Abstract
Maternal RNA/protein degradation and zygotic genome activation (ZGA), occurring during maternal-to-zygotic transition (MZT), are the first essential events for the development of pre-implantation embryos. Previously, we have shown the importance of the ubiquitin-proteasome system (UPS) for initiation of minor ZGA at the 1-cell stage of mouse embryos. However, little is known about the mechanism of involvement of the UPS-degraded maternal proteins in ZGA. In this study, we investigated the effect of inhibiting maternal protein degradation by the reversible proteasome inhibitor, MG132, on post-implantation development and ZGA regulation during early cleavage stages. Our study revealed that zygotic transcription by RNA polymerase II (Pol II) at the 1-cell stage was delayed and the full-term development was affected by transient proteasome inhibition during 1 to 9 h post-insemination (hpi). Furthermore, we found that the transient inhibition of proteasome activity at the 2-cell stage delayed the onset of transcription of some major ZGA genes. These results support the model hypothesizing the requirement of sequential degradation of maternal proteins by UPS for the proper onset of ZGA and normal progression of MZT in early mouse embryos.
Keywords: Maternal-to-zygotic transition, Ubiquitin-proteasome system, Zygotic genome activation
Oocytes, fertilized by a sperm, can acquire and maintain totipotency until a particular period of early embryonic development. Maternally inherited RNA/protein degradation and zygotic genome activation (ZGA) are essential events in establishing totipotency during the maternal-to-zygotic transition (MZT) [1, 2]. In mouse, ZGA consists of two waves: the minor ZGA at the late 1-cell stage, followed by the major ZGA at the 2-cell stage [3, 4]; each being strictly regulated by multiple mechanisms such as cell cycle, epigenetic modification, chromatin remodeling, and transcription factors activities [5]. Furthermore, the clearance of maternal RNAs/proteins and ZGA cooperatively progress through MZT toward embryogenesis [5]. Understanding these mechanisms will provide an important insight into the generation of a totipotent zygote.
In eukaryotic cells, autophagy and ubiquitin-proteasome system (UPS) are the major intracellular protein degradation pathways. The autophagy pathway is a bulk degradation system in which the cellular components, including macromolecules and organelles, are delivered to the lysosome for degradation [6]. On the other hand, UPS-dependent proteolysis is a selective degradation system in which the destruction of proteins is initiated by attaching multiple ubiquitin molecules to the target protein that is subsequently degraded by the 26S proteasome complex [7]. These two proteolytic pathways play a vital role in the maternal protein clearance during MZT [8,9,10]. In particular, UPS regulates transcription in multiple ways, both proteolytic and non-proteolytic, ranging from the level of chromatin structure to gene expression regulation [11,12,13].
Previously, we have shown that UPS is important for the onset of minor ZGA in early mouse embryos [14]. Under the selected optimal conditions, in terms of best efficiency and least side effects for development, we found that the onset of zygotic genes was delayed in both normally developed 2-cell embryos and arrested 1-cell embryos after a transient treatment with the reversible proteasome inhibitor MG132 [14]. We also identified a zygote-specific proteasome assembly chaperone (ZPAC), a unique proteasome assembly pathway specifically expressed in mouse gonads; expression of ZPAC was transiently increased at the mouse MZT [9, 15]. Furthermore, another group recently found that E3 ubiquitin ligase RNF114-mediated ubiquitination and degradation of TAB1 activates the NF-κB pathway during MZT [16]. Thus, the UPS-dependent proteolysis plays a crucial role in MZT. However, the mechanism by which the UPS-mediated maternal protein degradation is involved in full-term development and the onset of major ZGA in early mouse embryos is not yet understood.
In this study, we investigated the effect of transient inhibition of proteasome activity in early mouse embryos on their post-implantation development and the regulation of ZGA. We found that the fetal development rate was significantly reduced after the transient inhibition of proteasome activity in the early stage of fertilized eggs. In addition, the transient inhibition led to delayed recruitment of RNA polymerase II (Pol II) into pronuclei, delayed phosphorylation of Pol II as well as delayed DNA replication. Furthermore, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis of 2-cell embryos, with transient inhibition of proteasome activity, revealed a delay in the expression of some genes which are otherwise scheduled to be expressed during the major ZGA period. These findings imply that a transient inhibition of proteasome activity induces a delay in the onset of ZGA and affects full-term development in mice. Thus, our results suggest that a proper regulation of ZGA, via UPS-mediated degradation of maternal proteins, is required for normal progression of MZT in early mouse embryos.
Materials and Methods
Animals
For the collection of oocytes and in vitro fertilization, all mice (ICR strain) were purchased from Kiwa Laboratory Animals (Wakayama, Japan) at > 8 weeks of age and maintained in light-controlled, air-conditioned rooms. This study was carried out in strict accordance with the recommendations in the Guidelines of Kindai University for the Care and Use of Laboratory Animals. The protocol was approved by the Committee on the Ethics of Animal Experiments of Kindai University (Permit Number: KABT-26-002). All efforts were made to minimize the number of mice used in the study; all mice were killed by cervical dislocation, ensuring minimum suffering.
Collection of oocytes, in vitro fertilization and embryo culture
Collection of spermatozoa, oocytes, and fertilized embryos was performed as described [17]. In brief, spermatozoa were collected from the cauda epididymis of male mice. The sperm suspension was incubated in human tubal fluid (HTF) medium for 1.5 h to allow for capacitation at 37°C under 5% CO2. Oocytes were collected from the excised oviducts of female mice (2–3 months old) that had been super-ovulated with pregnant mare serum gonadotropin (PMSG; Serotropin, Teikoku Zoki, Tokyo, Japan) followed by human chorionic gonadotropin (hCG; Puberogen, Sankyo, Tokyo, Japan) 48 h later. Cumulus-oocyte complexes were recovered into pre-equilibrated HTF medium. The sperm suspension was added to the oocyte cultures, and morphologically normal fertilized oocytes were collected 1 h after insemination. The fertilized embryos were cultured in potassium simplex optimized medium (KSOM) at 37°C under 5% CO2.
