Abstract
Local structure and dynamics of lipid membranes play an important role in membrane function. The diffusion of small molecules, the curvature of lipids around a protein and the existence of cholesterol-rich lipid domains (rafts) are examples for the membrane to serve as a functional interface. The collective fluctuations of lipid tails, in particular, are relevant for diffusion of membrane constituents and small molecules in and across membranes, and for structure and formation of membrane domains. We studied the effect of aspirin (acetylsalicylic acid, ASA) on local structure and dynamics of membranes composed of dimyristoylphosphocholine (DMPC) and cholesterol. Aspirin is a common analgesic, but is also used in the treatment of cholesterol. Using coherent inelastic neutron scattering experiments and molecular dynamics (MD) simulations, we present evidence that ASA binds to liquid-ordered, raft-like domains and disturbs domain organization and dampens collective fluctuations. By hydrogen-bonding to lipid molecules, ASA forms ‘superfluid’ complexes with lipid molecules that can organize laterally in superlattices and suppress cholesterol’s ordering effect.
Keywords: lipid membranes, cholesterol, aspirin, liquid-ordered phase, membrane–drug interactions
1. Introduction
Membrane research progresses with ever-increasing levels of granularity. Initial models of the membrane as an inert, physical barrier were revised with the discovery of membrane-embedded proteins. Singer & Nicholson [1] devised their famous ‘fluid mosaic’ model to describe a system where proteins float in a featureless soup of various lipid types. Most recently, membrane research has focused on membrane details on the level of individual lipids and proteins. The lipid environment in the specific region around a protein, such as the hydrophobic thickness or spontaneous curvature of the lipid tails, is now believed to be crucial to proper protein function [2]. Lateral diffusion of membrane constituents [3–7] and transmembrane diffusion of small molecules are believed to involve the local, collective motion of lipid tails [8–11].
Lipid rafts are an example of function arising from nanoscale structure. Rafts are small lipid heterogeneities in plasma membranes [12,13]. Rafts are typically described as structures that are enriched in cholesterol and exist to chaperone proteins from the Golgi apparatus to the plasma membrane surface [14]. Although their existence in biological membranes is still a matter of debate [15], various raft-like structures have been observed in model membranes, and from studies on model membranes, plasma membrane rafts are believed to be manifestations of the so-called liquid-ordered (lo) phase [12,16–27]. The lo phase is unique in that it is characterized by elevated cholesterol concentrations as well as high lipid positional and chain order, however, at the same time low viscosity [28] and high surface tension. It is soft and stiff at the same time by being more rigid than a gel-state membrane, but less viscous than a fluid membrane.
The effect of drug molecules on lipid membranes is typically characterized by their effect on bulk membrane properties, such as mechanical properties and area per lipid head group [29–31], which affect, for instance, permeability. Drug–membrane interactions can also indirectly influence the function of membrane proteins through their membrane effects, and they also affect membrane heterogeneities [30,32,33]. For example, there are numerous reports that the common analgesic aspirin (acetylsalicylic acid, ASA) interacts with the lipid membrane and makes it softer and more fluid, and also impacts the formation of lipid raft structures [34–39]. Neutron scattering experiments recently presented evidence that aspirin creates local structural distortions in the lo phase in model membranes [30], suppressing the formation of cholesterol clustering.
In this paper, we performed inelastic neutron scattering experiments and molecular dynamics (MD) simulations on membranes containing cholesterol and aspirin. The cholesterol concentration was chosen such that cholesterol rafts form, but well below the solubility limit of cholesterol [40]. We observed that aspirin creates local increases in area per lipid, causing a decrease in positional order and a damping of collective lipid tail fluctuations. We find evidence for ‘superfluid’ ASA–lipid complexes that organize in the membranes and impact cholesterol’s effect on the bilayers. We also present evidence for a direct ASA–cholesterol interaction in membranes through hydrogen bonding of ASA to cholesterol molecules.
2. Results
2.1. Inelastic neutron scattering
Coherent, inelastic neutron scattering experiments were performed on oriented DMPC-d54 (1,2-dimyristoyl-sn-glycero-3-phosphocholine) bilayers containing cholesterol and ASA. The lipid chains were selectively deuterated, enhancing the contribution of collective tail dynamics to the signal. In addition, the bilayers were hydrated with D2O to reduce incoherent contributions. Two membrane systems were prepared: bilayers with 32.5 mol% cholesterol (CHOL sample), and bilayers with 29 mol% cholesterol and 10 mol% ASA (ASA sample). This cholesterol concentration has been shown to form rafts before and is well below the solubility limit of cholesterol in dimyristoylphosphocholine (DMPC) of approximately 40 mol% [40]. Membranes were prepared by dissolving DMPC, cholesterol and aspirin in a 1 : 1 solution of trifluoroethanol/chloroform, at the appropriate molar ratios, and depositing the solution on 1×1 cm2 silicon wafers. Following drying in vacuum and incubation at 100% relative humidity for 48 h, highly oriented bilayers are formed [30,34,36,41]. Twenty such wafers were prepared and aligned with respect to each other to create a ‘sandwich sample’ with a total mosaicity of less than 0.5° and a total mass (lipids, cholesterol and aspirin) of 34 mg.
Neutron measurements were performed on the IN12 cold triple-axis spectrometer at the high flux reactor of the Institut Laue-Langevin (ILL) in Grenoble, France. All experiments were conducted at a temperature of 30°C, in the fluid phase of the membranes. The scattering vector Q was placed in the plane of the membranes (q∥) to measure the static (S(q∥)) and dynamic structure factors (S(q∥,ω)). A sketch of the scattering geometry is shown in figure 1a.
Figure 1.
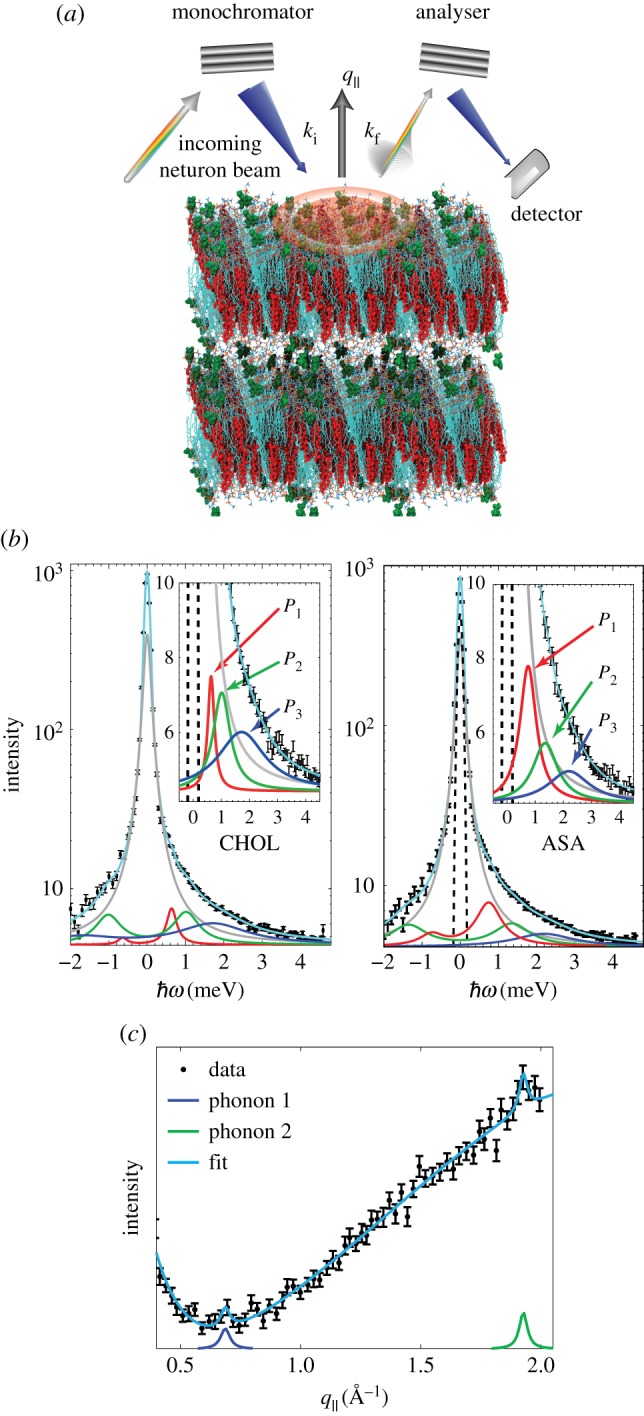
(a) Sketch of the scattering geometry. (b) Sample constant-q∥ scans for the CHOL sample at q∥=1.37 Å −1. Data were fit using a Gaussian instrumental resolution, a Lorentzian peak centred at q∥=0 to capture incoherent scattering contributions, and three phonon excitations. The Gaussian instrumental resolution was fixed to values based on resolutions calculated using the ResLib package. The position of the incoherent peak was fixed at ; all other parameters are free. The inset shows three phonon excitations (P1, P2 and P3) in more detail. (c) Constant-energy scan for the ASA sample at an energy of . All parameters are free in the constant-energy scans.
We note that IN12 was recently upgraded and was used in focusing mode, which significantly increases the neutron intensity at the sample position, however, at the cost of Q and energy resolution. When comparing our results to previous experiments in similar systems by Armstrong et al. [42] using a parallel beam configuration on IN12, signals in Q and energy, thus, appear slightly broader. In previous experiments, however, 400 mg of material was needed to conduct inelastic experiments, while only 34 mg was sufficient to see a clear signal in this paper.
Typical constant-q∥ scans, taken from the ASA and CHOL samples at q∥=1.37 Å −1 are shown in figure 1b. The constant-q∥ scans were well fit by analytical functions described by three phonon excitations, as described in the Material and methods.
