Abstract
Introduction of selectively chemical reactive groups at the cell surface enables site-specific cell surface labeling and modification opportunity, thus facilitating the capability to study the cell surface molecular structure and function and the molecular mechanism it underlies. Further, it offers the opportunity to change or improve a cell’s functionality for interest of choice. In this study, two chemical reactive anchor lipids, phosphatidylethanolamine–poly(ethylene glycol)–dibenzocyclooctyne (DSPE–PEG2000–DBCO) and cholesterol–PEG–dibenzocyclooctyne (CHOL–PEG2000–DBCO) were synthesized and their potential application for cell surface re-engineering via lipid fusion were assessed with RAW 264.7 cells as a model cell. Briefly, RAW 264.7 cells were incubated with anchor lipids under various concentrations and at different incubation times. The successful incorporation of the chemical reactive anchor lipids was confirmed by biotinylation via copper-free click chemistry, followed by streptavidin-fluorescein isothiocyanate binding. In comparison, the cholesterol-based anchor lipid afforded a higher cell membrane incorporation efficiency with less internalization than the phospholipid-based anchor lipid. Low cytotoxicity of both anchor lipids upon incorporation into the RAW 264.7 cells was observed. Further, the cell membrane residence time of the cholesterol-based anchor lipid was evaluated with confocal microscopy. This study suggests the potential cell surface re-engineering applications of the chemical reactive anchor lipids.
Introduction
The cell surface is made of a diversity of biomolecules that govern the biological processes of the cell, such as cell signaling, cell–cell adhesions, and other extracellular/intracellular communications. Cell surface re-engineering with biologically important molecules has scope for potential applications such as cell labeling,1 imaging,2 and functionalization.3,4 In general, direct addition of biological functionality onto live cell surfaces allows for the molecular level analysis of cell surface phenomena and manipulation of cell functions as well. In addition, introduction of chemical reactive groups at the cell surface enables rapid and site-specific cell surface labeling and modification opportunity, providing enormous capability to study the cell surface molecular structure and function and the molecular mechanism it underlies. Further, it provides potential opportunity to change or improve cell functionality for different interests. Due to the growing importance for cell surface re-engineering and its promising applications, several approaches, such as direct chemical modification,5 membrane fusion,6 and metabolic engineering methods,7 have been explored so far.
Macrophages play pivotal roles in both innate and adaptive immunity, most importantly in antigen processing and presenting processes. Therefore, macrophages have been explored widely as drug/antigen delivery targets,8−12 drug delivery carrier systems,13−15 and also in transplantation/grafting applications16 for the treatment of many disease conditions.17 Recently, macrophage-mediated programmed cell removal has been confirmed as an important mechanism in diseased and damaged cell elimination before programmed cell death.18 Based on this fact, a surface modification of macrophages with nucleic acid aptamers, so-called “eat-you” motifs, was proposed as it can bind to membrane proteins of cancer cells and capture the cell.19 It indicated that enhancing the selective adhesion of macrophage to cancer cells may be an effective macrophage-mediated anticancer therapy. In this study, we propose a cell surface re-engineering strategy of macrophages with bio-orthogonal functionality via lipid fusion aiming to chemoselectively label and modify the cell surface with biomolecules, paving a path for potential biomedical applications of macrophages.
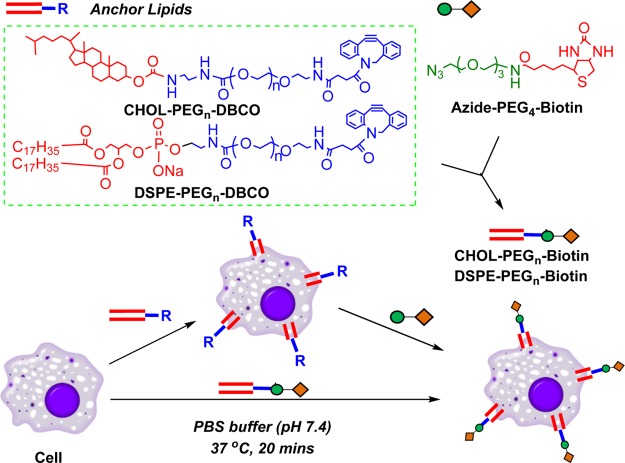
Direct chemical modification-based cell surface re-engineering has limited applications due to its low site selectivity. Metabolic engineering approaches often rely on the cell’s internal machinery and may inevitably perturb the cell’s physiology under investigation and thus have limited capability for cell surface re-engineering as well. Therefore, passive exogenous insertion of chemically defined structures into cellular membranes via lipid fusion is an attractive alternative approach for cell surface engineering.20−25 For example, lipid fusion has been a straightforward method for chemical glycocalyx engineering through passive insertion of lipid-anchored glycopolymers into the plasma membrane.26,27 In this study, two chemically reactive anchor lipids, phosphatidylethanolamine–poly(ethylene glycol)–dibenzocyclooctyne (DSPE–PEG2000–DBCO) and cholesterol–PEG–dibenzocyclooctyne (CHOL–PEG2000–DBCO) were synthesized and their potential applications for cell surface re-engineering with bio-orthogonal functionality were assessed using RAW 264.7 cells as model macrophages. Specifically, we systemically investigated the incorporation efficiency of anchor lipids under various concentrations and at different incubation times. The successful incorporation of the anchor lipids was confirmed by chemically selective biotinylation of the incorporated DBCO functionality via copper-free click chemistry (CFCC), targeting the biotin with streptavidin-fluorescein isothiocyanate (FITC) and analyzed by confocal microscopy and flow cytometry, respectively (Figure 1). Next, the cytotoxic effects of both anchor lipids upon incorporating onto the RAW 264.7 cells were assessed. Further, the cell membrane residence time of the anchored lipids was evaluated with confocal microscopy. This study suggests the possible use of these reactive anchor lipids for potential cell surface re-engineering applications of macrophages and other cells as well.
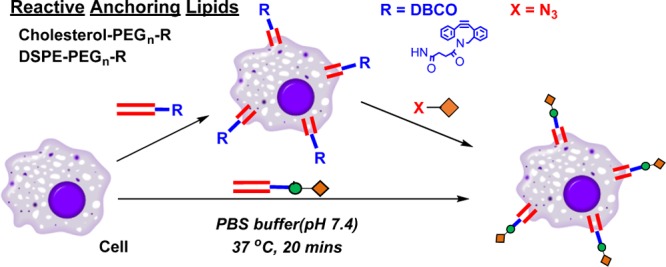
Figure 1.
Schematic illustration of cell surface re-engineering via CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO lipid anchoring approach and its bio-orthogonal modification via copper-free click chemistry.
