Abstract
Conventional methods for histopathologic tissue diagnosis are labor- and time-intensive and can delay decision-making during diagnostic and therapeutic procedures. We report the development of an automated and biocompatible handheld mass spectrometry device for rapid and nondestructive diagnosis of human cancer tissues. The device, named MasSpec Pen, enables controlled and automated delivery of a discrete water droplet to a tissue surface for efficient extraction of biomolecules. We used the MasSpec Pen for ex vivo molecular analysis of 20 human cancer thin tissue sections and 253 human patient tissue samples including normal and cancerous tissues from breast, lung, thyroid, and ovary. The mass spectra obtained presented rich molecular profiles characterized by a variety of potential cancer biomarkers identified as metabolites, lipids, and proteins. Statistical classifiers built from the histologically validated molecular database allowed cancer prediction with high sensitivity (96.4%), specificity (96.2%), and overall accuracy (96.3%), as well as prediction of benign and malignant thyroid tumors and different histologic subtypes of lung cancer. Notably, our classifier allowed accurate diagnosis of cancer in marginal tumor regions presenting mixed histologic composition. Last, we demonstrate that the MasSpec Pen is suited for in vivo cancer diagnosis during surgery performed in tumor-bearing mouse models, without causing any observable tissue harm or stress to the animal. Our results provide evidence that the MasSpec Pen could potentially be used as a clinical and intraoperative technology for ex vivo and in vivo cancer diagnosis.
INTRODUCTION
Tissue assessment and diagnosis are critical in the clinical management of cancer patients. Tissue diagnosis is particularly important during surgical excision of solid cancers for surgical margin evaluation. Many women diagnosed with breast cancer, for example, undergo breast-conserving surgery, which involves removing the lesion of interest with a rim of normal tissue and preserving the rest of the breast. One of the greatest challenges a breast cancer surgeon faces is determining the delicate boundary between cancerous and normal tissues to achieve negative margins for invasive and carcinoma in situ while optimizing aesthetic outcomes (1). Similarly, optimal surgical treatment of lung carcinomas includes complete local resection of the primary tumor (2) because adverse patient outcome is strongly associated with residual tumor at the bronchial resection margin (3). For high-grade serous ovarian cancer (HGSC) patients, postoperative residual disease after surgical debulking is also negatively associated with progression-free survival and response to adjuvant chemotherapy (4). Thus, accurate negative margin assessment and complete tumor excision are highly desirable across cancer surgeries because they offer the greatest potential for prolonged disease-free and overall survival (1, 5–7).
Intraoperative assessment of the extent of tumor involvement can be challenging through conventional histopathologic analysis of frozen sections. Frozen section preparation is time- and labor-intensive and requires skilled technicians and pathologists to produce and interpret the results. Moreover, freezing artifacts can negatively interfere with tissue structure and cell morphology, thus complicating pathological interpretation. Logistically, intraoperative frozen section analysis prolongs operative time, subjecting the patient to increased risks related to extended anesthesia. Therefore, margin specimens are frequently processed postoperatively as permanent specimens. However, when positive margins are found during the final pathologic evaluation, the patient is subjected to additional surgical procedures for re-excision of the involved margin, which increases health care costs and places the patient at risk for additional surgical complications, discomfort, and anxiety (1, 8).
Molecular analysis of cancer tissues offers the exciting opportunity to incorporate cancer-specific biomarkers into clinical decision-making for improved cancer detection and diagnosis. Several molecular imaging technologies have been developed and advanced to preclinical and clinical phases for ex vivo and in vivo tissue diagnosis. Immunohistochemistry protocols targeting protein biomarkers are routinely used in diagnostic pathology for postoperative evaluation of ex vivo tissue sections and typing of neoplasms (9). Gene sequencing technologies are powerful for postoperative identification of specific genetic mutations and chromosomal translocations in ex vivo tissue samples (10). Intraoperative real-time techniques including fluorescent probes that target tumor cells are currently being implemented for in vivo tumor and surgical margin visualization (11). Emerging optical technologies including Raman spectroscopy and stimulated Raman scattering microscopy have been recently applied for intraoperative diagnosis of brain cancers (12, 13). Mass spectrometry (MS) imaging approaches have been successfully applied for molecular imaging of cancer tissues (14–17).
Within the last decade, several ambient ionization MS techniques have been developed for rapid molecular diagnosis of cancer tissues and have shown exceptional potential for clinical use (18). Desorption electrospray ionization MS imaging (DESI-MSI), for example, has been used for ex vivo cancer diagnosis and surgical margin evaluation of tissue sections and tissue smears (19, 20). Yet, technical incompatibilities including the use of a spray of organic solvents, high-pressure nebulizing gas, and high voltage have prevented the use of DESI-MSI for fresh tissue and in vivo analyses. A few approaches for direct MS analysis of cancer tissue specimens have been developed (18). Rapid evaporative ionization MS, or the iKnife, combines an electrocauterization device with MS for direct tissue analysis and classification and has been successfully used intraoperatively for in vivo cancer diagnosis (21, 22). Ultraviolet and infrared lasers have also been coupled with MS for characterization of cancer tissues (23, 24). Although these approaches offer the advantage of incorporating common surgical methods into an MS-based diagnostic workflow, these technologies rely on tissue damage to produce molecular ions or are operationally constrained to a specific surgical modality.
Here, we describe the development and application of an automated, biocompatible, disposable handheld device, the MasSpec Pen, for direct, real-time nondestructive sampling and molecular diagnosis of tissues. We tested the MasSpec Pen for ex vivo molecular evaluation of human normal and cancerous tissue samples from 253 patients. The mass spectra obtained presented rich molecular information including diagnostic metabolites, lipids, and proteins. Statistical analysis using the least absolute shrinkage and selector operator (Lasso) technique allowed prediction of cancer with high sensitivity and specificity (25). In a tumor-bearing mouse model, we demonstrate that this technology is suited for in vivo use and diagnosis of breast cancer during surgery.
RESULTS
Optimization of the MasSpec Pen design and operation
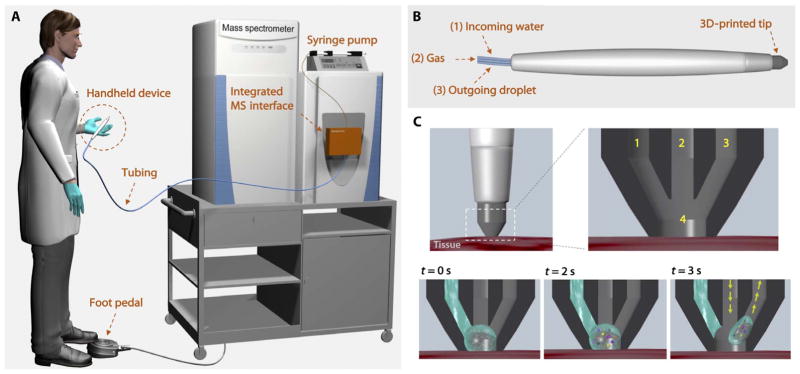
We designed the MasSpec Pen (Fig. 1A) as an automated and biocompatible handheld sampling probe that allows gentle and time- and volume-controlled extraction of molecules from a tissue sample using a discrete water droplet (movie S1). Several prototypes of the system were engineered with the goal of minimizing tissue damage, maximizing tissue-analyte extraction, and maximizing solvent transfer to the mass spectrometer. The optimized system contains three primary components: (i) a syringe pump that is programmed to deliver a defined water volume (4 to 10 μl) to the sampling probe; (ii) small diameter [inner diameter (ID), 800 μm] polytetrafluoroethylene (PTFE) tubing conduits, which are integrated to a fast (8 ms) two-way pinch valves for controlled solvent transport from the pump to the tissue and from the tissue to the mass spectrometer; and (iii) a handheld pen-sized probe for direct sampling of biological tissues.
Fig. 1. Schematic representation of the MasSpec Pen system and operational steps.
(A) The pen-sized handheld device is directly integrated into a laboratory-built MS interface through PTFE tubing. The integrated MS interface houses the pinch valves, microcontroller, and tubing to connect the system to the mass spectrometer inlet. The system is triggered by the user through a foot pedal. (B) The MasSpec Pen (handheld device) is designed with a PDMS tip and three PTFE conduits, which provide incoming water (1) and gas (2) to the tip and an outgoing conduit for the water droplet (3). (C) The tip contacts the tissue for analysis. Inset shows the three conduits (1 to 3) and solvent reservoir (4) within the tip. When the system is triggered (t = 0 s) by using the foot pedal, the syringe pump delivers a controlled volume of water to the reservoir. The discrete water droplet interacts with the tissue to extract the molecules (t = 2 s). After 3 s of extraction, the vacuum and the gas conduits are concomitantly opened (arrows) to transport the droplet from the MasSpec Pen to the mass spectrometer through the tubing system for molecular analysis.
