Abstract
Background:
Cutaneous melanoma is the most serious skin malignancy and new therapeutic strategies are needed for advanced melanoma. TP53 mutations are rare in cutaneous melanoma and hence activation of wild-type p53 is a potential therapeutic strategy in cutaneous melanoma. Here, we investigated the WIP1 inhibitor, GSK2830371, and MDM2–p53 binding antagonists (nutlin-3, RG7388 and HDM201) alone and in combination treatment in cutaneous melanoma cell lines and explored the mechanistic basis of these responses in relation to the genotype and induced gene expression profile of the cells.
Methods:
A panel of three p53WT (A375, WM35 and C8161) and three p53MUT (WM164, WM35-R and CHL-1) melanoma cell lines were used. The effects of MDM2 and WIP1 inhibition were evaluated by growth inhibition and clonogenic assays, immunoblotting, qRT–PCR gene expression profiling and flow cytometry.
Results:
GSK2830371, at doses (⩽10 μM) that alone had no growth-inhibitory or cytotoxic effects on the cells, nevertheless significantly potentiated the growth-inhibitory and clonogenic cell killing effects of MDM2 inhibitors in p53WT but not p53MUT melanoma cells, indicating the potentiation worked in a p53-dependent manner. The siRNA-mediated knockdown of p53 provided further evidence to support the p53 dependence. GSK2830371 increased p53 stabilisation through Ser15 phosphorylation and consequent Lys382 acetylation, and decreased ubiquitination and proteasome-dependent degradation when it was combined with MDM2 inhibitors. These changes were at least partly ATM mediated, shown by reversal with the ATM inhibitor (KU55933). GSK2830371 enhanced the induction of p53 transcriptional target genes, cell cycle arrest and apoptosis.
Conclusions:
GSK2830371, a WIP1 inhibitor, at doses with no growth-inhibitory activity alone, potentiated the growth-inhibitory and cytotoxic activity of MDM2 inhibitors by increasing phosphorylation, acetylation and stabilisation of p53 in cutaneous melanoma cells in a functional p53-dependent manner.
Keywords: cutaneous melanoma, p53, MDM2, WIP1, nutlin-3, RG7388, HDM201
Cutaneous melanoma is the most serious skin malignancy (Miller and Mihm, 2006) and one of the most common cancers in developed countries (Torre et al, 2015). The prognosis of melanoma patients depends on the stage at presentation and the survival outcomes of patients with advanced stage are extremely poor (Balch et al, 2009). Both the MAPK (RAS–RAF–ERK) (Demunter et al, 2001; Davies et al, 2002) and ARF–MDM2–p53 (Chin, 2003) pathways are critical in melanoma tumourigenesis and genetic alterations associated with them provide important druggable targets. The combination of BRAF and MEK inhibitors has become the standard treatment for unresectable or metastatic melanoma harbouring a BRAFV600 mutation (Larkin et al, 2014; Robert et al, 2015). However, a therapeutic strategy targeting the p53 network has not been established in melanoma and could be a good option for advanced melanoma patients harbouring no BRAFV600 mutation (Lu et al, 2014).
The tumour suppressor p53 is a transcription factor that regulates a number of genes with a broad range of functions, including DNA repair, metabolism, cell cycle arrest, apoptosis and senescence (Levine et al, 1991; Wade et al, 2010; Gannon and Jones, 2012). Mutation of TP53 results in loss of wild-type (WT) p53 tumour suppressor function and can also have dominant oncogenic functions (gain-of-function mutations) that are entirely independent of WT p53, causing cancer cell development, survival and proliferation (Muller and Vousden, 2013). However, melanomas have p53 mutations only in a minority of cases (∼20%). Furthermore, p53 mutations and loss of CKDN2A appear to be mutually exclusive, possibly reflecting p53 dependence (Hodis et al, 2012; Zhang et al, 2016). Therefore, a therapeutic strategy to rescue and reactivate p53 function can be envisioned in melanomas that retain WT p53.
The MDM2–p53 binding antagonists block the p53-binding pocket of MDM2 and stabilise p53 by preventing MDM2-mediated ubiquitylation and degradation. This results in activation of the p53 pathway in p53WT (p53 wild type) rather than p53MUT (p53 mutated) cancer cells, causing cell cycle arrest, apoptosis and growth inhibition of human cancer cells (Brown et al, 2009). Nutlin-3 was the first MDM2–p53 binding antagonist to be developed and shown to have efficacy in vitro and in vivo (Vassilev et al, 2004; Sachweh et al, 2013; Polanski et al, 2014). The RG7388 (Idasanutlin), an orally available second-generation MDM2–p53 binding antagonist, efficiently suppressed tumour growth in vivo (Ding et al, 2013); clinical trials of RG7388 are currently ongoing to investigate the clinical efficacy of MDM2 inhibitors in relapsed multiple myeloma (NCT02633059), relapsed or refractory acute myeloid leukaemia (NCT02670044, NCT02545283) and relapsed or refractory follicular lymphoma or diffuse large B-cell lymphoma (NCT02624986). The HDM201, a new-generation and highly potent and selective MDM2 inhibitor, is also under investigation in early clinical trials (Hyman et al, 2016).
PPM1D encodes wild-type p53-induced phosphatase 1 (WIP1) that is involved in homeostatic regulation of p53 function and stability by directly dephosphorylating p53 after cellular stress. PPM1D is also a direct transcriptional target of p53, thus forming a negative autoregulatory loop with the p53 network by dephosphorylating p53 (Ser15) and other signalling components (such as ATM (ataxia-telangiectasia mutated), ATR (ataxia telangiectasia and Rad3-related) and MDM2) involved in p53 post-translational regulation (Lu et al, 2007; Lowe et al, 2012). GSK2830371 allosterically inhibits the enzymatic activity of WIP1 protein and also enhances ubiquitin-mediated degradation of WIP1 (Gilmartin et al, 2014). Preclinical studies have shown that GSK2830371 can enhance p53-mediated tumour suppression by MDM2 inhibitors, nutlin-3 (Esfandiari et al, 2016; Pechackova et al, 2016), nutlin-3a (Sriraman et al, 2016) and RG7388 (Esfandiari et al, 2016) or by chemotherapy (Gilmartin et al, 2014; Pechackova et al, 2016).
The aim of the current study was to investigate WIP1 inhibition (WIP1i), GSK2830371 and MDM2–p53 inhibitors, nutlin-3, RG7388 and HDM201, alone and in combination treatment specifically in cutaneous melanoma cell lines and to explore the mechanistic basis of these responses in relation to the genotype and induced gene expression of the cells.
Materials and methods
Cell lines and reagents
Cutaneous melanoma cell lines were routinely cultured using Dulbecco’s modified Eagle’s medium (DMEM) medium containing 4.5 g l−1 glucose (Sigma-Aldrich, Gillingham, UK) and supplemented with 10% (v/v) foetal calf serum. All the cell lines were authenticated by serial tandem repeat (STR) profiling (NewGene, Newcastle, UK) and tested to confirm lack of mycoplasma infection. Nutlin-3 was purchased from NewChem (Newcastle, UK), RG7388 and HDM201 were obtained from Astex Pharmaceuticals (Cambridge, UK) and GSK2830371 was purchased from Sigma-Aldrich. All compounds were initially dissolved in DMSO (Sigma-Aldrich) and used to dose cells at a final concentration of 0.5% DMSO with minimal cytotoxic effects on cells.
Growth inhibition assay
Melanoma cells were seeded in 96-well plates 24 h before 72 h of treatment with nutlin-3 RG7388, HDM201, GSK2830371 or combinations. The cells were fixed using Carnoy’s fixative followed by Sulforhodamine B (SRB) assay (Skehan et al, 1990). The GI50 value, the concentration of a compound that can reduce the growth of the cell population by 50% compared with solvent control, was determined. The details of the calculation for growth inhibition are described in Supplementary Information.
Clonogenic assay
Melanoma cell lines were seeded in 6-well plates and left for 24 h before treatment with MDM2 inhibitors for 72 h, combined with or without WIPi. Fresh medium was replaced and the cells were fixed after 10–21 days depending on the growth rates of the cells. The LC50 value, the concentration of a compound that can reduce the number of colonies by 50% compared with solvent control, was determined.
Immunoblotting
Cell lysates were harvested by 2% SDS lysis buffer, heated and sonicated. Equal quantities of protein were loaded onto and separated by SDS–polyacrylamide gels (4–20% Mini-PROTEAN TGX Gel, BioRad, Watford, UK). The separated proteins were transferred and immobilised onto Amersham nitrocellulose membranes (GE Healthcare Life Science, Amersham, UK). Primary antibodies against p53 (DO-7) (M7001, Dako, Glostrup, Denmark), MDM2 (Ab-1) (OP46, Merck Millipore, Watford, UK), p21WAF1 (EA10) (OP64, Calbiochem, Merck Millipore), WIP1 (F-10) (sc-376257, Santa Cruz Biotechnology, Dallas, TX, USA), phospho-p53(Ser-15) (9284, Cell Signaling Technology, Danvers, MA, USA), acetyl-p53(Lys382) (2525, Cell Signaling Technology), BAX (2772, Cell Signaling Technology), GAPDH (14C10) (2118, Cell Signaling Technology) and secondary goat anti-mouse/rabbit horseradish peroxidase-conjugated antibodies (P0447/P0448, Dako) were used. All antibodies were diluted in 5% (w/v) non-fat milk or BSA in TBS–Tween. Proteins were visualised using enhanced chemiluminescence (GE Healthcare Life Sciences) and X-ray film (Fujifilm, Bedford, UK). Densitometry was carried out using ImageJ software (National Institutes of Health, Rockville, MD, USA).
