Abstract
Seasonal influenza is controlled through vaccination campaigns. Evolution of influenza virus antigens means that vaccines must be updated to match novel strains, and vaccine effectiveness depends on scientists’ ability to predict nearly a year in advance which influenza variants will dominate in upcoming seasons. In this review, we highlight a promising new surveillance tool: predictive models. Developed through data-sharing and close collaboration between the World Health Organization and academic scientists, these models use surveillance data to make quantitative predictions regarding influenza evolution. Predictive models demonstrate the potential of applied evolutionary biology to improve public health and disease control. We review the state of influenza predictive modeling and discuss next steps and recommendations to ensure that these models deliver upon their considerable biomedical promise.
Keywords: influenza, predictive evolution, predictive modeling, vaccine strain selection
1. The need to predict influenza evolution
Influenza viruses annually cause large numbers of emergency room visits, hospitalizations, ICU admissions and deaths worldwide [1–6], as well as billions of dollars in economic losses in the United States alone [2, 7]. Persistent, annual influenza epidemics are possible in the face of population immunity because influenza lineages gradually accumulate genetic changes that alter antigenic phenotype and allow reinfection of previously exposed individuals. Strong positive selection for these new antigenic variants produces antigenic drift (see Glossary).
In the face of antigenic drift, the influenza vaccine must be updated frequently to maintain its effectiveness. Because it takes at least 6–8 months to develop and produce an updated influenza vaccine, scientists must decide which influenza virus variants to include in the vaccine nearly a year in advance.
To aid these predictions and facilitate informed vaccine update decisions, the World Health Organization (WHO) coordinates a sweeping collaborative global effort to survey and characterize the diversity of influenza viruses circulating in humans [8–10] (see also WHO vaccine recommendation reportsiii). Surveillance produces a massive corpus of antigenic and genetic data, which must be analyzed rapidly and systematically. Researchers have developed a number of quantitative tools to aid these analyses.
Predictive models that forecast which influenza virus clades will predominate in future influenza seasons are a new class of quantitative tools at the cutting edge of influenza surveillance. These models could represent the first step toward an era of applied evolutionary biology, in which influenza surveillance not only aims to monitor the diversity of circulating viruses, but also to produce robust forecasts of future circulation [11]. Influenza also promises to serve as a model system for predictive evolutionary modeling in other areas. For example, tumors under therapy show a very similar mode of evolution, characterized by competing genetic clones [12, 13], high mutation rates, and strong selection pressure driven by the host immune response [14, 15]. Predictive evolutionary modeling also has the potential to provide crucial insights into the ongoing emergence of treatment resistance in pathogens, vectors and agricultural pests.
In this article, we review recent work in predictive modeling of influenza and discuss the current role of models in the influenza vaccine strain selection process. We then outline key next steps to ensure that predictive models provide robust insights for vaccine strain selection. We give particular attention to the importance of maintaining and strengthening the global research community that has made forecasting influenza evolution possible.
2. Influenza prediction and strain selection today
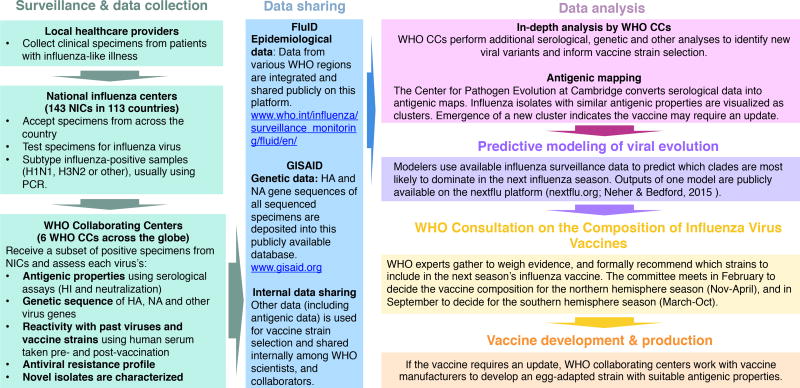
Influenza vaccine strain selection is a year-round, continuous process. Predictive models are most useful when they can be tightly integrated into the fast-paced influenza surveillance and vaccine development pipeline (Fig. 1, 3). In this section, we describe that pipeline and the modeling’s current role within it.
Figure 1.
Schematic of the influenza surveillance and vaccine strain selection process
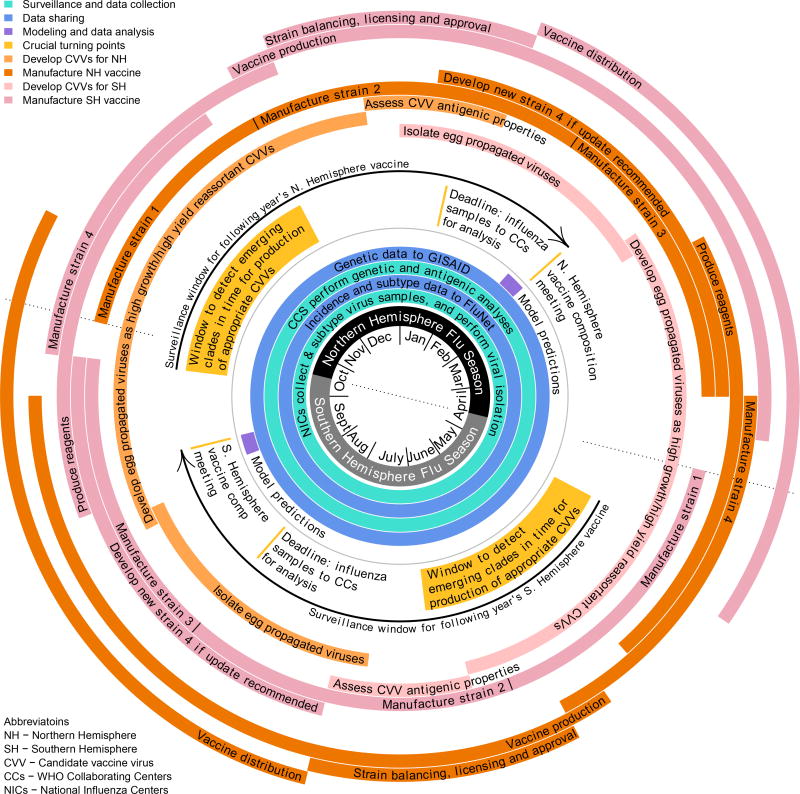
Figure 3.
Approximate calendar of the vaccine update process for the quadrivalent vaccine.
Vaccine strains are determined at meetings in February (for the Northern Hemisphere) and September (for the Southern Hemisphere), so that vaccine production and delivery can be completed prior to the onset of each hemisphere’s influenza season. Influenza surveillance, and vaccine development and production are underway year-round.
2.1. Vaccine strain selection
The WHO’s Global Influenza Surveillance and Response System (GISRS) coordinates influenza surveillance efforts (Fig. 1). Samples are collected from patients across the globe and sent to one of 143 National Influenza Centers (NICs). NICs identify influenza-positive samples to the type and subtype level, and then send a representative subset of samples to one of six WHO Collaborating Centers on influenza (WHOCCs) (Fig. 1).
WHOCCs sequence a large proportion of influenza virus specimens received (Fig. 2), and typically use hemagglutination inhibition (HI) assays to characterize of isolates’ antigenic phenotype. Some circulating influenza A (H3N2) variants have lost the ability to agglutinate red blood cells [16], so WHOCCs now use microneutralization assays to complement and corroborate HI titer data.
