Abstract
Background
Trajectories of complex neurocognitive phenotypes in preclinical aging may be produced differentially through selective and interactive combinations of genetic risk.
Objective
We organize three possible combinations into a “network” of genetic risk indices derived from polymorphisms associated with normal and impaired cognitive aging, as well as Alzheimer’s disease (AD). Specifically, we assemble and examine three genetic clusters relevant to non-demented cognitive trajectories: (1) Apolipoprotein E (APOE), (2) a Cognitive Aging Genetic Risk Score (CA-GRS; Catechol-O-methyltransferase + Brain-derived neurotrophic factor), and (3) an AD-Genetic Risk Score (AD-GRS; Clusterin + Complement receptor 1 + Phosphatidylinositol-binding clathrin assembly protein).
Method
We use an accelerated longitudinal design (n = 634; age range = 55–95 years) to test whether AD-GRS (low versus high) moderates the effect of increasing CA-GRS risk on executive function (EF) performance and change as stratified by APOE status (ε4+ versus ε4-).
Results
APOE ε4 carriers with high AD-GRS had poorer EF performance at the centering age (75 years) and steeper 9-year decline with increasing CA-GRS but this association was not present in APOE ε4 carriers with low AD-GRS.
Conclusions
APOE ε4 carriers with high AD-GRS are at elevated risk of cognitive decline when they also possess higher CA-GRS risk. Genetic risk from both common cognitive aging and AD-related indices may interact in intensification networks to differential predict (1) level and trajectories of EF decline and (2) potential selective vulnerability for transitions into impairment and dementia.
Keywords: Alzheimer’s disease, Cognitive Aging, Genetic Risk, Apolipoprotein E, Executive Function, Victoria Longitudinal Study
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease that accounts for approximately 60–80% of dementia cases worldwide [1–3]. The AD pathophysiological process includes the build-up of amyloid plaques and neurofibrillary tangles, which start years before the onset of clinical symptoms that lead to diagnosis [3,4]. Current theoretical models of AD suggest that the accumulation of amyloid-β (Aβ) peptide triggers a pathological cascade that leads to increased cerebrospinal tau production, brain atrophy, and cognitive impairment [5]. Primary clinical characteristics include memory loss, decline in global cognition, and early impairments in executive function (EF) [6]. Although age is a prominent factor in the incidence and prevalence of sporadic AD [7], studies have shown that genetic risk factors play a critical role in AD development [8]. Single AD risk genes may operate through (1) interactive effects with other AD genetic polymorphisms [9], (2) panel effects with other polymorphisms of similar structure and function [10], (3) aggregated or magnified associations with genetic risk for exacerbated but non-demented cognitive decline [11], and (4) networks of panels that reflect coordinated functions or canonical pathways [12]. Arguably, network-based approaches may lead to early identification of individual non-demented adults with elevated genetic risk for accelerated cognitive decline, Mild Cognitive Impairment (MCI) or AD, thus promoting timely and effective prevention programs [13]. In the present study, we examine a network approach assembling three clusters of genetic risk: (1) a prominent and commonly examined AD risk gene (i.e., Apolipoprotein (APOE; rs7412, rs429358)), (2) three AD risk genetic polymorphisms with different functions in the central nervous system (i.e., Clusterin (CLU; rs11136000), Complement receptor 1 (CR1; rs6656401), and Phosphatidylinositol-binding clathrin assembly protein (PICALM; rs3851179)), and (3) two commonly studied polymorphisms relevant to EF trajectories in non-demented aging (i.e., Catechol-O-methyltransferase (COMT; rs4680) and Brain-derived neurotrophic factor (BDNF; rs6265)). As we focus on testing a network of genetic risk for preclinical aging, we examine these three clusters for interactive predictions of longitudinal cognitive trajectories in non-demented older adults. We have selected the EF domain, which involves cognitive processes such as planning, goal-directed actions, and problem solving [14,15]. Recent studies have shown that changes in EF performance and decline can be detected prior to clinical diagnosis making EF an important and promising early marker for MCI and AD [16].
The APOE gene located on chromosome 19q13.2 has three isoforms, ε2, ε3, and ε4 [17]. The ε4 allele is consistently associated with increased risk for sporadic AD [18–20] and also observed in recent AD genome wide association studies (GWAS) [21,22]. The ε2 allele is shown to have a protective effect and the ε3 allele is considered neutral [17]. Approximately 40% of AD patients across major ethnic groups are carriers of at least one copy of the ε4 allele [17,23]. APOE is involved in regulating cholesterol levels which plays an important role in Aβ metabolism, aggregation, and deposition, leading to increased senile plaques and cerebral amyloid angiopathy in brains of APOE ε4 carriers diagnosed with AD. In healthy older adults, APOE ε4 homozygotes are associated with higher amyloid pathology, greater medial temporal lobe atrophy, elevated resting-state activity in the default mode network, neuroinflammation [17] and accelerated decline on cognitive tests [24,25].
Recent studies have identified additional single nucleotide polymorphisms (SNPs) that may play an important role in AD development [26]. A meta-analysis conducted by the AD Genetics Consortium confirmed that CLU, CR1, and PICALM [9,21,22] are associated with high AD risk susceptibility in independent samples [9]. These three SNPs have been further replicated for association with late-onset AD in European [27], North American [28], and Chinese [29] populations. The three SNPs represent varying functions in the brain where CLU is mainly involved with cholesterol metabolism, CR1 is associated with immune response, and PICALM with endocytosis [30]. We now summarize the three polymorphisms as related to risk for cognitive decline and AD.
CLU is located on chromosome 8p21.1 and differentially regulates lipid transport, Aβ clearance, brain atrophy, and apoptosis [30]. AD patients show higher CLU levels implying that CLU acts in response to poor neuronal functions [31]. In such cases, CLU may (1) act as an anti-apoptotic signal, (2) provide oxidative stress protection, (3) defend activated complement proteins as a result of inflammation, and (4) bind to partially unfolded proteins to prevent aggregation [31]. CLU allelic risk carriers (C+) are at 1.16 greater odds of developing AD compared to their low risk homozygotes (T/T) [32]. In healthy young adults, CLU allelic risk had lower white matter integrity than their counterparts [33] which may indicate an increased risk for developing dementia in old age. CLU is also similar in structure with the molten globule structure of APOE and they may influence each other in frontal lobe regions [10,34,35].
CR1 gene is located on chromosome 1q32. CR1 is a multifunctional glycoprotein expressed on many cells including dendritic cells [36]. The protein is involved in a number of functions including regulation of the complement cascade and clearance of immune complexes. In relation to AD, CR1 acts as a receptor for the Aβ-42 peptide removal from the brain and the circulatory system [22]. Thus, the CR1 SNP may be responsible for modifying the rate of Aβ-42 clearance in AD patients [37].
The PICALM gene is located on chromosome 11q14 is involved in the production of Aβ peptide and linked to the formation of amyloid plaques and Aβ metabolism [38]. The PICALM protein is involved in clathrin-mediated endocytosis. A recent study replicated GWAS findings with 2816 AD and 2706 control subjects in a European population [22] and confirmed that the T allele was associated with AD risk. PICALM risk (T/T) homozygotes have been associated with decreased cerebrospinal fluid Aβ-42 levels and therefore increased Aβ-42 levels in the brain [39]. A meta-analysis reported that PICALM interacts with APOE such that APOE ε4 risk carriers with the PICALM risk genotype are at increased AD risk [9]. Similarly, a recent study reported interaction effects for PICALM rs3851179 risk and APOE ε4+ leading to changes in brain atrophy and cognitive performance in AD [40].
The three AD-related SNPs have also independently been associated with accelerated cognitive decline in healthy older adults. Specifically, a faster rate of memory decline in older adults was observed for PICALM risk carriers [41], CLU risk carriers who eventually converted to MCI or AD [42], and CR1 risk carriers as mediated by amyloid plaque burden [43]. Although identified and confirmed in large GWAS and examined in single candidate gene association studies with normal cognitive performance [42,43], the neurobiological underpinnings for the synergistic effect of CLU, CR1, and PICALM on cognitive trajectories in non-demented aging has not yet been studied. CLU, CR1, and PICALM broadly represent three distinct processes (cholesterol metabolism, immune response, endocytosis) in the brain [30] and all been linked to Aβ metabolism and production, and cognitive decline in older adults at risk for dementia.