Site-directed mutagenesis and ectopic expression of mutant p53
The details of site-directed mutagenesis have been described in a previous study (Esfandiari et al, 2016). The WM35 was transfected with plasmid cDNA constructs encoding either wild-type or mutated p53 (S15A or S15D) using Lipofectamine 2000 (Thermo Fisher Scientific, Cramlington, UK) and incubated for 18 h to allow protein expression before collecting lysates for immunoblotting.
Denaturing immunoprecipitation
Cell lysates were collected by 2% SDS lysis buffer and aliquots were used as input. Non-SDS lysis buffer was used to dilute to the rest of lysates. A total of 2 μg of anti-p53 (sc-126, Santa Cruz Biotechnology), anti-Ub (sc-8017, Santa Cruz Biotechnology) or normal IgG (sc-2025, Santa Cruz Biotechnology) was incubated with lysates. Dynabeads (ThermoFisher Scientific) were used for immunoprecipitation according to the manufacturer’s protocol. Immunoblotting was performed as above and anti-mouse IgG VeriBlot for IP secondary antibody (HRP) (ab131368, Abcam, Cambridge, UK) was used.
RNA extraction and qRT–PCR
Total RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany). The RNA purity and concentration was estimated with an ND-1000 spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific). Complementary DNA was generated using the High-Capacity cDNA Reverse Transcription Kit (4368814, Thermo Fisher Scientific) as described by the manufacturer. The qRT–PCR was carried out using SYBR green RT–PCR master mix (Life Technologies, Thermo Fisher Scientific) as per the manufacturer’s guidelines and the following primers:
MDM2: F-5′-AGTAGCAGTGAATCTACAGGGA-3′ and R-5′-CTGATCCAACCAATCACCTGAAT-3′
CDKN1A: F-5′-TGTCCGTCAGAACCCATGC-3′ and R-5′-AAAGTCGAAGTTCCATCGCTC-3′
PUMA: F-5′-ACCTCAACGCACAGTACGA-3′ and R-5′-CTGGGTAAGGGCAGGAGTC-3′
TP53INP1: F-5′-TCTTGAGTGCTTGGCTGATACA-3′ and R-5′-GGTGGGGTGATAAACCAGCTC-3′
FAS: F-5′-AGATTGTGTGATGAAGGACATGG-3′ and R-5′-TGTTGCTGGTGAGTGTGCATT-3′
TNFRSF10B: F-5′-ATGGAACAACGGGGACAGAAC-3′ and R-5′-CTGCTGGGGAGCTAGGTCT-3′
BAX: F-5′-CCCGAGAGGTCTTTTTCCGAG-3′ and R-5′-CCAGCCCATGATGGTTCTGAT-3′
GAPDH: F-5′-CAATGACCCCTTCATTGACC-3′ and R-5′-GATCTCGCTCCTGGAAGAT-3′.
The qRT–PCR reactions using a total of 20 ng of the cDNA samples per 10 μl final reaction volume with the standard cycling parameters were performed and products detected in real time on an ABI 7900HT system (Applied Biosystems, Foster City, CA, USA) (Esfandiari et al, 2016). The GAPDH was used as endogenous control and samples of cells exposed to DMSO solvent control were used as the calibrator for each independent repeat, with the formula 2ΔΔCt used to calculate fold changes. Analysis was carried out using SDS 2.2 software (Applied Biosystems).
Reverse phase protein arrays
The reverse phase protein arrays (RPPA) detection of proteins was carried out by ArrayGen UK (Newcastle, UK) on a collaboration basis. The detailed experimental procedure and technical background are described in Supplementary Information (Voshol et al, 2009).
Fluorescence-activated cell sorting
After treatment, floating and adhered cells were pooled and fixed using 70% cold ethanol. Samples were incubated in 250 μl PBS with 40 μg ml−1 propidium iodide (Sigma-Aldrich) and 20 μg ml−1 RNAse A (Sigma-Aldrich) for 20 min in the dark at room temperature, and then were analysed on a fluorescence-activated cell sorting (FACS) Calibur flow cytometer using CellQuest Pro software (Becton Dickinson, Oxford, UK). Cell cycle distribution based on DNA content was determined using Cyflogic (CyFlo Ltd, Turku, Finland).
Caspase-3/7 activity assay
Melanoma cells were seeded in white 96-well plates and treated for 24 h. Caspase-3/7 enzymatic activities were measured using a FLUOstar Omega plate reader (BMG Labtech, Aylesbury, UK) after adding a 1 : 1 ratio of CaspaseGlo-3/7 reagent (Promega, Southampton, UK) to growth media and incubating for 30 min. All values were expressed as a ratio of signal relative to solvent control.
Transfection of siRNAs
A total of 40 nM siRNA duplex (Eurogentec, Liège, Belgium) against TP53 and control noncoding sequence was transfected by Lipofectamine 2000 (Thermo Fisher Scientific) in OptiMEM-glutamax serum free media (Thermo Fisher Scientific). The sequences were designed as follows: Control SiRNA (SiControl), sense: 5′-GCGCGCUUUGUAGGAUUCGdTdT-3′, antisense: 5′-CGAAUCCUACAAAGCGCGCdTdT-3′ two alternative TP53 targeted siRNAs (SiP53), SiP53 #1, sense: 5′-CCACCAUCCACUACAACUAdTdT-3′, antisense: 5′-UAGUUGUAGUGGAUGGUGGdTdT-3′, SiP53 #2, sense: 5′-CUGGAUGGAGAAUAUUUCAdTdT-3′, antisense: 5′-UGAAAUAUUCUCCAUCCAGdTdT-3′.
Statistical analysis
Data were presented as mean±s.e.m. unless otherwise stated. Statistical tests were carried out using GraphPad Prism 6 (GraphPad Software, La Jolla, CA, USA) and all P-values represent paired t-tests of at least three independent repeats. A P-value of <0.05 was considered as statistically significant.
Results
GSK2830371, at doses showing no growth inhibition alone, potentiated the effects of MDM2 inhibitors on A375 and WM35 cells
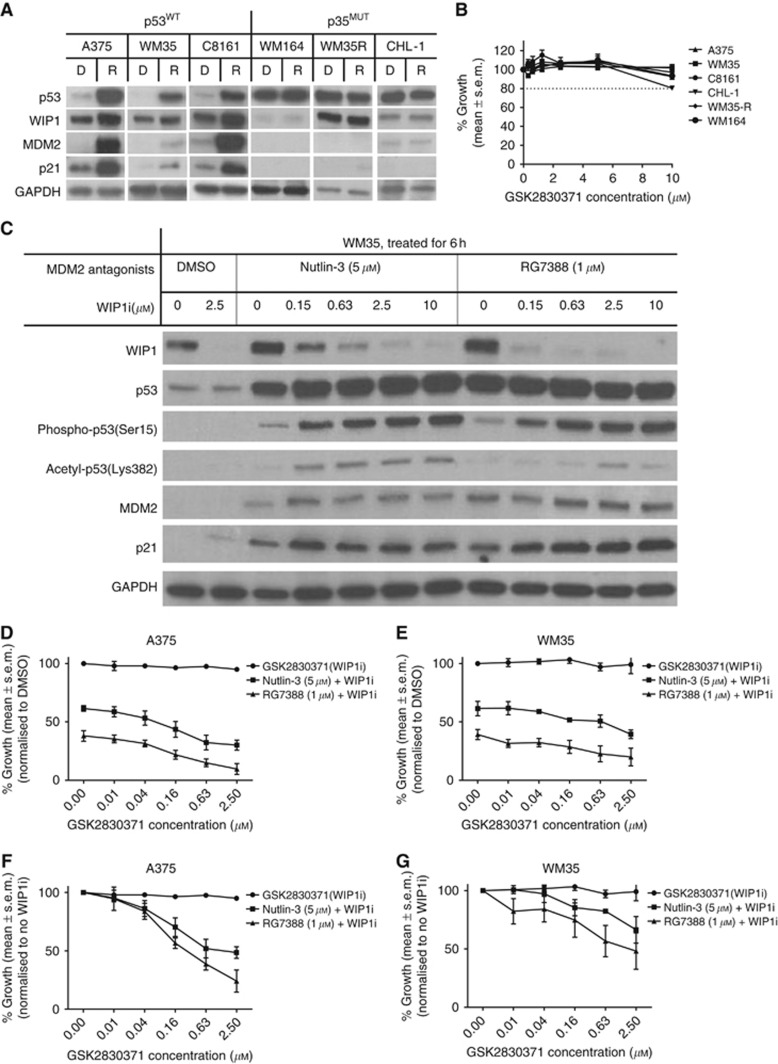
The expression of WIP1 protein after p53 activation was evaluated by immunoblotting, showing induction of WIP1 was associated with p53 stabilisation after RG7388 (0.2 μM) treatment for 6 h in p53WT but not p53MUT melanoma cell lines (Figure 1A). The WIP1i, GSK2830371, was used as a single agent in growth-inhibitory experiments. Growth inhibition measured by SRB assay showed that GSK2830371 at concentrations of ⩽10 μM had no or minimal growth-inhibitory effect on p53WT (A375, WM35, C8161) and p53MUT (WM164, WM35-R, CHL-1) melanoma cells (Figure 1B). Immunoblotting of WM35 cells treated by MDM2 inhibitor (either 5 μM nutlin-3 or 1 μM RG7388), combining with different concentrations of GSK2830371 (Figure 1C), illustrated that WIP1 protein decreased gradually with increased dosages of GSK2830371. Phospho-p53 (Ser15) and acetyl-p53 (Lys382) were probed because phospho-p53 (Ser15) is a direct substrate of WIP1 phosphatase and p53 can be acetylated by CBP/P300 that has been reported to be recruited after phosphorylation of p53 (Ser15) (Meek, 2015). Marked increases in phospho-p53 (Ser15) and acetyl-p53 (Lys382) as a consequence of WIP1 inhibition were observed. The transcriptional targets of p53, MDM2 and p21 increased modestly after combination treatment with GSK2830371.