A constant-energy scan at an energy of 2.5 meV is shown in figure 1c as an example. Constant-E scans were fit with an empirical exponential background, as well as a broad background Lorentzian peak at q∥∼2.5 Å −1. The background peak arises from convolution of dynamics at higher q∥ values (figure 10). The position of the phonon branches are determined by the positions of the Lorentzian signals. The positions of phonons observed in both constant-q∥ and constant-E scans were combined to determine entire dispersion relations, shown in figure 2.
Figure 10.
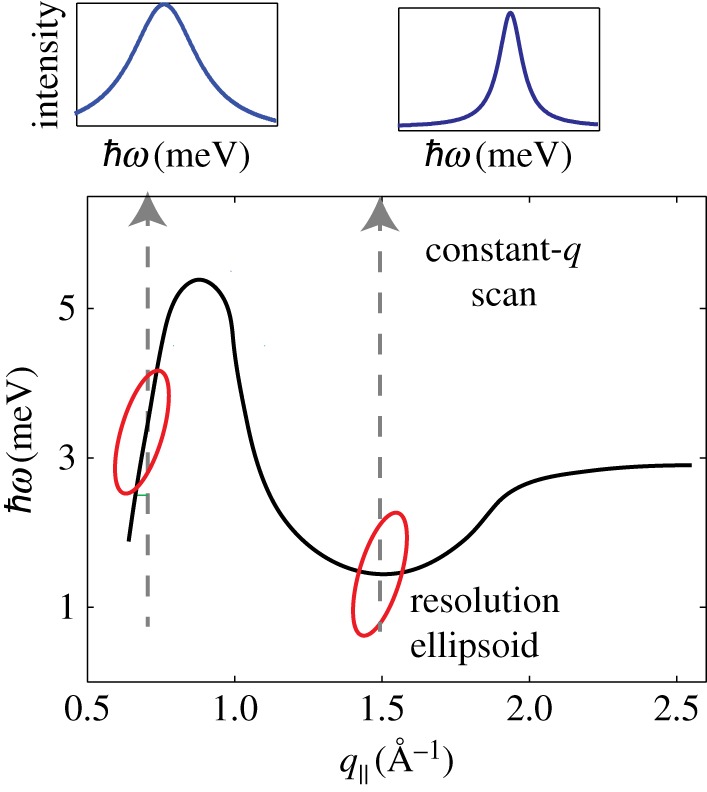
Lipid dispersion and instrumental resolution. The lipid dispersion is drawn schematically. During a constant-q∥ scan, the resolution ellipsoid is moved across the dispersion to detect the corresponding excitations. The smallest peak width in the spectra is observed when the dispersion is cut perpendicular, in the minimum of the dispersion relation.
Figure 2.
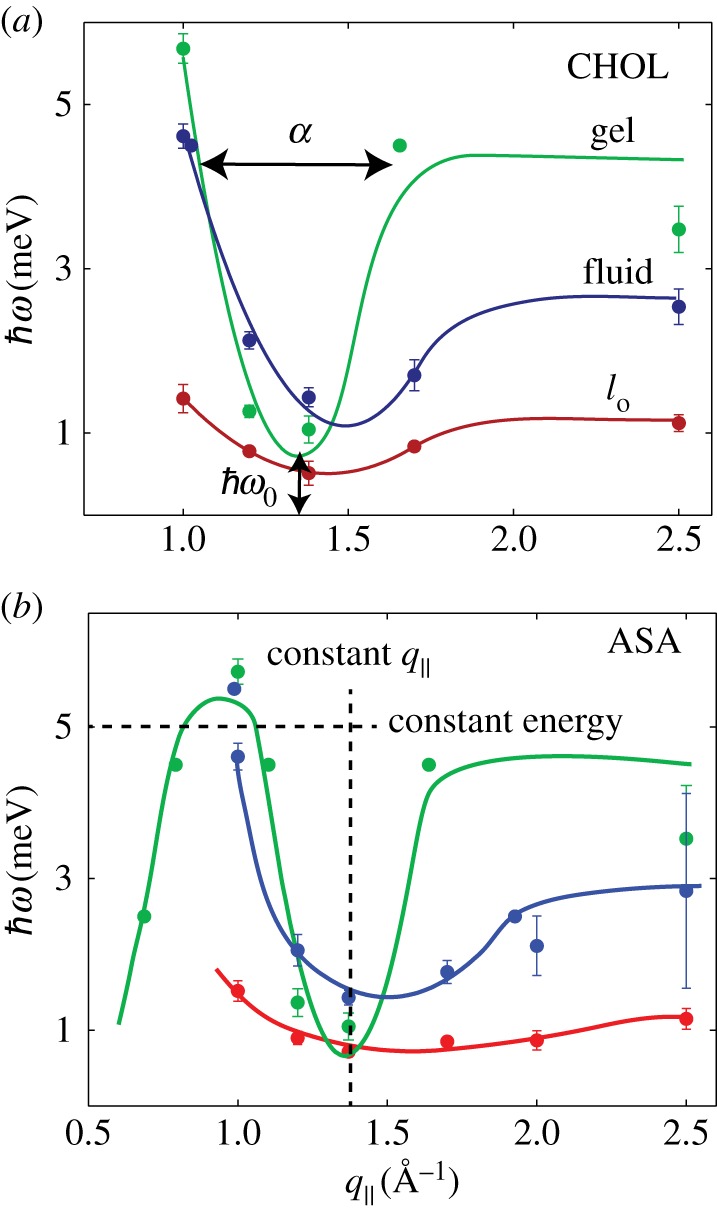
Dispersion relations for (a) the CHOL sample and (b) the ASA sample, as determined from constant-q∥ and constant-energy scans in figure 1. Three dispersion branches were observed, related to lipid molecules in gel (Pβ), fluid (Lα) and liquid-ordered (lo) patches. The CHOL dispersion in (a) is in qualitative agreement with previous reports on the collective short-wavelength dynamics in cholesterol-rich lipid membranes [42]. The meaning of the parameters α and in equation (2.1) are displayed in (a). The direction of constant-q∥ and constant-energy scans is shown in (b).
Lipid dispersions have been measured previously in inelastic X-ray and neutron scattering experiments [9–11,42–47] and using MD simulations [46,48,49]. All dispersions show similar features: at low q∥, the dispersion increases linearly with the slope related to the speed of sound. After reaching a maximum, the dispersion descends into a minimum at q∥=qT, where qT typically agrees well with the position of the chain correlation peak (the maximum in the static structure factor). Finally, the dispersion increases with increasing q∥ to form a plateau.
The dispersion in the region of the minima can empirically be described by a quadratic function [42]:
| 2.1 |
where is the minimum energy in the dispersion and α is the width of the dispersion; is related to lipid order, as a lower value indicates better-ordered, more ‘crystal-like’ bilayer; α is an empirical measure of the ‘softness’ of the bilayer [42].
As previously observed by Armstrong et al. [42], DMPC membranes containing cholesterol generate collective excitations belonging to three dispersions, related to coexisting gel, fluid and lo phases. Evidence for this phase coexistence was also observed in the long wavelength fluctuations from neutron spin-echo measurements [28]. Armstrong et al. and Toppozini et al. later used neutron diffraction and demonstrated that lo phases coexist in DMPC and DPPC bilayers with cholesterol [26,27,42,50,51].
Phonons were, therefore, assigned to three different phases. In the CHOL membranes shown in figure 2a, the gel phase (green) reaches at q∥=1 Å −1, then decreases to an energy minimum of at q∥∼1.4 Å −1. A second of the phases is observed, with a wider minimum at (blue), and is assigned to the fluid phase. Finally, a third dispersion is recorded at lower energies, with a minimum . Armstrong et al. assigned the dispersion at low energy, which is only observed in the presence of cholesterol, to the lo phase (red). The dispersion relations in the presence of ASA are shown in figure 2b.
With the phonons assigned to specific phases, the effect of ASA on the collective motions of each phase is examined. First of all, the damping of the phonons can be ascertained from the width of the phonon peak. As described in the Materials and methods, measurements along q∥=1.37 Å −1 represent the truest representation of the phonon widths. An increase in peak width, from approximately 0.14 meV to approximately 0.31 meV, in the lo phase is observed in the ASA sample. However, no statistically significant changes to the damping of the fluid or gel phases were observed, as displayed in figure 3a).
Figure 3.