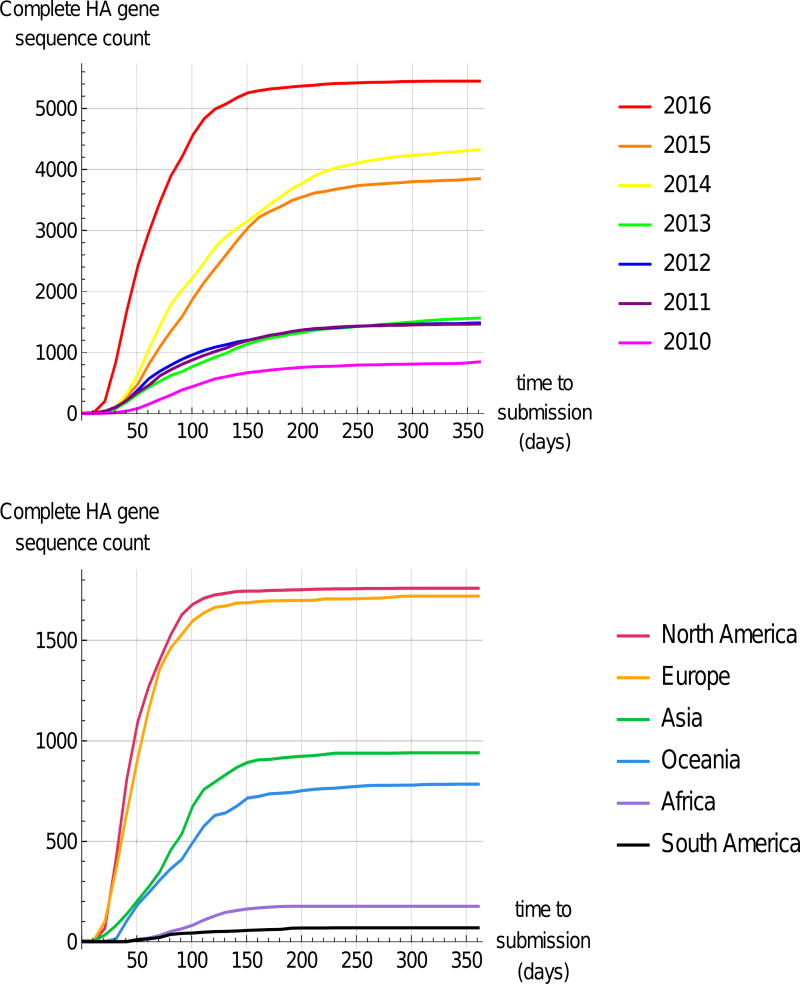
Figure 2.
Accumulation of Newly Sampled Complete HA Gene Sequences in GISAID EpiFlu™ over the Course of a March–March Year. Top: Cumulative count of sequences deposited in EpiFlu™ as a function of time to submission (difference between sampling date and submission date). Sequences are grouped by collection year. Bottom: Accumulation of sequence data for the year 2016 by geographic region. Plotted data was obtained by downloading sequence records from EpiFlu™ databasev (data downloaded on 31st March 2017), selecting only sequences that were submitted to and published directly in GISAID, and comparing collection date and submission date metadata. Abbreviation: HA, hemagglutinin.
Twice yearly, leaders of the WHOCCs, WHO Essential Regulatory Laboratories, regulators, and others meet to assess surveillance data and develop WHO recommendations for the composition of vaccines. Recommendations issued at these vaccine composition meetings (VCMs) inform vaccine campaigns during the Northern Hemisphere (Nov–April) and Southern Hemisphere (March–Oct) influenza seasons.
Vaccine updates are not possible unless a suitable candidate vaccine virus (CVV) has been developed well in advance of the VCM (Fig. 3). Emerging influenza variants with high probabilities of spread in upcoming seasons must therefore be identified as fit and developed into CVVs at least 9–12 months in advance of their possible inclusion in the vaccine.
Predictive models can systematically search through large surveillance datasets to help identify emerging influenza clades with high probabilities of expansion. The challenge now is to integrate the diverse array of existing models into the strain selection process in ways that fully leverage their predictive power to improve strain selection.
2.2. Predictive models
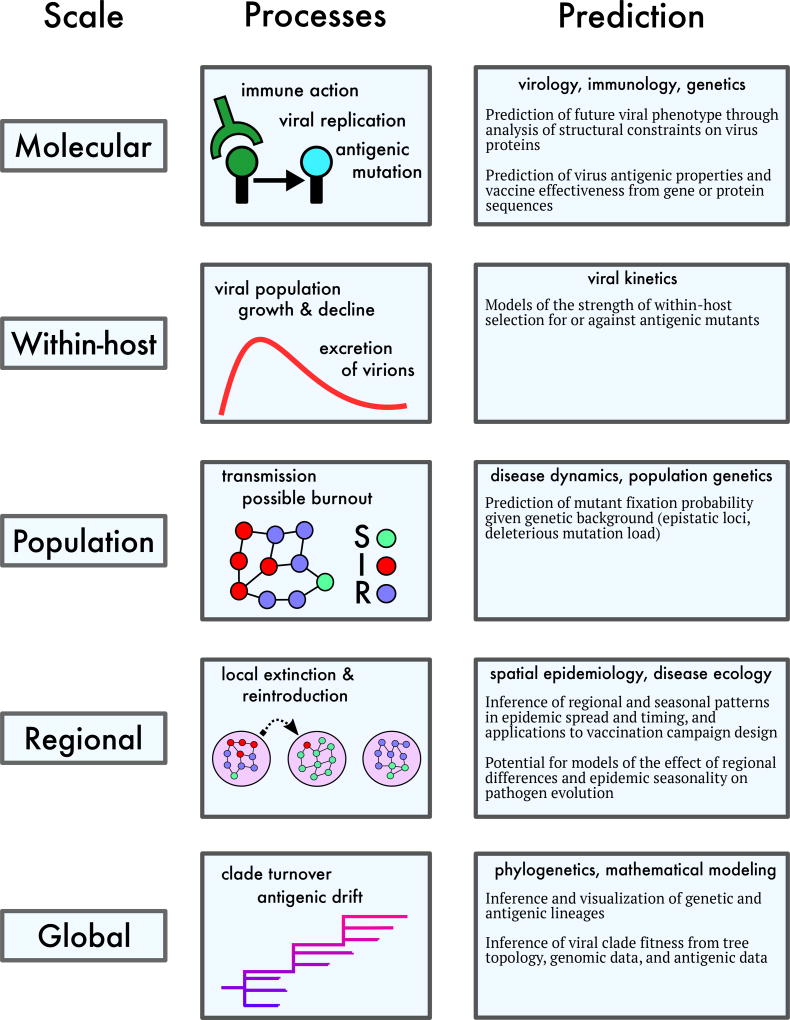
Advances in biomedicine have begun to elucidate the biological determinants of seasonal influenza’s evolution at a variety of scales (Fig. 4). The majority of predictive modeling studies focus on the gene segment that encodes the surface protein hemagglutinin (HA). The globular head of HA contains epitope antigenic sites – the primary targets for the human adaptive immune response. HA gene sequences for thousands of influenza A strains isolated over the last 40 years are publicly available [17, 18].
Figure 4.
The multi-scale process of influenza evolution, with patterns and possible sources of predictability at each scale.
This has made it possible to study influenza’s phylodynamics [19, 20] -– the interaction between its epidemiological and evolutionary processes. Predictive analysis can also build upon advances in influenza population genetics, in particular methods of inferring selection on virus proteins [21–25].
To predict influenza evolution, it is first necessary to identify viral sublineages or clades coexisting in the global population and to partition viral phylogenies inferred from sequence data into such clades. Changes in clade frequency can then be predicted by estimating the fitness differences between viral clades – that is, their expected relative growth rates.
We first describe predictive models with an evolutionary focus; these models predict properties of future viral populations based on past and present data (see Box 1).
Box 1: Predictive models.
Antigenicity-stability fitness model [26]
This model estimates the fitness (i.e. expected growth rate) of viral clades – ensembles of genetically related strains that descend from a single common ancestor. The original model [26] used only genetic data, but current iterations utilize both genetic and antigenic input data to estimate multiple components contributing to viral fitness. In the current model, antigenic changes (inferred from HI and neutralization data or from epitope mutations) increase fitness, while protein destabilization (inferred from non-epitope mutations) decreases fitness. The model predicts the frequency trajectory of clades for about one year into the future, and it allows for the early detection of new antigenic variants and inference of their genetic basis. It has been validated by historical predictions over two decades. To estimate the contribution of the individual model components several years of historical data are required.
Epitope clade growth [27]
This model ranks genetic HA clades seeded by epitope mutations by their recent growth inferred from a genealogical tree. By mapping HI data on the tree, the model estimates the antigenic differences of high-growth clades. It has been tested by limited historical predictions. Factors affecting prediction quality include sampling inhomogeneities and fitness effects outside epitope mutations.