Figure 1.
The WIP1 phosphatase induction by p53 activation with either nutlin-3 or RG7388 and inhibition by GSK2830371 (WIP1i). (A) Increase in WIP1 expression in p53WT but not p53MUT melanoma cell lines by p53 activation with RG7388 (0.2 μM) for 6 h (D, DMSO, R, RG7388). (B) Growth inhibition by SRB assay showed WIP1i at ⩽10 μM has no growth-inhibitory effect on p53WT or p53MUT melanoma cells. (C) Immunoblotting of A375 and WM35 cells treated by MDM2 antagonists (5 μM nutlin-3 or 1 μM RG7388) combined with different concentrations of WIP1i for 6 h. (D–G) Growth inhibition of A375 and WM35 by either 5 μM nutlin-3 or 1 μM RG7388, combined with different concentrations of WIP1i for 72 h. The % growth is shown either normalised to DMSO solvent control treatment (D and E) or to the fixed doses of MDM2 inhibitors (F and G).
To choose an optimal concentration of GSK2830371, growth inhibition assays were performed for A375 and WM35 treated with either MDM2 inhibitor (5 μM nutlin-3 or 1 μM RG7388) combined with different concentrations of GSK2830371 (Figure 1D–G). Dose-dependent growth inhibition by GSK2830371 was only found after p53 activation with nutlin-3 or RG7388. A GSK2830371 concentration of 2.5 μM displayed the best potentiation when combined with either MDM2 inhibitor and was therefore used for subsequent experiments to evaluate the role of WIP1 in p53 network responses.
Supplementary Figure S1A shows immunoblotting of A375, WM35 and C8161 cells treated with MDM2 inhibitor (5 μM nutlin-3 or 1 μM RG7388) combined with 2.5 μM GSK2830371 for 6 h. Supplementary Figure S1B shows immunoblotting of A375 and C8161 cells treated with HDM201 combined with 2.5 μM GSK2830371 for 6 and 24 h. The protein changes were similar to the findings in Figure 1C. The same treatment was performed on A375 and phospho-p53 (Ser15) was detected by RPPA. Significantly increased phospho-p53 (Ser15) was found after combination treatment with either nutlin-3 or RG7388 and GSK2830371 (Supplementary Figure S1C). The signals were suppressed by alkaline phosphatase, validating the antibody used in the RPPA to be specific for phosphorylated protein.
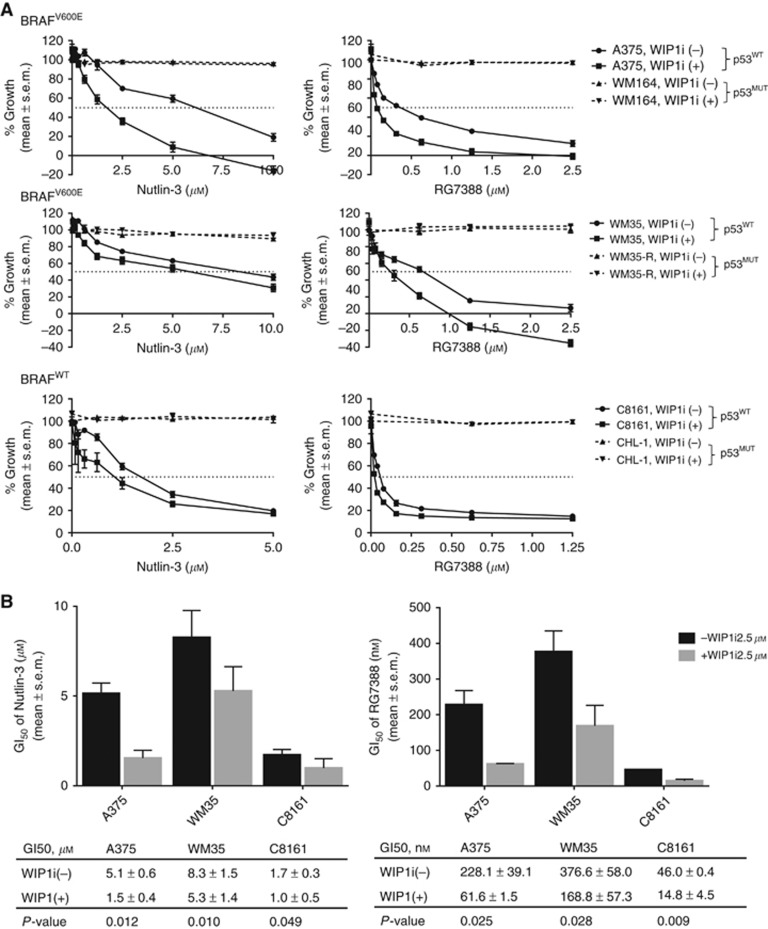
GSK2830371 potentiated the growth-inhibitory and cytotoxic activity of MDM2 inhibitors in a p53-dependent manner
Growth inhibition and clonogenic survival following treatment with MDM2 inhibitor (either nutlin-3, RG7388 or HDM201) with or without GSK2830371 (2.5 μM) combination was evaluated for a panel of p53WT (A375, WM35 and C8161) and p53MUT (WM164, WM35-R, CHL-1) melanoma cell lines (Figures 2 and 3 and Supplementary Figure S2). GSK2830371 enhanced the growth-inhibitory effects of MDM2 inhibitors in p53WT rather than p53MUT melanoma cells, showing that potentiation by GSK2830371 worked in a p53-dependent manner (Figure 2A and Supplementary Figure S2A). GSK2830371 significantly decreased the GI50 for nutlin-3, RG7388 or HDM201 compared with the GI50 for nutlin-3 or RG7388 alone in p53WT melanoma cells (Figure 2B and Supplementary Figure S2B).
Figure 2.
GSK2830371 (WIP1i) potentiated the growth-inhibitory effect of MDM2 antagonists in a p53-dependent manner. (A) Growth inhibition measured by SRB assay for p53WT (A375, WM35 and C8161) and p53MUT (WM164, WM35-R, CHL-1) melanoma cell lines treated with different concentrations of MDM2 antagonists (either nutlin-3 or RG7388), combined with or without WIP1i (2.5 μM) for 72 h. All % of growth was normalised to DMSO treatment. (B) Summary of GI50 (mean±s.e.m.) values for nutlin-3 or RG7388 with or without WIP1i in p53WT melanoma cells.
Figure 3.
The reduction in melanoma clonogenic cell survival by MDM2 antagonists was potentiated by GSK2830371 (WIP1i). (A) The p53-wild-type (WT) melanoma cell lines (A375, WM35 and C8161) were treated by different concentrations of MDM2 antagonists (nutlin-3 or RG7388), combined with or without WIP1i (2.5 μM) for 72 h. (B) Summary of the effect of WIP1 inhibition on LC50 values for nutlin-3 or RG7388 from at least three independent repeats. (C) Clonogenic survival of p53WT and p53 MT cells treated with either DMSO, WIP1i, nutlin-3, RG7388 or combination. (D) Clonogenic formation of C8161 cells following treatment with nutlin-3 (200 nM), RG7388 (20 nM), WIP1i (2.5 μM) or combination. *P<0.05, **P<0.005, ***P<0.0005.
