Abstract
Tomato is an important horticultural and economic crop cultivated worldwide. As Phytophthora infestans becomes a huge threat to tomato production, it is necessary to study the resistance mechanisms of tomato against P. infestans. Our previous research has found that miR482 might be involved in tomato–P. infestans interaction. In this study, miR482b precursor was cloned from Solanum pimpinellifolium “L3708” and miR482b was shown to decrease in abundance in tomato following P. infestans infection. Compared to wild-type tomato plants, tomato plants that overexpressed miR482b displayed more serious disease symptoms after P. infestans infection, with more necrotic cells, longer lesion diameters, and increased P. infestans abundance. Meanwhile, silencing of miR482b was performed by short tandem target mimic (STTM), resulting in enhancement of tomato resistance to P. infestans. Using miRNA and degradome data sets, NBS–LRR disease-resistance genes targeted by miR482b were validated. Negative correlation between the expression of miR482b and its target genes was found in all miR482b-overexpressing and -silencing tomato plants. Our results provide insight into tomato miR482b involved in the response to P. infestans infection, and demonstrate that miR482b–NBS–LRR is an important component in the network of tomato–P. infestans interaction.
Late blight resistance genes identified in tomato
Researchers in China have identified a genetic pathway in tomato involved in resistance to the late blight pathogen, Phytophthora infestans. A team at Dalian University of Technology led by Yushi Luan engineered tomatoes to express more or less miR482b and then infected them with late blight. Symptoms were more severe in plants with excess miR482b and attenuated in those with reduced miR482b. The researchers also predicted the targets of miR482b and found 30 defense-related genes known as NBS-LRRs. Sequencing revealed that four of these genes were degraded in tomatoes infected with late blight. In addition, the expression of all four decreased when miR482b was strongly expressed and increased when it was silenced. These findings identify miR482b-NSS-LRR as an important pathway in tomato’s response to P. infestans which may be a valuable target for molecule breeding.
Introduction
Tomato, as an important horticultural and economic crop cultivated worldwide, always suffers from the aggression of various pathogens1–3. Phytophthora infestans, the agent that causes late blight, has become one of the most devastated pathogens4 and can cause great losses in tomato production5. For example, in Inner Mongolia of China and the USA, late blight caused the loss of tomato production in early 2000s6,7. Up to now, the molecular mechanisms of tomato–P. infestans interaction are beyond understanding and the method for controlling tomato late blight is still not effective enough. It is necessary to study tomato resistance mechanisms against P. infestans and identify the key resistance genes which are used in disease-resistance breeding of tomato transgene.
MicroRNA (miRNA), a class of about 22 nt endogenous small noncoding RNA, have been discovered to play an important role in regulating endogenous gene expressions in plants8,9. MiRNAs attack their targets by sequence-specific binding to the targeting sites on the gene transcripts, leading to either target mRNA degradation or translational repression mediated by the miRNA-associated proteins10. Up to now, the major methods to study miRNA functions are to generate and analyze the transgenic lines overexpressing miRNAs11,12. For example, in our previous studies, overexpression of miR172 in tomato enhanced its susceptibility to P. infestans13. Meanwhile, the tomato resistance to P. infestans was reduced by the overexpression of miR191814. However, these approaches are insufficient to fully understand miRNA functions. Short tandem target mimic (STTM), which was invented based on an endogenous mechanism that modulates miR399 activity in Arabidopsis thaliana15, is a newly developed approach to explore miRNA functions by blocking miRNAs in plants16. In previous studies, STTM had been used to silence miR165/166, miR160, and miR159 in A. thaliana, cotton, soybean, tobacco and these miRNAs were found functioning as regulators of plant growth and development17–21. In tomato, blocking miR858 by overexpressing STTM858 resulted in high accumulation of anthocyanins, which demonstrated miR858 could negatively control the anthocyanin accumulation of tomato22.
Many miRNAs have been demonstrated to play important roles in stress responses. A number of plant miRNAs, such as miR159, miR160, miR166, miR169, miR172, and miR396, are involved in the response to drought, water deficit, and salt stresses23,24. More and more evidence have indicated that plant miRNAs also play an important role in plant–pathogen interaction25,26. For example, miR393 and miR472 can contribute to pathogen-associated molecular pattern-triggered immunity (PTI) in plants27,28. Other miRNAs, such as miR482, miR6019, miR6020, miR6022, miR5300, miR1507, and others, are thought to suppress nucleotide-binding site and leucine-rich repeat (NBS–LRR) defense genes, which play critical roles in effector-triggered immunity (ETI), also a crucial component of plant immune system29–33.
MiR482, a class of miRNAs found in various plants, acts in plant–pathogen interaction34. Previous studies have shown, miR482 is involved in the process of tomato, cotton, soybean, and peanut interacting with Potato spindle tuber viroid, Cucumber mosaic virus, Verticillium dahliae, soybean cyst nematode, and Ralstonia solanacearum35–39. MiR482 regulates defense mechanisms of plant via targeting conserved sequences encoding the P-loop of NBS–LRR resistance proteins38. In cotton, ~12% of cotton NBS–LRR genes were predicted targets by gra-miR482 family40. In Solanaceae, miR482 was also found targeted to NBS–LRR genes30,31,34. NBS–LRR genes were the most represented gene family of resistance (R) genes41. They can enhance plant resistance to various pathogens, such as nematodes, Magnaporthe oryzae, Xanthomonas oryzae, tomato mosaic virus (ToMV), and Pseudomonas syringae42–46. Previous study in potato found the increased susceptibility of miR482e-overexpressing plants to V. dahliae infection could be explained by the enhancement of miR482e-mediated silencing on NBS–LRR disease-resistance genes31.
Using high-throughput sequencing and homology-based computational research, we have previously identified a number of tomato miRNAs involved in tomato–P. infestans interaction including miR482, miR172, miR6024, miR6026, miR6027, etc.13,25,47,48. Our previous work showed that only miR482b was identified in tomato, Solanum pimpinellifolium “L3708” (L3708) inoculated with P. infestans and its transcripts per million clean tags was lower than wild-type (WT) tomato after analysis of the miRNA-Seq data. However, whether miR482b affected tomato resistance to P. infestans has not been determined. In this study, we analyzed the expression patterns of miR482b in tomato L3708 after P. infestans treatment. Then the function of miR482b was explored by overexpressing and silencing approach and its target genes were also identified. These studies will benefit not only understanding of the molecular mechanisms of tomato–P. infestans interaction but also future molecular breeding.