Results and Discussion
Lipid fusion has been reported as a useful cell surface re-engineering approach by using lipid anchors.20−25 In this study, we systemically investigated the incorporation efficiencies of two types of anchor lipids, CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO, and their further chemically selective modification potentials. The terminal DBCO moiety gives bio-orthogonal functionality for reacting with azide-containing biomolecules via copper-free click chemistry (CFCC). CFCC is a widely used biocompatible and bio-orthogonal technique employed for labeling and modification of biomolecules and cells. The reaction can be performed under mild conditions in aqueous buffers without a catalyst and in high yields under reasonable reaction times. Therefore, the incorporation of DBCO onto the cell surface is anticipated to be useful for the coupling of many other ligands onto the cell surface, facilitating many biomedical applications. Further, a poly(ethylene glycol) (PEG) molecule acts as the spacer between the lipid head group and the terminal DBCO functional group, which makes the anchor lipids water soluble and facilitates a longer lifetime of the biomolecules introduced on the cell surface.
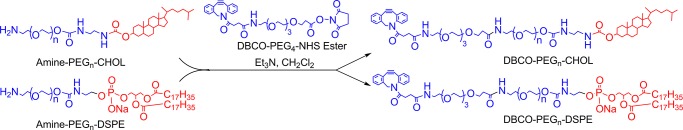
Syntheses of Anchor Lipids CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO for Cell Surface Re-engineering
The syntheses of these anchor lipids were carried out by an amidation reaction between commercially available DBCO–PEG4-N-hydroxysuccinimide (NHS) ester group and amine-terminated cholesterol and DSPE lipids (Scheme 1). First, monocholesteryl–PEG2000-amine was synthesized as per a previously described method.28 The amidation of monocholesteryl–PEG2000-amine with DBCO–PEG4-NHS ester in anhydrous CH2Cl2 (3 mL) afforded anchor lipids CHOL–PEG2000–DBCO in 72% high yield. Next, DSPE–PEG2000–PEG4–DBCO was synthesized by amidation of commercially available DSPE–PEG2000-amine with DBCO–PEG4-NHS ester in 72% high yield. The resultant anchor lipids were purified via silica gel column chromatography using a gradient mobile phase comprising CHCl3/MeOH. The obtained products were characterized by both 1H NMR and 13C NMR spectra data.
Scheme 1. Scheme for the Syntheses of CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO Anchor Lipids for Cell Surface Re-engineering Applications.
Incorporation Efficiency of Anchor Lipids into the Cell Surface
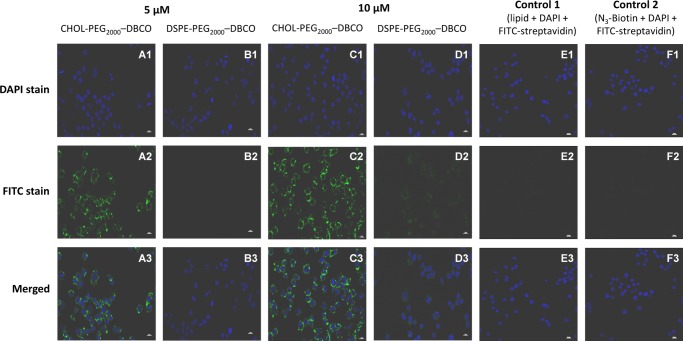
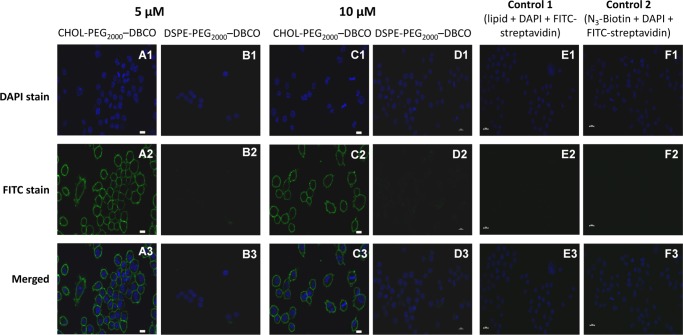
To investigate the anchoring ability of both cholesterol- and DSPE-based anchor lipids in the cell membranes, RAW 264.7 macrophage cells were directly incubated with different concentrations (5 and 10 μM) of aqueous solutions of anchor lipids (CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO) for 20 min at 37 °C. The incorporation of the anchor lipid into the cell membrane was detected by labeling with azide–PEG4–biotin and further detected by fluorescein-labeled streptavidin (streptavidin-FITC), which was confirmed by confocal microscopy and flow cytometry. As a result, apparent fluorescence signals were observed for RAW 264.7 cells treated with both cholesterol- and DSPE-based anchor lipids for 20 min in confocal microscopy images (Figure 2A2–D2). From the negative control experiments, it is clearly evident that the cells did not exhibit any fluorescence as that of both the cells treated with anchor lipids but without azide–PEG4–biotin and treated with azide–PEG4–biotin but without anchor lipids because there is no binding of FITC-streptavidin (Figure 2E2,F2). Apparently, the cholesterol anchor lipid showed higher fluorescence signals for RAW 264.7 cells (Figure 2A,C) compared to DSPE anchor lipid (Figure 2B,D).
Figure 2.
Confocal microscopy images of RAW 264.7 macrophage cells treated with anchor lipids for 20 min at 37 °C, in phosphate-buffered saline (PBS) buffer (pH 7.4) followed by biotinylation via CFCC and streptavidin-FITC labeling: RAW 264.7 macrophage cells treated with CHOL–PEG2000–DBCO (A, C) and DSPE–PEG2000–DBCO (B, D) with varying concentrations (5 and 10 μM) for 20 min at 37 °C in PBS (pH 7.4) followed by biotinylation and streptavidin-FITC labeling; RAW 264.7 macrophage cells treated with anchor lipids but without azide–PEG4–biotin (E) and treated with azide–PEG4–biotin but without anchor lipids (F).
