Abstract
The concept of antibody microarrays is one of the most versatile approaches within multiplexed immunoassay technologies. These types of arrays have increasingly become an attractive tool for the exploratory detection and study of protein abundance, function, pathways, and potential drug targets. Due to the properties of the antibody microarrays and their potential use in basic research and clinical analytics, various types of antibody microarrays have already been developed. In spite of the growing number of studies utilizing this technique, few reviews about antibody microarray technology have been presented to reflect the quality and future uses of the generated data. In this review, we provide a summary of the recent applications of antibody microarray techniques in basic biology and clinical studies, providing insights into the current trends and future of protein analysis.
Electronic supplementary material
The online version of this article (10.1186/s12014-018-9184-2) contains supplementary material, which is available to authorized users.
Keywords: Antibody microarray, Signalling, Drug mechanism, Clinical application, Systems biology, Technology advances
Background
Antibody microarrays are built on immobilizing antibodies for a parallel analysis of multiple targets in a given sample [1]. Today’s antibody and affinity reagent-engineering methods have helped to advance the methodology [2, 3]. Antibodies and a variety of antibody derivatives have been used to build arrays, including nanobodies, single-chain variable fragments (scFvs) and fragment antigen-binding (Fab)-fragments [4]. In addition, phage display [5] and ribosome display [6], combined with advanced materials and bioinformatics development have being driving forces in recent years [7].
The typical workflow of an antibody microarray is depicted in Fig. 1. Briefly, antibodies are immobilized onto a chemically functionalized or otherwise modified surface. After blocking the reactive groups of the surface, a sample containing soluble proteins of interest is incubated on the array, and the targeted proteins from the sample are captured by the antibodies. The resulting binding events are reported directly by fluorescent labelling of the sample or by the addition of a secondary detection reagent.
Fig. 1.
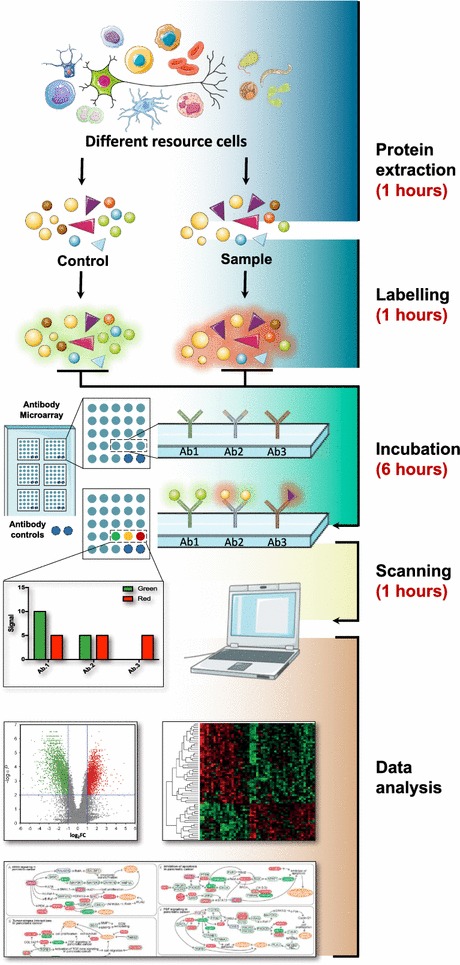
Review of planar antibody microarray technologies and their applications in the field of proteomics. Images were adopted from Servier Medical Art by Servier (http://www.servier.com/Powerpoint-image-bank) and modified by the authors under the following terms: CREATIVE COMMONS Attribution 3.0 Unported (CC BY 3.0)
The attractiveness of antibody microarrays is that they can be used to study a diverse number of biological processes [8] and have been used to investigate protein–protein interactions [9], signal pathway analysis [10], studies of post-translation modifications [11], and detection of toxins [12]. In the clinical context, arrays have enabled opportunities to identify novel disease biomarkers [13] as well as generating unique proteome signature by comparing healthy and disease states. This information will be of great value in the future, enabling better disease management through improved diagnostics and the ability to track disease status and therapeutic efficacy.
Antibody microarrays have demonstrated a number of advantages compared to traditional, single analyte methods of protein analysis, such as, enzyme-linked immunosorbent assays (ELISA) and Western blotting. Microarrays are high throughput, highly sensitivity, require small sample volumes, and more recently have become more standardized and user friendly experimental procedures. Compared with mainstream proteomics strategies, especially mass spectrometry (MS), the process of antibody microarray assays is fast and takes less than 24 h from sample preparation to data interpretation. Detailed comparison is shown Fig. 2.
Fig. 2.
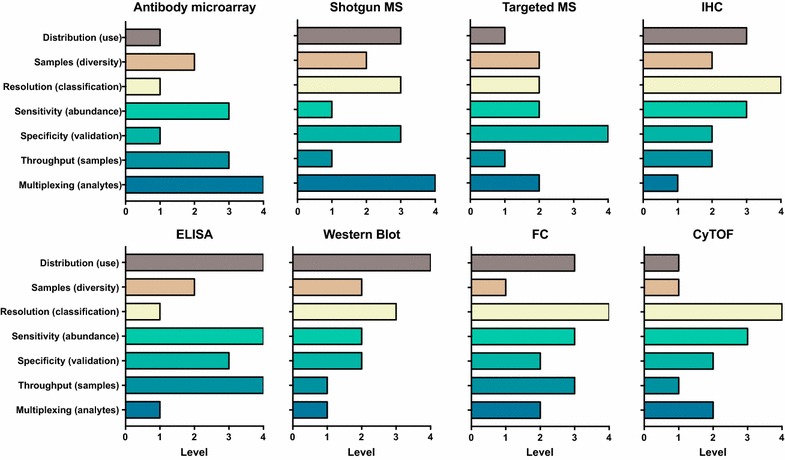
Comparison of different protein detection method following 7 categories: multiplexing (analytes), throughput (samples), specificity (validation), sensitivity (resolution), flexibility (content), samples (types), distribution (use). The scale number equal to high (4), medium (3), low (2), lower (1). The use of the scales as a comparative measure within each category. IHC immunohistochemistry, ELISA enzyme-linked immunosorbent assay, FC flow cytometry, CyTOF mass cytometry
In theory, like DNA microarrays, antibody microarrays can be designed to host a few to thousands, or even ten-thousands, of features. Currently, high features have been achieved by immobilizing proteins [14] or lysates [15, 16], and antibody microarrays are under active technological development and to-date operate at a few hundred features [17–19]. The arrays can be constructed either host many features per sample or be designed to compartmentalize the array into sets of arrays that allow many samples to be investigated simultaneously. Generally, the latter is more common, particularly if a large number of clinical samples are analysed in a given study. For the analysis of a large amount of samples, bead based systems have become attractive, as they can be processed in microtiter plates and by liquid handling devices using cytometry for instant data availability [20, 21]. Importantly, antibody microarrays generate high dimensional data that can be processed using already well-established DNA microarray software, since the images scanned from planar antibody microarrays are similar to those of DNA microarrays.
Antibody microarray experiments are most often used as the initial tool during the biomarker discovery process. These results continue down the validation pipeline using additional antibody-based assays, such as ELISA and Western blotting, or immune-capture MS to validate the findings clinical/pathophysiological relevance [22]. However, the cross-reactivity to off-target proteins that are more abundant than the target of interest is still a challenge for the single binder version of the assay. Thus, undesired reactions with other target proteins represent one of the main obstacles in establishing high performance and high specificity assays. The scope of this review is to outline the antibody microarray techniques that are being used today for biology studies and clinical research.
Recent applications (since 2011)
Due to unique capabilities of the antibody microarray and its applicability in a range of biomedical projects, a series of different antibody microarrays have been developed, of which some have become commercially available. Herein, we have collected some of the representative studies in the last 7 years (Table 1), and organized our overview into two parts depending on the focus of the projects: basic biology-driven studies and clinical research. As it remains practically impossible to cover all the recent applications of antibody microarray, we focus on those applications which we deemed are of high interest and findings of important clinical significance.
Table 1.
