Summary
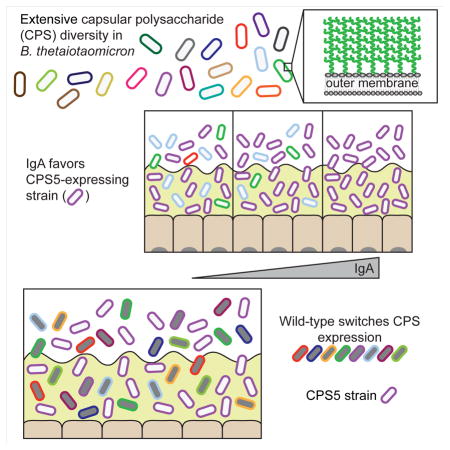
Capsular polysaccharides (CPS) play multiple roles in protecting bacteria from host and environmental factors, and many commensal bacteria can produce multiple capsule types. To better understand the roles of different CPS in competitive intestinal colonization, we individually expressed the eight different capsules of the human gut symbiont Bacteroides thetaiotaomicron. Certain CPS were most advantageous in vivo, and increased anti-CPS IgA correlated with increased fitness of a strain expressing one particular capsule CPS5, suggesting that it promotes avoidance of adaptive immunity. A strain with the ability to switch between multiple capsules was more competitive than those expressing any single capsule except CPS5. After antibiotic perturbation, only the wild-type, capsule-switching strain remained in the gut, shifting to prominent expression of CPS5 only in mice with intact adaptive immunity. These data suggest that different capsules equip mutualistic gut bacteria with the ability to thrive in various niches, including those influenced by immune responses and antibiotic perturbations.
eTOC Blurb
Symbiotic Bacteroides species encode for multiple capsular polysaccharides, the roles of which have remained poorly defined. Porter et al. identify one capsule involved in competitive colonization of the mouse gut that assists in evading adaptive immune responses. The genes for synthesizing this capsule are frequently identified in human gut samples.
Introduction
The microbial community (microbiota) residing in the human intestine encodes many functions that are not represented in the human genome. Some of these functions complement physiological deficits of humans, such as our inability to digest dietary fiber polysaccharides (Kaoutari et al., 2013). Other commonly observed functions serve less defined roles in shaping the membership and physiology of the microbiota. One example is the propensity of many individual human gut bacteria to synthesize a variety of different capsular polysaccharides (CPS) on their surface (Coyne and Comstock, 2008). The ability to cover their cell surfaces and even vary these coatings could protect bacteria from host immune responses (Peterson et al., 2007), other bacteria, bacteriophage or even allow them to alter host physiology (Mazmanian et al., 2005).
Bacteroides thetaiotaomicron (B. theta) is a model gut symbiont that degrades a wide variety of dietary, host and microbial glycans (Martens et al., 2011). The type strain of this species also dedicates a substantial portion of its genome (182 genes contained in 8 distinct loci) to CPS production, a typical phenomenon so far observed among members of the Bacteroides and Parabacteroides genera (Coyne and Comstock, 2008). These loci may vary widely between strains of the same species (Patrick et al., 2010), partly due to lateral gene transfer or other events (Martens et al., 2014). The Bacteroides have complex mechanisms for regulating expression of CPS loci, including invertible promoters that are recombinationally flipped between “on” and “off” positions (Coyne et al., 2003; Hickey et al., 2015), NusG-like antitermination factors (Chatzidaki-Livanis et al., 2009), and trans locus inhibitors (Chatzidaki-Livanis et al., 2010). It has been hypothesized that this regulation allows for one CPS locus to be expressed by an individual cell (Chatzidaki-Livanis et al., 2010), while equipping cells in the greater population with the ability to explore expression of different capsules. Such a strategy could pre-adapt subpopulations of B. theta and related symbionts to better compete in environmental conditions for which specific CPS are most advantageous.
There are multiple roles that CPS could play in the fitness of intestinal symbiotic microbes (reviewed in Porter and Martens, 2017). First, bacteria may optimize cell energy expenditures by synthesizing CPS similar in composition to sugars that the bacterium is metabolizing. Alternatively, as the capsule may be hundreds of nanometers thick (Martens et al., 2009) and acts as a barrier to acquisition of some nutrients (Cameron et al., 2014), conditional expression of some CPS may facilitate passage of various polysaccharides to the cell surface. In support of this hypothesis, B. theta CPS loci have previously been found to be coordinately regulated with genes involved in the degradation of some host glycans (Martens et al., 2009).
In addition to roles in nutrient acquisition, CPS may benefit gut mutualists in similar ways as they do pathogens, including: evasion of the complement system and/or phagocytosis (Avery and Dubos, 1931; Cunnion et al., 2003), shielding of the bacterium from detection through molecular mimicry (Cress et al., 2014), or directly modulating the immune system (Mazmanian et al., 2005). Indeed each of these roles has gained support in studies of just a few gut Bacteroides: B. fragilis CPS provide complement resistance (Coyne et al., 2008); CPS in B. fragilis and other species display molecular mimicry of fucosylated host glycoconjugates (Coyne et al., 2005); and zwitterionic CPS from B. fragilis and others modulates host cytokine levels (Neff et al., 2016; Round et al., 2011). Finally, production of antibodies against a dominantly expressed B. theta CPS (CPS4) results in down-regulation of CPS4 and switching to other CPS (Peterson et al., 2007), suggesting that these bacterial capsules are targeted by adaptive immunity and the ability to dynamically express alternatives is an advantage in evading this response.
Taken together, the ability to vary CPS may enable gut bacteria to evade host immune responses. However, since no collection of CPS from symbiotic gut bacteria have been systematically studied in a single species, it is unclear what general roles these prominent cellular features play, especially in the highly abundant gut-dwelling Bacteroides. In this study, we measured the contributions of individual CPS towards enabling B. theta to compete in the mammalian gut in the face of various selective pressures, including diet, changing immune responses, and antibiotic pressure. We reveal that specific CPS provide advantages in the gut environment, and that the abundance of strains expressing individual capsules can be shaped by adaptive immunity. Moreover, we determined that the ability of B. theta to dynamically express multiple CPS provides an advantage over expression of any single CPS, although antibiotic perturbation is needed to reveal this competitive phenotype. This study shows the importance of capsules within the quotidian functions of a single commensal bacterium and highlights the importance of these massively diversified structures in equipping bacteria for colonization of the mammalian intestinal tract.
Results
Long-term colonization of mice with B. theta reveals dynamic expression of CPS over time
Experiments with the well-studied type strain of Bacteroides thetaiotaomicron (B. theta) have shown that it varies expression of its 8 different CPS in different in vivo conditions (summarized in Figure S1G). A limitation of these experiments is that they were short-term colonizations of gnotobiotic mice, providing only a “snapshot” of CPS locus expression at a single time point generally 10–30 days after inoculation or diet change. Longer-term colonization of mice could select for different CPS than those dominantly expressed in vitro or may reveal dynamic changes in locus expression. Thus, we inoculated B. theta into C57BL/6 (herein, referred to as wild-type) germ-free mice and measured transcriptional changes in CPS expression over a period of approximately 5 months (Figure 1; combined data in Figure S1A–C). To determine if dietary conditions (Sonnenburg et al., 2005) or adaptive immunity (Peterson et al., 2007) play a role in long-term CPS expression, we also periodically fed one group of wild-type mice a low-fiber diet and colonized Rag1−/− mice (adaptive immune-deficient, fed a high-fiber diet).
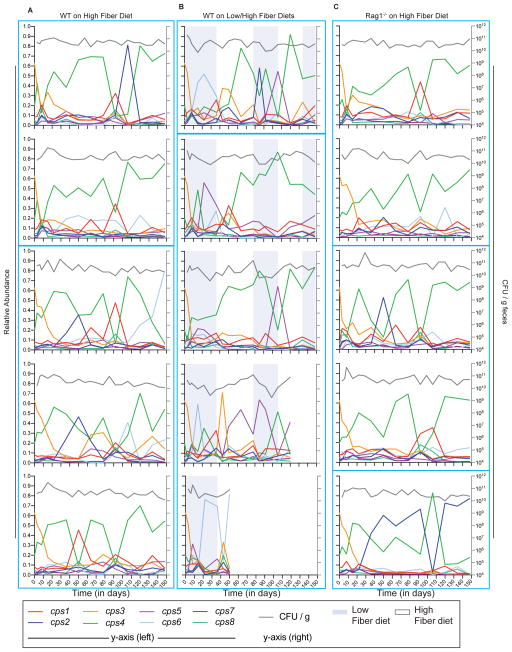
Figure 1.
B. theta dynamically alters expression of its cps loci over time in the mouse gut. Wild-type (WT) B. theta was inoculated into one of 3 groups (n=5 mice/group): A) wild-type mice fed a high-fiber diet; B) wild-type mice switched between low-fiber and high-fiber diets; or C) Rag1−/− mice fed a high-fiber diet. Shown are relative cps locus expression in RNA extracted from stool (left y-axis) and colony forming units per gram of stool (CFU/g) (right y-axis). Each panel represents an individual mouse. Blue boxes indicate groups of co-housed mice. Expression of the inoculum is represented as “Day 0”. Two mice in panel B died prematurely. See also Figure S1.
Quantitative PCR (qPCR)-based measurements of fecal bacterial community cps locus transcription at regular intervals revealed that CPS expression quickly changed from the inoculum, which was enriched for cps3 transcription (shown at day 0). However, expression proceeded to change dynamically in individual mice over time, ultimately exhibiting a unique profile of temporal expression of different CPS for each mouse (Figure 1). Dynamic changes in CPS expression occurred on both high- and low-fiber diets as well as in adaptive immune-deficient mice, suggesting that these factors were not exclusively responsible for driving expression changes. As previously observed in shorter-term experiments (Figure S1G), expression of CPS4 was most often highest in wild-type and Rag1−/− mice fed a high-fiber diet. However, there were periodic decreases in CPS4 expression (up to ~62% total change) that did not always coincide with changes in total B. theta abundance. Additionally, some decreases spanned multiple time points, collectively suggesting that these periodic events truly reflect large-scale changes in community capsule expression. CPS1 and CPS2, and to a far lesser degree CPS6, were the capsules that were expressed antagonistically with CPS4 when mice were fed a high-fiber diet. However, in most cases in which CPS4 was dominantly expressed prior to another capsule, the community resumed expression of CPS4 within 40 or fewer days. Interestingly, bacteria in stool taken from mice on the day of sacrifice could express a different CPS than in the cecum of the same mouse (Figure S1D–F).
Compared to wild-type or Rag1−/− mice fed a high-fiber diet, those fed a low-fiber diet exhibited different CPS expression. Consistent with previous studies of B. theta-colonized mice consuming low fiber (Bjursell et al., 2006; Sonnenburg et al., 2005), 4 of 5 mice showed dominant expression of CPS5 and/or CPS6 at some time during 35 days of low-fiber feeding and this trend was never observed in the same time frame for the high fiber-fed mice. As the low-fiber diet eventually leads to illness in some B. theta-monoassociated animals, we switched the low-fiber diet group to the high-fiber diet at day 35. Immediately following the diet switch, CPS3 expression increased transiently in most animals, but then gave way to a different dominant capsule (CPS4, CPS5 or CPS6). Additional diet switches may have triggered changes in CPS expression, although the occurrence of these is difficult to attribute to diet change alone given the different responses observed and the dynamic expression in constant feeding of the high-fiber diet. We utilized Dirichlet regression to more quantitatively compare changes in relative cps expression among the 3 experimental groups (see STAR Methods). While there is great variation among mice even within experimental groups, possibly masking some transcriptional changes, expression of CPS2 was found to be significantly lower when mice were fed a low-fiber diet (p = 0.0144). Significant decreases in CPS4 (p < 0.0001) and significant increases in CPS6 (p = 0.0002) were also observed in low-fiber fed mice but these changes were only associated with the first period of low-fiber feeding. Taken together, these changes in CPS expression suggest that specific capsules may be advantageous in vivo and that the corresponding selective forces are dynamic and influenced by diet.