Materials and methods
Plant material collection and P. infestans inoculation
Tomato L3708 was grown in a greenhouse at 25 ± 3 °C under 16 h of light per day. P. infestans strain “P12103” was cultured in oat medium in the dark at 20 °C. Tomato plants at the five to six leaf stage were inoculated with P. infestans spores suspension (106 zoospores mL−1) and placed at 20 ± 1 °C in a 100% relative humidity environment without light. Tomato plants treated with sterile water were used as controls, and they were kept under the same conditions. Leaf samples were collected at 0, 1, 2, 3, and 4 days post inoculation (dpi). All samples were quickly frozen in liquid nitrogen and stored at −80 °C until DNA and RNA isolation.
Cloning of pre-miR482b
MiR482b precursor (pre-miR482b) was cloned using tomato DNA as a template. The primers (miR482bF and miR482bR; Table S2) were designed according to the tomato genome sequences using the Primer Premier 5 software. The PCR product was cloned into T-Vector pMD 19 (Takara, Dalian, China), and the identity of the sequence was verified by DNA sequencing. Multiple nucleic acid sequence alignments were performed with ClustalX2.
Construction of silencing and overexpressing plasmid of miR482b and their transient overexpression in tomato
Silencing of miR482b with STTM was performed. The structure of STTM482b was design based on the description of Yan et al.17. It contained two copies of imperfect miR482b binding sites (22 nucleotides) linked with a 48 nt linker. A bulge contain three additional nucleotides (CTA) was designed around positions 10 to 11 of the miRNA to trap miR482b without being cleaved by it. The pre-miR482b and STTM482b inserts were then subcloned into BamHI–SacI cut pBI121 to generate the recombinant overexpressing plasmids pBI121-miR482b and silencing plasmid pBI121-STTM482b, in which the expression of miR482b and STTM482b are controlled by the strong constitutive CaMV35S promoter. Freeze-thaw method was used to introduce pBI121-miR482b and pBI121-STTM482b into Agrobacterium tumefaciens strain “GV3101”.
A sample from A. tumefaciens culture was centrifuged at 4000 × g for 10 min and the pellet was resuspended in infiltration medium (10 mM MgCl2, 10 mM MES and 20 μM acetosyringone; OD600 = 1.0)49. A. tumefaciens was introduced into tomato leaves by infiltration. A. tumefaciens with empty vector was used as a control. After 3 days, each infiltrated leaf region was inoculated with 20 μL of P. infestans (106 zoospores mL−1). The lesions were observed at the 5th day and the length of the lesion diameter was calculated.
Transformation and identification of transgenic tomato
Tomato transformation was performed based on the method of Li et al.50. The seeds of tomato S. lycopersicum “Zaofen No. 2” were cultured in 1/2 Murashige and Skoog (MS) medium at 25 ± 3 °C under 16 h of light per day after surface sterilized with 75% (v/v) ethanol and 2.5% (v/v) sodium hypochlorite. Cotyledon excised from 1-week-old seedlings were used as explants. Transgenic tomato plants were generated by A. tumefaciens-mediated leaf disk method. Putative transgenic plants were selected on 1/2 MS medium with 200 mg L−1 carbenicillin and 50 mg L−1 kanamycin. After obtaining T0 kanamycin-resistant plants, the presence of the transgene in the regenerating plantlets was further confirmed using PCR with a pair of specific primers of neomycin phosphoryltransferase II (nptII) (Table S2). The abundance of miR482b in the selected positive overexpressing and silencing lines were examined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
Analysis of transgenic tomato resistance against P. infestans
The detached leaves of WT and transgenic tomato plants were inoculated with 10 μL of 106 zoospore mL−1 zoospore suspension of P. infestans according to the method of Li et al.50. In whole-plant assays, a suspension of P. infestans zoospores (106 zoospores mL−1) was also used to spray plants to run off. The inoculated leaves or whole plants were maintained at 20 ± 1 °C under high humidity condition for 24 h without light, and then placed in a greenhouse at 20 ± 1 °C under 16 h of light per day. Necrotic regions and disease indices were recorded at 5 dpi. UV photographs were also taken at 5 dpi. Images of the leaves were taken and the diameters of lesions were measured. Trypan blue staining assays used to detect dead cells of harvested leaves was also based on the method described by Li et al.50. In addition, the transcript level of P. infestans Actin gene was used to indicate P. infestans growth in plant by qRT-PCR51. Categorization of disease grades (DG) was performed as described by Li et al.50, and the disease index (DI) was calculated according to the following formula:
where DGi is the value of DG, ni is the number of plants in each DG, and n is the total number of plants. Each experiment was carried out at least three times.
The leaves were also collected to measure the content of malonaldehyde (MDA) according to previously described method50.
Identification of target genes
The mature-miR482b sequence was input into psRNATarget (http://plantgrn.noble.org/psRNAtarget/) and WMD3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi) to predict its target genes in tomato. Tomato transcripts (cDNA library, version 2.4/2.3) were used as data sets. The degradome sequencing library of tomato inoculated with P. infestans was used to verify the targets. CleaveLand software was used to find the cleavage site of the miR482b targets52,53. T-plot figures were produced based on R language54.
qRT-PCR analysis
The abundance of miR482b in tomato was examined by qRT-PCR. The specific forward primer for miR482b was designed following the method described by Varkonyi-Gasic et al.55. Quantitative RT-PCR was performed using the SYBR PrimeScript miRNA RT-PCR kit (Takara, Dalian, China) with Corbett Rotor Gene 3000 qRT-PCR machine (Corbett Research, Mortlake, Vic., Australia). The primers for the target genes were designed following the manufacturer’s instructions for the SYBR Premix Ex Taq II kit (Takara, Dalian, China). The miR482b cleavage sites of target genes were ensured between the forward and reverse primers binding sites. Information on all primers are shown in Table S2. Actin was used as a reference control gene for the miRNA and target gene qRT-PCR analysis. Of the nine leaves sampled in each experiment, three leaves were pooled into one biological replicate, resulting in three biological replicates.
Statistical analysis
All statistical analysis of data was performed with Origin 8.5, and all data were expressed as means ± SEs from three independent experiments. Based on http://abacus.bates.edu/~ganderso/biology/resources/writing/HTW_Guide_Reporting_Statistics_3-7-2011.pdf, the capital letters were used over the error bars. Letters shared in common between or among the groups would indicate no significant difference.