Antibody microarray summary
| Lab/company | Area | Antibody microarray type | Abs number | PMID |
|---|---|---|---|---|
| Medsaic Pty | Colorectal cancer | DotScan™ CRC microarrays | 122 | 25445327 |
| Medsaic Pty | Systemic lupus erythematosus | DotScan™ antibody microarray | 82 | 23516448 |
| Medsaic Pty | Chronic lymphocytic leukemia | DotScan™ CLL antibody microarray | 182 | 24289109 |
| Medsaic Pty | Cancer | DotScan™ antibody microarrays | 144 | 27086589 |
| Clontech | Alzheimer’s disease | Antibody Microarray 500 | 500 | 22554416 |
| Clontech | Prostate cancer | Antibody Microarray 507 | 507 | 23280553 |
| Clontech | Glaucoma | Ab Microarray 500 | 508 | 22974818 |
| Fullmoon Biosystems Inc | Explorer Antibody Microarray | 656 | ||
| Fullmoon Biosystems Inc | Squamous cell lung and esophageal carcinoma | PEX100 | 1318 | 26040563 |
| Fullmoon Biosystems Inc | Prostate cancer | Phosphorylation-specific antibody microarray | 95 | 24009409 |
| Fullmoon Biosystems Inc | Customized antibody microarray | 248 | ||
| Kinexus Bioinformatics | Smoke | KAM-1.0 | 650 | 21627322 |
| Kinexus Bioinformatics | Breast cancer | KAM-1.1 | 650 | 21423216 |
| Kinexus Bioinformatics | A. pleuropneumoniae infection | KAM-850 | 854 | 26577697 |
| Kinexus Bioinformatics | hiPSC-CMs | KAM-850 | 854 | 25055963 |
| Sigma-Aldrich | Breast cancer | Cell Signaling Antibody Microarray | 224 | 21394501 |
| Lab Vision Corporation | Programmed cell death | TAA-001 Lab Vision Corporation | 720 | 23619569 |
| Abnova | Alzheimer’s disease | Master Antibody Microarray | 656 | 25428253 |
| Proteogen Inc | Drug mechanism | ProteoChip | 48 | 21558493 |
| Jörg D. Hoheisel | Non-muscle-invasive bladder cancer | Home made | 818 | 24610664 |
| Jörg D. Hoheisel | Glioblastoma | Home made | 724 | 26232108 |
| Jörg D. Hoheisel | Pancreatic cancer | Home made | 810 | 22579748 |
| Paul D. Lampe | Pancreas cancer | Home made | 4096 | 25589628 |
| Paul D. Lampe | Biomarker for cancer | Home made | 3600 | 24185138 |
| Paul D. Lampe | Autoantigen-autoantibody complexes | Home made | 3600 | 23541305 |
| Nicholas A. Shackel | Liver diseases | Home made | 60 | 22863037 |
| Nicholas A. Shackel | Hepatitis C virus | Home made | 90 | 25706280 |
| Christer Wingren | Pancreatitis | Home made | 121 | 22930578 |
| Diane M. Cibrik | Renal allograft rejection | Home made | 108 | 24323459 |
| Daniel Böhm | Breast cancer | Home made | 23 | 21885915 |
| Carl Borrebaeck | Pancreas cancer | Home made | 180 | 25196118 |
Review of planar antibody microarrays from recent studies. Lab/company, area of research, antibody microarray type and number of immobilized antibodies (abs) are summarized. The microarray with the highest number of antibodies belongs to Paul D. Lampe’s group and their home-made antibody microarray
Basic research
Signalling pathway related
Changes in signalling pathways is a hallmark of many disease states, including cancer [23, 24], diabetes [25, 26] and neurodegenerative disorders [27]. Building antibody arrays using capture reagents targeting proteins in the signalling cascade of interest has enabled researchers to investigate changes in protein profiles and modifications in signalling pathways in normal biological processes and disease states. Researchers have applied this concept to a range of biological sample materials and preparations, including cell lysates [28], tissue extracts [29], and plasma [30].
Calbindin-D28 k (CB), an important calcium-binding protein that acts as a calcium buffer and is found to be expressed at lower levels in the brains of mice and humans with Alzheimer’s disease but it is not known if these changes contribute to AD-related dysfunction (AD). Kook et al. generated a CB-deficient Alzheimer’s transgenic mouse (CBKOTg) to investigate in CB’s contribution to signaling pathways in AD [23]. Utilizing antibody microarrays to examine mouse brain tissue, they identified significant alterations in cell death pathways, synaptic transmission and MAPK signalling pathways upon CB deletion. These findings were verified by immunohistochemistry which showed increased apoptotic markers and increased neuronal death, demonstrating that antibody microarrays can provide novel information, enhancing our understanding of the role CB and its implication in AD pathophysiology.
Antibody microarrays have been used to uncover the physiological role in highly conserved proteins, including elongation factor 4. Gao et al. dissected the mechanism of quality-control factor mitochondrial Elongation factor 4 (mtEF4) in translation using a phospho-explorer antibody microarray [31]. By comparing cell lysates between mtEF4 knock-out and WT mice testis tissues, they found that the fold enrichment of mTOR was the highest among different signalling pathways. The authors showed that with mtEF4 deletion, the major feedback signal from the somatic cytoplasm is mTOR upregulation. This was accompanied by an increased cytoplasmic translation, thus indicating that mTOR plays a crucial role as a downstream effector compensating for mitochondrial translation deficiency [32]. Furthermore, the study linked previously undescribed cross-talk between mtEF4-dependent quality control in the mitochondria and the mTOR pathway in the cytoplasm.
Drug mechanism
With the help of antibody microarray assays, researchers can investigate drug mechanisms in a systematic and efficient manner. Among many possible applications, tumor-induced angiogenesis has been studied as it plays a pivotal role in cancer progression [33]. P11, a novel peptide ligand containing a PDZ-binding motif (Ser-Asp-Val) with high affinity to integrin αvβ3, was identified from a hexapeptide library (PS-SPCL) using an antibody microarray [34]. The pharmacological mechanism of P11, was elucidated using a specifically designed pharmacoproteomic microarray approach containing 48 cancer related antibodies [35]. The study revealed that P11 inhibits bFGF-induced human umbilical vein endothelial cell proliferation via mitogen-activated protein kinase and extracellular-signal regulated kinase inhibition. In addition, the microarray revealed P11 caused the upregulation of apoptotic marker p53, resulted in apoptosis induction via activation of the caspases system, which indicates P11 may play a key role in preventing tumor progression. The link between P11 and p53 presents clinical and basic cancer researchers will help to elucidate further clues about the role of P11 and its inhibitory target.
Lovastatin, a natural product derived from Aspergillus terreus or Monascus ruber, has been widely used as a cholesterol-lowing drug in the clinic. Yang et al. have used an antibody microarray containing 656 antibodies with a focus on different functional cell pathways, which suggested that lovastatin also has anti-cancer properties, through poorly defined mechanisms. There is a strong need to find effective cancer treatments and to understand their mechanisms of action. Using a microarray, breast cancer cells were studied under hypoxic conditions to investigate the molecular mechanisms through Lovastin exherts its effect. They showed 17 up-regulated proteins and 20 down-regulated proteins for lovastatin treated breast cancer cells, compared to the control, and the results were subsequently validated by real-time PCR [36]. The protein signature included proteins involved in apoptosis, cell proliferation and tumour metastasis, linking modulation of these pathways to the pharmacological action of lovastin.
The type 2 diabetes drug metformin, has also proposed to have anti-cancer properties. Lee et al. investigated the potency of novel metformin derivatives using an antibody microarray built to study cell cycle-related molecules. The authors showed that metformin-butyrate (MFB) had better anti-tumor efficacies than metformin-HCl, through greater neoplastic activity and better efficiency at impairing cell cycle progression during S and G2/M phases in addition to appearing to have preferential cytotoxic effects on breast cancer stem cell populations [37]. These findings highlight how antibody arrays can provide proteomic evidence supporting development of drugs with better efficacy.
For basic biology research, antibody microarrays can serve in detecting signalling proteins and proteins associated to a particular phenotype. Arrays with parallel protein detection create an opportunity to investigate multiple signalling pathways in a single experiment, providing insights into novel mechanisms, such as the study of disease progression, drug interaction, and response to infections.
Clinical research
Analysis of clinical samples at the protein level will enable improvement in diagnostics markers, and directing treatment options. Since proteins are involved in the majority of cellular processes, protein analysis may provide a biological disease signature, generating detailed insights about the current state of a cell, organ or system. Antibody microarrays can provide a real-time guide of current changes in biological processes during health and disease. In this section, we will exemplify how antibody microarrays have been used to investigate clinical cohorts, primarily analysing serum and plasma samples. It is not possible to cover all the aspects of antibody microarrays for clinical research, herein, we only focus on several of the most important diseases, i.e. autoimmune diseases, infectious diseases, cancer and neurodegenerative diseases.
Autoimmune diseases
The diagnosis of systemic lupus erythematosus (SLE) is challenging due to its heterogeneous clinical presentation and the lack of robust biomarkers to distinguish it from active and inactive disease as well as from other autoimmune diseases. Lin et al. [38] performed a DotScan™ antibody microarray screening of peripheral blood mononuclear cells from 60 SLE patients of varying disease activity, 25 rheumatoid arthritis patients, 28 other autoimmune disease samples, and 24 healthy controls. The antibody microarray profiles could distinguish active SLE patients from healthy controls. Using a leukocyte capture array improved the discriminative ability of conventional SLE diagnostics, by verifying serum anti-dsDNA, complements C3 and C4, the microarrays increased the capability of discrimination of semi-active and active SLE, information which will assist better disease management.
In another study, Carlsson et al. [39] constructed 135 human recombinant single-chain fragment variables (scFv) targeting immune proteins to build an in-house antibody microarray. In this study, they examined patients with systemic sclerosis (SSc), SLE and 15 healthy volunteers. SSc is an autoimmune disorder affecting connecting tissue, which can be challenging to differentially diagnose from SLE. The array identified 40 differentially expressed proteins creating a candidate proteome signature to delineate SLE and its severity from SSc. This protein signature was a better disease classification than single or even combinations of conventional clinical parameters, including, ANA, anti-DNA, SLEDAI-2 k, C1q, C3, C4 and CRP, illustrating the potential use for antibody microarrays to create new disease-signatures which will add clinical value to disease management.
Infectious diseases
Infections activate the immune system differently, and these mechanisms can be investigated using antibody arrays on a proteome-wide level. Helicobacter pylori is a pathogen that colonizes roughly one half of the world’s population which causes chronic gastritis. Sukri et al. [40] employed the DotScan™ antibody microarray to determine the tolerance of immune system toward tumor cells in gastric cancer using 144 CD antibodies to profile the distribution of CD markers between Helicobacter pylori infected and un-infected gastric adenocarcinoma cells. Interestingly, they found gastric adenocarcinoma cell line AGS infected by cagA + H. pylori showed increased CD27 expression, which is essential for maintenance of T cell population, and increased CD markers were also detected in H. pylori-infected gastric cancer patients. This study suggests not only the tolerance of the immune system toward gastric cancer, but also the immune response variations exploited by different H. pylori strains.