The main component of the handheld pen-sized probe is a three-dimensional (3D) printed polydimethylsiloxane (PDMS) tip (Fig. 1B). The probe tip is designed with three ports: an incoming port that delivers a single water droplet to the probe tip (conduit 1), a central port for gas (N2, CO2, or air) delivery (conduit 2), and an outgoing port to transport molecular constituents in the water droplet from the tissue to the mass spectrometer (conduit 3). At the probe tip, all ports combine into a small reservoir where a single water droplet is retained and exposed to the tissue sample for a controlled amount of time (3 s), allowing efficient analyte extraction. After the 3-s extraction period, the MasSpec Pen is removed from the tissue. Concomitantly, conduit 3 is opened, allowing vacuum extraction of the droplet to the mass spectrometer, whereas positive pressure from a low-pressure gas delivery (<10 psi) is provided through conduit 2 (Fig. 1C). The gas provided by the second tube does not participate in the extraction process but is used instead to prevent the collapse of the system due to the vacuum used and to assist solvent transport from the tissue to the mass spectrometer. A subsequent flush step cleans the system; this is not used for extraction of biomolecules from tissues because there is no contact with the tissue during this period. Conduit 3 is directly connected to the transfer tube of a high–mass resolution Orbitrap mass spectrometer so that the negative pressure of the mass spectrometer vacuum system drives the movement of the droplet from the reservoir to the mass spectrometer for ionization and mass analysis. This setup simplifies the operational steps and precludes the use of ionization sources, although various connection and ionization methods could be coupled to our system.
The diameter of the reservoir at the probe tip determines the volume of solvent exposed to the tissue and the spatial resolution of the device. Using current tooling, we have designed MasSpec Pen tips with sampling sizes ranging from 1.5 to 5.0 mm, which is determined by the reservoir diameter. At a 2.77-mm reservoir diameter, a solvent volume of 10 μl is retained in the reservoir and contacts the tissue sample for a defined time period, whereas 4.4 μl is retained in a reservoir with a 1.5-mm diameter. Contact times of 1, 3, and 5 s between the droplet and the tissue sample were evaluated (fig. S1). The 3-s contact time was selected for all the experiments because it allowed ease of operation by the user and yielded mass spectra with sufficient total ion intensity. A tube length of 1.5 m was used for all the conduits to allow free handheld use of the device by an operator without geometrical or spatial constraints.
The tip design using three conduit tubes and high-speed actuated pinch valves allowed precise control of droplet motion and showed excellent performance and robustness. The entire process from sampling to mass spectral acquisition is completed in 10 s or less and is automated using an Arduino microcontroller so that each acquisition and analysis is individually triggered through a one-step click using a foot pedal. System automation ensures that each solvent droplet is delivered separately to the inlet, yielding several mass spectra that are averaged for a final molecular profile of the sample. Controlled droplet delivery allowed the mass spectrometer to operate without any evident performance degradation. After each use, the MasSpec Pen was cleaned through a rapid and automated cleaning flush or by replacing the disposable tip. Contamination of the mass spectrometer was evaluated by installing commercially heated ESI source and acquiring mass spectra after the MasSpec Pen analysis. No lipid contamination was observed other than background peaks commonly observed in the mass spectra.
Molecular analysis of thin tissue sections using the MasSpec Pen
We tested the effectiveness of the MasSpec Pen by analyzing thin mouse and human tissue sections and pieces of tissue samples. First, 16-μm-thick tissue sections were analyzed on standard histologic glass slides following the operational steps described above for the MasSpec Pen, using pure water as the solvent. Several probe tips with different reservoir diameters of the MasSpec Pen were tested, yielding mass spectra presenting lipid species characteristic of mouse brain tissue gray matter, white matter, or mixed composition for larger sampling sizes (fig. S2) (26). Figure S3 shows a representative mass spectrum obtained in the negative ion mode using a 2.7-mm pen tip from the gray matter region of a mouse brain tissue section and a representative background mass spectrum obtained from a region of glass slide (no sample).
The negative ion mode mass spectrum obtained from the glass slide region presented background ions from a mass/charge ratio (m/z) of 120 to 400, corresponding to common solvent clusters and chemical contaminants present on the glass slide or within the mass spectrometer. The negative ion mode mass spectrum obtained from the gray matter region of the mouse brain tissue section presented rich molecular information, including a variety of ions corresponding to deprotonated or chloride adducts of lipid species commonly detected from biological tissues using solvent-based ambient ionization MS techniques (16). Peaks at high relative abundances were identified as fatty acids (FA) from m/z 120 to 350, sphingolipids such as sulfa-tides from m/z 700 to 1100 and chloride adducts of ceramides (Cer) from m/z 500 to 700, and glycerophospholipids (GL) such as glycerophosphoinositols (PI), glycerophosphoethanolamines (PE), glycerophosphoserines (PS), and doubly charged cardiolipins (CL) from m/z 700 to 1100. In the higher mass range from m/z 1100 to 1800, GL dimers and singly charged CL were observed. A variety of peaks tentatively identified as small metabolites including glutamine at m/z 145.061, glutamate at m/z 146.045, N-acetylaspartic acid at m/z 174.041, and chloride adduct of hexose at m/z 215.033 were detected in the lower mass range from m/z 120 to 250, based on high–mass accuracy measurements and tandem MS data (table S1). The negative ion mode mass spectra obtained from the gray matter from different tissue sections of the same mouse brain were reproducible [relative standard deviation (RSD) = 9.3%; n = 9], comparable to what has been reported using the same method for DESI-MSI (RSD = 8.0%; n = 5) (26).
In the positive ion mode, the mass spectrum obtained presented high relative abundances of commonly observed molecular species identified as diacylglycerols, PE, and glycerophosphocholines (fig. S4). Tentative assignments were performed using high–mass accuracy measurements, as well as tandem MS analysis when adequate intensity of fragment ions was achieved for structural interpretation (see Identification of molecular ions in the Supplementary Materials). Mass errors and the m/z of fragment ions obtained by tandem MS experiments are described for all the species identified throughout the study (tables S1 to S5). Because isomerism of the double bonds in the FA chains of complex lipids complicates precise structural assignment, FA chains were tentatively assigned for lipid species.
To evaluate the extraction efficiency and molecular profiles obtained, we compared the molecular species detected in the negative ion mode using the MasSpec Pen with those observed in DESI-MSI acquired from a serial tissue section of the same mouse brain using water as the solvent and under analogous experimental conditions. The mass spectra obtained using the MasSpec Pen and DESI-MSI were similar with a calculated cosine similarity of 0.9 (see Data analysis in the Supplementary Materials), sharing a large number of molecular species at comparable relative abundances and signal-to-noise ratios (fig. S5). Other solvent systems including mixtures of water with ethanol at various ratios were also explored as solvent systems for the MasSpec Pen. The mass spectra obtained presented similar lipid species to those observed in those obtained using pure water, with variations in their relative abundances (fig. S6). To ensure full biocompatibility of our device, we selected water as the solvent for all subsequent MasSpec Pen experiments performed.
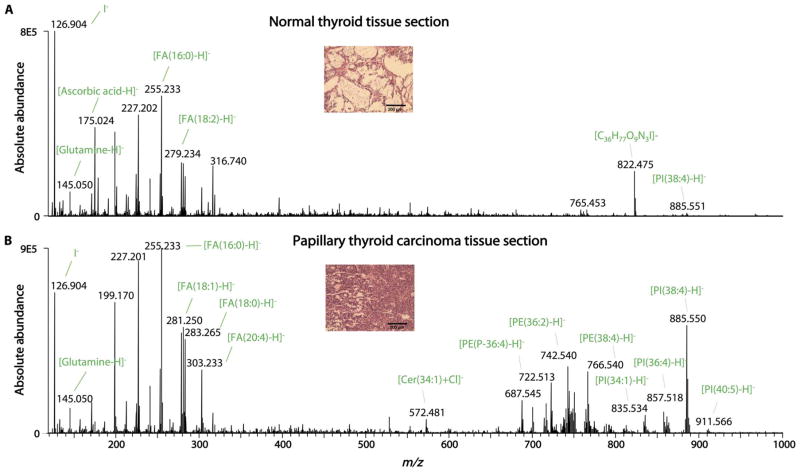
We then tested the capability of the MasSpec Pen to analyze 20 thin tissue sections of human breast tissues (n = 5 normal breast and 5 breast ductal carcinoma tissues) and thyroid tissues [n = 5 normal thyroid, 4 papillary thyroid carcinoma (PTC), and 1 follicular thyroid adenoma (FTA) tissues]. The mass spectra obtained in the negative ion mode for each tissue type presented a rich variety of molecular ions commonly observed from human tissues by DESI-MSI (18), with high relative abundances of metabolites, FA, and complex lipids. For example, the mass spectra obtained for PTC tissue sections presented lipid species previously identified as diagnostic markers by DESI-MSI (27), including a variety of doubly charged CL and other GL such as PI(38:4) (m/z 885.550), PI(36:4) (m/z 857.518), PE(38:4) (m/z 766.539), and PE (36:2) (m/z 742.539) (table S2). A distinct mass spectral profile was obtained for the normal thyroid tissue sections, presenting high relative abundances of iodine (m/z 126.904), glutamine (m/z 145.050), ascorbic acid (m/z 175.024), C36H78O9N3I (tentatively assigned; m/z 822.472), and PI(38:4) (m/z 885.551) (Fig. 2). A series of multiply charged molecular ions at different charge states (z) including m/z 991.091 (z = −5), m/z 1239.113 (z = −4), and m/z 1652.484 (z = −3) were detected in the mass spectra obtained from all tissue sections analyzed. These ions were tentatively identified as different charge states of the protein thymosin β-4 based on high–mass accuracy measurements (fig. S7 and table S1). Principal components analysis (PCA) performed on the data obtained from the human tissue sections showed separation between tumor and normal tissues (fig. S8).