Results
Cloning, tissue-specific expression and expression patterns of tomato miR482b under P. infestans infection
The pre-miR482b was cloned from tomato L3708 based on tomato sRNAs available in the miRBase and tomato genome. The result of multiple nucleic acid sequence alignments showed that the sequence of the pre-miR482b and mature-miR482b were identical to the miR482b from cultivated tomato S. lycopersicum in miRbase (Figure S1).
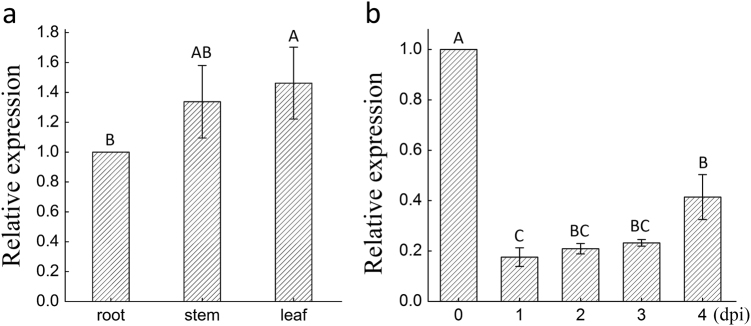
To analyze the distribution of miR482b within tomato L3708, qRT-PCR was used to measure the levels of miR482b transcript in the leaf, stem, and root of the plant. MiR482b was found in high abundance in leaf, whereas relatively low abundance was detected in root (Fig. 1a). To verify whether miR482b was involved in the response to P. infestans infection, the expression of miR482b in P. infestans-treated leaves at different dpi was measured. The result showed that after P. infestans treatment, the expression level of miR482b was lowest one day after the infection, but increased to a moderate level four days after the infection (Fig. 1b).
Fig. 1. Quantitative real-time PCR analysis of the expression pattern of miR482b in different tissues and in response to Phytophthora infestans.
a Tissue-specific expression of miR482b in tomato root, stem, and leaf. b Expression pattern of miR482b in tomato leaves at the indicated time after being sprayed with P. infestans. Data are the means ± SEs of three independent experiments. Letters indicate significant difference among samples, and letters shared in common between or among the groups indicate no significant difference at the P < 0.05 level
Overexpression and silencing of miR482b by agrobacterium infiltration
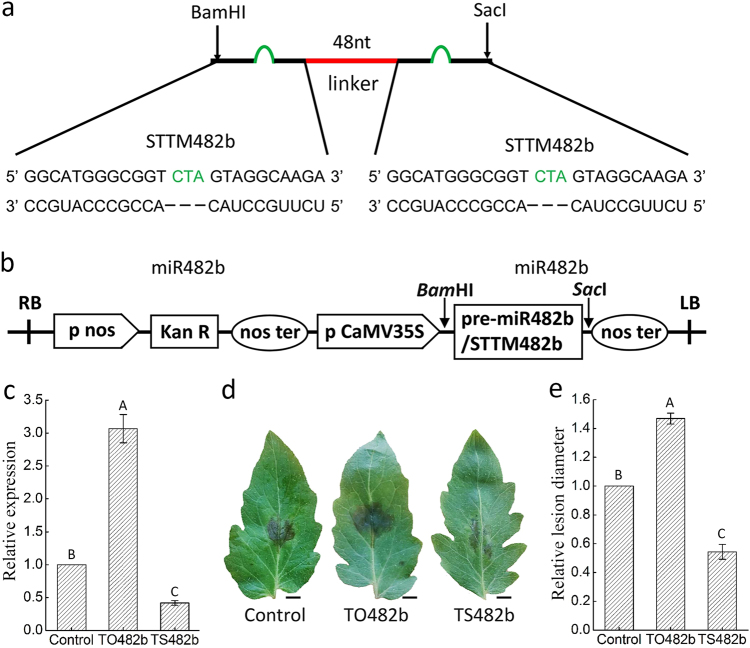
STTM482b sequence was constructed to silence miR482b, which contained two copies of imperfect miR482b binding sites with a 48 nt linker and each cope had a cleavage-preventive bulge contained three additional nucleotides (CTA) (Fig. 2a). Then the overexpressing and silencing plasmids, pBI121-miR482b and pBI121-STTM482b were constructed (Fig. 2b) and introduced into tomato leaf by agrobacterium infiltration. The abundance of miR482b was significantly increased in tomato plants that transiently overexpressed miR482b (TO482b) and significantly decreased in tomato plants transiently silenced miR482b (TS482b) (Fig. 2c). Compared to control plants, TO482b tomatoes displayed more severe disease condition following infection of P. infestans, with significantly larger lesions in the leaves, while TS482b tomato showed less severe disease condition after P. infestans infection (Fig. 2d, e). These results suggested that miR482b may have weakened the resistance of tomato against P. infestans.
Fig. 2. Transient overexpression and silencing of miR482b in tomato leaves.