Furthermore, antibody arrays can be useful in tracking a continuing change in the physiological environment, including disease status and response to therapies and interventions. After liver transplantation, 94% of patients were found with hepatitis C virus (HCV) induced histological damage, with some patients suffering severe disease recurrence [41]. There is an urgent need for predictive biomarkers to identify recurrent disease severity. Using a CD antibody microarray, peripheral blood from patients defined as pre-transplant, early, mid, and late-transplant groups were investigated to predict the severity of HCV recurrence after transplantation [42]. Serial blood samples taken from patients before and after liver transplant enabled tracking of the disease milieu, with its own internal controls. Five CD markers (CD27, CD182, CD260, CD41, and CD34) were significantly increased in severe recurrence compared to mild recurrence. This result shows that antibody arrays may assist in assessing recurrent HCV disease severity after liver transplantation.
Ellmark et al. [43] devised a single framework recombinant antibody (SinFabs) microarray containing 127 different antibodies against immune-regulatory antigens selected from the n-CoDeR library on gastric adenocarcinoma. They elucidated that distinct tumor- as well as infection-associated protein expression signatures, such as IL-9, IL-11, and MCP-4, could be identified from the plasma proteomes and serve as potential biomarkers. These findings may aid to improve the understanding of H-pylori induced cancer and may pave the way for refined diagnostics in the future.
Bead based antibody arrays have been performed to investigate differential protein profiles in plasma in children suffering from malaria and malaria-related complications [44]. From 1000 proteins screened, 41 proteins had differential expression between malaria infected children and community controls. Thirteen further proteins were linked to malaria-disease severity. These findings indicate the involvement of inflammation as well as unbalanced metabolism of glucose in severe forms of the disease and the biggest changes were seen in two of muscle proteins (carbonic anhydrase 3 and creatine kinase) pointed at muscle damage and lesions for children with cerebral malaria. These findings may aid the development of simple tests for classifying malaria-infected children into group showing a higher risk for the severe form of the disease.
Cancer
Antibody microarrays have been applied for cancer research mainly to study cancer progression and candidate proteins that may serve as diagnostic biomarkers [45, 46]. Pancreatic cancer is an aggressive disease with poor prognosis, and disease-specific biomarkers that offer early and accurate diagnosis are in urgent need. Using an in-house developed recombinant antibody microarray platform, Wingren et al. [47] screened sera from 148 patients with pancreatic cancer, chronic pancreatitis, autoimmune pancreatitis (AIP), and healthy controls. They identified a panel of 25 protein targets, including IL-2, IL-11, IL-12, TNF-, which contributed to distinguishing pancreatic cancer from healthy controls. This 25 protein signature exhibits a high diagnostic potential (AUC of 0.88). In addition, the group has recently extended their analysis of their panel to additional sample sets, including lymphoma [48], prostate cancer [49], and breast cancer in cell lysates [50] and plasma [51]. These findings are useful to aid the development of novel and multivariate diagnostics methods.
One of the most important questions in cancer biology, is determining the invasiveness of a tumor. One study developed a customized an antibody microarray platform containing 4096 features to interrogate plasma samples spanning pre-invasive and invasive diseases from a mouse model of pancreatic ductal adenocarcinoma (PDA) [18]. They found a protein signature, comprising of the differential expression of three proteins, ERBB2, TNC and ESR1, which could be used to improve the AUC from 0.86 (95% confidence interval [CI] 0.76–0.96) to 0.97 (95% CI 0.92–1.0) when the PDA marker CA19-9 included.
The majority of newly diagnosed cases of bladder cancer are low-stage, low-grade, non-muscle-invasive [52]. After standard transurethral resection, 50–70% of tumors recur, however 10–30% of the tumors will progress to muscle-invasive disease [53, 54]. Understanding the mechanism of tumor progression could provide more useful information to clinical scopes. Srinivasan et al. studied cell lysates with an antibody microarray containing 810 cancer related antibodies constructed by the Hoheisel lab [55]. The authors built a multivariate classifier containing 20 proteins, which facilitatated the prediction of recurrence with a sensitivity of 80% and a specificity of 100%. Interestingly, they found repression of the TGF-β signaling pathway in recurrent cancer. The signaling factors IFNG, TNF-α, and THBS1 were expressed less and the abundance of the inhibitor MAPK3 (also known as ERK1) was higher, while SMAD2, SMAD3, and SMAD4 were again significantly underrepresented [56]. The data indicated that TGF-β signaling pathway inhibitors may reduce bladder cancer recurrence [57].
Puig-Costa et al. [58] employed antibody microarrays to discover biomarkers for gastric cancer (GC). Notably, the antibody microarray contained antibodies targeting different well-known functional proteins that play a crucial role in cancer progression, including 120 cytokines, 43 angiogenic factors, 41 growth factors, 40 inflammatory factors and 10 metalloproteinases. Ingenuity pathway analysis confirmed that some biomarkers, such as ICAM-1 and angiogenin, indicated high inflammatory response in GC patients. Cellular movement and immune cell trafficking targets such as monocyte chemoattractant protein (MCP)-1 was found to be overrepresented within GC patients. The positive predictive and negative predictive values in this validation cohort were 75% (95% CI 53–90) and 80% (95% CI 56–94), respectively. Finally, antibody microarray analyses of the GC-associated inflammatory proteome identified a 21-protein inflammatory protein-driven gastric cancer signature (INPROGAS) that accurately discriminated GC from noncancerous gastric mucosa, and may provide new leads for the analysis of cancer progression.
In prostate cancer, Schwenk et al. [59] used antibody arrays on suspension bead arrays to compare plasma levels of proteins between different groups in order to find additional biomarkers alongside prostate-specific antigen (PSA). Besides classifying the patients based on PSA, they identified decreased plasma CNDP1 levels and in a subsequent larger studies this was shown to association in more aggressive forms of the disease. A sandwich immunoassay was developed to validate these findings in more than 1200 patients, and the association of the decreased CNDP1 to lymph node metastasis was further elucidated [60]. This demonstrates one of the very few studies that have successfully validated an initial indication through the antibody microarray pipeline and the development of a targeted assay. In a subsequent investigation of cancer cachexia, plasma levels of CNDP1 was found to be decreased in cachexic patients, hence pointing at a metabolic role of changes in CNDP1 levels [61].
In a study of small intestine neuroendocrine tumours, Darmanis et al. [62] investigated the plasma levels of two independent study set (77 and 132 samples) and a targeted bead array for 124 unique proteins. They could achieve classification accuracy of up to 85% using a panel of proteins and concluded with proposing novel candidates for classifying the tumors. Of the shortlisted candidates, such as for IGFBP2 and IGF1, the group performed ELISA assays to confirm the indications. These findings indicate a metabolic association of plasma proteins with cancer and support to consider these targets in future cancer studies.
Neurodegenerative diseases
Analysis of protein expression provides a possibility to extend the current knowledge on neurodegenerative pathophysiology [63]. Recent advances within the field of proteomics have offered the potential to search for novel biomarkers using antibody microarrays [64]. Advancement in protocol development have allowed researchers to study not only blood, but also cerebrospinal fluid (CSF) [69].
Within neuroproteomics, the Nilsson lab has conducted several studies by using antibodies form the Human Protein Atlas [65] together with the suspension bead array assays. Profiling CSF from patients with Multiple Sclerosis (MS), they found GAP43, a cytoplasmic protein involved in the formation, and regeneration of neurons, hence a promising biomarker diseases of the brain [66]. More recently, Remnestål et al. [67] compared protein levels in 441 CSF samples collected from different neurodegenerative disease sample sets as well as CSF collected post-mortem. Among 376 antibodies, the synaptic proteins GAP43 and NRGN were found to be associated to AD patients compared with controls.
Using plasma samples, CSF and brain tissue from patients suffering from MS, a large-scale screening was conducted that started from utilizing 4500 antibodies on bead arrays [68]. A set of proteins was found to be associated with MS subtypes in CSF and plasma. Utilizing some of the candidate antibodies raised against IRF8, IL7, and METTL14 also for immuno-fluorescence analysis of brain tissue showed staining of neurons in proximity of MS lesions. This indicates that antibodies selected from array-based assays for analysis of body fluids can also provide further evidence at the affected tissue.
Lastly, the bead arrays were used to profile plasma from patients suffering from amyotrophic lateral sclerosis (ALS) [69], where 367 ALS patients and 101 controls were analyzed for 278 proteins. The study concludes with proposing neurofilament medium polypeptide (NEFM), solute carrier family 25 (SLC25A20), and regulator of G-protein signalling 18 (RGS18) as valuable proteins because they are involved in processes related to disease pathophysiology, which warrant further validation in independent sample sets.
Current issues and solutions
We list commercial antibody microarrays in Table 2, and further details about these products can be found as supplementary informtion (Additional file 1: Table S1). Despite the rapid technological advances in recent years, there are still technical issues that need to be overcome to ensure high-specificity and reproducibility of antibody arrays, to ensure high impact data and meaningful conclusions.
Table 2.
Summary of commercial available antibody microarrays from different companies, Type G (Glass), M (Membrane)
| Provider | Ab microarray category | Type | Max number of abs | Major focus |
|---|---|---|---|---|
| Raybio | 215 | G/M | 400 | Immune response, cytokines, cancer, signalling |
| Full moon | 43 | G | 1358 | Signalling, cancer biomarker, cytokines |
| Abnova | 39 | G/M | 247 | Signalling, cytokines |
| R&D | 20 | M | 105 | Signalling, cytokines, functional assay, proteomic profile |
| Abcam | 14 | M | 400 | Signalling, cytokines, functional assay, biomarker |
| MyBiosource | 4 | G | 656 | Signalling, proteomic profile |
| Hypromatrix | 3 | M | 400 | Signalling, functional assay |
| Panorama | 2 | G | 112 | Gene regulation, signalling |
| Arrayit | 1 | G | 380 | Plasma protein |
| Kinex™ | 1 | G | 878 | Signalling |
There is a need for validated antibodies for antibody microarray applications and respective sample types. The data from such validation efforts should consequently be provided. The quality of validated antibodies, the standardization of data analysis, and data storage and sharing are three important challenges to be considered.