Fig. 2. MasSpec Pen analysis of PTC and normal thyroid tissue sections.
(A) A representative negative ion mode MasSpec Pen mass spectra obtained from a normal human thyroid tissue section (average of n = 5 mass spectra) and (B) a PTC tissue section (average of n = 4 mass spectra) are shown. Identification of the most abundant molecular ions is provided. Insets shows an optical image of the H&E-stained tissue sections evaluated by histopathology.
Nondestructive molecular analysis of tissue samples
The MasSpec Pen was designed to operate directly on tissue specimens independently of tissue stiffness and morphology. We tested the performance of the MasSpec Pen to analyze soft tissue samples (0.1 to 5 g) from different organs including mouse brain and human breast, thyroid, lung, and ovary tissues. Tissue analyses were performed under ambient conditions through a simple one-step experiment, following the same operational steps described previously. The MasSpec Pen tip was gently brought into contact with the surface of the tissue sample for a period of 3 s during which extraction took place. The mass spectra obtained for a region of gray matter probed from a piece of fresh mouse brain tissue were reproducible (RSD = 4.6%; n = 10) and highly similar to the mouse brain tissue section mass spectra (cosine similarity of 0.93), indicating that the extraction process at the tissue surface occurs independently of the tissue shape and rigidity (fig. S9). MasSpec Pen analyses of human tissue samples provided similar rich molecular information, especially of tissues composed of epithelial and cancerous cells. Noncancerous tissue specimens that were mostly composed of soft connective tissue such as stroma provided less abundant mass spectra. Many of the normal breast tissue samples analyzed presented abundant fat content, which is immiscible with water and thus yielded lower total ion counts in the mass spectra when compared to breast cancer tissues or normal breast cancer glands.
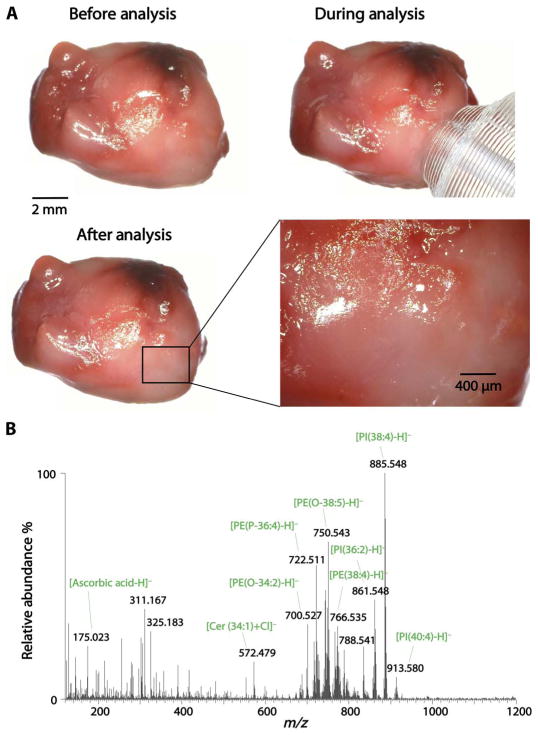
Visual and microscopic inspection of the tissue samples after the MasSpec Pen analysis revealed no detectable damage to the tissue morphology in the region probed. Figure 3A shows optical images obtained from a lung tissue sample before, during, and after the MasSpec Pen analysis. No observable damage to the tissue was seen at the region analyzed, and rich mass spectra were obtained (Fig. 3B). The automated and time-controlled operational steps of the MasSpec Pen prevent tissue damage because the tissue is only exposed to the small water droplet and not to the vacuum used to transport the droplet from the reservoir to the mass spectrometer. These results provide evidence that the MasSpec Pen can obtain rich molecular information from tissue samples nondestructively.
Fig. 3. Nondestructive molecular analysis of human tissue samples using the MasSpec Pen.
(A) Optical images show a lung adenocarcinoma tissue sample before, during, and after the MasSpec Pen analysis. A magnification of the tissue specimen (inset) shows no macroscopic damage to the tissue region analyzed by the MasSpec Pen. (B) Negative ion mode mass spectrum obtained for the tissue region analyzed including the identification of the most abundant molecular ions.
Molecular diagnosis and statistical prediction of cancer in human tissues
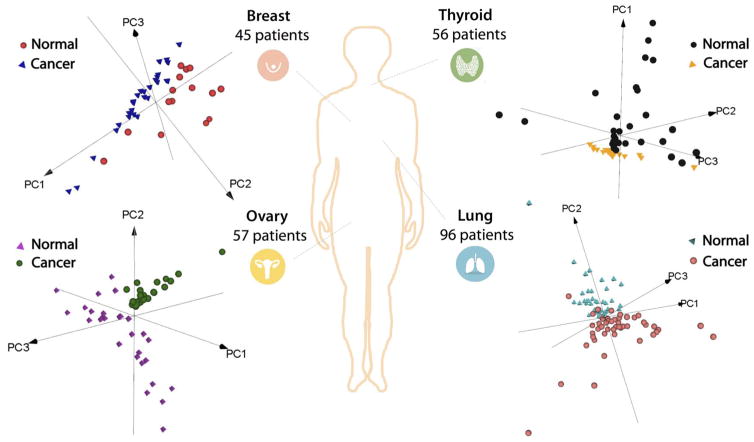
We next evaluated whether the molecular information obtained from human tissue samples using the MasSpec Pen was diagnostic and predictive of disease state. We analyzed a total of 253 human tissue specimens using the MasSpec Pen, including 95 lung samples (47 normal and 48 cancer samples including 17 adenocarcinoma, 17 squamous cell carcinoma, and 14 cancer samples of other histologic subtypes), 57 ovary samples (29 normal and 28 HGSC samples), 56 thyroid samples (27 normal, 11 FTA, and 18 PTC samples), and 45 breast samples (29 normal and 16 ductal carcinoma samples). Patient demographic information is provided in table S6. After the MasSpec Pen analysis, the region analyzed was demarcated and registered through a series of optical images. Parallel pieces of the samples were frozen, sectioned at the demarcated region, hematoxylin and eosin (H&E)–stained, and evaluated by histopathology to derive a diagnosis. Only samples with a predominant cell composition and clear diagnosis were used to build molecular databases. The histologically validated mass spectra obtained for the cancerous samples presented molecular species identified as several lipids and metabolites previously described as potential disease markers using ambient ionization MS techniques. For the lung cancer tissue, characteristic molecular markers such as PI(36:1) (as m/z 863.565), PG(36:2) (m/z 773.542), PG(34:1) (m/z 747.514), and FA(18:1) (m/z 281.249) were observed (Fig. 3B and table S4). For the normal lung, m/z 885.550, identified as PI(38:4), and m/z 744.552, identified as PE(36:1), were observed. The mass spectra obtained for breast cancer tissue presented diagnostic lipid markers previously described by DESI-MSI (28, 29), including m/z 885.550, identified as PI(38:4), m/z 863.565, identified as PI(36:1), m/z 773.542, identified as PG(36:2), and several FA such as m/z 303.233, identified as FA(20:4), and m/z 281.249, identified as FA(18:1) (table S5). PCA performed on the data obtained from the 253 human tissue samples analyzed showed separation between cancer and normal tissues for each organ (Fig. 4).
Fig. 4. PCA of the data obtained from human tissue samples using the MasSpec Pen.