Local tree shape [28]
In this model, the branching patterns of reconstructed genealogical trees are used to infer the relative growth rates of the sampled genetic sublineages, without an explicit modeling of the viral fitness. The growth rate is estimated with the local branching index, which is defined as the length of the tree, averaged with an exponential decreasing weight, in the proximity of an internal node. This model can predict the most likely strain to dominate in upcoming influenza seasons without using historical data. The validity of the prediction has been assessed both with simulated data and with extensive retrospective analysis on data from influenza surveillance. Temporal and geographical inhomogeneities of strain sampling limit the quality of the prediction.
The fitness model presented in [26] uses genetic clades as units of predictive analysis and proposes an approximate mapping between HA sequences and viral fitness. Mutations at epitope sites are assigned a positive fitness effect describing cross-immunity across multiple strains. Mutations outside the epitope region, which can cause protein destabilization, are assigned a fitness cost. This fitness model can predict the future evolutionary trajectories of viral sublineages, and can therefore be used to identify which viral clade is most likely to dominate in upcoming influenza seasons.
A second group of predictive clade frequency models are based primarily on genealogical tree data and work without explicitly modeling viral fitness. In [27], recent growth of HA clades seeded by epitope changes is extrapolated to identify future predominant clades, which are then weighed by their inferred antigenic differences. In contrast, the model of[28] evaluates recent clade growth only from the information contained in the local shape of the HA genealogical tree. Branching patterns of reconstructed phylogenies are used to infer the relative growth rate of the sampled viral sublineages. Rapidly growing populations undergo fast diversification and therefore show a high degree of local tree branching. A measure of fitness differences between clades can then be obtained by interpreting the inferred growth rate as fitness.
Other theoretical models do not aim to predict clade content of the future viral population but infer phenotypic properties of the current population (see Box 2). These include the expected effectiveness of the current vaccine strains against newly identified strains [29, 30] and amino acid substitutions in protein loci [31]. Some models can also be used for the identification of new antigenic variants at their early stage of expansion [26, 27, 29, 30, 32].
Box 2: Inference of antigenic characterization.
Linking antigenic properties and genetic data
Antigenic properties of influenza A viruses can be inferred from viral HA sequences using machine learning and other inference methods. Some methods use genetic distances computed from sequence alignments [29, 109]. Others apply phylogenetic methods [44, 45]. Structural and physicochemical properties of HA sequences can also be taken into account [32, 110]. These methods can be used to predict antigenic cluster membership [32, 110], to quantify the expected effectiveness of the current vaccine against circulating viral strains [30], or to assess the degree of antigenic change caused by particular mutations [44, 45, 109, 111].
Identification of proposed vaccine strains
The antigenicity-stability fitness model [26] uses minimization of cross-immunity with predicted future circulating strains as a criterion to identify candidate vaccine strains. The approach presented in [27] combines measures of strain proliferation rate with measures of strain antigenic novelty obtained from HI data to identify possible candidate strains for seasonal influenza vaccine updates; a similar approach has been recently integrated in a new bioinformatic pipeline (S.A. Schobel. PhD thesis, University of Maryland, College Park, 2015). The local branching index [28] can be used to identify vaccine candidate strains by proximity to high ranking tree nodes.
All the models are probabilistic, not least because stochastic events are important drivers of influenza evolution: from mutations in individual viral sequences to local burnout of epidemics (Fig. 4).
3. Next steps for modeling and the influenza prediction community
Each of the modeling approaches described above represents a unique combination of biological details, simplifying assumptions and informative data. The immediate challenges are to determine how different approaches and data sources can be combined, and how modeling results should be interpreted when informing strain selection decisions. In particular, there is a need (1) to develop best practices in data collection, management, and use, (2) to choose an appropriate level of biological detail for predictive modeling, and (3) to develop tools and strategies for comparing models and understanding their limitations (see Outstanding Questions). All three goals will require collaboration among scientists working at all levels of the surveillance and strain selection process, including modelers, laboratory scientists, epidemiologists and clinicians.
3.1. Best practices for data collection, management, and use
All predictive models of influenza evolution infer viral fitness using some combination of genetic and antigenic data. Here, we summarize cautions and challenges associated with each data stream. We then discuss key next steps.
3.1.1. Genetic data
Each season, WHOCCs characterize the whole genome sequence, or HA and NA gene sequences of thousands of the influenza viruses collected and submitted by NICs around the globe (Fig. 2). The WHO’s FluID databaseiii provides open access to the resulting epidemiological data, while WHOCCs deposit sequence data into the publicly available GISAID EpiFlu™ databasev, Fig. 1. The genetic data within EpiFlu™ has been instrumental to the development of all modeling approaches reviewed here, and has facilitated a wealth of other basic influenza research.
In recent years, the quantity of data deposited into GISAID has increased dramatically while delays between virus collection and sequence sharing have decreased (Fig. 2). This has given modelers a better dataset from which to make predictions in advance of VCMs. Visualization and prediction tools such as the public nextu web applicationvi [33] have leveraged these data to allow rapid and up-to-date exploratory analysis. WHO NICs and CCs should be commended for their efforts to curate and publish this wealth of data [17, 18, 34, 35].
In the past, lags between data collection and sequencing limited the amount of data available to inform model predictions prior to VCMs. But efforts to reduce delays between sample collection and sequence publication have been fruitful. In 2016, thousands of complete HA gene sequences were deposited within 50 days of sample collection; this accounts for more than one-third of that year’s deposited sequences (Fig. 2). Continuing to streamline this data deposition pipeline can increase robustness and decrease uncertainty in model predictions. For similar reasons, it is worth continuing to develop rapid data analysis pipelines so that models can make use of the large quantities of new data that may become available shortly before a VCM (Fig. 2). Another ongoing challenge is to address geographical disparity in surveillance data quantity and quality. Europe and North America are well-sampled, and data processing and deposition happens quickly, but the same is not true for other regions (Fig. 2).
One caution when using influenza sequence data is that clinical samples are often passaged in cell culture prior to sequencing and antigenic analysis. Passaging adaptations that arise when viral isolates are propagated in vitro can influence both antigenic [16, 36], and genetic characterizations of viral specimens [37]. It is possible to correct or exclude genetic sequences containing passaging adaptations before input into predictive models [37]. Helpful computational tools can be found onlinevii. Laboratories have also been adopting best practices to reduce passaging adaptations in sequenced isolates [16, 37, 38], or to sequence virus specimens without isolation.
However most sequenced isolates are still passaged at least once [37], and passaging is necessary to characterize an isolate’s antigenic phenotype. Thus, there is a need to standardize sequence annotation across laboratories to unambiguously communicate a sequence’s passage history to modelers and other data users. Metadata that helps modelers compare the passage histories of isolates used to generate antigenic data would also help map from phenotype to genotype.
3.1.2. Antigenic data
Panels of hemagglutination inhibition (HI) serological assays are the standard tool used to assess the antigenic properties of influenza viral isolates. In HI panels, antisera are obtained from ferrets infected with a single reference virus. To characterize pairwise similarities in antigenicity between viruses, each antiserum is systematically tested against each reference virus. A high titer indicates high antigenic similarity and strong potential for cross-reactivity between the virus-antiserum pair tested [39].
Integration of HI data into predictive models can be challenging for a number of reasons. First, some clades of H3N2 no longer react in HI assays [16], so neutralization assays are increasingly used to complement HI titer data. Integration of various types of assays requires specialized modeling approaches. Experiments with human sera have shown that HI titer and neutralization titer correlate strongly [40], so in principle it is possible to do a mixed analysis that employs both types of serological data. However, the complexity of interpreting antigenic data should not be underestimated [36].
A second challenge is that serological patterns obtained using ferret antisera provide a close, albeit imperfect[41, 42] proxy for patterns of antigenic cross-reactivity in humans. Thus, panels of post-vaccination huma sera have been used for many years by WHO to confirm the antigenic properties of circulating viruses. Nevertheless, an increased use of panels of human antisera and human monoclonal antibodies is likely to improve further serological analysis of circulating viruses and potential candidate vaccine viruses.
