Abstract
The road to producing an eye begins with the decision to commit a population of cells to adopting an eye tissue fate, the process of retinal determination. Over the past decade and a half, a network of transcription factors has been found to mediate this process in all seeing animals. This retinal determination network is known to regulate not only tissue fate but also cell proliferation, pattern formation, compartment boundary establishment, and even retinal cell specification. The compound eye of the fruit fly, Drosophila melanogaster, has proven to be an excellent experimental system to study the mechanisms by which this network regulates organogenesis and tissue patterning. In fact the founding members of most of the gene families that make up this network were first isolated in Drosophila based on loss-of-function phenotypes that affect the eye. This chapter will highlight the history of discovery of the retinal determination network and will draw attention to the molecular and biochemical mechanisms that underlie our understanding of how the fate of the retina is determined.
1. Introduction
The retinal determination network in Drosophila sets in motion the process by which approximately 20,000 undifferentiated cells are specified and incorporated into the several hundred unit eyes or ommatidia that comprise the adult retina. The genes that are included within the network work to coordinate cell proliferation rates, regulate the initiation and progression of the morphogenetic furrow, specify and maintain individual cell fates, as well as eliminate excess numbers by programmed cell death. The network is a central part of eye development from its beginnings during embryogenesis through its completion within the adult. As retinal precursor cells are initially set aside during embryogenesis (Cohen, 1993; Held, 2002), several members of the network begin the task of canalizing these cells toward adopting an eye fate. And as the morphogenetic furrow later patterns the retina (Lebovitz and Ready, 1986; Ready et al., 1976; Wolff and Ready, 1991), the retinal determination network plays critical roles in its initiation and progression. Later, as individual ommatidia are being assembled behind the furrow (Cagan and Ready, 1989a,b; Tomlinson and Ready, 1986, 1987a,b), a number of these factors play critical roles in the acquisition of photoreceptor neurons, lens-secreting cone, and optically insulating pigment cell fates. And finally, select retinal determination genes function in the adult retina to activate the expression of light-capturing rhodopsin genes (Sheng et al., 1997). This review will introduce the reader to the genes that comprise the retinal determination network in Drosophila and will highlight the role that these genes play during eye specification. It will also draw attention to the intricate molecular and biochemical relationships that exist between network members. Particular attention will be placed on emphasizing the spatial and temporal nature of these relationships within the retinal epithelium.
2. Structure and Development of the Drosophila Eye
Since its initial structural and developmental description by Ready and coworkers more than 30 years ago, the compound eye has served as an excellent model system for understanding a myriad of developmental processes including organogenesis, cell proliferation and apoptosis, compartment boundary establishment, pattern formation, cell fate specification, planar cell polarity and cell rotation, as well as axon projection and guidance. Its simple adult structure and stereotyped developmental history have allowed us to also study basic mechanisms of morphogen gradients, gene regulation and signal transduction, and gene regulatory networks. Despite three decades of exploration, the eye continues to provide fertile ground for the discovery of new and exciting cellular mechanisms. Indeed, each passing year brings more than a hundred new papers and this continues to fuel our growing understanding of the mechanisms that underlie the specification and patterning of this near perfect simple nervous system.
The adult retina consists of approximately 800 unit eyes. Each is a six-sided replica of its neighbors; thus the adult retina is a precise hexagonal display that has been described as a neurocrystalline lattice (Fig. 1.1A; Ready et al., 1976). Each ommatidium contains approximately 20 cells: 8 photoreceptor neurons and 12 lens-secreting cone cells and optically insulating pigment cells (Fig. 1.1B and C; Ready, 1989). The placement of the photoreceptors within the ommatidium is stereotyped and resembles that of an asymmetric trapezoid (Fig. 1.1D and E; Dietrich, 1909). Each cell within the ommatidium is specified through a combination of cell–cell interactions as well as short- and long-range signals during the final larval instar and pupal stage of the life cycle (Cagan and Ready, 1989a,b; Tomlinson and Ready, 1986, 1987a,b). The template for the pupal and adult retinas is the eye imaginal disk. Prior to the onset of pattern formation, all cells within the disk are unpatterned and undifferentiated. But during the third and final instar, a wave of differentiation initiates at the posterior margin of the disk and proceeds toward the anterior edge of the epithelium. The most anterior edge of this wave is called the morphogenetic furrow and transforms the sea of undifferentiated cells into a tiling of periodically spaced ommatidial rudiments (Fig. 1.2A–C; Ready et al., 1976; Wolff and Ready, 1991). Once the photoreceptors cells have been specified, they will undergo significant morphological changes including the elaboration of their rhabdomeres, which are the light sensitive organelles homologous to the outer segments of vertebrate photoreceptors (Kumar and Ready, 1995; Longley and Ready, 1995). These cells will go on to express rhodopsins, which are light-capturing photopigments, while the cone cells will secrete the overlying lens and the pigment cells will provide the bulk of the optically insulating pigment granules (reviewed in Wolff and Ready, 1993).
Figure 1.1.
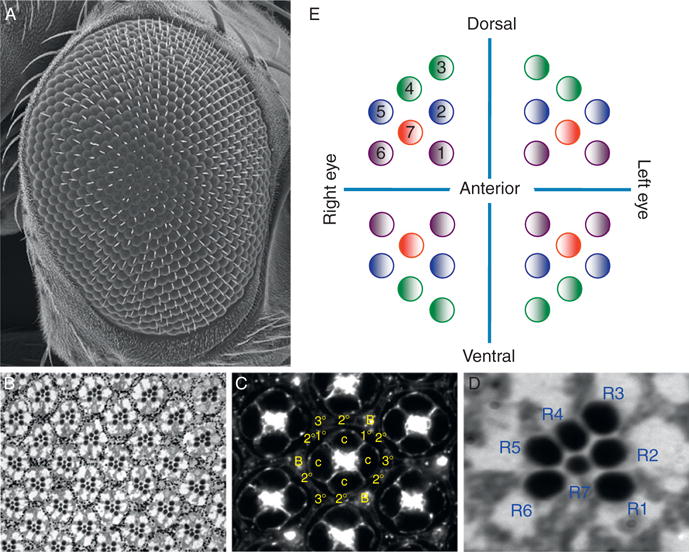
Structure of the adult compound eye. (A) Scanning electron micrograph of the adult eye. (B) Section of the adult retina showing the photoreceptor neurons. (C) Section of the pupal retina showing all cone, pigment, and bristle cells. (D) A high magnification of a single ommatidium from the adult retina. Note that each photoreceptor neuron is given a unique identifier number. (E) A schematic describing the orientation of ommatidia in the dorsal and ventral quadrants in the left and right compound eyes. Anterior is to the right in all images.
Figure 1.2.
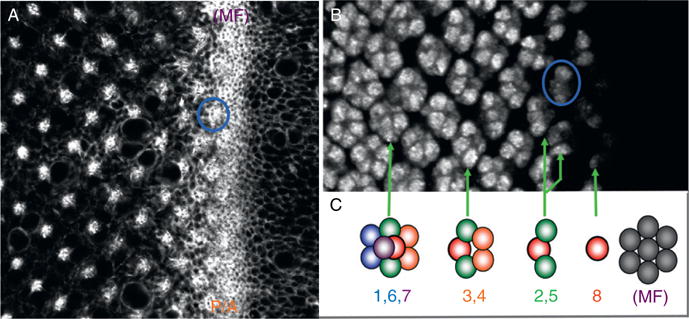
The morphogenetic furrow and ommatidial assembly. (A) A confocal section of a third instar larval eye disk that has been stained with phalloidin, which marks F-actin. Note that cells ahead of the furrow are unpatterned while those behind the furrow are organized into ommatidial rudiments. The blue circle marks one individual unit eye. (B) A confocal section of a third instar larval eye disk that has been stained with an antibody that is directed against the ELAV protein. The blue circle marks a unit eye that is in roughly the same position as the one in panel A. (C) A schematic drawing of the order of ommatidial assembly within the eye disk. Anterior is to the right in all images. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this chapter.)
3. The Retinal Determination Network: Membership Has Its Privileges
Membership within the retinal determination network currently stands at 14 genes, the vast majority of which code for DNA-binding proteins (Fig. 1.3A). These include eyeless (ey; Quiring et al., 1994), twin of eyeless (toy; Czerny et al., 1999), eyegone (eyg; Jun et al., 1998), twin of eyegone (toe; Aldaz et al., 2003), sine oculis (so; Cheyette et al., 1994; Serikaku and O’Tousa, 1994), optix (Seimiya and Gehring, 2000), teashirt (tsh: Pan and Rubin, 1998), tiptop (tio; Laugier et al., 2005), distal antenna (dan; Curtiss et al., 2007), distal antenna related (danr; Curtiss et al., 2007), dachshund (dac; Mardon et al., 1994), and homothorax (hth; Pai et al., 1997). The remaining two genes encode the eyes absent (eya) transcriptional coactivator/protein tyrosine phosphatase (Bonini et al., 1993) and the nemo (nmo) protein kinase (Braid and Verheyen, 2008; Choi and Benzer, 1994). While these factors were initially thought to function as purely selector genes, more recent evidence indicates that these genes play roles in the proliferation of progenitor cells, the differentiation of retinal precursors, and the specification and/or maintenance of photoreceptor neurons (Bessa et al., 2002; Lopes and Casares, 2009; Peng et al., 2009; Pignoni et al., 1997). Nearly every one of these genes is also represented in vertebrates, and many have been implicated in retinal disorders (Fig. 1.3A; Chi and Epstein, 2002; Hanson, 2001; Jean et al., 1998; Kozmik, 2005; Kumar, 2009a,2009b; Mansouri et al., 1999; Wawersik and Maas, 2000). Additionally, several of these factors also play crucial roles in the development of a broad range of nonretinal tissues and organs (Brodbeck and Englert, 2004; Chi and Epstein, 2002; Christensen et al., 2008; Dressler, 2006; Kawakami et al., 2000), thereby adding to their growing importance in both development and disease.