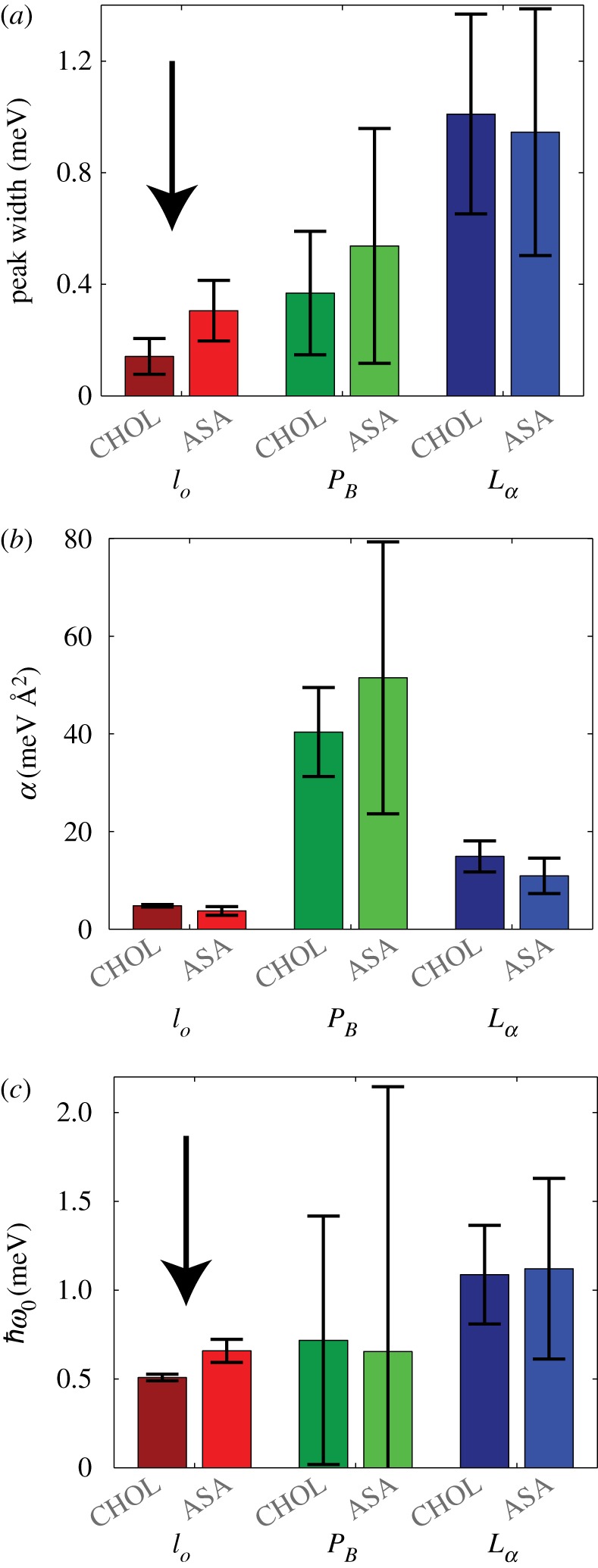
(a) Phonon widths for gel, fluid and lo phase in figure 2 from phonon lines measured at q∥=1.37 Å −1. (b) Parameters α and from equation (2.1) from fits to the dispersion curves. Statistically significant differences between CHOL and ASA samples were observed in the phonon width (a) and energy value of the minimum (c) in the lo phase, only. Statistically significant results are indicated by the arrow.
Equation (2.1) was fit to the phonon branches in the region 1.0 Å −1<q∥<2.0 Å −1, around the minimum. The fitting parameters are summarized in table 1 and also plotted in figure 3b,c. When comparing the dispersions from the ASA sample to the CHOL sample, two distinct points become important. First of all, for all structures, there is no statistically significant difference in bilayer softness, α. Secondly, we observe an increase in in the lo phase for the ASA sample, indicating a decrease in lipid order. The increase in phonon damping and the decrease in lipid order are compatible with the occurrence of defects in the structure and a fluidification of the lo phase, as will be discussed below.
Table 1.
Results of the fits to equation (2.1). We see that α is a metric of the ‘stiffness’ of the tails on nanoscale distances; ω0 can be related to the order in the bilayer. Results are compared to Armstrong et al. [42] for DMPC membranes containing 40 mol% cholesterol. Parameters for the CHOL and ASA samples are also plotted in figure 3.
|
α (meV/) |
(meV) |
|||||
|---|---|---|---|---|---|---|
| dispersion | Armstrong et al. (40 mol% CHOL) | cholesterol 30 mol% | ASA 10 mol% | Armstrong et al. (40 mol% CHOL) | cholesterol 30 mol% | ASA 10 mol% |
| gel | 30.3 | 41 ± 9 | 51.5 ± 28 | 1.80 | 0.72 ± 0.70 | 0.65 ± 1.49 |
| fluid | 10.7 | 14.9 ± 3 | 11.0 ± 4 | 2.63 | 1.09 ± 0.28 | 1.12 ± 0.51 |
| lo | 6.6 | 4.8 ± 0.3 | 3.7 ± 0.9 | 1.09 | 0.51 ± 0.02 | 0.66 ± 0.07 |
2.2. Molecular dynamics simulation
To gain an atomistic view of aspirin’s effect on the lo phase, MD simulations of membranes with 30 mol% cholesterol were performed. Simulations were performed on MacSim, a GPU-accelerated workstation, using parameters described previously [52] and also in the Material and methods. A united-atom DMPC + 30 mol% cholesterol bilayer, with 142 lipid + cholesterol molecules, was obtained from Hub et al. [53]. The number of water molecules per lipid was adjusted to 25, to mimic typical full-hydration conditions in agreement with the experiment, and the system was simulated for 200 ns. The membranes equilibrated to an area per lipid (DMPC and cholesterol) of AL=41.1 Å 2, which agrees closely with simulations by Hub et al. [53] and the experimentally determined area from Armstrong et al. [50]. In addition, the area per DMPC was determined using the GridMAT protocol to be 48.6±1 Å 2 [54], slightly smaller than the experimentally determined partial lipid area in membranes containing 32.5 mol% cholesterol [50].
Afterwards, the water in the system was removed and the aspirin was added to the now-dehydrated aqueous phase. Next, all water molecules (25 water molecules per lipid) were replaced and the system was equilibrated using the identical procedure as the pure bilayer. The equilibrated bilayer + aspirin system was then simulated for 200 ns. A snapshot of the simulation is shown in figure 4a. The molecules spontaneously embedded in the membrane within 10 ns. The density decomposition of the bilayer with ASA in figure 4b revealed that the molecules partitioned into the lipid head groups at z-values of |z|∼20 Å , in excellent agreement with past experiments [34,36]. To compare the MD simulations with the experiment, figure 4c shows the neutron scattering length density (SLD) with and without ASA. While the simulations were run in protonated membranes, the deuterium-labelled atoms were considered to be deuterated for the SLD calculation, as reported in [55]. The neutron scattering length agrees well with the experimentally determined neutron SLD shown figure 8c.
Figure 4.
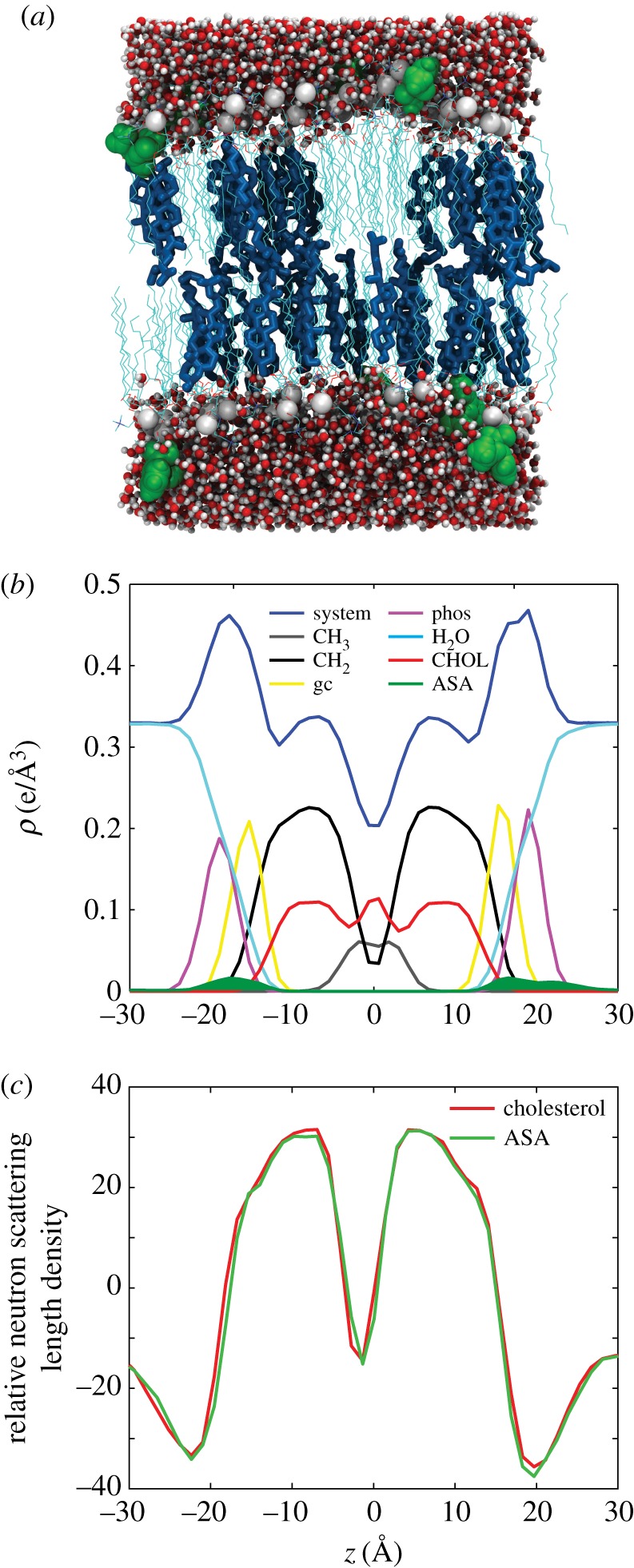
Results of the MD simulations. Simulations were performed for 30 mol% cholesterol in DMPC (CHOL), and the same system with 10 mol% ASA (ASA). (a) A snapshot of the ASA simulation. (b) An electron density breakdown for the components of the bilayer, indicating that the position of ASA (green) is in the head groups at |z|∼20 Å . (c) Neutron SLD from MD simulations. While the simulations were run in protonated membranes, the deuterium-labelled atoms were considered to be deuterated for the SLD calculation, as reported in [55].
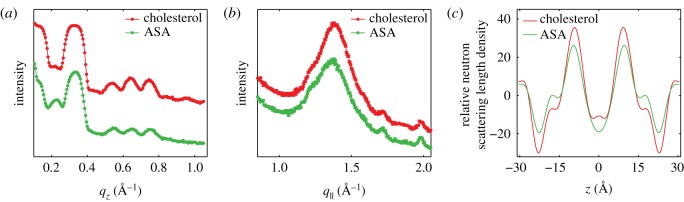
Figure 8.