A total of 253 patient tissue samples were analyzed including breast (n = 45), thyroid (n = 56), ovary (n = 57), and lung (n = 96) cancer and normal tissue samples. 3D PCA (PC1, PC2, and PC3) score plots are shown for each tissue type. The first three PCs explain the 77, 69, 51, and 87% of the total variance of breast, thyroid, lung, and ovarian data sets, respectively.
To evaluate whether the MasSpec Pen molecular signatures are predictive of cancer and normal tissues, we applied the Lasso method to build classification models using the histologically validated mass spectral database. The performance of the model was evaluated through a leave-one-patient-out cross-validation approach and was measured by sensitivity and specificity for cancer, as well as accuracy and the area under the curve (AUC) (Table 1). For breast cancer (n = 45), 87.5% sensitivity, 100% specificity (AUC = 1.0), and an overall accuracy of 95.6% were achieved, which is comparable to the results reported using DESI-MSI (98.2% accuracy; n = 126) (29), the iKnife (95.5% accuracy; n = 10) (22), and matrix-assisted laser desorption/ionization imaging of lipids and proteins (94.1% accuracy; n = 68) (30). For HGSC (n = 57), 100% sensitivity, 89.7% specificity, and 94.7% accuracy were achieved (AUC = 0.98), which is also similar to classification results obtained by DESI-MSI (97.1% accuracy; n = 31) (31). For lung cancer (n = 96), 97.9% sensitivity, 95.7% specificity, and 96.8% accuracy were achieved (AUC = 0.97). When predicting on the basis of lung cancer histologic subtypes, 93.8 and 92.2% accuracy was achieved for squamous cell carcinoma and adenocarcinoma, respectively. Thyroid tumor samples investigated included benign FTA and malignant PTC samples. A classifier for each was built, yielding 94.7% accuracy for FTA and 97.8% accuracy for PTC. Overall, 96.4% sensitivity, 96.2% specificity, and 96.3% accuracy were achieved for all four types of cancer investigated. These results demonstrate that the molecular information obtained from human tissue samples by the MasSpec Pen can be used to identify cancer and indicate that the statistical classifiers built are robust and may be used in an automated approach for rapid clinical diagnosis of tissue samples.
Table 1. Human tissue sample details and results obtained using the MasSpec Pen.
Pathological diagnosis, number of patient samples, and the Lasso prediction sensitivity, specificity, accuracy, and AUC obtained using a leave-one-out cross-validation approach are shown.
| Organ | Pathologic evaluation | Number of patients | Lasso prediction | ||||
|---|---|---|---|---|---|---|---|
| Diagnosis | Histologic type | Sensitivity | Specificity | Accuracy | AUC | ||
| Breast | Normal | 29 | 87.5% | 100.0% | 95.6% | 1.00 | |
| Cancer | Ductal carcinoma | 16 | |||||
| Lung* | Normal | 47 | 97.9% | 95.7% | 96.8% | 0.97 | |
| Cancer | Adenocarcinoma | 17 | 88.2% | 93.6% | 92.2% | 0.98 | |
| Squamous cell | 17 | 88.2% | 95.7% | 93.8% | 0.93 | ||
| Others | 14 | — | — | — | — | ||
| Ovary | Normal | 29 | 100.0% | 89.7% | 94.7% | 0.98 | |
| Cancer | High-grade serous | 28 | |||||
| Thyroid† | Normal | 27 | — | — | — | — | |
| Tumor | Papillary carcinoma | 18 | 94.4% | 100.0% | 97.8% | 0.99 | |
| Follicular adenoma | 11 | 90.9% | 96.3% | 94.7% | 0.93 | ||
Lasso prediction results for lung are shown for normal versus all cancer tissues (first row), followed by normal versus lung adenocarcinoma (middle row) and normal versus squamous cell carcinoma (last row).
Lasso prediction results for thyroid are shown for normal versus malignant papillary carcinoma and normal versus benign follicular adenoma.
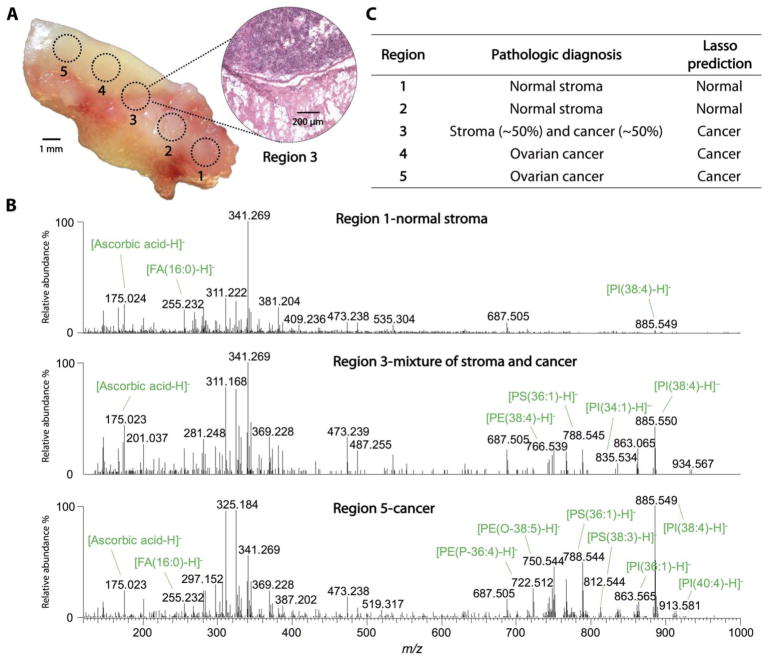
Intrasample analysis of histologically distinct and cancer margin tissue regions
We evaluated the ability of the MasSpec Pen to identify histologically distinct regions in a single human tissue sample that contained regions of HGSC adjacent to normal ovarian stroma tissue. Five regions of the tissue sample were analyzed consecutively using a MasSpec Pen with a 1.5-mm probe tip diameter, as demarcated in the optical image shown in Fig. 5A. A tissue section of the sample including the regions analyzed by the MasSpec Pen was subjected to H&E staining and evaluated by histopathology. Regions 1 and 2 were diagnosed by histopathology as normal stroma, whereas regions 4 and 5 were diagnosed as HGSC. Region 3 was in the margin between the cancer and normal stroma tissue regions, presenting ~50% tumor tissue and ~50% normal stroma tissue (Fig. 5A, inset). Figure 5B shows the mass spectra obtained for regions 1, 3, and 5. The spectra obtained for region 5, HGSC, presented characteristic lipid markers detected in the HGSC tissues analyzed ex vivo to build our statistical classifier (table S3). The mass spectra obtained for region 1, diagnosed as normal ovarian stroma tissue, presented less abundant molecular ions as also observed for the other stroma tissues analyzed ex vivo. Region 3 presented molecular profiles characteristic of HGSC with lower total abundance due to the contribution of normal stroma tissue present within the region analyzed. The mass spectra obtained for the five regions were then evaluated by our ovarian cancer molecular classifier as an independent validation set. Our classifier correctly identified regions 1 and 2 as normal and regions 3 to 5 as cancer (Fig. 5C). Similar results were obtained for a different tissue sample with histologically distinct regions (fig. S10). These results show that the molecular information obtained by the MasSpec Pen can be used to detect cancer in marginal regions with mixed composition of normal and cancer cells.
Fig. 5. MasSpec Pen analysis of an HGSC tissue sample with mixed histologic composition.
(A) Optical image shows the tissue sample that was analyzed at the demarcated regions (1 to 5) using a 1.5-mm-diameter MasSpec Pen. After the MasSpec Pen analysis, the tissue sample was frozen, sectioned, and H&E-stained. An optical image of the H&E-stained tissue section obtained at region 3 is shown (inset), presenting a mixed histologic composition including cancer and adjacent normal stroma tissue. (B) The MasSpec Pen negative ion mode mass spectra are shown for regions 1 (normal stroma; average of n = 3 mass spectra), 3 (mixture of normal stroma and cancer; average of n = 3 mass spectra), and 5 (cancer; average of n = 3 mass spectra). (C) Table listing the pathologic diagnosis of the five regions analyzed and the Lasso prediction results.
