top: Autoradiograph. Lower: Total protein and immunoglobulin G (IgG) immunoblots for loading. n = 3 independent experiments. (E and F) Coimmunoprecipitation (IP) immunoblots in HUVECs transfected with WT DNMT1, RBBP7, or HAT1 or their corresponding Ser-to-Ala mutants (DNMT1-S730A, HAT1-S190A, and RBBP7-S314A) and treated with AICAR or left untreated for 30 min (top) or 10 min (bottom). P-ACC (phospho–acetyl-CoA carboxylase), T-ACC (total acetyl-CoA carboxylase), T-RBBP7, T-DNMT1, and T-HAT1 immunoblotting was conducted with the input IP crude cell lysate. n = 4 independent experiments. Comp C, compound C. (G and H) Densitometry analysis of coimmunoprecipitation immunuoblots comparing coimmunoprecipitated protein to total protein. *P < 0.05. AU, arbitrary units.
AMPK phosphorylates DNMT1, RBBP7, and HAT1
To validate AMPK phosphorylation sites (Fig. 1A), we performed in vitro kinase assays using peptides (13-mers) containing the AMPK phosphorylation sequences with or without Ser-to-Ala substitution, mimicking the unphosphorylated state. SAMS peptide was used as a standard positive control for AMPK activity (Fig. 1C) (8). DNMT1 peptides were composed of residues flanking Ser730; RBBP7 peptides were composed of residues flanking Ser314, Ser145, Ser354, or Ser393; and HAT1 peptides were composed of residues flanking Ser190. Peptides containing Ser were phosphorylated more efficiently than the Ala-substituted counterparts (Fig. 1C, top). However, the phosphorylation of the RBBP7 peptides encompassing Ser145, Ser354, and Ser393 was not statistically significant (fig. S2, A and B). Kinase assays revealed that recombinant DNMT1, RBBP7, and HAT1 were phosphorylated by active AMPK (Fig. 1C, bottom). Furthermore, when immunoprecipitated from transfected human umbilical vein endothelial cells (HUVECs), the wild-type forms of His-tagged DNMT1, FLAG-tagged RBBP7, or His-tagged HAT1, but not their corresponding Ser-to-Ala mutants, were phosphorylated (Fig. 1D). Liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis results indicated that Ser730 in DNMT1 was phosphorylated (fig. S2, C and D). We cannot exclude the possibility that AMPK phosphorylates sites other than Ser314 on RBBP7, which could be ruled out by further LC-MS/MS analysis of cells using phosphosite mutants. As expected, the AMPK activator AICAR increased the amount of phosphoserine on the AMPK substrate H2B in HUVECs (fig. S3A) but did not change the abundance of DNMT1, RBBP7, or HAT1 (fig. S3B). Together, these results indicate that AMPK phosphorylated DNMT1-Ser730, RBBP7-Ser314, and HAT1-Ser190.
AMPK activation increased interactions of DNMT1, RBBP7, and HAT1
Because interaction between RBBP7 and HAT1 is required for acetyltransferase activity (17), we examined whether AMPK initiated protein-protein interactions among DNMT1, RBBP7, and HAT1. Coimmunoprecipitation analysis showed that AICAR increased the interaction between DNMT1 and RBBP7 at 30 min and between HAT1 and RBBP7 at 10 min in transfected HUVECs in a manner that was abolished by mutation of the phosphorylation sites in these proteins (Fig. 1, E to H). Temporal analysis indicated that RBBP7 and HAT1 transiently interacted within 10 min of AICAR treatment. At 30 min, RBBP7 interacted more robustly with DNMT1 than with HAT1 (fig. S3C). These results indicated that AMPK promoted the DNMT1-RBBP7 and HAT1-RBBP7 interactions.
AMPK inhibited DNMT1 through phosphorylation of Ser730 in DNMT1 and Ser314 in RBBP7
Next, we examined whether AMPK-mediated phosphorylation of DNMT1 affected its activity. AICAR or metformin decreased DNMT1 activity in HUVECs and AMPK+/+ mouse embryonic fibroblasts (MEFs) but not in AMPK−/− MEFs (Fig. 2, A and B, and fig. S3D). DNMT1 activity was also decreased in AMPK−/− MEFs infected with adenovirus encoding constitutively active AMPK (Ad-AMPK-CA) but not those infected with adenovirus encoding dominant-negative AMPK (Ad-AMPK-DN) (Fig. 2C and fig. S3E). AICAR or metformin decreased DNMT1 activity in HUVECs but not after knockdown of AMPK, PARP-1, or RBBP7 (Fig. 2D and fig. S3F). Because PARP-1 associates with and ADP ribosylates DNMT1 to inhibit DNA methylation (16), PARP-1 knockdown (fig. S3F) was used as a control. Furthermore, AICAR or metformin decreased DNMT1 activity in HUVECs expressing wild-type or phosphomimetic (Ser-to-Asp) forms of DNMT1 or RBBP7 but not the Ser-to-Ala forms of these proteins (Fig. 2E). Similar results were observed in HUVECs subjected to pulsatile shear stress (Fig. 2, F and G), which activated AMPK (fig. S3D) and increases mitochondrial biogenesis (18). These results suggested that DNMT1 inhibition depended on AMPK-mediated phosphorylation of DNMT1-Ser730 and RBBP7-Ser314.
Fig. 2. AMPK decreased DNMT1 activity.