Results of elastic neutron scattering. (a) Out-of-plane neutron reflectivities along qz. Both the cholesterol and ASA samples show a set of approximately seven evenly spaced Bragg peaks. (b) In-plane elastic scattering along q∥. A broad feature is observed at q∥∼1.37 Å −1. Additional reflections are associated with higher-order reflections of silicon. (c) Relative neutron SLDs assembled by Fourier analysis of the qz peaks.
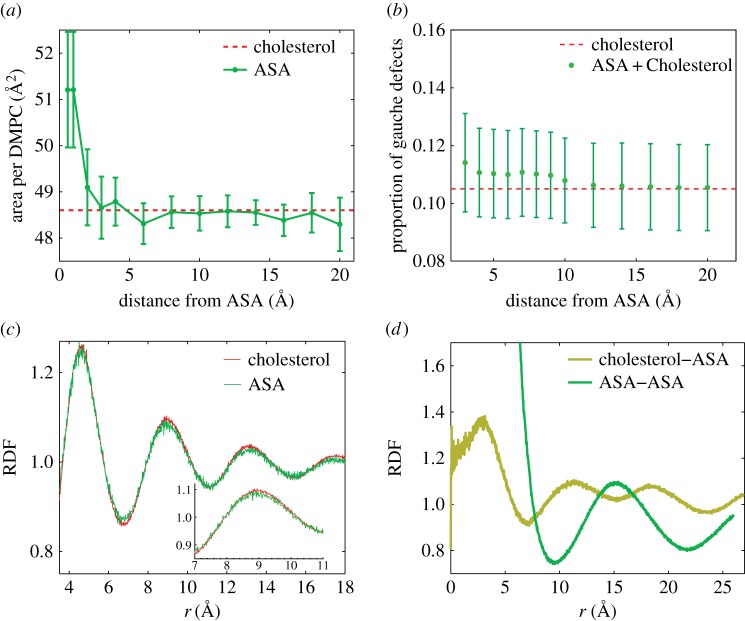
A local change in the area per lipid near ASA molecules was observed. As depicted in figure 5a, DMPC molecules within an approximately 5 Å radius around an ASA molecule have an area per lipid of more than 50 Å 2, a 5% increase. As the area per lipid is a common proxy for membrane fluidity [56], these results suggest a local increase in fluidity due to ASA. The proportion of gauche defects in the lipid chains was calculated in figure 5b. The presence of ASA did not result in a statistically significant change in the number of tail gauche defects. So while ASA was found to increase positional disorder, the proportion of gauche defects in the lipid tails was found to be unchanged, indicating that ASA did not influence chain segmental order [57].
Figure 5.
(a) Area per DMPC molecule at increasing distance from an ASA molecule. (b) Proportion of gauche defects in the DMPC molecules at increasing distance from ASA. (c) Radial distribution function (RDF) for DMPC–DMPC correlations, taken from both CHOL and ASA simulations. (d) RDF for cholesterol–ASA and ASA–ASA interactions.
Radial distribution functions (RDFs) were calculated for DMPC–DMPC correlations. The RDF in figure 5c is proportional to the probability of finding a DMPC tail at a distance r from another DMPC tail. A peak at r∼4.8 Å indicates a lipid tail separation of 4.8 Å, in agreement with diffraction experiments in bilayer systems [42]. The lipid–lipid distance appears to be unchanged in the presence of ASA.
The simulations can in particular be used to search for correlations between cholesterol and ASA and ASA–ASA molecules. The cholesterol–ASA RDF is plotted in figure 5d. This function probes potential organization between cholesterol and ASA molecules in the bilayers. Peaks appear at r∼11.5 Å and 19 Å, indicating a preferred separation between ASA and cholesterol. Peaks are observed at length scales larger than a lipid–lipid distance, suggesting superlattice ordering, as has been suggested previously [30]. This ordering is further supported by the ASA–ASA RDF, also plotted in figure 5d. A maximum is observed at 15 Å as is a second peak just outside the measurement frame at 25 Å. Both peaks are compatible with a superlattice of lipid–ASA complexes, as will be discussed in the next section.
We note that an additional peak in the cholesterol–ASA RDF at |r|∼3 Å was observed. This was an important finding as, on visual inspection of the simulation results, ASA molecules were found to transiently hydrogen-bond with cholesterol molecules, without embedding in the bilayer.
3. Discussion
Much literature exists showing that ASA has a disordering effect on membranes [30,34,37,58]. This disordering effect is, for instance, used to explain how aspirin disrupts proteins embedded in lipid rafts [59]. So far, studies of drug–membrane interactions have mainly focused on bulk, global changes in membrane properties due to drugs, such as changes in phase behaviour (a decrease or broadening in the gel–fluid transition), or an overall change in bilayer thickness. However, physiological membranes are dynamic and heterogeneous, and their nanoscale properties, such as lipid curvature near a protein or the presence of domains, matter to the membrane’s function. In this paper, we used neutron scattering and MD simulations to demonstrate that ASA preferably interacts with the cholesterol-rich lo phase and leads to local changes in the lipid arrangement and collective fluctuations.
ASA is known to partition into the lipid head groups and decrease lamellar spacing and widen the main transition in calorimetry experiments. Dispersion curves of the collective short-wavelength dynamics were measured in DMPC bilayers containing cholesterol and cholesterol and ASA. As was reported previously, the inelastic spectra present evidence for three dispersion branches, corresponding to lipids in their gel, fluid and liquid-ordered phase. By fitting an empirical function to the dispersion minimum, the degree of order and the softness of the corresponding phases were estimated from the depth of the minimum () and the width of the dispersion in the minimum (α). A statistically significant difference was observed in the energy value of the minimum. The increase in in the presence of ASA was assigned to a decrease in lipid order when ASA was present in the membranes. The decrease in order was accompanied by an increase in phonon line width, indicative of an increased phonon damping, probably related to the formation of defects.
Our experiments, therefore, present evidence that aspirin has the largest effect on lo phases, where it reduces the size of coherently coupled patches [4] and makes the lo phase overall more fluid. These observations are compatible with the idea that ASA forms superfluid complexes with lipid molecules that act as defects in the otherwise well-ordered lo phase, as has been proposed by Alsop et al. using neutron diffraction [30]. It has been suggested that these complexes can organize throughout the lo and form a superlattice-type structure in membranes with cholesterol and aspirin. The lattice spacing of this two-dimensional structure was reported as a=21.2 Å , b=18 Å , γ=103°. In addition, lipid–aspirin complexes were observed where aspirin caused local increases in the area per lipid. The authors proposed that ASA interacted with the lo phase to create fluid patches and that these patches organize in the membranes and suppress cholesterol raft formation.
In experiments using highly monochromatic beams, such as synchrotron X-rays, local structure much smaller than the coherence length will be averaged with the surrounding membrane [27,50,51,60]. In a membrane at high coherence length, non-raft regions dominate in a diffraction measurement. By shrinking the coherence length of the X-ray or neutron probe, the scattering signal becomes an incoherent sum of many smaller coherent averages, giving more weight to the smaller scale structure. The longitudinal coherence length of a neutron beam, ξ, is defined by ξ=λ2/Δλ [61]; ξ has been estimated as [50], where E is the energy of the neutron beam, and ΔE the energy resolution. In a typically monochromatic diffraction experiment, ΔE is small, making ξ large. Alsop et al. used a set-up with a low beam monochromaticity in their diffraction experiments to observe small, local structures in membranes with ASA and cholesterol with a coherence length, ξ, in the order of approximately 30 Å . The coherence length in our inelastic experiments is estimated as (with the energy of the incident neutrons of approx. 5 meV measuring an excitation at approx. 1.5 meV). This value is comparable to the above diffraction experiments and also other inelastic neutron experiments in membranes [4]. We, therefore, argue that inelastic neutron scattering experiments can provide information about local structures in membranes.
Details of the molecular structure are assessed using our MD simulations. Lipid molecules in close proximity to ASA molecules were found to be more fluid, showing an increasing area per lipid. ASA and cholesterol molecules are surprisingly well organized, with well-defined nearest-neighbour distances. A snapshot of the structure of the membranes containing cholesterol and ASA from MD simulations is shown in figure 6. Here, the ASA–lipid complexes indeed form superlattices in the membrane plane with dimensions of 15 Å ×25 Å , in very good agreement with the experimentally determined dimensions. The peaks in the cholesterol–ASA RDF at 11.5 Å and 19 Å are roughly positioned at the minima in the ASA–ASA RDF, indicating that the cholesterol prefers to be in the lattice at positions not occupied by ASA.
Figure 6.