a Scheme of the base pairing pattern of STTM482b and miR482b. b Schematic diagram of gene cassette used for the overexpression and silencing of miR482b in tomato. c Relative quantities of miR482b in tomato leaves transiently overexpressing and silencing miR482b. d Disease symptoms of tomato leaves transiently overexpressing and silencing miR482b 5 days after inoculation with P. infestans. Control, transient overexpression of the empty vector; TO482b, transient overexpression of pBI121-miR482b; TS482b, transient silencing of pBI121-STTM482b. Scale bars = 0.5 cm. e The relative lesion diameter. Data are the means ± SEs of three independent experiments. Letters indicate significant difference among samples, and letters shared in common between or among the groups indicate no significant difference at the P < 0.05 level
Identification of transgenic plants with high and low miR482b expression levels
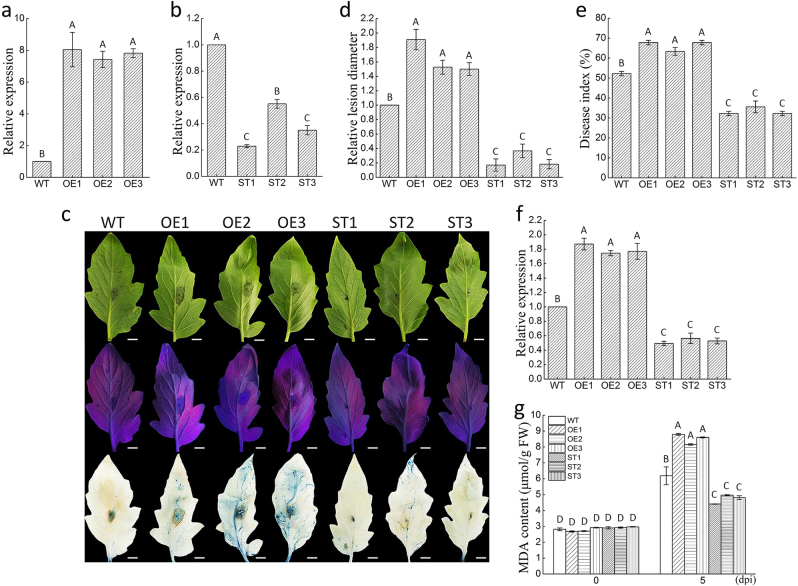
The transgenic tomatoes were constructed using pBI121-miR482b and pBI121-STTM482, respectively. Four-week-old tomato seedlings (WT and representative positive transgenic lines) were transferred to pots and grown for 2 weeks before they were subjected to qRT-PCR analysis to estimate the abundance of miR482b in their leaf tissue. MiR482b was more abundant in all overexpressing lines and less abundant in all silencing lines compared to WT lines (Fig. 3a, b) with the expression in three overexpressing lines (OE1, OE2, and OE3) being ~8-fold, 7.4-fold, and 7.8-fold, respectively, the expression in WT, while in three silencing lines (ST1, ST2, and ST3) the expression were 0.32-fold, 0.28-fold, 0.36-fold, respectively, the expression in WT. In addition, after genomic DNA-PCR detection based on a pair of specific primers of nptII, transgenic lines exhibited the expected transgene-specific band (395 bp), WT was not found (Figure S2).
Fig. 3. Effect of miR482b overexpression and silencing on the resistance of transgenic tomato to P. infestans.
a, b Quantitative real-time PCR analysis of the abundance of miR482b in WT, overexpressing (a) and silencing (b) tomato lines. c Detached-leaf inoculated with P. infestans showing disease signs (scale bars = 0.5 cm). Top: disease symptoms; middle: leaves were photographed in UV-light; bottom: trypan Blue staining for detection of dead cells. d The relative lesion diameter. e Whole-plant assay of P. infestans-infected tomato plants showing disease index of WT, overexpressing and silencing tomato lines. f Transcript accumulation of P. infestans Actin gene in these inoculated plants. g MDA content of the leaves from WT, and overexpressing and silencing tomato lines at 5 days after inoculation with P. infestans. Data are the means ± SEs from three independent experiments. Letters indicate significant difference among samples, and letters shared in common between or among the groups indicate no significant difference (P < 0.05)
MiR482b enhances tomato susceptibility to P. infestans
After the inoculation of P. infestans, the detached leaves were gradually darkened and the disease symptoms gradually appeared. Compared to WT, the disease symptoms were more severe on the leaves of miR482b-overexpressing lines and less severe on the leaves of miR482b-silencing lines (Fig. 3c). The dead cells of the treated leaves were examined by trypan blue staining. The darker blue marks in the leaves of overexpressing plants indicated that a greater number of dead cells occurred in miR482b-overexpressing leaves than WT, while the dark blue marks were reduced after miR482b silenced which demonstrated fewer dead cells were existed in miR482b-silencing leaves compared to WT (Fig. 3c). Furthermore, lesion diameters of miR482b-overexpressing and miR482b-silencing leaves were significantly longer and shorter than WT leaves, respectively (Fig. 3d).
In the whole-plant inoculation assay, the DI was found to be higher in miR482b-overexpressing lines than WT, which indicated that blight lesions obviously occupied more leaf area in tomato plants overexpressed miR482b. Conversely, compared to WT, the DI of silencing lines was significantly reduced (Fig. 3e). The transcript levels of P. infestans Actin gene measured by qRT-PCR were used to indicate P. infestans growth in plant and the result showed a significant increase and decrease in abundance of P. infestans in overexpressing and silencing lines compared to WT, respectively (Fig. 3f). In plant–pathogen interaction, plant cellular membranes were damaged because of late massive ROS generation56. MDA, which is taken as an indicator of membrane damage50, was detected in the overexpressing, silencing and WT plants inoculated with P. infestans. A significantly higher MDA level was found in overexpressing lines, suggesting that these plants suffered more severe membrane damage than WT (Fig. 3g). In silencing lines, the lower MDA level suggested that they suffer less severe membrane damage compared to WT plants (Fig. 3g). To some extent, these results also suggested that miR482b had led to the enhancement of tomato susceptibility to P. infestans.