Figure 1.3.
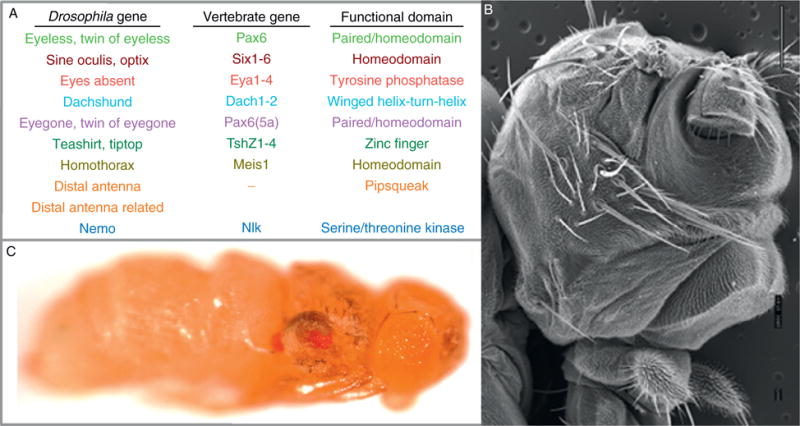
The retinal determination network: genes and phenotypes. (A) A list of the known retinal determination genes, the vertebrate homologs, and the known functional domains. (B) A scanning electron micrograph of a sine oculis loss-of-function mutant. Note that the compound eyes are missing and have been replaced by head cuticle. (C) A light microscope image of an animal in which the ey gene has been expressed in the wing and haltere disks using an ap-GAL4 driver. Anterior is to the right in all images.
Historically, the founding members of the retinal determination network were grouped together and considered part of a single regulatory system if they displayed two physical attributes. First, loss-of-function mutations needed to result in a strongly reduced or missing eye phenotype (Fig. 1.3B; Bonini et al., 1993; Cheyette et al., 1994; Dominguez et al., 2004; Jang et al., 2003; Mardon et al., 1994; Quiring et al., 1994; Serikaku and O’Tousa, 1994). Second, forced expression in nonretinal tissues had to be sufficient to force these cells into adopting a retinal fate (Fig. 1.3C; Bonini et al., 1997; Chen et al., 1997; Halder et al., 1995; Pignoni et al., 1997; Weasner et al., 2007). But as the number of genetic screens and genome/proteome-wide studies has grown, new genes have been included in the network based on combinations of criteria that had been expanded to include genetic, molecular, or biochemical interactions with existing pathway members (Bessa et al., 2009; Braid and Verheyen, 2008; Curtiss et al., 2007; Czerny et al., 1999; Datta et al., 2009; Pai et al., 1997; Pan and Rubin, 1998; Seimiya and Gehring, 2000; Yao et al., 2008). As of this writing, the number of genes that are universally accepted as being bona fide retinal determination genes stands at 14 (see above). This expanded list of genes has provided us with an opportunity to gain a much more sophisticated understanding of how early decisions in developing organs, particularly that of the eye, are executed. Our appreciation for how an eye is specified will only accelerate exponentially as new genes are identified as playing a role during early retinal differentiation.
4. The Molecular Biology and Biochemistry of Retinal Determination
The genes that govern eye specification are said to constitute a network rather than a cascade or pathway because the experimentally verified genetic, molecular, and biochemical interactions among the various members include several reinforcing positive feedback loops, mutually dampening negative interactions, and self-fortifying autoregulatory circuits (Fig. 1.4; Kumar, 2009a,2009b). A growing body of evidence is also suggesting that the transcriptional output of each retinal determination gene is controlled by the combined regulatory inputs of other network members (Niimi et al., 1999; Pappu et al., 2005; Pauli et al., 2005). Adding additional layers of regulatory complexity is the fact that numerous signaling pathways are known to integrate into the network: multiple pathways are used to activate/repress transcription of individual genes and each pathway appears to integrate into the network at multiple levels (Chen et al., 1999; Kenyon et al., 2003; Kumar and Moses, 2001a; Kurata et al., 2000). All these interactions (at least the ones that are relevant for eye specification) occur in two places: first, they occur throughout the eye disk prior to the initiation of the morphogenetic furrow and second, once pattern formation has initiated, they occur ahead of the advancing furrow. This section will highlight features of the retinal determination proteins themselves and will draw attention to the molecular and biochemical interactions that are known to exist between the network members.
Figure 1.4.
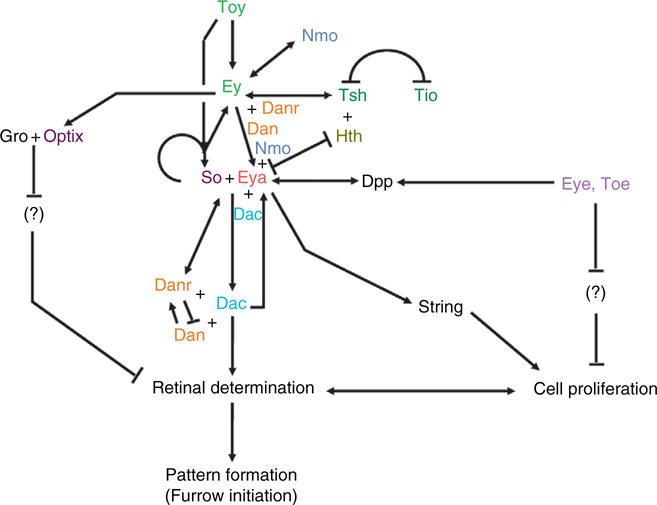
The workings of the retinal determination network. A schematic of all known interactions within the retinal determination network. Note that the cascade does not function as a linear pathway but rather contains autoregulatory circuits and feedback loops.
4.1. Eyeless/Pax6: King of kings
The founding member of the retinal determination network was ey. Mutations in this gene were discovered nearly 100 years ago based on their no-eye phenotype (Hoge, 1915). Its central role in early eye development was established by its description as a transcription factor with homology to human Pax6 and murine Small eye (Quiring et al., 1994), the demonstration that it could force cell populations in nonretinal tissues to adopt an eye fate (Halder et al., 1995), the revelation that it regulates a large number of target genes in the developing eye (Michaut et al., 2003; Niimi et al., 1999; Ostrin et al., 2006), and the identification of functional orthologs within nearly every phylum of the animal kingdom (Callaerts et al., 1997; Gehring, 1996; Gehring and Ikeo, 1999). Unlike vertebrates, the fly genome contains a second Pax6 gene, toy. Its expression precedes that of ey, and Toy protein can directly activate ey transcription during embryonic development (Czerny et al., 1999; Hauck et al., 1999; Kronhamn et al., 2002). This interaction appears to be unidirectional since Ey can neither feed backward to activate toy expression nor bind to its own eye-specific enhancer (Fig. 1.4; Czerny et al., 1999). These two Pax6 proteins then, either separately or together depending upon the target enhancer, activate the transcription of several other retinal determination genes including so, eya, dac, and optix (Fig. 1.4; Halder et al., 1998; Niimi et al., 1999; Ostrin et al., 2006; Pappu et al., 2005). During the first and second larval instars, these interactions take place throughout the entire eye disk. However, by the third larval instar, Toy and Ey are only able to activate their targets within specific regions of the epithelium (see below).
The two proteins cannot fully substitute for each other: expression of toy in an ey mutant background fails to promote retinal development (Czerny et al., 1999; Kronhamn et al., 2002). While forced expression of either Pax6 gene can direct ectopic eye formation, Ey appears to be able to induce retinal development in a broader topographical range than Toy (Czerny et al., 1999; Halder et al., 1995; Salzer and Kumar, 2010). These differences have been the subject of several molecular dissections (Clements et al., 2009; Punzo et al., 2001, 2004; Weasner et al., 2009), and results from these studies indicate that reported distinctions and disparities can be attributed to different transactivation potentials (Punzo et al., 2004; Weasner et al., 2009) as well as the unique presence of transcriptional repressive activity within Ey (Weasner et al., 2009). Additionally, it appears that the paired DNA-binding domain and the C-terminal segment (which contains the activation domain) are the only segments of either Ey or Toy that are required for the promoting of eye ectopic formation (Punzo et al., 2001; Weasner et al., 2009); this suggests that Pax6 proteins may in fact function in modular fashion with different DNA-binding domains and nonconserved segments being used in different molecular and developmental contexts. Immediately, after ey was initially cloned and shown capable of inducing ectopic eye formation, it was crowned as “the master control gene” for eye development. Ey was thought to assume its role as king when transcription of it (and its sister gene toy) initiated during embryogenesis (Callaerts et al., 1997; Halder et al., 1995; Quiring et al., 1994). Ey still rules the eye although it must share its throne with the other eye specification genes as they share many of the properties that once were the realm of Ey alone.