A clonogenic assay was performed to see whether GSK2830371 can enhance the cytotoxic activity of MDM2 inhibitors. The GSK2830371 further inhibited colony formation when it was added to either MDM2 inhibitor in p53WT melanoma cells (Figure 3A and Supplementary Figure S2C). GSK2830371 significantly decreased the LC50 for nutlin-3, RG7388 or HDM201 compared with the LC50 for MDM2 inhibitor alone for A375 or C8161 cells (P<0.05) and with a similar trend for WM35 (Figure 3B and Supplementary Figure S2D). Compared with control (DMSO), there was no significant reduction of colony formation on treatment by GSK2830371 (2.5 μM) as a single agent (Figure 3C and D). The potentiation by GSK2830371 in clonogenic assays was limited to p53WT and was not seen with p53MUT melanoma cells (Figure 3C).
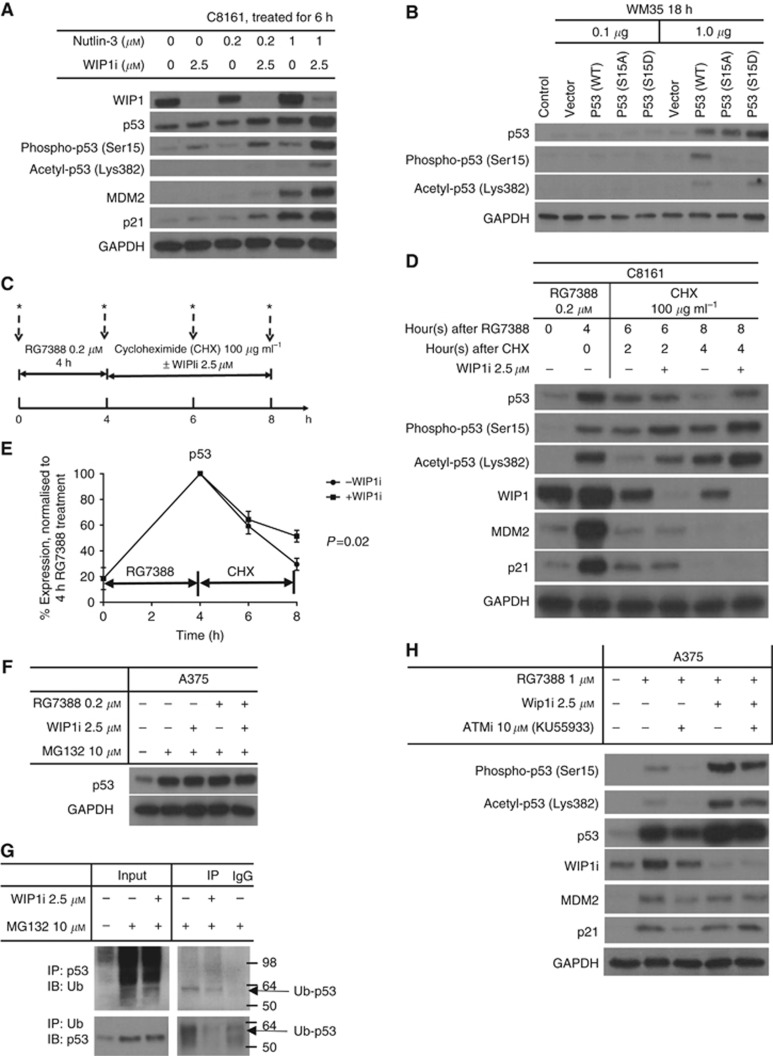
Increased p53 stabilisation by GSK2830371 through increased phosphorylation and acetylation when combined with MDM2 inhibitors could be reversed by ATM inhibition
Ubiquitylation and acetylation happen on the same lysine residues of the C-terminal region of p53 and both are mutually exclusive. Therefore, it was hypothesised that inhibition of WIP1 protein by GSK2830371 could enhance the stabilisation of p53 through increasing acetylation and decreasing ubiquitin-mediated degradation. The lack of obvious change in total p53 protein in the presence of WIP1 inhibition in Figure 1C was postulated to reflect saturation of p53 stabilisation following treated by 5 μM nutlin-3 and 1 μM RG7388. Therefore, lower concentrations of nutlin-3 and RG7388 were tested in combination with GSK2830371. The resulting immunoblots confirmed that combination of lower MDM2 inhibitor doses and WIP1i increased p53 stabilisation compared with the effect of MDM2 inhibitor as a single agent (Figure 4A and Supplementary Figures S1B and S3). Furthermore, this increase was associated with higher levels of phosphorylation (Ser15) and acetylation (Lys382) of p53, and higher expressions of MDM2 and p21.
Figure 4.
The p53 increase by GSK2830371(WIP1i) and MDM2 inhibitors was reversed by ATM inhibition (ATMi, KU55933). (A) Immunoblotting of C8161 cells treated with nutlin-3, WIP1i or combination of both for 6 h. (B) Effect of WM35 transfection with different p53 (Ser15) mutant plasmid constructs on acetyl-p53(Lys 382). (C) Design of a cycloheximide (CHX) experiment to measure protein turnover. (D) Immunoblotting for C8161 cells treated by RG7388 (0.2 μM) followed by CHX with or without 2.5 μM WIP1i. (E) The relative expression of p53 by densitometry, normalised to GAPDH; P-value shown for three independent repeats. (F) Immunoblotting for A375 cells treated by MG132±RG7388±WIP1i for 6 h. (G) The A375 was treated by MG132±WIP1i for 6 h. The Ub–p53 was immunoprecipitated with anti-p53 antibody and immunoblot with anti-Ub antibody and by reverse way. The last lane is the IgG pulldown negative control that was incubated with lysates after MG132 treatment. (H) Immunoblotting of A375 cells treated with R7388, WIP1i, ATMi and combinations for 6 h. *Lysates.
To examine whether the phosphorylation of p53 (Ser15) is a key residue for subsequent C-terminal acetylation of p53, WM35 cells were transfected with different p53 mutant plasmid constructs, including WT, S15A and S15D mutations (Figure 4B). After 18 h of transfection with 1 μg plasmid, increased p53 expression was detected for all three constructs. However, phospho-p53 (Ser15) could only be detected in cells after WT p53 cDNA transfection and not p53 mutant transfections, showing the antibody used was specific against WT p53 (Ser15) phosphorylation. Acetyl-p53 (Lys382) was detected in the cells after p53 (WT) and the S15D phosphomimetic mutant transfection but not when ser15 was mutated to the non-phosphorylatable alanine, indicating that Ser15 and its phosphorylation is an essential and sufficient residue for p53 acetylation.
To evaluate whether WIP1 inhibition by GSK2830371 stabilises p53 by slowing down its degradation, cycloheximide (CHX) was used to block de novo protein synthesis (Figure 4C). The C8161 was treated with 0.2 μM RG7388 for 4 h to make p53 accumulate. The cells were then treated with CHX (100 μg ml−1) to block protein synthesis, in the presence or absence of GSK2830371 for 2 and 4 h. Protein lysates were collected for immunoblotting at the indicated time points. During the 4 h CHX treatment, GSK2830371 slowed down p53 degradation, evidenced by significantly higher levels of p53 protein with concurrent GSK2830371 treatment than without GSK2830371 treatment (P=0.02) (Figure 4D and E). To test whether the degradation is proteasome dependent, A375 cells were treated with RG7388±WIP1i and MG132 for 6 h. The MG132 completely prevented the degradation of p53 and masked any difference in stabilisation of p53 by RG7388±WIP1i (Figure 4F). Co-immunoprecipitation showed GSK2830371 treatment decreased Ub–p53 by pulling down either p53 or ubiqutin (Figure 4G).
We next examined whether the p53 (Ser15) phosphorylation seen following combined treatment with MDM2 inhibitors and WIP1i is ATM mediated. The A375 cells were treated with 0.2 μM RG7388±WIP1i and±ATM inhibitor (KU55933 10 μM) concurrently for 6 h. The phosphorylation, acetylation and stabilisation of p53 were reversed by ATM inhibitor, indicating that these changes are at least in part mediated by basal ATM activity (Figure 4H). The KU55933 also decreased the growth-inhibitory effects of MDM2 inhibitors on A375 cells (Supplementary Figure S4).
Paired WM35 and WM35-R cells showed GSK2830371 increased p53 stabilisation in a p53-dependent manner
The WM35-R is an MDM2 inhibitor resistant cell line that we have selected from parental WM35 cells by culturing in medium containing 5 μM RG7388. Sanger sequencing revealed that WM35-R has a homozygous TP53 point mutation (1001G>T) resulting in a Gly334Val amino acid substitution in the p53 protein (Supplementary Figure S5), which is the hinge between β-strand (residues 326–333) and α-helix (residues 335–354) regions in the oligomerisation domain (Kawaguchi et al, 2005). Therefore, this pair of melanoma cells is useful for exploring p53-dependent pathway mechanisms in cutaneous melanoma.
Compared with MDM2 inhibitors alone, the combination of MDM2 inhibitor and GSK2830371 increased p53 phosphorylation (Ser15), acetylation (Lys382) and stabilisation in WM35 but did not increase p53 acetylation (Lys382) and stabilisation in WM35-R (Supplementary Figure S6A). A CHX experiment was performed in WM35 and WM35-R cells to investigate whether GSK2830371 inhibition slowed down the degradation of p53 only in WM35, because acetylation was not found in WM35-R (Supplementary Figure S6B). Interestingly, the mutant p53 in WM35-R cells was still subject to some degradation; however, after 4 -h of CHX treatment, GSK2830371 decreased the wt-p53 degradation in WM35 more than the mut-p53 in WM35-R (Supplementary Figure S6B).