Identification of miR482b targets
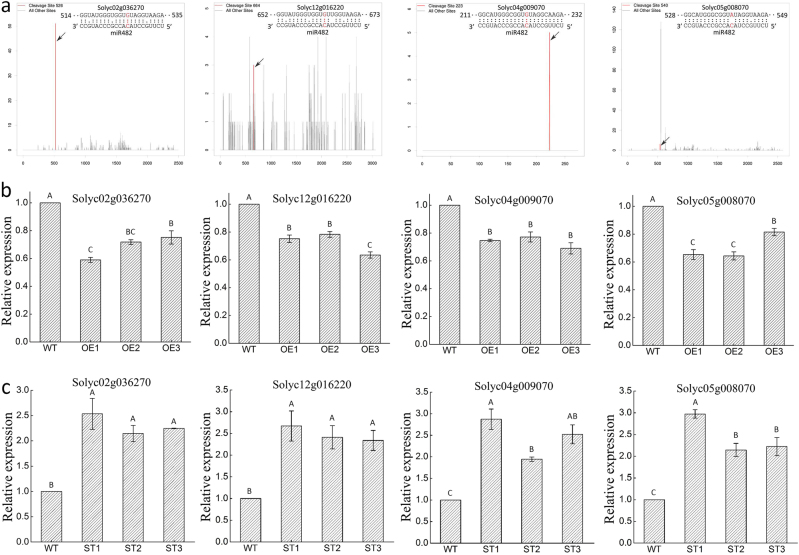
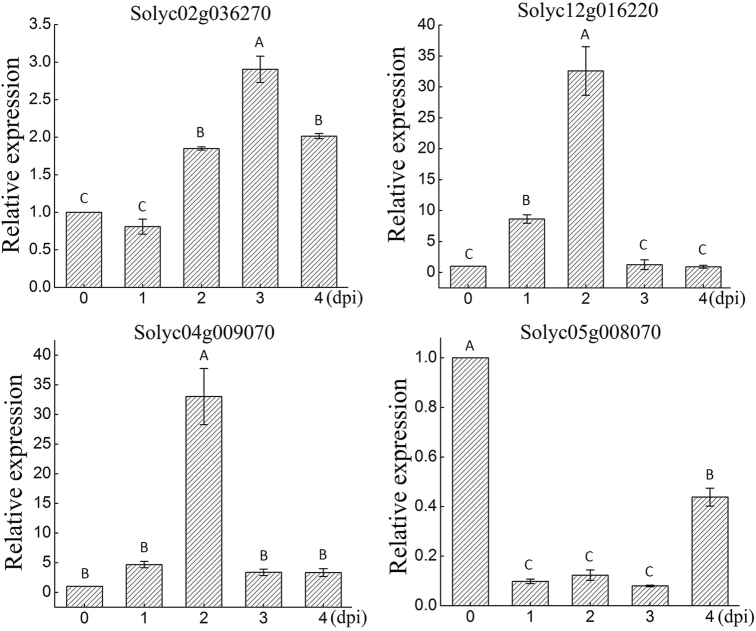
Identification of target genes is essential to show the regulatory networks of miRNA. Prediction of miR482b target genes using psRNATarget and WMD3 showed 30 NBS–LRR disease-resistance genes (Table S1). Four of these genes, Solyc02g036270.2, Solyc04g009070.1, Solyc12g016220.2, and Solyc05g008070.2, were found to have been cleaved at the miR482b-specific cleavage site according to the available tomato degradome sequencing data, indicating that they were the target genes of miR482b (Fig. 4a). Overexpression of miR482b led to significant reduction in the expression of its four target genes (Fig. 4b). On the contrary, these four target genes was upregulatedly expressed in miR482b-silencing tomatoes (Fig. 4c).
Fig. 4.
Identification of miR482b target genes in tomato. a T-plots showing the miR482b cleavage sites of target genes, Solyc02g036270.2, Solyc12g016220.2, Solyc04009070.1, and Solyc05g008070.2. The bar graph shows the number of sRNA reads mapped to each position on the transcript (x axis) and the sRNA raw read number (y axis). Arrow indicates cleavage sites. b, c Quantitative real-time PCR analysis of the expression levels of target genes in WT and the transgenic tomato overexpressed (b) and silenced (c) miR482b. Data are the means ± SEs of three independent experiments. Various letters indicate significant difference among samples, and letters shared in common between or among the groups indicate no significant difference (P < 0.05)
Upon infection by P. infestans, the expression levels of Solyc02g036270.2, Solyc04g009070.1, and Solyc12g016220.2 were initially increased, but then decreased (Fig. 5). Their expression trend was in contrast to that of miR482b, which further verified the relationship between miR482b and the three genes. However, the expression level of Solyc05g008070.2 was decreased at the first day after infection by P. infestans, but remained constant for the next two days, and then increased (Fig. 5), suggesting a lack of correlation with the expression trend of miR482b. According to the tomato degradome sequencing data, Solyc05g008070.2 was also found to have been cleaved by miR6024, which might be the reason why there was no correlation between the expression of miR482b and Solyc05g008070.2. These results therefore indicated that miR482b might play an important role in the regulation of NBS–LRR genes during tomato–P. infestans interaction.
Fig. 5. Expression patterns of target genes in tomato leaves at the indicated time after being sprayed with P. infestans.
Data are the means ± SEs of three independent experiments. Various letters indicate significant difference among samples, and letters shared in common between or among the groups indicate no significant difference (P < 0.05)
Discussion
To gain insight into the molecular mechanisms responsible for the late blight resistance, we previously used high-throughput sequencing to identify P. infestans-induced miRNAs in tomato, including miR169, miR398, miR482, miR6024, miR6026, and miR602725,47,48. Among these miRNA candidates, miR482 is one of the most important miRNAs involved in plant–pathogen interaction. In this study, we showed that the abundance of miR482b in tomato was reduced following P. infestans infection (Fig. 1b). Similar results have also been reported by other investigators, who found that decreased abundance of miR482 occurs in Solanaceae upon infection with different pathogens, including V. dahlia, Fusarium oxysporum, etc.30,31. Recently, it was found that RNA silencing suppressors from pathogens might suppress the accumulation of miRNA in plant–pathogen interaction57. For example, two effectors form the soybean pathogen P. sojae, named PsPSR1 and PsPSR2 suppressed transgene silencing by inhibiting the accumulation of plant sRNAs58,59. In addition, the accumulation of miRNA also was regulated by lncRNAs and transcription factors. For example, in tomato–TYLCV interaction, a tomato lncRNA, slylnc0195 acted as a target mimic to suppress the expression of miR166a60. Meanwhile, ripening inhibitor (RIN), a vital transcription factor acted though sequence-specific interactions with cis-regulatory DNA elements in the promoter of MIR172a to regulate its accumulation61.
Tomato L3708 is a resistant tomato to P. infestans. But, the miR482b sequences in tomato L3708 and cultivated tomato showed 100% sequence identity (Figure S1). To study the function of miR482b in tomato–P. infestans interaction, we introduced miR482b into cultivated tomato to constructed transgenic tomato. Three independent transgenic tomato lines were generated, which increased transcript amounts of miR482b. After the infection of P. infestans, more serious disease symptoms occurred in tomatoes overexpressed miR482b than WT, which suggested miR482b could enhance the susceptibility of tomato to P. infestans infection (Fig. 3). Similar results have also been reported in potato. The overexpression of miR482e in potato led plants to be sensitive to V. dahliae infection31. These results implicate a role for miR482b in the defense response of tomato against P. infestans infection.
As overexpression is not enough to fully understand miRNA functions and some consequence from overexpression studies may be caused by temporal or spatial misexpression62, miRNA-silencing technology should be used to explore functions of miRNAs. STTM, an effective tools to restrain miRNA activity, make it technically possible to silence miRNAs in various plants. In rice, transgenic STTM lines silencing 35 miRNA families (miR398, miR172, miR156, etc.) were produced as resources for functional studies and crop improvement62. STTM was also be used in tomato to silence miR396, which resulted in tomato flowers, sepals and fruits all obviously becoming bigger63. Meanwhile, effectively inhibiting the expression of miR858 by STTM induced anthocyanin accumulation in tomato22. In present study, we used STTM to silence miR482b to validate its function. In miR482-silencing lines, with the abundance of miR482b reduced significantly, plant resistance to P. infestans enhanced observably, which further proved miR482b could enhance the susceptibility of tomato to P. infestans.