Inhibitor treatment
Proteasome inhibitor MG132 (carbobenzoxy-L-leucyl-L-leucyl-L-leucinal) was purchased from Sigma-Aldrich (St. Louis, MO, USA; C2211). To inhibit the activity of proteasomes in early embryos, they were cultured in KSOM medium containing 5 µM MG132 [14]. In the control, same protocol was used without MG132. For inhibition of DNA replication, zygotes were treated with 1 µg/ml aphidicolin (Sigma-Aldrich; A0781). Zygotes were cultured in KSOM medium in presence of only 0.1% DMSO, for the control.
Embryo transfer
At 24 h post-insemination, morphologically normal 1-cell or 2-cell stage embryos were transferred into the oviducts of Day 1 pseudo-pregnant female mice (Jcl:MCH (ICR); CLEA Japan, Tokyo, Japan) for full-term development following standard procedures. In the controls, the same procedure was performed with MG132-untreated embryos. Autopsies of the recipient animals were performed on Day 19.5. The number of implantation sites and fetuses were examined. At least three independent experiments were performed for each group.
Proteasome activation assay
Peptidase activity of the specimens was measured with a fluorescent peptide substrate, N-succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (Suc-LLVY-AMC, Peptide Institute, Osaka, Japan; 3120-v), as described previously [18]. One hundred fresh embryos were used in each stage. At least three independent experiments were performed for each group.
Western blot analysis
The procedures were essentially performed as described previously [9, 15]. In brief, 30 cells of each embryonic stage, were subjected to sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis for the detection of polyubiquitinated proteins. The protein extracts were resolved in 7.5% running gel and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare, Little Chalfont, UK). The membranes were incubated in Block Ace (Dainippon Sumitomo Pharma, Osaka, Japan) at room temperature (RT) for 1 h, washed with phosphate-buffered saline containing 0.2% Tween 20 (PBST), and incubated at 4°C overnight with either anti-Ub antibody (final dilution, 1:300; Santa Cruz Biotechnology, Dallas, TX, USA; sc-9133) or anti-Actin antibody (final dilution, 1:50,000; Santa Cruz Biotechnology; sc-1616) as a loading control. The membranes were washed in PBST, incubated with donkey anti-rabbit IgG horseradish peroxidase (HRP) conjugate (final dilution, 1:50,000; Millipore, Billerica, MA, USA; AP182P) for anti-Ub or donkey anti-goat IgG HRP conjugate (final dilution, 1:50,000; Millipore; AP180P) for anti-Actin at RT for 1 h, washed again with PBST, and developed using ECL Prime Western Blotting detection reagent (GE Healthcare). At least three independent experiments were performed for each group.
Densitometric quantification analysis
Densitometric quantification analysis of the immunoblot bands was performed using an Image Studio Lite version 5.0.21 (LI-COR Biosciences, Lincoln, NE, USA).
Immunofluorescence analysis and microscopy
Subcellular localization of Pol II and its carboxyl-terminal domain (CTD) phosphorylation were determined by immunofluorescence analysis of early embryos, as described [19, 20]. Embryos were fixed in 4% paraformaldehyde (PFA; Nacalai Tesque, Kyoto, Japan) in PBS for 15 min at RT and the fixed samples were then incubated in PBS containing 0.2% Triton X-100 (Nacalai Tesque) for 1 h at RT. They were then incubated with monoclonal antibodies 8WG16, H14, and H5 (final dilution, 1:400; Covance, Berkeley, CA, USA; MMS-126R, MMS-134R, and MMS-129R), which recognize the CTD heptapeptide repeats and CTD phosphorylation at Ser-5 and Ser-2, respectively, in PBS containing 30 mg/ml bovine serum albumin at 4°C overnight. Thereafter, the cells were incubated with Alexa Fluor 488-labeled donkey anti-mouse IgG antibody (final dilution, 1:2,000; Invitrogen; A-21202) for anti-8WG16, and with Alexa Fluor 488-labeled goat anti-mouse IgM antibody (final dilution, 1:2,000; Invitrogen; A-21042) for anti-H14 and H5, all at RT for 1 h. Specimens were mounted on glass slides in VECTASHIELD mounting medium (Vector Laboratories, Burlingame, CA, USA) containing 3 µg/ml 4', 6-diamidino-2-phenylindole (DAPI) (Invitrogen; D1306). Finally, the slides were imaged using an Olympus BX51 microscope (Olympus, Tokyo, Japan) equipped with an Olympus DP 70 digital camera (Olympus). All experiments were performed at least in triplicates.
DNA replication analysis by EdU incorporation
The procedures were essentially performed using a commercial kit (Click-iT EdU Alexa Fluor 488 Imaging Kit, Life Technologies; C10337) as prescribed by the manufacturer. In brief, following in vitro fertilization, control and MG132-treated embryos were cultured in presence of 10 µM 5-ethynyl-2'-deoxyuridine (EdU) at indicated intervals. Embryos were fixed at the end of each interval and permeabilized with 0.2% Triton X-100 in PBS at RT for 1 h. Click-iT reaction cocktail was added for incubation at RT for 30 min. Specimens were mounted on glass slides in VECTASHIELD mounting medium containing 3 µg/ml DAPI. At least three independent experiments were performed for each group.
RT-qPCR analyses
Total RNA was isolated from 85 pooled embryos using the RNeasy Micro Kit (QIAGEN, Hilden, Germany; 74004) and from 10 embryos using PicoPure RNA Isolation Kit (Life Technologies; KIT0204). In brief, cDNA was synthesized from total RNA using Superscript III RT First-Strand Synthesis system (Life Technologies; 18080051). Prepared cDNA samples were amplified and analyzed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The primers used are described in Supplementary Table 1 (online only). Amplified products were run in a 7300 ABI Prism Sequence Detector (Applied Biosystems). At least three independent experiments were performed for each group.