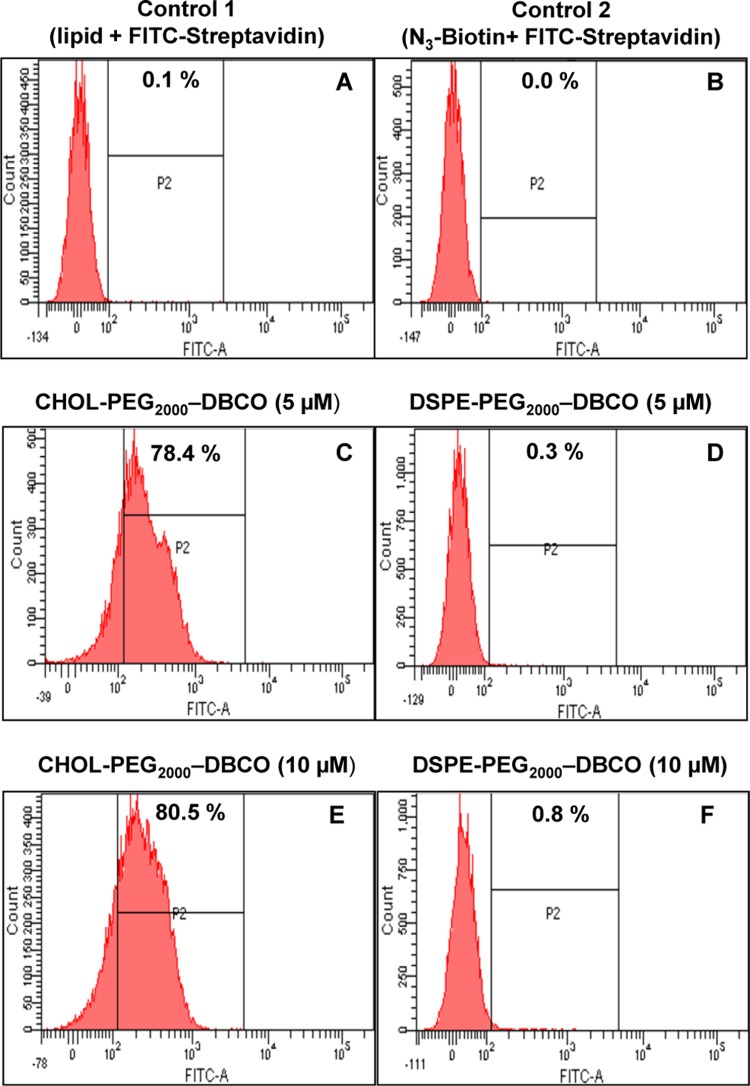
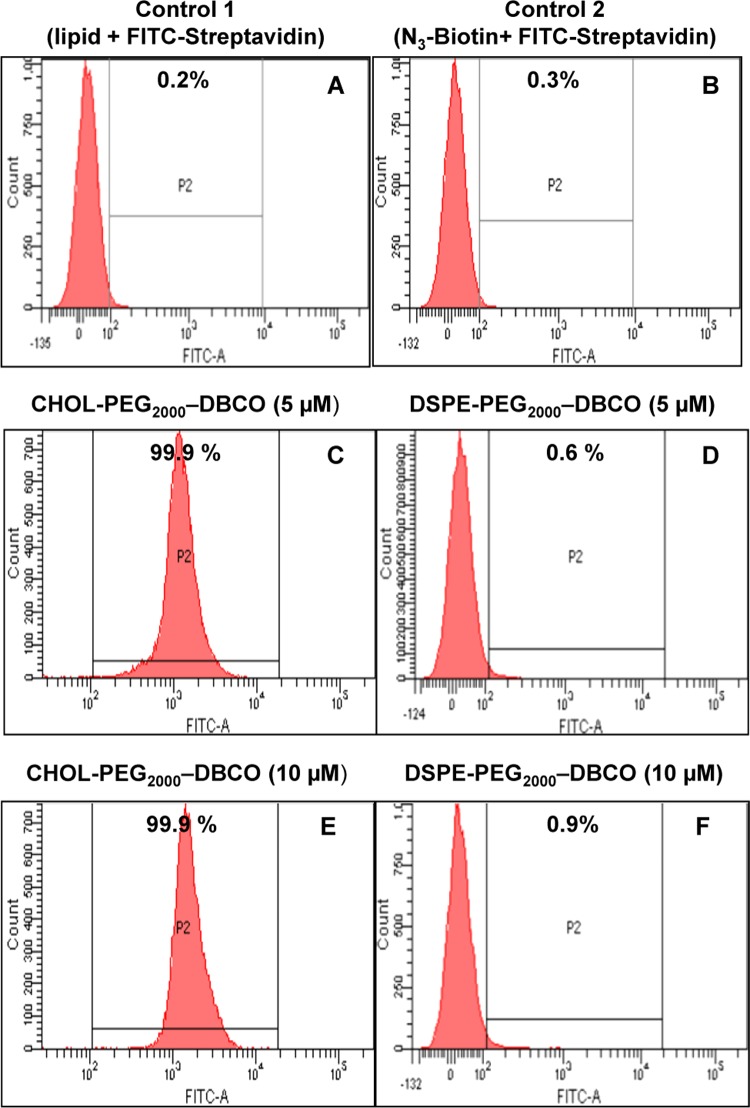
Further, flow cytometry analysis demonstrated higher fluorescence intensities for cells treated with CHOL–PEG2000–DBCO followed by biotinylation. Nearly, 78–81% of the cells were incorporated with CHOL–PEG2000–DBCO anchor lipids (Figure 3C,E). In comparison, relatively lower fluorescence signal was observed for the DSPE–PEG2000–DBCO anchor lipids indicating low amounts of incorporation (Figure 3D,F), whereas control experiments showed no fluorescence signals (Figure 3A,B). These results are well in agreement with the confocal data above, further indicating a higher incorporation efficiency of the cholesterol-based anchor lipids compared to DSPE-based anchor lipids. This may be a cell type-specific phenomenon. It may be associated with higher levels of raft-forming lipids in the membranes of the macrophage cells. It deserves a future study in different types of cells to clarify this cell type-specific phenomenon.
Figure 3.
Flow cytometry analysis of RAW 264.7 macrophage cells treated with CHOL-PEG-DBCO (C, E) and DSPE-PEG-DBCO anchor lipids (D, F) in varying concentrations (5 and 10 μM) for 20 min at 37 °C in PBS (pH 7.4) followed by biotinylation and FITC-streptavidin binding and controls (A, B).
Direct Anchoring of Biotinylated Lipid Conjugates into the Cell Surface
To further confirm the high incorporation efficiency of the cholesterol-based anchor lipid, we next investigate the direct anchoring of biotinylated lipid conjugates into the cell surface of RAW 264.7 cells. First, the biotin lipids are prepared by treating varying concentrations of anchor lipids (CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO) with azide–PEG4–biotin for 1 h at room temperature (RT), followed by direct incubation of the RAW 264.7 cells at different incubation times. Both confocal microscopy and flow cytometry results imply that most of the cells incubated with biotinylated CHOL–PEG2000–DBCO lipids exhibited much stronger fluorescence signals (Figures 4A,C and 5C,E) compared to biotinylated phospholipid (DSPE) anchor lipid, which showed almost no detectable signals (Figures 4B,D and 5D,F). These results indicate that there is uniform incorporation of the biotinylated cholesterol into the cell membrane with almost negligible signals from within the cytoplasm. As a comparison, the direct incubation of preconjugated biotinylated lipid (Figures 4 and 5) gave a higher insertion efficiency compared to the two-step click process (Figures 2 and 3) at the cell surface. Due to its highly efficient surface-anchoring ability, further rate of anchoring studies were conducted with CHOL–PEG2000–DBCO anchor lipids.