Competition of single CPS-expressing strains reveals differential success in the mouse gut
To better understand the role of individual CPS in the fitness of gut commensals, without the influence of the complex molecular regulation that is known to govern Bacteroides cps gene expression (Chatzidaki-Livanis et al., 2010), we employed a series of B. theta variants that each encodes and expresses one of its 8 CPS biosynthetic loci (cps loci) (Hickey et al., 2015; see STAR Methods for details on strain generation and validation). These strains allowed us to isolate the role of individual CPS in subsequent experiments. We first sought to test the hypothesis that expression of distinct CPS would be advantageous in specific immunological environments. We pooled the 8 single CPS-expressing strains, along with an acapsular strain, and inoculated these into individually housed germ-free mice of one of three genotypes: wild-type mice, mice with a deficient adaptive immune response (Rag1−/−), and mice with deficiencies in innate immune signaling (MyD88−/−/Trif−/−). All mice were fed a standard high-fiber diet, and relative levels of each strain in stool over time were determined via qPCR.
Although all strains were present in the inoculum at relatively equal levels, 3 strains outcompeted the others within a few days after inoculation (Figure 2 for average strain abundance over time; Figure S4 for individual mouse data). The strain expressing CPS5 (“cps5 strain”) predominated in the stool of 4 of the 5 wild-type mice, with the cps4 and cps6 strains found at lower levels or outcompeted by the end of the experiment. In one wild-type mouse, the cps5 and cps6 strains appeared to be codominant. However, in adaptive immune deficient (Rag1−/−) mice, a more stable competition occurred over time that reflected both higher abundance of the cps6 and cps4 strains at most time points compared to wild-type mice and less propensity for these strains to diminish at later time points, especially cps4 in some individual mice (Figure S4). Specifically, while the cps5 strain was most abundant in four of the five Rag1−/− mice by the end of the time course, the cps4 and cps6 strains persisted in all mice and maintained higher abundance over time, with a statistically significant lower abundance of the cps5 strain over the first 40 days. Even though they were still outcompeted, several other strains (cps1, cps2, cps3, and cps7) also maintained significantly higher levels in Rag1−/− mice in the first 20 days. This was especially apparent in one mouse, where the cps1 strain maintained a persistent population for several weeks before dropping below the limit of detection. The less stringent dominance of the cps5 strain in Rag1−/− mice reinforces that adaptive immune responses play a role in CPS-mediated fitness (Peterson et al., 2007) and suggests that CPS5 could be more resilient under these conditions.
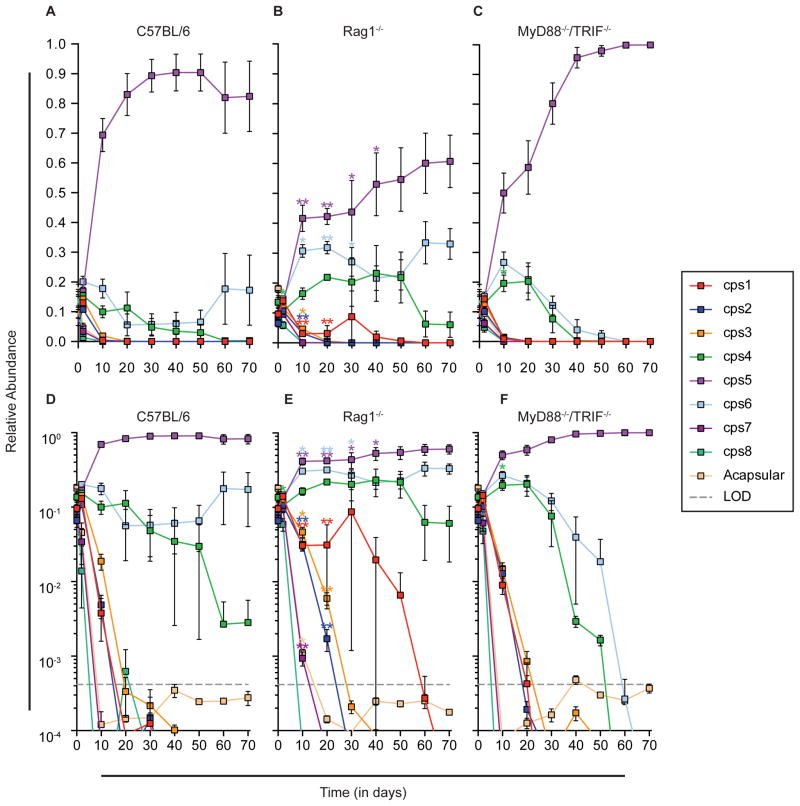
Figure 2.
Competition of single CPS-expressing strains in germ-free mice reveals distinct advantages conferred by specific CPS. Eight single CPS-expressing strains and an acapsular strain were pooled and inoculated into germ-free mice of 3 genotypes (n=5 mice/group): A, D) wild-type; B, E) MyD88−/−/Trif−/−; C, F) Rag1−/−. Panels A–C include a linear axis for relative abundance, whereas panels D–F use a logarithmic axis to reveal strains that have fallen below the limit of detection. All mice were fed a high-fiber diet. Relative abundance of each strain in stool was determined at regular intervals via qPCR. The relative abundance in the inoculum is represented as “Day 0”. Each mouse was individually caged for this experiment. Data are represented as mean ± SEM. Asterisks (in matching colors to bacterial strain) indicate significant differences (* p < .05, ** p < .01; Kruskal-Wallis test, with Benjamini-Hochberg correction) in relative strain abundance between wild-type mice and the mutant mouse genotype shown in that panel at each time point. See also Figures S2–6 and Table S3. LOD: Limit of detection.
Compared to Rag1−/− mice, an opposite trend was observed in MyD88−/−/Trif−/− mice (deficient in innate immunity). In all 5 mice, the cps5 strain became the most dominant strain by day 10. While the cps4 and cps6 strains maintained detectable populations for several weeks in individual mice, by the end of the experiment, the cps5 strain was the only strain remaining, with all other strains (including the cps6 and cps4 strains) outcompeted below the limit of detection. Despite the differences in strain abundance, all 3 groups of mice had similar bacterial load at the end of the experiment (Figure S4F). These data further suggest that CPS5 provides an advantage to B. theta in vivo, while deficiency in innate immunity provokes a more fierce competition between colonizing strains.
Besides our genome sequencing data demonstrating few mutations among the strains used in this study (Table S2), we sought to rule out secondary mutations by repeating the competition experiment described above with a newly constructed cps5 strain. Mirroring our previous results, the rederived cps5 strain outcompeted all other strains in two of three mice, and was codominant with the cps6 strain in the last mouse (Figure S4E). While some variation existed among the 4 replicates of this competition in wild-type mice (Figures 2, S4D–E, and S5D), including greater persistence of the cps2 strain over the cps4 strain in some cases, combined data from wild-type mice similarly exhibited significant differences in cps5, cps6, and other strain abundances compared to Rag1−/− mice (Figure S4). We also sought to determine if there were specific global gene expression profiles shared by the most competitive strains (cps4, cps5, and cps6; “winners”) compared to some of the strains that were consistently outcompeted (cps1, cps2, and acapsular; “losers”) by performing RNA-Seq on bacteria from monoassociated mice. When directly comparing all in vivo expression profiles for winners vs. losers (n=9 each) there were no significant changes in gene expression under the conditions used (see STAR Methods), although a few strain-specific changes in mostly genes of unknown function were observed (Table S3). From these data we conclude that CPS4, CPS5, and CPS6 provide a direct advantage to the bacterium in vivo that is not reflected in strong changes in global transcription.
Longitudinal sampling of the stool might not adequately represent geographical differences in population structure in the intestinal lumen vs. the mucus layer or along the intestinal length. Laser capture microdissection of bacteria from longitudinal (ileum, cecum, colon) or lateral (lumen, mucus layer) locations in the Rag1−/− mice from Figure 2 generally showed similar patterns among sites and stool (Figure S5A–B). Interestingly, the small intestine (ileum) was more permissive to strains other than cps4, cps5, and cps6 strains. A lack of geographical difference in cps transcript (Figure S5C) was confirmed in whole colon segments from mice monoassociated with wild-type B. theta, indicating that gross location within the intestinal tract, at least in the absence of a complex microbiota, is not a major driver of which CPS type is most advantageous.
Intestinal anti-CPS IgA levels correlate with increased competition among single CPS-expressing strains
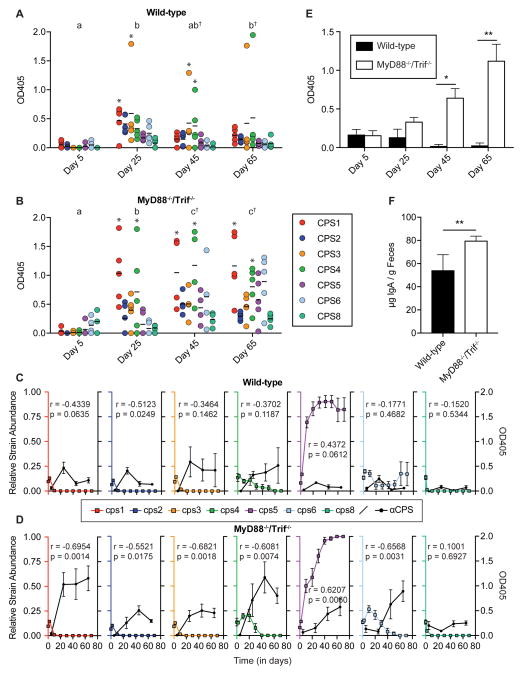
Each of the 3 groups of mice in our competition in Figure 2 exhibited a different competition profile. Whereas the Rag1−/− mice produce no intestinal IgA, the wild-type and MyD88−/−/Trif−/− mice produce adaptive immune responses. To determine whether differences in CPS-specific IgA levels between these two groups correlated with competition outcomes, we used ELISA with each purified CPS as antigen to quantify the magnitude of the CPS-specific IgA response in stool samples over time from the wild-type and MyD88−/−/Trif−/− mice shown in Figure 2. Consistent with development of adaptive immune responses during or after the first week of colonization, levels of anti-CPS IgA in wild-type mice at day 5 were low but significantly increased by day 25 (Figure 3A). At this time point, IgA levels were significantly higher against CPS1 and CPS3 than against CPS5, with significantly higher anti-CPS4 titers (compared to anti-CPS5 titers) emerging by day 45. This is noteworthy given that the intestinal community consisted mainly of the cps5, cps4, and cps6 strains, illustrating that the most competitive strain in these mice (the cps5 strain) is not also the most targeted by IgA. Interestingly, over the entire experiment, levels of anti-CPS IgA were generally higher in the innate immune-deficient (MyD88−/−/Trif−/−) mice (Figure 3B), which correlated with greater competition among strains and eventual elimination of the cps4 and cps6 strains (Figure 2). To measure the significance of this change in anti-CPS IgA production, we used a univariate mixed model with mouse genotype as a predictor, determining that levels of IgA in the MyD88−/−/Trif−/− mice were significantly higher (p < 0.001) than those of wild-type mice. A multivariate model including time showed no statistical difference in anti-CPS IgA levels between mouse genotypes at the first two time points, indicating that differences between mouse genotypes at days 45 (p < 0.001) and 65 (p < 0.001) drive the significance between groups—correlating with the outcompetition of both the cps4 and cps6 strains from the MyD88−/−/Trif−/− mice. We also directly correlated CPS-specific IgA titers with relative strain abundance data (Figure 3C–D). All capsules other than CPS5 were associated with a negative or non-significant correlation coefficient, whereas there was a strong positive relationship between anti-CPS5 IgA titers and cps5 strain abundance. While this positive relationship did not reach our level of significance (a = 0.05) in wild-type mice (r = 0.4372, p = 0.0612), it was significant in the MyD88−/−/Trif−/− mice (r = 0.6207, p = 0.006). The increased IgA response in MyD88−/−/Trif−/− mice was not limited to B. theta CPS, as ELISA using whole acapsular B. theta (Figure 3E) and quantification of total IgA (Figure 3F) both showed a correspondingly stronger response in MyD88−/−/Trif−/− mice. These data implicate the adaptive immune response (particularly, IgA levels) in driving changes in relative abundance seen in the three genotypes of mice tested.