Third, measured titer differences reect a combination of antigenic and non-antigenic effects [43–45]. Non-antigenic effects arise from differences in the receptor binding avidity of a particular reference virus, the potency of a particular antiserum, and avidity of the red blood cells being agglutinated [44]. Differences in assay protocol can also affect measured HI titer values [46]; standardizing protocols makes titer values obtained by different labs more directly comparable [47].
Finally, serological data are available for many, but not all of the viral sequences available in the EpiFlu™ database. Modelers have therefore developed tools that infer antigenic phenotype using known genetic sequences. The basic strategy is to gather antigenic data collected from the annual and interim reports of the WHOCC Londoniv and then to map these antigenic data onto inferred viral phylogenies [27, 44, 45, 48]. These genotype-to-phenotype mapping tools can be used as a substitute for missing antigenic data [44, 45, 48]. The challenge for predictive models is then to integrate this information in order to predict the future antigenic composition of influenza virus populations.
3.1.3. Next steps
Key next steps for improving data collection, management, and use include developing and enforcing clear data annotation standards, exploring the feasibility of an antigenic data repository, and using models to identify important gaps in surveillance.
Influenza research would benefit from clear, well-enforced data annotation standards. Robust model predictions require selecting input data based on its quality and collection date. This is only possible if metadata are unambiguous. Passaging adaptations can only be properly taken into account if isolates’ passage histories are documented in a consistent way. Evaluating the practical usefulness of model predictions for strain selection requires a record of what sequence and titer data was available for modeling in advance of a particular year’s VCM. Clear and consistent annotation also deepens the pool of researchers who can contribute to influenza prediction; it allows scientists not directly involved in data collection to be confident that their work’s assumptions are not violated by an undocumented aspect of the collection process.
Despite the challenges and new developments discussed, antigenic data remain a key part of influenza surveillance. To that end, we believe it is worth studying the feasibility of creating a public HI database to complement the genetic data available in EpiFlu™. Ideally, this database would also include data from neutralization assays. Presently, antigenic data are often unpublished, so the leaders of any database project would need to ensure that data are used responsibly, and that those collecting the data can receive due credit for their work. There is an ongoing discussion within biomedicine of how best to recognize – and incentivize – the collection, curation, and archiving of large, high-quality epidemiological datasets [49]. The influenza community should remain in the vanguard of efforts to share data equitably.
Finally, models may be able to aid surveillance efforts by identifying gaps or inefficiencies in existing surveillance data. Retrospective analysis of predictive error could help identify undersampled geographic areas or other factors that contribute to bias in surveillance data. Models could also help identify which data would most improve model informativeness if shared rapidly.
Modeling influenza evolution has been made possible through generous sharing of data. We advocate continued effort to facilitate and improve data sharing. This will allow predictive models to be better integrated into the strain selection process, increase the robustness and accuracy of existing models, and aid basic research on influenza.
3.2. Choosing an appropriate level of biological detail
An ongoing challenge for influenza forecasting will be to assess whether specific simplifying assumptions undermine the accuracy or robustness of model predictions. Here, we summarize key biological considerations that have not yet been explicitly included in predictive models.
3.2.1. Genetic background
Gene interactions and deleterious mutational load can influence which antigenic mutations are ultimately able to fix. Recent research has found evidence of epistasis between sites in the influenza HA gene [50] and of inter-gene epistasis between HA sites and NA sites [51]. Modeling work suggests that epistatic interactions could play a key role in driving patterns of influenza antigenic evolution [52]. A model that incorporates epistasis in antigenic phenotype can reproduce both the gradual antigenic changes that steadily accumulate within a broadly-cross-immunizing “antigenic cluster” [44, 53] and also the rarer cluster transitions that occur every few years [52].
Deleterious mutational load may also regulate the probability that antigenic mutants proliferate and reach fixation. Against a deleterious background, only large-effect antigenic mutants will experience strong positive selection; this is another possible explanation for the punctuated pattern of antigenic cluster transitions [54].
Taking account of the genetic background of newly-discovered antigenic variants could allow for more precise prediction of their likely fitness and their fixation probability.
3.2.2. How much of the influenza genome should models consider?
Despite evidence that substitutions at just a handful of key sites lead to large antigenic changes [21, 53, 55], recent analyses [44, 45] have shown that tracking substitutions at these key sites alone yields poor predictions of antigenic phenotype. Rather, the accumulation of small antigenic changes via substitutions at other sites may be a stronger driver of influenza virus evolution [44, 45] and can increase the accuracy of predicting antigenic phenotypes of emerging viruses from genotype [44].
HA sites outside the antigenic epitopes are also likely contribute to the total viral fitness, as many are under purifying selection [23, 26]. Moreover, high rates of amino-acid substitutions, possibly signaling positive selective pressure by host immunity, have been observed for HA sites outside the epitope region [56] and for genomic regions other than HA [57]. In particular, neuraminidase (NA), which drives the release and escape of new virions from the cell [58], has been shown to evolve under strong selection pressures imposed by the human immune system and antiviral drugs [22, 59, 60].
Incorporating genomic data from sites outside the HA epitope regions may thus lead to a better estimation of viral fitness, and therefore improve predictions. If loci on multiple influenza genome segments are key to determining strains’ relative fitness, models may also need to account for intra-lineage reassortment events [59]. Reassortment events have been shown to be biased towards certain segment combinations [61, 62], implying an associated fitness cost. As a result, the pool of observed new reassortant strains is restricted, and reassortment events may be followed by a number of subsequent compensatory mutations [63].
3.2.3. How important are mixed infections and within-host diversity?
The data deposited into GISAID’s EpiFlu™ database are either collected using Sanger sequencing, or are consensus sequences obtained from next-generation sequencing. Thus these data do not capture the fact that multiple viral variants coexist within an infected host. Deep sequencing studies show that mixed influenza infections involving multiple viral variants occur frequently in humans [64–69], and that minor variants can be transmitted alongside major variants [64, 65, 70]. Ongoing research now aims to determine how evolutionary forces within the host influence viral dynamics at the population level. To date, most deep sequencing studies at the within-host level suggest that de novo genetic diversification is limited [71], and that rare antigenic variants do not often rise to high frequencies within the host [67–69], despite the potential fitness advantages provided by their ability to evade host immunity. New evidence suggests these antigenic mutations may fail to rise in frequency because of clonal interference [69] or purifying selection on non-antigenic substitutions within the same viral genome [71]. The evolutionary trajectory of influenza within immunocompromised hosts can thus mirror global patterns of virus evolution [69]; clonal interference and purifying selection are also known to prevent some antigenic mutations from fixing at the global scale.
3.2.4. What predictive insights can be gained from observing influenza evolution in the lab?
Further empirical work on short-term influenza evolution could help improve our understanding of the selection pressures that act on the virus and the antigenic phenotypes that are likely to result. Evolutionary experiments in which researchers attempt to generate and select for antigenic mutants under laboratory conditions may also be valuable, and in theory such experiments could even be used to recreate or predict patterns of antigenic drift in vitro [72]. As discussed above, passaging adaptations can introduce spurious signals of selection and should wherever possible be avoided in experimental infections. Studying influenza infections in natural animal hosts – and development of associated assays and reagents – is also highly desirable where possible.
In practice, evolutionary experiments are often challenging. Circulating influenza viruses experience varied and complex selection pressures that are difficult to replicate in the laboratory, both within individual hosts and during transmission between hosts (see Fig. 4). In particular, the process of transmission remains a challenge to tackle experimentally at scale. In the face of these challenges, most recent studies have taken an observational approach, studying influenza’s evolutionary trajectories within human hosts [64, 65, 67–69, 71, 73].
But there is no substitute for experimental infections in testing the mechanistic assumptions of epidemiological models (see for instance [74]). As our mechanistic understanding of within-host evolution and our capacity to process and interpret deep sequencing data continue to improve, it may become possible to develop predictive models of within-host influenza evolution.
3.2.5. Regional differences and seasonality
Existing predictive models largely operate at the global scale, although modelers and users are aware of evidence for geographic structure in influenza dynamics [75–77]. Most influenza virus variants originate in and spread from East or Southeast Asia. Influenza B and A/H1N1 lineages then appear to persist regionally for longer periods of time than A/H3N2 [77]. Yet in practice, it has been difficult to balance the collection of surveillance data from across the globe. There are strong geographical biases in reported data [78]; Europe and North America are sampled more intensely than other regions (Fig. 2). A more sophisticated treatment of regional differences and virus migration patterns [79] could potentially make predictive models more accurate and robust.