Figure 4.
Confocal microscopy images of RAW 264.7 macrophage cells treated with biotinylated cholesterol and DSPE lipid for 20 min at 37 °C in PBS buffer (pH 7.4).
Figure 5.
Flow cytometry analysis of RAW 264.7 macrophage cells treated with biotinylated cholesterol (C and E) and biotinylated DSPE lipid (D and F) in varying concentrations for 20 min at 37 °C in PBS buffer (pH 7.4) followed by FITC-streptavidin binding and control (A and B).
Rate of Anchoring Biotinylated Lipid Conjugate (CHOL–PEG2000–biotin) onto the Cell Membrane
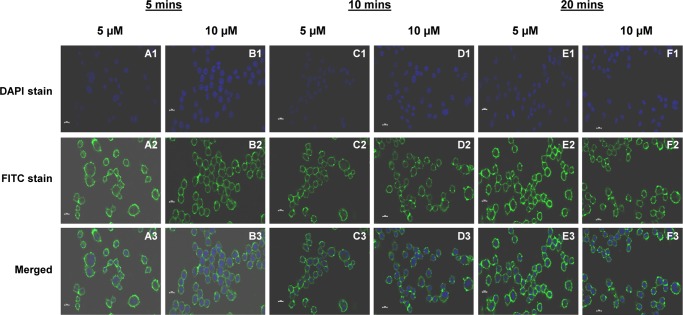
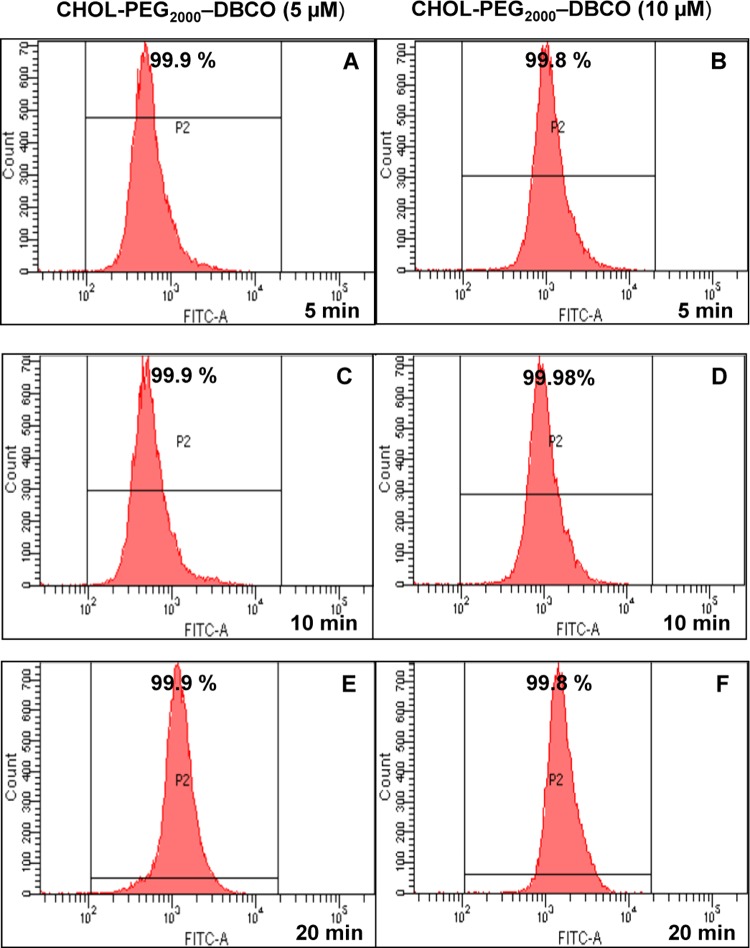
The rate of anchoring for the cholesterol-based anchor lipid into cell membranes was investigated at different periods of time. For this purpose, the cells were incubated with different concentrations of anchor lipid (5 and 10 μM) and for different incubation times (5, 10, and 20 min) at 37 °C in 1× PBS buffer (pH 7.4). As shown in the confocal microscopy image (Figure 6), the incorporation of cholesterol-based anchor lipid was observed within 5 min and a higher fluorescent intensity signal was observed at 20 min. Additionally, the rate of anchoring was also determined by flow cytometry analysis (Figure 7), which also resulted in high fluorescence intensities, thereby confirming that almost 99.9% of the cells were incorporated with the biotinylated cholesterol anchor lipids synthesized by the one-pot step procedure.
Figure 6.
Confocal images of direct anchoring biotinylated cholesterol lipid (5 and 10 μM) onto cell surface of RAW 264.7 cells at different time points at 37 °C in PBS (pH 7.4): (A, B) 5 min; (C, D) 10 min; and (E, F) 20 min.
Figure 7.
Flow cytometry analysis of RAW 264.7 cells treated with biotinylated cholesterol lipid at different concentrations (5 and 10 μM) and at varying incubation periods at 37 °C in PBS (pH 7.4): (A, B) 5 min; (C, D) 10 min; and (E, F) 20 min.
Retention Time of Biotinylated Lipid Conjugate (CHOL–PEG2000–biotin) into the Cell Membrane
The plasma membrane is a dynamic structure whose components are constantly being internalized and refreshed. The properties of the anchor lipids control the fate of the extra molecules introduced onto the cell surface, and the outcome of the desired cell surface re-engineering as well.29 As confirmed above, cholesterol-based anchor lipid afforded a higher cell membrane incorporation efficiency than the phospholipid-based anchor lipid. Furthermore, we investigated the cell membrane retention time of the cholesterol-based anchor lipid and its potential for cell surface re-engineering applications. The copper-free click chemistry was done in 1 h, during which the internalization may occur. Therefore, in this study, we used a biotinylated lipid conjugate (CHOL–PEG2000–biotin) instead of doing a click biotinylation on the cell membrane after anchor lipid incorporation. We probed the dynamic behavior of the cell-bound biotinylated cholesterol at higher and lower concentrations. Briefly, RAW 264.7 cells were incubated with 5 and 10 μM of CHOL–PEG2000–biotin for 30 min at 37 °C in 1× PBS (pH 7.4), followed by confocal imaging with fluorescently labeled streptavidin to track the remaining exogenously associated biotin overtime. Interestingly, at the concentration of both 5 and 10 μM of CHOL–PEG2000–biotin, the average fluorescent intensity underwent a sharp jump at the first 30 min and then a straightforward decrease (Figure 8). This phenomenon can be explained by either a high concentration of the cholesterol–PEG2000–biotin inserted on the cell surface, which may bind less streptavidin-FITC due to steric hindrances, multiple binding of streptavidin, or a fluorescent quench of the bound streptavidin-FITC. To confirm this stipulation, we investigated a low concentration of 2.5 μM of CHOL–PEG2000–biotin. Interestingly, the fluorescent intensity of bound streptavidin-FITC underwent a straightforward decreased without a high fluorescent signal jump, which indicates no steric hindrance nor fluorescent quench of streptavidin-FITC binding at the low concentration of CHOL–PEG2000–biotin inserted. Overall, we found concentration-dependent incorporation rates of anchor lipid. The retention half-life time of the cholesterol–PEG–biotin is about 1 h on macrophages cell surface, which is shorter than that from the cholesterol–PEG–glycopolymer in Jurkats cells reported by Bertozzi et al.27 This may be both lipid-conjugate and cell type-specific phenomenon.