Complex neurocognitive phenotypes observed in non-demented older adults may be a result of select combinations of genes associated with AD and those linked with cognitively normal aging. Network-based approach incorporating interactions between cognitive aging and AD genes may provide insight into specific AD disease mechanism and molecular interactions in preclinical decline [12]. Although inconsistent in their independent effects, select combinations of COMT and BDNF polymorphisms have been shown to play a magnifying role in predicting the extent of neurocognitive deficits observed among groups of non-demented older adults [44–46]. COMT homozygotes and carriers of the risk allele (G/G, G/A) have lower dopamine levels in the prefrontal cortex [47]. BDNF homozygotes and carriers of the risk allele (A/A, A/G) secrete lower levels of neurotrophic factors, particularly in the hippocampus [48]. In two previous studies [11,49], we established an additive (COMT + BDNF) Cognitive Aging Genetic Risk Score (CA-GRS). We observed a significant additive effect (and no interactive associations) between COMT and BDNF, where higher CA-GRS was associated with poorer EF decline. This implies additive pathways for cognitive aging SNPs (i.e., COMT and BDNF) where eliminating one risk factor does not reduce the risk associated with the other allelic risk. The CA-GRS effect was further modified by APOE genotype, where APOE ε4 carriers displayed poorer EF performance with increasing CA-GRS. EF risk associated with increasing CA-GRS may be especially magnified for older adults who are carriers of a notable genetic risk allele for cognitive impairment and AD [11].
Recent genetic reports on AD and cognitive impairment have involved (1) single candidate genes, (2) genetic risk scores, and (3) network of molecular and pathway analysis of genetic variants. AD genetic network based approaches focus on using large molecular networks [50] to identify and understand specific AD-related biological functions. For example, recent network approaches include (1) co-expression networks (gene-gene correlations), (2) genetic integration (protein-protein interactions), (3) tissue specificity (network for tissues associated with AD), (4) robustness (strength in patterns of gene co-expressions in specific regions) [51,52], (5) network-based stratification (protein-protein interaction to stratify patients and identify disease molecules within the network), (6) analysis of directed networks (predicts specific signals), and (7) disease state-specific networks (networks that are only significant in specific disease states) [12,51,52]. The present network analysis represents a combination of three genetic risk clusters integrated by elements of both network-based stratification and disease state-specific network approaches. Specifically, we examine the interaction between an Alzheimer’s disease-Genetic Risk Score (AD-GRS) and a CA-GRS as modified by APOE genotype. The target phenotype is EF performance (level and trajectories) in non-demented older adults. We examine select group of cognitive aging and AD risk alleles that may work in synergy to magnify cognitive decline in non-demented older adults. To date, previous work with additive risk panels [53,54] have been examined independently and lack integration with complex interactive networks between additive panels commonly linked to AD independently, in risk panels, and mechanisms related to cognitive impairment.
Genetic risk scores for AD and cognitive impairment risk have included varying number and types of polymorphisms. AD genetic risk scores have been associated with increased risk of late life cognitive impairment [55], AD risk [56], greater risk of conversion from MCI to AD [54,57,58], and discriminating an AD group from controls [59]. Research on genetic risk approaches have used several procedures for calculating risk scores. For example, prior studies have used an explained variance-weighted genetic risk score [60], an odds ratio weighted risk score [53,54], large number of SNPs to create a polygenic risk score [61,62], weighted sum of the top risk scores [56], and an additive allelic risk score [11,53,63].
Although previous studies have focused on a variety of methods to create AD genetic risk scores, the results have been incomplete with potential biases [12]. Previous reports have not examined interactive and modification effects with AD and cognitive aging genetic risk scores to predict non-demented cognitive change. In the present study, we focus on predicting actual trajectories of non-demented cognitive change, not the outcome of AD diagnosis. AD genetic risk is one important component, but not the only source, of genetic prediction of preclinical cognitive decline or impairment. We therefore design a test of a “network” approach that combines not only AD risk genes but also brain/cognitive decline risk genes that, in combination or interaction, may magnify cognitive decline and impairment in non-demented older adults. Accordingly, we examine interactions between sets (or panels) of genotypes that could intensify their effects on cognitive trajectories. Specifically, we select three components: (1) a genetic cognitive aging risk panel (COMT + BDNF), (2) a targeted set of relatively well-known AD risk genes for an AD genetic risk panel (CLU + CR1 + PICALM) and (3) the best-known and predominant AD risk gene (APOE) as modifier. We then test the interactions among these three genetic risk components, with APOE playing the special role of modifier. Regarding APOE, we elected to stratify APOE ε4− and ε4+ because AD and cognitive aging genes may differentially influence the mechanisms in these two groups. We select these specific genetic risk factors as previous research has accumulated considerable information on associated mechanisms (often as independent or candidate gene predictors) so that we could both (1) provide a strong test of potential genetic network effects and (2) propose mechanisms that underlie them and promote future research with this approach and on these mechanisms. In our own previous research [11], we have been investigating all of the genes (and their possible mechanisms and interactions) we examine in this research report, building to the point that we can now examine whether their influence on non-demented cognitive trajectories can be modelled as a network largely influenced by selected sets of AD-related and cognitive-aging-related risk genes.
In designing the present study, we acknowledged and adapted several important methodological and mechanistic contributions from both candidate gene approaches and basic gene x gene interaction approaches. Our network approach included a sequence of interactions between two clusters of aging and AD genetic risk, as modified by APOE, in the context of predicting differential cognitive trajectories over a 40-year band of aging. Specifically, we examined a sequence of interactions between two additive panels of genetic risk factors for AD (CLU + CR1 + PICALM) and accelerated cognitive decline (COMT + BDNF) as modified by APOE ε4 status. The sequence represents a network of genetic effects in cognitively normal older adults. To our knowledge, the present study is the first to examine a genetic network approach to examine (1) synergistic associations between clusters of AD (AD-GRS) and cognitive aging (CA-GRS) genes (2) as modified by APOE genotype (ε4− vs ε4+) (3) to predict EF trajectories (4) in non-demented older adults. We expect to observe that APOE ε4 carriers with interactive associations of higher CA-GRS and higher AD-GRS show poorer EF performance and steeper 9-year decline than their counterparts with lower CA-GRS and lower AD-GRS, and those who are in the APOE ε4− group.
Method
Participants
We used data from the Victoria Longitudinal Study (VLS), a large-scale, longitudinal sequential study examining biomedical, health, genetic, lifestyle, cognitive and other aspects of aging. General information on recruitment, methodological, and VLS characteristics are available elsewhere [64]. All volunteers in the VLS were cognitively normal (non-demented) and relatively healthy, with no reported brain-related serious conditions. They were enrolled through advertisements, and received a small honorarium for their participation. The VLS and all present data collection procedures are in full and certified compliance with prevailing human/institutional research ethics guidelines. Written informed consent was obtained from all participants. Approximately 99.2% of participants were White, not of Hispanic Origin. All had complete access to Canadian national health care. At baseline, all recruited VLS participants are cognitively normal (no dementia) and followed over time as they develop impairment or dementia. For the present study, we ensured a non-demented sample by applying a set of exclusionary criteria (including diagnosis of AD or neuropsychiatric disorder or Mini Mental State Exam (MMSE) score < 24). The present sample reflects the implementation of exclusionary criteria affecting individuals with (a) diagnosis of dementia, (b) anti-psychotic medication, (c) MMSE scores less than 24, (d) uncontrolled hypertension, (e) insulin-controlled diabetes, and (f) history of serious head injury (e.g., hospitalized). Accordingly, 634 participants (age range = 53–95 years, mean age = 70.58, SD = 8.65), including 423 females and 211 males with genetic data were included at baseline (Table 1). We followed an accelerated longitudinal design by assembling three partial samples (S; S1, S2, S3) from the VLS. The present Wave 1 (W1) and Wave 2 (W2) included participants from all three samples and Wave 3 (W3) included participants from S3. Specifically, throughout this report (a) W1 (n = 632) refers to S1W6, S2W4, and S3W1, (b) W2 (n = 517) refers to S1W7, S2W5, S3W2, and (c) W3 (n = 293) refers to S3W3. The average interval was 4.4 years between W1 and W2, and 4.5 years between W2 and W3. The retention rates for each wave interval ranged between 74% and 88%.