4.2. So–Eya: A workhorse complex in the early eye
While Ey has grabbed much of the spotlight over the years, several of the more downstream members of the network such as So, Eya, and Dac may in fact turn out to be just as influential in terms of regulating early eye development. Transcription of each of these three genes is responsive to the expression of ey (Halder et al., 1998; Michaut et al., 2003; Ostrin et al., 2006; Pappu et al., 2005; Punzo et al., 2002) with additional evidence supporting direct binding of both Ey and Toy to an eye-specific enhancer within the so transcriptional unit (Niimi et al., 1999). So, the founding member of the Six family is a homeobox containing transcription factor (Cheyette et al., 1994; Serikaku and O’Tousa, 1994) that on its own can bind to DNA but is unable to strongly activate transcription of its target. It forms a biochemical complex with the Eya transcriptional coactivator (Kenyon et al., 2005a,b; Pignoni et al., 1997), which also functions as a protein tyrosine phosphatase (Li et al., 2003; Rayapureddi et al., 2003; Tootle et al., 2003). The So–Eya complex is thought to influence eye specification by regulating the other members of the retinal determination network such as dac (Pappu et al., 2005) as well as by feeding back to autoregulate its own transcription and that of ey (Pauli et al., 2005). But the influence that So–Eya has on eye development does not stop at retinal determination. In fact, the complex regulates the transcription of genes that play critical roles in the initiation of the furrow (hedgehog; Pauli et al., 2005), the cell cycle (string; Jemc and Rebay, 2007), and cell fate decisions (lozenge; Tanaka-Matakatsu and Du, 2008; Yan et al., 2003; atonal, Zhang et al., 2006).
A second Six protein, Optix, is distributed in the eye and can induce ectopic eye formation (Seimiya and Gehring, 2000), but the lack of published loss-of-function phenotypes has made it difficult to define a role for optix in retinal development. Despite this, it has been possible to gain some hints into the molecular workings of Optix. Recent reports have indicated that the binding sites for all Six proteins in Drosophila are very similar, if not identical, to each other (Berger et al., 2008; Noyes et al., 2008). Thus, it is likely that Optix and So bind and regulate common target genes. However, rescue experiments have indicated that Optix cannot substitute for So in eye development (Weasner et al., 2007). This is due to cryptic functional motifs embedded within the nonconserved C-terminal (Weasner and Kumar, 2009) and differences in binding partner specificity (Kenyon et al., 2005a, b; Weasner et al., 2007). Of particular interest is the ability of So to toggle between activator and repressor by interacting with both Eya and the transcriptional corepressor Groucho (Gro; Kenyon et al., 2005a; Salzer and Kumar, 2009) while Optix appears to be a dedicated repressor as it can only interact with Gro (Kenyon et al., 2005a).
4.3. Dac: A transcription factor in search of a target
While a requirement for dac in eye specification and morphogenetic furrow initiation has been well documented (Chen et al., 1997; Mardon et al., 1994; Shen and Mardon, 1997), its molecular and biochemical roles in these processes have remained somewhat elusive, in part, because neither consensus binding sites nor transcriptional targets in the eye have been identified despite a significant body of evidence suggesting that it functions as a bona fide transcription factor. For example, sequence analysis suggests that Dac is related to members of the Ski/Sno family of proto-oncogenes (Hammond et al., 1998), contains a winged helix-turn-helix DNA-binding motif, and can physically interact with double-stranded nucleic acids (Kim et al., 2002). Further evidence indicates that Dac is not only capable of activating transcription of a reporter, on its own, in yeast but can also physically interact with Eya (Chen et al., 1997). Several reports indicate that the vertebrate homolog Dach1 switches between serving as an activator (Ikeda et al., 2002; Li et al., 2003) and functioning as a repressor (Li et al., 2002; Wu et al., 2003). The only potential targets known in the eye are the upstream factors so and eya, as loss of dac at the margin of the eye field leads to the loss of both of these genes (Salzer and Kumar, 2009).
4.4. Hth–Exd and Tsh: Suppressors of eye specification
In Drosophila, homeotic (Hox) genes regulate the development of body segments along the anterior–posterior (A/P) axis. They are expressed sequentially along the embryonic A/P axis and encode homeobox containing transcription factors. It was discovered early on that these proteins bind to very similar sequences (reviewed in Mann, 1995). It was thus proposed and later confirmed that Hox genes required cofactors to ensure binding specificity. One such cofactor is the product of the extradenticle (exd) gene. It also encodes a homeodomain and its binding to Hox proteins can alter their DNA-binding specificity (Chan and Mann, 1996; Chan et al., 1996). Interestingly, Exd is not located in the nucleus of all cells but rather is found both in the cytoplasm and the nucleus (Aspland and White, 1997). The Homothorax (Hth) protein, which shares extensive homology to the murine Meis1/2 proteins, contains a TALE class homeobox, binds to Exd, and translocates it to the nucleus where it can interact with Hox genes (Jaw et al., 2000; Pai et al., 1997; Rieckhof et al., 1997).
The Hth–Exd complex is important for eye development as loss-of-function mutations in either factor lead to the formation of ectopic eyes (Gonzales-Crespo et al., 1998; Pai et al., 1997; Pichaud and Casares, 2000; Rauskolb et al., 1995). This inhibition of normal eye development likely occurs through direct transcriptional repression of retinal determination genes as forced expression of hth can repress both eya and dac (but not ey). Presently, the Hth-binding site has not been determined but an effort to identify transcriptional targets has localized Hth to approximately 150 sites on Drosophila polytene chromosomes (Cohen and Salzberg, 2008). At the moment, the resolution of this map does not allow for the identification of individual genes but a scan of the listed cytological location suggests that Hth may not be occupying sites within eya or dac. This does not necessarily rule out a direct regulatory mechanism since the efficiency of repression is synergistically increased with the coexpression of both Ey and the zinc finger transcription factor Teashirt (Tsh), two factors that physically interact with Hth (Bessa et al., 2002). It is likely that some configuration of this complex is necessary to directly repress both eya and dac since Hth is a transcriptional activator (Inbal et al., 2001) while both Ey and Tsh can, depending upon the circumstance, function as transcriptional repressors (Bessa et al., 2009; Weasner et al., 2009).
Tiptop (Tio) is structurally related to Tsh (Laugier et al., 2005) and can promote eye development in forced expression assays (Bessa et al., 2009; Datta et al., 2009). In the eye, both genes are expressed in identical patterns and at approximately equal levels. Interestingly, tio loss-of-function mutants are completely viable and have no retinal defects. Similarly, tsh loss-of-function mutations have little to no effect on the eye (Laugier et al., 2005; Pan and Rubin, 1998). This suggests that these genes are functionally redundant. The loss of each gene leads to an upregulation of the other while elevated expression levels of each gene results in the downregulation of the other (Bessa et al., 2009). Both results provide further evidence that Tsh and also Tio function as transcriptional repressors. Does Tio function to also repress eya and dac transcription? At present, the answer is not clear as binding sites for neither gene have been identified, but such a mechanism would be consistent with the redundant role that Tio plays in the early eye.
4.5. Nemo: From ommatidial rotation to eye specification
nmo is a founding member of Nemo-like kinase family of proline-directed serine–threonine kinases and was initially studied for its role in ommatidial rotation (Choi and Benzer, 1994). But its expression pattern, which overlapped with several retinal determination genes, hinted of roles in early eye development. A recent report has demonstrated that nmo interacts genetically with ey and eya during normal retinal development as well as during the induction of ectopic eye formation (Fig. 1.4; Braid and Verheyen, 2008). Excitingly, forced expression of nmo is sufficient, on its own, to induce ectopic eye formation (Braid and Verheyen, 2008). To date, the only known substrate of Nemo is the Mothers against dpp (Mad) protein, which is a downstream component of the TGFβ signaling pathway (Zheng et al., 1995). A tantalizing model is that one or more retinal determination proteins are a substrate for phosphorylation by Nmo. As nmo genetically interacts with both eya and ey, these two factors would be the likely targets.
4.6. Dan and Danr: An antennal gene regulates the eye
The dan and danr genes code for Pipsqueak-class DNA-binding proteins and were initially identified as playing critical roles in antennal development (Emerald et al., 2003). Both genes appear to also play important roles in specifying the eye, in part, by participation within the retinal determination network (Curtiss et al., 2007). Both Dan and Danr contribute by regulating the expression of ey and eya. In turn, the So–Eya–Dac complex activates expression of both dan and danr (Fig. 1.4). Furthermore, dan and danr appear to regulate each other with Dan activating danr transcription and Danr repressing dan expression (Fig. 1.4; Curtiss et al., 2007). It is not yet known if any of these interactions are direct. In another twist, Dan and Danr form physical complexes with both Ey and Dac (Fig. 1.4; Curtiss et al., 2007). These multiple molecular and biochemical interactions suggest that both Dan and Danr play important roles in specifying the compound eye.
5. One Network yet Several Incarnations
As anyone who has read papers dealing with retinal determination can attest, one will usually find a depiction of the retinal determination network that is not much different than the one in Fig. 1.4, which is replete with activation steps, inhibitory, and autoregulatory loops. But closer inspections of expression patterns and mutant phenotypes indicate that all these interactions cannot and are not happening within the entire eye. In fact, several reports indicate that subsets of interactions are taking place in different geographical locations within the eye (Bessa et al., 2002; Lopes and Casares, 2009; Mardon et al., 1994; Peng et al., 2009; Pignoni et al., 1997; Salzer and Kumar, 2009); therefore, this section will place the aforementioned genetic, molecular, and biochemical interactions in temporal and spatial contexts.