Figure 8.

Retention time of directly anchored biotinylated cholesterol lipid onto cell surface of RAW 264.7 cells at different concentrations (2.5, 5, and 10 μM) in 1× PBS (pH 7.4) at room temperature. Error bars represent standard deviation, n = 3.
Cytotoxic Effect of the Anchor Lipids and upon Their Incorporation—3-(4, 5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay
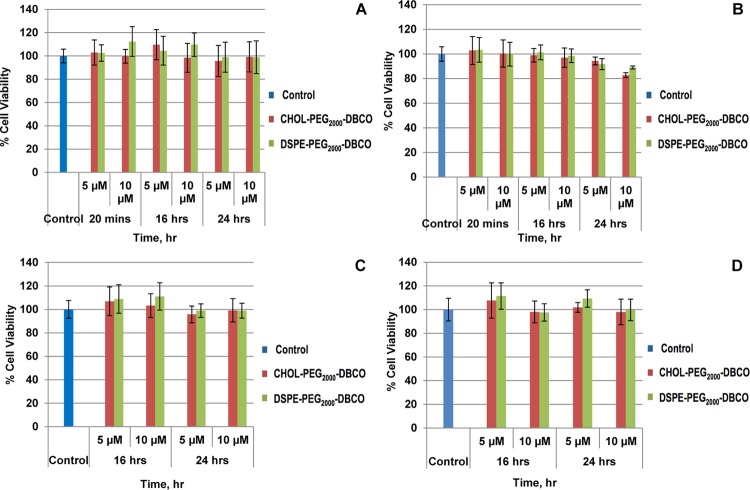
To evaluate the effect of the individual anchor lipid on cell viability upon and after incorporation, MTT assays were performed on the RAW 264.7 cell lines at different incubation times (20 min, 16, and 24 h) and at varying concentrations of anchor lipids. In addition, the effect after biotinylation on the cell viability was also explored for 20 min, 16, and 24 h, respectively. As a result, the anchor lipids did not show any significant cell toxicity during incorporation and after biotinylation for both two-step and direct biotinylation methods (Figure 9). This indicates that these reactive anchor lipids can be used for the potential biomedical applications.
Figure 9.
Viability of RAW 264.7 cells during DBCO-lipid conjugate treatment at varying time points (20 min, 16, and 24 h) at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) (A) and after biotinylation of DBCO-lipid conjugate treatment for 20 min at 37 °C in DMEM, followed by washing in buffer and incubation in DMEM for 16 and 24 h (B). Viability of RAW 264.7 cells during one-pot biotinylated lipid conjugate treatment at varying time points (20 min, 16, and 24 h) at 37 °C in DMEM (C) and after biotinylation of DBCO-lipid conjugate treatment for 20 min at 37 °C in DMEM, followed by washing in buffer and incubation in DMEM for 16 and 24 h (D). Cell viability was determined by the MTT assay. Error bars represent the standard deviation, n = 3.
Conclusions
In summary, the present study demonstrated the potentials of anchor lipids, DBCO-containing cholesterol- and DSPE-based anchor lipids for an efficient and chemoselective cell surface re-engineering application. The successful incorporation of the reactive anchor lipids and its selective modification was confirmed by biotinylation via copper-free click chemistry followed by specific interactions with streptavidin-FITC under confocal microscopy and flow cytometry analysis. The results indicated the successful incorporation of cholesterol-based anchor lipids with high fluorescence intensities from the cell membrane and with little or negligible internalization when compared to the DSPE-based anchor lipid. This difference in the incorporation efficiencies of the anchor lipids can be attributed to the difference in their structural characteristics and physiochemical properties. Furthermore, the cell viability assay (MTT) study also revealed that the incorporation of the cholesterol- and DSPE-based anchor lipids into the cell surface did not cause any significant toxicity to the modified RAW 264.7 cells. These results suggest the potential use of these reactive anchor lipids for cell surface re-engineering applications. In particular, the copper-free click chemistry can be used for conjugation of many important biomolecules such as peptides, proteins, and carbohydrates, which allows for various cell surface re-engineering applications.
Experimental Section
Materials and Reagents
All of the solvents and reagents were purchased from commercial sources and used as received, unless otherwise noted. Deionized water was used as a solvent in all of the procedures. N-Hydroxysuccinimide (NHS-ester) and cholesteryl chloroformate were purchased from Sigma-Aldrich (St. Louis, MO). 1,2-Dipalmitoyl-sn-glycero-3-phosphocholine, 1,2-distearoyl-sn-glycero-3-phosphoethanolamine–N-[amino (poly(ethylene glycol))-2000] (ammonium salt) (DSPE–PEG2000-NH2) were purchased from Avanti Polar Lipids, Inc (Alabaster, AL). DBCO–PEG4-NHS ester was purchased from Click Chemistry tools (Scottsdale, AZ). Azido–PEG4–biotin and streptavidin-fluorescein isothiocyanate (streptavidin-FITC) were purchased from Biolegend (San Diego, CA). Dulbecco’s modified Eagle’s medium (DMEM), (3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), and MTT reagent were purchased from Life Technologies (Grand Island, NY).