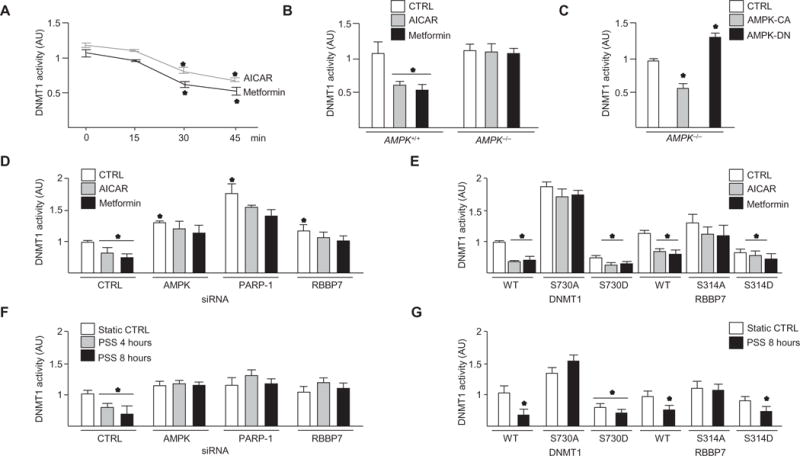
(A and B) DNMT1 activity in HUVECs (A) and AMPK+/+ or AMPK−/− MEFs (B) treated with AICAR or metformin or left untreated (CTRL). n = 4 independent experiments for (A) and (B). (C) DNMT1 activity in AMPK−/− MEFs infected with Ad-AMPK-CA or Ad-AMPK-DN [50 multiplicity of infection (MOI)]. n = 3 independent experiments. (D) DNMT1 activity in HUVECs transfected with control, AMPK, PARP-1, or RBBP7 small interfering RNA (siRNA) and treated as indicated. n = 3 independent experiments. (E) DNMT1 activity in cells transfected with the indicated forms of DNMT1 or RBBP7 and treated as indicated. n = 4 independent experiments. (F) DNMT1 activity in HUVECs transfected with control, AMPK, PARP-1, or RBBP7 siRNA and subjected to pulsatile shear stress (PSS). n = 3 independent experiments. (G) DNMT1 activity in HUVECs transfected with the indicated forms of DNMT1 or RBBP7 and subjected to PSS. n = 3 independent experiments. *P < 0.05.
AMPK activation decreased promoter methylation of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes
Promoter methylation analysis revealed that AICAR or metformin decreased promoter methylation of the PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 promoters in AMPK+/+ MEFs but not in AMPK−/− MEFs (Fig. 3A and fig. S4A). AICAR or metformin did not induce promoter hypomethylation in HUVECs upon PARP-1, AMPK, or RBBP7 knockdown (fig. S5, A to D). Furthermore, AICAR, metformin, or pulsatile shear stress decreased promoter methylation in HUVECs transfected with wild-type or DNMT1-S730D or RBBP7-S314D but not DNMT1-S730A or RBBP7-S314A (Fig. 3, B to D, and fig. S4, B to D).
Fig. 3. AMPK activation decreased promoter methylation of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes.
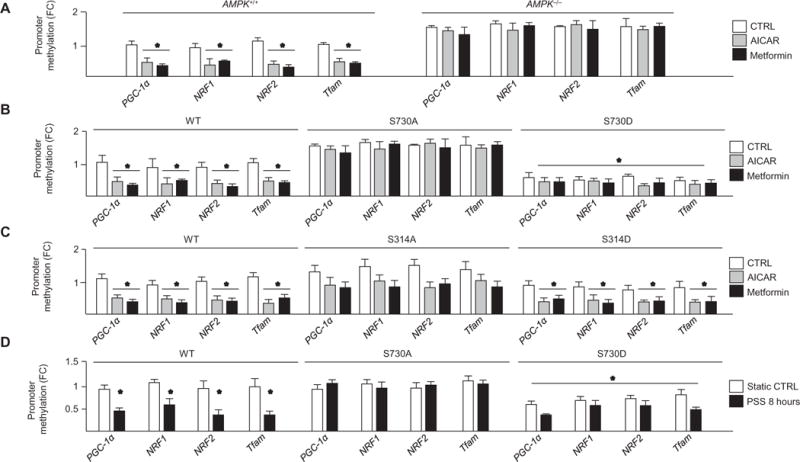
(A to C) Methylation-specific quantitative polymerase chain reaction (qPCR) analysis of promoter methylation in AICAR- or metformin-treated AMPK+/+ and AMPK−/− MEFs compared to nontreated AMPK+/+ group (A); HUVECs transfected with DNMT1-WT, DNMT1-S730A, or DNMT1-S730D (B); or HUVECs transfected with RBBP7-WT, RBBP7-S314A, or RBBP7-S314D (C). Cells were treated as indicated. n = 4 independent experiments for (A) to (C). (D) Promoter methylation status in HUVECs transfected with DNMT1-WT, DNMT1-S730A, or DNMT1-S730D and subjected to PSS. n = 3 independent experiments. *P < 0.05. FC, fold change.
AMPK increased HAT1 activity through phosphorylation of HAT1-Ser190 and RBBP7-Ser314
Because HAT1 and RBBP7 form an active acetyltransferase complex to acetylate histone 4 (H4) (acetyl-H4K5), which increases euchromatin (the decondensed chromatin state) (17, 19), we examined the effect of AMPK activation on HAT1 activity. AICAR or metformin increased HAT1 activity in AMPK+/+ MEFs but not in AMPK−/− MEFs (Fig. 4A). In addition, HAT1 was activated in AMPK−/− MEFs infected with Ad-AMPK-CA but not in those infected with Ad-AMPK-DN (Fig. 4B). AICAR, metformin, or pulsatile shear stress increased HAT1 activity, an effect that was attenuated by knockdown of AMPK, HAT1, or RBBP7 (Fig. 4, C and D). Furthermore, AICAR, metformin, or pulsatile shear stress did not increase HAT1 activity in HAT1-S190A– or RBBP7-S314A–transfected HUVECs (Fig. 4, E and F). These results suggested that HAT1 activity depended on AMPK-mediated phosphorylation of HAT1-Ser190 and RBBP7-Ser314.
Fig. 4. AMPK increased HAT1 activity.
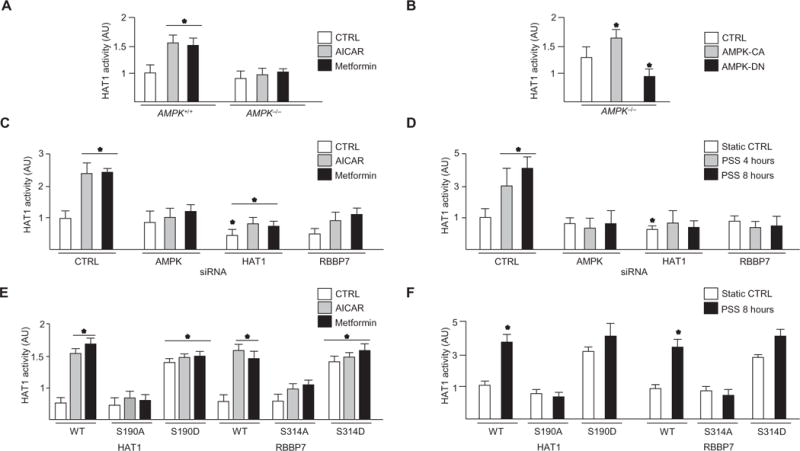
HAT1 activity was assessed in cells treated with AICAR or metformin or left untreated. (A) AMPK+/+ and AMPK−/− MEFs. (B) AMPK−/− MEFs were infected with Ad-AMPK-CA or Ad-AMPK-DN AMPK (50 MOI) before treatment. n = 3 independent experiments for (A) and (B). (C) HAT1 activity in HUVECs transfected with control, AMPK, HAT1, or RBBP7 siRNA before the indicated treatment. n = 4 independent experiments. (D) HAT1 activity in cells transfected with control, AMPK, HAT1, or RBBP7 siRNA and then subjected to PSS. n = 3 independent experiments. (E and F) HUVECs were transfected with the indicated forms of HAT1 or RBBP7 and treated as indicated. n = 4 independent experiments for (E) and n = 3 independent experiments for (F). *P < 0.05.