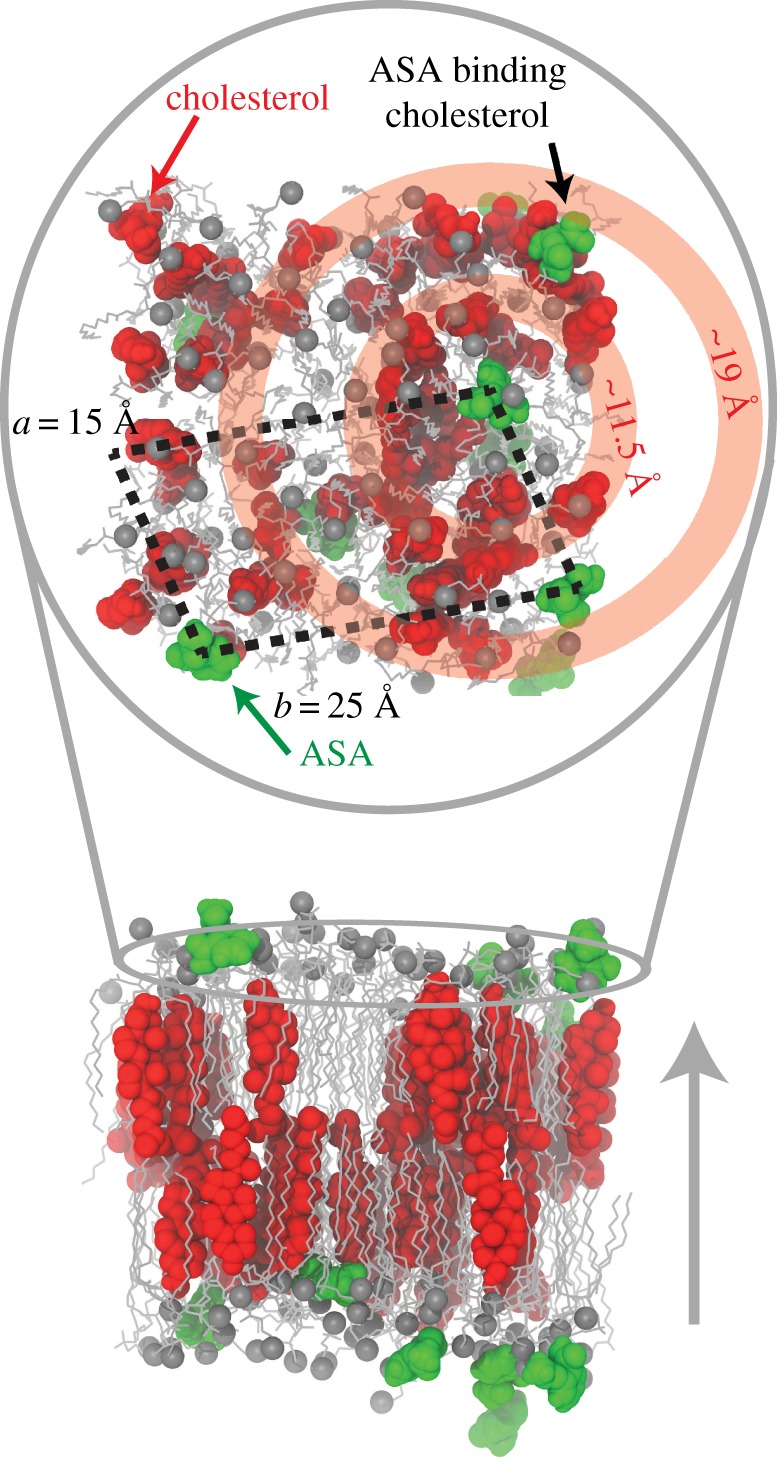
A simulation snapshot that captures the main points of the paper. The upper panel is an overhead view (along qz). The black dashed lines indicate the lateral separation of ASA into a lattice-like structure. The concentric red circles, centred on ASA, illustrate that cholesterol molecules prefer to be at regular distances from ASA.
An interesting question is why aspirin seems to be preferably attracted to lo phases. It has been shown that ASA increases the solubility of cholesterol in lipid membranes [36] and has an effect on cholesterol-rich lipid rafts [30]. Aspirin’s affinity to lo phases has recently also been demonstrated in human red blood cell plasma membranes [38]. This affinity could be entropy-driven, as ASA increases the fluidity of membranes and enhances lipid tail fluctuations. However, our results point to a free energy argument as the MD simulations also show evidence for ASA molecules hydrogen-bonding to cholesterol molecules. To the best of our knowledge, this is the first report of a direct interaction between ASA and cholesterol molecules in lipid bilayers.
4. Conclusion
By combining coherent, inelastic neutron scattering experiments and MD simulations, we investigated the effect of ASA on the liquid-ordered phase in phospholipid membranes. ASA was found to preferably bind to the liquid-ordered, raft-like domains. The dynamical experiments present evidence that ASA makes the lo phase more fluid by creating local defects. From MD simulations, there is evidence that the ASA molecules form superfluid complexes by hydrogen-bonding to lipid molecules, which organize laterally in superlattices and suppress cholesterol’s ordering effect. There is also evidence for ASA molecules directly hydrogen bonding to cholesterol molecules in the bilayers. Our results are in agreement with previous observations in the literature and provide a molecular mode of action for aspirin’s known effect on membrane cholesterol.
5. Material and methods
5.1. Membrane preparation
Highly oriented multi-lamellar stacks of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), cholesterol and ASA were prepared on 1×1 cm, 300 μm thick, single-side polished Si wafers. The coherent scattering of the lipid hydrocarbon chains was enhanced by using tail-deuterated DMPC-d54. A 20 mg ml−1 solution of DMPC-d54 1 : 1 in chloroform and 2,2,2-trifluoroethanol (TFE) was prepared. Solutions of chloroform and ASA were also prepared in TFE : chloroform. DMPC, cholesterol and ASA were then mixed to achieve the desired molecular concentrations.
The Si wafers were cleaned with 30 min sonications in dichloromethane (DCM) at 310 K to remove all organic contamination and leave the substrates in a hydrophobic state. The wafers were then thoroughly rinsed three times using alternating approximately 50 ml of ultrapure water and methanol. The cleaned wafers were placed on a heated sample preparation surface, which was kept at 40°C (313 K). This temperature is above the main phase transition of DMPC, thus the heated substrates ensured that the lipids were in the fluid phase during deposition and the self-assembly of the lipids. An 80 μl aliquot of the lipid solution was deposited on each Si wafer in a tilting incubator, which was set to a speed of 15 rev min−1 and tilt of 10°, such that the lipid solution spread evenly across the wafer. The temperature was kept at 313 K and the solvent was allowed to slowly evaporate for . The wafers were kept in vacuum overnight to remove all traces of the solvent and then incubated with heavy water, D2O, at 313 K for 48 h. Following this protocol, each wafer contained approximately 3000 highly oriented membranes totalling approximately 10 μm in thickness.
Eighteen sample-containing Si wafers were mounted in an aluminium sample holder fabricated to be inserted into IN12’s orange cryostat. A photo of the sample and the aluminium sample holder is shown in figure 7. Hydration of the lipid membranes from the vapour phase was achieved by a D2O reservoir in the bottom of the sample can. The samples were mounted vertically in the neutron beam such that the scattering vector (Q) could either be placed in the plane of the membrane (q∥), or perpendicular to the membrane (qz).
Figure 7.
Photographs of the silicon wafers with highly oriented membranes. Wafers were mounted in the sample holder, custom manufactured for the IN12 cryostat. The water reservoir in the bottom of the can was filled with D2O to ensure hydration of the membranes during the elastic and inelastic scans. Sample can was sealed and mounted in an orange cryostat for temperature control (±0.01°C).
5.2. Neutron scattering
Experiments were conducted using the cold triple-axis spectrometer IN12 at the high flux reactor at the Institut Laue-Langevin in Grenoble, France. Two instrumental set-ups were used: (i) a lower resolution set-up with kf=1.5 Å −1 and λ=4.2 Å , resulting in an energy resolution of approximately 0.3 meV and (ii) a higher resolution set-up with kf=1.1 Å −1 and λ=5.71 Å , resulting in an energy resolution of approximately 0.2 meV . The higher resolution set-up was implemented at q∥=1.37 Å −1 to verify the presence of three phonon peaks. The three axes of the spectrometers refer to the axes of rotation of the monochromator, the sample and the analyser. The incident and final neutron energies are defined by the Bragg reflections from pyrolytic graphite crystals. In-plane and out-of-plane structure can be measured simultaneously on a TAS by simply rotating the sample by 90°.
5.2.1. Elastic scattering
Elastic neutron scattering scans were taken along qz (q∥=0) and q∥ (qz=0) to measure the static structure of the bilayers. Scans along qz (figure 8a) observe a series of Bragg peaks, evenly separated by Δqz, indicating well-ordered and lamellar samples. The lamellar spacing is calculated by dz=2π/Δqz. We obtained dz=61.1 Å for the cholesterol sample and dz=60.6 Å for the ASA sample, which agrees with previous reports of a decrease in dz for membrane samples with ASA [34,36] (table 2).
Table 2.
Structural parameters determined from neutron diffraction.
| sample | dz (Å) | dHH (Å) | AT (Å2) |
|---|---|---|---|
| cholesterol | 61.1 | 46.0 | 23.9 |
| ASA | 60.6 | 45 | 24.4 |
By integrating the lamellar peaks, the neutron SLDs, ρz are calculated using:
| 5.1 |
where In are the integrated peak intensities, qn are the peak positions, and νn are the Fourier phases, which are ±1 for the stacked membrane systems. The phases for both samples are taken from Armstrong et al. as [1 −1 −1 −1 −1 1 1 −1 1] [42]. The resulting densities are shown in figure 8c on a relative scale. Unlike bilayer electron density profiles, neutron densities, for bilayers with deuterated chains, show a minima in the headgroups at |z|∼20 Å . The SLDs are maximum in the tails at |z|∼10 Å and at the edge of the bilayer, |z|=dz/2, where deuterated water and tails contribute. The position of the minima at |z|∼20 Å is used to determine the bilayer head–head spacing, dHH. In the cholesterol sample, dHH was determined to dHH=46 Å and reduced to 45 Å for the ASA sample.
Elastic scans along q∥ measure the in-plane lipid structure, and are displayed in figure 8b. For both samples, a single intense peak is observed, associated with lipid tail correlations, at q∥∼1.37 Å −1. Additional, smaller peaks are observed at q∥∼1.25 Å −1, q∥∼1.7 Å −1 and 1.95 Å −1 associated with higher-order scattering of silicon and aluminium [30].
While the area per lipid is not directly accessible by in-plane scattering, the area per tail is calculated from qT by . AT=23.9 Å 2 for the cholesterol sample and AT=24.2 Å 2 for the ASA sample, a slight increase.