Drivers of influenza seasonality in temperate versus tropical regions are still not fully understood [80, 81]. An improved understanding of influenza seasonality could improve model predictions of when new clades are likely to proliferate, and could help policymakers continue ongoing work to time vaccine campaigns optimally – especially in the tropics [82].
3.2.6. Immune history and the challenge of individual variation
HI assays presently assess an influenza isolate’s antigenicity using antisera from ferrets exposed to a single reference virus. Thus, these data do not capture realistic heterogeneity in human immune responses, which arises from differences in individual histories of exposure to multiple influenza strains over a lifetime. Individual histories of exposure can strongly and predictably alter which B cell responses are expressed against a given seasonal influenza challenge strain [72, 83–89], and childhood influenza exposures appear to play a particularly strong role in shaping an individual’s lifelong immune memory [83–85, 90, 91].
These immune history effects (reviewed in [90]) suggest a mechanistic basis for individual or birth-year-specific variation in vaccine effectiveness [88, 92], and there is a need to better understand when these effects should or should not be expected to cause meaningful differences between antigenic maps based on ferret sera and maps generated using human sera [41, 42]. Meanwhile, there is scope for modelers to incorporate serological data and known patterns of pre-existing population immunity into predictive models.
3.3. Comparing models and assessing the limits of model foresight
Improving and making practical use of model predictions will require a standard for what constitutes improvement and an understanding of predictive models’ theoretical and practical limitations.
3.3.1. Model assessment and comparison
One difficulty for decision-makers looking to use information from multiple predictive models is model heterogeneity. Models make different assumptions about virus evolution. They use overlapping but not identical datasets. When two predictions substantially differ, it must be determined whether differences stem from differences in assumptions, data, or both. Models also have different outputs, which often complicates direct comparisons of predictive success.
The scarcity of testing data compounds this difficulty. Only about 20 historical influenza seasons are available for rigorous testing of whether a new model outperforms existing ones, and overfitting of models is a serious concern.
The influenza prediction community should agree upon shared standards for success in viral forecasting so that outputs from different models can be more readily compared.
It may also be worth developing standard methods for meta-analysis. Differences in model assumptions may make some models more robust in certain contexts. Meta-analysis could help identify such patterns, making it easier to weigh evidence appropriately when different models disagree.
Finally, it will be important to determine what vaccine policy is optimal given an uncertain prediction. This will require determining the costs of different degrees of prediction failure given a certain policy.
3.3.2. What are the limits of model foresight?
Prudent model-based policymaking requires understanding the limits of models’ predictive capacity. Ongoing work aims to quantify both the timescales at which models can deliver accurate results and the uncertainty in existing model predictions [11].
At present, one temporal limitation is clear–existing models cannot predict which antigenic variants will emerge in the future. They can only assess the probability that an existing, observed antigenic variant will have high fitness and proliferate.
Virologists and evolutionary biologists have long hoped to overcome this barrier and predict the emergence of antigenic phenotypes that have not yet been observed [93]. Such prediction would provide a clear temporal advantage for selection and development of vaccine viruses. But whether such prediction is feasible will depend upon how truly constrained the antigenic trajectory of influenza is; evidence remains equivocal [94, 95].
The degree to which cross-reactivity between two arbitrary strains can be predicted from their amino-acid differences (what Gog and Grenfell termed the “geometry of the strain space”[96]) remains unclear, as does the degree to which this geometry can be captured by simple, reductive models [44, 45, 97, 98]. The virus’s path through this space of possible antigenic amino acid sequences may be further constrained by non-immunological fitness benefits, costs, and tradeoffs [54, 99, 100], as well as immune pressure acting on more conserved epitopes [101]. How measurable and constant these constraints are will help determine whether it is practical to attempt the prediction of unobserved viruses.
3.3.3. Universal influenza vaccines and evolutionary effects of vaccination
A recent epidemiological study provides direct evidence for the feedback of influenza vaccination on viral evolution [102]. This study compares viral isolates from cohorts of vaccinated and unvaccinated humans. The sequence isolates from vaccinated individuals are found to have a significantly increased distance from the vaccine strain but remain broadly distributed on the strain tree, indicating that vaccination tends to accelerate evolution. This is in accordance with the predictions on vaccination feedback from the antigenicity-stability fitness model [26]. In this model, vaccination introduces an additional artificial selection fitness cost on circulating strains, which depends on their cross-immunity with the vaccine strain. The vaccination fitness component accelerates evolution away from the vaccine strain and, hence, reduces the efficacy of vaccination.
Ongoing efforts to develop a universal influenza vaccine [103, 104] may eventually render twice-yearly vaccine update decisions obsolete. Universal influenza vaccines would stimulate immune memory against conserved influenza epitopes, thus providing broad protection against the full gamut of influenza types and subtypes (reviewed in [105] and [106]). Recent studies [107, 108] have suggested that universal vaccines could even slow the pace of antigenic evolution.
4. Concluding Remarks
The global program to control influenza produces great public health benefit under enormous time pressure, often with minimal recognition. The process is a continuous cycle (Fig. 3). This public health work is mapping out a new frontier in biomedicine: applied evolutionary biology.
The ongoing effort to improve influenza surveillance, prediction, and control has spurred important advances in areas ranging from the population-level modeling of viral evolution to the fine-scale virology and immunology of human influenza infections (Fig. 4). Perhaps most remarkably, influenza research has demonstrated that predictive evolutionary biology can have practical public health applications. This sets a valuable precedent for other medical fields. The increasing threat posed by antibiotic-resistant bacteria is but one example of the public health importance of anticipating pathogen evolution.
The advances in basic and applied science reviewed here have been made possible through close collaboration between specialists in various fields and through generous data-sharing.
Outstanding challenges include improving influenza surveillance data management and use, integrating relevant additional biological detail into predictive models, and developing methods for comparing models and assessing their limitations (see Outstanding Questions).
Our view is that addressing these challenges and making further advances will require continued commitment to a collaborative approach. There already exists an incipient influenza prediction community: virologists and immunologists who study immune escape and develop novel techniques for viral surveillance, modelers who aggregate surveillance data to make predictions, and epidemiologists who understand how best to use novel insights to control influenza. This community should be fostered; it promises to advance understanding of virus evolution and improve disease control.
Trends Box.
Seasonal influenza evolves to evade immune recognition, necessitating regular vaccine updates. The World Health Organization has collaborated with academic institutions and national public health organizations to build a global surveillance program for monitoring influenza evolution.
Scientists have built predictive models grounded in evolutionary theory that use surveillance data to forecast which viral strains or clades will predominate in the coming months.
Output from these models is already being used to inform influenza vaccine strain selection.
This modeling sheds light on basic science questions: the degree to which evolution is directed and the phylogenetic and genomic signatures of fitness.
This is a success story for large scale collaborative science.
Outstanding Questions.
What degree of biological detail makes influenza modeling most effective?
How can newer forms of surveillance data (e.g. microneutralization assays) be most effectively integrated into the modeling and strain-selection processes?
To what extent must surveillance and modeling efforts account for individual variation among hosts in order be effective? How can this variation best be incorporated into predictive models?
What currently unavailable surveillance data would be most informative about influenza antigenic evolution? Can models help the surveillance community determine this?
How should models take account of known biases and uncertainties in existing data (e.g. regional biases in sampling and imperfect external validity of lab assays)?
How should multiple models be compared and used for vaccine strain selection?
What methods should decision-makers use to integrate information from multiple models, particularly when those models disagree?
How should model performance be assessed? Developing agreed-upon standards for modeling success within the collaborative influenza community will ensure that new models are compared fairly against existing ones
What vaccine policy decision is optimal given an uncertain prediction? For a given policy, what are the costs of different degrees of prediction failure?
