Abstract
NG2 cells are a resident glial progenitor cell population that is uniformly distributed throughout the developing and mature mammalian CNS. Those in the postnatal CNS generate exclusively myelinating and non-myelinating oligodendrocytes and are thus equated with oligodendrocyte precursor cells. Prenatally, NG2 cells in the ventral gray matter of the forebrain generate protoplasmic astrocytes as well as oligodendrocytes. The fate conversion from NG2 cells into protoplasmic astrocytes is dependent on downregulation of the key oligodendrocyte transcription factor Olig2. We showed previously that constitutive deletion of Olig2 in NG2 cells converts NG2 cells in the neocortex into protoplasmic astrocytes at the expense of oligodendrocytes. In this study, we show that postnatal deletion of Olig2 caused NG2 cells in the neocortex but not in other gray matter regions to become protoplasmic astrocytes. However, NG2 cells in the neocortex became more resistant to astrocyte fate switch over the first 3 postnatal weeks. Fewer NG2 cells differentiated into astrocytes and did so with longer latency after Olig2 deletion at postnatal day 18 (P18) compared with deletion at P2. The high-mobility group transcription factor Sox10 was not downregulated for at least 1 month after Olig2 deletion at P18 despite an early transient upregulation of the astrocyte transcription factor NFIA. Furthermore, inhibiting cell proliferation in slice culture reduced astrocyte differentiation from Olig2-deleted perinatal NG2 cells, suggesting that cell division might facilitate nuclear reorganization needed for astrocyte transformation.
SIGNIFICANCE STATEMENT NG2 cells are glial progenitor cells that retain a certain degree of lineage plasticity. In the normal postnatal neocortex, they generate mostly oligodendrocyte lineage cells. When the oligodendrocyte transcription factor Olig2 is deleted in NG2 cells in the neocortex, they switch their fate to protoplasmic astrocytes. However, the efficiency of the fate switch decreases with age over the first 3 postnatal weeks and is reduced when cell proliferation is inhibited. As the neocortex matures, sustained expression of the oligodendrocyte lineage-specific key transcription factor Sox10 becomes less dependent on Olig2. Together, our findings suggest a gradual stabilization of the oligodendrocyte lineage genes and loss of lineage plasticity during the first 3 weeks after birth, possibly due to nuclear reorganization.
Keywords: astrocyte, lineage, NG2, Olig2, oligodendrocyte, Sox10
Introduction
NG2 cells, also called NG2 glia or polydendrocytes, are widely distributed throughout the developing and mature mammalian CNS (Nishiyama et al., 2009, 2016). Genetic fate-mapping studies have confirmed that, in the postnatal CNS, NG2 cells either self-renew or differentiate into myelinating and non-myelinating oligodendrocytes and thus they are commonly equated with oligodendrocyte precursor cells (OPCs). They express two OPC signature proteins, NG2 (Cspg4 gene product) and platelet-derived growth factor receptor α (Pdgfra), which are downregulated upon their terminal differentiation into postmitotic oligodendrocytes. NG2 cells remain proliferative throughout life, but the rate of their proliferation declines with age (Dawson et al., 2003; Psachoulia et al., 2009; Young et al., 2013).
Olig2 is a basic helix-loop-helix (bHLH) transcription factor that is broadly expressed in the germinal zones and is required for oligodendrocyte specification and maturation (Lu et al., 2002; Takebayashi et al., 2002; Zhou and Anderson, 2001; Maire et al., 2010; Wegener et al., 2015). During oligodendrocyte specification, Olig2 positively regulates transcription of Sox10, which belongs to the SoxE family of high-mobility group (HMG)-domain transcription factors and is expressed and functionally important throughout all stages of the oligodendrocyte lineage (Kuhlbrodt et al., 1998; Küspert et al., 2011; Stolt and Wegner, 2016). The onset of NG2 and Pdgfra expression immediately follows Sox10 induction in committed oligodendrocyte lineage cells as they emigrate from the germinal zone (for review, see Nishiyama et al., 2016).
A subpopulation of NG2 cells in the prenatal CNS also generate protoplasmic astrocytes in a region-specific manner (Zhu et al., 2008a,b; Ge et al., 2012; Huang et al., 2014). NG2 cells emerge in the telencephalon around embryonic day 14.5 (E14.5) (Zhu et al., 2008a). Subsequently, a subpopulation of them in the ventral forebrain downregulate Olig2 and become protoplasmic astrocytes, whereas other NG2 cells in the same region develop along the oligodendrocyte lineage. When Olig2 is constitutively deleted in NG2 cells, those in the neocortex switch their fate to become protoplasmic astrocytes at the expense of oligodendrocytes (Zhu et al., 2012), underscoring the critical role for Olig2 in maintaining NG2 cells in the oligodendrocyte lineage. In the normal adult CNS, NG2 cells do not produce astrocytes (Dimou et al., 2008; Rivers et al., 2008; Kang et al., 2010; Zhu et al., 2011), and deletion of Olig2 in the injured adult brain has yielded variable results (Dimou et al., 2008; Tatsumi et al., 2008; Komitova et al., 2011). To investigate the temporal changes in Olig2-dependent lineage plasticity of NG2 cells, we examined the fate of NG2 cells in the neocortex after deleting Olig2 at different postnatal ages. We show that there was an age-dependent delay and decline in astrocyte differentiation from NG2 cells after Olig2 deletion, which was correlated with delayed downregulation of Sox10. Furthermore, inhibiting cell division in slice cultures attenuated astrocyte differentiation from NG2 cells.
Materials and Methods
Generation of tamoxifen-inducible NG2 cell-specific Olig2 conditional knock-out mice.
Triple transgenic mouse line NG2creER:YFP:Olig2fl/fl (conditional knock-out, Cko) and heterozygous NG2creER:YFP:Olig2fl/+ (Ctr) were generated as follows. Mice that express tamoxifen-inducible Cre in NG2 cells (NG2creER; The Jackson Laboratory strain #008533, RRID:MGI_4819178) were crossed with the Cre reporter gtROSA-EYFP (The Jackson Laboratory strain #006148, RRID:MGI_2449038; YFP) as described previously (Zhu et al., 2011) and maintained as NG2creER:YFP double homozygotes. Mice with a conditional Olig2 allele (Olig2fl) were described previously (Yue et al., 2006; Zhu et al., 2012). NG2creER:YFP mice were mated with Olig2fl/fl mice to generate Olig2 Cko and Ctr mice. To obtain Olig2fl/− mice, Olig2fl/+ mice were first mated with EIIa-cre mice (The Jackson Laboratory strain #0003724, RRID:IMSR_JAX:003724), which causes recombination in the early embryo, leading to germline deletion of the floxed Olig2 allele. The resulting Olig2+/− mice were then crossed to obtain NG2creER:YFP:Olig2fl/− mice. Because Olig2 deletion efficiency was similar between NG2creER:YFP:Olig2fl/− and NG2creER:YFP:Olig2fl/fl. mice, we used the two strains interchangeably as Cko mice. Cre-mediated recombination was induced at postnatal day 2 (P2) or P18 by intraperitoneal injection of 0.1 mg/g body weight of 4-hydroxytamoxifen (4OHT; Sigma-Aldrich H7904) once daily over 4 consecutive days starting at P2 or P18. The stock solution of 10 mg/ml 4OHT was prepared in 19:1 mixture of canola oil and ethanol as described previously (Zhu et al., 2011). To examine cell proliferation, we injected 50 μg/g body weight of 5-ethynyl-2′-deoxyuridine (EdU; Invitrogen) into the mice twice at 4 and 2 h before perfusion. Both males and females were used for all the experiments. All animal procedures were approved by the Institutional Animal Care and Use Committee.
Tissue processing and immunohistochemistry.
On 14, 30, or 90 d after 4OHT injection [14, 30, or 90 d postinfection (dpi), respectively], mice were fixed by intracardiac perfusion of 4% paraformaldehyde-lysine-periodate solution and processed for immunohistochemistry as described previously (Zhu et al., 2008a). Mice were also perfused at 1 or 2 dpi to estimate Olig2 deletion efficiency after 4OHT injection into P2 or P18 mice. Fixed brains were cut into 50 μm vibratome sections or embedded in optimum cutting temperature solution (Fisher Scientific) after incubating overnight in 30% sucrose and 12 μm sections were cut on a Leica CM3050 S cryostat. Coronal, sagittal, and transverse sections were used for the brain, cerebellum, and spinal cord, respectively.