Syntheses of CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO Anchor Lipids
Synthesis of CHOL–PEG2000–DBCO
First, monocholesteryl–PEG2000-amine was synthesized as per a previously described method.28 Triethylamine (Et3N, 50 μL, 0.35 mmol) was added to the solution of monocholesteryl–PEG2000-amine (100 mg, 40.4 μmol) in anhydrous CH2Cl2 (3 mL) and stirred for 30 min at room temperature (RT) under argon gas atmosphere, followed by the addition of DBCO–PEG4-NHS ester (31 mg, 48.5 μmol, 1.2 equiv) dissolved in anhydrous CH2Cl2 (1 mL) to the above mixture and continuous stirring of the reaction solution for 48 h at RT. Thin-layer chromatography analysis was performed to monitor the reaction. Then, the reaction solution was concentrated to obtain a residue, which was purified by silica gel column chromatography eluted with a gradient solvent system (CHCl3/MeOH, 95:5 ∼ 80:20 v/v) to yield CHOL–PEG2000–PEG4–DBCO (92 mg, 70% yield). 1H NMR (CDCl3, 400 MHz, δ): 7.65 (d, 1H), 7.40–7.28 (d, 8H), 6.53 (br, 1H), 5.35 (br, 1H), 5.22 (br, 1H), 5.14 (d, 1H), 4.46 (br, 1H), 3.81 (t, 3H), 3.75–3.69 (m, 3H), 3.76–3.40 (m, 180 H), 3.36–3.27 (m, 4H), 3.26–3.21 (m, 2H), 2.87 (t, 3H), 2.80 (br, 6H), 2.60–2.20 (m, 10 H), 2.00–1.75 (m, 10 H), 1.50–0.95 (m, 30 H), 0.89 (d, 3H), 0.84 (dd, 6H), 0.65 (d, 3H) ppm. 13C NMR (CDCl3, 100 MHz, δ): 169.62, 168.69, 166.58, 164.36, 138.5, 129.75, 126.73, 126.22, 125.96, 125.89, 125.46, 124.84, 123.23, 120.16, 119.33, 105.41, 69.42, 68.34, 68.20, 68.11, 67.87, 63.36, 54.33, 53.12, 47.65, 39.95, 37.15, 34.52, 32.39, 29.79, 29.54, 29.29, 23.22, 20.46, 20.20, 16.35, 9.50, 9.38 ppm.
Synthesis of DSPE–PEG2000–DBCO
DSPE–PEG2000–PEG4–DBCO was synthesized by following the method from previous literature,30 and is commercially available from Avanti Polar Lipids (Alabaster, Alabama).
Cell Culture
The mouse macrophage cell line RAW 264.7 cells (ATCC) were cultured in DMEM supplemented with 10% fetal bovine serum, penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37 °C in 5% CO2 conditions. The cells were further subcultured by trypsinization with (0.25% trypsin, 0.02% ethylenediaminetetraacetic acid) after cells became confluent.
Preparation of Anchor Lipids Solution
A solution of 1 mg/mL anchor lipid in CHCl3 was prepared and then CHCl3 was removed by evaporation under a stream of N2 gas to obtain a thin film of lipids. The thin film was further kept under vacuum for overnight to ensure the complete removal of any residual solvent. The thin film was hydrated with PBS buffer (pH 7.4) with vigorous sonication and vortexing for about 3 h to form aqueous solutions. Serial dilutions from the above solution resulted in 2.5, 5, and 10 μM aqueous solutions of anchor lipids for further studies.
Cell Surface Re-engineering of RAW 264.7 Cells with CHOL–PEG–DBCO and DSPE–PEG–DBCO Anchor Lipids and Their Further Biotinylation via Copper-Free Click Chemistry
The RAW 264.7 cells were washed twice with 1× PBS buffer (pH 7.4) and incubated with different concentrations (5 and 10 μM) of CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO anchor lipids for different incubation times (5, 10, and 20 min) in 1× PBS buffer (pH 7.4) at 37 °C. At each time point, the lipid solution was gently aspirated, followed by washing three times with 1× PBS buffer (pH 7.4), the cells were treated with 750 μL of Azido–PEG4–biotin (3 equiv of anchor lipid) (50 mM stock solution, 1× PBS (pH 7.4) for 1 h at 37 °C). Then, the solution was gently aspirated and the cells were washed again with 1× PBS buffer (pH 7.4) and incubated with 750 μL FITC-streptavidin (1:800 dilution, 0.5 mg/mL stock solution) for 1 h in darkness at room temperature. The cell nuclei were stained by adding 3 μL of 4′,6-diamidino-2-phenylindole (DAPI) solution. The treated cells were washed, fixed by the addition of 4% PFE solution and set aside for 10 min, gently aspirated, washed three times with PBS buffer (pH 7.4), and finally mounted onto the glass slides (using Gold Anti-fade Solution) for viewing under a Nikon A1RSI Confocal Microscope with a 60x oil objective
Direct Anchoring of Biotinylated Lipid Conjugates into the Cell Surface
Biotin–PEG–lipids were synthesized by the incubation of aqueous lipid solutions of CHOL–PEG2000–DBCO and DSPE–PEG2000–DBCO with 2 equiv of azido–PEG4–biotin in Eppendorf tubes at RT for 1 h. Then, the RAW 264.7 cells were incubated with different concentrations of the resultant biotin–PEG–lipids (5 and 10 μM) at different incubation times (5, 10, and 20 min) in 1× PBS buffer (pH 7.4) at 37 °C. After each time point, the lipid solution was gently aspirated, followed by washing three times with 1× PBS buffer (pH 7.4). Finally, the RAW 264.7 cells with the incorporated biotin–PEG–lipids were incubated with 750 μL of streptavidin-FITC (1:800 dilution from 0.5 mg/mL stock solution, 1× PBS buffer, pH 7.4) for 1 h in darkness. The cell nuclei were stained by adding 3 μL of 4′,6-diamidino-2-phenylindole (DAPI) solution. Then, the buffer medium was gently aspirated, followed by washing three times with 1× PBS buffer (pH 7.4) between each step. The coverslips were mounted onto glass slides ProLong Gold Anti-fade reagent (Life Technologies). The slides were imaged using a Nikon A1RSI Confocal Microscope with a 60× oil objective.
Flow Cytometry Analysis of Streptavidin-FITC Labeled Biotinylated-Lipid Anchored Cells
RAW 264.7 cells (1 × 106 cells/test tube) were cultured overnight, washed three times with PBS buffer via centrifugation (1200 rpm, 5 min), treated with different concentrations of anchor lipids (500 μL/test tube, 5 and 10 μM) in 1× PBS buffer (pH 7.4), and incubated for different time intervals (5, 10, and 20 min) at 37 °C. After incubation, the cells were washed twice with PBS buffer (pH 7.4) via centrifugation, aspirated, and then treated with 750 μL of azido–PEG4–biotin (3 equiv of anchor lipid) (50 mM stock solution, 1× PBS, pH 7.4) for 1 h at 37 °C. The cells were washed and then incubated with streptavidin-FITC for 1 h at room temperature in darkness. Similarly, cells were treated with biotin–PEG–lipids solutions (5 and 10 μM) for different incubation times (20 min, 2 h, and 4 h) at 37 °C and followed by washing and incubation with streptavidin-FITC for 1 h at room temperature in darkness. After further washing steps, the fluorescence signal of 2 × 104 cells were measured using a flow cytometer (BD-Canto).