Acknowledgments
This paper originated at the workshop “Exploring Predictive Models for Improving Influenza Vaccine Virus Selection”, held in July 2016 at Princeton University. The workshop brought together WHO Collaborating Centres on Influenza and academic research groups. The organizers were Nancy Cox (CDC Atlanta), Bryan Grenfell (Princeton University), Jaqueline Katz (CDC Atlanta), Michael Lässig (Cologne University), John McCauley (Crick Worldwide Influenza Centre London), and Wenqing Zhang (WHO). We thank the WHO, Princeton University, and Deutsche Forschungsgemeinschaft for their support of the meeting.
We thank Wenqing Zhang for productive discussion of these ideas and two anonymous reviewers for helpful comments on the manuscript.
KMG is supported by the National Institute of Allergy and Infectious Disease of the National Institutes of Health (F31AI134017). SP and ML acknowledge support by Deutsche Forschungsgemeinschaft grant SFB 680. TB is a Pew Biomedical Scholar and is supported by NIH R35 GM119774-01 and NIH R01 AI127893-01. MŁ acknowledges support from NCI-NIH grant P01CA087497. BTG acknowledges support from the Bill & Melinda Gates Foundation (OPP1091919), the RAPIDD program of the Science and Technology Directorate, the Department of Homeland Security, and the Fogarty International Center, National Institutes of Health (NIH). JWM was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001030), the Medical Research Council (FC001030) and the Wellcome Trust (FC001030).
Glossary
- Antigenic drift
In antigenic drift, influenza antigens accumulate amino acid sequence changes over time. Mutations that alter the antigenic phenotype of circulating influenza viruses experience positive selection if they help variant viruses escape existing population immunity.
- Epistasis
An interaction between genetic loci in which phenotypic or fitness effects are non-additive. Positive epistasis denotes a combined effect that is greater than the sum of the individual effects. Negative epistasis denotes a combined effect that is less than the sum of the individual effects.
- GISAID EpiFlu™ database
A public database of influenza genetic sequences. Sequence data from global surveillance efforts collected by WHO NICs, by WHOCCs, and by others are all deposited into EpiFlu™.
- Hemagglutination inhibition (HI) assay
Influenza virus can typically bind red blood cells and cause them to agglutinate (clump into a lattice). The hemagglutination inhibition assay measures the maximum dilution at which antibody-containing serum prevents a particular influenza virus from agglutinating red blood cells. A high HI titer indicates that dilute serum (i.e. low antibody concentrations) can prevent agglutination.
- Hemagglutinin (HA)
An influenza surface protein that facilitates binding to cell surface receptors and cell entry. The globular head of hemagglutinin contains key antigenic epitopes recognized by the human adaptive immune system. Human antibodies that recognize the hemagglutinin stalk can also be generated.
- National Influenza Centre (NIC)
A laboratory or medical institution that coordinates influenza surveillance for a given county. NICs analyze virological samples collected from respiratory illness patients in their country and ship influenza-positive samples and viruses to a WHOCC for further analysis.
- Microneutralization assay
An assay that measures the dilution at which serum antibodies are able to prevent a particular reference virus from infecting tissue culture cells; (i.e. neutralize the virus). A high neutralization titer indicates that dilute serum (i.e. low antibody concentrations) can still neutralize the virus of interest.
- Passaging adaptation
Genetic changes that arise as lab-propagated influenza viruses adapt to grow and replicate efficiently in cell culture or eggs. Since lab work on sampled influenza viruses requires growing the virus in vitro, passaging adaptations accumulate in virus lineages.
- Phylodynamics
The simultaneous analysis of a virus’s evolution (especially its phylogenetics) and its epidemic dynamics. Necessary because many viruses, including human seasonal influenza, evolve rapidly enough that epidemics and substantial evolution occur on the same timescale.
- Vaccine composition meeting (VCM)
Twice-yearly meetings at which leaders of the WHOCCs, WHO Essential Regulatory Laboratories, regulators, and others meet to assess surveillance data and develop WHO recommendations for the composition of vaccines. Recommendations issued at these meetings inform vaccine campaigns during the Northern Hemisphere (November–April) and Southern Hemisphere (March–October) influenza seasons.
- WHO Collaborating Centres (WHOCCs)
Six facilities across the globe that perform laboratory analyses and genetic sequencing on influenza viruses collected by National Influenza Centres.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Resources
References
- 1.Ayscue P, et al. Influenza-associated intensive-care unit admissions and deaths – California, September 29, 2013–January 18, 2014. MMWR Morb. Mortal. Wkly. Rep. 2014;63:143–147. [PMC free article] [PubMed] [Google Scholar]
- 2.Young-Xu Y, et al. The annual burden of seasonal influenza in the US Veterans Affairs population. PLoS One. 2017;12:e0169344. doi: 10.1371/journal.pone.0169344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Descalzo MA, et al. Estimating the burden of influenza-associated hospitalizations and deaths in Central America. Influenza Other Respir. Viruses. 2016;10:340–345. doi: 10.1111/irv.12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi WS, et al. Disease burden of 2013–2014 seasonal influenza in adults in Korea. PLoS One. 2017;12:e0172012. doi: 10.1371/journal.pone.0172012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vestergaard L, et al. Excess all-cause and influenza-attributable mortality in Europe, December 2016 to February 2017. Eurosurveillance. 2017;22:30506. doi: 10.2807/1560-7917.ES.2017.22.14.30506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Charu V, et al. Mortality burden of the 2009–10 influenza pandemic in the United States: improving the timeliness of influenza severity estimates using inpatient mortality records. Influenza Other Respir. Viruses. 2013;7:863–871. doi: 10.1111/irv.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molinari NAM, et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine. 2007;25:5086–5096. doi: 10.1016/j.vaccine.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 8.Barr IG, et al. Epidemiological antigenic and genetic characteristics of seasonal influenza A(H1N1), A(H3N2) and B influenza viruses: Basis for the WHO recommendation on the composition of influenza vaccines for use in the 2009–2010 Northern Hemisphere season. Vaccine. 2010;28:1156–1167. doi: 10.1016/j.vaccine.2009.11.043. [DOI] [PubMed] [Google Scholar]
- 9.Klimov AI, et al. WHO recommendations for the viruses to be used in the 2012 Southern Hemisphere Influenza Vaccine: epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from February to September 2011. Vaccine. 2012;30:6461–6471. doi: 10.1016/j.vaccine.2012.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barr IG, et al. WHO recommendations for the viruses used in the 2013–2014 Northern Hemisphere influenza vaccine: epidemiology, antigenic and genetic characteristics of influenza A(H1N1)pdm09, A(H3N2) and B influenza viruses collected from October 2012 to January 2013. Vaccine. 2014;32:4713–4725. doi: 10.1016/j.vaccine.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Lässig M, et al. Predicting evolution. Nature Ecology & Evolution. 2017;1:0077. doi: 10.1038/s41559-017-0077. [DOI] [PubMed] [Google Scholar]
- 12.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, et al. Clonal evolution of glioblastoma under therapy. Nat. Genet. 2016;48:768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anagnostou V, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7:264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y, et al. The characteristics and antigenic properties of recently emerged subclade 3C.3a and 3C.2a human influenza A (H3N2) viruses passaged in MDCK cells. Influenza Other Respir. Viruses. 2017 doi: 10.1111/irv.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao Y, et al. The influenza virus resource at the national center for biotechnology information. J. irol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu Y, et al. GISAID: Global initiative on sharing all influenza data–from vision to reality. Eurosurveillance. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grenfell BT, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 20.Holmes EC, Grenfell BT. Discovering the phylodynamics of RNA viruses. PLoS Comput. Biol. 2009;5:e1000505. doi: 10.1371/journal.pcbi.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shih ACC, et al. Simultaneous amino acid substitutions at antigenic sites drive influenza A hemagglutinin evolution. Proc. Natl. Acad. Sci. U.S. A. 2007;104:6283–6288. doi: 10.1073/pnas.0701396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhatt S, et al. The genomic rate of molecular adaptation of the human influenza A virus. Mol. Biol. Evol. 2011;28:2443–2451. doi: 10.1093/molbev/msr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strelkowa N, Lässig M. Clonal interference in the evolution of influenza. Genetics. 2012;192:671–682. doi: 10.1534/genetics.112.143396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Illingworth CJ, Mustonen V. Components of selection in the evolution of the influenza virus: linkage effects beat inherent selection. PLoS Pathog. 2012;8:e1003091. doi: 10.1371/journal.ppat.1003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer AG, et al. Cross-species comparison of site-specific evolutionary-rate variation in influenza haemagglutinin. Phil. Trans. R. Soc. B. 2013;368:20120334. doi: 10.1098/rstb.2012.0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luksza M, Lässig M. A predictive fitness model for influenza. Nature. 2014;507:57–61. doi: 10.1038/nature13087. [DOI] [PubMed] [Google Scholar]