The source and dilution of the primary antibodies were as follows: mouse anti-aldehyde dehydrogenase 1 L1 (Aldh1L1, UC Davis/NIH NeuroMab catalog #75–140, RRID:AB_10673448, 1:100); chicken anti-GFP antibody (Aves Laboratories catalog #GFP-1020, RRID:AB_10000240, 1:1000); rabbit anti-glutathione S-transferase-π (Gst-π) antibody (MBL International catalog #312 RRID:AB_591792, 1:2000); mouse anti-glutamine synthetase (GS) antibody (Millipore catalog #MAB302 RRID:AB_2110656, 1:1000); rabbit anti-Id2 antibody (Cell Signaling Technology catalog #3431S RRID:AB_2122877, 1:1000), rabbit anti-NFIA antibody (gift from Dr. Deneen, Baylor College of Medicine, Houston, TX, 1:1000; currently available from Active Motif catalog #39329 RRID:AB_2314931); rabbit anti-NG2 antibody (Millipore catalog #AB5320 RRID:AB_91789, 1:500); mouse anti-Olig2 antibody (Millipore catalog #MABN50 RRID:AB_10807410, 1:1000); rabbit anti-Olig2 antibody (Novus catalog #NBP1–28667 RRID:AB_1914109, 1:500); rabbit anti-Sox9 antibody (Cell Signaling Technology catalog #82630 RRID:AB_2665492, 1:500; rabbit anti-Sox10 antibody (Millipore catalog #AB5727 RRID:AB_2195375, 1:1000); mouse anti-Sox10 antibody (Santa Cruz Biotechnology catalog #sc-365692, RRID:AB_10844002; 1:100) and guinea pig anti-Sox10 antibody (gift from Dr. Michael Wegner). Immunohistochemical analysis for GFP, GS, Gst-π, NG2, and Olig2 was performed as described previously (Zhu et al., 2008a). Additional antigen retrieval steps were used on some of the vibratome sections with rabbit Sox10, Id2, and NFIA antibodies following the method described by Jiao et al. (1999). For some nuclear antigens, sections were permeabilized with 1% Triton X-100 instead of 0.1%. Labeled sections were mounted in Vectashield mounting medium containing DAPI (Vector Laboratories) or Fluoro-Gel (Electron Microscopy Sciences) with 5 μg/ml Hoechst 33324. For EdU detection, samples were permeabilized in 0.5% Triton X-100/PBS, washed in 3% BSA (bovine serum albumin)/PBS twice for 5 min each and incubated for 30 min at room temperature in a reaction mixture that contained 150 mm NaCl, 100 mm TrisHCl, pH7.15, 4 mm CuSO4·5H2O, μg/ml Alexa Fluor-647 azide (Invitrogen), and 100 mm sodium ascorbate. After washes, the nuclei were labeled with Hoechst 33324 and mounted.
Microscopy and cell counts.
Images were acquired using a Zeiss Axiovert 200M equipped with Hamamatsu ORCA ER camera and Apotome or a Leica SP8 confocal microscope. Cells were counted on the Zeiss Axiovert 200M or from Leica SP8 confocal stacks in randomly selected fields in the neocortex at the level of anterior commissure. The proportion of cells displaying each phenotype was obtained by quantifying the percentage of YFP+Olig2− cells exhibiting the phenotype in Olig2 Cko mice and the percentage of YFP+Olig2+ cells exhibiting the phenotype in Ctr mice. To obtain the density of YFP+ astrocytes, numbers of YFP+GS+ astrocytes were obtained in four fields of view on Leica SP8 for each animal and the density per unit volume was calculated from the area of the field of view and the z-depth.
Slice culture.
Slice cultures were prepared from P4 Cko or Ctr mouse brains as described previously (Hill et al., 2013) after 4OHT injection on P2 and P3 to induce Cre recombination. Coronal slices were cultured on Millicell inserts (Millipore) for 5–6 d in the presence or absence of 20 μm PI3K inhibitor LY294002 (Sigma-Aldrich) or 5 μm DNA polymerase inhibitor aphidicolin. Culture medium was changed every other day. During the last 8 h of culture, 10 μm EdU was added. At the end of the culture period, slices were fixed in 4% paraformaldehyde and labeled for YFP, GFAP, and EdU. Slices were imaged on Leica SP8 confocal microscope or on Zeiss Axiovert 200M. The proportion of proliferating NG2 cells was determined by counting the number of YFP+ and YFP+EdU+ cells in randomly selected neocortical regions. The proportion of GFAP+ astrocytes among YFP+ cells was determined by counting the number of YFP+ and YFP+GFAP+ cells in randomly selected neocortical regions. All values are expressed as means ± SDs.
Results
We showed previously that constitutive deletion of Olig2 in NG2 cells led to an almost complete conversion of NG2 cells into protoplasmic astrocytes in the neocortex at the expense of oligodendrocytes and that deleting Olig2 at P2 caused 50% of the Olig2-deleted NG2 cells to become protoplasmic astrocytes in the neocortex (Zhu et al., 2012). In this study, we initially deleted Olig2 at P2, P18, and at P60 in Olig2 Cko mice (NG2creER:YFP:Olig2fl/fl) to determine whether loss of Olig2 from NG2 cells led to astrocyte differentiation at all postnatal ages. Because we did not observe astrocyte differentiation after deleting Olig2 at P60, we focused our analyses on mice in which Olig2 was deleted at P2 and P18 (Fig. 1A). We describe here our findings primarily in the neocortex, where we showed previously that NG2 cells do not generate protoplasmic astrocytes under normal conditions and that the effect of Olig2 deletion on astrocyte fate of NG2 cells is most prominent (Zhu et al., 2008a,b, 2012). We have also analyzed the gray matter of the spinal cord and cerebellum. By restricting our analysis to the gray matter, we have avoided the confounding phenomena of the requirement for Olig2 in astrocyte development in the corpus callosum (Cai et al., 2007) and our inability to perform long-term fate analysis in white matter due to replacement of Olig2-deleted NG2 cells by the more rapidly proliferating NG2 cells that had escaped Olig2 deletion.
Figure 1.
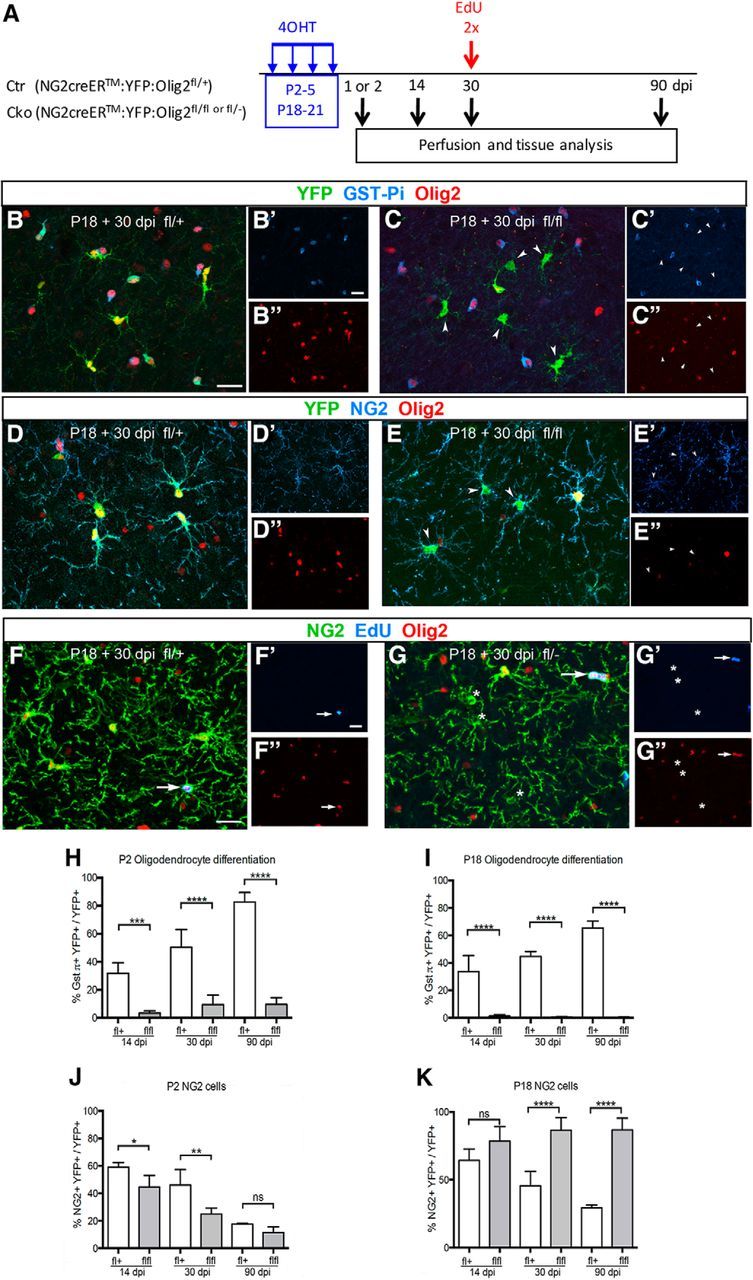
Compromised oligodendrocyte differentiation of Olig2-deleted cells. A, Schematic showing a timeline of the experiments. 4OHT was injected into Ctr or Cko mice at P2–P5 or P18–P21 and analyzed at 14, 30, or 90 dpi. A 1 or 2 dpi time point was taken to estimate the Olig2 deletion efficiency. EdU was injected twice 2 h apart before perfusion. B–E, Oligodendrocyte differentiation from NG2 cells in Ctr and Cko neocortex at P18 + 30 dpi. Neocortical sections from Ctr (B and D; fl/+) or Cko (C and E; fl/fl) mice immunolabeled for YFP (green), Olig2 (red) and Gst-π (blue in B and C) or NG2 (blue in D and E). YFP+ cells that lack Olig2 do not express Gst-π but are NG2+ (arrowheads in C and E). F, G, EdU incorporation into NG2 cells in Ctr (F) and Cko (G) neocortex at P18 + 30 dpi. Arrows indicate NG2+Olig2+ cells that are EdU+ in Ctr and Cko mice. EdU is not detected among NG2+ cells that lack Olig2 in Cko (asterisks in G). B′ and C′ are single channel images of Gst-pi immunofluorescence. D' and E' are single channel images of NG2 immunofluorescence. F′ and G′ show EdU labeled cells. B″, C″, D″, E″, F″, and G″ represent single channel images of Olig2 immunofluorescence. Scale bars, 20 μm. H–K, Quantification of oligodendrocytes and NG2 cells in Ctr and Cko neocortex. H, I, Oligodendrocyte differentiation after Cre activation at P2 (H) or P18 (I) showing the percentage of YFP+Gst-π+Olig2+ cells among YFP+Olig2+ cells in the neocortex of Ctr mice (open bars) and the percentage of YFP+Gst-π+Olig2− cells among YFP+Olig2− cells in Cko mice (gray bars). J, K, NG2 cells after Cre activation at P2 (J) or P18 (K) showing the percentage of YFP+NG2+Olig2+ cells among YFP+Olig2+ cells in the neocortex of Ctr mice (open bars) and the percentage of YFP+NG2+Olig2− cells among YFP+Olig2− cells in Cko mice (gray bars). The quantification in Cko mice was restricted in YFP+Olig2− cells (gray bars). Two-way ANOVA, Fisher's least significant difference test, n = 3. ns: not significant (p > 0.05); *0.01 < p < 0.05; **0.001 < p < 0.01; ***0.0001 < p < 0.001, ****p < 0.0001. Error bars indicate SD.