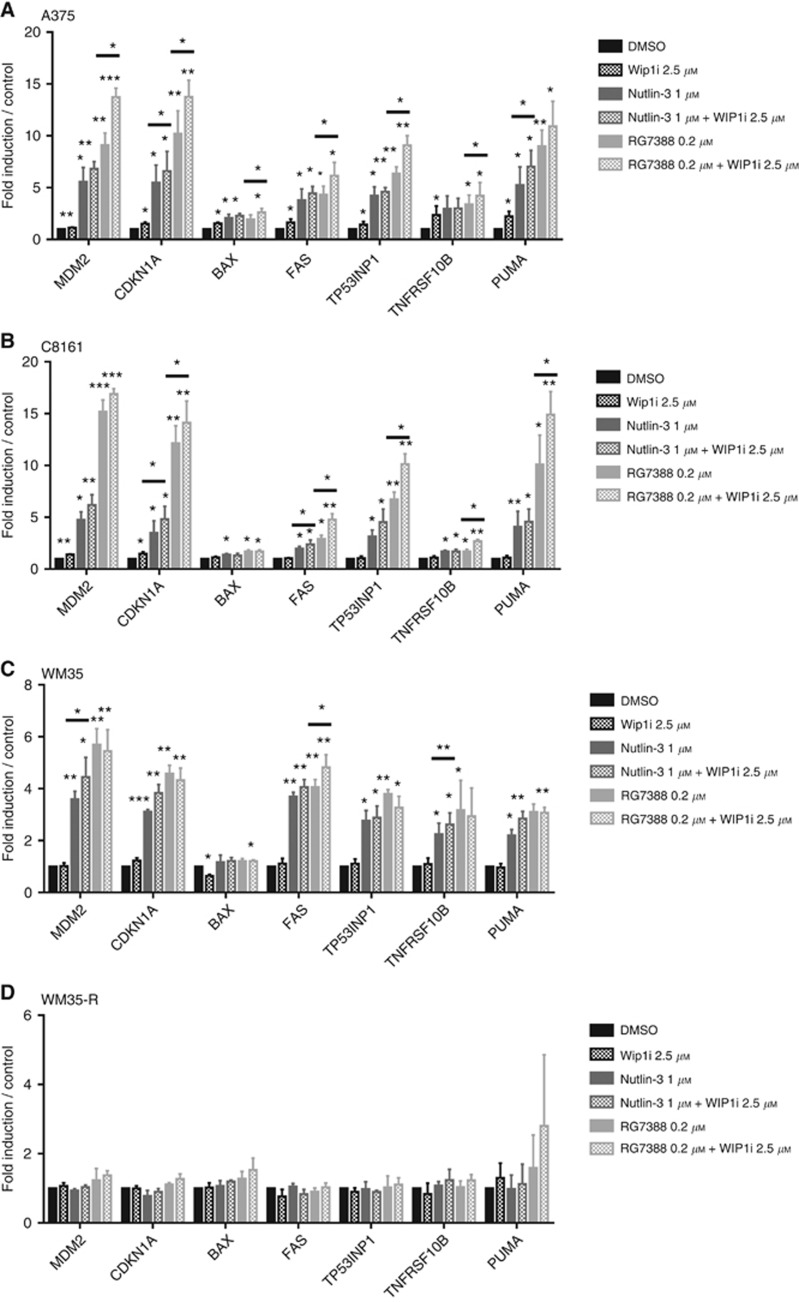
GSK2830371 increased the mRNA expression of p53 transcriptionally regulated genes when combined with MDM2 inhibitors
To test the hypothesis that GSK2830371 enhances the transcriptional activity of p53 in cutaneous melanoma cells treated by MDM2 inhibitors, mRNA expression of candidate genes related to cell cycle arrest and apoptosis were evaluated by quantitative real-time PCR (qRT–PCR). The p53WT cells (A375, C8161, WM35) and one p53MUT cell line, WM35-R, were treated with either 1 μM nutlin-3 or 0.2 μM RG7388, combined with or without GSK2830371 (2.5 μM) for 6 h.
Overall, nutlin-3 or RG7388 induced expression of candidate genes and GSK2830371 potentiated the induced genes in p53WT cells rather than WM35-R. The fold changes of mRNA in response to MDM2 inhibitors with or without WIP1i were less in WM35 than A375 and C8161 (Figure 5 and Supplementary Figure S7). Interestingly, WIP1i alone could significantly induce some of the p53 transcriptional target genes. However, the effects of GSK2830371 on transcript levels were modest, consistent with GSK2830371 alone having no or minimal growth-inhibitory effect in SRB and clonogenic assays (Figures 1B and 3C and D) and with the lack of obvious changes in p21 and MDM2 proteins in immunoblotting (Figure 1C and Supplementary Figure S1).
Figure 5.
The mRNA expression of p53 transcriptional target genes by qRT–PCR. The mRNA expression of p53 transcriptionally regulated genes in response to either 2.5 μM WIP1i, 1 μM Nutlin-3, 0.2 μM RG7388 or combinations for 6 h relative to DMSO solvent and GAPDH control in A375 (A), C8161 (B), WM35 (C) and WM35-R (D) melanoma cells. Statistical significance of differences (*P<0.05, **P<0.005, ***P<0.0005) is shown above each bar for each treatment compared with DMSO control. The significance of differences with or without WIP1i are indicated above the horizontal bars. Only the P-values of <0.05 are shown. Data are presented as mean±s.e.m. for three independent repeats.
Genes for cell cycle arrest, CDKN1A, and autoregulatory negative feedback, MDM2, were generally induced to a higher level by combination treatment with GSK2830371 and MDM2 inhibitors in p53WT cells, except for the RG7388 plus GSK2830371 combination treatment in WM35. Pro-apoptotic BAX, FAS, PUMA, TNFBSF10B and TP53INP1 genes were also evaluated. For BAX, the induction was modest even though some of the differences were statistically significant. This finding was consistent with another study that showed no significant increase of BAX expression in ovarian cancer cells treated by 5 μM nutlin-3 or 0.5 μM RG7388 for 6 h (Zanjirband et al, 2016). For FAS, PUMA, TNFBSF10B and TP53INP1 genes in A375 and C8161, GSK2830371 significantly potentiated the effect of RG7388 on the expression levels of these genes (P<0.05). GSK2830371 also showed the same trend for enhancing the effect of nutlin-3 on the expression of these genes, but the fold changes were mostly not statistically significant (P>0.05).
The WM35 had less fold changes of pro-apoptotic genes than in A375 and C8161. GSK2830371 appeared to increase the effect of nutlin-3 treatment in WM35 on the expression of pro-apoptotic genes, although some of these trends were not statistically significant (P>0.05). The poor potentiation of RG7388 by GSK2830371 found in WM35 was possibly because the transcripts of those genes were saturated by RG7388 treatment alone; WM35 treatment with 1 μM RG7388 did not increase more mRNA expression of MDM2, CDKN1A, PUMA, TNFBSF10B and TP53INP1 compared with 0.2 RG7388, either with or without WIP1i (Supplementary Figure S8).
The effect of combined MDM2 and WIP1 inhibition on cell cycle distribution and apoptosis
Given that GSK2830371 potentiated the growth inhibition and clonogenic reduction by MDM2 inhibitors, FACS analysis was carried out to investigate changes in cell cycle distribution. In addition, sub-G1 events detected by FACS analysis and caspase-3/7 catalytic activity were used as indicators of apoptosis. The responses to MDM2 and WIP1 inhibitors were cell line dependent.