Retention Time of Biotinylated Lipid Conjugate (CHOL–PEG2000–Biotin) on the Cell Membrane
RAW 264.7 cells were seeded into 6-well plates, with each well containing a glass coverslip. After overnight incubation, the cell medium was aspirated off and the cells were washed twice with 1× PBS buffer (pH 7.4). The cells were then incubated with a 2.5, 5, or 10 μM CHOL–PEG2000–biotin solution (total volume 1 mL), or 1× PBS buffer (pH 7.4) for control wells, for 30 min at 37 °C. The cholesterol solution was aspirated off and the cells were washed twice with 1× PBS buffer (pH 7.4). Fresh medium was added to each well and the cells were placed back into the incubator. After a set time point, the cells were removed from incubation, washed, and fixed. The time points were 0, 0.5, 1, 2, and 4 h after the removal of the cholesterol solution. The cells at 0 time point were used as control cells. At a given time point, the media was aspirated from specific wells and the cells were washed twice with 1× PBS buffer (pH 7.4). The cells were fixed using a 4% para-formaldehyde solution for 10 min at room temperature and then washed twice with 1× PBS buffer (pH 7.4). 300 μL of 0.625 μg/mL streptavidin-FITC solution was added to each coverslip and allowed to set for 1 h at room temperature. After the streptavidin incubation, the coverslips were washed by submerging them twice in 1× PBS buffer (pH 7.4) for 5 min. After washing, 300 μL of 300 nM DAPI solution was added to each coverslip and allowed to sit for 10 min at room temperature. After incubation, the coverslips were washed by submerging them thrice in 1× PBS buffer (pH 7.4) for 5 min. The coverslips were mounted on the slides by adding 30 μL of ProLong Gold Anti-fade reagent to each coverslip and allowed to set overnight. The slides were imaged using a Nikon A1RSI Confocal Microscope with a 60× oil objective.
Cytotoxic Effect of the Anchor Lipids and upon Their Incorporation—MTT Assay
The cell viability of RAW 264.7 cells upon anchor lipids incorporation was carried out using a MTT assay (Vybrant MTT Cell Proliferation Assay Kit). Briefly, RAW 264.7 cells were seeded at a density of 2.5 × 104 cells per well in 96-well plates overnight, then the medium was removed, and the cells were washed with 1× PBS buffer (pH 7.4). The cell viability during incorporation of the anchor lipid and its biotinylation process was assessed by the addition of FBS-free culture medium with anchor lipids and incubated for different time points (20 min, 16 h, 24 h) at 37 °C. In addition, the cell viability after biotinylation was investigated as well. After biotinylation, the cells were washed with 1× PBS buffer (pH 7.4) and placed back in the medium at 37 °C under 5% CO2, followed by the assessment of the cell viability at 16 and 24 h. Briefly, the medium was removed by gentle aspiration, to which 100 μL of fresh culture medium and 10 μL of 12 mM MTT solution were added and incubated for an additional 4 h. Subsequently, 100 μL of the sodium dodecyl sulfate–HCl solution was added (10% SDS in 0.01 M HCl) and incubated for another additional 3 h. Finally, the measurements were read at 570 nm using a UV–vis multiplate reader. Similar protocols were followed for cell viability assessments of biotin–PEG–lipids at different concentrations (5 and 10 μM) and incubation times.
Acknowledgments
This work was partially supported by grants from NIH (1R01HL102604-04, X.-L.S.), Faculty Research Fund from the Center for Gene Regulation in Health and Disease (GRHD) (X.-L.S.) as well as Dissertation Research Award (P.V.) at Cleveland State University. H.N. appreciates the China Oversea Scholar Award from China Scholarship Council. The authors acknowledge the NIH shared instrumentation grant NIH 1 S10 OD010381 for the acquisition of confocal microscope and NSF MRI grant for the acquisition of 400 MHz NMR at Cleveland State University (NSF CHE 1126384, X.-L.S.).
Author Contributions
The manuscript was written through contributions of all of the authors. All of the authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
References
- Hong V.; Steinmetz N. F.; Manchester M.; Finn M. G. Labeling live cells by copper-catalyzed alkyne-azide click chemistry. Bioconjugate Chem. 2010, 21, 1912–1916. 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin J. M.; Prescher J. A.; Laughlin S. T.; Agard N. J.; Chang P. V.; Miller I. A.; Lo A.; Codelli J. A.; Bertozzi C. R. Copper-free click chemistry for dynamic in vivo imaging. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 16793–16797. 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan M. T.; Irvine D. J. Enhancing Cell therapies from the Outside In: Cell Surface Engineering Using Synthetic Nanomaterials. Nano Today 2011, 6, 309–325. 10.1016/j.nantod.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkis C.; Gras G.; Sanchez F.; Mallet J.; Serguera C. Long term survival and limited migration of genetically modified monocytes/macrophages grafted into the mouse brain. J. Biomed. Sci. Eng. 2013, 6, 561–571. 10.4236/jbise.2013.65071. [DOI] [Google Scholar]
- Stabler C. L.; Sun X.-L.; Cui W.; Wilson J. T.; Haller C. A.; Chaikof E. L. Surface re-engineering of pancreatic islets with recombinant azido-thrombomodulin. Bioconjugate Chem. 2007, 18, 1713–1715. 10.1021/bc7002814. [DOI] [PubMed] [Google Scholar]
- Zope H. R.; Versluis F.; Ordas A.; Voskuhl J.; Spaink H. P.; Kros A. In vitro and in vivo supramolecular modification of biomembranes using a lipidated coiled-coil motif. Angew. Chem., Int. Ed. 2013, 52, 14247–14251. 10.1002/anie.201306033. [DOI] [PubMed] [Google Scholar]
- Kang S.-W.; Lee S.; Na J. H.; Yoon H. I.; Lee D.-E.; Koo H.; Cho Y. W.; Kim S. H.; Jeong S. Y.; Kwon I. C.; Choi K.; Kim K. Cell labeling and tracking method without distorted signals by phagocytosis of macrophages. Theranostics 2014, 4, 420–431. 10.7150/thno.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.; Rabinowitz S. Macrophages as targets for drug delivery. Adv. Drug Delivery Rev. 1989, 4, 27–47. 10.1016/0169-409X(89)90036-7. [DOI] [Google Scholar]