Proteomic investigations using antibody microarrays have provided valuable data that reveal the pathophysiological background of a disease [70]. But there are also issues to address. Besides the overall still relatively small number of studies, the number of samples analysed in each study is usually limited to less than a hundred. Typically, these experiments are performed in an “exploratory cohort” first, examining preferably 1000’s of different features. These microarray results need to be validated with an independent set of preferably larger number of specimens and by other methods.
Moreover, samples prepared from frozen or fresh samples may provide different profiles depending on the specific set and that could be referred to a “study markers”. Another challenge of applying antibody microarrays for further studies is the heterozygosity of specific diseases (e.g. relapse remitting multiple sclerosis versus non-relapse multiple sclerosis) requesting careful study designs, in-depth knowledge about the sample itself and the disease phenotype of the subject it was obtained from. Some of these considerations concerning the design of discovery assays for the analysis of plasma proteome have been recently addressed elsewhere [71–73].
Finally, for a better study applying antibody microarray. Control should be carefully taken into consideration. Sample controls, not only internalize control like beta-actin or GAPDH for each sample, but also positive (or negative) control that already confirmed by other method would help to set cut-off for further analysis.
The reproducibility challenge of the antibody microarray
One challenge for data derived from antibody microarrays is often linked to a lack of reproducibility. There are several experimental factors that may contribute to these problems, such as the surface chemistry and the mode of antibody immobilization, lack of sufficiently stringent processes for generation and validation of antibodies, as well as a bias from sample preparation, labelling, and batch effects from analysing samples across several slides or plates. We next focus on how to use general and novel methods to reduce systematic errors and potentially improve the reproducibility of antibody microarray experiment.
In the single-color approach, labels can be introduced at different efficacy in different samples due to chemistry dye binding stronger to certain amino acids. This might expand since the detection of proteins may reveal different fluorescent signal even though their quantity is the same. The samples could be labelled by a single fluorescent reporter dye, e.g. Cy3, Cy5, then probed one by one on antibody microarrays. In this circumstance, the slide-to-slide variation is unavoidable, thus reducing the reliability of the final microarray data and introducing systematic error. This systemic error can be attenuated by employing a dual-color labeling strategy inspired by DNA microarrays. Schroder et al. [74] reported an antibody microarray containing 741 cancer-related antibodies, employing dual-color mode. This approach has greatly improved the reproducibility, as reported in the study. The coefficients of variation (CV) of 89% features between 20 slides derived from the five production batches were under 15%, the CV of 96% features were under 20%. This dual-color mode can be applied in many other protein microarray experiments as long as they are comparing paired samples. This dual-color strategy will not only enhance the reproducibility of antibody microarrays, but may also improve the robustness of the assay.
The cross-reactivity of antibody microarray
The DNA microarray has rapidly been scaled up from 256 to 6.5 million features, but to date sensitive antibody microarray assays have only been scaled up to thousands of targets [75]. Cross-reactivity, commonly arising when multiple detection antibodies are mixed, is a known weakness of these assays that is mitigated by lengthy optimization [76].
In antibody co-localization microarray (ACM) described by Pla-Roca et al. [77], both capture and detection antibodies were physically spotted onto the same two-dimensional coordinate, which reduced cross-reactivity and improved antibody reproducibility. Specifically, after spotting of the capture antibodies, the chip was removed from the arrayer, incubated with the sample, and then placed back onto the arrayer to be spotted with detection antibodies. After 3 years, the same group developed the next generation ACM, employing an array containing 50 capture antibodies. Instead of spotting, they used another perfect matched array with pre-spotted detection antibodies to transfer detection antibodies onto the assay slide. This method employs a pair of antibodies to detect a single protein similar to ELISA, thus improving the accuracy of the test that sets the path for an improved reproducibility.
The availability of validated antibodies for antibody microarray
The primary power of antibody microarrays based on single binder assays lies in the capability of multiple protein detection. Theoretically, the power is only limited to the number of antibodies on the microarray. However, unlike DNA microarrays where the probes can easily be synthesized at large scale either off-chip or on-chip, antibodies have to be produced and validated one-by-one. Thus, the number of qualified antibodies for antibody microarrays is still limited and remains a challenge. To address this, high-throughput screening strategies such as phage display may hold the solution to this problem.
In addition, binding molecules of non-protein scaffolds have been described [78], yet the number remains low when compared to almost 3 million commercially available antibodies (http://www.antibodypedia.com/). However, the size of synthetic binder libraries can further be increased by introducing additional modifications to the scaffolds, such as aptamers [79]. To maximize the use and impact of affinity reagents, the EU FP7 programme AFFINOMICS (http://www.affinomics.org), together with the preceding EU programmes ProteomeBinders and AffinityProteome, aims to extend affinity proteomics research by generating a large-scale resource of validated protein-binding molecules for characterization of the human proteome [80].
Despite antibody use, a comprehensive scientific framework for the validation antibodies across research applications (i.e. immunohistochemistry, immuno-precipitation) is essential for the success of the antibody microarray platform. Uhlen et al. [81] have developed a five ‘pillars’ strategy for antibody validation: (1) genetic strategies, (2) orthogonal strategies, (3) independent antibody strategies, (4) expression of tagged proteins, and (5) immunocapture followed by MS for antibody validation. Immunoreagents, such as polyclonal, monoclonal antibodies, and other recombinant or synthetic binders are also suitable for this proposal. It will highly improve the quality and reproducibility for methods, such as antibody microarrays, given that the binders are validated in the intended application and for the sample type of interest.
For antibody microarrays, immuno-capture mass spectrometry or alternative assays such as dual-capture assays [82] offer similar assay formats for the target enrichment, as both require the immobilization of the binder. In the dual-capture system, antibodies are used to enrich their target proteins from a solution (e.g. plasma) and after labelling the proteins on the capture bead surface, the eluted proteins are detected by a multiplexed array consisting of several antibodies of interest. This allows two antibodies to bind a common target, hence presenting a sandwich assay based on consecutive binding. In comparison to MS, the antibodies used for read-out in affinity based assays need to be selected in relation to the target of the primary enrichment. Affinity assays are often still more sensitive and require less antibody and sample material then MS, however an MS based read-out will provide a wider view on which proteins have been enriched from a give sample, such as cell lysates [83] and more recently also plasma [84]. The breadth of MS data generated from such immuno-enrichment assessments requires though in order to carefully define specific peptides of interest from those that appear as unspecific and common contaminants [85].
Data processing, analysis and storage
Unlike the DNA microarray community, which has established standards for data submission and storage, such as minimum information about a microarray experiment (MIAME) [86] these guidelines have not yet been implemented for antibody microarrays. Most of the analysis tools in use for proteome-analysis have been adopted from the field of DNA microarrays. The data formats are similar between antibody microarrays and traditional DNA microarrays, the data analysis can also be subdivided into steps of image capture, data preprocessing, differential expression detection, clustering. There are now several R packages available to support some of these steps, such as Limma [87], Clusterprofiler [88], Qlucore Omics Explorer (http://www.qlucore.com) and IPA (http://www.ingenuity.com). DNA microarray-specific MIAME standards have been applied in Gene Expression Omnibus (GEO), which includes some data sets from antibody microarrays. Since diversity of antibody microarrays exceeds those of DNA microarrays due to diversified applications, a classification scheme that can include different types of antibody microarray data is needed.
In data acquisition and analysis, Ensink et al. [89] provided an advanced approach for locating signals in microarray image data, called segment and fit thresholding (SFT). This approach was optimized based on the initial settings by locating background and signal on the acquired antibody microarray image and immunofluorescence data. It was found that SFT performed well over multiple, diverse image characteristics without readjustment of settings, promoting full automation in microarray image analysis.
Image acquisition parameters such as the laser power and photomultiplier tube gain (PMT) during scanning can influence the readout of fluorescent intensities and thus may affect data quantitation. Gu et al. demonstrated an experimental approach using two fluorescent dyes to determine optimal settings of scan parameters for microarray experiments. Their efforts may facilitate the improvement of the accuracy of quantitative outcome in antibody microarray experiments [90].
Several main technical features and assay procedures remain to be improved. The handling of protein microarray data, i.e. the biostatistics parts, is one of the key features. Delfani et al. [91] have standardized the analytical workflow of their in-house designed recombinant antibody microarray platform and Hong et al. [92] have described an approach of handling data generated from different assay batches from suspension bead array assays. This is an important aspect when aiming to generate data for a larger number of samples addressing the remaining technical issues such as: antibody quality, array production, sample labelling, and selected assay conditions and biostatistics subjects.
Finally, to address the database issue, Xu et al. [93] constructed the protein microarray database (PMD), which is specifically designed for archiving and analyzing protein microarray data including antibody microarray data. In PMD, users can easily browse and search the entire database by experimental name, protein microarray type, and sample information. Additionally, PMD integrates several data analysis tools and provides an automated data analysis pipeline for users, who can obtain a comprehensive analysis report for their protein microarray data. Making antibody array data available to the community will be one of the important aspects to assess data quality and eventually improve the acceptance of microarray assay data.
Discussion
Antibody and affinity reagent libraries
Beyond antibodies, other types of affinity reagents may unlock the bright future of microarrays [78]. Fridy et al. have showed a robust, fast pipeline to produce nanobodies. Inspired by their previous studies of neutralizing HIV antibodies in human, they have developed a strategy for rapid discovery of nanobodies through computation. The nanobody discovery is based on MS identification of affinity-purified heavy-chain antibodies isolated from an individual llama, using a DNA sequence database generated from the same animal. Notably, the authors have provided an advanced technology to improve nanobody’s affinity by conforming dimerized nanobodies to specific antigens. Utilizing this method, phage display could be used to screen for interesting proteins and develop novel nanobody microarrays [94].