5.2.2. Fitting the inelastic spectra
The constant-q∥ scans were well fit by analytical functions described by three contributions: the energy resolution of the spectrometer described by a Gaussian peak centred at . The corresponding Gaussian width was calculated using the ResLib software using the geometry of the IN12 spectrometer [62]. Secondly, a Lorentzian peak, arising from incoherent molecular motions in the sample, centred at . The remaining scattering contributions were fit by three phonon peaks, positioned at . The width of these peaks is proportional to the damping experienced by the collective motions. The overall fitted analytic function also includes a linear background and can be written as
| 5.2 |
Fixed values were used for the position and width of the experimental resolution. The position of the incoherent peak was fixed to . The ratio of the amplitudes A+ and A− was determined by detailed balance.
A comparison between fits using one, two and three phonon excitations is shown in figure 9. Based on the determined χ2-value, the three-phonon fit was found to best describe the experimental spectra. However, the inclusion of additional parameters into a fit function may lead to an improved fit without statistical significance. The quality of the model fit was, therefore, also studied using an F-test. The data at a q∥-value of q∥=1.37 Å −1 were fitted using one, two and three phonon peaks successively. χ2 was determined for each fit as shown in figure 9. The Fχ-value was calculated for every fit [63]:
| 5.3 |
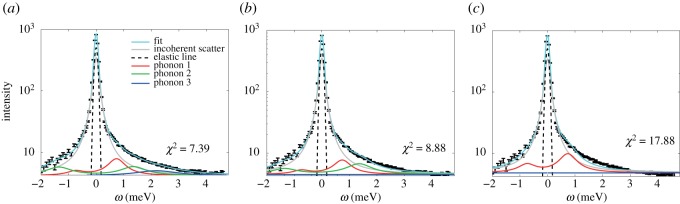
Figure 9.
Inelastic scans measured at a q∥-value of q∥=1.37 Å −1. The spectra can be fitted using different models: all models contain an elastic line, centred at energy transfer 0 (described by a Gaussian peak profile) and a Lorentzian peak also centred at ‘0’ to account for the incoherent scattering. (a) A fit including three phonon excitations, which shows a good agreement with the measured data. It was also attempted to fit the spectra with two excitations, and one excitation only (c). By comparing the resulting χ2-values, the fit with three phonons provides the best model to the data. This is also supported by the results of an F-test in table 3.
where m is the number of parameter used to fit N data points. The data included 108 data points. There are six parameters used to fit the background (two for the elastic peak, two for the inelastic peak and two for a linear background) and four parameters for every additional phonon signal. The calculated Fχ-values are listed in table 3. According to [63] the additional parameters are valid if the Fχ-value exceeds a critical F-value, which are listed in tables and were taken from [64]. Based on this test, the three-phonon fit indeed best describes the experimental data.
Table 3.
Fχ-values for for phonon fits when adding additional phonon peaks. The Fχ-values increase by including two and eventually three-phonon excitations and exceed the F-value indicating that the inclusion of the additional parameters improves description of the experimental data. F-values were taken from database [64].
| Fχ | F(2.5%) | |
|---|---|---|
| adding second phonon | 94.25 | 6.41 |
| adding third phonon | 17.94 | 6.04 |
5.2.3. Resolution considerations
The phonon peak width in the constant-q∥ energy scans is associated with phonon damping in the damped harmonic oscillator model. However, the width of the peak is not just a sample feature, but is convoluted with the instrumental resolution function. For a triple-axis spectrometer, this resolution function takes the form of a tilted elipsoid in the energy–momentum plane, where any excitation within the ellipsoid is recorded. A cartoon of the elipsoid on a lipid dispersion is in figure 10. When this ellipsoid intersects a region on the dispersion with high slope, the effect is to make peaks appear broader. Therefore, at the minima in the dispersion, where the slope is zero, a constant-q∥ scan would produce a peak width closest to the true width.
5.3. Molecular dynamics simulations
All simulations were run in-house on MacSim, a GPU-accelerated workstation with 20 physical Intel Xeon CPU cores and two GeForce GTX 1080 high-power graphics cards resulting in 5120 CUDA cores. This system produces about 180 ns per day of MD simulations in standard 128 lipid membrane patches in GROMACS.
A united-atom, DMPC + 30 mol% cholesterol bilayer with 142 lipid + cholesterol molecules, was obtained from Hub et al. [53]. Aspirin topology was obtained using the Automated Force Field Topology Builder (ATB) [65,66]. The SPC water model was used for system solvation [67]. All MD simulations were performed using the GROMACS 5.1.2 software package [68], implementing the GROMOS 54a7 force field [69] modified with Berger lipid parameters [70]. All simulations used a 2 fs time step, a periodic boundary condition applied to all directions, the particle-mesh Ewald to solve for long-range electrostatics [71], a short-range van der Waals cut-off of 1.2 nm and the LINCS algorithm to determine bond constraints [72]. Temperature coupling was controlled using a Nose-Hoover thermostat at 28°C (τ=0.5 ps) [73], and pressure was kept at 1.0 bar using Parrinello–Rahman semi-isotropic weak coupling (τ=1 ps) [74].
A total of two distinct simulations were conducted. First, the DMPC + cholesterol system was equilibrated with 25 waters per lipid for 200 ns. Next, the bilayer system with 10 mol% aspirin was prepared in a three-step process. First, all water in the system was removed, and the aspirin added to the now empty space outside the bilayer. Second, all water (25 per lipid) was replaced in the system. Finally, the system was re-equilibrated and then simulated for 200 ns. All analyses were performed with the final 50 ns of the simulations using GROMACS algorithms and simple scripts [75]. The electron density profiles were calculated for different constituents of the system. The function used calculates the relative distance along the bilayer normal of each atom within the specified index group, assigns a weighting based upon the number of electrons in each atom, and delivers an electron density as averaged over the specified time range.
The proportion of gauche dihedrals within a lipid system is commonly used as a measure of bilayer fluidity [76–78]. The proportion of gauche dihedrals as a function of increasing distance from aspirin was determined using dynamic scripting and GROMACS algorithms. A script was constructed to generate an index file containing only carbon chains belonging to lipids within the specified radius from the centre of mass of any aspirin molecule within the system every 50 frames. This index file specified the DMPC molecules whose carbons were to be used in calculation of the Ryckaert–Bellemans dihedral angles over that time interval. This was repeated over the final 50 ns of the simulation and averaged for each carbon position. Averaging across the SN1 and SN2 tails was then performed to generate the value shown in figure 5d, and the script was run successively to consider each new distance from aspirin.
Supplementary Material
Data accessibility
Inelastic neutron scattering data are provided as electronic supplementary material.
Authors' contributions
R.J.A. conceived and designed the study, prepared membrane samples for the neutron experiments, conducted the experiment, analysed the data, conducted and analysed the MD simulations, and wrote the manuscript. S.H. collected and analysed data. A.D. conducted the MD simulations. K.S. conceived of the study, coordinated and conducted the neutron experiment, and analysed data. M.C.R. conceived of the study, designed the study, coordinated the study, analysed data and wrote the manuscript. All the authors gave their final approval for publication.
Competing interests
The authors declare no competing interests.
Funding
This research was funded by the Natural Sciences and Engineering Research Council of Canada (NSERC), the National Research Council Canada (NRC), the Canada Foundation for Innovation (CFI) and the Ontario Ministry of Economic Development and Innovation. R.J.A. is the recipient of an NSERC PGS-D scholarship. M.C.R. is the recipient of an Early Researcher Award of the Province of Ontario and a University Scholar Award from McMaster University.
References
- 1.Singer SJ, Nicolson GL. 1972. The fluid mosaic model of the structure of cell membranes. Science 175, 720–731. [DOI] [PubMed] [Google Scholar]
- 2.Callan-Jones A, Sorre B, Bassereau P. 2011. Curvature-driven lipid sorting in biomembranes. Cold Spring Harb. Perspect. Biol. 3, a004648 (doi:10.1101/cshperspect.a004648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falck E, Róg T, Karttunen M, Vattulainen I. 2008. Lateral diffusion in lipid membranes through collective flows. J. Am. Chem. Soc. 130, 44–45. (doi:10.1021/ja7103558) [DOI] [PubMed] [Google Scholar]
- 4.Rheinstädter MC, Das J, Flenner EJ, Brüning B, Seydel T, Kosztin I. 2008. Motional coherence in fluid phospholipid membranes. Phys. Rev. Lett. 101, 248106 (doi:10.1103/PhysRevLett.101.248106) [DOI] [PubMed] [Google Scholar]
- 5.Busch S, Smuda C, Pardo LC, Unruh T. 2010. Molecular mechanism of long-range diffusion in phospholipid membranes studied by quasielastic neutron scattering. J. Am. Chem. Soc. 132, 3232–3233. (doi:10.1021/ja907581s) [DOI] [PubMed] [Google Scholar]
- 6.Armstrong CL, Trapp M, Peters J, Seydel T, Rheinstädter MC. 2011. Short range ballistic motion in fluid lipid bilayers studied by quasi-elastic neutron scattering. Soft Matter 7, 8358–8362. (doi:10.1039/c1sm05691c) [Google Scholar]
- 7.Armstrong CL, Toppozini L, Dies H, Faraone A, Nagao M, Rheinstädter MC. 2013. Incoherent neutron spin-echo spectroscopy as an option to study long-range lipid diffusion. ISRN Biophys. 2013, 439758 (doi:10.1155/2013/439758) [Google Scholar]
- 8.Paula S, Volkov AG, Van Hoek AN, Haines TH, Deamer DW. 1996. Permeation of protons, potassium ions, and small polar molecules through phospholipid bilayers as a function of membrane thickness. Biophys. J. 70, 339–348. (doi:10.1016/S0006-3495(96)79575-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rheinstädter MC, Ollinger C, Fragneto G, Demmel F, Salditt T. 2004. Collective dynamics of lipid membranes studied by inelastic neutron scattering. Phys. Rev. Lett. 93, 108107 (doi:10.1103/PhysRevLett.93.108107) [DOI] [PubMed] [Google Scholar]
- 10.Kaye MD, Schmalzl K, Nibali VC, Tarek M, Rheinstädter MC. 2011. Ethanol enhances collective dynamics of lipid membranes. Phys. Rev. E 83, 050907 (doi:10.1103/PhysRevE.83.050907) [DOI] [PubMed] [Google Scholar]
- 11.Zhernenkov M, Bolmatov D, Soloviov D, Zhernenkov K, Toperverg BP, Cunsolo A, Bosak A, Cai YQ. 2016. Revealing the mechanism of passive transport in lipid bilayers via phonon-mediated nanometre-scale density fluctuations. Nat. Commun. 7, 11575 (doi:10.1038/ncomms11575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alsop RJ, Rheinstädter MC. 2016. Lipid rafts in binary lipid/cholesterol bilayers. In Membrane organization and lipid rafts in the cell and artificial membranes (ed. A Catalá). Series on Cell Biology Research Progress, pp. 17–42. Hauppauge, NY: Nova. ISBN:978-1-63484-589-2.