5.1. Embryogenesis
The eye field initiates its development during embryogenesis as a simple cluster of approximately 20 cells (Cohen, 1993; Held, 2002). As these cells delaminate from the surface ectoderm, expression of the four Pax genes (ey, toy, eyg, and toe) and the Six gene optix is initiated (Aldaz et al., 2003; Czerny et al., 1999; Kronhamn et al., 2002; Kumar and Moses, 2001a; Quiring et al., 1994; Seimiya and Gehring, 2000). Very little is known about the molecular interactions that take place in the embryonic eye disk, save for the initial step in which toy expression precedes and presumably activates ey through binding at the eye-specific enhancer (Czerny et al., 1999; Hauck et al., 1999; Kronhamn et al., 2002).
5.2. Larval development
Upon hatching of the embryo into a larva, these cells continue to divide rapidly and are self-organized into a monolayer epithelium called the eye-antennal imaginal disk (Fig. 1.2). During the first two larval instars, the disk is primarily concerned with establishing dorsal–ventral compartment boundaries (Cho and Choi, 1998; Dominguez and de Celis, 1998; Papayannopoulos et al., 1998; Sato and Tomlinson, 2007), while rapidly generating the requisite number of cells that are needed for ommatidial assembly and with molecularly canalizing the epithelium toward an eye fate. This last task is achieved, in part, by the expression of the remaining retinal determination genes (Bonini et al., 1993; Braid and Verheyen, 2008; Curtiss et al., 2007; Kumar and Moses, 2001b; Mardon et al., 1994; Serikaku and O’Tousa, 1994; Singh et al., 2002).
As the larva enters the third and final instar stage, a wave of differentiation initiates at the posterior margin of the disk and proceeds toward the anterior border of the eye field (Ready et al., 1976). As this mobile compartment boundary progresses across the retinal field, the vast expanse of unpatterned and undifferentiated cells are transformed into a highly ordered display of periodically spaced unit eyes or ommatidia (Ready et al., 1976; Tomlinson and Ready, 1987a,1987b; Wolff and Ready, 1991). This moving current of differentiation can be visualized by an indentation in the tissue and is referred to as the morphogenetic furrow (Ready et al., 1976). As the furrow progresses across the eye field, expression of most retinal determination genes, which was once uniform throughout the entire disk, is now relegated to regions that remain undifferentiated and possibly undetermined (Bessa et al., 2009; Curtiss et al., 2007; Czerny et al., 1999; Datta et al., 2009; Dominguez et al., 2004; Jang et al., 2003; Pai et al., 1997; Pan and Rubin, 1998; Quiring et al., 1994; Seimiya and Gehring, 2000). A few genes such as so, eya, dac, and nmo continue to be expressed in cells that now lie behind the furrow and contribute to cell fate choices within developing ommatidia (Bonini et al., 1993; Cheyette et al., 1994; Choi and Benzer, 1994; Mardon et al., 1994; Serikaku and O’Tousa, 1994). The full complexity of retinal determination gene expression can most easily be observed and appreciated when the furrow has advanced roughly halfway across the eye field (Fig. 1.5).
Figure 1.5.
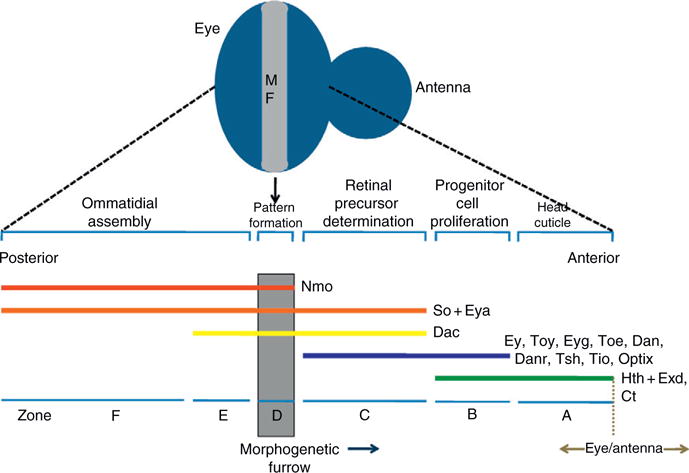
Expression patterns of retinal determination genes within the developing eye field. The eye disk is divided into six zones based on the expression patterns and functions of the known retinal determination genes. The zones are listed at the bottom of the figure. The expression patterns of each gene are represented by the colored horizontal lines. The ongoing developmental processes within each zone are listed at the top of the figure. The morphogenetic furrow is shown in gray. Anterior is to the right in this schematic diagram. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this chapter.)
Based on the expression patterns of genes that are known to regulate eye specification, furrow progression, and cell fate specification, the mid-third instar eye field can be divided into several broad zones. However, there is some disagreement as to the physical extent of each expression pattern and the role that each gene may play in the proliferation of progenitor cells versus their role in the specification of precursor cells. This has in turn resulted in differences in the labeling/nomenclature of individual zones (Bessa et al., 2002; Braid and Verheyen, 2008; Lopes and Casares, 2009; Pappu and Mardon, 2004; Silver and Rebay, 2005). Based on the published expression domains of each retinal determination gene and on loss-of-function phenotypes, we subdivide the eye into six zones (A–F; Fig. 1.5). We discuss each zone in turn starting with the region of the disk that borders the antennal segment.
The most anterior edge of the eye primordium (Zone A) actually will not yield retinal tissue at all but rather will give rise to head cuticle (Fig. 1.5; Haynie and Bryant, 1986). Cells within this zone appear not to express any retinal determination genes, save for one exception, hth, and its cofactor exd (Bessa et al., 2002; Pai et al., 1997; Pichaud and Casares, 2000). The Hth–Exd complex appears to normally block retinal development as loss-of-function clones of either gene result in ectopic eyes where head cuticle is normally produced (Gonzales-Crespo et al., 1998; Pai et al., 1997; Rauskolb et al., 1995). Similarly, simultaneously coexpressing both factors leads to an inhibition of normal eye development (Pai et al., 1997). In addition to the Hth–Exd complex, the antennal specifying factor cut (ct) is also expressed within Zone A thereby serving as an additional block on retinal development (Blochlinger et al., 1993; Dong et al., 2002). Thus, the combined efforts of these factors result in the proliferation of precursor cells that will be simultaneously blocked from adopting a retinal fate while being directed toward giving rise to head cuticle.
Directly adjacent to the head cuticle producing cells lie a swath of cells (Zone B) that are eventually slated to adopt an eye fate (Fig. 1.5). These cells, in addition to hth and exd, now begin to express most retinal determination genes including ey/toy (Czerny et al., 1999; Quiring et al., 1994), eyg/toe (Aldez et al., 2003), tsh/tio (Bessa et al., 2009; Datta et al., 2009; Pan and Rubin, 1998), dan/danr (Curtiss et al., 2007), and optix (Seimiya and Gehring, 2000). However, since the transcription of several key genes such as so, eya, and dac is not yet activated, the cells within this region of the disk will be held in a highly proliferative state while being temporarily blocked from being specified (Fig. 1.5; Lopes and Casares, 2009; Peng et al., 2009). There appears to be several simultaneously acting mechanisms for promoting cell proliferation. First, Notch signaling promotes growth through at least one Pax6(5a) variant Eyg (Chao et al., 2004; Dominguez et al., 2004). Second, a complex containing Hth and Yorki (Yki), the most downstream member of the Hippo tumor suppressor pathway, stimulates growth by targeting and activating transcription of the bantam microRNA which in turn results in the downregulation of the cell death gene head involution defective (hid: Peng et al., 2009). And finally, a transcription complex containing Hth, Tsh, and potentially Ey also contributes to cell proliferation rates in this region of the disk. Support for this last input comes from experiments showing that mutant hth clones rarely survive anterior to the furrow (Bessa et al., 2002) and that disruptions with Ey function via developmental pathway interference can be partially rescued by the expression of several cell cycle genes such as cyclin E (cycE) and myc (Jiao et al., 2001). It should be noted that this last complex is also thought to play a role in repressing so, eya, and dac, each of which is critical for the proper specification of the eye. The rare hth loss-of-function clones that do survive anterior to the furrow ectopically express these three genes in Zone B and this in turn promotes precocious eye development (Bessa et al., 2002; Lopes and Casares, 2009; Pai et al., 1997; Pichaud and Casares, 2000). Additionally, the coexpression of different combinations of these three genes (Tsh–Hth, Tsh–Ey, and Hth–Ey) is sufficient to inhibit expression of so, eya, and dac (Bessa et al., 2002).