AMPK activation increased the histone acetylation in the promoters of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes
To determine the functional consequence of AMPK activation on histone acetylation, we measured H4K5 acetylation in AMPK+/+ and AMPK−/− MEFs using chromatin immunoprecipitation (ChIP). AICAR or metformin increased H4K5 acetylation of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 promoters in AMPK+/+ MEFs (Fig. 5A and fig. S6B) but not in AMPK−/− MEFs; HUVECs expressing HAT1-S190A or RBBP7-S314A (Fig. 5, A to C, and fig. S6, C and D); or HUVECs with knockdown of AMPK, HAT1, or RBBP7 (fig. S6A). The dependence of promoter acetylation on AMPK was also evident in HUVECs subjected to pulsatile shear stress (Fig. 5D and fig. S7, A and B).
Fig. 5. AMPK activation decreased nucleosomal compaction of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes.
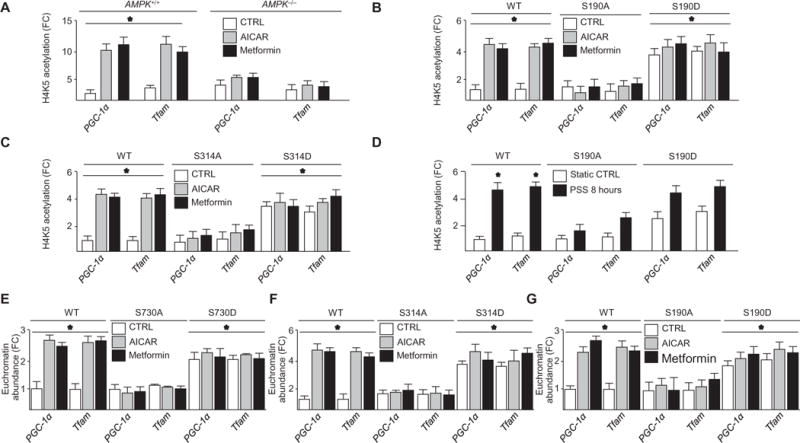
(A) H4K5 acetylation in AMPK+/+ or AMPK−/− MEFs treated with or without AICAR or metformin. n = 4 independent experiments. (B) H4K5 acetylation in HUVECs expressing indicated forms of HAT1 and treated as indicated. n = 4 independent experiments. (C) H4K5 acetylation in HUVECs expressing the indicated forms of RBBP7 and treated as indicated. n = 4 independent experiments. (D) H4K5 acetylation in cells expressing the indicated forms of HAT1 and then subjected to PSS. n = 3 independent experiments. (E to G) FAIRE analysis in HUVECs transfected with the indicated forms of DNMT1, RBBP7, or HAT1 and treated as indicated. n = 4 independent experiments. *P < 0.05.
AMPK activation decreased nucleosomal compaction of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes
We used formaldehyde-assisted isolation of regulatory elements (FAIRE) (20) to determine whether AMPK phosphorylation of the DNMT1-RBBP7-HAT1 epigenetic network increased euchromatin state and transcription. AICAR, metformin, or pulsatile shear stress increased the amount of euchromatin in PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 promoters in HUVECs transfected with wild-type, DNMT1-S730D, RBBP7-S314D, or HAT1-S190D but not DNMT1-S730A, RBBP7-S314A, or HAT1-S190A (Fig. 5, E to G, and figs. S8, A to F, and S9, A to C) or those with AMPK, RBBP7, DNMT1, or HAT1 knockdown (fig. S7, D and E). We also found that euchromatin was increased in AMPK+/+ MEFs but not in AMPK−/− MEFs (fig. S7C). In line with the increased euchromatin state, AICAR increased the protein abundance of PGC-1α, Tfam, NRF1, and NRF2 in HUVECs but not in HUVECs expressing DNMT1-S730A, RBBP7-S314A, or HAT1-S190A (fig. S8, G to I).
AMPK-mediated phosphorylation of DNMT1, RBBP7, and HAT1 increased membrane potential and biogenesis
We examined the consequence of AMPK-mediated phosphorylation of DNMT1, RBBP7, and HAT1 on mitochondrial biogenesis and function using HUVECs stably expressing wild-type DNMT1, RBBP7, HAT1, or their corresponding mutants (21). Fluorescence analysis of JC-1, a stain that shifts from green to red fluorescence upon an increase in mitochondrial membrane potential, indicated that AICAR failed to increase mitochondrial membrane potential in DNMT1-S730A–, RBBP7-S314A–, or HAT1-S190A–transfected HUVECs (Fig. 6A). MitoTracker staining indicated that AICAR increased mitochondrial mass in HUVECs expressing the wild-type DNMT1, RBBP7, or HAT1 genes but not those expressing the nonphosphorylatable forms (Fig. 6, B and C). Correlating with increased mitochondrial mass, AICAR increased mitochondrial DNA abundance and citrate synthase activity in HUVECs expressing wild-type or phosphomimetic mutants but not those expressing DNMT1-S730A, RBBP7-S314A, or HAT1-S190A (Fig. 6, D and E) (22). AICAR increased the activity of the first electron transporter in oxidative phosphorylation, complex I, and of the final electron transporter, complex IV, in HUVECs transfected with wild-type or phosphomimetic forms of DNMT1, RBBP7, or HAT1 compared to those transfected with DNMT1-S730A, RBBP7-S314A, or HAT1-S190A (Fig. 6, F and G). Correlating with mitochondrial activity, AICAR also increased complex V (ATP synthase) activity in HUVECs transfected with wild-type or phosphomimetic forms (Fig. 6H) (23). AICAR increased ATP production and decreased ROS generation in wild-type or phosphomimetic forms of DNMT1-, RBBP7-, or HAT1-transfected HUVECs but not in DNMT1-S730A–, RBBP7-S314A–, or HAT1-S190A–transfected HUVECs (Fig. 6, I and J). UCP2 and UCP3 are required to mitigate ROS production upon the onset of enhanced oxidative phosphorylation (24), and this compensatory response was eliminated upon DNMT1-S730A, RBBP7-S314A, or HAT1-S190A transfection (Fig. 6J). Our results suggest that epigenetic remodeling was an additional level of regulation enabling transcription factor access to promoters. To support this notion, we demonstrated that AICAR and metformin increased cyclic AMP response element–binding protein binding to the PGC-1α promoter, an AMPK substrate, and a PGC-1α transcriptional activator (fig. S3G) (25, 26). These results suggest that AMPK-mediated mitochondrial biogenesis and function occur partly through phosphorylation of DNMT1-Ser730, RBBP7-Ser314, and HAT1-Ser190.