Figure 3.
Fecal IgA levels in in vivo competition of single CPS-expressing strains. IgA was isolated from stool from A) wild-type and B) MyD88−/−/Trif−/− mice displayed in Figure 2 (n=5 mice/group). IgA was quantified via an ELISA using purified CPS as an antigen. Black bars indicate the mean optical density at 405 nm (OD405) against each purified CPS. Significance was determined using a series of linear mixed models. Distinct lower case letters denote significantly different time points within a host genotype (p < .05). Obelisks (†) denote significant differences between host genotypes at the same time point (p < .05). Asterisks (*) indicate samples within a time point with significantly higher OD405 values than the corresponding value for CPS5 (p < .05). C–D) Relative strain abundance (left y axis) and corresponding CPS-specific IgA titers (right y axis) are plotted over time for C) wild-type mice; D) MyD88−/−/Trif−/− mice. Pearson correlation coefficients (r) and p values for each correlation (p) are also indicated. E) Fecal IgA to the acapsular strain from wild-type and MyD88−/−/Trif−/− mice was quantified via ELISA. F) Total IgA from wild-type and MyD88−/−/Trif−/− mice at day 65 was quantified via ELISA. Significant differences between mouse genotypes in panels E–F were determined at each time point via Mann-Whitney test (* p < .05, ** p < .01). For panels C–F, data are represented as mean ± SEM.
Diet alterations do not have major impact on in vivo competition or on in vitro growth rate
Previously, we determined that activation of individual glycan degradation systems induced changes in CPS locus expression (Martens et al., 2009) and others have seen changes in CPS locus expression in vivo upon switching mouse diet (Bjursell et al., 2006; Sonnenburg et al., 2005). To investigate whether specific CPS conferred advantages in different dietary nutrient environments we used two approaches. First, we inoculated the same mixture of strains (8 single CPS-expressing strains and the acapsular strain) into wild-type mice fed one of 5 different diets (Figure S5D–H). While somewhat less stringent competition occurred compared to our previous experiment (Figure 2), the same strains (cps5, cps6, and cps4) predominated in high fiber fed mice. While the cps6 strain dominated for most of the time in low fiber-fed mice (similar to results in Figure 1), on the whole we observed little difference among groups of mice fed different diets. Second, we excluded the immune response as a factor by growing each single CPS-expressing strain in minimal medium containing one of 28 different monosaccharides or polysaccharides as the sole carbon source. Overall, we observed few significant differences in growth rate, which were mostly slower growth of some single CPS-expressing strains on monosaccharides (Figure S6). Taken together, our in vitro and in vivo data indicate that competition for nutrients, which must permeate each surface capsule to be utilized for growth (Cameron et al., 2014), is insufficient to explain the dominance of cps4, cps5 and cps6 strains in most conditions.
The ability to express multiple CPS confers an additional advantage in gut competition
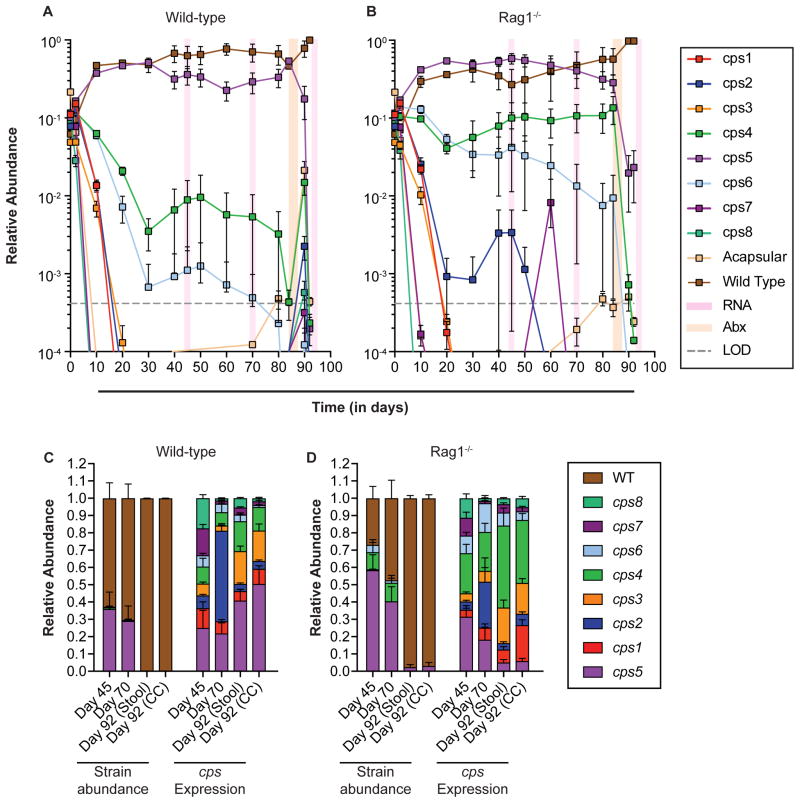
B. theta and its relatives have the potential to switch expression of their CPS loci to adapt to changing conditions (Coyne et al., 2003), including the development of an immune response (Peterson et al., 2007). While some single CPS-expressing strains of B. fragilis have been competed against an isogenic wild-type strain in vivo, studies have yielded conflicting results. In one, a strain expressing only one of its capsules (PSH) competed equally with the wild-type for 15 days (Coyne et al., 2008). However, in another report, 3 other single CPS-expressing strains (PSA, PSB, and PSC) were outcompeted by the wild-type strain over a 7-day period (Liu et al., 2008). Thus, it is unclear as to whether phase-variable expression of CPS loci provides an advantage over any individual CPS. Our collection allows us to test all 8 single CPS-expressing B. theta strains, which may compete differently with wild-type. Thus, we next tested the hypothesis that wild-type B. theta (WT) would exhibit increased fitness compared to the isogenic single CPS-expressing strains because it possesses the ability to dynamically adapt its CPS to thrive in potentially more diverse but unknown selective conditions in the gut. We repeated our previous in vivo competition by inoculating germ-free mice with the single CPS-expressing strain community, plus the acapsular strain, along with WT B. theta (Figures 4, S7). Similar to previous experiments, the CPS5 capsule was the most advantageous for the single CPS-expressing strains. Interestingly, the cps5 strain persisted at substantial levels alongside WT, although in wild-type mice (with adaptive immunity) WT B. theta competed slightly better at most later time points with the cps5 strain periodically dropping to low levels in some mice before regaining abundance. In contrast, in Rag1−/− mice, the cps5 strain competed better overall, with WT B. theta persisting at lower, dynamic levels in all but one mouse. As previously observed in Rag1−/− mice, the cps4 and cps6 strains were maintained at low levels, and other strains emerged at low levels in some individuals, corroborating our previous experiments (Figure S7B). Since WT and cps5 strains persisted together, we hypothesized that WT would also mainly express its cps5 locus, which generally provided the most advantage in our previous competitions. We sampled stool at two different time points to determine relative cps locus expression in fecal RNA. Opposite our expectations, we observed a variety of capsules expressed in the mixed community of fecal B. theta. As the cps5 strain can only express its cps5 locus, expression of CPS5 was higher than that of other loci at days 45 and 70 and similar to the abundance of the cps5 strain (Figure 4C). At day 45, each of the remaining seven capsules was expressed above ~5% total locus transcript, presumably by WT B. theta. At day 70, this relatively equal proportion of capsule expression was skewed in favor of CPS2 expression, one of the CPS favored in our long-term experiment (Figure 1). Strikingly, this same pattern was observed in the separate group of colonized Rag1−/− mice (Figure 4D), albeit with increased presence of CPS4 and CPS6 locus transcripts, presumably due to the higher abundance of the respective single CPS strains.
Figure 4.
In vivo competition of the single CPS-expressing strains with the acapsular and WT strains. 10 strains (8 single CPS-expressing strains, the acapsular strain, and the WT strain) were pooled and inoculated into germ-free mice of 2 genotypes (n=5 mice/group): A) wild-type (all co-housed); B) Rag1−/− (in two cages). All mice were fed a high-fiber diet. Relative abundance of each strain in stool was determined via qPCR. Ciprofloxacin was administered in the drinking water of all mice from days 84–88. C–D) Relative abundance of bacterial strains and relative expression of CPS loci in C) wild-type and D) Rag1−/− mouse stool at days 45, 70, and 92, and from cecal contents at mouse sacrifice (day 92). Data are represented as mean ± SEM. See also Figure S7 and Table S4. LOD: Limit of detection.
Given the ability of WT B. theta to persist alongside the cps5 strain (and others in Rag1−/− mice) by largely expressing a different capsule repertoire, we hypothesized that creating a perturbation to the community would reveal a differential advantage for one of these two populations. To test this, we treated the mice with ciprofloxacin, an antibiotic that kills Bacteroides but does not sterilize the gut (Lee et al., 2013) and to which the cps5 and WT B. theta strains are equally susceptible (Table S4). Administering antibiotics in water for 4 days (days 84–88) to both B. theta-colonized wild-type and Rag1−/− mice reduced the level of fecal bacteria by at least 7 orders of magnitude. We then allowed the respective populations to regrow in the presence or absence of a developed adaptive immune response. In support of our hypothesis, and despite 4/10 mice having higher cps5 populations prior to antibiotic treatment, the B. theta populations in all mice recovered in favor of the WT B. theta strain. This post-antibiotic dominance of the WT strain occurred in all wild-type mice, and in 3 of 5 Rag1−/− mice (2 mice harbored cps5 at 5–10%), suggesting that adaptive immunity explains a small part of this phenotype. The dominance of the WT strain was unlikely to be explained by differential growth on carbohydrates (Figure S6) or to killing mechanisms such as antimicrobial peptides (Table S4, Figure S7C) or complement (Figure S7D).