Cells that lie closest to the morphogenetic furrow (Zone C—also referred to as the preproneural [PPN] zone), while still proliferating, are on the cusp of being incorporated into the growing neurocrystalline lattice (Lebovitz and Ready, 1986). This transition, however, requires the presence of So, Eya, and Dac, as mutations in these genes affect the initiation and progression of the morphogenetic furrow (Mardon et al., 1994; Pignoni et al., 1997; Salzer and Kumar, 2009). These genes are activated by the downregulation of hth (Bessa et al., 2002) although the exact mechanism of hth transcriptional inhibition remains unknown. At this point in the disk, the activation of all retinal determination genes is sufficient to transition cells from being undifferentiated and undetermined to adopting cellular fates that are appropriate for the retina (Kumar and Moses, 2001b). As these cells prepare to be assembled into ommatidial units, several mechanisms for regulating the movement of the furrow and for selecting the first photoreceptor are initiated and are discussed in other chapters. Briefly, within Zone C, the bHLH protein Hairy (H) and the HLH protein Extramacrochaete (Emc) function together to prevent the pace of the patterning from outstripping the rate of cell proliferation ahead of the furrow (Brown et al., 1995). There is evidence now to indicate that the loss of emc, on its own, is sufficient to accelerate the pace of the furrow (C.M. Spratford and J.P. Kumar, unpublished data). Emc and its vertebrate homologs (inhibitors of differentiation) function by sequestering DNA-binding proteins away from target promoters (Benezra et al., 1990; Ellis et al., 1990; Garrell and Modolell, 1990). At present, the targets of sequestration by Emc ahead of the furrow are yet to be found. Their identification will be a huge advance in understanding how the rate of pattern formation in the eye is regulated. In addition to Emc holding the furrow to a sustainable pace across the disk, the Wingless (Wg) signaling pathway prevents pattern formation from initiating inappropriately from the lateral margins of the epithelium. Wg is expressed at the margins and its loss leads to the initiation of ectopic morphogenetic furrows (Ma and Moses, 1995; Treisman and Rubin, 1995).
Within the morphogenetic furrow itself (Zone D), cells are arrested in the G1 phase of the cell cycle. As they transition to positions behind the furrow, a subset of cells will exit the cell cycle and begin to initiate their development as the first five cells of the ommatidium (R8, R2, R5, R3, and R4) while the remainder will undergo one last round of synchronous cell division which will in turn lead to the production of the remaining photoreceptor neurons (R1, R6, and R7), the nonneuronal cone and pigment cells, as well as the cells of the bristle complex (Wolff and Ready, 1991). The G1 arrest within the furrow is established and maintained by high levels of the TGFβ ligand Decapentaplegic (Dpp: Fig. 1.5; Horsfield et al., 1998). In addition to affecting the cell cycle, Dpp signaling is also required for the initiation and progression of the furrow across the eye field (Chanut and Heberlein, 1997a,b; Heberlein et al., 1995). One attractive model for linking Dpp signaling (originating within the furrow) to cells just ahead of the furrow invokes a situation in which Dpp would regulate retinal determination gene expression, particularly that of so, eya, and dac. In fact, an eye-specific enhancer within the eighth intron of dac is responsive to Dpp signaling (Pappu et al., 2005), and loss of Dpp signaling leads to reductions in dac, eya, and so transcription (Curtiss and Mlodzik, 2000). Unfortunately, the regulation of these three retinal determination genes by Dpp appears to occur only at the margins of the disk during furrow initiation. Not only is expression of so, eya, and dac normal within internally located Mad clones but also the furrow progresses normally through this tissue (Curtiss and Mlodzik, 2000; Greenwood and Struhl, 1999). Thus, the molecular link between signaling pathway cells in the furrow (Zone D) and the retinal determination network in cells that lie just anterior (Zone C) remains unresolved. As a consequence, the morphological mechanism by which cells are canalized into the furrow also remains an open issue. Also, the exact roles of the retinal determination network in the furrow itself are not known with any degree of certainty.
The area behind the furrow can be divided into two sections (Zones E and F). The first encompasses cells that lie just posterior to the furrow (Zone E) and can be molecularly marked by the expression of four retinal determination genes, so, eya, dac, and nmo with the dac expression profile defining the most posterior edge of this zone (Fig. 1.5). The second (Zone F) begins where dac expression terminates and extends all the way to the posterior edge of the eye field. These cells continue to express only so, eya, and nmo (Fig. 1.5). The role played by the retinal determination genes in cells behind the furrow has not been extensively studied and is not well understood. However, an analysis of mutant phenotypes suggests that these factors contribute to at least two processes. The first involves a potential role for the so, eya, and dac genes in the acquisition and maintenance of individual cell fates within developing ommatidia. The loss of either so or eya within cells that are born in the second mitotic wave results in the loss of R1, R6, and R7 photoreceptors (Pignoni et al., 1997). It is not clear, however, if the So–Eya complex functions to specify these cell fates or to maintain cell identity. Also, a role for So–Eya in cells of the precluster (R8, R2, R5, R3, and R4) has not been described. Downstream of the So–Eya complex lies dac, so it is thought that it might play a role in photoreceptor cell specification and/or maintenance. However, the loss of dac in the middle of the eye field does not appear to inhibit the movement of the furrow or have a significant effect on the initial steps in photoreceptor specification (Salzer and Kumar, 2009).
In addition, each unit eye is executing dorsal–ventral patterning signals by assuming a chiral form that is appropriate for its location within either the dorsal or ventral half of the retina. Also, ommatidia in the two compartments must rotate in opposite directions. The combined effects of chirality and rotation result in ommatidia in the dorsal half being positioned in a mirror-image orientation to those in the ventral section (Fig. 1.5; reviewed in Fanto and McNeill, 2004; Mlodzik, 1999; Strutt and Strutt, 2003; Wolff and Ready, 1993). The Nmo kinase, while functioning to promote eye formation (Braid and Verheyen, 2008), also plays a major role in ommatidial rotation as loss-of-function mutations prevent ommatidia from rotating past the first 45° (Choi and Benzer, 1994; Fiehler and Wolff, 2008). It should be noted that the expression of all other retinal determination genes is extinguished at the furrow and thus do not appear to play roles in cell fate specification or ommatidial rotation.
6. Taking Instructions from Higher Authorities
The nuclear retinal determination network, while functioning as a unit, does not do so in isolation. Rather, it is a nexus point for integrating instructions that are being transmitted across the disk by diffusible morphogens and signal transduction pathways. These cascades are used reiteratively during eye development and intersect with the retinal determination network at multiple levels (reviewed in Kumar, 2001; Voas and Rebay, 2004). This final section will briefly bring to light the known interactions between signaling pathways and the eye specification network.
6.1. Notch and the EGF receptor: A role in sensory organ identity
As mentioned above, there is a unidirectional flow of information between toy and ey with the former residing molecularly upstream of the latter. Within this section of the network, there does not appear to exist any feedback loops or autoregulatory circuits. It raises the issue of how these genes are initially transcribed in the eye anlagen and how their expression patterns restricted are to just the eye field after being initiated throughout the entire eye-antennal epithelium. It appears that Notch signaling activates and maintains ey expression while in the second instar, the EGF receptor (EGFR) pathway functions to restrict ey transcription to the developing eye (Kumar and Moses, 2001b; Kurata et al., 2000). Alterations of either signaling cascade during this stage results in the transformation of the eye into an antenna (Kumar and Moses, 2001b). The latter half of the second instar appears to be a key developmental window for organogenesis, as manipulations of EGFR signaling within the wing disk during this same interval can convert the developing notum into wing tissue (Baonza et al., 2000).
A role for the EGFR in retinal determination does not stop with the regulation of ey expression. In fact, the pathway also goes on to regulate eya at multiple levels. This signaling cascade appears to regulate eya expression, possibly directly, as mutations in yan and pointed (pnt), which encode Ets type transcription factors, lead to up- and downregulation of eya transcription, respectively, in both the embryo and the developing eye (Salzer et al., 2010). The EGFR pathway also modulates the activity of Eya protein through phosphorylation by MAPK (Hsiao et al., 2001). This modification is critical for the ability of Eya to support normal and ectopic eye development.
6.2. Hedgehog and Dpp: Required for ectopic eye formation
Since the very first reports of ectopic eye formation, it has been noted that widespread expression of any retinal determination gene only transforms portions of some imaginal disks. A recent systematic effort to document this phenomenon identified nine cell populations that can support eye formation in the eye-antenna, leg, wing, and haltere disks (Salzer and Kumar, 2010). Several of these transformation “hot spots” appear to coincide geographically with several previously identified transdetermination weak points (Maves and Schubiger, 1998, 2003). These points seem to be under the control of several signaling pathways including Wg and Dpp. The coincidental location of the transdetermination weak points and the transformation hot spots suggests that the ability of the retinal determination network to support eye development within the hot spots and even in the normal eye may require that these cell populations be primed by the expression and activity of short- and long-range diffusible signals. Two efforts have attempted to confirm this hypothesis and uncover the identity of the morphogens. First, clones expressing retinal determination genes were induced in random locations within the wing disk. The only clones that contained ectopic retinal tissue were ones that were contained within the posterior compartment, which expresses the Hh morphogen (Kango-Singh et al., 2003). Second, coexpression of ey with dpp in cells that surround the developing wing pouch expanded the range of cells that can be transformed into retinal tissue (Chen et al., 1999). Together, these two reports suggest that the Hh and Dpp pathways are required in cells prior to the onset of the retinal determination network. While two pathways do play important roles in promoting eye development, the signaling requirements are likely to be more complicated, as several of the recently identified cell populations that can support eye development lie well outside the hh and dpp expression zones (Salzer and Kumar, 2010).