Fig. 6. AMPK activation increased mitochondrial biogenesis and function.
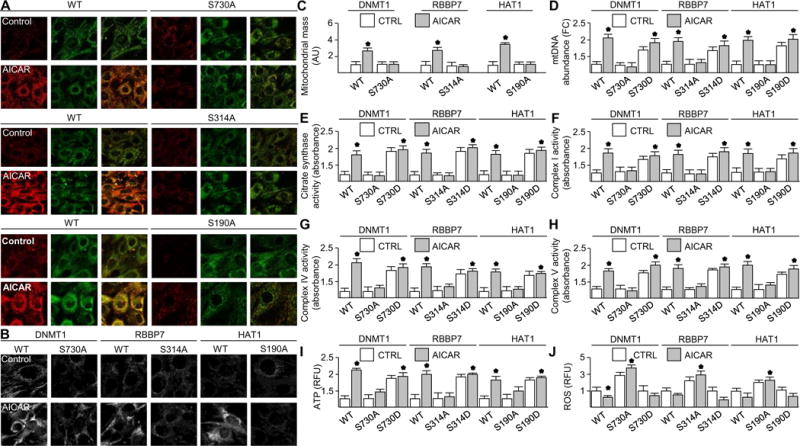
HUVECs expressing the indicated forms of DNMT1, RBBP7, or HAT1 were treated with AICAR or left untreated. (A) JC-1 staining. n = 6 independent experiments. (B) MitoTracker staining. n = 6 independent experiments. (C) Quantification of MitoTracker staining of six fields. n = 6 independent experiments. (D) Mitochondrial DNA (mtDNA) abundance. (E to H) Activity of citrate synthase (E), complex I (F), complex IV (G), and complex V (H). (I) ATP abundance. n = 3 independent experiments. RFU, relative fluorescence unit. (J) ROS abundance. n = 3 independent experiments for (D) to (J). *P < 0.05.
AMPK regulates the promoters of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes in vivo
To determine the effects of AMPK on the epigenetic regulation of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 in mouse aortas, we administered AICAR or metformin to AMPKα2+/+ mice and their AMPKα2−/− littermates. Consistent with our in vitro results, AICAR and metformin decreased DNMT1 and increased HAT1 binding to the respective promoters in AMPKα2+/+ mice (Fig. 7, A and B, and fig. S10, A and C). Likewise, euchromatin was increased; promoter methylation was decreased; and PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 mRNA abundance were increased in AMPKα2+/+ mouse aortas (Fig. 7, C to E, and fig. S10, B, D, and E). Conversely, AICAR and metformin had little or no effect on modulating these epigenetic or transcriptional events in AMPKα2−/− mice (Fig. 7, A to E, and fig. S10, A to E). Complex IV activity was also higher in AMPKα2+/+ mice with or without AICAR or metformin, compared to AMPKα2−/− mice (Fig. 7F). Collectively, these data support the role of AMPK in regulating mitochondrial functions through nucleosome remodeling in vivo.
Fig. 7. AICAR regulates nucleosome remodeling and gene expression through AMPKα2 in vivo.
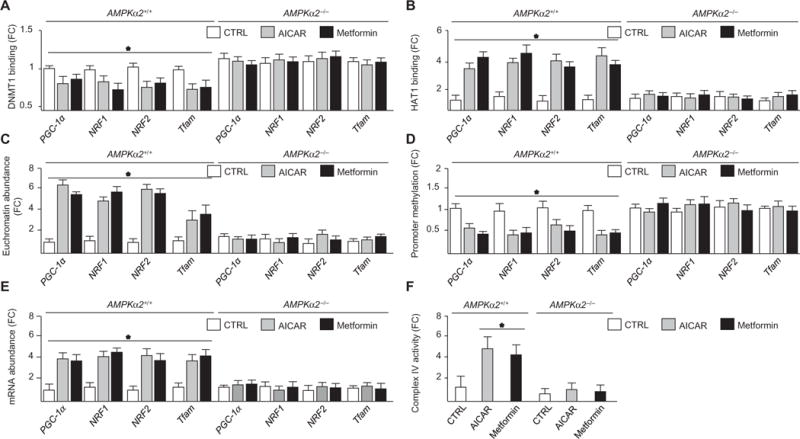
Aortas were isolated from AMPKα2+/+ and AMPKα2−/− mice administered AICAR or metformin. (A) DNMT1 binding to promoters. (B) HAT1 binding to promoters. (C) Euchromatin abundance. (D) Methylation status. (E) Abundance of the indicated mRNAs. (F) Complex IV activity. n = 12 mice per genotype and treatment. *P < 0.05.
DISCUSSION
This work enhances our understanding of AMPK’s role as an epigenetic regulator. Specifically, AMPK increased histone acetylation and decreased DNA methylation at PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 promoters through phosphorylation of DNMT1, RBBP7, and HAT1. DNMT1 is the major methyltransferase responsible for regulating cytosine methylation patterns and genome imprinting (27). By copying methylation patterns from parent to daughter DNA strands and being active only at replication forks, DNMT1 provides an inheritable mechanism of genetic regulation irrespective of DNA sequence heterogeneity (27). Throughout this process, DNMT1 activity is bimodal because it has both an active and autoinhibitory mechanism that ensures the high-fidelity preservation of DNMT1-mediated DNA methylation status. DNMT1 “self-regulates” to increase methylation at hemi-mCpG regions while structurally protecting unmethylated CpG sites, which associate with its CXXC domain and CXXC-BAH1 linker (28, 29). AMPK phosphorylation of Ser730, which is located near the autoinhibitory domain, may impart structural changes that cause constitutive association between unmethylated CpG regions and the CXXC domain, thus inhibiting methyl group transfer. Alternatively, the phosphorylation of Ser730 may induce conformational changes in DNMT1, thereby altering its protein-protein interactions. Because several proline residues flank Ser730 in the AMPK phosphorylation sequence, it is likely that AMPK-mediated phosphorylation of DNMT1 requires isomerization of these proline residues into their trans form before phosphorylation. Our results indicate that AMPK-mediated phosphorylation of DNMT1 had a direct inhibitory effect as well as an indirect inhibitory effect by affecting its interaction with RBBP7 (Figs. 1 and 2). Thus, it is possible that AMPK may also govern proline isomerases, such as peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, which also contains an AMPK phosphorylation sequence, to allow AMPK-mediated phosphorylation of DNMT1.