Statistical analysis
For statistical analysis, we used StatView version 5.0 (SAS Institute, Cary, NC, USA) and Microsoft Excel, and performed analysis of variance (ANOVA) with an α level of 0.05 to determine possible statistically significant differences between the means of groups.
Results
Proteasome inhibition after fertilization accumulates ubiquitinated proteins
MG132 is a reversible proteasome inhibitor, which is a substrate analogue and potent transition-state inhibitor, primarily of the chymotrypsin-like activity of the proteasome [21]. Under previously optimized experimental conditions, ensuring the best efficiency and least side effects in early mouse embryos for transient inhibition of proteasome activity with MG132 (Fig. 1A) [14], we investigated the kinetics of proteasome activity in the transient MG132-treated embryos. As an initial observation, we detected polyubiquitinated proteins, yet to be degraded, using western blot analysis (Fig. 1B). Compared to that in untreated embryos, polyubiquitinated proteins accumulated in MG132-treated embryos at 9 hpi and reduced by 21 hpi (Fig. 1B). To confirm whether the proteasome activity is inhibited by the transient treatment with MG132, and to understand the recovery of activity after the treatment, we measured the chymotrypsin-like activity of proteasome in MG132-treated embryos. In accordance with the above results (Fig. 1B), the proteasome activity significantly decreased at 9 hpi in MG132-treated embryos (Fig. 1C); it recovered gradually thereafter, between 12 to 24 hpi, to the same level as the untreated embryos (Fig. 1C). From these results, we confirmed that ubiquitinated proteins were accumulated in MG132-treated fertilized 1-cell zygotes due to the low activity of proteasomes. In contrast, the accumulated proteins were re-degraded in embryos cultured in MG132-free medium due to the recovery of proteasome activity.
Fig. 1.
Proteasome inhibition, after fertilization, causes accumulation of ubiquitinated proteins. (A) Schematic diagram of experiments using fertilized oocytes at 1 hpi. (B) Total protein extracts from untreated metaphase II oocytes (MII), untreated 1-cell embryos (1C), untreated 2-cell embryos (2C), MG132-treated (+) MII, and MG132-treated 1C embryos were immunoblotted with anti-Ub antibody. Actin was used as a loading control. Molecular masses (kDa) are shown on the right. (C) Effect of transient MG132 treatment (after fertilization) on the proteasome chymotrypsin-like activity in embryos. Proteasome chymotrypsin-like activity in crude lysate from 100 fresh untreated 1C, 2C, and MG132-treated 1C, were measured using Suc-LLVY-AMC as a substrate. This experiment was performed in triplicate. Statistically significant differences between untreated and MG132-treated embryos are shown (* P < 0.05). Bars represent the standard error of the mean. Untreated (white bars), Treated with MG132 (black bars).
Low proteasome activity in fertilized 1-cell zygotes relates to full-term mouse development
In our previous study, we showed that approximately 60% of embryos could develop into blastocysts, with a delay in 5 µM MG132-treated cases [14]. However, the effect of the transient inhibition in the fertilized 1-cell zygote on its post-implantation development is not clear. To this end, we performed an embryo transfer assay using MG132-treated 1- and 2-cell embryos. Since all MG132-treated embryos were delayed in the 1- and/or 2-cell embryos, we transferred all MG132-treated 1- and 2-cell embryos into pseudo-pregnant recipient mice. The implantation rate of the MG132-treated 1-cell embryos at 24 hpi was significantly lower than that of the untreated embryos (Table 1). However, no significant difference in the implantation rates were observed between the MG132-treated and untreated 2-cell embryos at 24 hpi (Table 1). The fetal rate of the MG132-treated 1-cell embryos at 24 hpi was significantly lower than that of untreated ones; however, the fetal rate of the MG132-treated 2-cell embryos at 24 hpi tended to be low. While 5% of the untreated embryos were embryonically lethal, 31% of the implanted embryos, derived from MG132-treated 2-cell embryos, were shown to be embryonically lethal (Table 1). These results indicate that proteasome activity deficiency in the early stages of 1-cell zygotes could affect full-term mouse development.
Table 1. Transient inhibition of proteasome activity after fertilization affects full-term development.
| MG132 | No. of embryos used | Morphological stage of embryos at 24 hpi | No. of embryos transferred | No. (%) of |
||
| Implantations * | Fetuses ** | Embryonically lethal *** | ||||
| Treated without MG132 (Control) | 154 | 2-cell | 154 | 114 (74) a | 108 (70) a | 6 (5) a |
| Treated with MG132 for 8 h (from 1 to 9 hpi) | 309 | 1-cell | 221 | 119 (54) b | 97 (44) b | 22 (18) a |
| 2-cell | 88 | 67 (76) a, b | 46 (52) a, b | 21 (31) a | ||
*The percentage of transferred embryos that resulted in implantations. ** The percentage of transferred embryos that resulted in a live birth. *** The percentage of implanted embryos that resulted in embryonically lethal. Superscripts indicate significant differences from the control (P < 0.05).