6.3. Wingless: A repressive signal
The eye imaginal disk gives rise to more than just the compound eye; the surrounding head cuticle is also derived from this epithelium (Haynie and Bryant, 1986). The bulk of retinal determination genes, save for hth, are not expressed within the tissue that will give rise to head cuticle (see above). So how does the epithelium subdivide itself in this way? The Wg signaling pathway is known to inhibit eye development by blocking ectopic morphogenetic furrow initiation (Ma and Moses, 1995; Treisman and Rubin, 1995). Further analysis of its working in the eye indicated that Wg prevents the expression of so, eya, and dac as clones of axin or armadillo (arm) lead to their upregulation and ectopic wg expression downregulates their transcription in the eye (Baonza and Freeman, 2002). This effect is not direct as the Wg pathway activates hth expression, which in turn blocks initiation of so, eya, and dac in conjunction with tsh (Bessa et al., 2002; Singh et al., 2002). So while the Notch, EGFR, Hh, and Dpp signaling cascades promote eye development, the Wg pathway acts to balance these effects and subdivide the eye imaginal disk into the eye proper and head cuticle.
7. Concluding Remarks
The past 30 years have seen remarkable advances in our understanding of how a simple nervous system, the insect compound eye, is specified and patterned. The retina has been an enduring model for studying a myriad of developmental processes including organogenesis. This review has attempted to summarize the known molecular and biochemical events that lead to the specification of the eye. In addition, special emphasis has been put on placing these interactions within temporal and spatial contexts of the developing eye field. Despite the enormous progress that has been made on understanding how the retina is determined, we still have a long way to go before we will have a complete understanding of how the eye is constructed. Based on past experience, it will not be long before new genes are identified as retinal determination factors and their roles in eye development are elucidated. The advent of new technologies, application of high throughput assays, and the implementation of creative genetic, molecular, and biochemical screens will only accelerate this process. In time, we will surely unravel the mystery of how gene regulatory networks function to coordinate the fates of large groups of undifferentiated cells and produce a unique organ or tissue.
Acknowledgments
I would like to thank all those who have worked on the developing Drosophila compound eye and apologize to those whose work is not cited here. I would also like to thank Bonnie M. Weasner and Carrie M. Spratford for comments and suggestions on this chapter as well as Claire L. Salzer for the image in Fig. 1.3B. Justin P. Kumar is supported by a grant from the National Eye Institute (R56 EY014863).
References
- Aldaz S, Morata G, Azpiazu N. The Pax-homeobox gene eyegone is involved in the subdivision of the thorax of Drosophila. Development. 2003;130:4473–4482. doi: 10.1242/dev.00643. [DOI] [PubMed] [Google Scholar]
- Aspland SE, White RA. Nucleocytoplasmic localisation of extradenticle protein is spatially regulated throughout development in Drosophila. Development. 1997;124:741–747. doi: 10.1242/dev.124.3.741. [DOI] [PubMed] [Google Scholar]
- Baonza A, Freeman M. Control of Drosophila eye specification by Wingless signalling. Development. 2002;129:5313–5322. doi: 10.1242/dev.00096. [DOI] [PubMed] [Google Scholar]
- Baonza A, Roch F, Martin-Blanco E. DER signaling restricts the boundaries of the wing field during Drosophila development. Proc Natl Acad Sci USA. 2000;97:7331–7335. doi: 10.1073/pnas.97.13.7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Gebelein B, Pichaud F, Casares F, Mann RS. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002;16:2415–2427. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessa J, Carmona L, Casares F. Zinc-finger paralogues tsh and tio are functionally equivalent during imaginal development in Drosophila and maintain their expression levels through auto- and cross-negative feedback loops. Dev Dyn. 2009;238:19–28. doi: 10.1002/dvdy.21808. [DOI] [PubMed] [Google Scholar]
- Blochlinger K, Jan LY, Jan YN. Postembryonic patterns of expression of cut, a locus regulating sensory organ identity in Drosophila. Development. 1993;117:441–450. doi: 10.1242/dev.117.2.441. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Leiserson WM, Benzer S. The eyes absent gene: Genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- Bonini NM, Bui QT, Gray-Board GL, Warrick JM. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- Braid LR, Verheyen EM. Drosophila nemo promotes eye specification directed by the retinal determination gene network. Genetics. 2008;180:283–299. doi: 10.1534/genetics.108.092155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodbeck S, Englert C. Genetic determination of nephrogenesis: The Pax/Eya/Six gene network. Pediatr Nephrol. 2004;19:249–255. doi: 10.1007/s00467-003-1374-z. [DOI] [PubMed] [Google Scholar]
- Brown NL, Sattler CA, Paddock SW, Carroll SB. Hairy and emc negatively regulate morphogenetic furrow progression in the Drosophila eye. Cell. 1995;80:879–887. doi: 10.1016/0092-8674(95)90291-0. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989a;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989b;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu Rev Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- Chan SK, Popperl H, Krumlauf R, Mann RS. An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. EMBO J. 1996;15:2476–2487. [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Brocchieri L, Shen WF, Largman C, Cleary ML. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol Cell Biol. 1996;16:1734–1745. doi: 10.1128/mcb.16.4.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanut F, Heberlein U. Retinal morphogenesis in Drosophila: Hints from an eye-specific decapentaplegic allele. Dev Genet. 1997a;20:197–207. doi: 10.1002/(SICI)1520-6408(1997)20:3<197::AID-DVG3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chanut F, Heberlein U. Role of decapentaplegic in initiation and progression of the morphogenetic furrow in the developing Drosophila retina. Development. 1997b;124:559–567. doi: 10.1242/dev.124.2.559. [DOI] [PubMed] [Google Scholar]
- Chao JL, Tsai YC, Chiu SJ, Sun YH. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development. 2004;131:3839–3847. doi: 10.1242/dev.01258. [DOI] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and Eyes Absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Chen R, Halder G, Zhang Z, Mardon G. Signaling by the TGF-b homolog decapentaplegic functions reiteratively within the network of genes controlling retinal cell fate determination in Drosophila. Development. 1999;126:935–943. doi: 10.1242/dev.126.5.935. [DOI] [PubMed] [Google Scholar]
- Cheyette BN, Green PJ, Martin K, Garren H, Hartenstein V, Zipursky SL. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- Chi N, Epstein JA. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/s0168-9525(01)02594-x. [DOI] [PubMed] [Google Scholar]
- Cho KO, Choi KW. Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature. 1998;396:272–276. doi: 10.1038/24394. [DOI] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–136. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- Clements J, Hens K, Merugu S, Dichtl B, de Couet HG, Callaerts P. Mutational analysis of the eyeless gene and phenotypic rescue reveal that an intact Eyeless protein is necessary for normal eye and brain development in Drosophila. Dev Biol. 2009;334:503–512. doi: 10.1016/j.ydbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM. Imaginal disc development. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 747–841. [Google Scholar]
- Cohen L, Salzberg A. Chromosomal binding sites of the homeotic cofactor Homothorax. Mol Genet Genomics. 2008;280:73–81. doi: 10.1007/s00438-008-0347-0. [DOI] [PubMed] [Google Scholar]
- Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: The roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–1336. doi: 10.1242/dev.127.6.1325. [DOI] [PubMed] [Google Scholar]
- Curtiss J, Burnett M, Mlodzik M. Distal antenna and distal antenna-related function in the retinal determination network during eye development in Drosophila. Dev Biol. 2007;306:685–702. doi: 10.1016/j.ydbio.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Halder G, Kloter U, Souabni A, Gehring WJ, Busslinger M. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Datta RR, Lurye JM, Kumar JP. Restriction of ectopic eye formation by Drosophila teashirt and tiptop to the developing antenna. Dev Dyn. 2009;238:2202–2210. doi: 10.1002/dvdy.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich W. Die Fazettenaugen der Dipteren. Z Wiss Zool. 1909;92:465–539. [Google Scholar]
- Dominguez M, de Celis JF. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature. 1998;396:276–278. doi: 10.1038/24402. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Ferres-Marco D, Gutierrez-Avino FJ, Speicher SA, Beneyto M. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat Genet. 2004;36:31–39. doi: 10.1038/ng1281. [DOI] [PubMed] [Google Scholar]
- Dong PD, Dicks JS, Panganiban G. Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development. 2002;129:1967–1974. doi: 10.1242/dev.129.8.1967. [DOI] [PubMed] [Google Scholar]
- Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Spann DR, Posakony JW. extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell. 1990;61:27–38. doi: 10.1016/0092-8674(90)90212-w. [DOI] [PubMed] [Google Scholar]