Drastic reduction of the cps5 and other strains after antibiotics provided the opportunity to measure which CPS were expressed by WT B. theta after it expanded to dominate colonization. Surprisingly, in wild-type mice (with 100% WT B. theta in all 5 mice), WT B. theta displayed a new pattern of cps expression that varied from its pre-antibiotic profile by exhibiting far less cps2 expression and prominent expression of cps5, followed by expression of cps3 and cps1, plus others (Figure 4C). This same pattern was observed in post-antibiotic-treated Rag1−/− mice (Figure 4D), with the notable exception that far less cps5 was expressed by the population (with most cps5 expression found in the two mice with low populations of the cps5 strain), which instead favored corresponding increases in cps1, cps4 and cps6. While it is possible that high cps2 expression at Day 70 is caused by the same dynamic changes in locus expression observed in Figure 1, it is unlikely that the observed changes in cps5 expression, exclusively found in all wild-type mice, is due to a similar phenomenon, as we never observed a large (>20%) expression change of cps5 in wild-type or Rag1−/− mice fed this diet. Collectively, our data indicate that the ability to shift expression of multiple CPS provides an advantage to B. theta that is realized most strongly after antibiotic perturbation and that CPS5 confers some advantage(s) in the face of the adaptive immune response.
Extensive CPS biosynthetic locus diversity in sequenced B. theta isolates
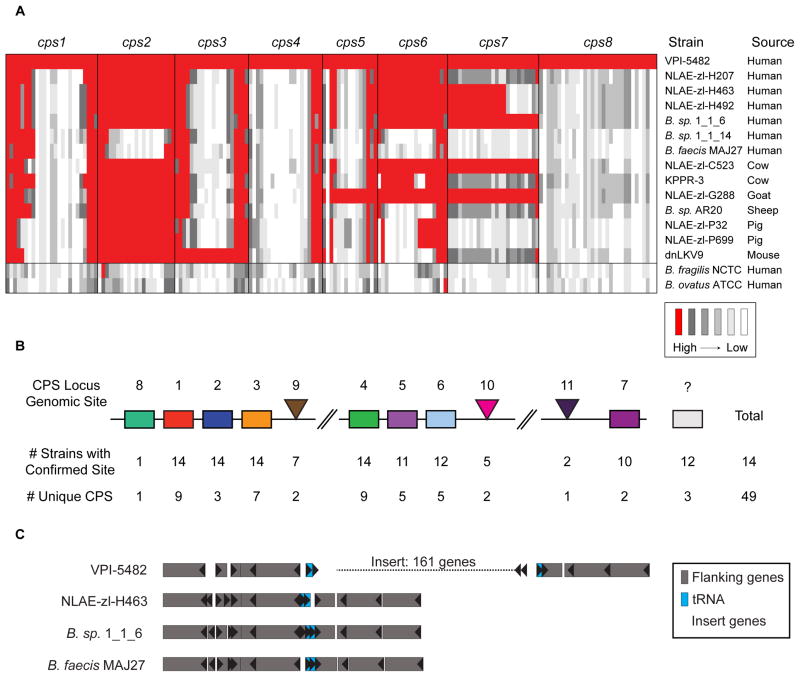
As Bacteroides fragilis strains harbor broad diversity in CPS biosynthetic loci (Patrick et al., 2010), we wondered whether the specific CPS that we determined to be advantageous in gnotobiotic mice would be prevalent in other B. theta strains and in humans. We probed the diversity of B. theta CPS loci in 14 sequenced members of this species (all with >98% 16S rDNA identity to the type strain). Interestingly, only 2 of the 8 CPS loci from the type strain (cps2 and cps6) are found in half or more of the strains (Figure 5A), raising the question of whether other strains encode different CPS loci and if they reside at the same genomic positions. Based on variations in gene content and homology, we identified a total of 49 unique cps loci among these 14 strains (Figure 5B). Strikingly, only one of these loci (cps8 from the VPI-5482 type strain) appeared in other non-B. theta strains (~45,000 genomes probed), suggesting that, while diverse, Bacteroides capsules are largely shared among closely related lineages. The cps8 locus is on a putative mobile element, which may explain its presence in more divergent bacteria (Figure 5C). Breaks in the draft genome assemblies around cps loci hindered the determination of the genomic site for several cps loci; however, when location could be determined, homologous loci occupied the same genomic site. Identical cps were not limited to isolates from a single host species, as examples from humans were also found in isolates from goats, cows, pigs and/or mice.
Figure 5.
Conservation of cps loci from B. theta VPI-5482 in other Bacteroidetes strains. A) The genomes of 14 B. theta strains (>98% full-length 16S identity to type strain VPI-5482) or closely-related strains were probed with genes from the 8 cps loci in the B. theta type strain. B. fragilis NCTC 9343 and B. ovatus ATCC 8483, Bacteroides species containing distantly related cps loci (Coyne and Comstock, 2008), are included as controls. Each gene corresponds to a column of boxes underneath its cognate cps locus (e.g. cps7). Red boxes indicate homology between the gene from the type strain and the genome of the probed strain. B) A simple schematic of the B. theta pangenome illustrates the relative location of the 49 unique cps loci identified in the 14 strains. Rectangular boxes indicate genomic sites utilized by the type strain, whereas triangular wedges indicate sites found only in other strains. Because of the frequency of genomic breaks at sites of cps loci, a “confirmed site” indicates the presence of any number of genes involved in CPS synthesis. C) The cps8 biosynthetic locus is encoded on a mobile genetic element. tRNA genes flanking the cps8 locus in VPI-5482 were identified, and 4–5 flanking genes on each side were used to identify the same genomic location in other closely related strains. These flanking genes were homologous in all loci. The 161-gene insert in VPI-5482 includes the cps8 locus and putative mobilization functions. See also Table S5.
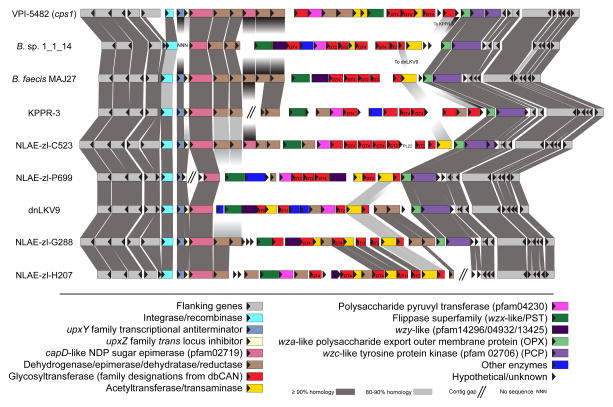
A more detailed inspection of a single genomic site, Site 1 (cps1 locus in the type strain) revealed that genes flanking the cps locus were well conserved, as were genes for regulation (upxY/upxZ-like) and export (wza-like/wzc-like), at the 5′ and 3′ ends (Figure 6). While a few strains had gene clusters at this site that shared homologous genes and even regions of local synteny, we identified a total of 9 unique loci in which most of the genes involved in polymerization or modification of the growing glycan (e.g., glycosyltransferases, acetyltransferases, epimerases) were not homologous. Additionally, glycosyltransferase (GT) content between these loci was different, in both combination and number, suggesting that each locus encodes the ability to produce a distinct glycan.
Figure 6.
Heterogeneity of cps loci at a single genomic site (Site 1). All of the genes present in 9 different cps locus variants found at the same site as the cps1 locus in B. theta VPI-5482 were analyzed for pairwise homology to each other. Gray boxes connecting genes indicate homology (>90% identity in dark grey, 80–90% identity in light grey). Genes are color-coded according to information found in the profile for each gene on the Integrated Microbial Genomes website. Annotations from the CAZy database are displayed within each gene as applicable.
CPS5 and CPS6 biosynthetic loci are prevalent in human gut metagenomic samples
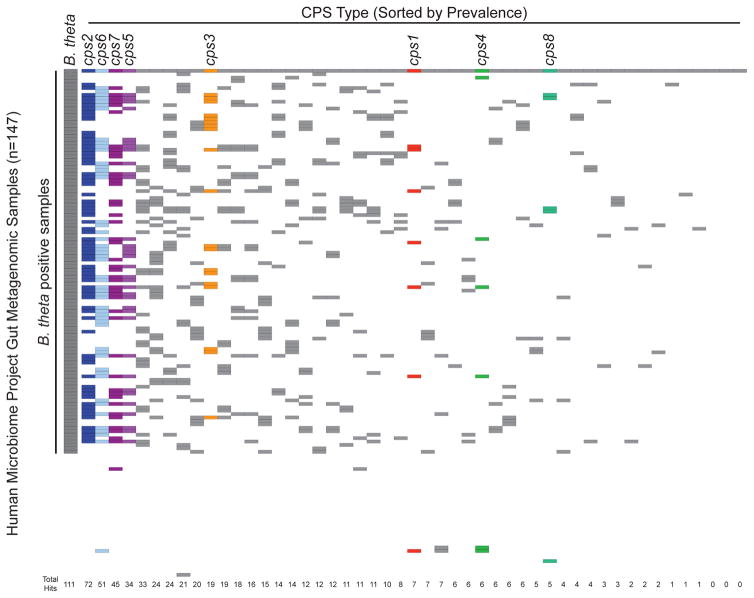
To more broadly survey the presence of B. theta CPS diversity we extended our analysis beyond cultured bacterial isolates to human fecal community samples. We probed metagenomic data from Human Microbiome Project stool samples (Markowitz et al., 2012) to determine the prevalence, in healthy adults, of the various capsule variants that we identified. Of the 48 B. theta-specific loci (excluding cps8), 45 were detected within at least one metagenomic sample (Figure 7). These ranged in prevalence from approximately 1–65% of all B. theta-positive samples. The 3 most prevalent loci in sequenced isolates (cps2, cps6 and cps7) were also the 3 loci most frequently identified in metagenomic samples (65, 46, and 41% prevalence in B. theta-positive samples, respectively), indicating that these loci may confer a competitive advantage in the human gut. Interestingly, while only found in 2 of the 14 sequenced B. theta isolates, the cps5 locus was the fourth most frequently identified cps locus in the metagenomic samples (31% of B. theta-positive samples). Taken together with our experimental data, CPS5 and other CPS undoubtedly provide unique, niche-specific advantages to the bacterium during competitive colonization of the gut. At least for CPS5, this may be due in part to its ability to facilitate evasion of the host immune response in mice and, based on the prevalence of the cps5 locus in the human metagenome, this advantage may extend to the human gut as well.
Figure 7.
Prevalence of B. theta cps biosynthetic loci in human metagenomic samples. The 147 gut metagenomic samples from the Human Microbiome Project were probed with genes from each of the 49 identified B. theta cps loci, as well as a set of genes specific to B. theta sequenced isolates. Each row represents a single metagenome, whereas each column represents a unique cps locus. Colored boxes indicate metagenomic samples with positive hits for the corresponding locus. cps loci are sorted by prevalence in metagenomic samples.
Discussion
Little has been uncovered about the specific roles that CPS play in bacterial survival in the gut and why members of this phylum encode so many variations of these surface coatings. With its full structure known and much of its immunomodulatory activity described (Mazmanian et al., 2008, 2005), by far the best-studied Bacteroides CPS is the zwitterionic polysaccharide A (PSA) from B. fragilis, one of 8 CPS produced by the type strain of this species. Deletion of only PSA abrogated these immunomodulatory changes and protective effects, indicating that the other 7 CPS in this strain likely play different roles.