In this study, four NBS–LRR genes were identified as the target genes of miR482b by degradome sequencing and qRT-PCR (Fig. 4). All these target genes showed more than 95% sequence identity and their binding regions of miR482b are completely identical between tomato L3708 and cultivated tomato. For example, the sequence identity of the target gene, Solyc02g036270.2 is 99% in these two tomato source (unpublished). That is to say, miR482b cleaves the same miR482b-specific cleavage site in tomato L3708 and cultivated tomato. NBS–LRR proteins have been shown to function as immune receptors that recognize specific pathogen effector proteins during plant–pathogen interaction, leading to the initiation of plant ETI25,28,64. The simplest model for its functioning is that NBS–LRR proteins recognize their corresponding effectors from various pathogens, and their interaction would lead to the activation of a series of defense responses65. Before the attack of a pathogen, the expression level of NBS–LRR genes is very low, but would rapidly increase at the stage of infection66–68. For example, VaRGA1, a grapevine NBS–LRR gene, is strongly induced by Plasmopara viticola at 12 hpi65. The expression of a novel peanut NBS–LRR gene, AhRRS5, is also increased in response to R. solanacearum infection, both in resistant and susceptible peanut cultivars69. Similar to these previous studies, the expression of Solyc02g036270.2, Solyc04g009070.1, and Solyc12g016220.2, three of the four NBS–LRRs genes identified as target genes of miR482b, were also elevated after P. infestans infection, which clearly demonstrated that these genes could respond to the infection of P. infestans in tomato (Fig. 5). In addition, previous studies also showed that overexpression of the NBS–LRR gene VaRGA1 in Nicotiana benthamiana would enhance its resistance to P. parasitica65, whereas silencing of a CC–NBS–LRR gene through VIGS in cotton dramatically increased its susceptibility to V. dahliae70. Similar results were obtained in this study. The suppression of NBS–LRRs, which was caused by the overexpression of miR482b (Fig. 4b), led to more disease symptoms in miR482b-overexpressing tomato lines (Fig. 3). In miR482b-silencing lines, the expression levels of NBS–LRRs was significantly upregulated (Fig. 4c) and the resistance of tomato plants to P. infestans enhanced (Fig. 3). Thus, these results provided additional evidence that these NBS–LRR genes were targeted by miR482b and they might play a role in the activation of tomato defense responses.
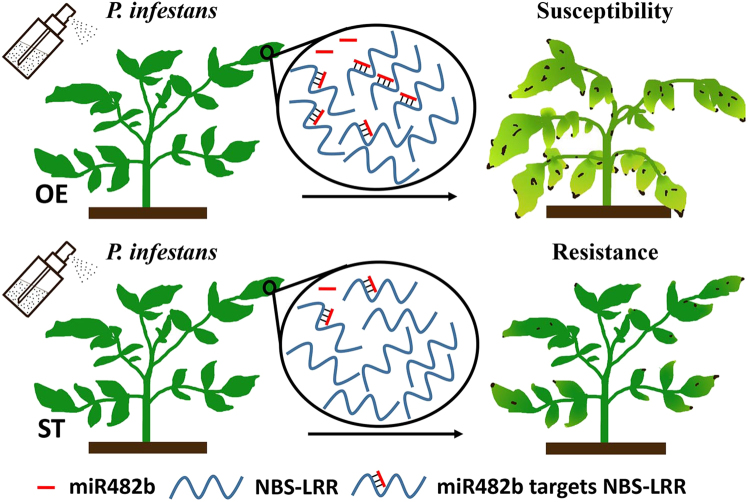
Based on these results, we propose a working model to explain how miR482b affects tomato resistance to P. infestans (Fig. 6). Following our results for degradome analysis, miR482b cleaves its target genes, the members of NBS–LRR family to suppress the abundance of NBS–LRRs (Fig. 4). When miR482b is overexpressed in tomato, the abundance of NBS–LRRs is decreased and more serious disease symptoms are shown after P. infestans infection. On the contrary, in miR482b-silencing tomato plants, more abundance of NBS–LRRs results in enhancement of tomato resistance to P. infestans. Our results provide insight into tomato miR482b–NBS–LRR module involved in the response to P. infestans infection and guidance for molecular breeding to improve P. infestans stress tolerance in the future.
Fig. 6. Model of miR482b-NBS-LRR regulation network involved in the tomato–P. infestans interaction.
Overexpression of miR482b in tomato negatively modulated P. infestans defense response by decreasing the abundance of NBS–LRRs, while the resistance was enhanced after miR482b silencing
Electronic supplementary material
Acknowledgements
This work was supported by Grants from the NFSC (Nos. 31471880 and 61472061).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s41438-018-0017-2.
Contributor Information
Jun Meng, Email: mengjun@dlut.edu.cn.
Yushi Luan, Email: ysluan@dlut.edu.cn.