Olig2 deletion efficiency
We first assessed the extent of Olig2 deletion in the neocortex of Olig2 Cko mice (NG2creER:YFP:Olig2fl/fl) and Ctr mice (NG2creER:YFP:Olig2fl/+) at different time points after Cre activation. In Ctr neocortex, Olig2 was expressed in the vast majority of YFP+ cells 30 d after Cre induction by 4OHT injection from P18 to P21 (P18 + 30 dpi) (Fig. 1B,D), similar to what we reported previously at 30 dpi after the last 4OHT injection from P2 to P5 (Zhu et al., 2012). After Cre activation at P2 in Cko mice, >50% of YFP+ cells in the neocortex lacked Olig2 expression at P2 + 14, 30, and 90 dpi (Table 1). After Cre activation at P18 in Cko mice, Olig2 was undetectable in >78% of YFP+ cells at 1, 14, and 30 dpi and in 43% of YFP+ cells at 90 dpi (Table 1). In addition to NG2creER:YFP:Olig2fl/fl mice, we have also used NG2creER:YFP:Olig2fl/− mice in which deletion of one allele of Olig2 was passed through the germline. Olig2 deletion efficiency in NG2creER:YFP:Olig2fl/− mice was assessed at P18 + 2 dpi and was 52% among YFP+ cells in the neocortex, 77% in the gray matter of the spinal cord, and 69% in the gray matter of the cerebellum. Because the Olig2 deletion efficiency was similar between the two strains of Cko mice, we have used them interchangeably in the analyses described below. Because of the incomplete deletion of Olig2 in NG2 cells, we assessed the phenotype of YFP+ cells exclusively among the Olig2-negative (Olig2−) cells in Cko mice throughout this study.
Table 1.
Olig2 deletion efficiency in the neocortex of Olig2 Cko mice after Cre activation at P2 or P18
| % YFP+Olig2− cells/YFP+ cells | |
|---|---|
| P2 + 1 dpi | 37.7 ± 5.1 |
| P2 + 14 dpi | 52.5 ± 4.0 |
| P2 + 30 dpi | 63.7 ± 11.5 |
| P2 + 90 dpi | 52.6 ± 4.1 |
| P18 + 1 dpi | 81.3 ± 2.0 |
| P18 + 14 dpi | 85.5 ± 1.7 |
| P18 + 30 dpi | 78.1 ± 6.1 |
| P18 + 90 dpi | 42.8 ± 2.7 |
Impaired oligodendrocyte differentiation after Olig2 deletion from NG2 cells in the neocortex
To examine the extent of oligodendrocyte differentiation in the neocortex of Olig2 Cko and Ctr mice, we analyzed the percentage of YFP+Olig2− cells that had become oligodendrocytes at 14, 30, and 90 dpi after Cre activation at P2 or P18. Triple labeling for YFP, Olig2 and Gst-π, which labels mature oligodendrocytes (Fig. 1B,C; Cammer et al., 1989; Hill et al., 2014), showed that, in Ctr mice, the production of oligodendrocytes from NG2 cells in the neocortex of P2- and P18-induced mice steadily increased from 14 to 90 dpi (Fig. 1H,I), reaching 83% and 65% at 90 dpi after Cre activation at P2 and P18, respectively. In contrast, the proportion of Gst-π+ oligodendrocytes among YFP+Olig2− cells in Cko mice remained <10% in P2-induced mice and <1.5% in P18-induced mice, suggesting a nearly complete abrogation of oligodendrocyte differentiation from Olig2-deleted NG2 cells (Fig. 1C,H,I).
Next, we examined the proportion of induced cells that remained as NG2 cells (Fig. 1D,E). In Ctr mice induced at P2 or P18, there was a progressive decline in the proportion of YFP+NG2+ cells from 14 to 90 dpi, suggesting that NG2 cells with normal Olig2 levels differentiated into Gst-π+ oligodendrocytes over a period of 90 d (Fig. 1J,K). In the Ctr, oligodendrocytes and NG2 cells accounted for 85–100% of the YFP+ cells. In Cko mice in which Olig2 was deleted at P18, the majority of the YFP+Olig2− cells were NG2 cells (Fig. 1K), suggesting that without Olig2, NG2 cells in P18 neocortex did not differentiate into oligodendrocytes but remained as NG2 cells. In contrast, when Olig2 was deleted at P2, the proportion of YFP+NG2+ did not remain elevated but declined from 14–90 dpi (Fig. 1J). This suggests that a significant fraction of NG2 cells in P2 neocortex had differentiated into another cell type after Olig2 deletion. In Cko mice, the proportion of oligodendrocytes and NG2 cells among YFP+Olig2+ cells, which represent cells that had escaped Olig2 deletion, was similar to that of Olig2+ cells in Ctr mice (data not shown).
The sustained high proportion of YFP+NG2+ cells among YFP+ cells in mice in which Olig2 deletion was induced at P18 could be either due to increased proliferation of NG2 cells in Cko or due to an accumulation of NG2 cells that were incapable of differentiating into oligodendrocytes or astrocytes. To examine these possibilities, we performed pulse labeling with EdU at P18 + 30 dpi before perfusion and compared the extent of EdU incorporation into NG2+Olig2+ cells in Ctr and into NG2+Olig2− cells in Cko cortex. EdU was incorporated into 2% of NG2+Olig2+ cells in both Ctr and Cko mice (Fig. 1F,G, arrows). In contrast, we did not detect any NG2+Olig2− cells that were EdU+ in Cko cortex. Therefore, it is likely that the sustained high proportion of NG2+ cells among YFP+ cells in Cko cortex reflects an accumulation of NG2 cells that are not differentiating into oligodendrocytes or astrocytes.
Delayed astrocyte differentiation after Olig2 deletion in NG2 cells in the neocortex at P18
We reported previously that when Cre is induced in Olig2 Cko mice at P2, 23% and 55% of Olig2-deleted NG2 cells (YFP+Olig2− cells) become protoplasmic astrocytes after 14 and 30 dpi, respectively (Zhu et al., 2012). To determine whether NG2 cells in P18 neocortex were also capable of switching their fate to generate astrocytes upon deletion of Olig2, we injected 4OHT in Ctr and Cko mice at P18 and determined the fraction of YFP+Olig2− cells that were astrocytes. We initially used glutamine synthetase (GS) to identify astrocytes in our experiments as described previously (Zhu et al., 2012).
In Ctr neocortex, YFP+ cells expressed Olig2, exhibited the typical morphology of NG2 cells (polydendrocytes) or oligodendrocytes, and lacked GS immunoreactivity (Fig. 2A,C) at all time points, indicating that NG2 cells were restricted to the oligodendrocyte lineage in the presence of at least one allele of Olig2. Unlike mice in which Cre was activated perinatally or prenatally (Zhu et al., 2012), we did not detect NG2 cell-derived astrocytes in Cko neocortex at P18 + 14 dpi or P18 + 30 dpi (Fig. 2B,H). Surprisingly, at P18 + 90 dpi, we found YFP+ cells with bushy processes typical of protoplasmic astrocytes (Fig. 2D, arrows). These cells expressed GS, lacked Olig2, and comprised 26% of YFP+Olig2− cells (Fig. 2H). Thus, approximately half of the YFP+Olig2− cells became astrocytes when Cre was activated at P2. The density of YFP+GS+ NG2 cell-derived astrocytes at 30 and 90 dpi (Fig. 2I) declined from P2 + 30 dpi to P2 + 90 dpi even though the percentage of YFP+ cells that were astrocytes remained similar, likely due to a decline in the number of total YFP+ cells. We have confirmed the astrocyte phenotype of the bushy YFP+ cells by using an antibody to aldehyde dehydrogenase family 1 L1 (Aldh1L1), which positively labeled bushy YFP+ NG2 cell-derived astrocytes in Cko neocortex (Fig. 2E). These observations suggest that NG2 cells in P18 neocortex still retained the ability to follow an astrocyte fate after Olig2 deletion, but they did so with a more protracted time course and reduced efficiency compared with those in P2 neocortex.
Figure 2.