Table 1.
Participant characteristics by genotype.
| COMT | BDNF | APOE | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| A/A | A/G | G/G | p-value | G/G | A/G | A/A | p-value | ε4- | ε4+ | p-value | |
| n | 146 | 336 | 150 | -- | 416 | 189 | 27 | -- | 453 | 149 | -- |
| Age (years) | 70.15 (8.86) | 70.82 (8.69) | 70.40 (8.39) | 0.709 | 70.30 (8.82) | 71.45 (8.52) | 68.54 (6.32) | 0.145 | 70.89 (8.84) | 69.86 (8.27) | 0.212 |
| Education (years) | 14.92 (3.11) | 15.36 (2.80) | 15.36 (3.15) | 0.294 | 15.29 (2.96) | 15.13 (2.99) | 15.72 (2.70) | 0.585 | 15.19 (2.97) | 15.55 (3.07) | 0.208 |
| Sex (F/M) | 101/45 | 225/111 | 96/54 | 0.637 | 276/140 | 128/61 | 18/9 | 0.946 | 304/149 | 93/56 | 0.295 |
| MMSE | 28.72 (1.20) | 28.72 (1.20) | 28.56 (1.32) | 0.390 | 28.62 (1.28) | 28.74 (1.15) | 29.15 (0.77) | 0.071 | 28.66 (1.24) | 28.68 (1.25) | 0.914 |
|
| |||||||||||
| CLU | CR1 | PICALM | Total | ||||||||
|
| |||||||||||
| T/T | C+ | p-value | G/G | A+ | p-value | C/C | T+ | p-value | |||
|
| |||||||||||
| n | 103 | 527 | -- | 213 | 417 | -- | 151 | 478 | -- | 632 | |
| Age (years) | 70.65 (8.11) | 70.55 (8.75) | 0.982 | 70.30 (8.90) | 70.70 (8.52) | 0.854 | 69.42 (8.69) | 70.95 (8.59) | 0.129 | 70.56 (8.65) | |
| Education (years) | 14.92 (2.80) | 15.33 (2.98) | 0.182 | 15.41 (3.02) | 15.19 (2.92) | 0.282 | 15.52 (2.93) | 15.18 (2.97) | 0.310 | 15.26 (2.96) | |
| Sex (F/M) | 65/38 | 355/172 | 0.428 | 137/76 | 283/134 | 0.408 | 96/55 | 323/155 | 0.314 | 422/210 | |
| MMSE | 28.71 (1.12) | 28.68 (1.25) | 0.387 | 28.76 (1.27) | 28.65 (1.20) | 0.222 | 28.69 (1.24) | 28.68 (1.22) | 0.356 | 28.68 (1.23) | |
Abbreviations: n = total number; COMT = Catechol-O-methyl transferase; BDNF = Brain-derived neurotrophic factor; APOE = Apolipoprotein E; CLU = Clusterin; CR1 = Complement receptor 1; PICALM = Phosphatidylinositol-binding clathrin assembly protein; p < .05; MMSE = Mini-Mental State Exam; Standard deviations are in parentheses.
DNA Extraction and Genotyping
Saliva was collected according to standard procedures from Oragene DNA Genotek and stored at room temperature in Oragene® disks until DNA extraction. DNA was manually extracted from 0.8 ml of saliva sample mix using the manufacturer’s protocol with adjusted reagent volumes. Genotyping was carried out by using a polymerase chain reaction-restriction fragment length polymorphism strategy to analyze the allele status for CLU (rs11136000), CR1 (rs6656401), PICALM (rs3851179), BDNF (rs6265), COMT (rs4680), and APOE (rs7412, rs429358). Genotyping was successful for the targeted SNPs for all present participants. The genotype frequencies did not differ significantly from Hardy-Weinberg equilibrium for: BDNF (χ2 = 0.868, p = 0.35), COMT (χ2 = 2.909, p = 0.08), APOE (χ2 = 0.189, p = 0.66), and CLU (χ2 = 0.710, p = 0.40). We note that the CR1 (χ2 = 6.219, p = 0.01) and PICALM (χ2 = 36.955, p = 0.00) genotype frequencies were not in Hardy-Weinberg equilibrium.
Executive Function Measures
Two dimensions of EF (inhibition, shifting) were each measured by two standard and frequently used tests for cognitive, clinical, and neurobiological studies in older adults [14,49,65,66].
Hayling Sentence Completion (Inhibition)
This test [67] consists of two sections, each comprising 15 sentences. The standardized scores are based on errors from the second of two sections and the speed of each response from both sections, which are then combined to obtain the final score (1 = very low to 10 = very high).
Stroop (Inhibition)
This test [68] consists of the standard three parts (Parts A, B and C), with the measures based on latencies. The score is the standardized Stroop interference index ([Part C− Part A]/Part A), with a lower index reflecting better performance.
Brixton Spatial Anticipation (Shifting)
This test [67] consists of 10 different circles, one being blue, whereas the rest are colorless. Participants are asked to guess where the blue colored circle will appear on subsequent pages. The total number of incorrect guesses are measured and the final scores are calculated (1 = very low to 10 = very high).
Color Trails (Shifting)
This test [69] comprises two different sections in which participants connect different attributes, such as numbered and colored circles. Latency scores in the second of two sections were computed and used in the final analyses. Lower scores reflected better performance.
Statistical Analyses
Structural equation modeling (SEM) was used for all analyses with Mplus Version 7 [70]. All missing values for cognitive measures were assumed to be missing at random and estimated using maximum likelihood. Cases with missing predictor values were removed using list-wise deletion in Mplus 7. Age was centered at 75 years because aging related changes in many cognitive domains are not visible until 75 years [71]. Only two participants with missing measures on all four EF tasks were lost due to list-wise deletion. Preliminary analyses were examined to obtain EF factor scores and the best latent growth model (see supplementary materials).
The APOE ε4− group was coded as 0 (lower risk) and APOE ε4+ group as 1 (higher risk). All ε2/ε4 carriers (n = 30) were deleted [10,11]. APOE ε4− and ε4+ groups were separated and examined for effect modification by APOE. The AD-GRS [53,54,72] was calculated in three steps. First, we dichotomized CLU (risk: C/C, C/T; no risk: T/T), CR1 (risk: A/A, A/G; no risk: G/G), and PICALM (risk: T/T, T/C; no risk: C/C) into no risk (0) and risk (1) groups. Second, we summed across CLU, CR1, and PICALM to obtain a score for each adult ranging from 0–3. Third, we performed a median split [53] for this score and grouped the CLU + CR1 + PICALM allelic risk score by low (0–1 risk allele) and high (2–3 risk allele) genetic risk. The CA-GRS was calculated using all three allelic combinations (A/A, A/G, and G/G) of COMT and BDNF. Both SNPs were coded from 1 to 3 (3 = highest risk) and summed across COMT and BDNF to obtain the CA-GRS ranging from 2–6.
EF was regressed on CA-GRS as moderated by low and high AD-GRS. This analysis was performed twice, for the APOE ε4− group and the APOE ε4+ group. We expected that sex differences may modify the effects of genetic risk on EF performance and decline [73]. Therefore, sex was included as a covariate in both group analyses. We accounted for variability associated with age by directly incorporating age as the metric of change in our analyses. For model fit statistics and significant results, we examined the regression estimate and p < .05, and −2 log likelihood (−2LL), Akaike information criteria (AIC), and Bayesian information criteria (BIC) values with lower values indicating better model fit (see Table 2).
Table 2.