References
- 1.Baysal-Gurel E, Li R, Ling KS, Miller SA. First report of tomato chlorotic spot virus infecting tomatoes in ohio. Plant Dis. 2015;99:163–164. doi: 10.1094/PDIS-06-14-0639-PDN. [DOI] [PubMed] [Google Scholar]
- 2.Balmant KM, et al. Redox proteomics of tomato in response to Pseudomonas syringae infection. Hortic. Res. 2015;2:15043. doi: 10.1038/hortres.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattarai K, Louws FJ, Williamson JD, Panthee DR. Differential response of tomato genotypes to Xanthomonas-specific pathogen-associated molecular patterns and correlation with bacterial spot (Xanthomonas perforans) resistance. Hortic. Res. 2016;3:16035. doi: 10.1038/hortres.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang CZ, Liu L, Wang XX, et al. The Ph-3 gene from Solanum pimpinellifolium encodes CC-NBS-LRR protein conferring resistance to Phytophthora infestans. Theor. Appl. Genet. 2014;127:1353–1364. doi: 10.1007/s00122-014-2303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fry WE, Birch PR, Judelson HS, et al. Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology. 2015;105:966–981. doi: 10.1094/PHYTO-01-15-0005-FI. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C, Liu L, Zheng Z, et al. Fine mapping of the Ph-3 gene conferring resistance to late blight (Phytophthora infestans) in tomato. Theor. Appl. Genet. 2013;126:2643–2653. doi: 10.1007/s00122-013-2162-1. [DOI] [PubMed] [Google Scholar]
- 7.Nowicki M, Fooled MR, Nowakowska M, Kozik EU. Potato and tomato late blight caused by Phytophthora infestans: an overview of pathology and resistance breeding. Plant Dis. 2012;96:4–17. doi: 10.1094/PDIS-05-11-0458. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 10.Tang G, Tang X. Short tandem target mimic: a long journey to the engineered molecular landmine for selective destruction/blockage of microRNAs in plants and animals. J. Genet. Genom. 2013;40:291–296. doi: 10.1016/j.jgg.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, Zou Z, Gong P, et al. Over-expression of microRNA169 confers enhanced drought tolerance to tomato. Biotechnol. Lett. 2011;33:403–409. doi: 10.1007/s10529-010-0436-0. [DOI] [PubMed] [Google Scholar]
- 12.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005;17:1360–1375. doi: 10.1105/tpc.105.031716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luan, Y. et al. Effective enhancement of resistance to Phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum. Planta247, 127–138 (2018). [DOI] [PubMed]
- 14.Luan Y, Cui J, Wang W, Meng J. MiR1918 enhances tomato sensitivity to Phytophthora infestans infection. Sci. Rep. 2016;6:35858. doi: 10.1038/srep35858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franco-Zorrilla JM, Valli A, Todesco M, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- 16.Tang G, Yan J, Gu Y, et al. Construction of short tandem target mimic (STTM) to block the functions of plant and animal microRNAs. Methods. 2012;58:118–125. doi: 10.1016/j.ymeth.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan J, Gu Y, Jia X, et al. Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell. 2012;24:415–427. doi: 10.1105/tpc.111.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia X, Ding N, Fan W, et al. Functional plasticity of miR165/166 in plant development revealed by small tandem target mimic. Plant Sci. 2015;233:11–21. doi: 10.1016/j.plantsci.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 19.Gu Z, Huang C, Li F, Zhou X. A versatile system for functional analysis of genes and microRNAs in cotton. Plant Biotechnol. J. 2014;12:638–649. doi: 10.1111/pbi.12169. [DOI] [PubMed] [Google Scholar]
- 20.Nizampatnam NR, Schreier SJ, Damodaran S, Adhikari S, Subramanian S. microRNA160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant J. 2015;84:140–153. doi: 10.1111/tpj.12965. [DOI] [PubMed] [Google Scholar]
- 21.Zhao J, Liu Q, Hu P, et al. An efficient Potato virus X-based microRNA silencing in Nicotiana benthamiana. Sci. Rep. 2016;6:20573. doi: 10.1038/srep20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia X, Shen J, et al. Small tandem target mimic-mediated blockage of microRNA858 induces anthocyanin accumulation in tomato. Planta. 2015;242:283–293. doi: 10.1007/s00425-015-2305-5. [DOI] [PubMed] [Google Scholar]
- 23.Akdogan G, Tufekci ED, Uranbey S, Unver T. miRNA-based drought regulation in wheat. Funct. Integr. Genom. 2016;16:221–233. doi: 10.1007/s10142-015-0452-1. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Wang T, Zhang Y, Li Y. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J. Exp. Bot. 2016;67:175–194. doi: 10.1093/jxb/erv450. [DOI] [PubMed] [Google Scholar]
- 25.Luan Y, et al. High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans. Planta. 2015;241:1405–1416. doi: 10.1007/s00425-015-2267-7. [DOI] [PubMed] [Google Scholar]
- 26.Jin H. Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 2008;582:2679–2684. doi: 10.1016/j.febslet.2008.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro L, et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006;312:436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 28.Boccara M, et al. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014;10:e1003883. doi: 10.1371/journal.ppat.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Pignatta D, Bendix C, et al. MicroRNA regulation of plant innate immune receptors. Proc. Natl Acad. Sci. USA. 2012;109:1790–1795. doi: 10.1073/pnas.1118282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang S, et al. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014;10:e1004464. doi: 10.1371/journal.ppat.1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, et al. Overexpression of potato miR482e enhanced plant sensitivity to Verticillium dahliae infection. J. Integr. Plant Biol. 2015;57:1078–1088. doi: 10.1111/jipb.12348. [DOI] [PubMed] [Google Scholar]
- 32.Li W, Xu YP, Cai XZ. Transcriptional and posttranscriptional regulation of the tomato leaf mould disease resistance gene Cf-9. Biochem. Biophys. Res. Commun. 2016;470:163–167. doi: 10.1016/j.bbrc.2016.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Bao, D. et al. Downregulation of genes coding for core RNAi components and disease resistance proteins via corresponding microRNAs might be correlated with successful SMV infection in soybean. Mol. Plant Pathol. 10.1111/mpp.12581 (2017). [DOI] [PMC free article] [PubMed]
- 34.Shivaprasad PV, et al. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsushima D, Adkar-Purushothama CR, Taneda A, Sano T. Changes in relative expression levels of viroid-specific small RNAs and microRNAs in tomato plants infected with severe and mild symptom-inducing isolates of Potato spindle tuber viroid. J. Gen. Plant Pathol. 2015;81:49–62. doi: 10.1007/s10327-014-0566-7. [DOI] [Google Scholar]