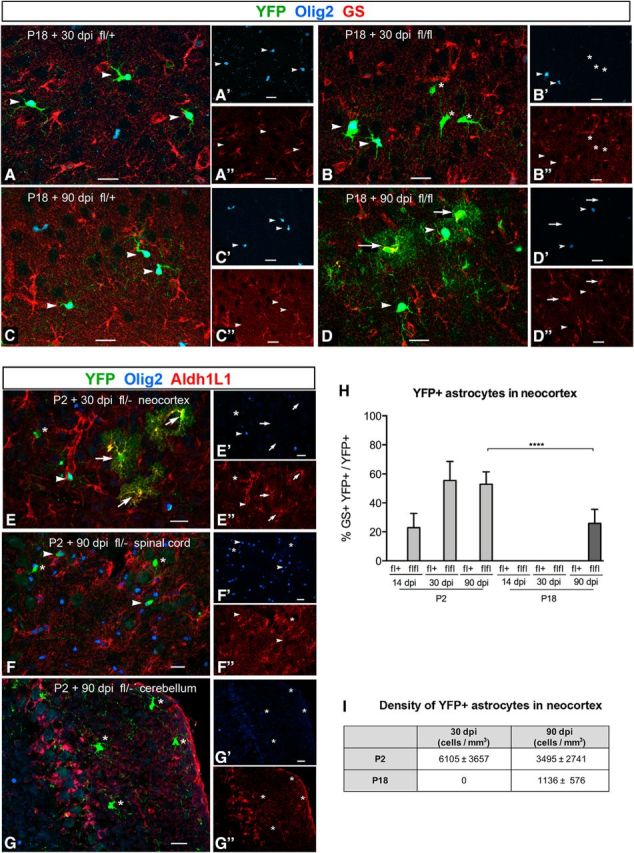
Astrocyte differentiation from NG2 cells in Cko neocortex. A–D, NG2 cell-derived astrocytes in Ctr (A, C, fl/+) and Cko mice (B, D, fl/fl) at P18 + 30 dpi (A, B) and 90 dpi (B, D). In Ctr neocortex, YFP+ cells that express Olig2 have the morphology of oligodendrocyte lineage cells and lack GS (arrowheads in A and C). In Cko neocortex, YFP+ cells that retain Olig2 lack GS and have the morphology of oligodendrocyte lineage cells as in Ctr (arrowheads in B and D). In Cko cortex at P18 + 90 dpi, there are clusters of YFP+ cells with bushy astrocyte morphology that express GS (arrows in D). Some Olig2− deleted cells at P18 + 30 dpi exhibit oligodendrocyte lineage morphology and lack GS (asterisks in B). E, Labeling for YFP, Olig2, and Aldh1L1 in Cko neocortex at P2 + 30 dpi confirming the astrocyte identify of bushy YFP+ cells (arrows). Some Olig2-deleted YFP+ cells (asterisks) and YFP+ cells that escaped Olig2 deletion (arrowheads) lack Aldh1L1 and have the morphology of oligodendrocyte lineage cells. F, G, In the spinal cord (F) and cerebellar cortex (G) of P2 + 90 dpi Cko mice, Olig2-deleted YFP+ cells do not express Aldh1L1 and exhibit the morphology of oligodendrocyte lineage cells (asterisks). Arrowheads denote YFP+ cells that retain Olig2. A′–G′: Olig2 immunofluorescence. A″–D″: GS immunofluorescence. E″–G″ Aldh1L1 immunofluorescence. Scale bars, 20 μm. H, Quantification of NG2 cell-derived astrocytes in Ctr and Cko neocortex. The proportion of YFP+GS+ astrocytes among YFP+ cells at 14, 30, and 90 dpi after Olig2 deletion at P2 (light gray bars) or P18 (dark gray bars) in the neocortex of Ctr (fl/+) and Cko (fl/fl) mice. Two-way ANOVA, Fisher's LSD (least significant difference) test, n = 3. ****p < 0.0001. Error bars indicate SD. I, Density of YFP+GS+ astrocytes at 30 and 90 dpi after Olig2 deletion at P2 or P18. The density of YFP+ astrocytes declines in the neocortex from P2 + 30 to 90 dpi and is lower at P18 + 90 dpi compared with that in P2 + 90 dpi neocortex.
There was variability in the proportion YFP+ astrocytes detected at 90 dpi after Olig2 deletion at P18, ranging from 14% to 33%, which is reflected in the variable proportion of YFP+NG2+ cells at P18 + 90 dpi ranging from 76% to 93% (Fig. 1). After Olig2 deletion at P2, Gst-π+ oligodendrocytes, NG2 cells, and astrocytes accounted for 65–85% of the YFP+ cells. It is possible that some of the YFP+ cells could be CC1+ immature oligodendrocytes because Gst-π is a relatively late antigen in the oligodendrocyte lineage, with a similar onset of expression to PLP-DsRed fluorescence (Hill et al., 2014).
We next investigated whether Olig2 deletion caused NG2 cells in other gray matter regions to differentiate into astrocytes. In the ventral gray matter of the forebrain, we did not detect YFP+ cells with bushy astrocyte morphology that were GS+ or Aldh1L1+. Nor did we see any YFP+ astrocytes in the gray matter of the spinal cord induced at P2 or P18 and analyzed at 30 and 90 dpi in the same animals in which we detected numerous YFP+ astrocytes in the neocortex (Fig. 2F) despite similar Olig2 deletion efficiency in the spinal cord and neocortex. We also examined the cerebellar cortex and did not find any YFP+ astrocytes in the molecular layer, Purkinje cell layer, or granule cell layer (Fig. 2G). These observations suggest that NG2 cells in the postnatal neocortex differ from those in other gray matter regions and are unique in their ability to switch their fate to astrocytes upon Olig2 deletion.
Delayed downregulation of Sox10 after Olig2 deletion at P18
In the CNS, Sox10 is expressed exclusively in the oligodendrocyte lineage and is present at all stages of oligodendrocyte development (Küspert et al., 2011; Küspert and Wegner, 2016). We showed previously that, in NG2 cells that are transitioning from NG2 cells into protoplasmic astrocytes around P4 after constitutive Olig2 deletion in NG2 cells, Sox10 is downregulated as the Olig2-deleted cells acquire astrocyte antigens (Zhu et al., 2012). To determine whether the delay in astrocyte fate switch from NG2 cells after Olig2 deletion at P18 could be attributed to delayed downregulation of Sox10 or delayed initiation of the astrocyte differentiation program after an early downregulation of Sox10, we examined the expression of Sox10 at different time points after Olig2 deletion at P18. In Ctr mice, Sox10 was expressed in all YFP+ cells in the neocortex at P18 + 14, 30, and 90 dpi. The morphology of these YFP+Sox10+ cells resembled that of NG2 cells or oligodendrocytes (Fig. 3A,C, arrowheads). In Cko mice at P18 + 30 dpi, Sox10 was detected in the nuclei of all YFP+Olig2+ cells and in 98.7 ± 2.2% of YFP+Olig2− cells (Fig. 3B, asterisks). By P18 + 90 dpi, Sox10 had become undetectable in YFP+Olig2− cells that exhibited bushy astrocyte morphology (Fig. 3D, arrows), whereas it was detected robustly in YFP+ cells that lacked Olig2, had the morphology of oligodendrocyte lineage cells, and apparently had not switched to an astrocyte fate (Fig. 3D, asterisks). This suggests that loss of Sox10 does not immediately follow the loss of Olig2 in most NG2 cells at P18 and that NG2 cells that eventually downregulate Sox10 expression become astrocytes.
Figure 3.
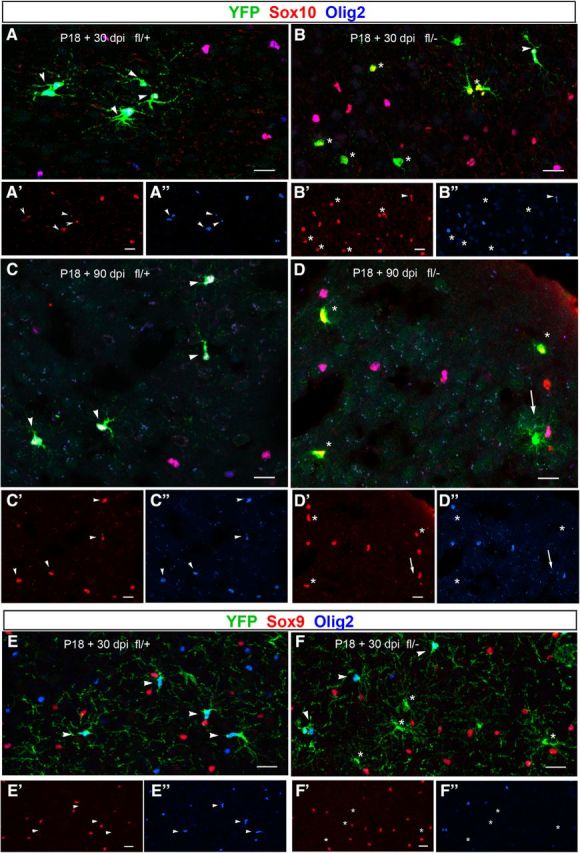
Delayed downregulation of Sox10 after Olig2 deletion at P18. A–D, Sox10 expression in Ctr (A, C) and Cko mice (B, D) at P18 + 30 dpi (A, B) and 90 dpi (C, D). Sections were triple labeled for Sox10, Olig2, and YFP. Sox10 is detected in YFP+Olig2+ cells in Ctr (fl/+) and Cko (fl/−) mice at 30 and 90 dpi (A, C, arrowheads) and in YFP+Olig2− cells in Cko (fl/−) mice at 30 dpi (B, asterisks) and in a subpopulation of YFP+Olig2− cells in Cko mice at 90 dpi with oligodendrocyte lineage morphology (D, asterisks). Sox10 immunoreactivity is not detectable in YFP+ cells with bushy protoplasmic astrocyte morphology found in Cko cortex at 90 dpi (D, arrows). E, F, Mutually exclusive expression of Sox9 and YFP+ cells. At P18 + 30 dpi, Sox9 is not detected in YFP+Olig2+ cells in the neocortex of Ctr (fl/+, E) and Cko (fl/−, F) mice (arrowheads) or YFP+Olig2− cells in Cko mice (F, asterisks). A′–D′: Sox10, A″–D″ Olig2. E′–F′: Sox9, E″–F″: Olig2. Scale bars, 20 μm.