Cognitive Aging-Genetic Risk Score (CA-GRS) effect model estimates for low and high Alzheimer’s disease-Genetic Risk Score (AD-GRS) as separated by APOE ε4− and ε4+ groups.
| Intercept | Slope | Model Fit Statistics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| β | SE | p | β | SE | p | H0 value | Free Parameters | −2LL | AIC | BIC | |
| APOE ε4− group | −1432.04 | 32 | 2864.08 | 2928.08 | 3059.58 | ||||||
|
| |||||||||||
| Low AD-GRS: CA-GRS | 0.248 | 0.166 | 0.135 | 0.012 | 0.011 | 0.279 | |||||
| Low AD-GRS: Sex | 0.031 | 0.284 | 0.912 | −0.004 | 0.017 | 0.797 | |||||
|
| |||||||||||
| High AD-GRS: CA-GRS | 0.006 | 0.058 | 0.919 | 0.002 | 0.003 | 0.599 | |||||
| High AD-GRS: Sex | −0.207 | 0.118 | 0.080 | −0.011 | 0.006 | 0.050 | |||||
|
| |||||||||||
| APOE ε4+ group | −490.72 | 32 | 981.44 | 1045.43 | 1141.56 | ||||||
|
| |||||||||||
| Low AD-GRS: CA-GRS | 0.300 | 3.393 | 0.930 | 0.001 | 0.122 | 0.994 | |||||
| Low AD-GRS: Sex | 0.792 | 2.495 | 0.751 | 0.017 | 0.114 | 0.883 | |||||
|
| |||||||||||
| High AD-GRS: CA-GRS | −0.396 | 0.151 | 0.009 | −0.015 | 0.007 | 0.045 | |||||
| High AD-GRS: Sex | 0.058 | 0.239 | 0.808 | 0.002 | 0.012 | 0.853 | |||||
Abbreviations: Est. = regression estimate; SE = standard error; H0 = log likelihood; −2LL = −2 log likelihood; AIC = Akaike Information Criteria; BIC = Bayesian Information Criteria; COMT = Catechol-O-methyltransferase; BDNF = Brain-derived neurotrophic factor; APOE = Apolipoprotein E; CLU = Clusterin; CR1 = Complement receptor 1; PICALM = Phosphatidylinositol-binding clathrin assembly protein. CA-GRS = COMT + BDNF; AD-GRS = CLU + CR1 + PICALM.
Results
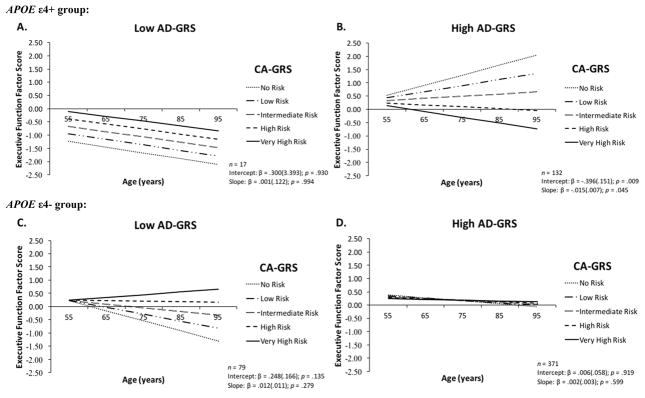
In our preliminary analyses and results (see supplementary materials) for the EF factor analysis and growth modeling, we established that the one-factor parsimonious model of EF provided the best fit to the data [74] and met partial scalar longitudinal invariance (see Supplementary Table 1). To obtain partial scalar invariance, we first need to meet configural invariance (all four indicators load on to the EF factor) and metric invariance (unstandardized EF factor loadings at all three waves are equal to each other). Partial scalar invariance is obtained when two out of the four EF indicator intercept were constrained to be equal across all three waves. The best latent growth model was obtained with the random intercept and random slope model (see Supplementary Table 2). This model shows that adults vary in their EF intercept (at age 75 years) and EF slope over time (9 years). In the present study, we extended our previous findings [11] and observed two novel genetic network associations with cognitive aging and AD genes. We observed that poorer EF performance at age 75 years with increasing CA-GRS in APOE ε4 carriers is (1) moderated by AD-GRS and (2) significant EF decline over 9 years is observed only when AD-GRS is included in this complex genetic network (Figure 1). Specifically, APOE ε4 carriers in the high AD-GRS group showed poorer EF performance at age 75 years (β = −0.396; SE = 0.151; p = .009) and steeper 9-year decline (β = −0.015; SE = 0.007; p = .045) with increasing CA-GRS (Figure 1B). Poorer EF performance at age 75 years or steeper 9-year decline was not observed in: (1) APOE ε4 carriers with low AD-GRS (Figure 1A); (2) the APOE ε4− group with high AD-GRS (Figure 1D); and (3) the APOE ε4− group with low AD-GRS (Figure 1C). We did not observe that sex (as a covariate) influenced the CA-GRS and AD-GRS interactive association on EF performance and change in either the APOE ε4− or APOE ε4+ group. CA-GRS effect on EF performance and change was selective and only present in APOE ε4 carriers with high AD-GRS.
Figure 1.
Increasing risk associated with Cognitive Aging-Genetic Risk Score [CA-GRS: Catechol-O-methyltransferase + Brain-derived neurotrophic factor] was magnified by high Alzheimer’s disease-Genetic Risk Score [AD-GRS: Clusterin + Complement receptor 1 + Phosphatidylinositol-binding clathrin assembly protein] selectively in Apolipoprotein E (APOE) ε4 carriers. (A) APOE ε4 risk carriers with low AD-GRS showed overall poorer EF performance and same rate of 9-year decline regardless of CA-GRS; (B) APOE ε4 risk carriers with high AD-GRS showed poorer EF performance at age 75 years and steeper 9-year decline with increasing CA-GRS; (C) APOE ε4− group with low AD-GRS did not show poorer EF performance at age 75 years or steeper 9-year decline with higher CA-GRS; (D) APOE ε4− group with high AD-GRS showed similar EF performance at age 75 years and 9-year decline for all levels in the CA-GRS.
Discussion
We examined a network approach to test AD and cognitive aging genetic effects on cognitive trajectories in preclinical aging. We tested interactive and effect modification of cognitive aging genetic risk factors [11] and clinically pertinent clusters of AD genetic risk factors [30] associated with lipid transport, inflammation, and endocytosis important in neuronal and synaptic changes in the brain. Specifically, whether an AD-GRS interacts with a CA-GRS as stratified by APOE risk status to differentially predict EF performance and change across a 40-year age band in non-demented older adults. We observed that the cumulative effect of high allelic risk in the AD-GRS magnifies the risk associated with increasing CA-GRS selectively for APOE ε4 carriers. This is the first study to show interactive effect of select AD and cognitive aging SNPs on EF trajectories as modified by APOE ε4 status. This finding advances the field on genetic and neurocognitive associations in preclinical aging by showing that network-related genetic associations may (1) lead to early detection of older adults most vulnerable or at high risk of cognitive impairment or dementia and (2) promote our understanding of the underlying genetic and molecular networks involved in preclinical cognitive impairment and potential risk of AD disease pathogenesis.
In a previous cognitive aging genetic risk panel study [11], we established a CA-GRS using COMT and BDNF. Both have been implicated in normal cognitive aging [46] and are thought to influence each other through basal ganglia-thalamocortical loops [75]. We observed poorer EF performance at centering age 75 and no significant 9-year change with increasing CA-GRS in APOE ε4 carriers. In the present study, we extended these results by identifying a complex interactive genetic network with AD-GRS and CA-GRS as modified by APOE ε4 genotype to predict EF performance and change. Our key finding is that the effect of APOE ε4 allelic risk on EF trajectories in non-demented aging is exacerbated via interactive effects of high AD-GRS and high CA-GRS. Specifically, APOE ε4 carriers in the high AD-GRS group had poorer EF performance at age 75 years and significantly steeper 9-year decline with increasing CA-GRS (Figure 1B). Compared to the high AD-GRS group, APOE ε4 carriers with low AD-GRS had poorer EF performance and were declining overall irrespective of their CA-GRS (Figure 1A). Whereas APOE ε4− group with high or low AD-GRS (Figure 1C–1D) did not show the same vulnerable pattern of poorer EF performance and steeper 9-year decline with increasing CA-GRS. Future studies focusing on the molecular underpinnings of cognitive decline and impairment should consider interactive effects of cognitive aging genes in conjunction with AD genes to detect subtle changes and intricate genetic networks effects in preclinical phenotypes.