- 13.Lingwood D, Simons K. 2010. Lipid rafts as a membrane-organizing principle. Science 327, 46–50. (doi:10.1126/science.1174621) [DOI] [PubMed] [Google Scholar]
- 14.Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387, 569–572. (doi:10.1038/42408) [DOI] [PubMed] [Google Scholar]
- 15.Simons K, Gerl MJ. 2010. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11, 688–699. (doi:10.1038/nrm2977) [DOI] [PubMed] [Google Scholar]
- 16.Yeagle PL. 1985. Lanosterol and cholesterol have different effects on phospholipid acyl chain ordering. Biochim. Biophys. Acta (BBA) 815, 33–36. (doi:10.1016/0005-2736(85)90470-5) [DOI] [PubMed] [Google Scholar]
- 17.Mouritsen OG, Jørgensen K. 1994. Dynamical order and disorder in lipid bilayers. Chem. Phys. Lipids 73, 3–25. (doi:10.1016/0009-3084(94)90171-6) [DOI] [PubMed] [Google Scholar]
- 18.Mouritsen OG, Jørgensen K. 1997. Small-scale lipid-membrane structure: simulation versus experiment. Curr. Opin. Struct. Biol. 7, 518–527. (doi:10.1016/S0959-440X(97)80116-9) [DOI] [PubMed] [Google Scholar]
- 19.Pitman MC, Suits F, MacKerell AD, Feller SE. 2004. Molecular-level organization of saturated and polyunsaturated fatty acids in a phosphatidylcholine bilayer containing cholesterol. Biochemistry 43, 15 318–15 328. (doi:10.1021/bi048231w) [DOI] [PubMed] [Google Scholar]
- 20.Mouritsen OG, Zuckermann MJ. 2004. What’s so special about cholesterol? Lipids 39, 1101–1113. (doi:10.1007/s11745-004-1336-x) [DOI] [PubMed] [Google Scholar]
- 21.Jacobson K, Mouritsen OG, Anderson RGW. 2007. Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 9, 7–14. (doi:10.1038/ncb0107-7) [DOI] [PubMed] [Google Scholar]
- 22.Róg T, Pasenkiewicz-Gierula M, Vattulainen I, Karttunen M. 2009. Ordering effects of cholesterol and its analogues. Biochim. Biophys. Acta (BBA) 1788, 97–121. (doi:10.1016/j.bbamem.2008.08.022) [DOI] [PubMed] [Google Scholar]
- 23.Pike LJ. 2009. The challenge of lipid rafts. J. Lipid Res. 50, S323–S328. (doi:10.1194/jlr.R800040-JLR200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eggeling C. et al. 2009. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature 457, 1159–1162. (doi:10.1038/nature07596) [DOI] [PubMed] [Google Scholar]
- 25.Mouritsen OG. 2010. The liquid-ordered state comes of age. Biochim. Biophys. Acta (BBA) 1798, 1286–1288. (doi:10.1016/j.bbamem.2010.02.017) [DOI] [PubMed] [Google Scholar]
- 26.Rheinstädter MC, Mouritsen OG. 2013. Small-scale structure in fluid cholesterol–lipid bilayers. Curr. Opin. Colloid Interface Sci. 18, 440–447. (doi:10.1016/j.cocis.2013.07.001) [Google Scholar]
- 27.Toppozini L, Meinhardt S, Armstrong CL, Yamani Z, Kučerka N, Schmid F, Rheinstädter MC. 2014. Structure of cholesterol in lipid rafts. Phys. Rev. Lett. 113, 228101 (doi:10.1103/PhysRevLett.113.228101) [DOI] [PubMed] [Google Scholar]
- 28.Armstrong CL, Häußler W, Seydel T, Katsaras J, Rheinstädter MC. 2014. Nanosecond lipid dynamics in membranes containing cholesterol. Soft Matter 10, 2600–2611. (doi:10.1039/c3sm51757h) [DOI] [PubMed] [Google Scholar]
- 29.Nunes C, Brezesinski G, Pereira-Leite C, Lima JLFC, Reis S, Lúcio M. 2011. NSAIDs interactions with membranes: a biophysical approach. Langmuir 27, 10 847–10 858. (doi:10.1021/la201600y) [DOI] [PubMed] [Google Scholar]
- 30.Alsop RJ, Toppozini L, Marquardt D, Kučerka N, Harroun TA, Rheinstädter MC. 2015. Aspirin inhibits formation of cholesterol rafts in fluid lipid membranes. Biochim. Biophys. Acta (BBA) 1848, 805–812. (doi:10.1016/j.bbamem.2014.11.023) [DOI] [PubMed] [Google Scholar]
- 31.Dies H, Cheung B, Tang J, Rheinstädter MC. 2015. The organization of melatonin in lipid membranes. Biochim. Biophys. Acta (BBA) 1848, 1032–1040. (doi:10.1016/j.bbamem.2015.01.006) [DOI] [PubMed] [Google Scholar]
- 32.Schmidt A, Lenzig P, Oslender-Bujotzek A, Kusch J, Lucas SD, Gründer S, Wiemuth D. 2014. The bile acid-sensitive ion channel (BASIC) is activated by alterations of its membrane environment. PloS One 9, e111549 (doi:10.1371/journal.pone.0111549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundbæk JA, Koeppe RE, Andersen OS. 2010. Amphiphile regulation of ion channel function by changes in the bilayer spring constant. Proc. Natl Acad. Sci. USA 107, 15 427–15 430. (doi:10.1073/pnas.1007455107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett MA, Zheng S, Roshankar G, Alsop RJ, Belanger RKR, Huynh C, Kučerka N, Rheinstädter MC. 2012. Interaction of aspirin (acetylsalicylic acid) with lipid membranes. PLoS ONE 7, e34357 (doi:10.1371/journal.pone.0034357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasedkin A, Davidsson J, Kumpugdee-Vollrath M. 2013. Determination of nanostructure of liposomes containing two model drugs by X-ray scattering from a synchrotron source. J. Synchrotron. Radiat. 20, 721–728. (doi:10.1107/S0909049513020074) [DOI] [PubMed] [Google Scholar]
- 36.Alsop RJ, Barrett MA, Zheng S, Dies H, Rheinstädter MC. 2014. Acetylsalicylic acid (ASA) increases the solubility of cholesterol when incorporated in lipid membranes. Soft Matter 10, 4275–4286. (doi:10.1039/C4SM00372A) [DOI] [PubMed] [Google Scholar]
- 37.Lichtenberger LM, Zhou Y, Dial EJ, Raphael RM. 2006. NSAID injury to the gastrointestinal tract: evidence that NSAIDs interact with phospholipids to weaken the hydrophobic surface barrier and induce the formation of unstable pores in membranes. J. Pharmacy Pharmacol. 58, 1421–1428. (doi:10.1211/jpp.58.10.0001) [DOI] [PubMed] [Google Scholar]
- 38.Himbert S. et al. 2017. The molecular structure of human red blood cell membranes from highly oriented, solid supported multi-lamellar membranes. Sci. Rep. 7, 39661 (doi:10.1038/srep39661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma VK, Mamontov E, Ohl M, Tyagi M. 2017. Incorporation of aspirin modulates the dynamical and phase behavior of the phospholipid membrane. Phys. Chem. Chem. Phys. 19, 2514–2524. (doi:10.1039/C6CP06202D) [DOI] [PubMed] [Google Scholar]
- 40.Barrett MA, Zheng S, Toppozini LA, Alsop RJ, Dies H, Wang A, Jago N, Moore M, Rheinstädter MC. 2013. Solubility of cholesterol in lipid membranes and the formation of immiscible cholesterol plaques at high cholesterol concentrations. Soft Matter 9, 9342–9351. (doi:10.1039/c3sm50700a) [Google Scholar]
- 41.Alsop RJ, Armstrong CL, Maqbool A, Toppozini L, Dies H, Rheinstädter MC. 2015. Cholesterol expels ibuprofen from the hydrophobic membrane core and stabilizes lamellar phases in lipid membranes containing ibuprofen. Soft Matter 11, 4756–4767. (doi:10.1039/C5SM00597C) [DOI] [PubMed] [Google Scholar]
- 42.Armstrong CL, Barrett MA, Hiess A, Salditt T, Katsaras J, Shi A-C, Rheinstädter MC. 2012. Effect of cholesterol on the lateral nanoscale dynamics of fluid membranes. Eur. Biophys. J. 41, 901–913. (doi:10.1007/s00249-012-0826-4) [DOI] [PubMed] [Google Scholar]
- 43.Chen SH, Liao CY, Huang HW, Weiss TM, Bellisent-Funel MC, Sette F. 2001. Collective dynamics in fully hydrated phospholipid bilayers studied by inelastic X-ray scattering. Phys. Rev. Lett. 86, 740 (doi:10.1103/PhysRevLett.86.740) [DOI] [PubMed] [Google Scholar]
- 44.Weiss TM, Chen P-J, Sinn H, Alp EE, Chen SH, Huang HW. 2003. Collective chain dynamics in lipid bilayers by inelastic X-ray scattering. Biophys. J. 84, 3767–3776. (doi:10.1016/S0006-3495(03)75105-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen P-J, Liu Y, Weiss TM, Huang HW, Sinn H, Alp EE, Alatas A, Said A, Chen S-H. 2003. Studies of short-wavelength collective molecular motions in lipid bilayers using high resolution inelastic X-ray scattering. Biophys. Chem. 105, 721–741. (doi:10.1016/S0301-4622(03)00099-1) [DOI] [PubMed] [Google Scholar]
