Abstract
Activating Ras mutations are associated with ∼30% of all human cancers and the four Ras isoforms are highly attractive targets for anticancer drug discovery. However, Ras proteins are challenging targets for conventional drug discovery because they function through intracellular protein–protein interactions and their surfaces lack major pockets for small molecules to bind. Over the past few years, researchers have explored a variety of approaches and modalities, with the aim of specifically targeting oncogenic Ras mutants for anticancer treatment. This perspective will provide an overview of the efforts on developing “macromolecular” inhibitors against Ras proteins, including peptides, macrocycles, antibodies, nonimmunoglobulin proteins, and nucleic acids.
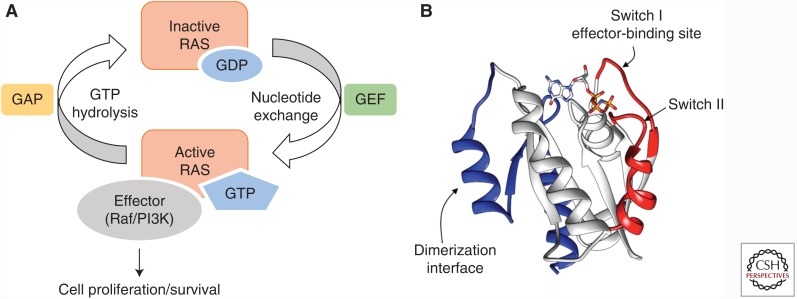
Ras is a small GTPase, acting as a molecular switch in many signaling pathways and regulating cell proliferation, differentiation, and survival, among other functions (Young et al. 2009). Its four isoforms, H-Ras, N-Ras, K-Ras4A, and K-Ras4B, are identical within the amino-terminal 85 amino acids and differ primarily in the carboxyl-termini (amino acids 165–189). Wild-type Ras oscillates between the inactive guanosine diphosphate (GDP)-bound form (Ras-GDP) and the active guanosine triphosphate (GTP)-bound form (Ras-GTP) (Fig. 1A). Ras-GTP interacts with and activates multiple effector proteins, including kinases Raf and phosphoinositide 3-kinase (PI3K), turning cells on for proliferation and survival. Somatic mutations at Gly-12, Gly-13, or Gln-61, which are all located within the GTPase active site, impair GTP hydrolysis, resulting in an excessive Ras-GTP population leading to uncontrolled cell growth. Ras mutations are found in ∼30% of all human cancers and are well-established cancer drivers (Prior et al. 2012; Singh et al. 2015). Mutations in K-Ras are particularly prevalent in some of the most deadly cancers, including pancreatic (90% prevalence), colon (35% prevalence), and lung cancers (16% prevalence). Disruption of Ras function genetically (i.e., by gene mutations or small-interfering RNA [siRNA]) inhibits the proliferation of Ras-mutant cancer cells and induces apoptosis, validating Ras as one of the most compelling cancer drug targets (Gupta et al. 2007; Singh et al. 2009; Castellano et al. 2013; Khvalevsky et al. 2013).
Figure 1.
Ras structure and function. (A) Interconversion between inactive Ras-guanosine diphosphate (GDP) and active Ras-guanosine triphosphate (GTP) mediated by guanine nucleotide exchange factor (GEF) and GTPase-activating protein (GAP). The active Ras-GTP interacts with effector proteins and activates downstream cell signaling. (B) Structure of K-Ras bound with GppNp, a nonhydrolyzable analog of GTP. The switch I/II regions are shown in red, the dimerization interface in blue, and the bound nucleotide analog is shown as sticks. (Figure was recreated from PDB data [pdb5p21].)