Recent studies are starting to combine omics data into multi-scale models to examine networks associated with AD-related risk factors [12]. A recent study applied a systems biology approach to examine common pathways and molecular networks among AD related genes. Three inter-connected pathway modules (neuronal and metabolic; cell growth/survival and neuroendocrine; immunological cluster) were identified [50]. Identifying genes involved in the same pathways will contribute to our understanding of how select clusters of genes operate to influence neurocognitive phenotypes associated with cognitive impairment and AD. Although the molecular pathways underlying the genetic network in our study require detailed mechanistic studies, we propose a potential process through which the three genetic combinations (APOE, AD-GRS, CA-GRS) may work together to magnify cognitive decline in APOE ε4 carriers with the high AD-GRS and high CA-GRS. First, APOE ε4 carriers may have decreased synaptic function, axonal growth, Aβ clearance, hippocampal dendrites, cholesterol metabolism, and mitochondrial function. They also may show greater medial temporal lobe atrophy, mostly around the hippocampus, accelerated loss in cortical thickness and hippocampal volume that is correlated with cognitive decline [17]. Second, increases in CA-GRS (COMT + BDNF allelic risk panel) are associated with lower dopamine levels in the prefrontal cortex [47] or lower neurotrophic factors [48] in the hippocampus that may influence each other through the basal ganglia thalamocortical loops [75]. This implies that higher CA-GRS may lead to poorer EF performance and steeper decline in non-demented aging [11] due to lower dopamine levels or lower neurotrophic factors.
Third, the additive effect of all three polymorphisms in the AD-GRS (CLU + CR1 + PICALM allelic risk panel) and their respective pathologies may lead to magnified risk for reductions in amyloid clearance and increases in brain atrophy, both of which are typically observed in older adults with cognitive impairment or dementia. Briefly, CLU is involved in cholesterol transport and binds to Aβ. The CR1 protein plays a role in inflammation especially in AD patients. PICALM influences neurotransmitter release at presynaptic terminals [30,76]. Furthermore, CLU protein levels are also higher in the frontal cortex and hippocampus of postmortem AD patients [77]. For example, older adults with low AD-GRS may be at a decreased risk of amyloid deposition, and lower dopamine levels (COMT risk allele in the CA-GRS) may not lead to poorer EF performance in the APOE ε4− group. Whereas, in the APOE ε4+ group, lower dopamine levels may result in steeper EF decline with risk intensification from high AD-GRS. Fourth, APOE and CLU show similar physiological and functional properties (e.g., molten globule structures and differences in CLU levels in the frontal cortex among APOE ε4 carriers versus non-carriers) [34] and this may partially explain the moderation observed among APOE ε4 carriers for low and high AD-GRS. In addition, reduced APOE levels in the hippocampus and frontal cortex and increased CLU levels are associated with increasing APOE ε4 allele dose [78].
In the present study, we observed a network-based interactive effect between two sets of genetic risk scores (CA-GRS and AD-GRS) on EF trajectories selectively for APOE ε4 carriers. APOE isoforms have previously been associated with the timing, location, and amount of amyloid beta deposition and clearance in the brain parenchyma and vasculature which may trigger a chain of pathological events resulting in AD [79]. Overall, poor central nervous system functioning (for APOE ε4 carriers) as moderated by rate of amyloid clearance, efficient synaptic transmission, and amount of brain atrophy (CLU + CR1+ PICALM), may change the influence of lower dopamine levels or neurotrophic factors (COMT + BDNF) on EF performance in non-demented aging. Future research may use this and similar network approaches (e.g., [12]) to (1) test other dementia and cognitive aging polymorphisms using varying combinations of interactive, additive, and effect modifications, (2) extend the networks to include other modalities of AD biomarkers (e.g., molecular, brain), and (3) replicate our findings to identify subpopulations of specific risk profiles that may be targeted for precision clinical interventions to delay cognitive impairment. In addition, this work suggests that some future AD clinical trials may benefit from including not only APOE [80], the most prominent AD genetic risk factor, but also a selected subset of other AD risk genes (such as CLU, CR1, and PICALM) to (1) improve precision in the determination of risk for cognitive decline and dementia and (2) promote efficient and effective evaluation of intervention outcomes.
We note several limitations of the present study. First, at present the VLS database does not include all SNPs linked with either AD or cognitive aging. Some recent and important ones are not available (e.g., Bridging Integrator 1, Triggering Receptor Expressed on Myeloid Cells 2). Future studies may benefit from genetic risk scores (AD-GRS and CA-GRS) that include contributions from additional SNPs. Second, the subset of VLS data we assembled did not feature full representation of three waves for all participants. Some were in data collection samples that have not been completed. However, we used maximum likelihood to estimate missing EF factor scores. Maximum likelihood uses all available data to identify and generate population parameter estimates that have the highest probability (log likelihood) [81]. In addition, some other features of the design were corresponding strengths, including the sample size (n = 634) and the coverage of a 40-year band of non-demented aging. Third, we focused on EF, to the exclusion of other important domains of cognition. Although this was reasonable, given the domain-specificity of many genetic-cognition associations in aging, future studies should validate this approach and these results with other cognitive domains (i.e., episodic memory, speed). Fourth, the present network approach is based on genetic markers, but could be extended to multi-modal biomarkers, such as cerebrospinal fluid-based and neuroimaging markers in similar designs. Fifth, although sex as covariate did not influence our models, future studies should consider examining sex differences for differential patterns in other complex multimodal network associations with preclinical phenotypes [82]. We note two additional strengths. First, the longitudinal design and analyses featured age as the metric of change. This allowed us to incorporate chronological age directly into our analyses [11] to account for EF variability associated with age. This approach is better than if age were included as a covariate and allowed us to examine EF change across a 40-year band of aging. Second, we used four standard neuropsychological tests that contributed to one EF latent variable. The latent modeling approach is representative of the overall EF construct and accounts for measurement error commonly present with single cognitive tests.
APOE ε4 carriers who were at additional AD risk (due to higher scores on the AD-GRS) and also had higher risk for cognitive decline (indicated by the CA-GRS) performed selectively worse on an EF latent variable at the intercept (age 75) and produced steeper 9-year longitudinal decline. This result was interpreted as both biologically feasible and selective, in that it did not appear for other tested networks of genetic risk. SNPs identified in AD GWAS studies may (1) work via select clusters of additive risk scores (AD-GRS), (2) in interaction with cognitive aging risk scores (CA-GRS), and (3) as stratified by APOE ε4 status to alter preclinical cognitive aging. Future research should examine the underlying molecular pathways involved in this or similar complex genetic networks as they predict cognitive trajectories and outcomes in non-demented aging. Such network analyses may lead to precision approaches for early detection and individualized intervention programs to identify older adults at higher and lower risk for accelerated decline or dementia.
Supplementary Material
Acknowledgments
The present research is supported by grants to Roger A. Dixon from (a) the National Institutes of Health (National Institute on Aging; R01 AG008235), (b) Alberta Health Services (University Hospital Foundation) (to David Westaway, Jack Jhamandas, and Roger Dixon), and (c) the Canadian Consortium on Neurodegeneration in Aging (CCNA) (with funding from the Canadian Institutes of Health Research and partners, including SANOFI-AVENTIS R&D). RAD is also supported by the Canada Research Chairs program. SS is also supported by the CCNA/Alzheimer Society of Canada Postdoctoral Fellowship. The funding sources did not have a role in the study design, data collection, statistical analysis, results interpretation, report writing, or submission decisions. SS is now at Sunnybrook Health Sciences Centre.
We thank the volunteer participants and the VLS staff for their many contributions. More information about the VLS may be found at: http://www.ualberta.ca/~vlslab/.