Several signal transduction pathways influence DNMT1 activity. For example, CTCF (CCCTC-binding factor) promotes PARP-1–mediated PARylation and inhibition of DNMT1 (16) to preserve the DNA methylation status, particularly at unmethylated regions of daughter cells (16, 27). This cascade may be governed by AMPK (8). We demonstrated here that AMPK decreased DNMT1 activity at 30 min (Fig. 2A), but promoter methylation status was not decreased until 4 hours (Fig. 3). Although DNMT1 copies methylation patterns during DNA replication, this delay may not be correlated to cell cycle because AMPK activation induces cell cycle arrest (30). Alternatively, our data indicated that AMPK-dependent nucleosome remodeling, which is required before transactivation, is likely to require various types of epigenetic regulation that promote gene transcription. This mechanism may explain the epigenetic mechanisms by which AMPK and pulsatile shear stress influence cellular lineage differentiation in the endothelium (30–32). Association with RBBP7 decreased the ability of DNMT1 to methylate or increased its protection of unmethylated CpG regions, allowing access by transcriptional activating machinery.
HAT1, initially classified as a type B histone acetyltransferase because of its cytosolic location and acetylation of free, newly synthesized histones (19, 33–35), is now understood to also play a role in chaperoning histones into the nucleus for nucleosomal integration (19, 36). HAT1 forms an active complex with RBBP7 and H4 (36). Because the interaction between RBBP7 occurs in helix 1 of the histone fold of H4, a region that is not accessible when H4 is in chromatin, it is likely that this interaction serves as a histone chaperone complex. Our data supported the notion that this mechanism may initiate the removal of histones from specific gene elements while integrating acetylated histones into neighboring genomic regions to facilitate transcriptional activation (Fig. 5, A to D, and figs. S6 and S7, A and B). Ultimately, this interaction results in the neutralization of the positive charge on acetylated histones that attenuates the electrostatic interaction between DNA and histones to create a relaxed nucleosome structure (31, 33).
Although AMPK phosphorylates H2B and indirectly regulates it through phosphorylation of O-linked-N-acetylglucosamine transferase, the phosphoproteome that AMPK governs is currently unknown (12, 37). Here, we demonstrated that AMPK-mediated phosphorylation of DNMT1, RBBP7, and HAT1 led to increased mitochondrial biogenesis through PGC-1α, NRF1, NRF2, and Tfam transactivation and control of mitochondrial membrane potential through transactivation of UCP2 and UCP3. However, the mechanism we define here does not mutually exclude other molecular events regulated by AMPK that facilitate mitochondrial biogenesis and function. Kinetically, histone modifications likely precede alterations in methylation patterns that facilitate nucleosome remodeling, a requirement for transcription factor access and gene transcription. PGC-1α is coactivated by AMPK phosphorylation and sirtuin 1 (SIRT1) deacetylation, and AMPK primes the nucleosome for PGC-1α binding to PGC-1α, NRF1, and NRF2 promoters to increase mitochondrial biogenesis (10). Nucleosome remodeling also likely precedes mitochondrial fission factor (MFF) transactivation, enabling phosphorylation of MFF by AMPK and mitochondrial fission and improved mitochondrial function (38). Independently of mitochondrial biogenesis, AMPK increases catabolic pathway activity, increasing electron flux through the electron transport chain. Chromatin remodeling plays an important role in priming mitochondria for increased oxidative phosphorylation, impairment of which results in increased ROS production (Fig. 6J). Nonetheless, these molecular events would ultimately improve ATP production and control of mitochondrial ROS production, possibly through complex I function (39, 40).
We present in vivo correlational data demonstrating that AMPKα2−/− results in alterations in DNA methylation patterns as well as inhibition of nucleosome remodeling. In vivo, AMPKα2 deficiency results in altered mitochondrial structure, decreased oxidative capacity, and hyperphysiological ROS (38, 39). Further, inhibition of DNMT1 in vivo by 5-aza-2′-deoxycytidine blocks endothelial inflammation and attenuates atherosclerotic lesion formation (32). Consistent with these results, enhanced DNA methylation patterns cause endothelial cell senescence (32). Thus, because of the fundamental importance of DNMT1 and HAT1, it is likely that the effects of AMPK regulation of DNMT1, RBBP7, and HAT1 extend beyond mitochondrial homeostasis to vascular function.
Our previous work indicates that AMPK is also likely to phosphorylate charged multivesicular body protein 1B (CHMP1B) and Zuotin-related factor 1 (ZRF1) (13). CHMP1B plays a role in gene regulation through chromatin structural maintenance, and ZRF1 facilitates histone 2A Lys119 ubiquitination, leading to chromatin remodeling and transcription activation. Therefore, these two molecules, together with HAT1, DNMT1, and RBBP7, may promote AMPK-mediated nucleosome remodeling on genome-wide scale, providing potential to uncover new treatment approaches to metabolic and vascular pathologies.
MATERIALS AND METHODS
Data mining
EntroSolve (EntroSolve.com) was recruited to mine databases composed of epigenetic factors that contained an AMPK phosphorylation sequence (βΦβXXXS/TXXXΦ,, where β represents basic residues H, K, or R; and Φ represents hydrophobic residues F, L, M, I, V, or A) or a variant sequence lacking the basic residues at the +4 and +6 positions (14).
Cell culture
HUVECs were cultured in M199 medium, and all other cell types were cultured in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, catalog #D6429-500M) supplemented with 10% fetal bovine serum as previously described (8).
Transfection
Transfection was conducted with Lipofectamine LTX (catalog #15338100) according to the manufacturer’s instructions, and then transfected cells were selected for and enriched as previously described (20).
In vitro kinase assay
Kinase assays were conducted as previously described (8) in 50 mM Hepes (pH 7.4), 1 mM AMP, 1 mM [32P]-γ-ATP, 5 mM MgCl2, 1 pM AMPKα2, and 1 nM recombinant protein in a 50-μl reaction volume at 37°C for 1 hour. Radioactive counts were detected by scintillation (8).
