To the Editor: The development of gut microbiota in the early years of life is essential for immune function and protection against pathogens in humans.[1,2] Stool sample is often the first choice for analysis of the gut microbiota. The dominant five phyla observed in stool samples have been Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Fusobacteria, ranked in the order of abundance. However, it might be difficult to obtain the fresh stool sample from infants and young children, especially when they are diseased. In this study, we aimed to evaluate if the rectal swab sample collected from children with hand, foot, and mouth disease (HFMD) can be an alternative for microbiome study using 16S rRNA gene sequencing technology.
This study was approved by the Medical Ethics Committee of Beijing You'an Hospital, Capital Medical University, Beijing, China. The study was conducted from May 2015 to June 2016. During this period, 42 patients with diagnosed HFMD were recruited at the Department of Infectious Diseases, Beijing You'an Hospital. Clinical diagnosis of HFMD was made by an experienced physician according to the HFMD Diagnosis and Treatment Guidelines (2010) issued by National Health and Family Planning Commission of People's Republic of China. Thirty patients were also confirmed by the detection of enteroviruses in their nasopharyngeal swab or stool samples by polymerase chain reaction (PCR) and/or culture. The patient age ranged from 3 months to 4.4 years (median: 2.0 years; male/female: 22:20). All patients were Chinese Han nationality. Patients were excluded if they had symptoms of other respiratory infection, digestive tract disease, or had a history of treatment with antibiotics or anti-inflammatory agents in the past 2 weeks.
Fifty swabs were collected from 42 patients. Eight patients also had paired swabs collected in acute and convalescent phases (sampling interval: >7 days between the two samples). Rectal swab was collected in a sterile flask container by perianal stimulation. To minimize the possible effect of sampling and sequencing, three parallel swabs were obtained from each patient and were mixed for DNA extraction. Samples were transferred to the laboratory within 30 min and stored at −80°C.
Genome DNAs were extracted using the Qiagen Stool Mini Kit according to manufacturer's instructions (Hilden, Germany) and diluted to 1 ng/μl using sterile water. The V3–V5 regions were amplified using the specific primers with the barcode, sequenced, and analyzed to define composition of the bacterial community. Sequencing libraries were generated using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) and index codes were added. The library was sequenced on an Illumina HiSeq 2500 platform and 250 bp paired-end reads were generated. Sequences with more than 97% similarity were assigned to the same operational taxonomic units (OTUs). Representative sequence for each OTU was screened for further annotation. Abundance of OTUs was normalized using a standard sequence number corresponding to the sample with the least sequences.
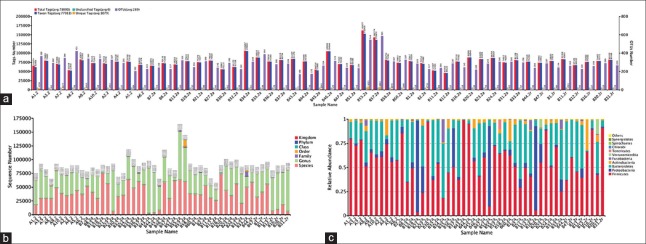
Figure 1 shows OTUs cluster and species annotation, sequence numbers classified into different level taxa and comparison of bacterial community composition at the phylum. A total of 3,904,495 tags were generated and clustered into 14,438 OTUs. Of them, 97.3% of the tags were classified into genus-level taxa and 48.0% into species-level taxa [Figure 1b]. For each sample, 78,090 tags were generated. Of them, 77,011 taxon tags were clustered into an average of 289 OTUs [Figure 1a]. Further, all (100%) of the OTUs could be classified and the average unique tags identified was 1079.
Figure 1.
Composition of bacterial microbiome identified in 50 rectal swabs collected from Chinese children with diagnosed hand, foot, and mouth disease. (a) OTUs cluster and species annotation. Total tags (red column) represented the total number of effective tags for OTUs clustered, taxon tags (blue column) the tags number for OTUs construction and annotation, unclassified tags (green column) the tags number for unannotated OTUs, unique tags (yellow column) the tags numbers for unclustered OTUs, and OTUs numbers (the purple column) were acquired OTUs number for each sample. Horizontal axis indicated the individual sample. (b) Sequence numbers classified into different level taxa. Horizontal axis indicated the individual sample. Vertical axis indicated the sequence numbers obtained. Different color indicated classified different taxa. (c) Comparison of microbial community composition at the phylum for each sample. Only the top 10 most abundant phylum are shown for demonstration and clarity. Horizontal axis indicated the individual sample. OTUs: Operational taxonomic units.