Proteasome inhibition delays Pol II recruitment in pronuclei
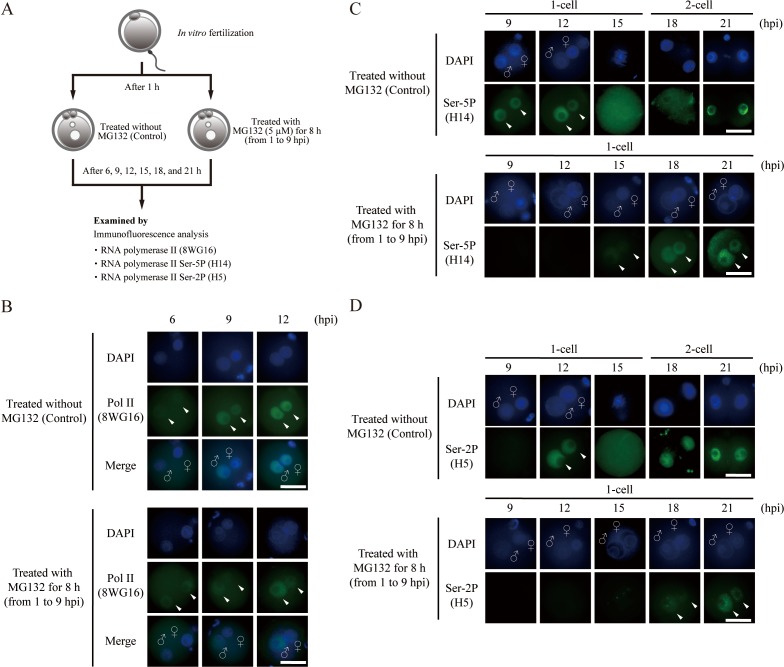
We next focused on the involvement of UPS-dependent proteolysis of maternal proteins in the Pol II mediated-gene transcription in 1-cell zygotes. In mouse embryos, the nuclear localization of Pol II is confirmed in 1-cell zygotes, when the minor ZGA initiates [20, 22, 23]. To investigate the effect of transient inhibition of proteasome activity on subcellular localization of Pol II in 1- and 2-cell embryos, we performed a localization assay using Pol II marker 8WG16 and Pol II phosphorylation markers H14 and H5 for Ser-5 (Ser-5P) and Ser-2 (Ser-2P), respectively (Fig. 2A). As shown in Fig. 2B, Pol II signals from 8WG16 stain were detected in pronuclei at 6 hpi in both MG132-treated and untreated embryos, and those signals intensified until 12 hpi. No difference of Pol II signals were observed in the MG132-treated 1-cell zygotes and the untreated embryos. We have previously shown that the onset of ZGA, tested with several ZGA marker genes, was delayed after the MG132 treatment in both normally developed 2-cell and arrested 1-cell embryos [14], suggesting that the transient inhibition of proteasome activity may have an effect beyond the recruitment of Pol II in nuclei.
Fig. 2.
Proteasome inhibition affects Pol II localization. (A) Schematic representation of the experimental procedures. (B) Immunofluorescence images of Pol II (green) in untreated and MG132-treated embryos. Representative images of embryos stained for Pol II with anti-8WG16 antibody are shown (arrowheads). (C) Immunofluorescence images of phosphorylation at serine residue 5 (Ser-5P) of Pol II with anti-H14 antibody in untreated and MG132-treated embryos (arrowheads). (D) Immunofluorescence images of phosphorylation at serine residue 2 (Ser-2P) of Pol II with anti-H5 antibody in untreated and MG132-treated embryos (arrowheads). All nuclei were stained by DAPI (blue). Merge images show all images combined with DAPI; ♀, female pronucleus; ♂, male pronucleus; hpi, hours post-insemination; scale bar = 50 µm.
To investigate the localization of CTD phosphorylation in proteasome activity-inhibited 1-cell zygotes, we performed immunofluorescence staining assay using Ser-5P and Ser-2P, which correspond to the transcriptional initiation and elongation forms, respectively (Fig. 2C, 2D). In the untreated embryos, signals for Ser-5P and Ser-2P were observed in both pronuclei at 9 hpi (Fig. 2C) and 12 hpi (Fig. 2D), respectively. Interestingly, no Ser-5P and Ser-2P signals could be observed in MG132-treated embryos at the same time points. In MG132-treated embryos, Ser-5P signals at 15 hpi and Ser-2P signals at 18 hpi were observed; the intensity of the signal was shown to increase till 21 hpi (Fig. 2C, 2D). These results demonstrate that MG132 treatment introduces a delay in transcriptional initiation and elongation due to the delayed recruitment of Pol II in the pronuclei.
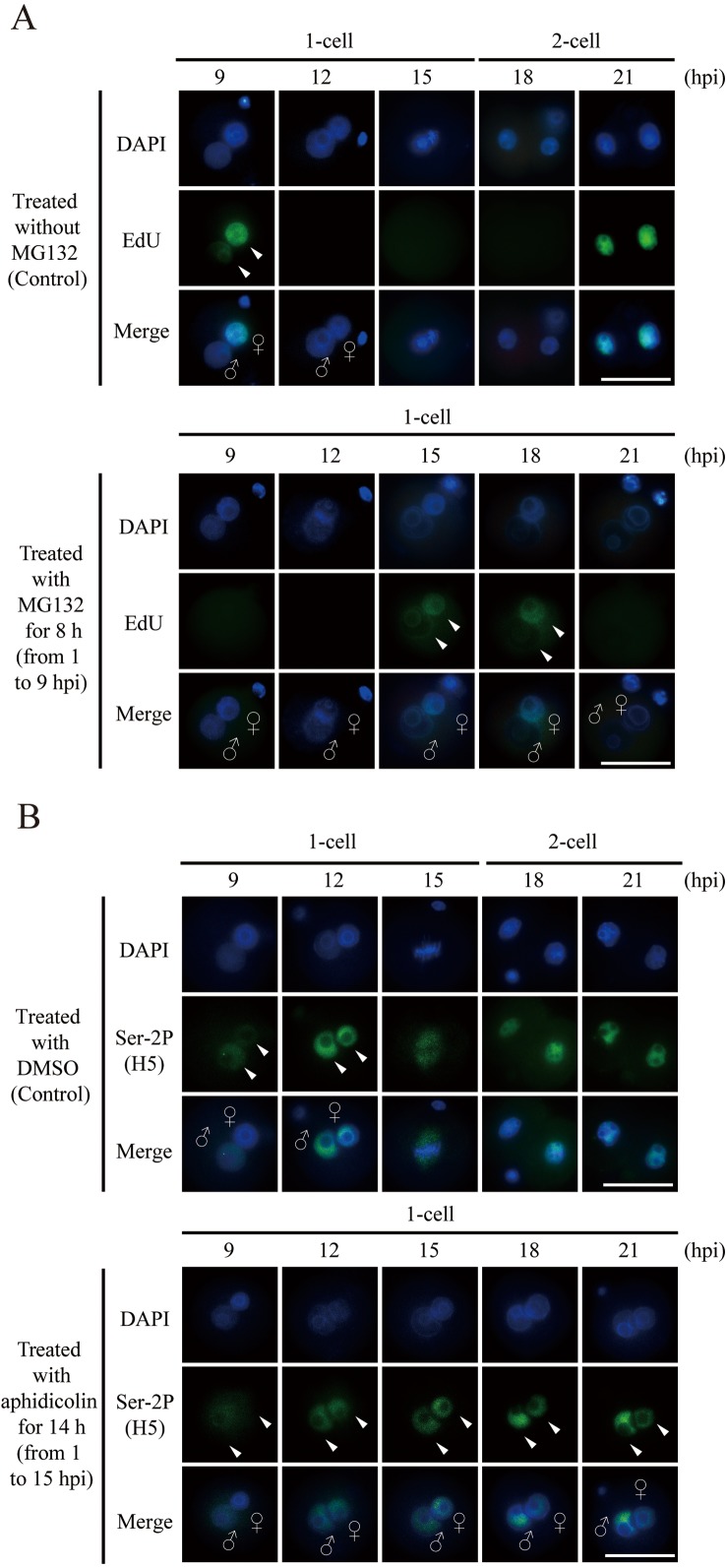