During embryonic development in the spinal cord, Sox9, another member of the SoxE family of HMG transcription factors that is closely related to Sox10, is critical for the appearance of Sox10+ cells and persists for some time in NG2 cells, but is downregulated upon terminal oligodendrocyte differentiation (Stolt et al., 2003; Finzsch et al., 2008). We investigated whether Sox9 might be upregulated in Olig2-deleted NG2 cells in Cko cortex and could play a role in retaining Sox10. In the neocortex at P18 + 2 dpi, ∼16% of Pdgfra+ Olig2+ NG2 cells and 2.7% of Pdgfra+ Olig2− NG2 cells expressed Sox9. However, at P18 + 30 dpi, none of the YFP+ cells expressed Sox9 regardless of whether they expressed Olig2 (Fig. 3E,F). The lack of Sox9 expression in Olig2-deleted YFP+ cells (Fig. 3F, asterisks) suggests that reexpression of Sox9 in Olig2-deleted NG2 cells does not play a role in retaining Sox10 expression after Olig2 deletion in NG2 cells at P18.
Early upregulation of nuclear factor I-A (NFIA) expression after Olig2 deletion
Next, we assessed the expression pattern of NFIA in Ctr and Cko neocortex at 14, 30, and 90 d after Olig2 deletion at P2 or P18. NFIA is a transcription factor that is critical for astrocyte fate determination (Deneen et al., 2006; Kang et al., 2012). In Ctr, NFIA immunoreactivity was detected throughout the cortical layers, but YFP+ cells exhibited little detectable levels of NFIA, indicating that little NFIA was expressed in Olig2+ oligodendrocyte lineage cells (Fig. 4A,C,E, arrowheads). In the neocortex of Cko mice, YFP+ cells that retained Olig2 expressed little detectable NFIA (Fig. 4B,D,F, arrowheads). Occasionally, faint NFIA immunofluorescence could be visible in some YFP+Olig2+ cells (Fig. 4C,D, arrowheads) and in neuron-like cells, but strong NFIA immunofluorescence was not detected in YFP+Olig2+ cells. In a subpopulation of YFP+Olig2− cells that exhibited oligodendrocyte morphology, we readily detected NFIA (Fig. 4B,D,F, asterisks). The intensity of NFIA immunofluorescence in these YFP+Olig2− cells was weaker than that of some YFP− cells that strongly exhibited NFIA immunofluorescence. The YFP− cells that exhibited strong NFIA immunoreactivity in the nucleus were likely to be astrocytes because they expressed GS (Fig. 4G, pink arrows). YFP+NFIA+ cells became less abundant at 30 dpi compared with 14 dpi (Fig. 4H). By 90 dpi, NFIA expression was barely detectable in YFP+Olig2− cells that were not astrocytes (Fig. 4F, asterisks). In contrast, NFIA was strongly detected in the nucleus of YFP+Olig2− cells that exhibited the morphology of bushy protoplasmic astrocytes (Fig. 4B,F, arrows). The intensity of NFIA in most of the YFP+Olig2− astrocytes was comparable to that in YFP− astrocytes. After Olig2 deletion at P2, 35.7 ± 7.5% of YFP+Olig2− cells were NFIA+ at 14 dpi and this fraction decreased to 15.9 ± 2.9% by 30 dpi (Fig. 4H). When Olig2 was deleted at P18, 15.1 ± 3.4% of YFP+Olig2− cells were NFIA+ at 14 dpi and this fraction decreased to 4.9 ± 2.1% by 30 dpi. These findings suggest that NFIA was induced in NG2 cells shortly after Olig2 deletion and was further upregulated as they transformed into astrocytes, whereas it was shut down in those that aborted the astrocyte fate.
Figure 4.
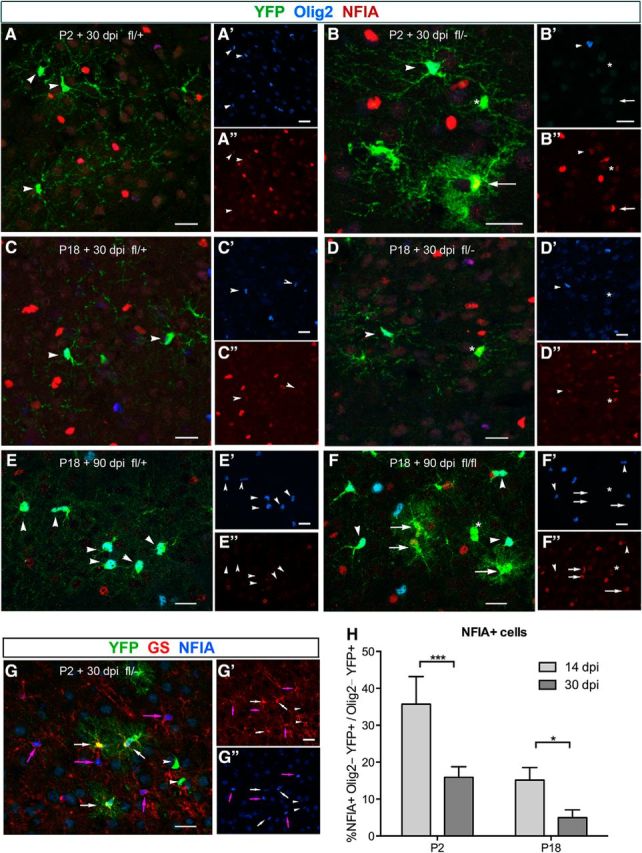
Early upregulation of NFIA expression in Olig2-deleted cells. A–F, Immunolabeling of neocortical sections from Ctr (fl/+; A, C, E) and Cko (fl/−; B, D, F) mice for YFP, Olig2, and NFIA. A, B, P2 + 30 dpi. In Ctr (A), YFP+ cells are Olig2+ (arrowheads) and have low levels or little detectable NFIA. In Cko (B), an Olig2+ YFP+ cell has a low level of NFIA and an Olig2− YFP+ cell with bushy astrocyte morphology (arrow) has a higher level of NFIA. Another YFP+ cell with an oligodendrocyte lineage cell morphology (asterisk) lacks Olig2 and has an intermediate level of NFIA. C, D, P18 + 30 dpi. In Ctr (C) and Cko (D), YFP+ cells that are Olig2+ (arrowheads) have low or no detectable NFIA. In Cko, one YFP+ cell that lacks Olig2 has a higher level of NFIA than its neighboring Olig2+ cells (asterisk). E, F, P18 + 90 dpi. In Ctr (E), YFP+ cells are Olig2+ (arrowheads) and have little detectable NFIA. In Cko (F), YFP+Olig2+ cells (arrowheads) have little detectable NFIA as in Ctr, whereas YFP+ Olig2− cells with bushy astrocyte morphology (arrows) have readily detectable NFIA. Asterisks indicate a YFP+Olig2− cell that does not have astrocyte morphology. G, Labeling for YFP, NFIA, and GS shows that cells that are strongly labeled for NFIA are GS+ astrocytes (pink arrows). Bushy YFP+ astrocytes derived from NG2 cells also express GS and NFIA (white arrows), whereas YFP+Olig2+ cells express neither GS nor NFIA (arrowheads). A′–F′: single channel images showing Olig2 immunofluorescence. A″–F″: single channel images showing NFIA immunofluorescence. G′: single channel image showing GS immunofluorescence. G″: single channel image showing NFIA immunofluorescence. Scale bars, 20 μm. H, Quantification of YFP+NFIA+ cells in the neocortex of Cko mice. Graph shows the proportion of YFP+Olig2− cells at 14 dpi (light gray bars) and 30 dpi (dark gray bars) after Olig2 deletion at P2 or P18. Fisher's least significant difference test, n = 3. *0.01 < p < 0.05, ***0.0001 < p < 0.001.
Astrocyte differentiation is inhibited by proliferation arrest in NG2 cells
Why does astrocyte fate conversion from NG2 cells in the P18 neocortex occur over a more prolonged period than that in the P2 neocortex? During the first postnatal 3 weeks, histone H3 acetylation gradually decreases and the class I histone deacetylases HDAC1 and HDAC2 are required for oligodendrocyte maturation and myelination (Marin-Husstege et al., 2002; Shen et al., 2005; Ye et al., 2009). To determine whether greater HDAC occupancy at astrocyte genes at P18 made these genes resistant to transcriptional activation upon Olig2 removal, we attempted to inhibit HDACs by administering the HDAC inhibitor suberanilohydroxamic acid (SAHA, also known as vorinostat; Guan et al., 2009) into P5 Cko mice. However, after 2 weeks of treatment, we did not observe any detectable changes in the level of total acetylated histone H3 or H3 acetylated on lysine 14 by Western blotting (data not shown) and therefore did not test the effects of SAHA on NG2 cell fate. Because HDACs 1 and 2 inhibit Wnt/β-catenin signaling and consequently transcription of inhibitory HLH factors such as Id2, which inhibit oligodendrocyte differentiation and promotes the astrocyte fate (Wang et al., 2001; Samanta and Kessler, 2004; Ye et al., 2009), we examined the expression of Id2 after Olig2 deletion at P18. Although we detected Id2 in YFP+ mature oligodendrocytes, we did not detect Id2 in YFP+ cells with polydendrocytes morphology at P18 + 14 and 30 dpi until the YFP+ cells differentiated into astrocytes at P18 + 90 dpi, when they also exhibited nuclear Id2 immunoreactivity (data not shown).