- 27.Steinbrück L, et al. Computational prediction of vaccine strains for human influenza A (H3N2) viruses. J. Virol. 2014;88:12123–12132. doi: 10.1128/JVI.01861-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neher RA, et al. Predicting evolution from the shape of genealogical trees. eLife. 2014;3:e03568. doi: 10.7554/eLife.03568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deem MW, Lee HY. Sequence space localization in the immune system response to vaccination and disease. Phys. Rev. Lett. 2003;91:068101. doi: 10.1103/PhysRevLett.91.068101. [DOI] [PubMed] [Google Scholar]
- 30.Gupta V, et al. Quantifying influenza vaccine efficacy and antigenic distance. Vaccine. 2006;24:3881–3888. doi: 10.1016/j.vaccine.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito K, et al. Gnarled-trunk evolutionary model of influenza A virus hemagglutinin. PLoS One. 2011;6:e25953. doi: 10.1371/journal.pone.0025953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du X, et al. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat. Commun. 2012;3:709. doi: 10.1038/ncomms1710. [DOI] [PubMed] [Google Scholar]
- 33.Neher RA, Bedford T. nextu: real-time tracking of seasonal influenza virus evolution in humans. Bioinformatics. 2015;31:3546. doi: 10.1093/bioinformatics/btv381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bogner P, et al. A global initiative on sharing avian u data. Nature. 2006;442:981–981. [Google Scholar]
- 35.McCauley JW. Viruses: Model to accelerate epidemic responses. Nature. 2017;542:414–414. doi: 10.1038/542414b. [DOI] [PubMed] [Google Scholar]
- 36.Lin YP, et al. Neuraminidase receptor binding variants of human influenza A (H3N2) viruses resulting from substitution of aspartic acid 151 in the catalytic site: a role in virus attachment? J. Virol. 2010;84:6769–6781. doi: 10.1128/JVI.00458-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McWhite CD, et al. Sequence amplification via cell passaging creates spurious signals of positive adaptation in influenza virus H3N2 hemagglutinin. Virus Evol. 2016;2:vew026. doi: 10.1093/ve/vew026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oh DY, et al. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. J. Clin. Microbiol. 2008;46:2189–2194. doi: 10.1128/JCM.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katz JM, et al. Serologic assays for influenza surveillance, diagnosis and vaccine evaluatio. Expert Rev. Anti Infect. Ther. 2011;9:669–683. doi: 10.1586/eri.11.51. [DOI] [PubMed] [Google Scholar]
- 40.Truelove S, et al. A comparison of hemagglutination inhibition and neutralization assays for characterizing immunity to seasonal influenza A. Influenza Other Respir. Viruses. 2016;10:518–524. doi: 10.1111/irv.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie H, et al. H3N2 mismatch of 2014–15 northern hemisphere influenza vaccines and head-to-head comparison between human and ferret antisera derived antigenic maps. Sci. Rep. 2015;5 doi: 10.1038/srep15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fonville JM, et al. Antigenic maps of influenza a (H3N2) produced with human antisera obtained after primary infection. J. Infect. Dis. 2015;213:31–38. doi: 10.1093/infdis/jiv367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ndifon W. New methods for analyzing serological data with applications to influenza surveillance. Influenza Other Respir. Viruses. 2011;5:206–212. doi: 10.1111/j.1750-2659.2010.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harvey WT, et al. Identification of low-and high-impact hemagglutinin amino acid substitutions that drive antigenic drift of influenza A (H1N1) viruses. PLoS Pathog. 2016;12:e1005526. doi: 10.1371/journal.ppat.1005526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neher RA, et al. Prediction dynamics, and visualization of antigenic phenotypes of seasonal influenza viruses. Proc. Natl. Acad. SciU.SA. 2016;113:E1701–E1709. doi: 10.1073/pnas.1525578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ampofo WK, et al. Improving influenza vaccine virus selection: Report of a WHO informal consultation held at WHO headquarters, geneva, switzerland, 14–16 June 2010. Influenza Other Respir. Viruses. 2011;6:142–152. doi: 10.1111/j.1750-2659.2011.00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zacour M, et al. Standardization of hemagglutination inhibition assay for influenza serology allows for high reproducibility between laboratories. Clin. Vaccine Immunol. 2016;23:236–242. doi: 10.1128/CVI.00613-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinbrück L, McHardy AC. Inference of genotype–phenotype relationships in the antigenic evolution of human influenza A (H3N2) viruses. PLoS Comput. Biol. 2012;8:e1002492. doi: 10.1371/journal.pcbi.1002492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roche DG, et al. Troubleshooting public data archiving: suggestions to increase participation. PLoS Biol. 2014;12:1–5. doi: 10.1371/journal.pbio.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kryazhimskiy S, et al. Prevalence of epistasis in the evolution of influenza A surface proteins. PLos Genet. 2011;7:e1001301. doi: 10.1371/journal.pgen.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neverov AD, et al. Coordinated Evolution of Influenza A Surface Proteins. PLoS Genet. 2015;11:e1005404. doi: 10.1371/journal.pgen.1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tria F, et al. Dynamically correlated mutations drive human influenza A evolution. Sci. Rep. 2013;3:2705. doi: 10.1038/srep02705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koel BF, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science. 2013;342:976–979. doi: 10.1126/science.1244730. [DOI] [PubMed] [Google Scholar]
- 54.Koelle K, Rasmussen DA. The effects of a deleterious mutation load on patterns of influenza A/H3N2's antigenic evolution in humans. eLife. 2015;4:e07361. doi: 10.7554/eLife.07361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muñoz ET, Deem MW. Epitope analysis for influenza vaccine design. Vaccine. 2005;23:1144–1148. doi: 10.1016/j.vaccine.2004.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meyer AG, Wilke CO. Geometric constraints dominate the antigenic evolution of influenza H3N2 hemagglutinin. PLoS Pathog. 2015;11:e1004940. doi: 10.1371/journal.ppat.1004940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki Y. Natural selection on the influenza virus genome. Mol. Biol. Evol. 2006;23:1902–1911. doi: 10.1093/molbev/msl050. [DOI] [PubMed] [Google Scholar]
- 58.Palese P, Compans R. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2, 3-dehydro-n-triuoroacetylneuraminic acid (FANA): mechanism of action. J. Gen. Virol. 1976;33:159–163. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- 59.Nelson MI, Holmes EC. The evolution of epidemic influenza. Nat. Rev. Genet. 2007;8:196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- 60.Bloom JD, et al. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science. 2010;328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lubeck MD, et al. Nonrandom association of parental genes in influenza A virus recombinants. Virology. 1979;95:269–274. doi: 10.1016/0042-6822(79)90430-6. [DOI] [PubMed] [Google Scholar]