Aptamers are single-stranded oligonucleotides, utilized as affinity reagents. Depending on their sequence, the temperature, pH and the presence of certain ions, the aptamers fold into defined three-dimensional (3D) structures [95]. Properly folded aptamers are able to bind other molecules with high affinity and specificity [96]. High-throughput DNA engineering methods can be applied to generate high-diversity libraries to screen for target proteins, which can be utilized in microarray applications. The use of aptamers selected for a slow off-rate has indeed lead to building high-throughput assays for plasma profiling [97], nowadays expanding into the analysis of the plasma proteome in thousands of samples [98]. Recent studies point at an extended use of this platform for studying thousands of proteins and samples. Other affinity reagents, such as designed ankyrin repeat proteins (DARPins) [99] and single-chain variable fragment (scFV) [100, 101] also hold the promise to further boost the advancement and application of affinity reagent microarrays [102].
Combine with other technologies
Flow cytometry
There are further developments and novel applications of antibody microarrays. Sharivkin et al. [103] showed a novel method of combining antibody microarrays with flow cytometry to isolate specific cell types from differentiating stem cell populations. An array of antibodies against cell-surface antigens was printed on a hydrogel coated glass slide. Live cells isolated by different markers using flow cytometry. And then captured on specific antibody spots by interaction between their surface antigens and the printed antibodies. This allowed identifying additional markers that further refined different subpopulation from flow sorting [104].
CRISPR/Cas9
The bacterial clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system for genome editing has greatly expanded the toolbox for mammalian genetics, enabling the rapid generation of isogenic cell lines and mice with modified alleles. Wang et al. [105] generated a knockout pool for two human cell lines, and this loss-of-function genetic screening approach was suitable for both positive and negative selection. With 1000’s of genes to manipulate in one experiment, the antibody microarray has a great opportunity to be combined with this pool and the CRISPR/Cas9 technology holds promise to create a whole human proteome knock out library, which had the potential to revolutionise how we study proteins and their interactions in health and disease.
Advance materials
Chinnasamy et al. presented a novel lateral flow microarray-based device using a novel dually labelled gold nanoparticle-strategy for rapid and sensitive detection of clinical serum samples. Each gold nanoparticle was conjugated to an optimized ratio of HRP and anti-IgE, allowing a significant improvement of assay sensitivity as compared to commercially available detection reagents. Also, due to the rapid and simple procedure, inexpensive materials and read-out by means of a consumer flatbed scanner, the presented assay may provide a sensitive and low-cost platform for multiple fast testing which could facilitate antibody microarray in translation medicine area [106].
Antibody microarrays have been established alongside the advancement of materials. Hu et al. [107] developed a hierarchically nanostructured organic–inorganic hybrid substrate, comprising of randomly oriented ZnO nanorods on glass slides with coaxially tethered dense polymer brush, which highly improved the limit of detection (LOD) to 100 fg/mL. Huang et al. [108] have invented a glycol-gold nanoparticle-based antibody microarray, with potential to amplify a signal that could be detected by the naked eye.
A perspective on the future applications of antibody microarrays
As outlined above, applications of antibody arrays can be found across various areas of biology and diseases. Some of the concepts have entered clinical testing, while a wide range of content and applications are still being developed or improved. The future of antibody arrays will, among many other factors, depend on three main aspects as eluted to below: (1) validation, (2) continued technical advances, and (3) translation and dissemination.
With currently ongoing discussions about the validation of antibodies used for the generation of affinity proteomics assays data, a key element for a continuous use of antibody microarrays will relate to how experimental results from these multiplexed systems can be validated and confirmed by targeted assays. Validation refers to confirming and certifying specificity and reproducibility of the assay across batches of experiments, as well as to describe possible sources of interference and variance. With more and more affinity reagents on the market—currently almost 3,000,000 antibody products can be purchased covering 94% of all human genes (19,155 genes) according to Anitbodypedia (http://www.antibodypedia.org)—not all of these will be functional for antibody microarray assays. Hence, careful upfront qualification and selection of binding molecules is required. This process will require additional, orthogonal and multiplexed assays and read-out systems that resemble the conditions of the array-based assays. An important readout with be the correlation with data from mass spectrometry, as this technology is capable of delivering absolute identifications of target molecules.
Analytical sensitivity and degrees of multiplexing are important areas of improvement but these are already common (popular) features that have been addressed ever since. Advances made through interactions with alternative assay concepts and read-out systems are often driven by other areas of life science and engineering. These have and will continue to advance array technologies and its derivate systems. This will also lead to new content being integrated into the current and coming platforms.
Lastly, disseminating the method’s utility and data will be key for a continued awareness about the technology. Making antibody array data and assays more available will subsequently enable its use beyond those labs developing technologies to meet and even larger group of users. The success of antibody arrays will then be dependent on how robust and translatable the generated information is. Only if the data provides new leads to advance research projects or supports clinical decision making for an improved precision, then we will see a continued use of antibody arrays.
Conclusion
In summary, various forms of antibody microarrays have gradually evolved as analytical tools for proteomics research. These microarrays have facilitated advances in basic biology and discoveries towards potential tools of clinical use. With the progressive development, standardization of the experimental workflow and data interpretation, increasing numbers of binding reagents as well as innovative concepts are arising and will provide further advances in the foreseeable future. Antibody microarrays and concepts based upon, hold the promises to contribute to the advancement of research and even diagnostic applications, but there is a strong need to apply the appropriate validation schemes and follow-up studies to confirm the presented indications for a possible clinical decision making.
Additional file
Additional file 1. Detailed information about the home-made and commercially available antibody microarrays.
Acknowledgments
Authors’ contributions
ZC and TC contributed to the writing of the manuscript, and had a significant input from both authors. JS and ST revised critically had a major input in the writing of the paper. All authors read and approved the final manuscript.
Acknowledgements
We thank Philippa Pettingill’s inspired comments for this review.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was funded in part by the National Key Research and Development Program of China (2016YFA0500601), and by National Natural Science Foundation of China Grants (31370813 and 31670831). This work was supported by grants for KTH-SJTU seed-funding program and Science for Life Laboratory. The KTH Center for Applied Precision Medicine (KCAP) funded by the Erling-Persson Family Foundation, and the Human Protein Atlas funded by The Knut and Alice Wallenberg Foundation are acknowledged for their support. We also thank Philippa Pettingill for her valuable input on the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- scFvs
single-chain variable fragments
- Fab
fragment antigen-binding
- CD
cluster of differentiation
- ELISA
enzyme-linked immunosorbent assays
- DNA
deoxyribonucleic acid
- MS
mass spectrometry
- CB
calbindin-D28 k
- AD
Alzheimer’s disease
- mtEF4
mitochondrial elongation factor 4
- mTOR
mammalian target of rapamycin-
- PDZ
ser-asp-val amino acid
- PS-SPCL
positional scanning-synthetic peptide combinatorial libraries
- bFGF
basic fibroblast growth factor
- PCR
polymerase chain reaction
- TNFR1
tumor necrosis factor receptor 1
- DD
death domain
- PARP
poly ADP-ribose polymerase
- MFB
metformin-butyrate
- SLE
systemic lupus erythematosus
- SSc
systemic sclerosis
- ANA
a positive antinuclear antibody
- SLEDAI-2k
systemic lupus erythematosus disease activity index 2000
- CRP
C-reactive protein
- cagA
cytotoxin-associated gene A
- HCV
hepatitis C virus
- SinFabs
single framework recombinant antibody
- IL-9
interleukin-9
- IL-11
interleukin-11
- MCP-1
monocyte chemoattractant protein 1
- MCP-4
monocyte chemotactic protein-4
- CA3
carbonic anhydrase III
- CK
creatine kinase
- AIP
autoimmune pancreatitis
- TNF-α
tumor necrosis factor α
- AUC
area under the curve
- PDA
pancreatic ductal adenocarcinoma
- CI
confidence interval
- TGF-β
transforming growth factor beta
- IFNG
interferon gamma
- THBS1
thrombospondin 1
- MAPK3
mitogen-activated protein kinase 3
- ERK1
extracellular signal-regulated kinase 1
- GC
gastric cancer
- ICAM-1
intercellular adhesion molecule 1
- INPROGAS
inflammatory protein-driven gastric cancer signature
- CNDP1
carnosine dipeptidase 1
- IGFBP2
insulin like growth factor binding protein 2
- IGF1
insulin like growth factor 1
- CSF
cerebrospinal fluid
- POLG
DNA polymerase gamma
- MGMT
O-6-methylguanine-DNA methyltransferase
- ApoD
apolipoprotein D
- GAP43
growth associated protein 43
- NRGN
neurogranin
- IRF8
interferon regulatory factor 8
- METTL14
methyltransferase like 14
- ALS
amyotrophic lateral sclerosis
- NEFM
neurofilament medium polypeptide
- RGS18
regulator of G-protein signalling 18
- SLC25A20
solute carrier family 25
- CV
coefficients of variation
- ACM
antibody co-localization microarray
- DARPins
designed ankyrin repeat proteins
- MIAME
minimum information about a microarray experiment
- GEO
gene expressiono
- SFT
segment and fit thresholding
- PMT
photomultiplier tube gain
- PMD
protein microarray database
- CRISPR -Cas9
clustered regularly interspaced short palindromic repeats
- HRP
horseradish peroxidase
- IgE
immunoglobulin E
- LOD
limit of detection
- IHC
immunohistochemistry
- FC
flow cytometry
- CyTOF
mass cytometr
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12014-018-9184-2) contains supplementary material, which is available to authorized users.
Contributor Information
Jochen M. Schwenk, Email: jochen.schwenk@scilifelab.se
Sheng-ce Tao, Email: taosc@sjtu.edu.cn.