- 46.Hub JS, Salditt T, Rheinstädter MC, De Groot BL. 2007. Short-range order and collective dynamics of dmpc bilayers: a comparison between molecular dynamics simulations, X-ray, and neutron scattering experiments. Biophys. J. 93, 3156–3168. (doi:10.1529/biophysj.107.104885) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brüning B, Rheinstädter MC, Hiess A, Weinhausen B, Reusch T, Aeffner S, Salditt T. 2010. Influence of cholesterol on the collective dynamics of the phospholipid acyl chains in model membranes. Eur. Phys. J. E: Soft Matter Biol. Phys. 31, 419–428. (doi:10.1140/epje/i2010-10574-6) [DOI] [PubMed] [Google Scholar]
- 48.Tarek M, Tobias DJ, Chen S-H, Klein ML. 2001. Short wavelength collective dynamics in phospholipid bilayers: a molecular dynamics study. Phys. Rev. Lett. 87, 238101 (doi:10.1103/PhysRevLett.87.238101) [DOI] [PubMed] [Google Scholar]
- 49.Nibali VC, D’Angelo G, Tarek M. 2014. Molecular dynamics simulation of short-wavelength collective dynamics of phospholipid membranes. Phys. Rev. E 89, 050301 (doi:10.1103/PhysRevE.89.050301) [DOI] [PubMed] [Google Scholar]
- 50.Armstrong CL, Marquardt D, Dies H, Kučerka N, Yamani Z, Harroun TA, Katsaras J, Shi A-C, Rheinstädter MC. 2013. The observation of highly ordered domains in membranes with cholesterol. PLoS One 8, e66162 (doi:10.1371/journal.pone.0066162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Armstrong CL, Barrett MA, Toppozini L, Kučerka N, Yamani Z, Katsaras J, Fragneto G, Rheinstädter MC. 2012. Co-existence of gel and fluid lipid domains in single-component phospholipid membranes. Soft Matter 8, 4687–4694. (doi:10.1039/c2sm07158d) [Google Scholar]
- 52.Khondker A, Dhaliwal A, Alsop RJ, Tang J, Backholm M, Shi A-C, Rheinstädter MC. 2017. Partitioning of caffeine in lipid bilayers reduces membrane fluidity and increases membrane thickness. Phys. Chem. Chem. Phys. 19, 7101–7111. (doi:10.1039/c6cp08104e) [DOI] [PubMed] [Google Scholar]
- 53.Wennberg CL, Van Der Spoel D, Hub JS. 2012. Large influence of cholesterol on solute partitioning into lipid membranes. J. Am. Chem. Soc. 134, 5351–5361. (doi:10.1021/ja211929h) [DOI] [PubMed] [Google Scholar]
- 54.Allen WJ, Lemkul JA, Bevan DR. 2009. Gridmat-md: A grid-based membrane analysis tool for use with molecular dynamics. J. Comput. Chem. 30, 1952–1958. (doi:10.1002/jcc.21172) [DOI] [PubMed] [Google Scholar]
- 55.Vogel A, Roark M, Feller SE. 2012. A reinterpretation of neutron scattering experiments on a lipidated Ras peptide using replica exchange molecular dynamics. Biochim. Biophys. Acta (BBA) 1818, 219–224. (doi:10.1016/j.bbamem.2011.08.016) [DOI] [PubMed] [Google Scholar]
- 56.Kučerka N, Gallová J, Uhríková D, Balgavỳ P, Bulacu M, Marrink S-J, Katsaras J. 2009. Areas of monounsaturated diacylphosphatidylcholines. Biophys. J. 97, 1926–1932. (doi:10.1016/j.bpj.2009.06.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hofsäß C, Lindahl E, Edholm O. 2003. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys. J. 84, 2192–2206. (doi:10.1016/S0006-3495(03)75025-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suwalsky M, Belmar J, Villena F, Gallardo MJ, Jemiola-Rzeminska M, Strzalka K. 2013. Acetylsalicylic acid (aspirin) and salicylic acid interaction with the human erythrocyte membrane bilayer induce in vitro changes in the morphology of erythrocytes. Arch. Biochem. Biophys. 539, 9–19. (doi:10.1016/j.abb.2013.09.006) [DOI] [PubMed] [Google Scholar]
- 59.Zhou Y, Cho K-J, Plowman SJ, Hancock JF. 2012. Nonsteroidal anti-inflammatory drugs alter the spatiotemporal organization of Ras proteins on the plasma membrane. J. Biol. Chem. 287, 16 586–16 595. (doi:10.1074/jbc.M112.348490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Majkrzak CF, Metting C, Maranville BB, Dura JA, Satija S, Udovic T, Berk NF. 2014. Determination of the effective transverse coherence of the neutron wave packet as employed in reflectivity investigations of condensed-matter structures. I. Measurements. Phys. Rev. A 89, 033851. [Google Scholar]
- 61.Rauch H. 1993. Reality in neutron interference experiments. Found. Phys. 23, 7–36. (doi:10.1007/BF01883988) [DOI] [PubMed] [Google Scholar]
- 62.Zheludev A. 2007. Reslib 3.4: 3-axis resolution library for Matlab. In Neutron Scattering Sciences Division, vol. 37831. Oak Ridge, TN: Oak Ridge National Laboratory. http://www.neutron.ethz.ch/research/resources/reslib.
- 63.Bevington P, Robinson DK. 2002. Data reduction and error analysis for the physical sciences. New York, NY: Mcgraw Hill Book Co; See http://www.ebook.de/de/product/3078650/philip_bevington_d_keith_robinson_data_reduction_and_error_analysis_for_the_physical_sciences.html. [Google Scholar]
- 64.Statistics Online Computational Resource (SOCR). 2017. F distribution tables. See http://www.socr.ucla.edu/applets.dir/f_table.html#FTable0.025 (Online; accessed 19 December 2017).
- 65.Koziara KB, Stroet M, Malde AK, Mark AE. 2014. Testing and validation of the automated topology builder (ATB) version 2.0: prediction of hydration free enthalpies. J. Comput. Aided Mol. Des. 28, 221–233. (doi:10.1007/s10822-014-9713-7) [DOI] [PubMed] [Google Scholar]
- 66.Malde AK, Zuo L, Breeze M, Stroet M, Poger D, Nair PC, Oostenbrink C, Mark AE. 2011. An automated force field topology builder (ATB) and repository: version 1.0. J. Chem. Theory Comput. 7, 4026–4037. (doi:10.1021/ct200196m) [DOI] [PubMed] [Google Scholar]
- 67.Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. 1981. Interaction models for water in relation to protein hydration. In Intermolecular forces, pp. 331–342. Dordrecht, The Netherlands: Springer.
- 68.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E. 2015. Gromacs: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25. (doi:10.1016/j.softx.2015.06.001) [Google Scholar]
- 69.Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, van Gunsteren WF. 2011. Definition and testing of the GROMOS force-field versions 54a7 and 54b7. Eur. Biophys. J. 40, 843–856. (doi:10.1007/s00249-011-0700-9) [DOI] [PubMed] [Google Scholar]
- 70.Berger O, Edholm O, Jähnig F. 1997. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72, 2002–2013. (doi:10.1016/S0006-3495(97)78845-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Darden T, York D, Pedersen L. 1993. Particle mesh Ewald: an method for Ewald sums in large systems. J. Chem. Phys. 98, 10 089–10 092. (doi:10.1063/1.464397) [Google Scholar]
- 72.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. 1997. Lincs: a linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472. (doi:10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H) [Google Scholar]
- 73.Evans DJ, Holian BL. 1985. The Nose–Hoover thermostat. J. Chem. Phys. 83, 4069–4074. (doi:10.1063/1.449071) [Google Scholar]
- 74.Parrinello M, Rahman A. 1981. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190. (doi:10.1063/1.328693) [Google Scholar]
- 75.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. 2005. Gromacs: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718. (doi:10.1002/jcc.20291) [DOI] [PubMed] [Google Scholar]
- 76.Saito H, Shinoda W. 2011. Cholesterol effect on water permeability through DPPC and PSM lipid bilayers: a molecular dynamics study. J. Phys. Chem. B 115, 15 241–15 250. (doi:10.1021/jp201611p) [DOI] [PubMed] [Google Scholar]
- 77.Hofsäß C, Lindahl E, Edholm O. 2003. Molecular dynamics simulations of phospholipid bilayers with cholesterol. Biophys. J. 84, 2192–2206. (doi:10.1016/S0006-3495(03)75025-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Venable RM, Brooks BR, Pastor RW. 2000. Molecular dynamics simulations of gel (LβI) phase lipid bilayers in constant pressure and constant surface area ensembles. J. Chem. Phys. 112, 4822–4832. (doi:10.1063/1.481085) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Inelastic neutron scattering data are provided as electronic supplementary material.