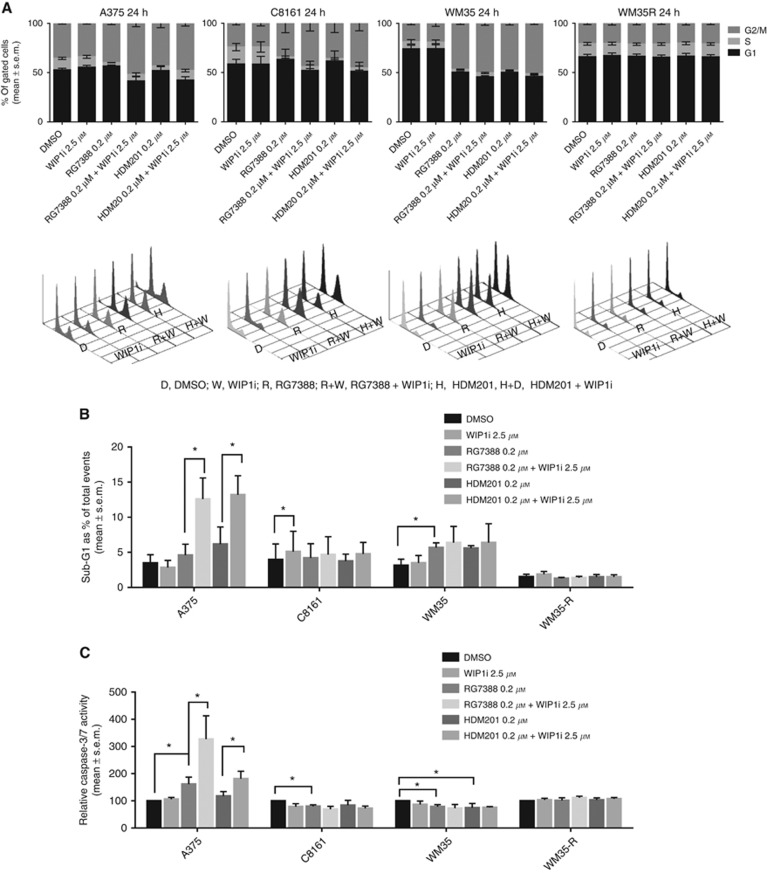
In all the cell lines, 2.5 μM GSK2830371 alone did not affect the cell cycle distribution through 72 h of treatment (Figure 6 and Supplementary Figures S9 and S10) In general, RG7388 (0.2 μM) or HDM201 (0.2 μM) treatment of p53WT cells (A375, WM35, C8161) decreased the proportion of cells in S phase that was accompanied by G1-phase increases, with or without G2-phase increases. Treatment with GSK2830371 further enhanced the changes in cell cycle distribution markedly when it was combined with MDM2 inhibitors.
Figure 6.
The effect of 24 h treatment with combinations of MDM2 and WIP1 inhibitors on cell cycle distribution and apoptosis. Melanoma cells were treated by either 2.5 μM WIP1i, 0.2 μM RG7388, 0.2 μM HDM201 or combinations for 24 h. (A) Cell cycle distribution changes by FACS analysis. (B) Sub-G1 events by FACS and (C) caspase-3/7 activity were used as indicators of apoptosis. Statistically significant P-values (*P<0.05) are indicated. Data are presented as mean±s.e.m. for three independent repeats.
Apoptotic responses, shown by increased FACS sub-G1 signals and caspase-3/7 activity after RG7388 or HDM201 treatment, was evident for A375 cells but not for WM35 or C8161 cells. The combination of MDM2 and WIP1 inhibitors significantly increased the apoptotic response of A375 cells. Cell cycle distribution was not affected in TP53 mutant WM35-R cells regardless of the treatment conditions, demonstrating that the changes in cell cycle distribution observed are p53 dependent.
SiRNA-mediated p53 knockdown
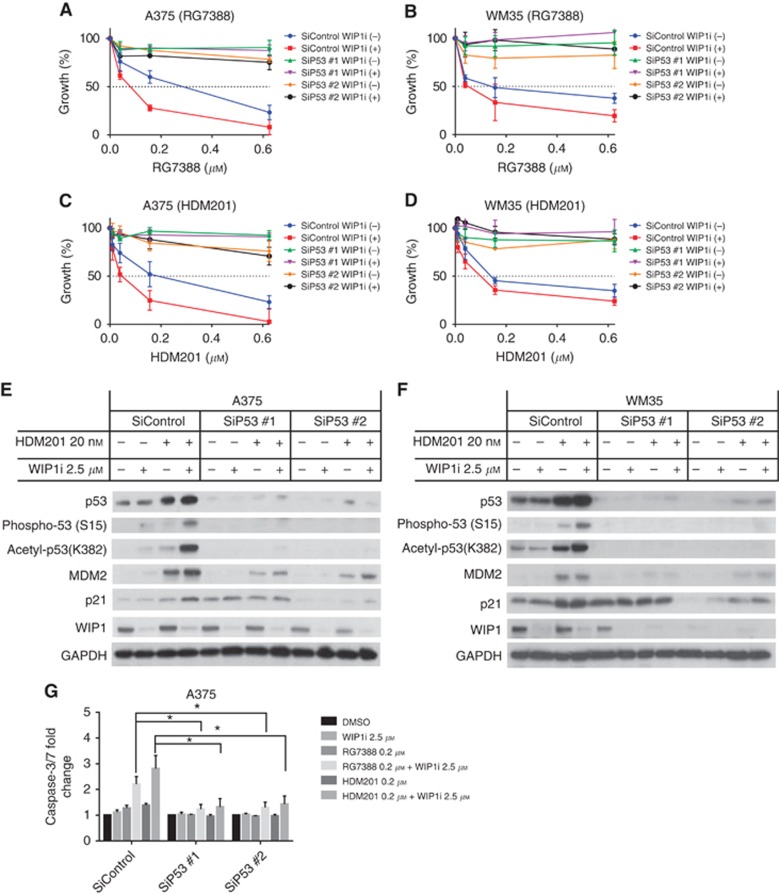
To further investigate the p53 dependence of MDM2 and WIP1 inhibition in cutaneous melanoma, siRNA-mediated p53 knockdown was performed in two p53WT melanoma cells, A375 and WM35. The cells were treated with siRNA for 24 h followed by RG7388 or HDM201±GSK2830371 treatment. The potentiation of growth inhibition by GSK2830371 on RG7388 or HDM201 was suppressed by the p53 knockdown (Figure 7).
Figure 7.
The combination of WIP1 and MDM2 inhibitions is p53 dependent. The A375 and WM35 were subjected to siRNA-mediated knockdown of p53 for 24 h followed by RG7388 and HDM201 treatments. (A–D) Growth inhibition of A375 (A and C) and WM35 (B and D) treated with siRNA followed by 72 h RG7388 (A and B)/HDM201 (C and D) with or without GSK2830371 (WIP1i). (E and F) Immunoblotting of A375, WM35 treated with siRNA followed by HDM201±WIP1i. (G) Caspase-3/7 activity of A375 treated with siRNA followed by RG7388/HDM201±WIP1i for 24 h. Statistically significant P-values (*P<0.05) are indicated. Data are presented as mean±s.e.m. for at least three independent repeats.
Discussion
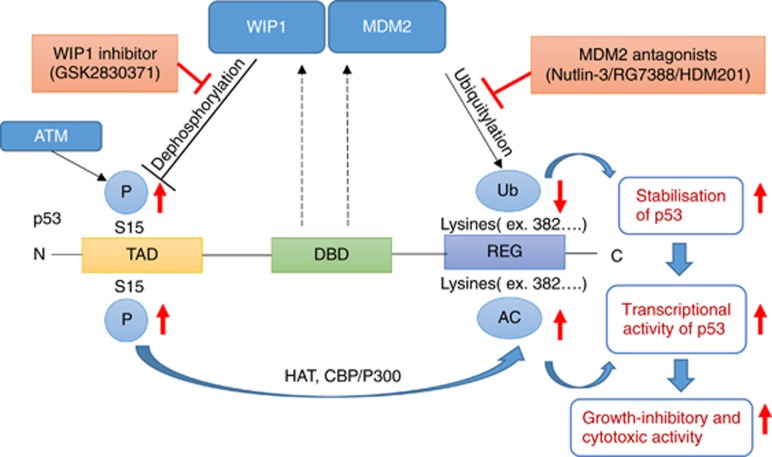
In the current study, we demonstrated that doses of the GSK2830371, WIP1 inhibitor, with no growth-inhibitory activity nevertheless potentiated the growth inhibition and cell killing of wild-type p53 cutaneous melanoma cells by MDM2 inhibitors, nutlin-3, RG7388 and HDM201. Mechanistic studies linked this potentiation to increased phosphorylation, acetylation and decreased ubiquitylation. This has a twofold effect, resulting in not only increased stabilisation of p53 but also increased functional activation by the increased post-translational phosphorylation and acetylation (Figure 8).
Figure 8.
Proposed model for GSK2830371 (WIP1i) potentiation of the growth-inhibitory and cytotoxic effects of MDM2 antagonists. After activation of p53 by MDM2 antagonists (nutlin-3/RG7388/HDM201), WIP1 inhibition (WIP1i) increases phospho-p53 (Ser 15) through allosteric inhibition of WIP1 phosphatase. Phospho-p53 (Ser 15) recruits histone acetyl transferases (HATs, e.g., CBP/P300) that acetylate lysine residues in the C-terminal region of p53. Acetylation of p53 increases the transcriptional activity of p53 directly, and also indirectly by stabilising p53. Dashed lines indicate p53 transcriptional upregulation of the corresponding genes for MDM2 and WIP1. TAD, transcriptional activation domain; DBD, DNA binding domain; REG, C-terminal regulatory region.