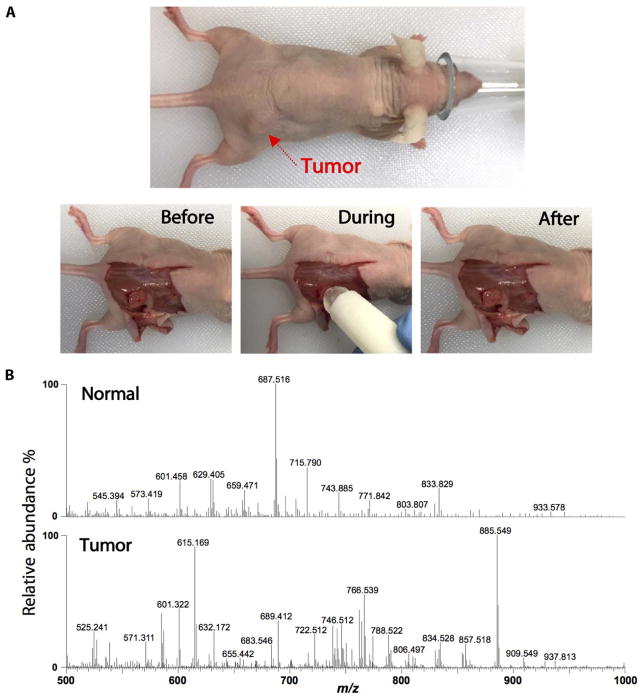
In vivo analysis of a murine model of human breast cancer during surgery
The MasSpec Pen was designed with biocompatible materials to ensure full compatibility as an in vivo molecular diagnostic tool. We tested the MasSpec Pen for in vivo tissue analysis using a murine model of human breast cancer. BT474 human epidermal growth factor receptor 2–positive (HER2+) breast cancer cells were implanted subcutaneously in nude athymic mice (n = 3). The tumors were grown to an average of 250 mm3 over a period of 4 weeks. Under anesthesia, the skin overlying the tumors was dissected away, and several tissue regions were analyzed following the same automated experimental steps described previously, including multiple positions of the top of the tumor, the core of the tumor after partial tumor resection, and adjacent normal soft connective tissue. Figure 6A shows an optical image of the animal under anesthesia before initiation of surgery, before analysis (after surgical removal of the skin), during the MasSpec Pen analysis, and after the analysis. The mass spectra obtained for the tumor regions presented many molecular species observed in human breast tissue, with a distinctive profile from what was obtained for adjacent normal soft connective tissue regions (Fig. 6B). Using optical microscopy, no observable macroscopic or microscopic damage to the tissue regions analyzed were detected due to MasSpec Pen analyses, as evident from the optical images obtained of the H&E-stained tissue sections (fig. S11). Further, no apparent effects to the health of the animals were observed due to the MasSpec Pen analysis during surgery. After in vivo analysis, freshly excised tumor specimens were also analyzed ex vivo, yielding mass spectra with common lipid species to those observed during in vivo analysis despite variations in relative abundances, which are likely due to the reanalysis process of the same tissue region (fig. S12). These results suggest that the MasSpec Pen is suitable for in vivo molecular evaluation and cancer diagnosis.
Fig. 6. Intraoperative analysis of tumor and normal tissues in a murine model.
(A) Experiments were performed in vivo in mice under anesthesia. Optical images show the animal under anesthesia and before, during, and after the MasSpec Pen analysis. (B) Representative negative ion mode mass spectra show distinct molecular profiles from normal (average of n = 3 mass spectra) and tumor (average of n = 3 mass spectra) tissues.
DISCUSSION
We developed the MasSpec Pen as an automated and biocompatible handheld sampling probe that allows gentle and time- and volume-controlled extraction of molecules from a tissue sample using a discrete water droplet. Our results provide evidence that the MasSpec Pen is suited for rapid ex vivo and in vivo cancer diagnosis of tissue samples. The mass spectra obtained from the analysis of 20 thin tissue sections and 253 human tissue samples presented rich molecular information that is diagnostic of disease state. The chemical extraction process used is gentle so that the tissue is undamaged after molecular analysis. Statistical classifiers built from the mass spectra obtained provided high sensitivity and specificity for cancer detection (>96%), including prediction of histologic subtypes of lung cancer and benign and malignant thyroid tumors. Experiments performed in animal models demonstrate that this technology is suitable for in vivo molecular evaluation of cancer and normal tissues without causing observable tissue harm or evident stress to the animal.
The MasSpec Pen provides rich mass spectra from biological samples characterized by a variety of singly and doubly charged ions of lipids and metabolites and multiply charged protein ions, similar to those obtained by DESI-MSI and ESI methods. A liquid-solid chemical extraction process is used to sample molecules from the tissue using a water droplet, without assistance from a gas or a strong electric field. Our results show that by depositing a discrete water droplet onto a tissue sample for a determined amount of time, efficient extraction of biomolecules is achieved while tissue integrity is preserved (Figs. 3 and 6). The extraction process is similar to that reported for liquid extraction surface analysis (32), liquid microjunction surface sampling probe (33), and nanoDESI (34), although the latter two techniques use a continuous flow of solvent onto a sample surface, whereas the MasSpec Pen uses a single water droplet for extraction. In our current system, the water droplet containing analytes is transported as a liquid sample to the mass spectrometer and directly introduced to an extended transfer tube, which is connected to the heated inlet. Vaporization and ionization occur in the inlet region of the mass spectrometer, similar to what has been observed in solvent-assisted inlet ionization (35). Additional means of ionization such as electric field or laser assistance were not applied, although various connection methods and ionization sources (ESI, atmospheric pressure chemical ionization, atmospheric pressure photoionization, and others) could be adapted for our system.
Biocompatibility has been recognized as a key functional requirement of next-generation medical devices (36). The MasSpec Pen was designed as a simple, disposable device made with biocompatible materials and chemicals for contact with living tissues. The probe tip material, the conduit tubes, and the chemical used were PDMS, PTFE, and water, respectively. PDMS and PTFE are widely recognized as biocompatible materials and have a long history of utilization in medical devices including catheters and long-term implants (37). Our experiments demonstrate that the low volume (10 μl or less) of high-purity water used caused no effect to the tissues analyzed in vivo and ex vivo. The MasSpec Pen has shown good compatibility for contact with living tissues, which should enable in vivo use.
We designed the MasSpec Pen as an automatic and user-friendly device that could be used during routine medical diagnosis (movie S1). A drawback of many ambient ionization MS methods is the need for geometrical optimization and alignment between the source, the sample, and the mass spectrometer inlet to achieve good ion transmission. On the other hand, the MasSpec Pen operates in a geometry-free manner independently of sample stiffness and morphology and does not require optimization because all of the components required for solvent delivery, tissue sampling, and solvent retrieval are incorporated within the tip. The 1.5-m tube transport systems enable free motion and ease of use to the operator. The high-speed electronically and time-controlled pinch valves allowed precise control of droplet motion and showed excellent performance, robustness, and reproducibility in the results obtained.
Our results suggest that the MasSpec Pen may provide the performance required for near real-time nondestructive molecular diagnosis of tissues in vivo and ex vivo. Intraoperative diagnosis and surgical margin evaluation continue to be research focuses of clinical ambient ionization MS, with efforts used to develop devices for in vivo diagnosis (18). For example, a sampling probe using a DESI-MSI source to produce a continuous high-pressure spray of microdroplets has been proposed for tissue analysis (38), but it has not been demonstrated for cancer diagnosis or in vivo use. Ultraviolet and infrared laser approaches have been reported for tissue analysis through ablation processes (23, 24). A resonant infrared laser ablation system was used to analyze a cancer tissue sample ex vivo and human finger skin in vivo (24). To date, the iKnife has been the most successful MS-based technique demonstrated for in vivo, intraoperative diagnosis (21, 22). Yet, the electrocauterization process used by the iKnife is necessary for ion generation for MS analysis, resulting in thermal and mechanical damage of the analyzed tissue. Further, surgical modalities other than electrocauterization are used in oncologic surgeries, such as ultrasonic surgical aspiration, cold knife and mechanical stapled excisions, and laser surgery. Different from laser and electrocauterization approaches, an advantage of the proposed water-based MasSpec Pen technology is its nondestructive nature, which allows diagnosis of cancerous tissues without damage to normal tissues. Because the MasSpec Pen performs molecular diagnosis independently of any dissection tools, it has the potential to be used in many surgical modalities. Intraoperatively, we envision the MasSpec Pen to be used in conjunction with a surgical resection tool, depending on the oncologic surgery and the preference of the medical professional. Because surgeons routinely exchange handheld surgical instruments during surgery, with assistance from surgical technologists, we expect the MasSpec Pen to be adapted into routine surgical workflows. Clinically, the MasSpec Pen could be suitable for pre- and postsurgical procedures that require diagnosis of ex vivo samples (fresh tissues, tissue sections, or biopsies) commonly examined by pathologists.