We next explored the possibility that NG2 cell reprogramming into astrocytes after Olig2 deletion required cell division and that it took longer for NG2 cells to become astrocytes after Olig2 deletion at P18 because the cell cycle time increases from <2 d at P2 to >18 d at P18 (Psachoulia et al., 2009; Young et al., 2013). We used coronal forebrain slice cultures from P4 Cko mice to test whether pharmacological inhibition of cell proliferation reduced the proportion of NG2 cells that differentiated into astrocytes after Olig2 deletion. We first injected Cko mice with 4OHT to activate Cre in vivo at P2–P3 and subsequently prepared slice cultures at P4 (Fig. 5A). In the first set of experiments, we incubated the slices for 6 d in vehicle or in 20 μm LY294002, which is a PI3K inhibitor and inhibits basal and PDGF AA-induced NG2 cell proliferation (Hill et al., 2013). Proliferation was assayed by quantifying the number of YFP+ cells that had incorporated EdU during the last 8 h of culture. In slice cultures treated with vehicle, we detected YFP+GFAP− cells with typical oligodendrocyte lineage cell morphology (Fig. 5B) and YFP+GFAP+ cells with protoplasmic astrocyte morphology (Fig. 5C,D). In slice cultures treated with LY294002, the density of YFP+ cells was reduced, and the majority of the YFP+ cells were GFAP− and had the morphology of polydendrocytes or immature oligodendrocytes (Fig. 5E,F). Quantification revealed that LY294002 treatment led to a 5-fold reduction in the proportion of YFP+ cells that had incorporated EdU (Fig. 5G). Under the same conditions, LY294002 treatment resulted in a 6.7-fold decrease in the proportion of YFP+ cells that were GFAP+ astrocytes (Fig. 5H). Because it is possible that LY294002 had inhibited a critical pathway required for astrocyte fate conversion from NG2 cells, we next incubated slice cultures with 0.5 μm aphidicolin, which is an inhibitor of DNA polymerase (Ikegami et al., 1978) and should block NG2 cell proliferation. Treatment of slice cultures with 0.5 μm aphidicolin completely suppressed incorporation of EdU into YFP+ cells and resulted in a concomitant 5-fold decrease in the proportion of YFP+ cells that were also GFAP+ (Fig. 5I). These observations suggest that astrocyte differentiation from Olig2-deleted NG2 cells is facilitated by cell division.
Figure 5.
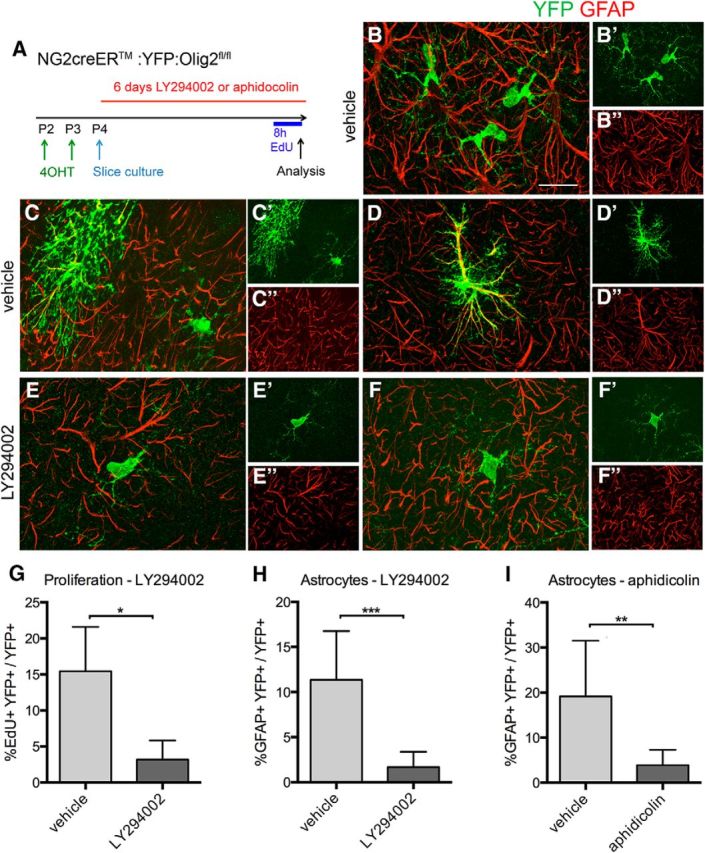
Astrocyte differentiation from Olig2-deleted NG2 cells in slice culture after inhibiting cell proliferation. A, Experimental setup. Slice cultures were prepared from P4 Cko mice after 4OHT injection in vivo at P2–P3. B–F, Slice cultures immunolabeled for YFP and GFAP. B, Vehicle-treated slice showing a cluster of YFP+GFAP− cells with polydendrocytes morphology. C, Vehicle-treated slice showing a YFP+GFAP+ cell with protoplasmic astrocyte morphology (left) and a YFP+GFAP− cell with polydendrocyte morphology (right). D, Vehicle-treated slice showing a YFP+GFAP+ astrocyte with strong GFAP immunoreactivity. E, F. LY29004-treated slice showing a YFP+GFAP− cell with the morphology of a polydendrocyte (E) or immature oligodendrocyte (F). A′, B′, C′, D′, E′, and F′ are single channel images showing YFP+ cells, and A″, B″, C″, D″, E″, and F″ are single channel images showing GFAP immunofluorescence. Scale bar, 20 μm. G, Proportion of EdU+ among YFP+ cells in slice cultures treated with vehicle (light gray bar) or LY294002 (dark gray bars). n = 4. H, Proportion of GFAP+ astrocytes among YFP+ cells in slice cultures treated with vehicle (light gray bar) or LY294002 (dark gray bar). n = 9. I, Proportion of GFAP+ astrocytes among YFP+ cells in slice cultures treated with vehicle (light gray bar) or aphidicolin (dark gray bar). n = 3. *p < 0.05, **0.01 < p < 0.05, ***0.001 < p < 0.01 Student's t test. Error bars indicate SD.
Discussion
We have shown that NG2 cells in the neocortex but not those in the spinal cord or cerebellar cortex differentiated into protoplasmic astrocytes after deletion of Olig2 during the first 3 postnatal weeks, but with increasing latency and reduced efficiency with age. Sox10 persisted for at least 30 d after Olig2 deletion at P18 and finally disappeared when the cells became astrocytes at 90 dpi. NFIA was transiently upregulated by 14 or 30 dpi after Olig2 deletion and more robustly in P2 than in P18 mice, correlating with the efficiency of astrocyte differentiation. Furthermore, conditions that inhibited proliferation of perinatal neocortical NG2 cells in slice cultures also reduced their astrocyte differentiation.
Role of Olig2 in the oligodendrocyte lineage
Olig2 is essential for oligodendrocyte specification (Zhou and Anderson, 2001; Lu et al., 2002; Takebayashi et al., 2002). Deletion of Olig2 in neural progenitor cells (Yue et al., 2006; Cai et al., 2007) or oligodendrocytes at different maturation stages (Mei et al., 2013) have yielded varying results, revealing the complexity of the function of Olig2 at different development stages. The critical role for Olig2 in oligodendrocyte differentiation was unequivocally shown by Olig2 gain of function (Liu et al., 2007; Maire et al., 2010), particularly within the Sox10+ oligodendrocyte lineage cells (Wegener et al., 2015). Consistently, we found that mature oligodendrocytes were severely reduced in the neocortex after Olig2 deletion in NG2 cells, further corroborating the essential role for Olig2 in oligodendrocyte maturation during the first 3 postnatal weeks.
Lineage plasticity of NG2 cells
Two recent studies support our observations on oligodendrocyte–astrocyte fate plasticity. In NG2-CreERT2 knock-in mice, NG2 cells differentiate into protoplasmic astrocytes when Cre is induced at E17.5 but not after birth (Huang et al., 2014), consistent with our previous observations using bacterial artificial chromosome transgenic mice (Zhu et al., 2011). Loss of the class I histone deacetylase HDAC3 in NG2 cells causes loss of Olig2 and a fate switch from oligodendrocytes to astrocytes, suggesting that HDAC3 is critical for sustaining Olig2 expression and maintaining cells in the oligodendrocyte lineage (Zhang et al., 2016). The lack of evidence for neuronal differentiation from Olig2-deleted NG2 cells suggests that neuronal genes may be more permanently repressed than astrocyte genes in NG2 cells.
Astrocyte differentiation from Olig2-deleted NG2 cells was unique to the neocortex and was not seen in the spinal cord or cerebellum, consistent with our previous finding that constitutive Olig2 deletion in NG2 cells in the spinal cord fails to convert them into astrocytes (Zhu et al., 2012). This could be a unique property of NG2 cells that arise in the Emx1 domain of the pallial germinal zone (Kessaris et al., 2006). In addition, because NG2 cells in the neocortex are the last cohort of oligodendrocyte lineage cells to be born, their plasticity could reflect chronological immaturity of NG2 cells or the relatively immature state of astrocytes such as their density. It is noteworthy that NG2 cells in anatomical regions that generate astrocytes during normal development, such as the ventral forebrain and spinal cord (Zhu et al., 2008a,b, 2012), do not undergo further fate switch into astrocytes when Olig2 is deleted.