Among the three p53WT melanomas investigated in the current study, we found A375 to be particularly primed to undergo apoptosis in response to p53 activation rather than the other two cell lines. This was evidenced by significant increases in sub-G1 signals on FACS cell cycle analysis and caspase-3/7 activity after combination treatment. The p53-dependent transcription profiles possibly explain some differences between the A375 and WM35 responses to MDM2 and WIP1 inhibition (Figure 5 and Supplementary Figure S7). Although p53 expression has generally been associated with the levels of transcriptional target genes responsible for cell cycle arrest and apoptosis, apoptosis has been reported only to proceed in certain conditions, such as achieving a transcriptional threshold by p53 reactivation or suppression of its negative regulators (Kracikova et al, 2013; Lu et al, 2013; Khoo et al, 2014). Therefore, two distinct subgroups of response are identifiable, depending on whether cells undergo apoptosis or not after p53 reactivation by MDM2 inhibitors. Irreversible apoptosis only occurs with some cell types and reversible growth suppression or senescence without apoptosis is observed with other cell types (Tseng et al, 2010; Lu et al, 2013). In the current study, inhibition of WIP1, a negative regulator of p53, potentiated the activation of p53 in all three p53WT melanoma cell lines through post-translation modification, but only enhanced apoptosis in A375 cells and did not shift cell cycle arrest to apoptosis in the cells not primed for apoptosis (WM35 and C8161 in the current study).
Post-translational modification of p53 has critical effects on its stability and function (Kruse and Gu, 2009). A key post-translational modification is the phosphorylation of p53 at Ser15, which is mediated by the ATM and ATR protein kinases, and has been considered as an initiating event in p53 activation (Saito et al, 2002) by promoting the acetylation of lysine residues of the p53 C-terminal region, through recruitment of p300/CBP histone acetyltransferase (Gu and Roeder, 1997; Lambert et al, 1998; Dumaz and Meek, 1999; Reed and Quelle, 2014). The MDM2 can ubiquitinate the same lysine residues of p53 (Brooks and Gu, 2011; Reed and Quelle, 2014), and acetylation and ubiquitination are mutually exclusive. Therefore, p53 acetylation increases its stability by inhibiting MDM2-mediated ubiquitination of p53 and can stimulate its sequence-specific DNA binding activity (Gu and Roeder, 1997; Li et al, 2002).
The WIP1 phosphatase and ATM kinase work antagonistically on the same p53-related substrates, ATM (Ser1981), MDM2 (Ser395) and p53 (Ser15), to maintain a fine balance of these proteins in order to regulate the function and stability of p53. For example, autophosphorylation of ATM (Ser1981) after irradiation has been shown to stabilise ATM at DNA damage sites and phosphorylate p53 (Ser15) (So et al, 2009). Ser1981 of ATM is also a substrate of WIP1 phosphatase (Shreeram et al, 2006) and WIP1 inhibition by GSK2830371 can increase phospho-ATM (Ser1981) in a non-genotoxic manner (Gilmartin et al, 2014). Following DNA damage stress, ATM is able to phosphorylate MDM2 at Ser395 that attenuates degradation of p53 (Khosravi et al, 1999; Maya et al, 2001). The WIP1 can stabilise MDM2 in an ATM-dependent manner and also inhibit ubiquitination of MDM2 (Lu et al, 2007). Therefore, GSK2830371 is an effective non-genotoxic way of increasing activation of the ATM-mediated network by inhibition of WIP1 to maintain the phosphorylated state of key proteins. This is particularly useful when the p53-dependent WIP phosphatase is induced by non-genotoxic MDM2 inhibitors.
GSK2830371 as a single agent selectively inhibits the growth of MCF-7 and MX-1 cells, PPM1D amplified breast cancer cells and a subset of p53WT haematological cancer cell lines (Gilmartin et al, 2014). In one previous report using cells with different PPM1D gene alterations (gain-of-function mutations, amplification or copy number gain), all cells except MCF-7 cell were resistant to GSK2830371 (GI50>10 μM) as a single agent treatment (Esfandiari et al, 2016). Consistent with this report, GSK2830371 alone had no obvious growth-inhibitory activity (GI50>10 μM) in all of the melanoma cell lines we investigated in the current study (Figure 1A). Actually, modest phosphorylation of p53 (Ser15) following treatment with single agent GSK2830371 was detected by immunoblotting (Figure 4A) and RPPA (Supplementary Figure S1B) that resulted in a statistically significant but small increase in transcript level of some genes (Figure 6) and slight induction of p21 protein (Figures 1C and 4A). However, the transactivation of genes by this small level of p53 phosphorylation was insufficient to have a significant effect on the growth of the cutaneous melanoma cells.
Previous studies with genotoxic modalities such as ionising or UV irradiation indicated a model in which resultant phosphorylation of p53 on Ser15 recruits histone acetyl transferases (HATs) to acetylate lysine residues in the p53 C-terminal domain (Saito et al, 2002). In the current study, this model was more specifically tested and shown to be applicable to non-genotoxic treatment with MDM2–p53 binding antagonists and WIP1 inhibitor, providing a mechanistic basis for the potentiation. Consistent with a recent report (Sriraman et al, 2016) using nutlin-3a and GSK2830371, our results showed that enhanced phosphorylation (Ser15) and acetylation (Lys382) of p53 were found after non-genotoxic nutlin-3 and GSK2830371 treatment. Here we extended these observations to more specific and potent MDM2 inhibitors that are progressing through clinical trials. In addition, we transfected p53 (Ser15) mutants to prove the Ser15 is the essential and sufficient residue for p53 acetylation. Furthermore, we investigated the p53 stability after combination treatment in the current study.
The CHX treatment was used to investigate the effect of acetylation of p53 after GSK2830371 treatment on p53 half-life and showed p53 levels were increased by slowing down degradation. The MG132 reversed the change in p53 expression, indicating that GSK2830371 decreased the proteasomal degradation of p53. Both the p53 acetylation leading to inhibition of its ubiquitination by MDM2 (Li et al, 2002) and WIP1 inhibition by siRNA leading to decrease in p53 ubiquitination (Lu et al, 2007) are consistent with our finding that WIP1 inhibition by GSK2830371 stabilised p53 as a result of the decreased MDM2-mediated ubiquitination shown by co-immunoprecipitation. When the cells were treated by the lower concentrations of MDM2 inhibitors, the MDM2 was only partially inhibited by MDM2 inhibitors, hence p53 acetylation then played a critical role in p53 stability by antagonising ubiquitiylation (Figure 4).
GSK2830371 potentiated MDM2 inhibitors to amplify the expression of p53 transcriptionally targeted genes that are responsible for cell cycle arrest and apoptosis in p53WT melanoma cells, but the degree of potentiation was cell line dependent. Greater potentiation by GSK2830371 was found with A375 and C8161 cells than for WM35 by qRT–PCR, immunoblotting, growth inhibition and clonogenic assay. Although combination treatment stabilised p53 by decreasing degradation in WM35 cells, the increases in p53-target gene mRNAs and proteins in this cell line were modest (Figure 5). Consequently, the fold changes in GI50 and LC50 values were more limited. We also checked basal expressions of MDMX and WIP1 (Supplementary Figure S11) and showed there was no clear correlation between basal expression levels and responses to MDM2 and WIP1 inhibition, as MDM4 was overexpressed in ∼65% melanoma cases (Gembarska et al, 2012). The results of the current study indicate that the responsiveness of melanoma cell lines to MDM2 inhibitors and WIP1i depends to a large part on the fold differences between the basal and maximal relative transcript levels of downstream p53 transcriptional target genes, and is also likely to be modulated by the downstream status of pro-survival and anti-apoptotic pathways (Supplementary Figure S8).
In summary, we report the non-genotoxic potentiation of nutlin-3 and clinically relevant second-generation MDM2–p53 binding antagonists, RG7388 and HDM201, by the WIP1 phosphatase inhibitor GSK280371, through p53 phosphorylation, acetylation, stabilisation and increased transcription of p53 regulated genes in a panel of p53WT cutaneous melanoma cell lines. This combination treatment activated the ATM-mediated response without DNA damage, highlighting a novel non-genotoxic therapeutic strategy to further explore for melanoma.