Our single CPS-expressing B. theta strains provide a means to test the functional roles of CPS in nutrition and other functions. Our current work fails to reveal broad support for the hypothesis that CPS expression is tightly tuned to specific polysaccharides. Instead, our findings, combined with others’ previous results, support a critical role for Bacteroidetes CPS in coping with aspects of the adaptive immune response and recovery from antibiotic challenge. Specifically, while CPS4, CPS5, and CPS6 are advantageous in vivo regardless of host immune proficiency, CPS4 and CPS6 are clearly less advantageous than CPS5 in the face of an adaptive immune response, especially one that is strengthened by removing MyD88/Trif (Figures 2, 4A–B). However, removing host adaptive immunity neither eliminates dynamic expression of capsules by WT B. theta in vivo (Figure 1) nor equilibrates levels of competing single CPS-expressing strains, suggesting that other selective pressures are also important.
In confirmation of a previous study in B. fragilis (Coyne et al., 2008), a strain expressing a single CPS (CPS5) co-colonizes to high levels with WT B. theta especially under stable conditions (e.g. consistent diet, lack of adaptive immunity). It is intriguing that, during coexistence with the cps5 strain, WT B. theta produces multiple CPS, which appear to mostly exclude CPS5. At least two scenarios may account for this phenomenon. First, the complex cps locus regulatory hierarchy employed in B. fragilis (Chatzidaki-Livanis et al., 2010; Coyne et al., 2003) likely applies to B. theta as well. As CPS3 and CPS4 are the most dominantly expressed in WT B. theta in vitro (see inoculum at “Day 0” in Figure 1), regulatory factors encoded in these CPS synthesis loci may actively downregulate the expression of other loci (including CPS5) in the absence of strong selective pressures. Second, having the facultative ability to shift production to different CPS may provide the bacteria with an advantage to fill some niches better than the cps5 strain. In light of these possibilities, it is even more remarkable that, although the WT strain is equally susceptible to antibiotic inhibition in vitro, the WT strain prevails as the dominant colonizer after antibiotic challenge in both adaptive immune proficient and deficient mice, but resorts to dominant expression of CPS5 only in wild-type mice with intact adaptive immunity. This observation supports a model in which CPS5 provides B. theta with the greatest advantage to occupy a particular niche in the gut that is influenced by adaptive immunity. The “size” of this hypothetical niche must be relatively large based on the >50% abundance of the CPS5 strain in single strain competitions. After antibiotic treatment, WT B. theta prevails, perhaps because a capsule other than CPS5 promotes better outgrowth or access to an antibiotic-resistant niche. But after outgrowth in wild-type mice, a subpopulation of WT B. theta adjusts its CPS expression and fills the CPS5 niche, which may either not exist or be smaller in adaptive immune-deficient mice. It is unclear whether WT B. theta would continue to maintain high levels of cps5 expression in adaptive immune-proficient mice, or whether expression would eventually decrease to levels similar to those in Rag1−/− mice. Further studies investigating community perturbations could provide more evidence for specific niches favoring CPS5 or other capsule types.
Such a conditionally advantageous niche exists for B. fragilis, which possesses a gene cluster (ccfA-E, for crypt colonization factor) that allows it to colonize the lumen of colonic crypts. Wild-type B. fragilis is more resistant to antibiotic perturbation than a ccf mutant unable to colonize deep within crypts (Lee et al., 2013). Several homologous loci exist in B. theta, which was recently shown to also colonize colonic crypts (Whitaker et al., 2017), and one of these loci (BT0865-BT0867) is upregulated approximately 5-fold in the cps4 strain in mice (Table S3). However, the cps4 strain is also outcompeted during antibiotic perturbation, suggesting that a more complex interplay between expression of specific CPS loci and niche-specific colonization factors may be involved. Additionally, we determined that relative abundance of the single CPS-expressing strains in our previous experiment (Figures 2, S5) did not vary along the length of the colon or between the mucus layer and lumen of the gut. Thus, the niches implied in our study may not refer to grossly geographical habitats but rather niches that could be based on any number or combination of other undefined factors that are not necessarily exclusive of each other. A complex, constantly-shifting environment will likely dynamically change the size of many of these niches, necessitating phase-variable expression of fitness factors, such as capsules, to pre-adapt some part of the population to thrive in the most important niches. In experiments involving single CPS-expressing strains, CPS5 is most often dominant but this is not an exclusive phenomenon. The presence of other abundant strains that may be even more dominant in individual mice, indicates that capsules such as CPS4 and CPS6 also provide substantial advantages either by promoting access to other competing niches or by promoting good access to the same niche as CPS5.
Finally, we identify numerous, genetically diversified CPS loci in sequenced B. theta isolates and human gut samples. Similar broad diversity has been found in other species of gut Bacteroidetes (Coyne and Comstock, 2008; Patrick et al., 2010) as well as a more phylogenetically diverse set of genomes from the human gut microbiome (Donia et al., 2014), underscoring the importance of CPS diversity to the fitness of bacteria that inhabit this niche. Given the extensive diversification of gut bacterial CPS and the currently limited knowledge of individual CPS function, we believe that future work into the roles of these CPS will provide a much greater understanding of the ways in which symbiotic bacteria interact with both their hosts and the complex and competitive microbiota in which they live.
STAR * METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources may be directed to and will be fulfilled by the Lead Contact Eric C. Martens (emartens@umich.edu; address: University of Michigan Medical School, Ann Arbor, Michigan 48109, USA).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Strains and culture conditions
Bacterial strains used in this study are listed in Table S1. Strains were routinely grown in TYG medium (Holdeman, 1977; 10 g/L tryptone, 5 g/L yeast extract, 4 g/L glucose, 100 mM KH2PO4 (pH 7.2), 15 mM NaCl, 8.5 mM (NH4)2SO4, 4 mM L-cysteine, 200 μM L-histidine, 100 μM MgCl2, 50 μM CaCl2, 1.9 μM hematin, 1.4 μM FeSO4•7H2O, 1 μg/ml vitamin K3, and 5 ng/ml vitamin B12), minimal medium (Martens et al., 2008; 100 mM KH2PO4 (pH 7.2), 15 mM NaCl, 8.5 mM (NH4)2SO4, 4 mM L-cysteine, 200 μM L-histidine, 100 μM MgCl2, 50 μM CaCl2, 1.9 μM hematin, 1.4 μM FeSO4•7H2O, 1 μg/ml vitamin K3, 5 ng/ml vitamin B12, and 0.5% glucose or other carbohydrate as indicated), or on brain heart infusion agar plates containing 10% horse blood (Quad Five, Rygate, Montana) (BHI-blood plates). Antibiotics were added as needed (gentamicin: 200 μg/ml; erythromycin: 25 μg/ml; 5-fluoro-2′-deoxyuridine: 200 μg/ml). Bacteria were routinely grown at 37 °C in an anaerobic chamber (Coy Laboratory Products Inc, Grass Lake, Michigan) or in borosilicate tubes using the NaHCO3/pyrogallol method for anaerobiosis (Holdeman, 1977; after placing a sterile cotton ball in the top of a borosilicate tube, burning the cotton ball and pushing it midway into the tube, 200 μl of saturated NaHCO3 solution and 200 μl of 35% pyrogallol solution were pipetted onto the cotton ball, a rubber stopper was used to seal the tubes, and the tubes were incubated at 37 °C.).
Locus deletions and promoter locking were performed in the B. thetaiotaomicron VPI-5482 tdk− background as described previously (Koropatkin et al., 2008). E. coli S17-1 λpir cells (carrying the pExchange plasmid, containing genomic flanks of the locus of interest that have been ligated together) were mated on BHI-blood plates aerobically at 37 °C with an approximately equal number of B. thetaiotaomicron recipient cells. After one day, selection for merodiploid B. thetaiotaomicron cells was carried out by plating on BHI-blood plates containing gentamicin and erythromycin and incubating for 2 days anaerobically. Resistant colonies were restreaked and regrown under the same conditions for 2 days, and then 10 resistant colonies were grown anaerobically overnight in TYG medium. Counterselection was carried out by pooling the overnight cultures and plating on BHI-blood plates containing 5-fluoro-2′-deoxyuridine. After 2 days of anaerobic growth, colonies were restreaked and regrown under the same conditions. Individual colonies were then picked and screened for loss of the locus or sequence of interest. Deletion constructs for cps loci were described in Rogers et al., 2013 (see also Table S1), and primers for promoter locking are listed in Table S1. cps3 promoter locking data in Figure S2C was previously shown in Hickey et al., 2015.
Gnotobiotic mouse experiments
All animal experiments were performed with approval from the University Committee on Use and Care of Animals at the University of Michigan. Mice were randomly assigned into groups by a technician unfamiliar with the project. Animal numbers for each experiment were chosen based on the minimum numbers required in previous studies to observe significant changes in the readouts measured in each experiment (gene expression, community member changes, antibody titers, etc). Germ-free mice of a C57BL/6 background (wild-type, MyD88−/−/TRIF−/−, or Rag1−/−, as indicated in each experiment) of varying ages and genders were used, dependent on availability, and none of these were involved in any previous experiments. Mixed gender groups with a wide age range (20–59 days old at inoculation, see Figure S4A–C) were used in the experiment for Figure 2, whereas all other experiments involved a single gender common to all groups within that experiment. Groups within an experiment were age matched to the greatest extent possible. For Figure 1, male mice were inoculated at 50–84 days of age. For Figure S4D–E, female mice age 35–47 days were used. For Figure S6, female mice age 64–80 days were used. For Figure 4, male mice 27–36 days of age were used. For the RNA-Seq experiment performed for Table S3, female mice 45–56 days of age were used.
Mice were inoculated with 100 μl of bacterial culture (individual strain for monocolonization experiments; or for competition experiments, strains pooled in equal volumes at approximately OD600 1.0, with the acapsular strain volume doubled). Mice were routinely fed a standard, autoclaved high-fiber diet (LabDiet 5013, LabDiet). For experiments involving other diets [low-fiber diet TD.130343 (Harlan Laboratories, Madison, WI) (Larsbrink et al., 2014), prebiotic diet (TD.130342, Harlan) (Desai et al., 2016), bread-based diet (TD.130682, Harlan) (Cuskin et al., 2015), or high-fat diet (TD.96132, Harlan) (Reed et al., 2000)], mice were pre-fed the corresponding diet for 7 days prior to inoculation with Bacteroides thetaiotaomicron (B. theta) strains. To reduce bacterial load in vivo in Figure 4, mice were switched to water containing 0.625 mg/ml ciprofloxacin for 4 days, prior to switching back to regular water. Reduction of bacterial load was confirmed via spot plating of stool samples.