Timing of astrocyte differentiation from Olig2-deleted NG2 cells
We showed previously that when Olig2 is deleted constitutively from NG2 cells in the neocortex, Olig2-deleted NG2 cells develop to normal density until P0, but by P4, they begin to downregulate Sox10 and Pdgfra and upregulate astrocyte antigens, and by P21 there is an almost complete conversion of NG2 cells into protoplasmic astrocytes at the expense of oligodendrocytes (Zhu et al., 2012). If Olig2 is downregulated in NG2 cells in the neocortical parenchyma by E16.5–E17.5, there would be a lag period of 7–8 d before Olig2-deleted NG2 cells begin to exhibit overt astrocyte phenotype. In postnatal neocortex, we detected NG2 cell-derived astrocytes by 14 dpi after Olig2 deletion at P2, but we did not detect NG2 cell-derived astrocytes until 90 dpi after Olig2 deletion at P18. Therefore, the latency between Olig2 deletion and astrocyte differentiation increased with the age of the animal.
Sox10 is activated directly by Olig2 at its U2 enhancer in the embryonic spinal cord at the time of oligodendrocyte specification (Küspert et al., 2011). Because Sox10 is critical for reprogramming neural progenitor cells or embryonic fibroblasts into oligodendrocyte lineage cells (Najm et al., 2013; Yang et al., 2013; Wang et al., 2014), successful conversion of NG2 cells into astrocytes requires an obligatory step of Sox10 downregulation. When Olig2 was deleted at P18, Sox10 expression persisted for at least 30 d despite efficient Olig2 deletion at 1 dpi, and its delayed downregulation was correlated with delayed astrocyte differentiation. Therefore, Sox10 expression in NG2 cells seemed to become less dependent on Olig2 with postnatal maturation. Interestingly, Brg1 (Brahma-related gene product 1), a subunit of the ATP-dependent chromatin remodeling complex, is required for Sox10 induction during oligodendrocyte specification (Yu et al., 2013), but becomes less critical for Sox10 expression in committed NG2 cells (Bischof et al., 2015), suggesting that differential mechanisms regulate Sox10 expression at different developmental stages.
NFIA is critical for astrocyte differentiation during embryonic development (Deneen et al., 2006; Namihira et al., 2009; Kang et al., 2012). Sox10 and NFIA interact and antagonize each other in the induction of myelin and astrocyte genes, respectively, and Olig2 facilitates the formation of the NFIA–Sox10 complex (Glasgow et al., 2014). In Olig2 Cko mice, NFIA was upregulated in NG2 cells early, by 14 dpi after Olig2 deletion at P2 or P18, when Sox10 was still expressed. Failure to extinguish Sox10 expression after Olig2 deletion at P18 could counter the effect of NFIA. Interestingly, deletion of HDAC3, which causes loss of Olig2, upregulates NFIA and its target astrocyte genes and downregulates Sox10 (Zhang et al., 2016).
Sox9, a transcription factor closely related to Sox10, interacts with NFIA to promote astrocyte differentiation and is highly expressed in postnatal neocortical astrocytes (Glasgow et al., 2014). Sox9 is also critical for oligodendrocyte specification and Sox10 induction during embryonic spinal cord development (Stolt et al., 2003; Finzsch et al., 2008). We did not detect Sox9 in Olig2-deleted YFP+ cells at P18 + 30 dpi. Therefore, it is unlikely that Sox9 is involved in sustaining Sox10 expression after Olig2 deletion at P18. Although Sox9 is expressed on all NG2 cells during embryonic development (Stolt et al., 2003), 81% of NG2 cells had lost Sox9 expression by P18 + 2 dpi, and no Sox9 was detected in NG2 cells at P18 + 30 dpi. The presence of Sox9 in younger NG2 cells could facilitate the fate switch by competing with Sox10 for NFIA binding (Glasgow et al., 2014). Other Sox family transcription factors such as Sox8 and Sox5/6 or developmental changes in Sox10 degradation could also play a role in sustaining Sox10 expression in Olig2-deleted NG2 cells (Stolt et al., 2004,2006; Lv et al., 2015; for review, see Weider and Wegner, 2017).
Role of NG2 cell proliferation in astrocyte fate switch
During development, progenitors generate diverse neuron and glial subtypes through temporal patterning, whereby the ability of progenitor cells to generate different cell types becomes restricted with age. Studies on the development of layer II/III neurons in mammals and neuroblast fate in Drosophila suggest that cell division could play a critical role in age-dependent fate restriction (McConnell and Kaznowski, 1991; Kohwi et al., 2013).
We reported previously that there is a finite temporal window after NG2 cell division in the postnatal brain during which the fate of NG2 cells is modulated by their microenvironment (Hill et al., 2014). Cell division could accelerate the astrocyte fate switch from NG2 cells after Olig2 removal by facilitating reorganization of the nuclear structure. Extinction of Sox10 expression upon Olig2 deletion might also require similar nuclear reorganization facilitated by cell division and possibly changes in Sox10-binding partners. The increase in the latency from Olig2 deletion to astrocyte differentiation could reflect the time it takes for the Olig2-deleted NG2 cells to undergo cell division because the cell cycle time increases with age (Psachoulia et al., 2009; Young et al., 2013). Consistently, pharmacological inhibition of cell proliferation in slice cultures significantly reduced the extent of astrocyte differentiation from Olig2-deleted NG2 cells. Proliferation of NG2 cells seems to become increasingly dependent on Olig2 with age. When Olig2 is deleted constitutively in NG2 cells, they expand normally until shortly after birth (Zhu et al., 2012). In contrast, there was reduced EdU incorporation into Olig2-deleted NG2 cells at P18 + 30 dpi. Therefore, the lower proliferative rate in Olig2-deleted NG2 cells in the more mature cortex could further contribute to the reduced efficiency of their astrocyte differentiation.
Although the mechanism for the age-dependent loss of NG2 cell lineage plasticity is currently unknown, changes in the chromatin landscape and nuclear organization could lead to progressive silencing of astrocyte genes, concomitant with progressive Olig2-independent activation of oligodendrocyte lineage genes. Reversal of these lineage-restrictive nuclear events would have to occur if reprogramming of NG2 cells into other cell types were to be achieved in the mature brain to promote lesion repair.
Footnotes
This work was supported by the National Institutes of Health (Grants R01NS049267, R01 NS074870, and R01 NS073425 to A.N.; Grants R01NS072427 and R01NS075243 to Q.R.L.) and the Connecticut Stem Cell Program (Grant 06SCB03 to A.N.). The Leica SP8 confocal microscope was purchased using National Institutes of Health Shared Instrumentation Grant S10OD016435 (principal investigator, A.N.) and is maintained by Dr. Chris O'Connell, Director of Advanced Microscopy Facility. We thank Youfen Sun for maintaining the transgenic mouse colony, Dr. Ben Deneen (Baylor College of Medicine) for the rabbit anti-NFIA antibody, and Dr. Michael Wegner (Erlangen University) for the guinea pig anti-Sox10 antibody.
The authors declare no competing financial interests.