SDS-PAGE and in-gel digestion
Proteins were separated using SDS–polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie blue and then in-gel–reduced with dithiothreitol, alkylated with iodoacetamide, and digested overnight with trypsin (Promega) at 37°C. Peptides were extracted from the gels with 5% acetic acid in H2O and then with 5% acetic acid in CH3CN/H2O (1:1, v/v).
LC-MS/MS analysis
LC-MS/MS analysis was performed with an LTQ-Orbitrap Velos mass spectrometer coupled to EASY-nLC II high-performance liquid chromatography (HPLC) system and a nanoelectrospray ionization source (Thermo Fisher Scientific). The sample injection, enrichment, desalting, and HPLC separation were carried out automatically on a custom-built trapping column (150 μm × 50 mm) and a separation column (75 μm × 120 mm) packed with ReproSil-Pur C18-AQ resin (5-μm particle size and 300 Å pore size from Dr. Maisch HPLC GmbH). The LTQ-Orbitrap Velos mass spectrometer was operated in positive-ion mode and spray voltage of 1.8 kV. MS/MS spectra were acquired in a data-dependent scan mode where 1 full MS scan was followed with 20 MS/MS scans. To obtain high-quality MS/MS, the mass spectrometer was set up in selected ion monitoring mode, and fragmentations of [M + 2H]2+ ion of the phosphorylated DNMT1 peptide containing residues 714 to 732 was monitored.
Data processing
The LC-MS/MS data were used for the identification of phosphorylation sites using Mascot Server 2.2 (Matrix Science, Boston, MA) by searching the mass/charge ratio data files against the National Center for Biotechnology Information database. The maximum number of miss-cleavages for trypsin was set at 2 per peptide. The mass tolerances for MS and MS/MS were 50 parts per million and 0.6 Da, respectively. Cys carbamidomethylation and Ser/Thr phosphorylation were set as fixed and variable modifications, respectively. Peptides identified with individual scores at or above the Mascot-assigned homology score, P < 0.01, and individual peptide score of >30 were considered specific peptide sequences. The false discovery rates (>0.95) were determined by decoy database search.
Immunoprecipitation and blotting
Immunoprecipitation was conducted according to the standard protocols outlined by Abcam under nondenaturing conditions. Immunoblotting was conducted as previously described (8).
Nuclear extraction
Nuclear extracts were prepared with the Active Motif Nuclear Extract Kit (catalog #40010) according to the manufacturer’s specifications.
DNMT1 activity assay
DNMT1 activity was quantified from nuclear extracts according to the manufacturer’s instructions (Active Motif, catalog #55006).
HAT1 activity assay
HAT1 activity was measured with a Histone Acetyltransferase Assay Kit by Active Motif (cat #56100) according to the manufacturer’s instructions.
Genomic DNA isolation
DNA was isolated with UltraPure Phenol/Chloroform/Isoamyl Alcohol (25:24:1, v/v) (Thermo Fisher Scientific, catalog #15593031) according to the manufacturer’s instructions.
Promoter methylation assay
Methylated and hemimethylated DNA was analyzed with the EpiMark 5-hmC and 5-mC Analysis Kit by New England Biolabs (catalog #E33175) according to the manufacturer’s guidelines quantified using qPCR and primers (table S2).
FAIRE analysis
Isolation of transcriptionally active euchromatin was achieved using FAIRE analysis as previously described (19) using the same promoter primers.
mRNA quantification
RNA was purified using TRIzol reagent from Life Technologies and converted to complementary DNA (cDNA) with Promega reverse transcriptase according to the manufacturer’s instructions. cDNA was quantified using qPCR with iTaq Universal SYBR Green Supermix from Bio-Rad. Results were calculated using the 2−ΔΔCt method. Primers used for qPCR analysis are listed in table S2.
ChIP assay
ChIP assays were performed as outlined by Abcam (8). Resulting DNA was purified using Qiagen PCR purification kit before qPCR analysis. The same primers used for promoter methylation analysis were used for ChIP analysis.
Mitochondrial biogenesis and mitochondrial membrane potential
Cells were grown in chambered glass bottom wells, treated, and stained with MitoTracker or JC-1 dye (Invitrogen, catalog #M7514 or Cayman Chemical, catalog #10009172, respectively) according to the manufacturer’s instructions. Imaging was conducted using a Leica SP5 inverted confocal microscope. Mitochondrial intensity was quantified using IMARIS imaging software.
Mitochondrial DNA isolation and mitochondrial enzyme activity
Mitochondria were isolated with the use of the MitoCheck Mitochondrial Isolation Kit (Cayman Chemical, catalog #701010). Complexes I, IV, and V or citrate synthase activity was monitored with the MitoCheck Complex I Activity Assay Kit (Cayman Chemical, catalog #700930), MitoCheck Complex IV Activity Assay Kit (Cayman Chemical, catalog #700990), MitoCheck Complex V Activity Assay Kit (Cayman Chemical, catalog #701000), or MitoCheck Citrate Synthase Activity Assay Kit (Cayman Chemical, catalog #701040) according to the manufacturer’s specifications.
Mitochondrial DNA quantification
Mitochondrial DNA was quantified as previously described (41).
ATP and ROS measurements
ATP abundance was measured using ATP Assay Kit (Colorimetric/Fluorometric) (Abcam, catalog #ab83355) according to the manufacturer’s instructions. ROS was measured with the Cellular ROS Detection Assay Kit (Abcam, catalog #ab113851) according to the manufacturer’s instructions. Data were obtained as relative fluorescence units.
Pulsatile shear stress
Glass slides were coated with collagen (Sigma-Aldrich, catalog #C7661-5MG) before seeding HUVECs, which were then loaded into flow channels and subjected to pulsatile shear stress at 12 ± −4 dyne/cm2 or left under standard culture conditions.
Animal protocol
Animals were administered AICAR (200 mg/kg; Sigma-Aldrich, catalog #A9978-25MG) or metformin (Sigma-Aldrich, catalog #D150959-5G) through intraperitoneal injection 4 hours before sacrificing.
Statistical analysis
Data are means ± SEM of at least three independent experiments. Tests used for or nonparametric data included Kruskal-Wallis test with Tukey’s post hoc test and Mann-Whitney U test. Parametric data were analyzed using analysis of variance (ANOVA) with post hoc Bonferroni. Unless otherwise indicated, P values <0.05 are considered statistically significant.