METHOD DETAILS
Resequencing of single CPS-expressing strains
For strain genomic sequencing, pellets from 35 ml overnight cultures were resuspended in 15 ml TE and incubated with 0.5 ml 20% SDS and 200 μl of 20 mg/ml proteinase K at approximately 50 °C for 2 hours. Samples were extracted twice with 1:1 phenol:chloroform, followed by two extractions with pure chloroform. DNA was precipitated using sodium acetate and isopropanol and centrifuged to form a pellet. After washing in 70% ethanol and drying, the pellet was resuspended in 5 ml TE overnight at 4 °C. Samples were treated with RNase prior to extractions with phenol, 1:1 phenol:chloroform, and two extractions with chloroform. DNA was precipitated with sodium acetate and ethanol, washed in 70% ethanol, dried, and resuspended in TE. 700 μl of 100 ng/μl DNA was fragmented in a Sonics Vibra-cell GE505 ultrasonicator at 25% amplitude for 2 minutes (30 seconds on, 30 seconds off). DNA libraries were prepared using the TruSeq DNA LT Sample Prep Kit (Illumina Inc., San Diego, California). Libraries were pooled and run in 1 lane of 50 base pair single read sequencing on an Illumina HiSeq 2500 at Michigan State University. Genomes were assembled in SeqMan NGen (DNASTAR Inc., Madison, Wisconsin) and SNPs against the sequenced wild type strain (tdk− + Tag1) were identified in SeqMan Pro (DNASTAR Inc.) (see Table S2). CPS locus deletions were also validated from sequencing assemblies. RNA isolation and quantitative PCR (qPCR) for relative cps expression. For in vitro samples, strains were streaked onto agar media and grown anaerobically for 2 days. 3 individual colonies were selected for each strain and inoculated into TYG medium overnight, then were subcultured 1:50 into minimal medium containing 0.5% glucose and grown using the NaHCO3/pyrogallol method. Upon reaching OD600 0.6–0.8, the supernatant was removed, and 1.5 ml of each culture was resuspended in 500 μl RNAprotect (Qiagen) and then handled according to the manufacturer’s instructions. RNA was extracted via the RNeasy Mini Kit, treated with the TURBO DNA-free Kit (Ambion) and purified again with the RNeasy Mini Kit.
RNA was extracted from flash frozen mouse stool or cecal contents via bead beating (Ley et al., 2005) for 5 minutes in 500 μl buffer (200 mM NaCl, 200 mM Tris, 20 mM EDTA at pH 8) with 210 μl of 20% SDS and 500 μl phenol:chloroform:isoamyl alcohol (125:24:1, pH 4.3, Fisher Scientific), followed by ethanol precipitation, purification using the RNeasy Mini Kit, treatment with TURBO DNA-free Kit, and repurification with the RNeasy Mini Kit. For measuring relative cps locus expression from inocula, the inoculum was passaged into minimal medium and handled as for other in vitro samples above.
cDNA was prepared with up to 1 ug RNA using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer’s instructions.Quantitative PCR (qPCR). qPCR analyses were performed using a Mastercycler ep realplex instrument (Eppendorf) and KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems, Wilmington, MA) with 500 nM primers and 10 ng DNA/cDNA as appropriate. Primers are listed in Table S1. For quantification of barcoded strains, a touchdown protocol was used: 40 cycles of 95 °C for 3 seconds, annealing at a variable temperature for 20 seconds, and 72 °C for 8 seconds. For the first 6 cycles annealing temperature started at 68 °C and then dropped one °C each for the subsequent 5 cycles. The annealing temperature was 62 °C for the remaining 34 cycles. Samples were normalized to a DNA standard curve for each strain. For cps locus gene expression, qPCR was performed for 40 cycles of 95 °C for 3 seconds, 55 °C for 20 seconds, and 72 °C for 8 seconds. Samples were normalized to a DNA standard curve of DNA from the wild-type strain. Both protocols were followed by a melting curve analysis for determination of amplicon purity. Relative abundance of strains/cps locus genes was then calculated.
Preparation of purified CPS and monosaccharide analysis
Purified CPS were prepared via the hot water phenol method (Martens et al., 2009). Briefly, 10 ml overnight cultures of each strain grown in TYG medium were inoculated into 4 L of minimal medium containing 0.5% glucose as the sole carbohydrate source for 2 days. Stationary phase cultures were centrifuged, flash frozen in liquid nitrogen, and stored at −80 °C until use. Cells were resuspended in 100 ml hot water prior to the addition of an equal volume of phenol. The mixture was stirred for 1 hour at 65 °C and allowed to cool at 4 °C overnight. Preparations were then centrifuged and the aqueous phase was dialyzed exhaustively against deionized distilled water (12–14 kDa cutoff). Preps were lyophilized to dryness and resuspended in buffer containing 20 mM Tris HCl (pH 7.4), 2.5 mM MgCl2, and 0.5 mM CaCl2. Both RNase A and DNase I were added and samples were rotated gently overnight at 37 °C. Proteinase K was then added and samples were incubated overnight at 65 °C. Phenol was added to the sample prior to vortexing and centrifuging for phase separation, and the aqueous phase was removed. Samples were again dialyzed exhaustively, prior to lyophilization to dryness. The cps7 strain did not produce sufficient product for downstream analysis, likely because of lower cps7 transcription and thus lower CPS production. Glycogen contamination in the samples was assayed using a quantitative iodine assay (Xiao et al., 2006): 50 ul of 5 mg/ml sample was added to a 96-well microtiter plate, and 50 ul of iodine reagent (5 mM I2 + 5 mM KI) was added. Color development was measured at 580 nm and compared to a standard curve of glycogen to determine glycogen levels in each sample. Samples displayed variable amounts of bacterial glycogen contamination (Figure S3H), which is also evident in relative abundance of glucose in purified CPS preparations (Figure S3A–G).
For monosaccharide composition analysis, purified CPS were analyzed via high pH anion exchange chromatography using pulsed amperometric detection (HPAEC-PAD) following acid hydrolysis at the UCSD Glycotechnology Core, using a uronic acid profile and authentic sugar standards (glucose, galactose, mannose, fucose, N-acetyl-glucuronic acid, N-acetyl-galacturonic acid, glucuronic acid, galacturonic acid, iduronic acid, guluronic acid, and mannuronic acid). Single CPS-expressing strains were confirmed to have different monosaccharide profiles (see Figure S3).
Single CPS-expressing strain characterization and validation
A set of B. theta strains that each encodes one of the type strain’s 8 CPS biosynthetic loci were previously generated (Hickey et al., 2015). These were produced by sequential deletion of all but one of the type strain’s 8 cps loci in parallel mutant lines (Figure S2A). Invertible promoters that flip between “on” and “off” orientations (Krinos et al., 2001) precede 5 of the 8 cps loci. To eliminate this as a confounding variable, the “outside” inverted terminal repeat region (i.e., lying upstream of the expressed promoter in its “on” state) of these 5 promoters was deleted to lock them in the expressed orientation (Figure S2B–C). To enable identification of each strain in a mixture, a unique 24bp barcode sequence was inserted into each of these 10 strains at one of the two NBU2 (tRNAser) sites (Koropatkin et al., 2008). As the panel of CPS-expressing strains is crucial for many of the experiments in this work, each strain was extensively validated. Genomic sequencing of each strain identified few mutations compared to the other strains (Table S2), and expected deletions, promoter locking, and barcode vector integration were confirmed in genome assemblies. Furthermore, for experiments in Figure S4E, the cps5 strain was rederived from an intermediate strain harboring just 4 cps loci (cps5, cps6, cps7, and cps8), with the reasoning that any secondary mutations found in this ancestral strain would be found in multiple strains, including other strains that competed both dominantly and poorly in vivo. Expression of individual cps loci in the 8 single CPS-expressing strains was confirmed by qPCR. As expected, each of the 8 single cps-encoding strains expressed only its respective cps, with all but cps7 being transcribed to similarly high levels. In contrast, a population of the wild-type (tdk−) parent strain was able to express all of the loci (Figure S2D). Unlike other strains possessing CPS biosynthetic genes, the CPS7-expressing strain (“cps7 strain”) that exhibited low expression sank to the bottom of the tube when grown in static liquid culture, similar to the acapsular strain of B. theta. This phenomenon has also been reported for acapsular B. fragilis (Coyne et al., 2008) and, together with low transcription, suggests that CPS7 is not fully functional. To measure differences in the chemical composition of the capsules, the CPS was extracted from each single CPS-expressing strain and quantitative sugar analysis was performed as described above. When grown in a defined medium containing glucose as the sole carbohydrate source, each strain produced CPS composed of different sugars or different stoichiometric abundances (Figure S3, note that CPS7 was not assayed due to low abundance of extractable CPS). These data indicate that each of these single CPS-expressing strains only produces a single CPS, allowing isolation of the role of individual CPS in experiments in this work.
Relative cps expression from previous published datasets
Microarray or RNA-Seq datasets measuring gene expression in samples containing B. thetaiotaomicron VPI-5482 were selected from the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/; see Table S6 for information on each sample set). In order to control for inter-experimental or inter-group variation, expression data for each of the genes in a cps biosynthetic locus was averaged within each individual sample, and the relative abundance of expression of each of the cps loci was calculated. Trends highlighted in some of the original studies (such as increased expression of the cps5 locus (Sonnenburg et al., 2005)) were still evident in the current analysis.
Quantification of fecal IgA via ELISA
IgA was isolated from mouse stool by preparing a 10% solution in PBS-BSA (1% BSA), vortexing for 20 minutes, centrifuging for 20 minutes at 14,000 rpm (18,400 rcf), and collecting the supernatant. For quantification of anti-CPS or anti-acapsular strain IgA responses, plates were coated with 50 μl of a 0.2 mg/ml solution of purified CPS in bicarbonate buffer (15 mM sodium carbonate, 35 mM sodium bicarbonate, 3 mM sodium azide, pH 8.5) and incubated for 2 hours at room temperature. Plates were then washed 3 times in PBS-T (0.05% Tween 20), then blocked with PBS-BSA (30 min at room temperature), and again washed with PBS-T. The fecal supernatant containing IgA was diluted 1:10 in PBS-BSA and added to the plate (50 μl per well) before an overnight incubation at 4 °C. Wells were washed 3 times with PBS-T prior to the addition of 50 μl of horseradish peroxidase (HRP)- conjugated goat anti-mouse IgA (Southern Biotech; diluted 1:1000 in PBS-BSA). After a 2 hour incubation, samples were washed three times with PBS-T, and 50 μl of biotinyl tyramide solution (diluted 1:500 in 0.1 M sodium borate buffer, pH 8, supplemented with 0.03% H2O2) was added to each well. Samples were incubated for 15 minutes at room temperature and washed 3 times in PBS-T; 50 μl of Neutralite Avidin-HRP (Southern Biotech; diluted 1:2000 in PBS-BSA) was added and incubated at room temperature for 2 hours. Samples were then washed 3 times in PBS-T and revealed using 100 μl of 1 mM 2,2′-azino-di-[3-ethylbenzthiazoline]-sulfonate solution (ABTS, Roche) in citrate buffer (100 mM citric acid, 50 mM sodium phosphate, pH 4.2), containing 0.03% H2O2. Samples were incubated for 15 minutes at room temperature prior to measuring absorbance at 405 nanometers. As possible (depending on amount of IgA isolated), samples were run in duplicate. For quantification of total IgA from stool, plates were coated with 50 μl of a 1:1000 dilution of goat anti-mouse IgA (Southern Biotech) in bicarbonate buffer and incubated for 2 hours at room temperature. Plates were then washed 3 times in PBS-T, then blocked with PBS-BSA (30 minutes at room temperature). Serial dilutions of the fecal supernatant were added to the plate (50 μl per well), starting with a 1:10 dilution, followed by serial 1:4 dilutions in BSA-PSA. Plates were incubated at room temperature for 2 hours. Wells were washed 3 times with PBS-T prior to the addition of 50 μl of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgA (diluted 1:1000 in PBS-BSA). After a 2 hour incubation, samples were washed three times with PBS-T, and 100 μl of 1 mM ABTS solution in citrate buffer supplemented with 0.03% H2O2 was added to each well, prior to measuring absorbance at 405 nanometers. A standard curve of mouse purified IgA (Southern Biotech) was used to calculate IgA titers from each sample.