- 62.Greenbaum BD, et al. Viral reassortment as an information exchange between viral segments. Proc. Natl. Acad. SciU.SA. 2012;109:3341–3346. doi: 10.1073/pnas.1113300109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neverov AD, et al. Intrasubtype reassortments cause adaptive amino acid replacements in H3N2 influenza genes. PLoS Genet. 2014;10:e1004037. doi: 10.1371/journal.pgen.1004037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poon LLM, et al. Quantifying influenza virus diversity and transmission in humans. Nat. Genet. 2016;48:195–200. doi: 10.1038/ng.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghedin E, et al. Presence of oseltamivir-resistant pandemic A/H1N1 minor variants before drug therapy with subsequent selection and transmission. J. Infect. Dis. 2012;206:1504–1511. doi: 10.1093/infdis/jis571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Illingworth CJ, et al. Identifying selection in the within-host evolution of influenza using viral sequence data. PLoS computational biology. 2014;10:e1003755. doi: 10.1371/journal.pcbi.1003755. doi: https://doi.org/10.1371/journal.pcbi.1003755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dinis JM, et al. Deep sequencing reveals potential antigenic variants at low frequencies in influenza a virus-infected humans. J. Virol. 2016;90:3355–3365. doi: 10.1128/JVI.03248-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Debbink K, et al. Vaccination has minimal impact on the intrahost diversity of H3N2 influenza viruses. PLoS Pathog. 2017;13:e1006194. doi: 10.1371/journal.ppat.1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xue KS, et al. Parallel evolution of influenza across multiple spatiotemporal scales. eLife. 2017;6:e26875. doi: 10.7554/eLife.26875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stack JC, et al. Inferring the inter-host transmission of influenza A virus using patterns of intra-host genetic variation. Proc. R. Soc. Lond. B: Biol. Sci. 2013;280:20122173. doi: 10.1098/rspb.2012.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leonard AS, et al. Deep sequencing of influenza A virus from a human challenge study reveals a selective bottleneck and only limited intrahost genetic diversification. J. Virol. 2016;90:11247–11258. doi: 10.1128/JVI.01657-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang KYA, et al. Focused antibody response to influenza linked to antigenic drift. J. Clin. Invest. 2015;125:2631–2645. doi: 10.1172/JCI81104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leonard AS, et al. The effective rate of influenza reassortment is limited during human infection. PLoS Pathog. 2017;13:e1006203. doi: 10.1371/journal.ppat.1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park AW, et al. Quantifying the impact of immune escape on transmission dynamics of influenza. Science. 2009;326:726–728. doi: 10.1126/science.1175980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nelson MI, et al. Stochastic processes are key determinants of short-term evolution in influenza A virus. PLoS Pathog. 2006;2:e125. doi: 10.1371/journal.ppat.0020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson MI, et al. Phylogenetic analysis reveals the global migration of seasonal influenza A viruses. PLoS Pathog. 2007;3:e131. doi: 10.1371/journal.ppat.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bedford T, et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature. 2015;523:217–220. doi: 10.1038/nature14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pereyaslov D, et al. Improving the representativeness of influenza viruses shared within the WHO Global Influenza Surveillance and Response System. Influenza Other Respir. Viruses. 2016;10:68–75. doi: 10.1111/irv.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lemey P, et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 2014;10:e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Viboud C, et al. Influenza in tropical regions. PLoS Med. 2006;3:e89. doi: 10.1371/journal.pmed.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tamerius J, et al. Global influenza seasonality: reconciling patterns across temperate and tropical regions. Environ. Health Perspect. 2011;119:439. doi: 10.1289/ehp.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hirve S, et al. Influenza seasonality in the tropics and subtropics–when to vaccinate? PLoS One. 2016;11:e0153003. doi: 10.1371/journal.pone.0153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Francis T. On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 1960;104:572–578. [Google Scholar]
- 84.Lessler J, et al. Evidence for antigenic seniority in influenza A (H3N2) antibody responses in southern China. PLoS Pathog. 2012;8:e1002802. doi: 10.1371/journal.ppat.1002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fonville JM, et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346:996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andrews SF, et al. Immune history profoundly affects broadly protective B cell responses to influenza. Sci. Transl. Med. 2015;7:316ra192–316ra192. doi: 10.1126/scitranslmed.aad0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y, et al. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J. Exp. Med. 2013;210:1493–1500. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Linderman SL, et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc. Natl. Acad. Sci. U.S. A. 2014;111:15798–15803. doi: 10.1073/pnas.1409171111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Linderman SL, Hensley SE. Antibodies with ‘original antigenic sin’ properties are valuable components of secondary immune responses to influenza viruses. PLoS Pathog. 2016;12:e1005806. doi: 10.1371/journal.ppat.1005806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cobey S, Hensley SE. Immune history and influenza virus susceptibility. Curr. Opin. Virol. 2017;22:105–111. doi: 10.1016/j.coviro.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gostic KM, et al. Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science. 2016;354:722–726. doi: 10.1126/science.aag1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li Y, et al. Immune history shapes specificity of pandemic h1n1 influenza antibody responses. Journal of Experimental Medicine. 2013:jem–20130212. doi: 10.1084/jem.20130212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fazekas de St. Groth S. Evolution and hierarchy of influenza viruses. Arch. Environ. Health. 1970;21:293–303. doi: 10.1080/00039896.1970.10667241. [DOI] [PubMed] [Google Scholar]
- 94.Wagner A. A genotype network reveals homoplastic cycles of convergent evolution in influenza A (H3N2) haemagglutinin. Proc. R. Soc. Lond. B: Biol. Sci. 2014;281:20132763. doi: 10.1098/rspb.2013.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doud MB, Bloom JD. Accurate measurement of the effects of all amino-acid mutations on influenza hemagglutinin. Viruses. 2016;8:155. doi: 10.3390/v8060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gog JR, Grenfell BT. Dynamics and selection of many-strain pathogens. Proc. Natl. Acad. SciU.SA. 2002;99:17209–17214. doi: 10.1073/pnas.252512799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kryazhimskiy S, et al. On state-space reduction in multi-strain pathogen models, with an application to antigenic drift in influenza A. PLoS Comput. Biol. 2007;3:e159. doi: 10.1371/journal.pcbi.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Pan K, Deem MW. Comment on Ndifon et al, “on the use of hemagglutination-inhibition for influenza surveillance: Surveillance data are predictive of influenza vaccine effectiveness”. Vaccine. 2009;27:5033. doi: 10.1016/j.vaccine.2009.05.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gog JR. The impact of evolutionary constraints on influenza dynamics. Vaccine. 2008;26:C15–C24. doi: 10.1016/j.vaccine.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 100.Kucharski A, Gog JR. Influenza emergence in the face of evolutionary constraints. Proc. R. Soc. Lond. B: Biol. Sci. 2011 doi: 10.1098/rspb.2011.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wikramaratna PS, et al. The antigenic evolution of influenza: drift or thrift? Phil. Trans. R. Soc. B. 2013;368:20120200. doi: 10.1098/rstb.2012.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chong Y, Ikematsu H. Effect of seasonal vaccination on the selection of influenza A/H3N2 epidemic variants. Vaccine. 2017;35:255–263. doi: 10.1016/j.vaccine.2016.11.084. [DOI] [PubMed] [Google Scholar]
- 103.Impagliazzo A, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349:1301–1306. doi: 10.1126/science.aac7263. [DOI] [PubMed] [Google Scholar]
- 104.Krammer F, et al. Influenza Pathogenesis and Control-Volume II. Springer; 2014. Advances in universal influenza virus vaccine design and antibody mediated therapies based on conserved regions of the hemagglutinin; pp. 301–321. [DOI] [PubMed] [Google Scholar]
- 105.Cho A, Wrammert J. Implications of broadly neutralizing antibodies in the development of a universal influenza vaccine. Curr. Opin. Virol. 2016;17:110–115. doi: 10.1016/j.coviro.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pica N, Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu. Rev. Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 107.Subramanian R, et al. Universal or specific? a modeling-based comparison of broad-spectrum influenza vaccines against conventional, strain-matched vaccines. PLoS Comput. Biol. 2016;12:1–17. doi: 10.1371/journal.pcbi.1005204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Arinaminpathy N, et al. Impact of cross-protective vaccines on epidemiological and evolutionary dynamics of influenza. Proc. Natl. Acad. Sci. U.S. A. 2012;109:3173–3177. doi: 10.1073/pnas.1113342109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun H, et al. Using sequence data to infer the antigenicity of influenza virus. MBio. 2013;4:e00230–13. doi: 10.1128/mBio.00230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Peng Y, et al. Predac-h3: a user-friendly platform for antigenic surveillance of human influenza a (h3n2) virus based on hemagglutinin sequences. Bioinformatics. 2016;32:2526–2527. doi: 10.1093/bioinformatics/btw185. [DOI] [PubMed] [Google Scholar]
- 111.Yang J, et al. Sequence-based antigenic change prediction by a sparse learning method incorporating co-evolutionary information. PLoS One. 2014;9:e106660. doi: 10.1371/journal.pone.0106660. [DOI] [PMC free article] [PubMed] [Google Scholar]