- 36.Feng J, Liu S, Wang M, Lang Q, Jin C. Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta. 2014;240:1335–1352. doi: 10.1007/s00425-014-2158-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, et al. Identification of miRNAs and their targets in cotton inoculated with Verticillium dahliae by high-throughput sequencing and degradome analysis. Int J. Mol. Sci. 2015;16:14749–14768. doi: 10.3390/ijms160714749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, et al. Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE. 2012;7:e39650. doi: 10.1371/journal.pone.0039650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao CZ, Xia H, Cao TJ, et al. Small RNA and degradome deep sequencing reveals peanut microRNA roles in response to pathogen infection. Plant Mol. Biol. Rep. 2015;33:1013–1029. doi: 10.1007/s11105-014-0806-1. [DOI] [Google Scholar]
- 40.Zhu QH, et al. MiR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS ONE. 2013;8:e84390. doi: 10.1371/journal.pone.0084390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marone D, Russo MA, Laido G, De Leonardis AM, Mastrangelo AM. Plant nucleotide binding site–leucine-rich repeat (NBS-LRR) genes: active guardians in host defense responses. Int. J. Mol. Sci. 2013;14:7302–7326. doi: 10.3390/ijms14047302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao ZC, et al. Cloning and functional analyses of pepper CaRKNR involved in Meloidogyne incognita resistance. Euphytica. 2015;205:903–913. doi: 10.1007/s10681-015-1438-8. [DOI] [Google Scholar]
- 43.Ma J, et al. Pi64, encoding a novel CC-NBS-LRR protein, confers resistance to leaf and neck blast in rice. Mol. Plant Microbe Interact. 2015;28:558–568. doi: 10.1094/MPMI-11-14-0367-R. [DOI] [PubMed] [Google Scholar]
- 44.Wang XM, et al. Characterization of a novel NBS-LRR gene involved in bacterial blight resistance in rice. Plant Mol. Biol. Rep. 2013;31:649–656. doi: 10.1007/s11105-012-0537-0. [DOI] [Google Scholar]
- 45.Lanfermeijer FC, Dijkhuis J, Sturre MJG, de Haan P, Hille J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-22 from Lycopersicon esculentum. Plant Mol. Biol. 2003;52:1039–1051. doi: 10.1023/A:1025434519282. [DOI] [PubMed] [Google Scholar]
- 46.Gutierrez JR, et al. Prf immune complexes of tomato are oligomeric and contain multiple Pto-like kinases that diversify effector recognition. Plant J. 2010;61:507–518. doi: 10.1111/j.1365-313X.2009.04078.x. [DOI] [PubMed] [Google Scholar]
- 47.Luan Y, Wang W, Liu P. Identification and functional analysis of novel and conserved microRNAs in tomato. Mol. Biol. Rep. 2014;41:5385–5394. doi: 10.1007/s11033-014-3410-4. [DOI] [PubMed] [Google Scholar]
- 48.Sun G, Luan Y, Cui J. Mining and characterization of miRNAs closely associated with the pathogenicity in tomato. Yi Chuan. 2014;36:69–76. doi: 10.3724/SP.J.1005.2014.00069. [DOI] [PubMed] [Google Scholar]
- 49.Cui J, Luan Y, Jiang N, Bao H, Meng J. Comparative transcriptome analysis between resistant and susceptible tomato allows the identification of lncRNA16397 conferring resistance to Phytophthora infestans by co-expressing glutaredoxin. Plant J. 2017;89:577–589. doi: 10.1111/tpj.13408. [DOI] [PubMed] [Google Scholar]
- 50.Li J, Luan Y, Liu Z. SpWRKY1 mediates resistance to Phytophthora infestans and tolerance to salt and drought stress by modulating reactive oxygen species homeostasis and expression of defense-related genes in tomato. Plant Cell Tissue Organ Cult. 2015;123:67–81. doi: 10.1007/s11240-015-0815-2. [DOI] [Google Scholar]
- 51.Jiang Y, et al. Overexpression of poplar PtrWRKY89 in transgenic Arabidopsis leads to a reduction of disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. PLoS ONE. 2016;11:e0149137. doi: 10.1371/journal.pone.0149137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fan C, Wei Q, Hao Z, Li G. Prediction and functional analysis of lincRNAs targeted by miRNAs. Yi Chuan. 2014;36:1226–1234. doi: 10.3724/SP.J.1005.2014.1226. [DOI] [PubMed] [Google Scholar]
- 55.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wi SJ, Ji NR, Park KY. Synergistic biosynthesis of biphasic ethylene and reactive oxygen species in response to hemibiotrophic Phytophthora parasitica in tobacco plants. Plant Physiol. 2012;159:251–265. doi: 10.1104/pp.112.194654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye W, Ma W. Filamentous pathogen effectors interfering with small RNA silencing in plant hosts. Curr. Opin. Microbiol. 2016;32:1–6. doi: 10.1016/j.mib.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Qiao Y, Liu L, Xiong Q, et al. Oomycete pathogens encode RNA silencing suppressors. Nat. Genet. 2013;45:330–333. doi: 10.1038/ng.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiao Y, Shi J, Zhai Y, Hou Y, Ma W. Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proc. Natl Acad. Sci. USA. 2015;112:5850–5855. doi: 10.1073/pnas.1421475112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, et al. Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 2015;5:16946. doi: 10.1038/srep16946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao C, et al. MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol. J. 2015;13:370–382. doi: 10.1111/pbi.12297. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H, et al. Short tandem target mimic rice lines uncover functions of miRNAs in regulating important agronomic traits. Proc. Natl Acad. Sci. USA. 2017;114:5277–5282. doi: 10.1073/pnas.1703752114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao D, et al. Regulations on growth and development in tomato cotyledon, flower and fruit via destruction of miR396 with short tandem target mimic. Plant Sci. 2016;247:1–12. doi: 10.1016/j.plantsci.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 64.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 65.Li X, Zhang Y, Yin L, Lu J. Overexpression of pathogen-induced grapevine TIR-NB-LRR gene VaRGA1 enhances disease resistance and drought and salt tolerance in Nicotiana benthamiana. Protoplasma. 2017;254:957–969. doi: 10.1007/s00709-016-1005-8. [DOI] [PubMed] [Google Scholar]
- 66.Gu K, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- 67.Radwan O, et al. Genetic diversity and genomic distribution of homologs encoding NBS-LRR disease resistance proteins in sunflower. Mol. Genet. Genom. 2008;280:111–125. doi: 10.1007/s00438-008-0346-1. [DOI] [PubMed] [Google Scholar]
- 68.Li X, et al. Comparative transcriptome analysis reveals defense-related genes and pathways against downy mildew in Vitis amurensis grapevine. Plant Physiol. Biochem. 2015;95:1–14. doi: 10.1016/j.plaphy.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 69.Zhang C, et al. Overexpression of a novel peanut NBS-LRR gene AhRRS5 enhances disease resistance to Ralstonia solanacearum in tobacco. Plant Biotechnol. J. 2017;15:39–55. doi: 10.1111/pbi.12589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang J, et al. Molecular cloning and functional analysis of GbRVd, a gene in Gossypium barbadense that plays an important role in conferring resistance to Verticillium wilt. Gene. 2016;575:687–694. doi: 10.1016/j.gene.2015.09.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.