RNA-Seq analysis on bacteria monoassociated in germ-free mice
Groups of 3 C57BL/6 wild-type mice were colonized with a single strain of B. thetaiotaomicron VPI-5482 (the cps1, cps2, cps4, cps5, cps6, or acapsular strains) for 4 weeks. Mice were fed a standard high-fiber diet (LabDiet 5013, LabDiet). RNA was extracted as described above and then treated with Ribo-Zero rRNA Removal Kit (Illumina Inc.) and concentrated using RNA Clean and Concentrator -5 kit (Zymo Research Corp, Irvine, CA). Sequencing libraries were prepared using TruSeq barcoding adaptors (Illumina Inc.), and 24 samples were multiplexed and sequenced with 50 base pair single end reads in one lane of an Illumina HiSeq instrument at the University of Michigan Sequencing Core. Demultiplexed samples were analyzed via Arraystar software (DNASTAR, Inc.) using RPKM normalization and the default parameters. Gene expression for each of the 5 CPS-expressing strains was normalized to the acapsular strain, and gene expression significantly upregulated or downregulated (5-fold cutoff, and significant by a moderated t test with Benjamini-Hochberg correction) was included in Table S3.
Bacterial growth rate analysis
Growth rate analysis was essentially as described previously (Martens et al., 2011). Briefly, strains were grown overnight in TYG medium, and were subcultured and grown overnight in minimal medium containing 0.5% glucose. Cells were washed in double strength minimal medium containing no carbohydrate, prior to resuspension in the same medium. 100 μl aliquots of the cell suspensions were then inoculated into 96-well plates containing an equal volume of 1.0% solution of the desired carbohydrate (2.0% for O-glycans or rhamnogalacturonan I) and grown for 4 days anaerobically. Absorbance values were measured using an automated plate reading device (BioTek Instruments). Specific rate [ (max OD600 − min OD600)/time ] was calculated using Microsoft Excel.
Antibiotic and antimicrobial susceptibility assays
Inhibition of bacterial growth by ciprofloxacin, polymyxin B, and human beta-defensin 3 were measured in a broth-based assay as described for antimicrobial peptide resistance (Cullen et al., 2015). Cultures were diluted to approximately OD600 0.05 in TYG medium, and 100 μl diluted culture was added to serially diluted concentrations of antibiotic/antimicrobial peptide. All 3 assays were set up anaerobically. For the assay containing human beta-defensin 3, plates were then incubated aerobically for 90 minutes before being placed back in an anaerobic environment. Growth was continuously monitored in an automated plate reading device and was compared to wells in which no compound was added; if wells achieved less than half the absorbance of the control at late log (~OD600 0.8), growth was considered to be inhibited at that concentration. For the complement-mediated killing assay (Coyne et al., 2008), 1 ml overnight culture of bacteria was diluted to OD600 0.2, washed, and resuspended in 1 ml PBS containing 5 mM MgCl2. 10 μl of normal human complement serum or freshly inactivated serum (incubated in a 56 °C water bath for 30 minutes) was added to 3 wells containing 90 μl of the bacterial resuspension. Wells were incubated anaerobically for 1 hour prior to serial dilution and plating on solid media. Each of the 3 replicates were normalized to counts for the heat inactivated complement [(inactive serum-treated counts – serum-treated counts)/inactive serum-treated counts] and then averaged and expressed as percent survival.
Laser capture microdissection and DNA extraction
Methanol Carnoy’s fixed intestinal segments were paraffin embedded prior to microscope slide generation. Slides were deparaffinized in xylene, twice incubated in 100% ethanol, and retreated with xylene prior to drying briefly and placing in a dessicator for approximately 1 hour. Mucus layer (~30 μm immediately adjacent to epithelium) or lumen-associated (middle of lumen) samples were taken for each intestinal segment (ileum, cecum, and distal colon) of the Rag1−/− mice using an Arcturus Veritas microdissection instrument (Applied Biosystems). DNA was extracted from each sample with the PicoPure DNA extraction kit (Applied Biosystems). Prior to qPCR for quantifying relative abundance of each strain, a PCR for non-specific amplification of the barcoding vector was employed (primers are listed in Table S1). 10 ng of extracted DNA were amplified in duplicate for each sample with Platinum Pfx (Invitrogen) with the following protocol: 95 °C for 5 minutes; 20 cycles of: 95 °C for 30 seconds, 58 °C for 30 seconds, and 72 °C for 30 seconds; final extension at 72 °C for 10 minutes. After PCR, duplicate samples were pooled and purified using Qiagen MinElute PCR Purification Kit and diluted (ileum diluted 1:10, colon segments diluted 1:100) prior to use in qPCR.
Identification of cps biosynthetic loci in sequenced isolates and in metagenomes
Strains of the B. theta genomic clique in the Integrated Microbial Genomes (IMG) database (Markowitz et al., 2014), as well as Bacteroides faecis MAJ27 (each with a >98% identity hit to the VPI-5482 16S rDNA) were probed with genes from the 8 cps loci from the VPI-5482 strain using the IMG “Genome Gene Best Homologs” tool with an initial percent identity cutoff of 20%. Strains with an identified homolog meeting identity cutoffs (90%, 80%, 60%, 40%, 20%, 0%) were shaded according to the color scale indicated in Figure 5A (from “high”/90% to “low”/0% homology). Type strains of B. ovatus (ATCC 8483) and B. fragilis (NCTC 9343) were included as controls. To identify other cps loci within each genome, multiple strategies were undertaken. First, homologs of flanking genes on both sides of known cps loci were investigated for the presence of neighboring, novel loci in other strains. Then, a blastp search with an E-value cutoff of 1e-5 was performed for conserved regulatory genes (upxY and upxZ family genes from the cps5 locus, BT1656 and BT1655) and for conserved export machinery genes (BT0398 from cps1 and BT1654 from cps5). To identify novel cps loci, each of the 14 strains was used as a reference strain in the “Genome Gene Best Homologs” tool and compared against the other 13 strains. A sufficient length of contiguous genome assembly was required to either identify the genomic location and/or compare gene content and homology to other loci. Although, gaps in assembled genomes prohibited full locus comparison in several cases. Genes in homologous loci were typically >98% similar to one another at the amino acid level. 3 genes within each locus were identified via this method that appeared to be highly conserved only among those strains that encoded the entire locus. A blastn search (E-value cutoff 1e-5) of all isolate genomes in IMG was performed, and each gene was confirmed to be specific to its cognate locus and not found within other genomes. Only the cps8 locus, which is found on a mobile genetic element, was found within other Bacteroides species (these were confirmed to contain a locus homologous to cps8). Genes from each locus were not found to be present outside of the Bacteroides genus. 49 unique cps biosynthetic loci were thus identified.
The 3 unique genes from each locus were also used to probe for each of the loci in the 147 Human Microbiome Project gut metagenomic samples found on the IMG website. Each sample was probed with the 3 genes from each of the 49 unique loci via blastn with more stringent criteria (E-value cutoff 1e-50, >100 bp hit, 95% identity). A sample with at least 2 out of 3 genes present was considered positive for the corresponding locus. 3 genes conserved within all 14 isolate genomes (but not found within other isolate genomes on IMG) were used to determine the presence of B. theta within each metagenomic sample. Genes used are listed in Table S5.
For cps locus synteny analysis (Figure 6), representative loci from each of the 9 unique cps locus types at genomic Site 1 (location of cps1) were compared using the IMG “Genome Gene Best Homologs” tool. Especially for some conserved gene families (e.g. upxY/upxZ) the closest homolog for a gene in one strain was found in a cps locus at a different genomic site. For these genes, Blastp (E-value cutoff 1e-2) was performed. Cutoffs of 80% or 90% identity were used to label genes as homologous.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
None of the investigators were blinded to sample identity during any of the described experiments and no samples or measurements were excluded from analysis. For each experiment, the total number of biological replicates “n” is indicated in the corresponding legend along with a description of what is considered a replicate (mice, replicate cultures, antibody titers, etc.). Statistical analyses (other than Dirichlet regression and linear mixed models) were performed in Prism software (GraphPad Software, Inc., La Jolla, CA). As data did not generally follow a normal distribution, nonparametric tests (Mann-Whitney/Kruskal-Wallis) were used to determine statistical significance. Unless otherwise indicated, statistical significance is indicated as follows: * p < 0.05; ** p < 0.01; *** p < 0.001; and **** p < 0.0001. Number of animals used (n) and statistical tests used are indicated in each figure legend. For comparing relative abundance of cps locus expression (Figure 1) between pairs of experimental groups, Dirichlet regression was performed in R using the alternate parameterization in the “DirichletReg” package (version 0.6–3; Maier, 2014). The parameters in this representation of the Dirichlet distribution are the relative proportions of cps locus expression and the total expression, which is a measure of precision. The parameterization models the logarithm of the ratio of the relative expression of locus cpsc (referring to CPS loci cps1, cps2, cps3, cps4, cps5, cps6, or cps8) to the relative proportion of the reference cps7 locus as a linear combination of the given variables of interest. The variables of interest were diet and low-fiber diet interval for comparison of the wild-type, high-fiber fed mice with the wild-type mice whose diets oscillated between intervals of low- and high-fiber diets. The variable of interest for comparison of the wild-type, high-fiber fed mice with the Rag1−/−, high-fiber fed mice, on the other hand, was mouse genotype. The precision for a given model was allowed to vary by group in cases where a likelihood ratio test indicated at significance level p < 0.05 that this model was superior to a corresponding model with constant precision.
For the IgA titer assay in Figure 3, it was necessary to use the entire fecal sample to provide enough IgA for the assay. Thus, to correlate specific anti-CPS IgA titers with strain abundance, a good estimation of the relative strain abundance at the time of the IgA titer data was generated by averaging the two closest relative strain abundances to the IgA time point (e.g. for a time point on day 25, the Day 20 and Day 30 abundances were averaged). A Pearson correlation was then performed.
Linear mixed models for Figure 3 were created and run in R using the package “lme4” (version 1.1–12, Bates et al., 2015). First, to examine the effect of mouse genotype on anti-CPS fecal IgA levels, a univariate model dependent only on mouse genotype as a predictor was employed: where i denotes an individual mouse, j denotes time, k denotes CPS type, Ri is the random effect of the mouse and eijk is the error term. The variable IgAijk refers to the normalized OD405 values shown in Figure 3 for subject i at time j for CPS type k and Genotypei is an indicator variable of the mouse genotype. This means that wild-type has value 0 and MyD88−/−/Trif−/− has value 1. Next, the contributions of both mouse genotype and day were examined in a multivariate model: with notation as described above. Dayd,j are indicator variables for days 25, 45, and 65. Furthermore, comparisons of anti-CPS5 titers to other anti-CPS titers at each time point were carried out using another model incorporating identity of each targeted CPS: where Typetk are indicator variables for CPS types CPS1, CPS2, CPS3, CPS4, CPS6, and CPS8, with other notation as described above. Linear combinations of coefficients in the mixed models represent differences between groups and between time points for each group. Significance was determined for each difference by testing the null hypothesis that the linear combination of coefficients representing the given difference is 0.