Footnotes
Conflict of Interest/Disclosure Statement
The authors have no conflict of interest to report.
All research has been approved continuously by relevant institutional review boards. Certificates are available and on file in the University of Alberta Research Services Office and the US National Institutes of Health. All participants have completed and signed informed consent forms.
References
- 1.Barnes DE, Yaffe K. The projected effect of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol. 2011;10:819–828. doi: 10.1016/S1474-4422(11)70072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brookmeyer R, Johnson E, Ziegler-Graham K, Arrighi HM. Forecasting the global burden of Alzheimer’s disease. Alzheimer’s Dement. 2007;3:186–191. doi: 10.1016/j.jalz.2007.04.381. [DOI] [PubMed] [Google Scholar]
- 3.Holtzman DM, Morris JC, Goate AM. Alzheimer’s Disease: The Challenge of the Second Century. Sci Transl Med. 2011;3:77sr1–77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison-Bogorad M, Wagster MV, Phelps CH. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jack CR, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, Shaw LM, Vemuri P, Wiste HJ, Weigand SD, Lesnick TG, Pankratz VS, Donohue MC, Trojanowski JQ. Update on hypothetical model of Alzheimer’s disease biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allain P, Etcharry-Bouyx F, Verny C. Executive functions in clinical and preclinical Alzheimer’s disease. Revue Neurologique. 2013:695–708. doi: 10.1016/j.neurol.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 7.Bekris LM, Yu C-E, Bird TD, Tsuang DW. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol. 2010;23:213–27. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of Genes and Environments for Explaining Alzheimer Disease. Arch Gen Psychiatry. 2006;63:168. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 9.Jun G, Naj AC, Beecham GW, Wang L-S, Buros J, Gallins PJ, Buxbaum JD, Ertekin-Taner N, Fallin MD, Friedland R, Inzelberg R, Kramer P, Rogaeva E, St George-Hyslop P, Cantwell LB, Dombroski BA, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Lunetta KL, Martin ER, Montine TJ, Goate AM, Blacker D, Tsuang DW, Beekly D, Cupples LA, Hakonarson H, Kukull W, Foroud TM, Haines J, Mayeux R, Farrer LA, Pericak-Vance MA, Schellenberg GD Alzheimer’s Disease Genetics Consortium. Meta-analysis Confirms CR1, CLU, and PICALM as Alzheimer Disease Risk Loci and Reveals Interactions With APOE Genotypes. Arch Neurol. 2010;67:1473. doi: 10.1001/archneurol.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McFall GP, Sapkota S, McDermott KL, Dixon RA. Risk-reducing Apolipoprotein E and Clusterin genotypes protect against the consequences of poor vascular health on executive function performance and change in nondemented older adults. Neurobiol Aging. 2016;42:91–100. doi: 10.1016/j.neurobiolaging.2016.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sapkota S, Bäckman L, Dixon RA. Executive function performance and change in aging is predicted by apolipoprotein E, intensified by catechol-O-methyltransferase and brain-derived neurotrophic factor, and moderated by age and lifestyle. Neurobiol Aging. 2017;52:81–89. doi: 10.1016/j.neurobiolaging.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaiteri C, Mostafavi S, Honey CJ, De Jager PL, Bennett DA. Genetic variants in Alzheimer disease — molecular and brain network approaches. Nat Publ Gr. 2016;12:413–427. doi: 10.1038/nrneurol.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Frias CM, Dixon RA, Strauss E. Structure of four executive functioning tests in healthy older adults. Neuropsychology. 2006;20:206–214. doi: 10.1037/0894-4105.20.2.206. [DOI] [PubMed] [Google Scholar]
- 15.Luszcz M. Executive function and cognitive aging. In: Schaie K, Willis S, editors. Handbook of the psychology of aging. Elsevier, Inc; 2011. pp. 59–72. [Google Scholar]
- 16.McFall GP, Sapkota S, Thibeau S, Dixon RA. Executive function in normal aging and neurodegenerative diseases: Trajectories and modifiers. In: Wiebe SA, Karbach J, editors. Executive Function: Development Across the Life Span. Taylor & Francis; Oxford, UK: 2017. p. 296. [Google Scholar]
- 17.Liu C-C, Liu C-C, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–18. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–72. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 19.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–3. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 20.Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S. Apolipoprotein E polymorphism and Alzheimer’s disease. Lancet (London, England) 1993;342:697–9. doi: 10.1016/0140-6736(93)91705-q. [DOI] [PubMed] [Google Scholar]
- 21.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schürmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frölich L, Hampel H, Hüll M, Rujescu D, Goate AM, Kauwe JSK, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Mühleisen TW, Nöthen MM, Moebus S, Jöckel K-H, Klopp N, Wichmann H-E, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambert J-C, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fiévet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossù P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanché H, Dartigues J-F, Tzourio C, Gut I, Van Broeckhoven C, Alpérovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 23.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–56. [PubMed] [Google Scholar]
- 24.Caselli RJ, Dueck AC, Locke DEC, Hoffman-Snyder CR, Woodruff BK, Rapcsak SZ, Reiman EM. Longitudinal modeling of frontal cognition in APOE ε4 homozygotes, heterozygotes, and noncarriers. Neurology. 2011;76:1383–8. doi: 10.1212/WNL.0b013e3182167147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vivot A, Glymour MM, Tzourio C, Amouyel P, Ch?ne G, Dufouil C. Association of Alzheimer?s related genotypes with cognitive decline in multiple domains: results from the Three-City Dijon study. Mol Psychiatry. 2015;20:1173–1178. doi: 10.1038/mp.2015.62. [DOI] [PubMed] [Google Scholar]
- 26.Bertram L, Tanzi RE. Genome-wide association studies in Alzheimer’s disease. Hum Mol Genet. 2009;18:R137–45. doi: 10.1093/hmg/ddp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff-Radford NR, Petersen RC, Morgan K, Younkin SG. Replication of CLU, CR1, and PICALM Associations With Alzheimer Disease. Arch Neurol. 2010;67:2687–2702. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corneveaux JJ, Myers AJ, Allen AN, Pruzin JJ, Ramirez M, Engel A, Nalls MA, Chen K, Lee W, Chewning K, Villa SE, Meechoovet HB, Gerber JD, Frost D, Benson HL, O’Reilly S, Chibnik LB, Shulman JM, Singleton AB, Craig DW, Van Keuren-Jensen KR, Dunckley T, Bennett DA, De Jager PL, Heward C, Hardy J, Reiman EM, Huentelman MJ. Association of CR1, CLU and PICALM with Alzheimer’s disease in a cohort of clinically characterized and neuropathologically verified individuals. Hum Mol Genet. 2010;19:3295–3301. doi: 10.1093/hmg/ddq221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen LH, Ping Kao PY, Fan YH, Yin Ho DT, Chan SY, Yip PY, Tung Ha JC, Chu LW, Song YQ. Polymorphisms of CR1, CLU and PICALM Confer Susceptibility of Alzheimer?s Disease in Southern Chinese Population. Alzheimer’s Dement. 2011;7:S181. doi: 10.1016/j.neurobiolaging.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Karch CM, Goate AM. Alzheimer’s Disease Risk Genes and Mechanisms of Disease Pathogenesis. Biol Psychiatry. 2015;77:43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calero M, Rostagno A, Matsubara E, Zlokovic B, Frangione B, Ghiso J. Apolipoprotein J (clusterin) and Alzheimer’s disease. Microsc Res Tech. 2000;50:305–315. doi: 10.1002/1097-0029(20000815)50:4<305::AID-JEMT10>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 32.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 33.Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW, Thompson PM. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci. 2011;31:6764–70. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu ZC, Yu JT, Li Y, Tan L. Clusterin in Alzheimer’s disease. Adv Clin Chem. 2012;56:155–173. doi: 10.1016/b978-0-12-394317-0.00011-x. [DOI] [PubMed] [Google Scholar]