At the phylum level, Firmicutes (mean relative abundance, 58.6%), Bacteroidetes (29.2%), Proteobacteria (6.1%), Actinobacteria (4.2%), and Fusobacteria (0.9%) comprised the vast majority (99.0%) of gut microbiota in the study participants [Figure 1c]. At family level, the gut microbiota was generally dominated by Clostridiales family XI Incertae sedis (mean relative abundance 21.8%, with representative genus Ezakiella and Finegoldia), Enterococcaceae (16.6%, representative genus Enterococcus), Prevotellaceae (15.2%, representative genus Prevotella), Bacteroidaceae (6.8%, representative genus Bacteroides), and Enterobacteriaceae (3.8%, representative genus Escherichia/Shigella).
Understanding the link between the intestinal microbiota and health has the potential to improve patient care in the context of infection prevention.[1,2] Optimizing study feasibility without altering results is critical for research on the gut microbiota in both out- and inpatients. The ongoing project “Impact of gut microbiota on severity of HFMD in children” provided us a unique opportunity to evaluate whether the rectal swabbing from child patients can be an alternative for microbiome study using 16S rRNA gene sequencing technology.
Feces have been widely used for the analysis of intestinal microbiome. Rectal swabbing is a relatively simple means to collect fecal sample among patients hospitalized or at home. It requires no special preparation and can be transported easily from bedside to laboratory. For children and pediatric patients, rectal swabs are more convenient to be collected by parents or nurses. Bassis et al.[3] recently performed the comparison of stool versus rectal swab samples for bacterial community profiles using 16S rRNA gene sequencing in eight adult patients and found that the stool and rectal swab microbiotas from the same participant were highly similar. Kuang et al.[4] used stool samples to study gut microbiota in 29 healthy Chinese infants and obtained a total of 6529 OTUs, with an average of 225 OTUs per sample. The dominant four phyla (Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria) identified comprised over 99% of bacterial community. Using stool samples, Qi et al.[5] recently compared gut microbiota between Chinese children with type 1 diabetes and healthy children (n = 15 for each group). The average OTUs obtained was 132.60 ± 32.29 for controls and 121.53 ± 36.06 for patients. There are certain limitations in this study. First, the number of patients included was limited. Second, parallel stool sample was not available. Third, no sample was obtained from healthy children. In our study, however, we found that both number of (average) OTUs and composition of bacterial communities identified in rectal swabs were rather similar to those reported in stool and rectal swab samples. Our preliminary result suggests that rectal swabs are an acceptable and practical proxy for the collection of fecal specimens for analysis of the intestinal microbiome in children. Further studies in which a large number of participants with parallel stool and rectal swabs are needed to confirm the finding.
Declaration of patient consent
The author certifies that they have obtained all appreciate patient consent forms. In the form, the patient/patient's guardians have given their consent for their images and other clinical information to be reported in the journal. The patient/patient's guardians understand that their names and initials will not be published and due efforts will be made to conceal their identity, but anonymity cannot be guaranteed.
Financial support and sponsorship
This study was supported by the grants from the Basic-clinical Research Cooperation Funding (No. 15JL13), the Excellent Youth Teacher Exchange Program (No. 0700-1160950157) of Capital Medical University, the Collaborative Innovation Center of Infectious Diseases, Capital Medical University (No. PXM 2016-014226-000052) and the Beijing Excellent Researcher Award Program (No. 2016000020124G105).
Conflicts of interest
There are no conflicts of interest.
Footnotes
Edited by: Ning-Ning Wang
REFERENCES
- 1.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–79. doi: 10.1056/NEJMra1600266. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 2.Zhang N, He QS. Commensal microbiome promotes resistance to local and systemic infections. Chin Med J. 2015;128:2250–5. doi: 10.4103/0366-6999.162502. doi: 10.4103/0366-6999.162502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassis CM, Moore NM, Lolans K, Seekatz AM, Weinstein RA, Young VB, et al. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017;17:78. doi: 10.1186/s12866-017-0983-9. doi: 10.1186/s12866-017-0983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuang YS, Li SH, Guo Y, Lu JH, He JR, Luo BJ, et al. Composition of gut microbiota in infants in China and global comparison. Sci Rep. 2016;6:36666. doi: 10.1038/srep36666. doi: 10.1038/srep36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi CJ, Zhang Q, Yu M, Xu JP, Zheng J, Wang T, et al. Imbalance of fecal microbiota at newly diagnosed type 1 diabetes in Chinese children. Chin Med J. 2016;129:1298–304. doi: 10.4103/0366-6999.182841. doi: 10.4103/0366-6999.182841. [DOI] [PMC free article] [PubMed] [Google Scholar]