DATA AND SOFTWARE AVAILABILITY
Genome sequencing and RNA-Seq data from this study have been deposited in the NCBI Short-Read Archive (SRA) under the BioProject ID PRJNA395217 (BioSample IDs for genomes: SAMN07373223-SAMN07373232; BioSample IDs for RNA-Seq data: SAMN07374632-SAMN07374649).
Supplementary Material
Strains, vectors, and oligonucleotides used in this study. Related to STAR Methods.
Single nucleotide polymorphisms (SNPs) from each of the 10 strains competed in this study. Related to STAR Methods. Genomic DNA from the WT strain, the acapsular strain, and each of the 8 single CPS-expressing strains was sequenced on an Illumina HiSeq and genomes were assembled in DNASTAR SeqMan NGen. SNPs not found in the WT strain (or in all but the WT strain) are displayed. Deletion of cps loci, locking of invertible promoters, and inclusion of unique barcode sequences were all verified but not included in this table.
Fold changes in bacterial genes in vivo: single CPS-expressing strains relative to the acapsular strain. Related to Figure 2. Locus tag numbers highlighted in yellow correspond to cps locus genes. Positive expression changes are highlighted in green while negative expression changes are in red.
Minimum inhibitory concentrations of ciprofloxacin, polymyxin B, and human beta-defensin 3 (μg/ml). Related to Figure 4. Mean ± SEM displayed for each strain incubated with each antimicrobial compound. Kruskal-Wallis tests for each antimicrobial compound for each strain versus wild-type were not significant at p = 0.05. ND: Not determined.
Strains, genes, and capsular polysaccharide synthesis loci in comparative genomic and metagenomic analyses. Related to Figure 5.
Additional information on relative cps expression from previously published datasets. Related to STAR Methods.
Highlights.
B. thetaiotaomicron dynamically changes its capsular polysaccharides (CPS) in vivo
One capsule, CPS5, provides the most advantage in the presence of adaptive immunity
The ability to express multiple CPS types promotes survival during antibiotic stress
CPS5 is one of the most prevalent B. thetaiotaomicron CPS loci in human samples
Acknowledgments
N.T.P. was supported by the NIH Cellular Biotechnology Training Program (T32GM008353). This work was supported by NIH grant numbers GM124136, DK096023, and DK034933. We thank the Germ Free Mouse Facility at University of Michigan for expert assistance with in vivo experiments and the University of California San Diego Glycotechnology Core for monosaccharide analysis of purified CPS. The Consulting for Statistics, Computing, and Analytics Research service (CSCAR) at University of Michigan assisted with linear mixed model and Dirichlet regression analysis.
Footnotes
Author Contributions
N.T.P., D.A.P., and E.C.M. conceived and designed the experiments. N.T.P. and P.C. performed experiments. N.T.P., D.A.P., P.C., and E.C.M. analyzed data. N.T.P. prepared figures. N.T.P and E.C.M. primarily wrote the manuscript. All authors reviewed the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avery OT, Dubos R. The protective action of a specific enzyme against Type III Pneumococcus infection in mice. J Exp Med. 1931;54:73–89. doi: 10.1084/jem.54.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting Linear Mixed-Effects Models Using lme4. J Stat Softw. 2015;67:51. [Google Scholar]
- Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- Cameron EA, Kwiatkowski KJ, Lee BH, Hamaker BR, Koropatkin NM, Martens EC. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. MBio. 2014;5:e01441–14. doi: 10.1128/mBio.01441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzidaki-Livanis M, Coyne MJ, Comstock LE. A family of transcriptional antitermination factors necessary for synthesis of the capsular polysaccharides of Bacteroides fragilis. J Bacteriol. 2009;191:7288–7295. doi: 10.1128/JB.00500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzidaki-Livanis M, Weinacht KG, Comstock LE. Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc Natl Acad Sci U S A. 2010;107:11976–11980. doi: 10.1073/pnas.1005039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne MJ, Comstock LE. Niche-specific features of the intestinal Bacteroidales. J Bacteriol. 2008;190:736–742. doi: 10.1128/JB.01559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne MJ, Weinacht KG, Krinos CM, Comstock LE. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc Natl Acad Sci U S A. 2003;100:10446–10451. doi: 10.1073/pnas.1832655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- Coyne MJ, Chatzidaki-Livanis M, Paoletti LC, Comstock LE. Role of glycan synthesis in colonization of the mammalian gut by the bacterial symbiont Bacteroides fragilis. Proc Natl Acad Sci U S A. 2008;105:13099–13104. doi: 10.1073/pnas.0804220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress BF, Englaender JA, He W, Kasper D, Linhardt RJ, Koffas MaG. Masquerading microbial pathogens: capsular polysaccharides mimic host-tissue molecules. FEMS Microbiol Rev. 2014;38:660–697. doi: 10.1111/1574-6976.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Schofield WB, Barry NA, Putnam EE, Rundell EA, Trent MS, Degnan PH, Booth CJ, Yu H, Goodman AL. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–175. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnion KM, Zhang HM, Frank MM. Availability of complement bound to Staphylococcus aureus to interact with membrane complement receptors influences efficiency of phagocytosis. Infect Immun. 2003;71:656–662. doi: 10.1128/IAI.71.2.656-662.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167:1339–1353. e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MSS, Cimermancic P, Schulze CJJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RGG, Fischbach MAA, Wieland Brown LC, et al. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CA, Kuhn KA, Donermeyer DL, Porter NT, Jin C, Cameron EA, Jung H, Kaiko GE, Wegorzewska M, Malvin NP, et al. Colitogenic Bacteroides thetaiotaomicron antigens access host immune cells in a sulfatase-dependent manner via outer membrane vesicles. Cell Host Microbe. 2015;17:672–680. doi: 10.1016/j.chom.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdeman LVE. Anaerobe Laboratory Manual. Blacksburg: Anaerobe Laboratory Virginia Polytechnic Institute & State University; 1977. [Google Scholar]
- Horwitz MA, Silverstein SC. Influence of the Escherichia coli capsule on complement fixation and on phagocytosis and killing by human phagocytes. J Clin Invest. 1980;65:82–94. doi: 10.1172/JCI109663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelacic TM, Chabot DJ, Bozue JA, Tobery SA, West MW, Moody K, Yang D, Oppenheim JJ, Friedlander AM. Exposure to Bacillus anthracis capsule results in suppression of human monocyte-derived dendritic cells. Infect Immun. 2014;82:3405–3416. doi: 10.1128/IAI.01857-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B, El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:3–8. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone Ca, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, Lee SM, VanLare JM, Kasper DL, Mazmanian SK. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci U S A. 2008;105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier MJ. DirichletReg: Dirichlet Regression for Compositional Data in R. 2014. p. 13. [Google Scholar]
- Markowitz VM, Chen IMA, Chu K, Szeto E, Palaniappan K, Jacob B, Ratner A, Liolios K, Pagani I, Huntemann M, et al. IMG/M-HMP: a metagenome comparative analysis system for the Human Microbiome Project. PLoS One. 2012;7:e40151. doi: 10.1371/journal.pone.0040151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Pillay M, Ratner A, Huang J, Woyke T, Huntemann M, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Kelly AG, Tauzin AS, Brumer H. The devil lies in the details: How variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. 2014;426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Mazmanian SKSSK, Liu C, Tzianabos AOA, Kasper DLD, Cui HL, Tzianabos AOA, Kasper DLD. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Neff CP, Rhodes ME, Arnolds KL, Collins CB, Donnelly J, Nusbacher N, Jedlicka P, Schneider JM, McCarter MD, Shaffer M, et al. Diverse intestinal bacteria contain putative zwitterionic capsular polysaccharides with anti-inflammatory properties. Cell Host Microbe. 2016;20:1–13. doi: 10.1016/j.chom.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick S, Blakely GWG, Houston S, Moore J, Abratt VR, Bertalan M, Cerdeño-Tárraga AM, Quail MA, Corton N, Corton C, et al. Twenty-eight divergent polysaccharide loci specifying within- and amongst-strain capsule diversity in three strains of Bacteroides fragilis. Microbiology. 2010;156:3255–3269. doi: 10.1099/mic.0.042978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–339. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- Porter NT, Martens EC. The Critical Roles of Polysaccharides in Gut Microbial Ecology and Physiology. Annu Rev Microbiol. 2017;71 doi: 10.1146/annurev-micro-102215-095316. annurev-micro-102215–095316. [DOI] [PubMed] [Google Scholar]
- Reed MJ, Meszaros K, Entes LJ, Claypool MD, Pinkett JG, Gadbois TM, Reaven GM. A new rat model of type 2 diabetes: the fat-fed, streptozotocin-treated rat. Metabolism. 2000;49:1390–1394. doi: 10.1053/meta.2000.17721. [DOI] [PubMed] [Google Scholar]
- Rogers TE, Pudlo NA, Koropatkin NM, Bell JSK, Moya Balasch M, Jasker K, Martens EC. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol. 2013;88:1–15. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- Thurlow LR, Thomas VC, Fleming SD, Hancock LE. Enterococcus faecalis capsular polysaccharide serotypes C and D and their contributions to host innate immune evasion. Infect Immun. 2009;77:5551–5557. doi: 10.1128/IAI.00576-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli A, Retini C, Monari C, Tascini C, Bistoni F, Kozel TR. Purified capsular polysaccharide of Cryptococcus neoformans induces interleukin-10 secretion by human monocytes. Infect Immun. 1996;64:2846–2849. doi: 10.1128/iai.64.7.2846-2849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker WR, Shepherd ES, Sonnenburg JL. Tunable Expression Tools Enable Single-Cell Strain Distinction in the Gut Microbiome. Cell. 2017;169:538–546. e12. doi: 10.1016/j.cell.2017.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Storms R, Tsang A. A quantitative starch–iodine method for measuring alpha-amylase and glucoamylase activities. Anal Biochem. 2006;351:146–148. doi: 10.1016/j.ab.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Strains, vectors, and oligonucleotides used in this study. Related to STAR Methods.
Single nucleotide polymorphisms (SNPs) from each of the 10 strains competed in this study. Related to STAR Methods. Genomic DNA from the WT strain, the acapsular strain, and each of the 8 single CPS-expressing strains was sequenced on an Illumina HiSeq and genomes were assembled in DNASTAR SeqMan NGen. SNPs not found in the WT strain (or in all but the WT strain) are displayed. Deletion of cps loci, locking of invertible promoters, and inclusion of unique barcode sequences were all verified but not included in this table.
Fold changes in bacterial genes in vivo: single CPS-expressing strains relative to the acapsular strain. Related to Figure 2. Locus tag numbers highlighted in yellow correspond to cps locus genes. Positive expression changes are highlighted in green while negative expression changes are in red.
Minimum inhibitory concentrations of ciprofloxacin, polymyxin B, and human beta-defensin 3 (μg/ml). Related to Figure 4. Mean ± SEM displayed for each strain incubated with each antimicrobial compound. Kruskal-Wallis tests for each antimicrobial compound for each strain versus wild-type were not significant at p = 0.05. ND: Not determined.
Strains, genes, and capsular polysaccharide synthesis loci in comparative genomic and metagenomic analyses. Related to Figure 5.
Additional information on relative cps expression from previously published datasets. Related to STAR Methods.