- Emerald BS, Curtiss J, Mlodzik M, Cohen SM. Distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs. Development. 2003;130(6):1171–1180. doi: 10.1242/dev.00323. [DOI] [PubMed] [Google Scholar]
- Fanto M, McNeill H. Planar polarity from flies to vertebrates. J Cell Sci. 2004;117:527–533. doi: 10.1242/jcs.00973. [DOI] [PubMed] [Google Scholar]
- Fiehler RW, Wolff T. Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev Biol. 2008;313:533–544. doi: 10.1016/j.ydbio.2007.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrell J, Modolell J. The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell. 1990;61:39–48. doi: 10.1016/0092-8674(90)90213-x. [DOI] [PubMed] [Google Scholar]
- Gehring WJ. The master control gene for morphogenesis and evolution of the eye. Genes Cells. 1996;1:11–15. doi: 10.1046/j.1365-2443.1996.11011.x. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax 6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Gonzales-Crespo S, Abu-Shaar M, Torres M, Martinez-A C, Mann RS, Morata G. Antagonism between extradenticle function and hedgehog signalling in the developing limb. Nature. 1998;394:196–200. doi: 10.1038/28197. [DOI] [PubMed] [Google Scholar]
- Greenwood S, Struhl G. Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development. 1999;126:5795–5808. doi: 10.1242/dev.126.24.5795. [DOI] [PubMed] [Google Scholar]
- Hoge MA. Another gene in the fourth chromosome of Drosophila. Am Nat. 1915;49:47–49. [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by target expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Flister S, Walldorf U, Kloter U, Gegring WJ. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–2191. doi: 10.1242/dev.125.12.2181. [DOI] [PubMed] [Google Scholar]
- Hammond KL, Hanson IM, Brown AG, Lettice LA, Hill RE. Mammalian and Drosophila dachshund genes are related to the Ski proto-oncogene and are expressed in eye and limb. Mech Dev. 1998;74:121–131. doi: 10.1016/s0925-4773(98)00071-9. [DOI] [PubMed] [Google Scholar]
- Hanson IM. Mammalian homologues of the Drosophila eye specification genes. Semin Cell Dev Biol. 2001;12:475–484. doi: 10.1006/scdb.2001.0271. [DOI] [PubMed] [Google Scholar]
- Hauck B, Gehring WJ, Walldorf U. Functional analysis of an eye specific enhancer of the eyeless gene in Drosophila. Proc Natl Acad Sci USA. 1999;96:564–569. doi: 10.1073/pnas.96.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie JL, Bryant PJ. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J Exp Zool. 1986;237:293–308. doi: 10.1002/jez.1402370302. [DOI] [PubMed] [Google Scholar]
- Heberlein U, Singh CM, Luk AY, Donohoe TJ. Growth and differentiation in the Drosophila eye coordinated by hedgehog. Nature. 1995;373:709–711. doi: 10.1038/373709a0. [DOI] [PubMed] [Google Scholar]
- Held LI. Imaginal discs: The genetic and cellular logic of pattern formation. Vol. 39. Cambridge Press; Cambridge: 2002. p. 460. (Developmental and Cell Biology Series). [Google Scholar]
- Horsfield J, Penton A, Secombe J, Hoffman FM, Richardson H. Decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development. 1998;125:5069–5078. doi: 10.1242/dev.125.24.5069. [DOI] [PubMed] [Google Scholar]
- Hsiao FC, Williams A, Davies EL, Rebay I. Eyes Absent mediates crosstalk between retinal determination genes and the receptor tyrosine kinase signaling pathway. Dev Cell. 2001;1:51–61. doi: 10.1016/s1534-5807(01)00011-9. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Watanabe Y, Ohto H, Kawakami K. Molecular interaction and synergistic activation of a promoter by Six, Eya, and Dach proteins mediated through CREB binding protein. Mol Cell Biol. 2002;22:6759–6766. doi: 10.1128/MCB.22.19.6759-6766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbal A, Halachmi N, Dibner C, Frank D, Salzberg A. Genetic evidence for the transcriptional-activating function of Homothorax during adult fly development. Development. 2001;128:3405–3413. doi: 10.1242/dev.128.18.3405. [DOI] [PubMed] [Google Scholar]
- Jang CC, Chao JL, Jones N, Yao LC, Bessarab DA, Kuo YM, Jun S, Desplan C, Beckendorf SK, Sun YH. Two Pax genes, eye gone and eyeless, act cooperatively in promoting Drosophila eye development. Development. 2003;130:2939–2951. doi: 10.1242/dev.00522. [DOI] [PubMed] [Google Scholar]
- Jaw TJ, You LR, Knoepfler PS, Yao LC, Pai CY, Tang CY, Chang LP, Berthelsen J, Blasi F, Kamps MP, Sun YH. Direct interaction of two homeoproteins, homothorax and extradenticle, is essential for EXD nuclear localization and function. Mech Dev. 2000;91:279–291. doi: 10.1016/s0925-4773(99)00316-0. [DOI] [PubMed] [Google Scholar]
- Jean D, Ewan K, Gruss P. Molecular regulators involved in vertebrate eye development. Mech Dev. 1998;76:3–18. doi: 10.1016/s0925-4773(98)00117-8. [DOI] [PubMed] [Google Scholar]
- Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev Biol. 2007;310:416–429. doi: 10.1016/j.ydbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao R, Daube M, Duan H, Zou Y, Frei E, Noll M. Headless flies generated by developmental pathway interference. Development. 2001;128:3307–3319. doi: 10.1242/dev.128.17.3307. [DOI] [PubMed] [Google Scholar]
- Jun S, Wallen RV, Goriely A, Kalionis B, Desplan C. Lune/eye gone, a Pax-like protein, uses a partial paired domain and a homeodomain for DNA recognition. Proc Natl Acad Sci USA. 1998;95:13720–13725. doi: 10.1073/pnas.95.23.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M, Singh A, Henry Sun Y. Eyeless collaborates with Hedgehog and Decapentaplegic signaling in Drosophila eye induction. Dev Biol. 2003;256:49–60. doi: 10.1016/s0012-1606(02)00123-9. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Sato S, Ozaki H, Ikeda K. Six family genes–structure and function as transcription factors and their roles in development. Bioessays. 2000;22:616–626. doi: 10.1002/1521-1878(200007)22:7<616::AID-BIES4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Ranade SS, Curtiss J, Mlodzik M, Pignoni F. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev Cell. 2003;5:403–414. doi: 10.1016/s1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Li DJ, Clouser C, Tran S, Pignoni F. Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev Dyn. 2005a;234:497–504. doi: 10.1002/dvdy.20442. [DOI] [PubMed] [Google Scholar]
- Kenyon KL, Yang-Zhou D, Cai CQ, Tran S, Clouser C, Decene G, Ranade S, Pignoni F. Partner specificity is essential for proper function of the SIX-type homeodomain proteins Sine oculis and Optix during fly eye development. Dev Biol. 2005b;286:158–168. doi: 10.1016/j.ydbio.2005.07.017. [DOI] [PubMed] [Google Scholar]
- Kim SS, Zhang RG, Braunstein SE, Joachimiak A, Cvekl A, Hegde RS. Structure of the retinal determination protein Dachshund reveals a DNA binding motif. Structure (Camb) 2002;10:787–795. doi: 10.1016/s0969-2126(02)00769-4. [DOI] [PubMed] [Google Scholar]
- Kozmik Z. Pax genes in eye development and evolution. Curr Opin Genet Dev. 2005;15:430–438. doi: 10.1016/j.gde.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Kronhamn J, Frei E, Daube M, Jiao R, Shi Y, Noll M, Rasmuson-Lestander A. Headless flies produced by mutations in the paralogous Pax6 genes eyeless and twin of eyeless. Development. 2002;129:1015–1026. doi: 10.1242/dev.129.4.1015. [DOI] [PubMed] [Google Scholar]
- Kumar JP. Signalling pathways in Drosophila and vertebrate retinal development. Nat Rev Genet. 2001;2:846–857. doi: 10.1038/35098564. [DOI] [PubMed] [Google Scholar]
- Kumar JP. The molecular circuitry governing retinal determination. Biochim Biophys Acta. 2009a;1789:306–314. doi: 10.1016/j.bbagrm.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009b;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Moses K. EGF receptor and Notch signaling act upstream of Eyeless/Pax6 to control eye specification. Cell. 2001a;104:687–697. doi: 10.1016/s0092-8674(01)00265-3. [DOI] [PubMed] [Google Scholar]
- Kumar JP, Moses K. Expression of evolutionarily conserved eye specification genes during Drosophila embryogenesis. Dev Genes Evol. 2001b;211:406–414. doi: 10.1007/s004270100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar JP, Ready DF. Rhodopsin plays an essential structural role in Drosophila photoreceptor development. Development. 1995;121:4359–4370. doi: 10.1242/dev.121.12.4359. [DOI] [PubMed] [Google Scholar]
- Kurata S, Go MJ, Artavanis-Tsakonas S, Gehring WJ. Notch signaling and the determination of appendage identity. Proc Natl Acad Sci USA. 2000;97:2117–2122. doi: 10.1073/pnas.040556497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugier E, Yang Z, Fasano L, Kerridge S, Vola C. A critical role of teashirt for patterning the ventral epidermis is masked by ectopic expression of tiptop, a paralog of teashirt in Drosophila. Dev Biol. 2005;283:446–458. doi: 10.1016/j.ydbio.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Lebovitz RM, Ready DF. Ommatidial development in Drosophila eye disc fragments. Dev Biol. 1986;117:663–671. doi: 10.1016/0012-1606(86)90335-0. [DOI] [PubMed] [Google Scholar]
- Li X, Perissi V, Liu F, Rose DW, Rosenfeld MG. Tissue-specific regulation of retinal and pituitary precursor cell proliferation. Science. 2002;297:1180–1183. doi: 10.1126/science.1073263. [DOI] [PubMed] [Google Scholar]
- Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, Nigam SK, Aggarwal AK, Maas R, Rose DW, Rosenfeld MG. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- Longley RL, Jr, Ready DF. Integrins and the development of three-dimensional structure in the Drosophila compound eye. Dev Biol. 1995;171:415–433. doi: 10.1006/dbio.1995.1292. [DOI] [PubMed] [Google Scholar]
- Lopes CS, Casares F. hth maintains the pool of eye progenitors and its downregulation by Dpp and Hh couples retinal fate acquisition with cell cycle exit. Dev Biol. 2009;339:78–88. doi: 10.1016/j.ydbio.2009.12.020. [DOI] [PubMed] [Google Scholar]