References
- 1.Tu S, Jiang HW, Liu CX, Zhou SM, Tao SC. Protein microarrays for studies of drug mechanisms and biomarker discovery in the era of systems biology. Curr Pharm Des. 2014;20(1):49–55. doi: 10.2174/138161282001140113123707. [DOI] [PubMed] [Google Scholar]
- 2.Kim SJ, Park Y, Hong HJ. Antibody engineering for the development of therapeutic antibodies. Mol Cells. 2005;20(1):17–29. [PubMed] [Google Scholar]
- 3.Maynard J, Georgiou G. Antibody engineering. Annu Rev Biomed Eng. 2000;2:339–376. doi: 10.1146/annurev.bioeng.2.1.339. [DOI] [PubMed] [Google Scholar]
- 4.Nelson AL. Antibody fragments: hope and hype. MAbs. 2010;2(1):77–83. doi: 10.4161/mabs.2.1.10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 6.Zahnd C, Amstutz P, Pluckthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Methods. 2007;4(3):269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 7.Chandra H, Reddy PJ, Srivastava S. Protein microarrays and novel detection platforms. Expert Rev Proteomics. 2011;8(1):61–79. doi: 10.1586/epr.10.99. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Guo S, Li Y, Zhou S, Tao S. Protein microarrays for systems biology. Acta Biochim Biophys Sin (Shanghai) 2011;43(3):161–171. doi: 10.1093/abbs/gmq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergsma D, Chen S, Buchweitz J, Gerszten R, Haab BB. Antibody-array interaction mapping, a new method to detect protein complexes applied to the discovery and study of serum amyloid P interactions with kininogen in human plasma. Mol Cell Proteomics. 2010;9(3):446–456. doi: 10.1074/mcp.M900418-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He HJ, Zong Y, Bernier M, Wang L. Sensing the insulin signaling pathway with an antibody array. Proteomics Clin Appl. 2009;3(12):1440–1450. doi: 10.1002/prca.200900020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivanov SS, Chung AS, Yuan ZL, Guan YJ, Sachs KV, Reichner JS, Chin YE. Antibodies immobilized as arrays to profile protein post-translational modifications in mammalian cells. Mol Cell Proteomics. 2004;3(8):788–795. doi: 10.1074/mcp.M300130-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Rucker VC, Havenstrite KL, Herr AE. Antibody microarrays for native toxin detection. Anal Biochem. 2005;339(2):262–270. doi: 10.1016/j.ab.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Carbayo M. Antibody microarrays as tools for biomarker discovery. Methods Mol Biol. 2011;785:159–182. doi: 10.1007/978-1-61779-286-1_11. [DOI] [PubMed] [Google Scholar]
- 14.Sutandy FX, Qian J, Chen CS, Zhu H. Overview of protein microarrays. Curr Protoc Protein Sci. 2013;27:1. doi: 10.1002/0471140864.ps2701s72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Zhao W, Akbani R, Liu W, Ju Z, Ling S, Vellano CP, Roebuck P, Yu Q, Eterovic AK, et al. Characterization of human cancer cell lines by reverse-phase protein arrays. Cancer Cell. 2017;31(2):225–239. doi: 10.1016/j.ccell.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gujral TS, Karp RL, Finski A, Chan M, Schwartz PE, MacBeath G, Sorger P. Profiling phospho-signaling networks in breast cancer using reverse-phase protein arrays. Oncogene. 2013;32(29):3470–3476. doi: 10.1038/onc.2012.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berrade L, Garcia AE, Camarero JA. Protein microarrays: novel developments and applications. Pharm Res. 2011;28(7):1480–1499. doi: 10.1007/s11095-010-0325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mirus JE, Zhang Y, Li CI, Lokshin AE, Prentice RL, Hingorani SR, Lampe PD. Cross-species antibody microarray interrogation identifies a 3-protein panel of plasma biomarkers for early diagnosis of pancreas cancer. Clin Cancer Res. 2015;21(7):1764–1771. doi: 10.1158/1078-0432.CCR-13-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borrebaeck CA, Sturfelt G, Wingren C. Recombinant antibody microarray for profiling the serum proteome of SLE. Methods Mol Biol. 2014;1134:67–78. doi: 10.1007/978-1-4939-0326-9_6. [DOI] [PubMed] [Google Scholar]
- 20.Drobin K, Nilsson P, Schwenk JM. Highly multiplexed antibody suspension bead arrays for plasma protein profiling. Methods Mol Biol. 2013;1023:137–145. doi: 10.1007/978-1-4614-7209-4_8. [DOI] [PubMed] [Google Scholar]
- 21.Schwenk JM, Gry M, Rimini R, Uhlen M, Nilsson P. Antibody suspension bead arrays within serum proteomics. J Proteome Res. 2008;7(8):3168–3179. doi: 10.1021/pr700890b. [DOI] [PubMed] [Google Scholar]
- 22.Neiman M, Fredolini C, Johansson H, Lehtio J, Nygren PA, Uhlen M, Nilsson P, Schwenk JM. Selectivity analysis of single binder assays used in plasma protein profiling. Proteomics. 2013;13(23–24):3406–3410. doi: 10.1002/pmic.201300030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168(6):960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT. The PI3 K pathway in human disease. Cell. 2017;170(4):605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brannmark C, Nyman E, Fagerholm S, Bergenholm L, Ekstrand EM, Cedersund G, Stralfors P. Insulin signaling in type 2 diabetes: experimental and modeling analyses reveal mechanisms of insulin resistance in human adipocytes. J Biol Chem. 2013;288(14):9867–9880. doi: 10.1074/jbc.M112.432062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boucher J, Kleinridders A, Kahn CR: Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol 2014, 6(1). [DOI] [PMC free article] [PubMed]
- 27.Bezprozvanny I. Calcium signaling and neurodegenerative diseases. Trends Mol Med. 2009;15(3):89–100. doi: 10.1016/j.molmed.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Lian S, Meng L, Qu L, Shou C. Antibody array revealed PRL-3 affects protein phosphorylation and cytokine secretion. PLoS ONE. 2017;12(1):e0169665. doi: 10.1371/journal.pone.0169665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dexlin-Mellby L, Sandstrom A, Antberg L, Gunnarsson J, Hansson SR, Borrebaeck CA, Wingren C. Design of recombinant antibody microarrays for membrane protein profiling of cell lysates and tissue extracts. Proteomics. 2011;11(8):1550–1554. doi: 10.1002/pmic.200900808. [DOI] [PubMed] [Google Scholar]
- 30.Mahlknecht P, Stemberger S, Sprenger F, Rainer J, Hametner E, Kirchmair R, Grabmer C, Scherfler C, Wenning GK, Seppi K, et al. An antibody microarray analysis of serum cytokines in neurodegenerative Parkinsonian syndromes. Proteome Sci. 2012;10(1):71. doi: 10.1186/1477-5956-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Qin Y. The paradox of elongation factor 4: Highly conserved, yet of no physiological significance? Biochem J. 2013;452(2):173–181. doi: 10.1042/BJ20121792. [DOI] [PubMed] [Google Scholar]
- 32.Gao Y, Bai X, Zhang D, Han C, Yuan J, Liu W, Cao X, Chen Z, Shangguan F, Zhu Z, et al. Mammalian elongation factor 4 regulates mitochondrial translation essential for spermatogenesis. Nat Struct Mol Biol. 2016;23(5):441–449. doi: 10.1038/nsmb.3206. [DOI] [PubMed] [Google Scholar]
- 33.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3(6):401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 34.Lee Y, Kang DK, Chang SI, Han MH, Kang IC. High-throughput screening of novel peptide inhibitors of an integrin receptor from the hexapeptide library by using a protein microarray chip. J Biomol Screen. 2004;9(8):687–694. doi: 10.1177/1087057104268125. [DOI] [PubMed] [Google Scholar]
- 35.Bang JY, Kim EY, Kang DK, Chang SI, Han MH, Baek KH, Kang IC. Pharmacoproteomic analysis of a novel cell-permeable peptide inhibitor of tumor-induced angiogenesis. Mol Cell Proteomics. 2011;10(8):M110.005264. doi: 10.1074/mcp.M110.005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang T, Yao H, He G, Song L, Liu N, Wang Y, Yang Y, Keller ET, Deng X. Effects of lovastatin on MDA-MB-231 breast cancer cells: an antibody microarray analysis. J Cancer. 2016;7(2):192–199. doi: 10.7150/jca.13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee KM, Lee M, Lee J, Kim SW, Moon HG, Noh DY, Han W. Enhanced anti-tumor activity and cytotoxic effect on cancer stem cell population of metformin-butyrate compared with metformin HCl in breast cancer. Oncotarget. 2016;7(25):38500–38512. doi: 10.18632/oncotarget.9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin MW, Ho JW, Harrison LC, dos Remedios CG, Adelstein S. An antibody-based leukocyte-capture microarray for the diagnosis of systemic lupus erythematosus. PLoS ONE. 2013;8(3):e58199. doi: 10.1371/journal.pone.0058199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlsson A, Wuttge DM, Ingvarsson J, Bengtsson AA, Sturfelt G, Borrebaeck CA, Wingren C. Serum protein profiling of systemic lupus erythematosus and systemic sclerosis using recombinant antibody microarrays. Mol Cell Proteomics MCP. 2011;10(5):M110.005033. doi: 10.1074/mcp.M110.005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sukri A, Hanafiah A, Kosai NR, Mohamed Taher M, Mohamed Rose I. Surface antigen profiling of helicobacter pylori-infected and -uninfected gastric cancer cells using antibody microarray. Helicobacter. 2016;21:417–427. doi: 10.1111/hel.12295. [DOI] [PubMed] [Google Scholar]