Supplementary Material
Fig. S1. Predicted AMPK-regulated epigenetic network.
Fig. S2. Phosphorylation of RBBP7 and DNMT1 by AMPK.
Fig. S3. Confirmation of pharmacologically induced AMPK activation and siRNA efficacy and time course analysis of RBBP7 interaction with DNMT1 and HAT1.
Fig. S4. AMPK activation decreased UCP2 and UCP3 promoter methylation.
Fig. S5. AMPK activation decreased promoter methylation in an AMPK- and RBBP7-dependent manner.
Fig. S6. AMPK activation enhanced NRF1, NRF2, UCP2, and UCP3 promoter histone acetylation through HAT1 and RBBP7.
Fig. S7. AMPK activation decreased nucleosomal compaction of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes.
Fig. S8. AMPK activation decreased nucleosomal compaction in promoters of NRF1, NRF2, UCP2, and UCP3, resulting in increased translation.
Fig. S9. Pulsatile shear stress decreased nucleosomal compaction of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes.
Fig. S10. AICAR regulated nucleosome remodeling and increased gene expression of UCP2 and UCP3 through AMPKα2 in vivo.
Table S1. AMPK phosphorylation sequence containing proteins predicted to be involved in epigenetic regulation.
Table S2. Primers used in methylation, FAIRE, ChIP, and mRNA quantification.
Acknowledgments
We thank G. Rosete for data management and S. Miller for numerous discussions about programming. Funding: This work was funded by Loma Linda University Grants for Research and School Partnerships (699330 to T.L.M.) and NIH (HL108735 to S.C., S.S., and J.Y.-J.S.).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencesignaling.org/cgi/content/full/10/464/eaaf7478/DC1
Author contributions: T.L.M. and B.G. conceived, designed and drafted the manuscript, and performed molecular assays, bioinformatics analysis, and statistical analysis. F.Z. performed MS analysis. M.M. and H.X. performed molecular assays. D.A.J., Y.W., S.S., S.C., and J.Y.-J.S. participated in design, coordination, and drafting the manuscript.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Cheng Z, Ristow M. Mitochondria and metabolic homeostasis. Antioxid Redox Signal. 2013;19:240–242. doi: 10.1089/ars.2013.5255. [DOI] [PubMed] [Google Scholar]
- 2.Jornayvaz FR, Shulman GI. Regulation of mitochondrial biogenesis. Essays Biochem. 2010;47:69–84. doi: 10.1042/bse0470069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lizcano JM, Göransson O, Toth R, Deak M, Morrice NA, Boudeau J, Hawley SA, Udd L, Mäkelä TP, Hardie DG, Alessi DR. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23:833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hawley SA, Pan DA, Mustard KJ, Ross L, Bain J, Edelman AM, Frenguelli BG, Hardie DG. Calmodulin-dependent protein kinase kinase-β is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2:9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Mungai PT, Waypa GB, Jairaman A, Prakriya M, Dokic D, Ball MK, Schumacker PT. Hypoxia triggers AMPK activation through reactive oxygen species-mediated activation of calcium release-activated calcium channels. Mol Cell Biol. 2011;31:3531–3545. doi: 10.1128/MCB.05124-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun W, Lee TS, Zhu M, Gu C, Wang Y, Zhu Y, Shyy JYJ. Statins activate AMP-activated protein kinase in vitro and in vivo. Circulation. 2006;114:2655–2662. doi: 10.1161/CIRCULATIONAHA.106.630194. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Wang Y, Bao C, Xu Y, Shen H, Chen J, Yan J, Chen Y. Metformin interacts with AMPK through binding to γ subunit. Mol Cell Biochem. 2012;368:69–76. doi: 10.1007/s11010-012-1344-5. [DOI] [PubMed] [Google Scholar]
- 8.Gongol B, Marin T, Peng IC, Woo B, Martin M, King S, Sun W, Johnson DA, Chien S, Shyy JYJ. AMPKα2 exerts its anti-inflammatory effects through PARP-1 and Bcl-6. Proc Natl Acad Sci USA. 2013;110:3161–3166. doi: 10.1073/pnas.1222051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calvert JWI, Gundewar S, Jha S, Greer JJ, Bestermann WH, Tian R, Lefer DJ. Acute metformin therapy confers cardioprotection against myocardial infarction via AMPK-eNOS–mediated signaling. Diabetes. 2008;57:696–705. doi: 10.2337/db07-1098. [DOI] [PubMed] [Google Scholar]
- 10.Cantó C, Auwerx J. PGC-1α, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr Opin Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: The central role of PGC-1α. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 12.Bungard D, Fuerth BJ, Zeng PY, Faubert B, Maas NL, Viollet B, Carling D, Thompson CB, Jones RG, Berger SL. Signaling kinase AMPK activates stress-promoted transcription via histone H2B phosphorylation. Science. 2010;329:1201–1205. doi: 10.1126/science.1191241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marin TL, Gongol B, Martin M, King SJ, Smith L, Johnson DA, Subramaniam S, Chien S, Shyy JYJ. Identification of AMP-activated protein kinase targets by a consensus sequence search of the proteome. BMC Syst Biol. 2015;9:13. doi: 10.1186/s12918-015-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weekes J, Ball KL, Caudwell FB, Hardie DG. Specificity determinants for the AMP-activated protein kinase and its plant homologue analysed using synthetic peptides. FEBS Lett. 1993;334:335–339. doi: 10.1016/0014-5793(93)80706-z. [DOI] [PubMed] [Google Scholar]
- 15.Shannon PT, Reiss DJ, Bonneau R, Baliga NS. The Gaggle: An open-source software system for integrating bioinformatics software and data sources. BMC Bioinformatics. 2006;7:176. doi: 10.1186/1471-2105-7-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zampieri M, Guastafierro T, Calabrese R, Ciccarone F, Bacalini MG, Reale A, Perilli M, Passananti C, Caiafa P. ADP-ribose polymers localized on Ctcf–Parp1–Dnmt1 complex prevent methylation of Ctcf target sites. Biochem J. 2012;441:645–652. doi: 10.1042/BJ20111417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klingberg R, Jost JO, Schümann M, Gelato KA, Fischle W, Krause E, Schwarzer D. Analysis of phosphorylation-dependent protein–protein interactions of histone H3. ACS Chem Biol. 2015;10:138–145. doi: 10.1021/cb500563n. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Peng IC, Cui X, Li YS, Chien S, Shyy JYJ. Shear stress, SIRT1, and vascular homeostasis. Proc Natl Acad Sci USA. 2010;107:10268–10273. doi: 10.1073/pnas.1003833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Han J, Kang B, Burgess R, Zhang Z. Human histone acetyltransferase 1 protein preferentially acetylates H4 histone molecules in H3.1-H4 over H3.3-H4. J Biol Chem. 2012;287:6573–6581. doi: 10.1074/jbc.M111.312637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu LS, Wei DH, Tang CK, Wang GX, Zhang SC, Yin WD, Yang YZ, Legrand AP, Guidoin R. A HUVEC line with a stable expression of the VEGF121 gene to achieve complete endothelialization of blood conduits. Artif Cells Blood Substit Immobil Biotechnol. 2007;35:319–331. doi: 10.1080/10731190701378642. [DOI] [PubMed] [Google Scholar]