- Ma C, Moses K. Wingless and patched are negative regulators of the morphogenetic furrow and can affect tissue polarity in the developing Drosophila compound eye. Development. 1995;121:2279–2289. doi: 10.1242/dev.121.8.2279. [DOI] [PubMed] [Google Scholar]
- Mann RS. The specificity of homeotic gene function. Bioessays. 1995;17:855–863. doi: 10.1002/bies.950171007. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Goudreau G, Gruss P. Pax genes and their role in organogenesis. Cancer Res. 1999;59:1707s–1709s. discussion 1709s–1710s. [PubMed] [Google Scholar]
- Mardon G, Solomon NM, Rubin GM. Dachshund encodes a nuclear protein required for normal eye and leg development in Drosophila. Development. 1994;120:3473–3486. doi: 10.1242/dev.120.12.3473. [DOI] [PubMed] [Google Scholar]
- Maves L, Schubiger G. A molecular basis for transdetermination in Drosophila imaginal discs: Interactions between wingless and decapentaplegic signaling. Development. 1998;125:115–124. doi: 10.1242/dev.125.1.115. [DOI] [PubMed] [Google Scholar]
- Maves L, Schubiger G. Transdetermination in Drosophila imaginal discs: A model for understanding pluripotency and selector gene maintenance. Curr Opin Genet Dev. 2003;13:472–479. doi: 10.1016/j.gde.2003.08.006. [DOI] [PubMed] [Google Scholar]
- Michaut L, Flister S, Neeb M, White KP, Certa U, Gehring WJ. Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc Natl Acad Sci USA. 2003;100:4024–4029. doi: 10.1073/pnas.0630561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M. Planar polarity in the Drosophila eye: A multifaceted view of signaling specificity and cross-talk. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrin EJ, Li Y, Hoffman K, Liu J, Wang K, Zhang L, Mardon G, Chen R. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–476. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Kuo TS, Jaw TJ, Kurant E, Chen CT, Bessarab DA, Salzberg A, Sun YH. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1997;12:435–446. doi: 10.1101/gad.12.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Rubin GM. Targeted expression of teashirt induces ectopic eyes in Drosophila. Proc Natl Acad Sci USA. 1998;95:15508–15512. doi: 10.1073/pnas.95.26.15508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papayannopoulos V, Tomlinson A, Panin VM, Rauskolb C, Irvine KD. Dorsal–ventral signaling in the Drosophila eye. Science. 1998;281:2031–2034. doi: 10.1126/science.281.5385.2031. [DOI] [PubMed] [Google Scholar]
- Pappu KS, Mardon G. Genetic control of retinal specification and determination in Drosophila. Int J Dev Biol. 2004;48:913–924. doi: 10.1387/ijdb.041875kp. [DOI] [PubMed] [Google Scholar]
- Pappu KS, Ostrin EJ, Middlebrooks BW, Sili BT, Chen R, Atkins MR, Gibbs R, Mardon G. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132:2895–2905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- Pauli T, Seimiya M, Blanco J, Gehring WJ. Identification of functional sine oculis motifs in the autoregulatory element of its own gene, in the eyeless enhancer and in the signalling gene hedgehog. Development. 2005;132:2771–2782. doi: 10.1242/dev.01841. [DOI] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: Homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud F, Casares F. Homothorax and iroquois-C genes are required for the establishment of territories within the developing eye disc. Mech Dev. 2000;96:15–25. doi: 10.1016/s0925-4773(00)00372-5. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Punzo C, Kurata S, Gehring WJ. The eyeless homeodomain is dispensable for eye development in Drosophila. Genes Dev. 2001;15:1716–1723. doi: 10.1101/gad.196401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo C, Seimiya M, Flister S, Gehring WJ, Plaza S. Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development. 2002;129:625–634. doi: 10.1242/dev.129.3.625. [DOI] [PubMed] [Google Scholar]
- Punzo C, Plaza S, Seimiya M, Schnupf P, Kurata S, Jaeger J, Gehring WJ. Functional divergence between eyeless and twin of eyeless in Drosophila melanogaster. Development. 2004;131:3943–3953. doi: 10.1242/dev.01278. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. (see comments). [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Smith KM, Peifer M, Wieschaus E. Extradenticle determines segmental identities throughout Drosophila development. Development. 1995;121:3663–3673. doi: 10.1242/dev.121.11.3663. [DOI] [PubMed] [Google Scholar]
- Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- Ready DF. A multifaceted approach to neural development. Trends Neurosci. 1989;12:102–110. doi: 10.1016/0166-2236(89)90166-5. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Salzer CL, Kumar JP. Position dependent responses to discontinuities in the retinal determination network. Dev Biol. 2009;326:121–130. doi: 10.1016/j.ydbio.2008.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer CL, Kumar JP. Identification of retinal transformation hot spots in developing Drosophila epithelia. PLoS ONE. 2010;5:e8510. doi: 10.1371/journal.pone.0008510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer CL, Elias Y, Kumar JP. The retinal determination gene eyes absent is regulated by the EGF receptor pathway throughout development in Drosophila. Genetics. 2010;184:185–197. doi: 10.1534/genetics.109.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Tomlinson A. Dorsal–ventral midline signaling in the developing Drosophila eye. Development. 2007;134:659–667. doi: 10.1242/dev.02786. [DOI] [PubMed] [Google Scholar]
- Seimiya M, Gehring WJ. The Drosophila homeobox gene optix is capable of inducing ectopic eyes by an eyeless-independent mechanism. Development. 2000;127:1879–1886. doi: 10.1242/dev.127.9.1879. [DOI] [PubMed] [Google Scholar]
- Serikaku MA, O’Tousa JE. Sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. doi: 10.1242/dev.124.1.45. [DOI] [PubMed] [Google Scholar]
- Sheng G, Thouvenot E, Schmucker D, Wilson DS, Desplan C. Direct regulation of rhodopsin 1 by Pax-6/eyeless in Drosophila: Evidence for a conserved function in photoreceptors. Genes Dev. 1997;11:1122–1131. doi: 10.1101/gad.11.9.1122. [DOI] [PubMed] [Google Scholar]
- Silver SJ, Rebay I. Signaling circuitries in development: Insights from the retinal determination gene network. Development. 2005;132:3–13. doi: 10.1242/dev.01539. [DOI] [PubMed] [Google Scholar]
- Singh A, Kango-Singh M, Sun YH. Eye suppression, a novel function of teashirt, requires Wingless signaling. Development. 2002;129:4271–4280. doi: 10.1242/dev.129.18.4271. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D. EGF signaling and ommatidial rotation in the Drosophila eye. Curr Biol. 2003;13:1451–1457. doi: 10.1016/s0960-9822(03)00545-1. [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Du W. Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev Biol. 2008;313:787–801. doi: 10.1016/j.ydbio.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Sevenless: A cell specific homeotic mutation of the Drosophila eye. Science. 1986;231:400–402. doi: 10.1126/science.231.4736.400. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Cell fate in the Drosophila ommatidium. Dev Biol. 1987a;123:264–275. doi: 10.1016/0012-1606(87)90448-9. [DOI] [PubMed] [Google Scholar]
- Tomlinson A, Ready DF. Neuronal differentiation in the Drosophila ommatidium. Dev Biol. 1987b;120:366–376. doi: 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- Treisman JE, Rubin GM. Wingless inhibits morphogenetic furrow movement in the Drosophila eye disc. Development. 1995;121:3519–3527. doi: 10.1242/dev.121.11.3519. [DOI] [PubMed] [Google Scholar]
- Voas MG, Rebay I. Signal integration during development: Insights from the Drosophila eye. Dev Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- Weasner BP, Kumar JP. The non-conserved C-terminal segments of Sine Oculis Homeobox (SIX) proteins confer functional specificity. Genesis. 2009;47:514–523. doi: 10.1002/dvg.20517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weasner B, Salzer C, Kumar JP. Sine oculis, a member of the SIX family of transcription factors, directs eye formation. Dev Biol. 2007;303:756–771. doi: 10.1016/j.ydbio.2006.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weasner BM, Weasner B, Deyoung SM, Michaels SD, Kumar JP. Transcriptional activities of the Pax6 gene eyeless regulate tissue specificity of ectopic eye formation in Drosophila. Dev Biol. 2009;334:492–502. doi: 10.1016/j.ydbio.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: The morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formation in the Drosophila retina. In: Bate M, Martinez Arias A, editors. The Development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 1277–1326. [Google Scholar]
- Wu K, Yang Y, Wang C, Davoli MA, D’Amico M, Li A, Cveklova K, Kozmik Z, Lisanti MP, Russell RG, Cvekl A, Pestell RG. DACH1 inhibits transforming growth factor-beta signaling through binding Smad4. J Biol Chem. 2003;278:51673–51684. doi: 10.1074/jbc.M310021200. [DOI] [PubMed] [Google Scholar]
- Yan H, Canon J, Banerjee U. A transcriptional chain linking eye specification to terminal determination of cone cells in the Drosophila eye. Dev Biol. 2003;263:323–329. doi: 10.1016/j.ydbio.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Yao JG, Weasner BM, Wang LH, Jang CC, Weasner B, Tang CY, Salzer CL, Chen CH, Hay B, Sun YH, Kumar JP. Differential requirements for the Pax6(5a) genes eyegone and twin of eyegone during eye development in Drosophila. Dev Biol. 2008;315:535–551. doi: 10.1016/j.ydbio.2007.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: Regulation of atonal by Ey and So. Development. 2006;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]