- 35.Sapkota S, Wiebe SA, Small BJ, Dixon RA. Apolipoprotein e and Clusterin can magnify effects of personality vulnerability on declarative memory performance in non-demented older adults. Int J Geriatr Psychiatry. 2016;31:502–509. doi: 10.1002/gps.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khera R, Das N. Complement Receptor 1: Disease associations and therapeutic implications. Mol Immunol. 2009;46:761–772. doi: 10.1016/j.molimm.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crehan H, Holton P, Wray S, Pocock J, Guerreiro R, Hardy J. Complement receptor 1 (CR1) and Alzheimer’s disease. Immunobiology. 2012;217:244–250. doi: 10.1016/j.imbio.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 38.Xiao Q, Gil S-C, Yan P, Wang Y, Han S, Gonzales E, Perez R, Cirrito JR, Lee J-M. Role of Phosphatidylinositol Clathrin Assembly Lymphoid-Myeloid Leukemia (PICALM) in Intracellular Amyloid Precursor Protein (APP) Processing and Amyloid Plaque Pathogenesis. J Biol Chem. 2012;287:21279–21289. doi: 10.1074/jbc.M111.338376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schjeide B-MM, Schnack C, Lambert J-C, Lill CM, Kirchheiner J, Tumani H, Otto M, Tanzi RE, Lehrach H, Amouyel P, von Arnim CAF, Bertram L. The Role of Clusterin, Complement Receptor 1, and Phosphatidylinositol Binding Clathrin Assembly Protein in Alzheimer Disease Risk and Cerebrospinal Fluid Biomarker Levels. Arch Gen Psychiatry. 2011;68:207. doi: 10.1001/archgenpsychiatry.2010.196. [DOI] [PubMed] [Google Scholar]
- 40.Morgen K, Ramirez A, Frölich L, Tost H, Plichta MM, Kölsch H, Rakebrandt F, Rienhoff O, Jessen F, Peters O, Jahn H, Luckhaus C, Hüll M, Gertz H, Schröder J, Hampel H, Teipel SJ, Pantel J, Heuser I, Wiltfang J, Rüther E, Kornhuber J, Maier W, Meyer-Lindenberg A. Genetic interaction of PICALM and APOE is associated with brain atrophy and cognitive impairment in Alzheimer’s disease. Alzheimers Dement. 2014;10:1–8. doi: 10.1016/j.jalz.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Barral S, Bird T, Goate A, Farlow MR, Diaz-Arrastia R, Bennett DA, Graff-Radford N, Boeve BF, Sweet RA, Stern Y, Wilson RS, Foroud T, Ott J, Mayeux R. Genotype patterns at PICALM, CR1, BIN1, CLU, and APOE genes are associated with episodic memory. Neurology. 2012;78:1464–1471. doi: 10.1212/WNL.0b013e3182553c48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thambisetty M, An Y, Nalls M, Sojkova J, Swaminathan S, Zhou Y, Singleton AB, Wong DF, Ferrucci L, Saykin AJ, Resnick SM. Effect of complement CR1 on brain amyloid burden during aging and its modification by APOE genotype. Biol Psychiatry. 2013;73:422–428. doi: 10.1016/j.biopsych.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chibnik LB, Shulman JM, Leurgans SE, Schneider JA, Wilson RS, Tran D, Aubin C, Buchman AS, Heward CB, Myers AJ, Hardy JA, Huentelman MJ, Corneveaux JJ, Reiman EM, Evans DA, Bennett DA, De Jager PL. CR1 is associated with amyloid plaque burden and age-related cognitive decline. Ann Neurol. 2011;69:560–569. doi: 10.1002/ana.22277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris SE, Deary IJ. The genetics of cognitive ability and cognitive ageing in healthy older people. Trends Cogn Sci. 2011;15:388–94. doi: 10.1016/j.tics.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 45.Mandelman SD, Grigorenko EL. BDNF Val66Met and cognition: all, none, or some? A meta-analysis of the genetic association. Genes, Brain Behav. 2012;11:127–136. doi: 10.1111/j.1601-183X.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagel IE, Chicherio C, Li S-C, von Oertzen T, Sander T, Villringer A, Heekeren HR, Bäckman L, Lindenberger U. Human aging magnifies genetic effects on executive functioning and working memory. Front Hum Neurosci. 2008;2:1. doi: 10.3389/neuro.09.001.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bilder RM, Volavka J, Lachman HM, Grace AA. The Catechol-O-Methyltransferase Polymorphism: Relations to the Tonic? Phasic Dopamine Hypothesis and Neuropsychiatric Phenotypes. Neuropsychopharmacology. 2004;29:1943–1961. doi: 10.1038/sj.npp.1300542. [DOI] [PubMed] [Google Scholar]
- 48.Savitz J, Solms M, Ramesar R. The molecular genetics of cognition: dopamine, COMT and BDNF. Genes, Brain Behav. 2006;5:311–328. doi: 10.1111/j.1601-183X.2005.00163.x. [DOI] [PubMed] [Google Scholar]
- 49.Sapkota S, Vergote D, Westaway D, Jhamandas J, Dixon RA. Synergistic associations of catechol-O-methyltransferase and brain-derived neurotrophic factor with executive function in agingare selective and modified by apolipoprotein E. Neurobiol Aging. 2015;36:249–256. doi: 10.1016/j.neurobiolaging.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Y, Xin J, Hu Y, Zhang L, Wang J. Analyzing the genes related to Alzheimer ‘ s disease via a network and pathway-based approach. 2017:1–15. doi: 10.1186/s13195-017-0252-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, Zhang C, Xie T, Tran L, Dobrin R, Fluder E, Clurman B, Melquist S, Narayanan M, Suver C, Shah H, Mahajan M, Gillis T, Mysore J, MacDonald ME, Lamb JR, Bennett DA, Molony C, Stone DJ, Gudnason V, Myers AJ, Schadt EE, Neumann H, Zhu J, Emilsson V. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song A, Yan J, Kim S, Risacher SL, Wong AK, Saykin AJ, Shen L, Greene CS. Network-based analysis of genetic variants associated with hippocampal volume in Alzheimer’s disease: a study of ADNI cohorts. BioData Min. 2016;9:3. doi: 10.1186/s13040-016-0082-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McFall GP, Wiebe SA, Vergote D, Anstey KJ, Dixon RA. Alzheimer’s genetic risk intensifies neurocognitive slowing associated with diabetes in nondemented older adults. Alzheimer’s Dement Diagnosis, Assess Dis Monit. 2015;1:395–402. doi: 10.1016/j.dadm.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez-Rodriguez E, Sanchez-Juan P, Vazquez-Higuera JL, Mateo I, Pozueta A, Berciano J, Cervantes S, Alcolea D, Mart?nez-Lage P, Clarim?n J, Lle? A, Pastor P, Combarros O. Genetic risk score predicting accelerated progression from mild cognitive impairment to Alzheimer’s disease. J Neural Transm. 2013;120:807–812. doi: 10.1007/s00702-012-0920-x. [DOI] [PubMed] [Google Scholar]