- 41.Neumann UP, Berg T, Fau-Bahra M, Bahra M, Fau-Puhl G, Puhl G, Fau-Guckelberger O, Guckelberger O, Fau-Langrehr JM, Langrehr JM, Fau-Neuhaus P, Neuhaus P. Long-term outcome of liver transplants for chronic hepatitis C: a 10-year follow-up. Transplantation. 2004;77(2):226–231. doi: 10.1097/01.TP.0000101738.27552.9D. [DOI] [PubMed] [Google Scholar]
- 42.Rahman W, Tu T, Budzinska M, Huang P, Belov L, Chrisp JS, Christopherson RI, Warner FJ, Bowden DS, Thompson AJ, et al. Analysis of post-liver transplant hepatitis C virus recurrence using serial cluster of differentiation antibody microarrays. Transplantation. 2015;99(9):e120–e126. doi: 10.1097/TP.0000000000000617. [DOI] [PubMed] [Google Scholar]
- 43.Ellmark P, Ingvarsson J, Carlsson A, Lundin BS, Wingren C, Borrebaeck CA. Identification of protein expression signatures associated with Helicobacter pylori infection and gastric adenocarcinoma using recombinant antibody microarrays. Mol Cell Proteomics. 2006;5(9):1638–1646. doi: 10.1074/mcp.M600170-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Bachmann J, Burte F, Pramana S, Conte I, Brown BJ, Orimadegun AE, Ajetunmobi WA, Afolabi NK, Akinkunmi F, Omokhodion S, et al. Affinity proteomics reveals elevated muscle proteins in plasma of children with cerebral malaria. PLoS Pathog. 2014;10(4):e1004038. doi: 10.1371/journal.ppat.1004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingvarsson J, Wingren C, Carlsson A, Ellmark P, Wahren B, Engstrom G, Harmenberg U, Krogh M, Peterson C, Borrebaeck CA. Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics. 2008;8(11):2211–2219. doi: 10.1002/pmic.200701167. [DOI] [PubMed] [Google Scholar]
- 46.Haab BB. Methods and applications of antibody microarrays in cancer research. Proteomics. 2003;3(11):2116–2122. doi: 10.1002/pmic.200300595. [DOI] [PubMed] [Google Scholar]
- 47.Wingren C, Sandstrom A, Segersvard R, Carlsson A, Andersson R, Lohr M, Borrebaeck CA. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012;72(10):2481–2490. doi: 10.1158/0008-5472.CAN-11-2883. [DOI] [PubMed] [Google Scholar]
- 48.Pauly F, Smedby KE, Jerkeman M, Hjalgrim H, Ohlsson M, Rosenquist R, Borrebaeck CA, Wingren C. Identification of B-cell lymphoma subsets by plasma protein profiling using recombinant antibody microarrays. Leuk Res. 2014;38(6):682–690. doi: 10.1016/j.leukres.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Nordstrom M, Wingren C, Rose C, Bjartell A, Becker C, Lilja H, Borrebaeck CA. Identification of plasma protein profiles associated with risk groups of prostate cancer patients. Proteomics Clin Appl. 2014;8(11–12):951–962. doi: 10.1002/prca.201300059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skoog P, Ohlsson M, Ferno M, Ryden L, Borrebaeck CAK, Wingren C. Tumor tissue protein signatures reflect histological grade of breast cancer. PLoS ONE. 2017;12(6):e0179775. doi: 10.1371/journal.pone.0179775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlsson A, Wingren C, Kristensson M, Rose C, Ferno M, Olsson H, Jernstrom H, Ek S, Gustavsson E, Ingvar C, et al. Molecular serum portraits in patients with primary breast cancer predict the development of distant metastases. Proc Natl Acad Sci USA. 2011;108(34):14252–14257. doi: 10.1073/pnas.1103125108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobs BL, Lee CT, Montie JE. Bladder cancer in 2010: how far have we come? CA Cancer J Clin. 2010;60(4):244–272. doi: 10.3322/caac.20077. [DOI] [PubMed] [Google Scholar]
- 53.Saad A, Hanbury DC, Fau-McNicholas TA, McNicholas TA, Fau-Boustead GB, Gb Boustead, Fau-Morgan S, Morgan S, Fau-Woodman AC, Woodman AC. A study comparing various noninvasive methods of detecting bladder cancer in urine. BJU Int. 2002;89(4):369–373. doi: 10.1046/j.1464-4096.2001.01699.x. [DOI] [PubMed] [Google Scholar]
- 54.Soloway MS, Sofer M, Fau-Vaidya A, Vaidya A. Contemporary management of stage T1 transitional cell carcinoma of the bladder. J Urol. 2002;167(4):1573–1583. [PubMed] [Google Scholar]
- 55.Schroder C, Jacob A, Tonack S, Radon TP, Sill M, Zucknick M, Ruffer S, Costello E, Neoptolemos JP, Crnogorac-Jurcevic T, et al. Dual-color proteomic profiling of complex samples with a microarray of 810 cancer-related antibodies. Mol Cell Proteomics. 2010;9(6):1271–1280. doi: 10.1074/mcp.M900419-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srinivasan H, Allory Y, Sill M, Vordos D, Alhamdani MS, Radvanyi F, Hoheisel JD, Schroder C. Prediction of recurrence of non muscle-invasive bladder cancer by means of a protein signature identified by antibody microarray analyses. Proteomics. 2014;14(11):1333–1342. doi: 10.1002/pmic.201300320. [DOI] [PubMed] [Google Scholar]
- 57.Liang Y, Zhu F, Zhang H, Chen D, Zhang X, Gao Q, Li Y. Conditional ablation of TGF-beta signaling inhibits tumor progression and invasion in an induced mouse bladder cancer model. Sci Rep. 2016;6:29479. doi: 10.1038/srep29479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puig-Costa M, Codina-Cazador A, Cortes-Pastoret E, Oliveras-Ferraros C, Cufi S, Flaquer S, Llopis-Puigmarti F, Pujol-Amado E, Corominas-Faja B, Cuyas E, et al. Discovery and validation of an inflammatory protein-driven gastric cancer signature (INPROGAS) using antibody microarray-based oncoproteomics. Oncotarget. 2014;5(7):1942–1954. doi: 10.18632/oncotarget.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schwenk JM, Igel U, Neiman M, Langen H, Becker C, Bjartell A, Ponten F, Wiklund F, Gronberg H, Nilsson P, et al. Toward next generation plasma profiling via heat-induced epitope retrieval and array-based assays. Mol Cell Proteomics MCP. 2010;9(11):2497–2507. doi: 10.1074/mcp.M110.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qundos U, Johannesson H, Fredolini C, O’Hurley G, Branca R, Uhlén M, Wiklund F, Bjartell A, Nilsson P, Schwenk JM. Analysis of plasma from prostate cancer patients links decreased carnosine dipeptidase 1 levels to lymph node metastasis. Transl Proteomics. 2014;2:14–24. [Google Scholar]
- 61.Arner P, Henjes F, Schwenk JM, Darmanis S, Dahlman I, Iresjo BM, Naredi P, Agustsson T, Lundholm K, Nilsson P, et al. Circulating carnosine dipeptidase 1 associates with weight loss and poor prognosis in gastrointestinal cancer. PLoS ONE. 2015;10(4):e0123566. doi: 10.1371/journal.pone.0123566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Darmanis S, Cui T, Drobin K, Li SC, Oberg K, Nilsson P, Schwenk JM, Giandomenico V. Identification of candidate serum proteins for classifying well-differentiated small intestinal neuroendocrine tumors. PLoS ONE. 2013;8(11):e81712. doi: 10.1371/journal.pone.0081712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humpel C. Identifying and validating biomarkers for Alzheimer’s disease. Trends Biotechnol. 2011;29(1):26–32. doi: 10.1016/j.tibtech.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haggmark A, Schwenk JM, Nilsson P. Neuroproteomic profiling of human body fluids. Proteomics Clin Appl. 2016;10(4):485–502. doi: 10.1002/prca.201500065. [DOI] [PubMed] [Google Scholar]
- 65.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 66.Haggmark A, Bystrom S, Ayoglu B, Qundos U, Uhlen M, Khademi M, Olsson T, Schwenk JM, Nilsson P. Antibody-based profiling of cerebrospinal fluid within multiple sclerosis. Proteomics. 2013;13(15):2256–2267. doi: 10.1002/pmic.201200580. [DOI] [PubMed] [Google Scholar]
- 67.Remnestal J, Just D, Mitsios N, Fredolini C, Mulder J, Schwenk JM, Uhlen M, Kultima K, Ingelsson M, Kilander L, et al. CSF profiling of the human brain enriched proteome reveals associations of neuromodulin and neurogranin to Alzheimer’s disease. Proteomics Clin Appl. 2016;10(12):1242–1253. doi: 10.1002/prca.201500150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bystrom S, Ayoglu B, Haggmark A, Mitsios N, Hong MG, Drobin K, Forsstrom B, Fredolini C, Khademi M, Amor S, et al. Affinity proteomic profiling of plasma, cerebrospinal fluid, and brain tissue within multiple sclerosis. J Proteome Res. 2014;13(11):4607–4619. doi: 10.1021/pr500609e. [DOI] [PubMed] [Google Scholar]