Our results show that the molecular information obtained using the MasSpec Pen is diagnostic of disease and can be used for rapid tissue classification, cancer diagnosis, and subtyping. We tested the MasSpec Pen using 253 patient tissue samples including the normal and tumorous breast, ovary, lung, and thyroid tissues. The mass spectra presented rich molecular profiles characterized by a variety of potential cancer biomarkers. The statistical classifiers built using machine learning algorithms and the histologically validated molecular information provided an overall sensitivity of 96.4%, specificity of 96.2%, and accuracy of 96.3% for cancer. Our classifiers allowed the identification of different histologic types of lung cancers and benign and malignant thyroid tumors when compared to normal tissue samples and correctly predicted ovarian cancer diagnosis in an independent test sample with mixed histologic features. Yet, as with any technology that relies on machine learning and statistical modeling of large data sets to provide predictive diagnosis, larger sample sets are needed to increase the training set size, improve the predictive accuracy of our classifiers including information on tumor cell concentration, and expand the histologic and molecular subtypes of neoplasms and normal tissues. Rigorous validation studies of the statistical results using large independent held-out test sets are necessary to determine possible overfitting of our statistical models and to more conclusively determine the overall performance of the method.
Here, we demonstrated that the MasSpec Pen is effective for in vivo diagnosis of cancer regions during murine oncological surgery. The entire procedure from triggering the system to data analysis is performed under 10 s, and further improvements are envisioned. Compared with the time necessary for intraoperative pathologic frozen section analysis of excised species (~30 min) or postoperative final pathologic evaluation (several days), the time required for the MasSpec Pen analysis could expedite surgical procedures, diagnosis, and treatment. Our results demonstrate that the molecular information obtained using a 1.5-mm sampling size allows accurate identification of cancer in marginal tumor regions of mixed histologic composition (Fig. 5), although further validation of these results with larger independent sample sets is needed. We are currently exploring other machining methods to increase sampling resolution; however, the 1.5-mm sampling size relates well with the degree of precision achieved during surgical resection.
We envision that the MasSpec Pen will become a valuable clinical technology for near real-time in vivo and ex vivo cancer diagnosis. Emerging technologies including fluorescence-guided surgery (11), Raman spectroscopy (12), optical coherence tomography (39), reflectance spectroscopy (40), and stimulated Raman scattering microscopy (13) have been proposed for cancer diagnosis and surgical guidance. Although powerful, many of these methods rely on injection of exogenous labels that target specific cell types for tumor visualization, require tissue excision and processing for analysis, suffer from moderate tissue specificity, or provide limited biochemical information. The approach we propose here leverages on the unparalleled sensitivity and specificity provided by MS for untargeted molecular evaluation and on its biocompatibility for disease diagnosis and clinical use. Yet, many challenges exist in translating and integrating new technologies into the workflow of a complex surgical and clinical environment (41). Integration of the MasSpec Pen into laparoscopic and robotic surgicalsystems for minimally invasive procedures and improved communication and visualization tools will facilitate its inclusion in clinical workflows. Similarly, size- and cost-effective mass spectrometers with sufficient analytical performance for molecular evaluation are needed for dedicated surgical use. Advances in building and validating databases for tissue identification, as well as automated computational methods for real-time output of predictive results, are also necessary for broad use. Further work will require careful evaluation of the long-term benefits to patients to determine the value of new technologies in clinical practice. We expect that the excellent performance and simple design and operation of the MasSpec Pen, combined with its clinically desirable features, may enable its translation to the clinic for routine medical use, improving patient care and treatment.
MATERIALS AND METHODS
Study design
The objective of this study was to evaluate the potential of an MS-based probe to nondestructively analyze and diagnose cancer in human tissue samples. Here, we investigated the molecular profiles of human tissue samples obtained from 281 patients including normal and cancer breast, lung, thyroid, and ovary tissues. All patient samples were obtained from the CHTN (Cooperative Human Tissue Network), Asterand Bioscience, and the Baylor College of Medicine Tissue Bank under approved Institutional Review Board protocol. The mass spectra obtained using the MasSpec Pen in tissue samples were normalized, background-subtracted, and analyzed using a statistical technique to build classification models. Expert, board-certified pathologists (J.L., W.Y., and C.N.) evaluated the H&E-stained tissue sections obtained from the tissue samples analyzed. The pathologists were blind to any information about the acquisition from MS analysis. Samples were excluded from statistical analysis if they were determined by the pathologist to have substantial heterogeneity in cell composition, which included 28 samples, resulting in a final sample set of 253 samples. The in vivo animal model experiments were conducted under approved Institutional Animal Care and Use Committee protocol.
Design and engineering of the MasSpec Pen
A 3D printer (Model uPrint SE Plus) was used to print the key component, a PDMS (Dow Corning) probe tip. The pen tips were fabricated by casting an elastomer from a negative mold designed using SolidWorks computer-aided design software and then dissolving the mold away. PTFE tubing (ID, 1/32 inch; outer diameter, 1/16 inch; Cole-Parmer) was directly inserted into the probe tip for experiments. For more information, see the Supplementary Materials.
MS data acquisition
All experiments were performed on a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific). Full scan was carried out at the range of m/z 120 to 1800, using a resolving power of 140,000, a capillary temperature of 350°C, and an S-lens radio frequency level of 100. Wild-type mouse brains were purchased from BioreclamationIVT. Human tissue samples were obtained frozen and stored in a −80°C freezer until analysis, when they were thawed at room temperature. The tissues were placed on a glass slide and analyzed by the MasSpec Pen using the experimental steps described. After experiments, the tissue regions analyzed were annotated and frozen, and 16-μm tissue sections were prepared using a CryoStar NX50 cryostat. Additional tissue sections at different regions of the tissue piece were obtained for the MasSpec Pen analysis. Tissue sections were H&E-stained and evaluated by histopathology after analysis.
In vivo mouse experiments
BT474 HER2+ cells were grown in improved minimum essential medium (IMEM) (Invitrogen) supplemented with 10% fetal bovine serum, 1% L-glutamine, and 1% insulin to a confluency of 80 to 90% in 5% O2 and 37°C. Cells were counted via a hemocytometer and trypan blue dye exclusion. Nude athymic female mice (n = 3) were subcutaneously implanted with a 17β-estradiol pellet (0.72 mg, 60-day release; Innovative Research of America) in the nape of the neck. About 24 hours later, BT474 breast cancer cells (107) in serum-free IMEM with 20% growth factor–reduced Matrigel were injected subcutaneously into the right flank of the mouse (total injection of 100 μl). Tumors were monitored weekly for growth until they reached 0.7 to 1.0 cm in diameter (average of 250 mm3). In vivo experiments were performed during surgical resection of tumors using murine animal models while the mice were under anesthesia (2% isoflurane and 98% O2). A surgical blade was used to open a flap of skin, leaving an estimated space of 1 to 2 cm around the tumors, and then, the skin flap was dissected from the surface of the tumor. The skin was flapped to expose the tumor and adjacent normal tissues, which were analyzed in several regions using the MasSpec Pen. Pieces of the tumor were then resected using a scalpel and analyzed ex vivo. Tumor tissue pieces analyzed by the MasSpec Pen were annotated, flash-frozen, sectioned, and subjected to H&E staining for diagnosis.
Statistical analysis
Averages of three mass spectra obtained during each 10-s MasSpec Pen analysis were used to build molecular databases. The Xcalibur raw data were converted to Microsoft Excel spreadsheet format. The full mass range of the spectra was partitioned into bins by rounding m/z values to the nearest hundredth. All mass spectra were first normalized according to total ion count or to the absolute intensity of m/z 885.55 to account for slight fluctuations in signal intensities that may occur between experiments. Then, background peaks and peaks not appearing in at least 10% of the samples analyzed were excluded to reduce random noise. For each tissue section (breast or thyroid), four representative mass spectra for each tissue section analyzed were imported to MetaboAnalyst (www.metaboanalyst.ca/) for PCA using the web-site built-in function. Score and loading plots were generated through the website for each tissue type. For each soft tissue sample type (breast, thyroid, lung, and ovary), the data were imported to R programming language. PCA was performed by centering the preprocessed data to mean zero and computing principal components using the prcomp function in R. The first three principal components were visualized with the rgl and pca3d packages for R. For tissue classification, the Lasso method was applied using the glmnet package in the CRAN R language library. Models generated using the Lasso are simpler to interpret than other regularization methods because it yields “sparse” models (models that involve only a subset of the features). A mathematical weight for each statistically informative feature is calculated by the Lasso depending on the importance that the mass spectral feature has in characterizing a certain class (cancer versus normal or a cancer subtype versus normal). Classification was performed using a leave-one-out cross-validation approach to assess the predictive accuracy within the training set. Performance of trained classifiers was measured by sensitivity, specificity, accuracy, and AUC.
Supplementary Material
Movie S1. Simulation demonstrating the use of the MasSpec Pen for routine intraoperative diagnosis.
Fig. S1. Effect of MasSpec Pen contact time on the mass spectra obtained.
Fig. S2. Effect of MasSpec Pen tip diameter on the mass spectra obtained.