Our previous report has shown that MG132-treated embryos are delayed in the first cleavage and a certain fraction of embryos remains at the 1-cell stage even at 24 hpi [14], suggesting that the transient MG132 treatment in early embryos presumably affects cell cycle progression, including DNA replication. Indeed, the ubiquitin-mediated proteasome system has also been reported to be crucial for the normal progression of DNA replication [24, 25]. Furthermore, inhibiting the first round of DNA replication by aphidicolin does not prevent the transcription initiation in the 1-cell embryo [26]. From these observations, we hypothesized that the MG132 treatment in this study affects the timing of DNA replication in 1-cell zygotes, but the timing of DNA replication does not affect the initiation of ZGA. To test this hypothesis, we initially examined the duration of DNA replication by the EdU incorporation assay in MG132-treated embryos. In untreated embryos, EdU was incorporated at 9 hpi while in the MG132-treated embryos, the signals were not observed at the same time; rather, most EdU-positive embryos were found from 15 to 18 hpi in 1-cell embryos (Fig. 3A). These results indicate that the DNA replication occurs from 15 to 18 hpi in the MG132-treated embryos and the cell cycle progression is recovered within at least 6 h from release of the transient proteasome activity inhibition, indicating that the transient inhibition actually delays the initiation of DNA replication.
Fig. 3.
Delayed DNA replication, due to a transient inhibition of proteasome activity, is unlikely the cause of delayed transcriptional initiation. (A) EdU incorporation images stained with EdU (green, arrowheads) in untreated (upper panel) and MG132-treated embryos (lower panel). (B) Immunofluorescence images of phosphorylation at serine residue 2 (Ser-2P) of Pol II (green) in untreated and aphidicolin-treated embryos. Representative images of embryos stained with anti-H5 antibody (green, lower panel) for Ser-2P (arrowheads). Merge images show all images combined with. ♀, female pronucleus; ♂, male pronucleus; hpi, hours post-insemination; scale bar = 50 µm.
To investigate whether the delay in transcriptional initiation is caused by the delay of DNA replication, we performed immunofluorescence staining using an anti-Ser-2P antibody in aphidicolin-treated 1-cell embryos from 1 to 15 hpi, which is the same duration as the delay of DNA replication observed in the transient MG132-treated embryos (Fig. 3A). At first, to confirm the time at which DNA replication occurs in embryos treated with aphidicolin for 14 h, aphidicolin-treated embryos were subjected to EdU labeling. As a result, DNA replication in the embryos were found to occur at 18 hpi, indicating that DNA replication is induced within at least 3 h after release of the aphidicolin treatment (Supplementary Fig. 1: online only). We then detected the subcellular localization of Ser-2P from 1 to 15 hpi in the embryos treated with aphidicolin for 14 h. In the aphidicolin-treated embryos, the Ser-2P signal was seen in pronuclei at 9 hpi, similar to that in the untreated embryos (Fig. 3B). Taken together, these results indicate that the delay of the DNA replication, induced by the transient inhibition of proteasome activity, may not be the main cause of the delayed onset of minor ZGA, consistent with the previous finding which reports that artificial inhibition of first round of DNA replication does not completely eliminate ZGA [27].