- 69.Haggmark A, Mikus M, Mohsenchian A, Hong MG, Forsstrom B, Gajewska B, Baranczyk-Kuzma A, Uhlen M, Schwenk JM, Kuzma-Kozakiewicz M, et al. Plasma profiling reveals three proteins associated to amyotrophic lateral sclerosis. Ann Clin Transl Neurol. 2014;1(8):544–553. doi: 10.1002/acn3.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu H, Qian J. Applications of functional protein microarrays in basic and clinical research. Adv Genet. 2012;79:123–155. doi: 10.1016/B978-0-12-394395-8.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwenk JM, Omenn GS, Sun Z, Campbell DS, Baker MS, Overall CM, Aebersold R, Moritz RL, Deutsch EW. The human plasma proteome draft of 2017: building on the human plasma PeptideAtlas from mass spectrometry and complementary assays. J Proteome Res. 2017;16(12):4299–4310. doi: 10.1021/acs.jproteome.7b00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith JG, Gerszten RE. Emerging affinity-based proteomic technologies for large-scale plasma profiling in cardiovascular disease. Circulation. 2017;135(17):1651–1664. doi: 10.1161/CIRCULATIONAHA.116.025446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geyer PE, Holdt LM, Teupser D, Mann M. Revisiting biomarker discovery by plasma proteomics. Mol Syst Biol. 2017;13(9):942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schroder C, Alhamdani MS, Fellenberg K, Bauer A, Jacob A, Hoheisel JD. Robust protein profiling with complex antibody microarrays in a dual-colour mode. Methods Mol Biol. 2011;785:203–221. doi: 10.1007/978-1-61779-286-1_14. [DOI] [PubMed] [Google Scholar]
- 75.Juncker D, Bergeron S, Laforte V, Li H. Cross-reactivity in antibody microarrays and multiplexed sandwich assays: shedding light on the dark side of multiplexing. Curr Opin Chem Biol. 2014;18:29–37. doi: 10.1016/j.cbpa.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 76.Poetz O, Ostendorp R, Brocks B, Schwenk JM, Stoll D, Joos TO, Templin MF. Protein microarrays for antibody profiling: specificity and affinity determination on a chip. Proteomics. 2005;5(9):2402–2411. doi: 10.1002/pmic.200401299. [DOI] [PubMed] [Google Scholar]
- 77.Pla-Roca M, Leulmi RF, Tourekhanova S, Bergeron S, Laforte V, Moreau E, Gosline SJ, Bertos N, Hallett M, Park M, et al. Antibody colocalization microarray: a scalable technology for multiplex protein analysis in complex samples. Mol Cell Proteomics. 2012;11(4):Antibody colocalization microarray: a scalable technology for multiplex protein analysis in complex samples. doi: 10.1074/mcp.M111.011460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lollo B, Steele F, Gold L. Beyond antibodies: new affinity reagents to unlock the proteome. Proteomics. 2014;14(6):638–644. doi: 10.1002/pmic.201300187. [DOI] [PubMed] [Google Scholar]
- 79.Gawande BN, Rohloff JC, Carter JD, von Carlowitz I, Zhang C, Schneider DJ, Janjic N. Selection of DNA aptamers with two modified bases. Proc Natl Acad Sci USA. 2017;114(11):2898–2903. doi: 10.1073/pnas.1615475114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stoevesandt O, Taussig MJ. European and international collaboration in affinity proteomics. N Biotechnol. 2012;29(5):511–514. doi: 10.1016/j.nbt.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 81.Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, Rimm DL, Rodriguez H, Hiltke T, Snyder M, et al. A proposal for validation of antibodies. Nat Methods. 2016;13:823–827. doi: 10.1038/nmeth.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ayoglu B, Birgersson E, Mezger A, Nilsson M, Uhlen M, Nilsson P, Schwenk JM. Multiplexed protein profiling by sequential affinity capture. Proteomics. 2016;16(8):1251–1256. doi: 10.1002/pmic.201500398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marcon E, Jain H, Bhattacharya A, Guo H, Phanse S, Pu S, Byram G, Collins BC, Dowdell E, Fenner M, et al. Assessment of a method to characterize antibody selectivity and specificity for use in immunoprecipitation. Nat Methods. 2015;12(8):725–731. doi: 10.1038/nmeth.3472. [DOI] [PubMed] [Google Scholar]
- 84.Fredolini C, Bystrom S, Sanchez-Rivera L, Ioannou M, Tamburro D, Branca RM, Nilsson P, Lehtio J, Schwenk JM. Utilizing a resource of enrichment profiles in plasma for the systematic assessment of antibody selectivity. bioRxiv. 2017;158022. 10.1101/158022. [DOI] [PMC free article] [PubMed]
- 85.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10(8):730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 87.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ensink E, Sinha J, Sinha A, Tang H, Calderone HM, Hostetter G, Winter J, Cherba D, Brand RE, Allen PJ, et al. Segment and fit thresholding: a new method for image analysis applied to microarray and immunofluorescence data. Anal Chem. 2015;87(19):9715–9721. doi: 10.1021/acs.analchem.5b03159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gu Q, Sivanandam TM. Optimizing scan parameters for antibody microarray experiments: accelerating robust systems diagnostics for life sciences. OMICS. 2014;18(6):385–398. doi: 10.1089/omi.2013.0095. [DOI] [PubMed] [Google Scholar]
- 91.Delfani P, Dexlin Mellby L, Nordstrom M, Holmer A, Ohlsson M, Borrebaeck CA, Wingren C. Technical advances of the recombinant antibody microarray technology platform for clinical immunoproteomics. PLoS ONE. 2016;11(7):e0159138. doi: 10.1371/journal.pone.0159138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hong MG, Lee W, Nilsson P, Pawitan Y, Schwenk JM. Multidimensional normalization to minimize plate effects of suspension bead array data. J Proteome Res. 2016;15(10):3473–3480. doi: 10.1021/acs.jproteome.5b01131. [DOI] [PubMed] [Google Scholar]
- 93.Xu Z, Huang L, Zhang H, Li Y, Guo S, Wang N, Wang SH, Chen Z, Wang J, Tao SC. PMD: a resource for archiving and analyzing protein microarray data. Sci Rep. 2016;6:2045–2322. doi: 10.1038/srep19956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fridy PC, Li Y, Keegan S, Thompson MK, Nudelman I, Scheid JF, Oeffinger M, Nussenzweig MC, Fenyo D, Chait BT, et al. A robust pipeline for rapid production of versatile nanobody repertoires. Nat Methods. 2014;11(12):1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reverdatto S, Burz DS, Shekhtman A. Peptide aptamers: development and applications. Curr Top Med Chem. 2015;15(12):1082–1101. doi: 10.2174/1568026615666150413153143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Witt M, Walter JG, Stahl F. Aptamer microarrays-current status and future prospects. Microarrays (Basel) 2015;4(2):115–132. doi: 10.3390/microarrays4020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE. 2010;5(12):e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, Burgess S, Jiang T, Paige E, Surendran P, Oliver-Williams C, Kamat MA, Prins BP, Wilcox SK, Zimmerman ES, Chi A, Bansal N, Spain SL, Wood AM, Morrell NW, Bradley JR, Janjic N, Roberts DJ, Ouwehand WH, Todd JA, Soranzo N, Suhre K, Paul DS, Fox CS, Plenge RM, Danesh J, Runz H, Butterworth AS. Consequences of natural perturbations in the human plasma proteome. bioRxiv. 2017;134551. 10.1101/134551.
- 99.Pluckthun A. Designed ankyrin repeat proteins (DARPins): binding proteins for research, diagnostics, and therapy. Annu Rev Pharmacol Toxicol. 2015;55:489–511. doi: 10.1146/annurev-pharmtox-010611-134654. [DOI] [PubMed] [Google Scholar]
- 100.Babel I, Barderas R, Pelaez-Garcia A, Casal JI. Antibodies on demand: a fast method for the production of human scFvs with minimal amounts of antigen. BMC Biotechnol. 2011;11:61. doi: 10.1186/1472-6750-11-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Seurynck-Servoss SL, Baird CL, Miller KD, Pefaur NB, Gonzalez RM, Apiyo DO, Engelmann HE, Srivastava S, Kagan J, Rodland KD, et al. Immobilization strategies for single-chain antibody microarrays. Proteomics. 2008;8(11):2199–2210. doi: 10.1002/pmic.200701036. [DOI] [PubMed] [Google Scholar]
- 102.Taussig MJ, Schmidt R, Cook EA, Stoevesandt O. Development of proteome-wide binding reagents for research and diagnostics. Proteomics Clin Appl. 2013;7(11–12):756–766. doi: 10.1002/prca.201300060. [DOI] [PubMed] [Google Scholar]
- 103.Sharivkin R, Walker MD, Soen Y. Functional proteomics screen enables enrichment of distinct cell types from human pancreatic islets. PLoS ONE. 2015;10(2):e0115100. doi: 10.1371/journal.pone.0115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sharivkin R, Walker MD, Soen Y. Proteomics-based dissection of human endoderm progenitors by differential cell capture on antibody array. Mol Cell Proteomics. 2012;11(9):586–595. doi: 10.1074/mcp.M111.016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chinnasamy T, Segerink LI, Nystrand M, Gantelius J, Svahn HA. A lateral flow paper microarray for rapid allergy point of care diagnostics. Analyst. 2014;139(10):2348–2354. doi: 10.1039/c3an01806g. [DOI] [PubMed] [Google Scholar]
- 107.Hu W, Liu Y, Chen T, Liu Y, Li CM. Hybrid ZnO nanorod-polymer brush hierarchically nanostructured substrate for sensitive antibody microarrays. Adv Mater. 2015;27(1):181–185. doi: 10.1002/adma.201403712. [DOI] [PubMed] [Google Scholar]
- 108.Huang LD, Adak AK, Yu CC, Hsiao WC, Lin HJ, Chen ML, Lin CC. Fabrication of highly stable glyco-gold nanoparticles and development of a glyco-gold nanoparticle-based oriented immobilized antibody microarray for lectin (GOAL) assay. Chemistry. 2015;21(10):3956–3967. doi: 10.1002/chem.201405747. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Detailed information about the home-made and commercially available antibody microarrays.
Data Availability Statement
All data generated or analysed during this study are included in this published article.