- 22.Cormio A, Guerra F, Cormio G, Pesce V, Fracasso F, Loizzi V, Cantatore P, Selvaggi L, Gadaleta MN. The PGC-1α-dependent pathway of mitochondrial biogenesis is upregulated in type I endometrial cancer. Biochem Biophys Res Commun. 2009;390:1182–1185. doi: 10.1016/j.bbrc.2009.10.114. [DOI] [PubMed] [Google Scholar]
- 23.Larsen S, Nielsen J, Hansen CN, Nielsen LB, Wibrand F, Stride N, Schroder HD, Boushel R, Helge JW, Dela F, Hey-Mogensen M. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J Physiol. 2007;590:3349–3360. doi: 10.1113/jphysiol.2012.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mailloux RJ, Harper ME. Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic Biol Med. 2011;51:1106–1115. doi: 10.1016/j.freeradbiomed.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am J Clin Nutr. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomson DM, Herway ST, Fillmore N, Kim H, Brown JD, Barrow JR, Winder WW. AMP-activated protein kinase phosphorylates transcription factors of the CREB family. J Appl Physiol. 2008;104:429–438. doi: 10.1152/japplphysiol.00900.2007. [DOI] [PubMed] [Google Scholar]
- 27.Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- 29.Song J, Rechkoblit O, Bestor TH, Patel DJ. Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation. Science. 2011;331:1036–1040. doi: 10.1126/science.1195380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motoshima H, Goldstein BJ, Igata M, Araki E. AMPK and cell proliferation—AMPK as a therapeutic target for atherosclerosis and cancer. J Physiol. 2006;574:63–71. doi: 10.1113/jphysiol.2006.108324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim EK, Lim S, Park JM, Seo JK, Kim JH, Kim KT, Ryu SH, Suh PG. Human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by AMP-activated protein kinase. J Cell Physiol. 2012;227:1680–1687. doi: 10.1002/jcp.22892. [DOI] [PubMed] [Google Scholar]
- 32.Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, Jang I, Son DJ, Kim D, Pan C, Fan Y, Jordan IK, Jo H. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J Clin Invest. 2014;124:3187–3199. doi: 10.1172/JCI74792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu H, Moshkina N, Min J, Zeng H, Joshua J, Zhou MM, Plotnikov AN. Structural basis for substrate specificity and catalysis of human histone acetyltransferase 1. Proc Natl Acad Sci USA. 2012;109:8925–8930. doi: 10.1073/pnas.1114117109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo WHW, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keck KM, Pemberton LF. Histone chaperones link histone nuclear import and chromatin assembly. Biochim Biophys Acta. 2012;1819:277–289. doi: 10.1016/j.bbagrm.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Q, Yang C, Du Y, Chen Y, Liu H, Deng M, Zhang H, Zhang L, Liu T, Liu Q, Wang L, Lou Z, Pei H. AMPK regulates histone H2B O-GlcNAcylation. Nucleic Acids Res. 2014;42:5594–5604. doi: 10.1093/nar/gku236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toyam EQ, Herzig S, Courchet J, Lewis TL, Jr, Losón OC, Hellberg K, Young NP, Chen H, Polleux F, Chan DC, Shaw RJ. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science. 2016;351:275–281. doi: 10.1126/science.aab4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Athéa Y, Viollet B, Mateo P, Rousseau D, Novotova M, Garnier A, Vaulont S, Wilding JR, Grynberg A, Veksler V, Hoerter J, Ventura-Clapier R. AMP-activated protein kinase α2 deficiency affects cardiac cardiolipin homeostasis and mitochondrial function. Diabetes. 2007;56:786–794. doi: 10.2337/db06-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang S, Zhang M, Liang B, Xu J, Xie Z, Liu C, Viollet B, Yan D, Zou MH. AMPKα2 deletion causes aberrant expression and activation of NAD(P)H oxidase and consequent endothelial dysfunction in vivo: Role of 26S proteasomes. Circ Res. 2010;106:1117–1128. doi: 10.1161/CIRCRESAHA.109.212530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rooney JP, Ryde IT, Sanders LH, Howlett EH, Colton MD, Germ KE, Mayer GD, Greenamyre JT, Meyer JN. PCR based determination of mitochondrial DNA copy number in multiple species. Methods Mol Biol. 2015;1241:23–38. doi: 10.1007/978-1-4939-1875-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Predicted AMPK-regulated epigenetic network.
Fig. S2. Phosphorylation of RBBP7 and DNMT1 by AMPK.
Fig. S3. Confirmation of pharmacologically induced AMPK activation and siRNA efficacy and time course analysis of RBBP7 interaction with DNMT1 and HAT1.
Fig. S4. AMPK activation decreased UCP2 and UCP3 promoter methylation.
Fig. S5. AMPK activation decreased promoter methylation in an AMPK- and RBBP7-dependent manner.
Fig. S6. AMPK activation enhanced NRF1, NRF2, UCP2, and UCP3 promoter histone acetylation through HAT1 and RBBP7.
Fig. S7. AMPK activation decreased nucleosomal compaction of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes.
Fig. S8. AMPK activation decreased nucleosomal compaction in promoters of NRF1, NRF2, UCP2, and UCP3, resulting in increased translation.
Fig. S9. Pulsatile shear stress decreased nucleosomal compaction of PGC-1α, NRF1, NRF2, Tfam, UCP2, and UCP3 genes.
Fig. S10. AICAR regulated nucleosome remodeling and increased gene expression of UCP2 and UCP3 through AMPKα2 in vivo.
Table S1. AMPK phosphorylation sequence containing proteins predicted to be involved in epigenetic regulation.
Table S2. Primers used in methylation, FAIRE, ChIP, and mRNA quantification.