Proteasome inhibition affects initiation of major ZGA
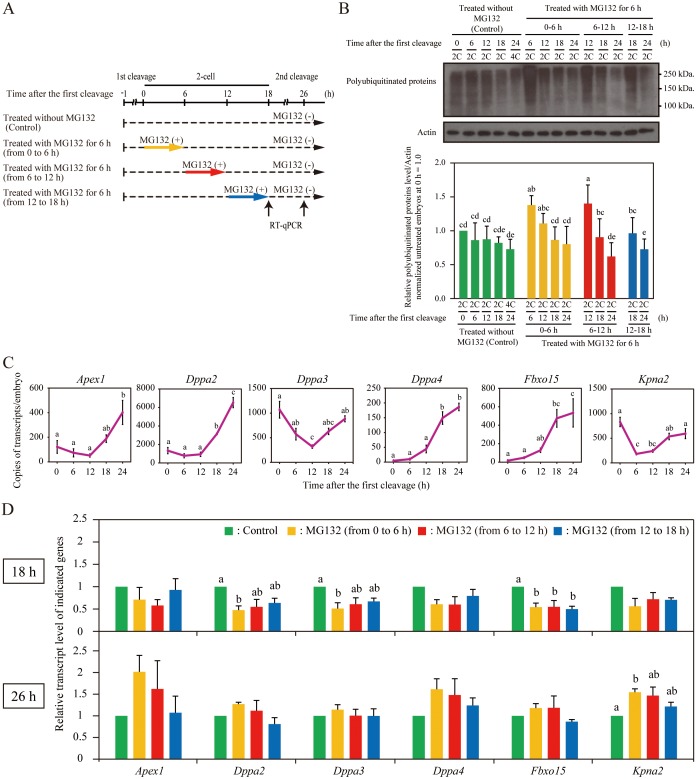
Recently, fundamental differences in chromatin state of minor ZGA from that of major ZGA were indicated [28, 29]. This led us to examine whether proteasome functions in 2-cell embryos are important for major ZGA. In order to confirm the optimal experimental conditions without side effects (as were determined for 1-cell embryos in our previous reports [14]), we divided the MG132 treatment into three groups based on the time after the first cleavage: the early, middle, and late 2-cell stages (0 to 6 h, 6 to 12 h, 12 to 18 h, respectively); all three groups were treated with 5 µM MG132 (Fig. 4A). As depicted in Fig. 4B, accumulation of polyubiquitinated proteins were confirmed by western blot analysis using anti-Ub antibody in each group. Furthermore, no arrest of pre-implantation embryonic development (after release of MG132 treatment) was observed in any group (Table 2; from 0 to 6 h, 6 to 12 h, 12 to 18 h). These results indicated that the temporal 5 µM MG132 treatment for 6 h in 2-cell embryos did not affect pre-implantation embryonic development. Hence, we chose this experimental condition for optimal transient treatment in the following experiments.
Fig. 4.
Delay of major ZGA by proteasome inhibition. (A) Schematic diagram of experiments in the 2-cell stage. (B) Effect of transient MG132 treatment in 2-cell embryos on the accumulation of polyubiquitinated proteins in embryos. Total proteins isolated from untreated 2-cell (2C), 4-cell (4C), and MG132-treated 2C embryos were immunoblotted with anti-Ub antibody (upper panel). Actin was used as a loading control. Molecular masses (kDa) are shown on the right. Densitometric quantification analysis of the immunoblot bands of polyubiquitinated proteins (lower panel). (C) Expression profile of indicated genes in 2-cell embryos (at 0, 6, 12, and 18 h) and early 4-cell embryos (at 24 h) was confirmed by RT-qPCR. (D) Expression levels in the untreated and MG132-treated embryos at 18 h (upper panel) and 26 h (lower panel). The mRNA levels of the untreated embryos were defined as 1. Untreated (green bars), MG132-treated (from 0 to 6 h) (yellow bars), MG132-treated (from 6 to 12 h) (red bars) and MG132-treated (from 12 to 18 h) (blue bars) embryos; h, time after the first cleavage. Different letters indicate statistical significances (P < 0.05). Bars represent standard error of the mean.
Table 2. Development of mouse embryos treated with 5 µM MG132 for 6 h.
| Duration of MG132 treatment (5 µM) | No. of embryos used | No. (%) of embryos developed to |
||||
| 3 or 4-cell embryos | 5–8-cell embryos | Morulae | Blastocysts | Degenerated | ||
| Untreated | 54 | 54 (100) | 54 (100) | 54 (100) | 52 (96) | 2 (4) |
| 0–6 h (16.5–22.5 hpi) | 61 | 60 (98) | 60 (98) | 59 (97) | 57 (93) | 6 (10) |
| 6–12 h (22.5–28.5 hpi) | 61 | 60 (98) | 60 (98) | 60 (98) | 60 (98) | 2 (3) |
| 12–18 h (28.5–34.5 hpi) | 63 | 63 (100) | 62 (98) | 60 (95) | 56 (89) | 9 (14) |
We examined whether the transient inhibition of proteasome activity in 2-cell embryos, using the same strategies as in Fig. 4A, affects the expression of marker genes of major ZGA such as Apex1, Dppa2, Dppa3, Dppa4, Fbxo15, and Kpna2 [3]. Consistent with the previous report [3], we observed that all these gene expressions were significantly increased in 2-cell embryos (Fig. 4C). In each transient MG132-treated embryo group, expression levels of all tested genes tended to be lower than those in untreated control embryos at 18 h (Fig. 4D, upper figure). Data were not normalized to any housekeeping gene, since the two housekeeping genes tested, namely Gapdh and Actb, showed differential expression. At 18 h, especially in the MG132-treated embryos from 0 to 6 h, the expression levels of Dppa2 and Dppa3 genes were significantly lower when compared to those in the untreated embryos. Moreover, the expression level of Fbxo15 was also significantly lower in all MG132-treated groups (Fig. 4D, upper figure). At 26 h after the treatment, gene expression levels in each MG132-treated embryo group were almost the same or higher than those in the untreated embryos (Fig. 4D, lower figure). These results show that the transient MG132 treatment in 2-cell embryos results in the overall low expression of some major ZGA genes at 18 h, but their expression was re-activated at 26 h, consistent with the recovered proteasome activity after release of MG132 treatment (Fig. 4B). We also examined whether the MG132 treatment affects the expression of maternal RNA, which is known to be degraded by the 2-cell stage. However, proper down-regulation of maternal RNA was not observed after MG132 treatment (data not shown). Collectively, these results indicate that the transient inhibition of proteasome activity in 2-cell embryos leads to a delay in the onset of some major ZGA genes.