Fig. S3. MasSpec Pen analysis of a mouse brain tissue section.
Fig. S4. Positive ion mode analysis using the MasSpec Pen.
Fig. S5. Comparison between MasSpec Pen and DESI-MSI mass spectra.
Fig. S6. Effect of MasSpec Pen solvent systems on the mass spectra obtained.
Fig. S7. MasSpec Pen detection of protein ions.
Fig. S8. PCA of the data obtained for the human tissue sections including normal and tumor thyroid and breast tissue sections.
Fig. S9. Comparison between the MasSpec Pen mass spectra obtained from the tissue section and fresh tissue piece.
Fig. S10. MasSpec Pen analysis of an HGSC tissue sample with mixed histologic composition.
Fig. S11. Intraoperative analysis of tumor and normal tissues in a murine model.
Fig. S12. MasSpec Pen analysis of the same tissue sample in vivo and ex vivo.
Table S1. Data obtained for the identification of selected negative ion mode molecular ions from mouse brain tissue.
Table S2. Data obtained for the identification of selected negative ion mode molecular ions from human thyroid tissue.
Table S3. Data obtained for the identification of selected negative ion mode molecular ions from human ovarian tissue.
Table S4. Data obtained for the identification of selected negative ion mode molecular ions from human lung tissue.
Table S5. Data obtained for the identification of selected negative ion mode molecular ions from human breast tissue.
Table S6. Patient demographics of the 253 human tissue samples used in this study.
Acknowledgments
We thank the Eberlin and Milner laboratory members for valuable discussions. L.S.E. thanks J. Brodbelt and R. Crooks (University of Texas at Austin) for reviewing the manuscript and their mentoring. We are grateful to R. Tibshirani (Stanford University) for the assistance with the statistical analysis. We also thank T. Hooper for his help with the hardware design and E. Que for the use of microscope. Tissue samples were provided by the Cooperative Human Tissue Network, a National Cancer Institute supported resource, and the Baylor College of Medicine Tissue Bank.
Funding: This work was supported by the National Cancer Institute of the NIH under award R00CA190783 to L.S.E., the startup funds provided to L.S.E. by the University of Texas at Austin, and the Cancer Prevention Research Institute of Texas to A.G.S. (CPRIT RR160005) and to T.E.M. (CPRIT DP 150102).
Footnotes
Author contributions: L.S.E., T.E.M., J.Z., J.Q.L., and J.R. designed the experiments. L.S.E, T.E.M., J.Z., J.R., J.Q.L., N.K., and A.Z. designed the MasSpec Pen. J.Z., J.R., J.Q.L., M.S., N.G., A.B., R.J.D., K.Y.G., B.L., L.S.E., A.G.S., and A.S. acquired the data. L.S.E., J.Z., T.E.M., M.S., and J.S. performed the data analysis and interpretation. W.Y., C.N., and J.L. performed the pathologic evaluation. J.H.Y., J.Z., J.Q.L., and L.S.E. performed the statistical analysis and interpretation. L.S.E., J.Z., and T.E.M. wrote and revised the manuscript. L.S.E. conceived and supervised the study.
Competing interests: L.S.E., T.E.M., J.Z., J.L., J.R., N.K., and A.Z. are inventors on a provisional patent application 62/462,524 owned by the Board of Regents of the University of Texas System that relates to a handheld probe and MS system, such as described in this study. All other authors declare that they have no competing interests.
Data and materials availability: The data for this study have been deposited in Dataverse, which can be found at https://dataverse.harvard.edu/dataset.xhtml?persistentId=doi:10.7910/DVN/XBVHGM.
REFERENCES AND NOTES
- 1.Buchholz TA, Somerfield MR, Griggs JJ, El-Eid S, Hammond MEH, Lyman GH, Mason G, Newman LA. Margins for breast-conserving surgery with whole-breast irradiation in stage I and II invasive breast cancer: American society of clinical oncology endorsement of the society of surgical oncology/american society for radiation oncology consensus guideline. J Clin Oncol. 2014;32:1502–1506. doi: 10.1200/JCO.2014.55.1572. [DOI] [PubMed] [Google Scholar]
- 2.Maygarden SJ, Detterbeck FC, Funkhouser WK. Bronchial margins in lung cancer resection specimens: Utility of frozen section and gross evaluation. Mod Pathol. 2004;17:1080–1086. doi: 10.1038/modpathol.3800154. [DOI] [PubMed] [Google Scholar]
- 3.Massard G, Doddoli C, Gasser B, Ducrocq X, Kessler R, Schumacher C, Jung GM, Wihlm JM. Prognostic implications of a positive bronchial resection margin. Eur J Cardiothorac Surg. 2000;17:557–565. doi: 10.1016/s1010-7940(00)00384-5. [DOI] [PubMed] [Google Scholar]
- 4.Nick AM, Coleman RL, Ramirez PT, Sood AK. A framework for a personalized surgical approach to ovarian cancer. Nat Rev Clin Oncol. 2015;12:239–245. doi: 10.1038/nrclinonc.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han SS, Jang JY, Kim SW, Kim WH, Lee KU, Park YH. Analysis of long-term survivors after surgical resection for pancreatic cancer. Pancreas. 2006;32:271–275. doi: 10.1097/01.mpa.0000202953.87740.93. [DOI] [PubMed] [Google Scholar]
- 6.Zhang M, Li ZG, Ma Y, Zhu GY, Zhang HF, Xue YW. Prognostic predictors of patients with carcinoma of the gastric cardia. Hepatogastroenterology. 2012;59:930–933. doi: 10.5754/hge09356. [DOI] [PubMed] [Google Scholar]
- 7.Sanai N, Berger MS. Glioma extent of resection and its impact on patient outcome. Neurosurgery. 2008;62:753–764. doi: 10.1227/01.neu.0000318159.21731.cf. [DOI] [PubMed] [Google Scholar]
- 8.Macario A. What does one minute of operating room time cost? J Clin Anesth. 2010;22:233–236. doi: 10.1016/j.jclinane.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Mino-Kenudson M, Chirieac LR, Law K, Hornick JL, Lindeman N, Mark EJ, Cohen DW, Johnson BE, Janne PA, Iafrate AJ, Rodig SJ. A novel highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res. 2010;16:1561–1571. doi: 10.1158/1078-0432.CCR-09-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones S, Anagnostou V, Lytle K, Parpart-Li S, Nesselbush M, Riley DR, Shukla M, Chesnick B, Kadan M, Papp E, Galens KG, Murphy D, Zhang T, Kann L, Sausen M, Angiuoli SV, Diaz LA, Jr, Velculescu VE. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stummer W, Pichlmeier U, Meinel T, Wiestler OD, Zanella F, Reulen H-J ALA-Glioma Study Group. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: A randomised controlled multicentre phase III trial. Lancet Oncol. 2006;7:392–401. doi: 10.1016/S1470-2045(06)70665-9. [DOI] [PubMed] [Google Scholar]
- 12.Jermyn M, Mok K, Mercier J, Desroches J, Pichette J, Saint-Arnaud K, Bernstein L, Guiot MC, Petrecca K, Leblond F. Intraoperative brain cancer detection with Raman spectroscopy in humans. Sci Transl Med. 2015;7:274ra19. doi: 10.1126/scitranslmed.aaa2384. [DOI] [PubMed] [Google Scholar]
- 13.Orringer DA, Pandian B, Niknafs YS, Hollon TC, Boyle J, Lewis S, Garrard M, Hervey-Jumper SL, Garton HJL, Maher CO, Heth JA, Sagher O, Wilkinson DA, Snuderl M, Venneti S, Ramkissoon SH, McFadden KA, Fisher-Hubbard A, Lieberman AP, Johnson TD, Xie XS, Trautman JK, Freudiger CW, Camelo-Piragua S. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng. 2017;1:0027. doi: 10.1038/s41551-016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chughtai K, Heeren RMA. Mass spectrometric imaging for biomedical tissue analysis. Chem Rev. 2010;110:3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu CC, Dorrestein PC. Visualizing life with ambient mass spectrometry. Curr Opin Biotechnol. 2015;31:24–34. doi: 10.1016/j.copbio.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu CP, Dill AL, Eberlin LS, Cooks RG, Ifa DR. Mass spectrometry imaging under ambient conditions. Mass Spectrom Rev. 2013;32:218–243. doi: 10.1002/mas.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norris JL, Caprioli RM. Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem Rev. 2013;113:2309–2342. doi: 10.1021/cr3004295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ifa DR, Eberlin LS. Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin Chem. 2016;62:111–123. doi: 10.1373/clinchem.2014.237172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberlin LS, Norton I, Orringer D, Dunn IF, Liu XH, Ide JL, Jarmusch AK, Ligon KL, Jolesz FA, Golby AJ, Santagata S, Agar NYR, Cooks RG. Ambient mass spectrometry for the intraoperative molecular diagnosis of human brain tumors. Proc Natl Acad Sci USA. 2013;110:1611–1616. doi: 10.1073/pnas.1215687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarmusch AK, Pirro V, Baird Z, Hattab EM, Cohen-Gadol AA, Cooks RG. Lipid and metabolite profiles of human brain tumors by desorption electrospray ionization-MS. Proc Natl Acad Sci USA. 2016;113:1486–1491. doi: 10.1073/pnas.1523306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schäfer K-C, Dénes J, Albrecht K, Szaniszló T, Balog J, Skoumal R, Katona M, Tóth M, Balogh L, Takáts Z. In vivo, in situ tissue analysis using rapid evaporative ionization mass spectrometry. Angew Chem Int Ed. 2009;48:8240–8242. doi: 10.1002/anie.200902546. [DOI] [PubMed] [Google Scholar]