- 55.Wollam ME, Weinstein AM, Saxton JA, Morrow L, Snitz B, Fowler NR, Suever Erickson BL, Roecklein KA, Erickson KI. Genetic Risk Score Predicts Late-Life Cognitive Impairment. J Aging Res. 2015;2015:1–8. doi: 10.1155/2015/267062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chouraki V, Reitz C, Maury F, Bis JC, Bellenguez C, Yu L, Jakobsdottir J, Mukherjee S, Adams HH, Choi SH, Larson EB, Fitzpatrick A, Uitterlinden AG, de Jager PL, Hofman A, Gudnason V, Vardarajan B, Ibrahim-Verbaas C, van der Lee SJ, Lopez O, Dartigues J-F, Berr C, Amouyel P, Bennett DA, van Duijn C, DeStefano AL, Launer LJ, Ikram MA, Crane PK, Lambert J-C, Mayeux R, Seshadri S International Genomics of Alzheimer’s Project. Evaluation of a Genetic Risk Score to Improve Risk Prediction for Alzheimer?s Disease. J Alzheimer’s Dis. 2016;53:921–932. doi: 10.3233/JAD-150749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lacour A, Espinosa A, Louwersheimer E, Heilmann S, Hernández I, Wolfsgruber S, Fernández V, Wagner H, Rosende-Roca M, Mauleón A, Moreno-Grau S, Vargas L, Pijnenburg YAL, Koene T, Rodríguez-Gómez O, Ortega G, Ruiz S, Holstege H, Sotolongo-Grau O, Kornhuber J, Peters O, Frölich L, Hüll M, Rüther E, Wiltfang J, Scherer M, Riedel-Heller S, Alegret M, Nöthen MM, Scheltens P, Wagner M, Tárraga L, Jessen F, Boada M, Maier W, van der Flier WM, Becker T, Ramirez A, Ruiz A. Genome-wide significant risk factors for Alzheimer’s disease: role in progression to dementia due to Alzheimer’s disease among subjects with mild cognitive impairment. Mol Psychiatry. 2017;22:153–160. doi: 10.1038/mp.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Adams HHH, de Bruijn RFAG, Hofman A, Uitterlinden AG, van Duijn CM, Vernooij MW, Koudstaal PJ, Ikram MA. Genetic risk of neurodegenerative diseases is associated with mild cognitive impairment and conversion to dementia. Alzheimer’s Dement. 2015;11:1277–1285. doi: 10.1016/j.jalz.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 59.Sleegers K, Bettens K, De Roeck A, Van Cauwenberghe C, Cuyvers E, Verheijen J, Struyfs H, Van Dongen J, Vermeulen S, Engelborghs S, Vandenbulcke M, Vandenberghe R, De Deyn PP, Van Broeckhoven C. A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid A?42. Alzheimer’s Dement. 2015;11:1452–1460. doi: 10.1016/j.jalz.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 60.Andrews SJ, Eramudugolla R, Velez JI, Cherbuin N, Easteal S, Anstey KJ. Validating the role of the Australian National University Alzheimer’s Disease Risk Index (ANU-ADRI) and a genetic risk score in progression to cognitive impairment in a population-based cohort of older adults followed for 12 years. Alzheimers Res Ther. 2017;9:16. doi: 10.1186/s13195-017-0240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, Morgan K, Passmore P, Holmes C, Powell J, Brayne C, Gill M, Mead S, Goate A, Cruchaga C, Lambert J-C, van Duijn C, Maier W, Ramirez A, Holmans P, Jones L, Hardy J, Seshadri S, Schellenberg GD, Amouyel P, Williams J, Amouyel P, Williams J. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain. 2015;138:3673–3684. doi: 10.1093/brain/awv268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marioni RE, Campbell A, Hagenaars SP, Nagy R, Amador C, Hayward C, Porteous DJ, Visscher PM, Deary IJ. Genetic Stratification to Identify Risk Groups for Alzheimer’s Disease. J Alzheimer’s Dis. 2017;57:275–283. doi: 10.3233/JAD-161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferencz B, Laukka EJ, Welmer A-K, Kalpouzos G, Angleman S, Keller L, Graff C, Lövdén M, Bäckman L. The benefits of staying active in old age: Physical activity counteracts the negative influence of PICALM, BIN1, and CLU risk alleles on episodic memory functioning. Psychol Aging. 2014;29:440–9. doi: 10.1037/a0035465. [DOI] [PubMed] [Google Scholar]
- 64.Dixon Ra, De Frias CM. The Victoria Longitudinal Study: From Characterizing Cognitive Aging to Illustrating Changes in Memory Compensation. Aging, Neuropsychol Cogn. 2004;11:346–376. [Google Scholar]
- 65.Bielak AAM, Mansueti L, Strauss E, Dixon RA. Performance on the Hayling and Brixton tests in older adults: Norms and correlates. Arch Clin Neuropsychol. 2006;21:141–149. doi: 10.1016/j.acn.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 66.McFall GP, Wiebe SA, Vergote D, Jhamandas J, Westaway D, Dixon Ra. IDE (rs6583817) polymorphism and pulse pressure are independently and interactively associated with level and change in executive function in older adults. Psychol Aging. 2014;29:418–430. doi: 10.1037/a0034656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burgess P, Shallice T. The Hayling and Brixton Tests. Thames Val. Test Company; Bury St. Edmunds, U.K: 1997. [Google Scholar]
- 68.Taylor SF, Kornblum S, Lauber EJ, Minoshima S, Koeppe RA. Isolation of Specific Interference Processing in the Stroop Task: PET Activation Studies. Neuroimage. 1997;6:81–92. doi: 10.1006/nimg.1997.0285. [DOI] [PubMed] [Google Scholar]
- 69.D’elia L, Satz P, Uchiyama C, Uchiyama C, White T. Color Trails Test Professional Manual. Florida: Psychological Assessment Resources; 1996. [Google Scholar]
- 70.Muthén L, Muthén B. Mplus user’s guide. 7. Muthén, L Muthén, B; Los Angeles, CA: 1998. [Google Scholar]
- 71.Small BJ, Dixon RA, McArdle JJ. Tracking Cognition-Health Changes From 55 to 95 Years of Age. Journals Gerontol Ser B Psychol Sci Soc Sci. 2011;66B:i153–i161. doi: 10.1093/geronb/gbq093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harrison TM, Mahmood Z, Lau EP, Karacozoff AM, Burggren AC, Small GW, Bookheimer SY. An Alzheimers Disease Genetic Risk Score Predicts Longitudinal Thinning of Hippocampal Complex Subregions in Healthy Older Adults. eNeuro. 2016:3. doi: 10.1523/ENEURO.0098-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Altmann A, Tian L, Henderson VW, Greicius MD. Sex modifies the APOE -related risk of developing Alzheimer disease. Ann Neurol. 2014;75:563–573. doi: 10.1002/ana.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kline RB. Principles and practice of structural equation modeling. Guilford Press; New York, NY: 2010. [Google Scholar]
- 75.Alexander GE, DeLong MR, Strick PL. Parallel Organization of Functionally Segregated Circuits Linking Basal Ganglia and Cortex. Annu Rev Neurosci. 1986;9:357–381. doi: 10.1146/annurev.ne.09.030186.002041. [DOI] [PubMed] [Google Scholar]
- 76.Ferrari R, Moreno JH, Minhajuddin AT, O’Bryant SE, Reisch JS, Barber RC, Momeni P. Implication of common and disease specific variants in CLU, CR1, and PICALM. Neurobiol Aging. 2012;33:1846.e7–1846.e18. doi: 10.1016/j.neurobiolaging.2012.01.110. [DOI] [PubMed] [Google Scholar]
- 77.Yu J-T, Tan L. The Role of Clusterin in Alzheimer?s Disease: Pathways, Pathogenesis, and Therapy. Mol Neurobiol. 2012;45:314–326. doi: 10.1007/s12035-012-8237-1. [DOI] [PubMed] [Google Scholar]
- 78.Bertrand P, Poirier J, Oda T, Finch CE, Pasinetti GM. Association of apolipoprotein E genotype with brain levels of apolipoprotein E and apolipoprotein J (clusterin) in Alzheimer disease. Mol Brain Res. 1995;33:174–178. doi: 10.1016/0169-328x(95)00097-c. [DOI] [PubMed] [Google Scholar]
- 79.Kim J, Basak JM, Holtzman DM. The Role of Apolipoprotein E in Alzheimer’s Disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kennedy RE, Cutter GR, Schneider LS. Effect of APOE genotype status on targeted clinical trials outcomes and efficiency in dementia and mild cognitive impairment resulting from Alzheimer’s disease. Alzheimer’s Dement. 2014;10:349–359. doi: 10.1016/j.jalz.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Enders CK. Analyzing longitudinal data with missing values. Rehabil Psychol. 2011;56:267–288. doi: 10.1037/a0025579. [DOI] [PubMed] [Google Scholar]
- 82.Koran MEI, Wagener M, Hohman TJ. Sex differences in the association between AD biomarkers and cognitive decline. Brain Imaging Behav. 2017;11:205–213. doi: 10.1007/s11682-016-9523-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.