Acknowledgments
We thank and gratefully acknowledge Newcastle University/Astex Pharmaceuticals Alliance and CRUK who funded the Drug Development Programme at the Newcastle University Northern Institute for Cancer research for their support and encouragement, including help with sourcing the HDM201 and RG7388 compounds by custom synthesis. This work was supported by Grants (CMRPG3C1521 to C-EW) from Chang Gung Memorial Hospital, Taiwan.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, Eggermont AM, Flaherty KT, Gimotty PA, Kirkwood JM, McMasters KM, Mihm MC Jr, Morton DL, Ross MI, Sober AJ, Sondak VK (2009) Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 27(36): 6199–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Gu W (2011) The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2(6): 456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP (2009) Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 9(12): 862–873. [DOI] [PubMed] [Google Scholar]
- Chin L (2003) The genetics of malignant melanoma: lessons from mouse and man. Nat Rev Cancer 3(8): 559–570. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA (2002) Mutations of the BRAF gene in human cancer. Nature 417(6892): 949–954. [DOI] [PubMed] [Google Scholar]
- Demunter A, Stas M, Degreef H, De Wolf-Peeters C, van den Oord JJ (2001) Analysis of N- and K-ras mutations in the distinctive tumor progression phases of melanoma. J Invest Dermatol 117(6): 1483–1489. [DOI] [PubMed] [Google Scholar]
- Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, Chu XJ, Bartkovitz D, Podlaski F, Janson C, Tovar C, Filipovic ZM, Higgins B, Glenn K, Packman K, Vassilev LT, Graves B (2013) Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem 56(14): 5979–5983. [DOI] [PubMed] [Google Scholar]
- Dumaz N, Meek DW (1999) Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J 18(24): 7002–7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandiari A, Hawthorne TA, Nakjang S, Lunec J (2016) Chemical inhibition of wild-type p53-induced phosphatase 1 (WIP1/PPM1D) by GSK2830371 potentiates the sensitivity to MDM2 inhibitors in a p53-dependent manner. Mol Cancer Ther 15(3): 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon HS, Jones SN (2012) Using mouse models to explore MDM-p53 signaling in development, cell growth, and tumorigenesis. Genes Cancer 3(3-4): 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de Lange J, Yip D, Goydos J, Haigh JJ, Haupt Y, Larue L, Jochemsen A, Shi H, Moriceau G, Lo RS, Ghanem G, Shackleton M, Bernal F, Marine JC (2012) MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 18(8): 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin AG, Faitg TH, Richter M, Groy A, Seefeld MA, Darcy MG, Peng X, Federowicz K, Yang J, Zhang SY, Minthorn E, Jaworski JP, Schaber M, Martens S, McNulty DE, Sinnamon RH, Zhang H, Kirkpatrick RB, Nevins N, Cui G, Pietrak B, Diaz E, Jones A, Brandt M, Schwartz B, Heerding DA, Kumar R (2014) Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nat Chem Biol 10(3): 181–187. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90(4): 595–606. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L (2012) A landscape of driver mutations in melanoma. Cell 150(2): 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman D, Chatterjee M, Langenberg MHG, Lin CC, Suarez C, Tai D, Cassier P, Yamamoto N, De Weger VA, Jeay S, Meille C, Halilovic E, Mariconti L, Guerreiro N, Kumar A, Wuerthner JU, Bauer S (2016) Dose- and regimen-finding phase I study of NVP-HDM201 in patients (pts) with TP53 wild-type (wt) advanced tumors. Eur J Cancer 69: S128–S129. [Google Scholar]
- Kawaguchi T, Kato S, Otsuka K, Watanabe G, Kumabe T, Tominaga T, Yoshimoto T, Ishioka C (2005) The relationship among p53 oligomer formation, structure and transcriptional activity using a comprehensive missense mutation library. Oncogene 24(46): 6976–6981. [DOI] [PubMed] [Google Scholar]
- Khoo KH, Verma CS, Lane DP (2014) Drugging the p53 pathway: understanding the route to clinical efficacy. Nat Rev Drug Discov 13(3): 217–236. [DOI] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D (1999) Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA 96(26): 14973–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kracikova M, Akiri G, George A, Sachidanandam R, Aaronson SA (2013) A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ 20(4): 576–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W (2009) Modes of p53 regulation. Cell 137(4): 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN (1998) Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem 273(49): 33048–33053. [DOI] [PubMed] [Google Scholar]
- Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, Mandala M, Demidov L, Stroyakovskiy D, Thomas L, de la Cruz-Merino L, Dutriaux C, Garbe C, Sovak MA, Chang I, Choong N, Hack SP, McArthur GA, Ribas A (2014) Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 371(20): 1867–1876. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Momand J, Finlay CA (1991) The p53 tumour suppressor gene. Nature 351(6326): 453–456. [DOI] [PubMed] [Google Scholar]
- Li M, Luo J, Brooks CL, Gu W (2002) Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem 277(52): 50607–50611. [DOI] [PubMed] [Google Scholar]
- Lowe J, Cha H, Lee MO, Mazur SJ, Appella E, Fornace AJ Jr (2012) Regulation of the Wip1 phosphatase and its effects on the stress response. Front Biosci (Landmark Ed) 17: 1480–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Breyssens H, Salter V, Zhong S, Hu Y, Baer C, Ratnayaka I, Sullivan A, Brown NR, Endicott J, Knapp S, Kessler BM, Middleton MR, Siebold C, Jones EY, Sviderskaya EV, Cebon J, John T, Caballero OL, Goding CR, Lu X (2013) Restoring p53 function in human melanoma cells by inhibiting MDM2 and cyclin B1/CDK1-phosphorylated nuclear iASPP. Cancer Cell 23(5): 618–633. [DOI] [PubMed] [Google Scholar]
- Lu M, Miller P, Lu X (2014) Restoring the tumour suppressive function of p53 as a parallel strategy in melanoma therapy. FEBS Lett 588(16): 2616–2621. [DOI] [PubMed] [Google Scholar]
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA (2007) The Wip1 Phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell 12(4): 342–354. [DOI] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, Kastan MB, Katzir E, Oren M (2001) ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 15(9): 1067–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW (2015) Regulation of the p53 response and its relationship to cancer. Biochem J 469(3): 325–346. [DOI] [PubMed] [Google Scholar]
- Miller AJ, Mihm Jr MC (2006) Melanoma. N Engl J Med 355(1): 51–65. [DOI] [PubMed] [Google Scholar]
- Muller PA, Vousden KH (2013) p53 mutations in cancer. Nat Cell Biol 15(1): 2–8. [DOI] [PubMed] [Google Scholar]
- Pechackova S, Burdova K, Benada J, Kleiblova P, Jenikova G, Macurek L (2016) Inhibition of WIP1 phosphatase sensitizes breast cancer cells to genotoxic stress and to MDM2 antagonist nutlin-3. Oncotarget 7(12): 14458–14475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanski R, Noon AP, Blaydes J, Phillips A, Rubbi CP, Parsons K, Vlatkovic N, Boyd MT (2014) Senescence induction in renal carcinoma cells by Nutlin-3: a potential therapeutic strategy based on MDM2 antagonism. Cancer Lett 353(2): 211–219. [DOI] [PubMed] [Google Scholar]
- Reed SM, Quelle DE (2014) p53 acetylation: regulation and consequences. Cancers (Basel) 7(1): 30–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, Lichinitser M, Dummer R, Grange F, Mortier L, Chiarion-Sileni V, Drucis K, Krajsova I, Hauschild A, Lorigan P, Wolter P, Long GV, Flaherty K, Nathan P, Ribas A, Martin AM, Sun P, Crist W, Legos J, Rubin SD, Little SM, Schadendorf D (2015) Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 372(1): 30–39. [DOI] [PubMed] [Google Scholar]
- Sachweh MC, Drummond CJ, Higgins M, Campbell J, Lain S (2013) Incompatible effects of p53 and HDAC inhibition on p21 expression and cell cycle progression. Cell Death Dis 4: e533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Goodarzi AA, Higashimoto Y, Noda Y, Lees-Miller SP, Appella E, Anderson CW (2002) ATM mediates phosphorylation at multiple p53 sites, including Ser(46), in response to ionizing radiation. J Biol Chem 277(15): 12491–12494. [DOI] [PubMed] [Google Scholar]
- Shreeram S, Demidov ON, Hee WK, Yamaguchi H, Onishi N, Kek C, Timofeev ON, Dudgeon C, Fornace AJ, Anderson CW, Minami Y, Appella E, Bulavin DV (2006) Wip1 phosphatase modulates ATM-dependent signaling pathways. Mol Cell 23(5): 757–764. [DOI] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR (1990) New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 82(13): 1107–1112. [DOI] [PubMed] [Google Scholar]
- So S, Davis AJ, Chen DJ (2009) Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J Cell Biol 187(7): 977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman A, Radovanovic M, Wienken M, Najafova Z, Li Y, Dobbelstein M (2016) Cooperation of Nutlin-3a and a Wip1 inhibitor to induce p53 activity. Oncotarget 7(22): 31623–31638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A (2015) Global cancer statistics, 2012. CA Cancer J Clin 65(2): 87–108. [DOI] [PubMed] [Google Scholar]
- Tseng HY, Jiang CC, Croft A, Tay KH, Thorne RF, Yang F, Liu H, Hersey P, Zhang XD (2010) Contrasting effects of nutlin-3 on TRAIL- and docetaxel-induced apoptosis due to upregulation of TRAIL-R2 and Mcl-1 in human melanoma cells. Mol Cancer Ther 9(12): 3363–3374. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303(5659): 844–848. [DOI] [PubMed] [Google Scholar]
- Voshol H, Ehrat M, Traenkle J, Bertrand E, van Oostrum J (2009) Antibody-based proteomics: analysis of signaling networks using reverse protein arrays. FEBS J 276(23): 6871–6879. [DOI] [PubMed] [Google Scholar]
- Wade M, Wang YV, Wahl GM (2010) The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 20(5): 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanjirband M, Edmondson RJ, Lunec J (2016) Pre-clinical efficacy and synergistic potential of the MDM2-p53 antagonists, Nutlin-3 and RG7388, as single agents and in combined treatment with cisplatin in ovarian cancer. Oncotarget 7(26): 40115–40134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Dutton-Regester K, Brown KM, Hayward NK (2016) The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res 29(3): 266–283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.