- Jain N. K.; Mishra V.; Mehra N. K. Targeted drug delivery to macrophages. Expert Opin. Drug Delivery 2013, 10, 353–367. 10.1517/17425247.2013.751370. [DOI] [PubMed] [Google Scholar]
- Kojima N.; Ishii M.; Kawauchi Y.; Takagi H. Oligomannose-coated liposome as a novel adjuvant for the induction of cellular immune responses to control disease status. BioMed Res. Int. 2013, 2013, 562924 10.1155/2013/562924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki N.; Vela J. L.; Nycholat C. M.; Rademacher C.; Khurana A.; van Rooijen N.; Crocker P. R.; Kronenberg M.; Paulson J. C. Targeted delivery of lipid antigen to macrophages via the CD169/sialoadhesin endocytic pathway induces robust invariant natural killer T cell activation. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 7826–7831. 10.1073/pnas.1219888110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. C.; Kawasaki N.; Nycholat C. M.; Han S.; Pilotte J.; Crocker P. R.; Paulson J. C. Antigen delivery to macrophages using liposomal nanoparticles targeting sialoadhesin/CD169. PLoS One 2012, 7, e39039 10.1371/journal.pone.0039039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehara Y.; Niwa T.; Biao L.; Ikehara S. K.; Ohashi N.; Kobayashi T.; Shimizu Y.; Kojima N.; Nakanishi H. A carbohydrate recognition-based drug delivery and controlled release system using intraperitoneal macrophages as a cellular vehicle. Cancer Res. 2006, 66, 8740–8748. 10.1158/0008-5472.CAN-06-0470. [DOI] [PubMed] [Google Scholar]
- Haney M. J.; Zhao Y.; Harrison E. B.; Mahajan V.; Ahmed S.; He Z.; Suresh P.; Hingtgen S. D.; Klyachko N. L.; Mosley R. L.; Gendelman H. E.; Kabanov A. V.; Batrakova E. V. Specific transfection of inflamed brain by macrophages: a new therapeutic strategy for neurodegenerative diseases. PLoS One 2013, 8, e61852 10.1371/journal.pone.0061852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Klyachko N. L.; Haney M. J.; Zhao Y.; Manickam D. S.; Mahajan V.; Suresh P.; Hingtgen S. D.; Mosley R. L.; Gendelman H. E.; Kabanov A. V.; Batrakova E. V. Macrophages offer a paradigm switch for CNS delivery of therapeutic proteins. Nanomedicine 2014, 9, 1403–1422. 10.2217/nnm.13.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori H.; Suzuki S.; Okazaki Y.; Suzuki N.; Kuwana M. Intracranial transplantation of monocyte-derived multipotential cells enhances recovery after ischemic stroke in rats. J. Neurosci. Res. 2012, 90, 479–488. 10.1002/jnr.22755. [DOI] [PubMed] [Google Scholar]
- Kelly C.; Jefferies C.; Cryan S.-A. J. Targeted liposomal drug delivery to monocytes and macrophages. Drug Delivery 2010, 2011, 1–11. 10.1155/2011/727241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M.; Chen J. Y.; Weissman-Tsukamoto R.; Volkmer J.-P.; Ho P. Y.; McKenna K. M.; Cheshier S.; Zhang M.; Guo N.; Gip P.; Mitra S. S.; Weissman I. L. Macrophages eat cancer cells using their own calreticulin as a guide: Roles of TLR and Btk. Proc. Natl. Acad. Sci. U.S.A. 2015, 112, 2145–2150. 10.1073/pnas.1424907112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto S.; Moriyama R.; Mori T.; Iwasaki Y. Surface engineering of macrophages with nucleic acid aptamers for the capture of circulating tumor cells. Chem. Commun. 2015, 51, 17428–17430. 10.1039/C5CC06211J. [DOI] [PubMed] [Google Scholar]
- Miura S.; Teramura Y.; Iwata H. Encapsulation of islets with ultra-thin polyion complex membrane through poly(ethylene glycol)-phospholipids anchored to cell membrane. Biomaterials 2006, 27, 5828–5835. 10.1016/j.biomaterials.2006.07.039. [DOI] [PubMed] [Google Scholar]
- Kato K.; Itoh C.; Yasukouchi T.; Nagamune T. Rapid protein anchoring into the membranes of Mammalian cells using oleyl chain and poly(ethylene glycol) derivatives. Biotechnol. Prog. 2004, 20, 897–904. 10.1021/bp0342093. [DOI] [PubMed] [Google Scholar]
- Takemoto N.; Teramura Y.; Iwata H. Islet surface modification with urokinase through DNA hybridization. Bioconjugate Chem. 2011, 22, 673–678. 10.1021/bc100453r. [DOI] [PubMed] [Google Scholar]
- Chung H. A.; Tajima K.; Kato K.; Matsumoto N.; Yamamoto K.; Nagamune T. Modulating the actions of NK cell-mediated cytotoxicity using lipid-PEG (n) and inhibitory receptor-specific antagonistic peptide conjugates. Biotechnol. Prog. 2005, 21, 1226–1230. 10.1021/bp049646b. [DOI] [PubMed] [Google Scholar]
- Teramura Y.; Iwata H. Islet encapsulation with living cells for improvement of biocompatibility. Biomaterials 2009, 30, 2270–2275. 10.1016/j.biomaterials.2009.01.036. [DOI] [PubMed] [Google Scholar]
- Teramura Y.; Kaneda Y.; Iwata H. Islet-encapsulation in ultra-thin layer-by-layer membranes of poly (vinyl alcohol) anchored to poly (ethylene glycol)-lipids in the cell membrane. Biomaterials 2007, 28, 4818–4825. 10.1016/j.biomaterials.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Rabuka D.; Forstner M. B.; Groves J. T.; Bertozzi C. R. Noncovalent cell surface engineering: incorporation of bioactive synthetic glycopolymers into cellular membranes. J. Am. Chem. Soc. 2008, 130, 5947–5953. 10.1021/ja710644g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods E. C.; Yee N. A.; Shen J.; Bertozzi C. R. Glycocalyx engineering with a recycling glycopolymer that increases cell survival in vivo. Angew. Chem., Int. Ed. 2015, 54, 15782–15788. 10.1002/anie.201508783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X.; Wu G.; Yang W.; Barth R. F.; Tjarks W.; Lee R. J. Synthesis of cetuximab-immunoliposomes via a cholesterol-based membrane anchor for targeting of EGFR. Bioconjugate Chem. 2007, 18, 101–108. 10.1021/bc060174r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. L.; Purcell S. C.; Verespy S.; Wang Y.; Godula K. Glycocalyx scaffolding with synthetic nanoscale glycomaterials. Biomater. Sci. 2017, 5, 1537–1540. 10.1039/C7BM00289K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.; Weingart J.; Jiang R.; Peng J.; Wu Q.; Sun X.-L. Bio-inspired liposomal thrombomodulin conjugate through bio-orthogonal chemistry. Bioconjugate Chem. 2013, 24, 550–559. 10.1021/bc300399f. [DOI] [PMC free article] [PubMed] [Google Scholar]