References
- Bischof M, Weider M, Küspert M, Nave KA, Wegner M (2015) Brg1-dependent chromatin remodelling is not essentially required during oligodendroglial differentiation. J Neurosci 35:21–35. 10.1523/JNEUROSCI.1468-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Chen Y, Cai WH, Hurlock EC, Wu H, Kernie SG, Parada LF, Lu QR (2007) A crucial role for Olig2 in white matter astrocyte development. Development 134:1887–1899. 10.1242/dev.02847 [DOI] [PubMed] [Google Scholar]
- Cammer W, Tansey F, Abramovitz M, Ishigaki S, Listowsky I (1989) Differential localization of glutathione-S-transferase yp and yb subunits in oligodendrocytes and astrocytes of rat brain. J Neurochem 52:876–883. 10.1111/j.1471-4159.1989.tb02536.x [DOI] [PubMed] [Google Scholar]
- Dawson MR, Polito A, Levine JM, Reynolds R (2003) NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci 24:476–488. 10.1016/S1044-7431(03)00210-0 [DOI] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ (2006) The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 52:953–968. 10.1016/j.neuron.2006.11.019 [DOI] [PubMed] [Google Scholar]
- Dimou L, Simon C, Kirchhoff F, Takebayashi H, Götz M (2008) Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. J Neurosci 28:10434–10442. 10.1523/JNEUROSCI.2831-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzsch M, Stolt CC, Lommes P, Wegner M (2008) Sox9 and sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating pdgf receptor alpha expression. Development 135:637–646. 10.1242/dev.010454 [DOI] [PubMed] [Google Scholar]
- Ge WP, Miyawaki A, Gage FH, Jan YN, Jan LY (2012) Local generation of glia is a major astrocyte source in postnatal cortex. Nature 484:376–380. 10.1038/nature10959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Zhu W, Stolt CC, Huang TW, Chen F, LoTurco JJ, Neul JL, Wegner M, Mohila C, Deneen B (2014) Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci 17:1322–1329. 10.1038/nn.3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH (2009) HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459:55–60. 10.1038/nature07925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RA, Patel KD, Medved J, Reiss AM, Nishiyama A (2013) NG2 cells in white matter but not gray matter proliferate in response to PDGF. J Neurosci 33:14558–14566. 10.1523/JNEUROSCI.2001-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RA, Patel KD, Goncalves CM, Grutzendler J, Nishiyama A (2014) Modulation of oligodendrocyte generation during a critical temporal window after NG2 cell division. Nat Neurosci 17:1518–1527. 10.1038/nn.3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Zhao N, Bai X, Karram K, Trotter J, Goebbels S, Scheller A, Kirchhoff F (2014) Novel NG2-CreERT2 knock-in mice demonstrate heterogeneous differentiation potential of NG2 glia during development. Glia 62:896–913. 10.1002/glia.22648 [DOI] [PubMed] [Google Scholar]
- Ikegami S, Taguchi T, Ohashi M, Oguro M, Nagano H, Mano Y (1978) Aphidicolin prevents mitotic cell division by interfering with the activity of DNA polymerase-alpha. Nature 275:458–460. 10.1038/275458a0 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Sun Z, Lee T, Fusco FR, Kimble TD, Meade CA, Cuthbertson S, Reiner A (1999) A simple and sensitive antigen retrieval method for free-floating and slide-mounted tissue sections. J Neurosci Methods 93:149–162. 10.1016/S0165-0270(99)00142-9 [DOI] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, Deneen B (2012) Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron 74:79–94. 10.1016/j.neuron.2012.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Fukaya M, Yang JK, Rothstein JD, Bergles DE (2010) NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron 68:668–681. 10.1016/j.neuron.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD (2006) Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci 9:173–179. 10.1038/nn1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi M, Lupton JR, Lai SL, Miller MR, Doe CQ (2013) Developmentally regulated subnuclear genome reorganization restricts neural progenitor competence in Drosophila. Cell 152:97–108. 10.1016/j.cell.2012.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komitova M, Serwanski DR, Lu QR, Nishiyama A (2011) NG2 cells are not a major source of reactive astrocytes after neocortical stab wound injury. Glia 59:800–809. 10.1002/glia.21152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M (1998) Sox10, a novel transcriptional modulator in glial cells. J Neurosci 18:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küspert M, Wegner M (2016) SomethiNG 2 talk about-transcriptional regulation in embryonic and adult oligodendrocyte precursors. Brain Res 1638:167–182. 10.1016/j.brainres.2015.07.024 [DOI] [PubMed] [Google Scholar]
- Küspert M, Hammer A, Bösl MR, Wegner M (2011) Olig2 regulates Sox10 expression in oligodendrocyte precursors through an evolutionary conserved distal enhancer. Nucleic Acids Res 39:1280–1293. 10.1093/nar/gkq951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Hu X, Cai J, Liu B, Peng X, Wegner M, Qiu M (2007) Induction of oligodendrocyte differentiation by Olig2 and Sox10: evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol 302:683–693. 10.1016/j.ydbio.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH (2002) Common developmental requirement for olig function indicates a motor neuron/oligodendrocyte connection. Cell 109:75–86. 10.1016/S0092-8674(02)00678-5 [DOI] [PubMed] [Google Scholar]
- Lv XB, Wu W, Tang X, Wu Y, Zhu Y, Liu Y, Cui X, Chu J, Hu P, Li J, Guo Q, Cai Z, Wu J, Hu K, Ouyang N (2015) Regulation of sox10 stability via ubiquitination-mediated degradation by fbxw7alpha modulates melanoma cell migration. Oncotarget 6:36370–36382. 10.18632/oncotarget.5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire CL, Wegener A, Kerninon C, Nait Oumesmar B (2010) Gain-of-function of olig transcription factors enhances oligodendrogenesis and myelination. Stem Cells 28:1611–1622. 10.1002/stem.480 [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P (2002) Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci 22:10333–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SK, Kaznowski CE (1991) Cell cycle dependence of laminar determination in developing neocortex. Science 254:282–285. 10.1126/science.1925583 [DOI] [PubMed] [Google Scholar]
- Mei F, Wang H, Liu S, Niu J, Wang L, He Y, Etxeberria A, Chan JR, Xiao L (2013) Stage-specific deletion of Olig2 conveys opposing functions on differentiation and maturation of oligodendrocytes. J Neurosci 33:8454–8462. 10.1523/JNEUROSCI.2453-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Lager AM, Zaremba A, Wyatt K, Caprariello AV, Factor DC, Karl RT, Maeda T, Miller RH, Tesar PJ (2013) Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat Biotechnol 31:426–433. 10.1038/nbt.2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K (2009) Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell 16:245–255. 10.1016/j.devcel.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Komitova M, Suzuki R, Zhu X (2009) Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci 10:9–22. 10.1038/nrn2495 [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Boshans L, Goncalves CM, Wegrzyn J, Patel KD (2016) Lineage, fate, and fate potential of NG2-glia. Brain Res 1638:116–128. 10.1016/j.brainres.2015.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psachoulia K, Jamen F, Young KM, Richardson WD (2009) Cell cycle dynamics of NG2 cells in the postnatal and ageing brain. Neuron Glia Biol 5:57–67. 10.1017/S1740925X09990354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivers LE, Young KM, Rizzi M, Jamen F, Psachoulia K, Wade A, Kessaris N, Richardson WD (2008) PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci 11:1392–1401. 10.1038/nn.2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler JA (2004) Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development 131:4131–4142. 10.1242/dev.01273 [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P (2005) Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol 169:577–589. 10.1083/jcb.200412101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Wegner M (2016) Schwann cells and their transcriptional network: evolution of key regulators of peripheral myelination. Brain Res 1641:101–110. 10.1016/j.brainres.2015.09.025 [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M (2003) The sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 17:1677–1689. 10.1101/gad.259003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M (2004) Transcription factors sox8 and sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development 131:2349–2358. 10.1242/dev.01114 [DOI] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgärtner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, Wegner M (2006) SoxD proteins influence multiple stages of oligodendrocyte development and modulate soxe protein function. Dev Cell 11:697–709. 10.1016/j.devcel.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y (2002) The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol 12:1157–1163. 10.1016/S0960-9822(02)00926-0 [DOI] [PubMed] [Google Scholar]
- Tatsumi K, Takebayashi H, Manabe T, Tanaka KF, Makinodan M, Yamauchi T, Makinodan E, Matsuyoshi H, Okuda H, Ikenaka K, Wanaka A (2008) Genetic fate mapping of Olig2 progenitors in the injured adult cerebral cortex reveals preferential differentiation into astrocytes. J Neurosci Res 86:3494–3502. 10.1002/jnr.21862 [DOI] [PubMed] [Google Scholar]
- Wang J, Pol SU, Haberman AK, Wang C, O'Bara MA, Sim FJ (2014) Transcription factor induction of human oligodendrocyte progenitor fate and differentiation. Proc Natl Acad Sci U S A 111:E2885–E2894. 10.1073/pnas.1408295111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA (2001) A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron 29:603–614. 10.1016/S0896-6273(01)00237-9 [DOI] [PubMed] [Google Scholar]
- Wegener A, Deboux C, Bachelin C, Frah M, Kerninon C, Seilhean D, Weider M, Wegner M, Nait-Oumesmar B (2015) Gain of Olig2 function in oligodendrocyte progenitors promotes remyelination. Brain 138:120–135. 10.1093/brain/awu375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weider M, Wegner M (2017) SoxE factors: transcriptional regulators of neural differentiation and nervous system development. Semin Cell Dev Biol 63:35–42. 10.1016/j.semcdb.2016.08.013 [DOI] [PubMed] [Google Scholar]
- Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, Hawkins JS, Geissler R, Barres BA, Wernig M (2013) Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol 31:434–439. 10.1038/nbt.2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, Hsieh J, Bassel-Duby R, Olson EN, Lu QR (2009) HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci 12:829–838. 10.1038/nn.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KM, Psachoulia K, Tripathi RB, Dunn SJ, Cossell L, Attwell D, Tohyama K, Richardson WD (2013) Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron 77:873–885. 10.1016/j.neuron.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, Chan JR, Wu J, Lu QR (2013) Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 152:248–261. 10.1016/j.cell.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T, Xian K, Hurlock E, Xin M, Kernie SG, Parada LF, Lu QR (2006) A critical role for dorsal progenitors in cortical myelination. J Neurosci 26:1275–1280. 10.1523/JNEUROSCI.4717-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, He X, Liu L, Jiang M, Zhao C, Wang H, He D, Zheng T, Zhou X, Hassan A, Ma Z, Xin M, Sun Z, Lazar MA, Goldman SA, Olson EN, Lu QR (2016) Hdac3 interaction with p300 histone acetyltransferase regulates the oligodendrocyte and astrocyte lineage fate switch. Dev Cell 36:316–330. 10.1016/j.devcel.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Choi G, Anderson DJ (2001) The bHLH transcription factor Olig2 promotes oligodendrocyte differentiation in collaboration with Nkx2.2. Neuron 31:791–807. 10.1016/S0896-6273(01)00414-7 [DOI] [PubMed] [Google Scholar]
- Zhu X, Bergles DE, Nishiyama A (2008a) NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development 135:145–157. 10.1242/dev.004895 [DOI] [PubMed] [Google Scholar]
- Zhu X, Hill RA, Nishiyama A (2008b) Ng2 cells generate oligodendrocytes and gray matter astrocytes in the spinal cord. Neuron Glia Biol 4:19–26. 10.1017/S1740925X09000015 [DOI] [PubMed] [Google Scholar]
- Zhu X, Hill RA, Dietrich D, Komitova M, Suzuki R, Nishiyama A (2011) Age-dependent fate and lineage restriction of single NG2 cells. Development 138:745–753. 10.1242/dev.047951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zuo H, Maher BJ, Serwanski DR, LoTurco JJ, Lu QR, Nishiyama A (2012) Olig2-dependent developmental fate switch of NG2 cells. Development 139:2299–2307. 10.1242/dev.078873 [DOI] [PMC free article] [PubMed] [Google Scholar]