Discussion
During MZT in mice, including the early 1-cell stage, a series of dynamic molecular events, such as epigenetic changes and highly coordinated chromatin remodeling regulated by maternal RNAs/proteins, triggers extensive reprogramming to establish a totipotent zygote [1, 2, 5]. The degradation of maternal proteins during MZT is especially regulated by UPS, with a role in turnover and quality control of proteins [10, 30]. Our data indicate that the transient inhibition of proteasome activity at the early 1-cell stage, 1 to 9 hpi, delays the onset of minor ZGA, primarily caused by the delayed recruitment of Pol II, and further affects full-term mouse development. This delayed onset of minor ZGA seems not to be directly connected to the delayed DNA replication. These findings explain the importance of the proper degradation of maternal proteins by UPS during mouse MZT for enabling full-term development of mouse totipotent zygotes. In case of somatic cell nuclear transfer (SCNT), high percentage of embryos gradually become embryonically lethal [31, 32]. The common cause of this failure has been suggested to be the incomplete nuclear reprogramming after SCNT [32]. Therefore, proper UPS function at the early 1-cell stage seems to cause an adequate reprogramming of the fertilized oocyte into a totipotent zygote. It is also noteworthy that autophagic activity regulates early embryonic development [8]. Therefore, it is possible that a portion of the embryos that developed to fetuses, after MG132 treatment, were rescued by the autophagy pathway instead of UPS. Indeed, the pathways are complementary, such that polyubiquitinated proteins are accumulated in the autophagy-defective neural cells [33].
As shown in Fig. 2, the delay in recruitment and phosphorylation of Pol II in transient MG132-treated 1-cell embryos are resultants of improper chromatin remodeling for Pol II recruitment. In general, UPS regulates gene expression and interacts with chromatin at multiple steps [11]. Transcriptional activators cannot translocate into nucleus or bind to DNA due to the failure of degrading their repressive regulators; a large number of transcription factors and cofactors have been reported to be regulated by UPS, indicating that UPS-mediated proteolysis of those factors is inherently linked to the way they stimulate transcription [11]. For example, in Caenorhabditis elegans, zinc-finger proteins OMA-1/2 bind to TBP-associated factor-4 (TAF-4), which is a component of the transcription factor-II D (TFIID) and pre-initiation complex of Pol II, sequestering TAF-4 into the cytoplasm during 1- and 2-cell stages. After the cell division, OMA-1/2 is degraded by UPS, TAF-4 is translocated into the nucleus, and is involved in TFIID-mediated transcriptional activation of the zygotic genome [34]. Furthermore, many histone modification enzymes, which are related to transcriptional silencing and activation, such as histone deacetylases (HDACs), methyltransferases (HMTs), and demethyltransferases (HDMs) are regulated by ubiquitin-mediated proteasomal degradation [35, 36]. At present, the molecular mechanisms of UPS governing minor ZGA in 1-cell embryos remain to be determined. Further study is needed to clarify which factors, degraded by UPS, can possibly participate in modulating chromatin remodeling and histone modifications, and in activating the transcription factors.
In this study, our results indicate that proteasome activity inhibition in 2-cell embryos leads to a delay in the major ZGA and exhibits differences in individual gene expression (Fig. 4). This is in contrast to the overall delay in minor ZGA in proteasome activity-inhibited 1-cell zygote [14]. A large group of SCF (Skp, Cullin, and F-box proteins) complex and E3 ubiquitin ligases that target substrate proteins were identified in mouse oocytes and zygotes [37, 38], indicating that UPS could be a major proteolytic system during MZT. We have previously reported that polyubiquitinated proteins start accumulating from mature oocytes to early 2-cell embryo stage, and then rapidly decrease after late 2-cell stage, finally disappearing between 24 and 36 hpi, when proteasomal chymotrypsin-like activity in 2-cell embryos was significantly up-regulated [9]. More recently, a global permissive chromatin state has been demonstrated, that causes promiscuous transcription in minor ZGA, and is converted to a global repressive chromatin state with the activities of cis-regulatory elements in major ZGA [28]. Therefore, the involvement of UPS in regulating the onset of minor and major ZGA for specific degradation of maternal proteins in 1-cell and 2-cell stages, respectively, could be explained by different mechanisms.
Taken together, our findings suggest that the maternal protein degradation by UPS during MZT is a basic requirement for a normal pre- and post-implantation embryonic development. Understanding the detailed mechanisms involved in the degradation of maternal proteins at early cleavage stages is essential to reveal how MZT is modulated.
Supplementary
Acknowledgments
We thank Ms N Backes Kamimura and Mr J Horvat for support with the manuscript and editing.
This study was supported, in part, by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science 25292189 (to KMa), and a grant from the INAMORI Foundation (to KMa). KMi is supported by Human Frontier Science Program (RGP0021/2016), JSPS KAKENHI Grant Numbers JP16H01321, JP16H01222, Grant for Basic Science Research Projects from The Sumitomo Foundation (150810), and by Kindai University Research Grant (15-I-2).