- 22.Balog J, Sasi-Szabó L, Kinross J, Lewis MR, Muirhead LJ, Veselkov K, Mirnezami R, Dezs B, Damjanovich L, Darzi A, Nicholson JK, Takáts Z. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci Transl Med. 2013;5:194ra93. doi: 10.1126/scitranslmed.3005623. [DOI] [PubMed] [Google Scholar]
- 23.Schäfer K-C, Szaniszló T, Günther S, Balog J, Dénes J, Keserű M, Dezső B, Tóth M, Spengler B, Takáts Z. In situ, real-time identification of biological tissues by ultraviolet and infrared laser desorption ionization mass spectrometry. Anal Chem. 2011;83:1632–1640. doi: 10.1021/ac102613m. [DOI] [PubMed] [Google Scholar]
- 24.Fatou B, Saudemont P, Leblanc E, Vinatier D, Mesdag V, Wisztorski M, Focsa C, Salzet M, Ziskind M, Fournier I. In vivo real-time mass spectrometry for guided surgery application. Sci Rep. 2016;6:25919. doi: 10.1038/srep25919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Series B Stat Methodol. 1996;58:267–288. [Google Scholar]
- 26.Dill AL, Eberlin LS, Costa AB, Ifa DR, Cooks RG. Data quality in tissue analysis using desorption electrospray ionization. Anal Bioanal Chem. 2011;401:1949–1961. doi: 10.1007/s00216-011-5249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang J, Yu W, Ryu SW, Lin J, Buentello G, Tibshirani R, Suliburk JW, Eberlin LS. Cardiolipins are molecular markers of mitochondria-rich thyroid oncocytic tumors. Cancer Res. 2016;76:6588–6597. doi: 10.1158/0008-5472.CAN-16-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calligaris D, Caragacianu D, Liu X, Norton I, Thompson CJ, Richardson AL, Golshan M, Easterling ML, Santagata S, Dillon DA, Jolesz FA, Agar NYR. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci USA. 2014;111:15184–15189. doi: 10.1073/pnas.1408129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenther S, Muirhead LJ, Speller AVM, Golf O, Strittmatter N, Ramakrishnan R, Goldin RD, Jones E, Veselkov K, Nicholson J, Darzi A, Takats Z. Spatially resolved metabolic phenotyping of breast cancer by desorption electrospray ionization mass spectrometry. Cancer Res. 2015;75:1828–1837. doi: 10.1158/0008-5472.CAN-14-2258. [DOI] [PubMed] [Google Scholar]
- 30.Kang HS, Lee SC, Park YS, Jeon YE, Lee JH, Jung SY, Park IH, Jang SH, Park HM, Yoo CW, Park SH, Han SY, Kim KP, Kim YH, Ro J, Kim HK. Protein and lipid MALDI profiles classify breast cancers according to the intrinsic subtype. BMC Cancer. 2011;11:465. doi: 10.1186/1471-2407-11-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sans M, Gharpure K, Tibshirani R, Zhang J, Liang L, Liu J, Young JH, Dood RL, Sood AK, Eberlin LS. Metabolic markers and statistical prediction of serous ovarian cancer aggressiveness by ambient ionization mass spectrometry imaging. Cancer Res. 2017;77:2903–2913. doi: 10.1158/0008-5472.CAN-16-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kertesz V, Van Berkel GJ. Fully automated liquid extraction-based surface sampling and ionization using a chip-based robotic nanoelectrospray platform. J Mass Spectrom. 2010;45:252–260. doi: 10.1002/jms.1709. [DOI] [PubMed] [Google Scholar]
- 33.Kertesz V, Ford MJ, Van Berkel GJ. Automation of a surface sampling probe/electrospray mass spectrometry system. Anal Chem. 2005;77:7183–7189. doi: 10.1021/ac0510742. [DOI] [PubMed] [Google Scholar]
- 34.Laskin J, Heath BS, Roach PJ, Cazares L, Semmes OJ. Tissue imaging using nanospray desorption electrospray ionization mass spectrometry. Anal Chem. 2012;84:141–148. doi: 10.1021/ac2021322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pagnotti VS, Chubatyi ND, McEwen CN. Solvent assisted inlet ionization: An ultrasensitive new liquid introduction ionization method for mass spectrometry. Anal Chem. 2011;83:3981–3985. doi: 10.1021/ac200556z. [DOI] [PubMed] [Google Scholar]
- 36.Helmus MN, Gibbons DF, Cebon D. Biocompatibility: Meeting a key functional requirement of next-generation medical devices. Toxicol Pathol. 2008;36:70–80. doi: 10.1177/0192623307310949. [DOI] [PubMed] [Google Scholar]
- 37.Mokkaphan J, Banlunara W, Palaga T, Sombuntham P, Wanichwecharungruang S. Silicone surface with drug nanodepots for medical devices. ACS Appl Mater Interfaces. 2014;6:20188–20196. doi: 10.1021/am505566m. [DOI] [PubMed] [Google Scholar]
- 38.Chen CH, Lin Z, Garimella S, Zheng L, Shi R, Cooks RG, Ouyang Z. Development of a mass spectrometry sampling probe for chemical analysis in surgical and endoscopic procedures. Anal Chem. 2013;85:11843–11850. doi: 10.1021/ac4025279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kut C, Chaichana KL, Xi J, Raza SM, Ye X, McVeigh ER, Rodriguez FJ, Quiñones-Hinojosa A, Li X. Detection of human brain cancer infiltration ex vivo and in vivo using quantitative optical coherence tomography. Sci Transl Med. 2015;7:292ra100. doi: 10.1126/scitranslmed.3010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajaram N, Aramil TJ, Lee K, Reichenberg JS, Nguyen TH, Tunnell JW. Design and validation of a clinical instrument for spectral diagnosis of cutaneous malignancy. Appl Optics. 2010;49:142–152. doi: 10.1364/AO.49.000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J, Yu W, Suliburk J, Eberlin LS. Will ambient ionization mass spectrometry become an integral technology in the operating room of the future? Clin Chem. 2016;62:1172–1174. doi: 10.1373/clinchem.2016.258723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1. Simulation demonstrating the use of the MasSpec Pen for routine intraoperative diagnosis.
Fig. S1. Effect of MasSpec Pen contact time on the mass spectra obtained.
Fig. S2. Effect of MasSpec Pen tip diameter on the mass spectra obtained.
Fig. S3. MasSpec Pen analysis of a mouse brain tissue section.
Fig. S4. Positive ion mode analysis using the MasSpec Pen.
Fig. S5. Comparison between MasSpec Pen and DESI-MSI mass spectra.
Fig. S6. Effect of MasSpec Pen solvent systems on the mass spectra obtained.
Fig. S7. MasSpec Pen detection of protein ions.
Fig. S8. PCA of the data obtained for the human tissue sections including normal and tumor thyroid and breast tissue sections.
Fig. S9. Comparison between the MasSpec Pen mass spectra obtained from the tissue section and fresh tissue piece.
Fig. S10. MasSpec Pen analysis of an HGSC tissue sample with mixed histologic composition.
Fig. S11. Intraoperative analysis of tumor and normal tissues in a murine model.
Fig. S12. MasSpec Pen analysis of the same tissue sample in vivo and ex vivo.
Table S1. Data obtained for the identification of selected negative ion mode molecular ions from mouse brain tissue.
Table S2. Data obtained for the identification of selected negative ion mode molecular ions from human thyroid tissue.
Table S3. Data obtained for the identification of selected negative ion mode molecular ions from human ovarian tissue.
Table S4. Data obtained for the identification of selected negative ion mode molecular ions from human lung tissue.
Table S5. Data obtained for the identification of selected negative ion mode molecular ions from human breast tissue.
Table S6. Patient demographics of the 253 human tissue samples used in this study.