References
- 1.Schier AF. The maternal-zygotic transition: death and birth of RNAs. Science 2007; 316: 406–407. [DOI] [PubMed] [Google Scholar]
- 2.Zhou LQ, Dean J. Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol 2015; 25: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 2004; 6: 117–131. [DOI] [PubMed] [Google Scholar]
- 4.Park SJ, Komata M, Inoue F, Yamada K, Nakai K, Ohsugi M, Shirahige K. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev 2013; 27: 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee MT, Bonneau AR, Giraldez AJ. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 2014; 30: 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizushima N. Autophagy: process and function. Genes Dev 2007; 21: 2861–2873. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka K. The proteasome: overview of structure and functions. Proc Jpn Acad, Ser B, Phys Biol Sci 2009; 85: 12–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N. Autophagy is essential for preimplantation development of mouse embryos. Science 2008; 321: 117–120. [DOI] [PubMed] [Google Scholar]
- 9.Shin SW, Shimizu N, Tokoro M, Nishikawa S, Hatanaka Y, Anzai M, Hamazaki J, Kishigami S, Saeki K, Hosoi Y, Iritani A, Murata S, Matsumoto K. Mouse zygote-specific proteasome assembly chaperone important for maternal-to-zygotic transition. Biol Open 2013; 2: 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeRenzo C, Seydoux G. A clean start: degradation of maternal proteins at the oocyte-to-embryo transition. Trends Cell Biol 2004; 14: 420–426. [DOI] [PubMed] [Google Scholar]
- 11.Geng F, Wenzel S, Tansey WP. Ubiquitin and proteasomes in transcription. Annu Rev Biochem 2012; 81: 177–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard GC, Tansey WP. Interaction of Gcn4 with target gene chromatin is modulated by proteasome function. Mol Biol Cell 2016; 27: 2735–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen R, Ferdoush J, Kaja A, Bhaumik SR. Fine-tuning of FACT by the ubiquitin proteasome system in regulation of transcriptional elongation. Mol Cell Biol 2016; 36: 1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin SW, Tokoro M, Nishikawa S, Lee HH, Hatanaka Y, Nishihara T, Amano T, Anzai M, Kato H, Mitani T, Kishigami S, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Inhibition of the ubiquitin-proteasome system leads to delay of the onset of ZGA gene expression. J Reprod Dev 2010; 56: 655–663. [DOI] [PubMed] [Google Scholar]
- 15.Shimizu N, Ueno K, Kurita E, Shin SW, Nishihara T, Amano T, Anzai M, Kishigami S, Kato H, Mitani T, Hosoi Y, Matsumoto K. Possible role of ZPAC, zygote-specific proteasome assembly chaperone, during spermatogenesis in the mouse. J Reprod Dev 2014; 60: 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Zhou C, Wang Y, Liu W, Liu C, Wang L, Liu Y, Shang Y, Li M, Zhou S, Wang Y, Zeng W, Zhou J, Huo R, Li W. The E3 ubiquitin ligase RNF114 and TAB1 degradation are required for maternal-to-zygotic transition. EMBO Rep 2017; 18: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuno S, Sono Y, Matsuoka T, Matsumoto K, Saeki K, Hosoi Y, Fukuda A, Morimoto Y, Iritani A. Expression and subcellular localization of GSE protein in germ cells and preimplantation embryos. J Reprod Dev 2006; 52: 429–438. [DOI] [PubMed] [Google Scholar]
- 18.Hirano Y, Hayashi H, Iemura S, Hendil KB, Niwa S, Kishimoto T, Kasahara M, Natsume T, Tanaka K, Murata S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol Cell 2006; 24: 977–984. [DOI] [PubMed] [Google Scholar]
- 19.Hatanaka Y, Shimizu N, Nishikawa S, Tokoro M, Shin SW, Nishihara T, Amano T, Anzai M, Kato H, Mitani T, Hosoi Y, Kishigami S, Matsumoto K. GSE is a maternal factor involved in active DNA demethylation in zygotes. PLoS ONE 2013; 8: e60205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokoro M, Shin SW, Nishikawa S, Lee HH, Hatanaka Y, Amano T, Mitani T, Kato H, Anzai M, Kishigami S, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Deposition of acetylated histones by RNAP II promoter clearance may occur at onset of zygotic gene activation in preimplantation mouse embryos. J Reprod Dev 2010; 56: 607–615. [DOI] [PubMed] [Google Scholar]
- 21.Lee DH, Goldberg AL. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol 1998; 8: 397–403. [DOI] [PubMed] [Google Scholar]
- 22.Zurita M, Reynaud E, Aguilar-Fuentes J. From the beginning: the basal transcription machinery and onset of transcription in the early animal embryo. Cell Mol Life Sci 2008; 65: 212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogolyubova IO, Bogolyubov DS. Nuclear distribution of RNA polymerase II and mRNA processing machinery in early mammalian embryos. Biomed Res Int 2014; 2014: 681596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamaguchi R, Dutta A. Proteasome inhibitors alter the orderly progression of DNA synthesis during S-phase in HeLa cells and lead to rereplication of DNA. Exp Cell Res 2000; 261: 271–283. [DOI] [PubMed] [Google Scholar]
- 25.Kawahara H, Philipova R, Yokosawa H, Patel R, Tanaka K, Whitaker M. Inhibiting proteasome activity causes overreplication of DNA and blocks entry into mitosis in sea urchin embryos. J Cell Sci 2000; 113: 2659–2670. [DOI] [PubMed] [Google Scholar]
- 26.Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181: 296–307. [DOI] [PubMed] [Google Scholar]
- 27.Davis W, Jr, Schultz RM. Role of the first round of DNA replication in reprogramming gene expression in the preimplantation mouse embryo. Mol Reprod Dev 1997; 47: 430–434. [DOI] [PubMed] [Google Scholar]
- 28.Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, Li W, Li Y, Ma J, Peng X, Zheng H, Ming J, Zhang W, Zhang J, Tian G, Xu F, Chang Z, Na J, Yang X, Xie W. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 2016; 534: 652–657. [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto R, Aoki F. A unique mechanism regulating gene expression in 1-cell embryos. J Reprod Dev 2017; 63: 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta 2014; 1843: 182–196. [DOI] [PubMed] [Google Scholar]
- 31.Wakayama T, Perry ACF, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 1998; 394: 369–374. [DOI] [PubMed] [Google Scholar]
- 32.Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci 2013; 368: 20110329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441: 885–889. [DOI] [PubMed] [Google Scholar]
- 34.Guven-Ozkan T, Nishi Y, Robertson SM, Lin R. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell 2008; 135: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bach SV, Hegde AN. The proteasome and epigenetics: zooming in on histone modifications. Biomol Concepts 2016; 7: 215–227. [DOI] [PubMed] [Google Scholar]
- 36.Zou C, Mallampalli RK. Regulation of histone modifying enzymes by the ubiquitin-proteasome system. Biochim Biophys Acta 2014; 1843: 694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Kou Z, Jing Z, Zhang Y, Guo X, Dong M, Wilmut I, Gao S. Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA 2010; 107: 17639–17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Ni X, Guo Y, Guo X, Wang Y, Zhou Z, Huo R, Sha J. Proteomic-based identification of maternal proteins in mature mouse oocytes. BMC Genomics 2009; 10: 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.