Over the past three decades, researchers have explored several approaches to inhibit Ras function (Wang et al. 2012; Spiegel et al. 2014; Stephen et al. 2014). The ideal approach would be direct binding to the GTPase active site by a small molecule. However, this approach was quickly found to be impractical, because Ras binds to GTP (and GDP) with pm affinity and GTP is present in the cytosol at mm concentrations. The next-best option is to block the interaction between Ras and its partner proteins and/or its nucleotide exchange activity. Unfortunately, Ras has been a very challenging target for direct inhibition, because its surface has no obvious pocket for small molecules to bind. Consequently, most of the early efforts were focused on targeting the signaling steps upstream or downstream of Ras. Small-molecule inhibitors against the posttranslational modification of Ras proteins (e.g., inhibitors against farnesyl and geranylgeranyl transferases) ultimately proved to be ineffective clinically, because of either a lack of efficacy or unacceptable toxicity (Whyte et al. 1997; Konstantinopoulos et al. 2007; Berndt et al. 2011). Another indirect approach is inhibition of the protein kinases downstream of Ras. Several MEK and PI3K inhibitors are currently undergoing clinical evaluation, although the clinical effectiveness of this approach remains to be seen (Engelman et al. 2008). It appears that simultaneous inhibition of at least two different pathways (e.g., by both MEK and PI3K inhibitors) is necessary to effectively block Ras signaling in Ras-mutant tumors (Britten et al. 2013); however, blocking multiple signaling pathways may result in extensive toxicity.
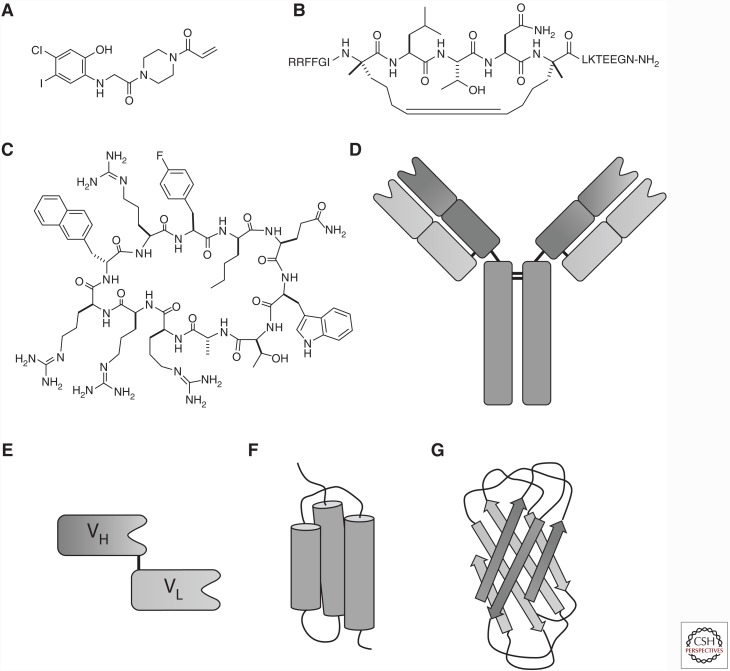
Driven by both urgent unmet medical needs and remarkable technological breakthroughs, there has been a recent “renaissance” of the direct Ras inhibition approach (Ledford 2015), which attacks Ras-driven cancers at the source and provides an opportunity for selective inhibition of Ras-mutant cancers. Development of direct Ras inhibitors is challenging because Ras performs its biological functions by engaging in intracellular protein–protein interactions (PPIs). PPIs are in general a challenging class of drug targets for conventional small molecules (defined as molecules of ≤500 molecular weight), because small molecules typically do not make a sufficient number of contacts with the large flat PPI interfaces to impart high affinity or specificity. To effectively compete with the protein partners, a PPI inhibitor should also possess a binding surface comparable in size to that of the partner proteins. Recognizing this, researchers have increasingly turned their attention to larger molecules, including peptides, macrocycles, and proteins, as PPI inhibitors. Powerful combinatorial library technologies for discovering peptide—especially macrocyclic peptide—ligands and novel approaches to improving their membrane permeability have recently been developed (Dougherty et al. 2017). Application of these technologies to Ras has resulted in a variety of Ras inhibitors, ranging from small molecules, peptides, macrocycles, proteins, to nucleic acids over the past few years (Fig. 2). In this review, we provide a summary of the “macromolecular” approaches to Ras inhibitor development (as opposed to the small-molecule approach). For an updated account on small-molecule Ras inhibitors, readers are referred to Zhou et al. (2017).
Figure 2.
Molecular modalities that have been used as direct Ras inhibitors. (A) Small molecules (example shown is the K-RasG12C-selective inhibitor by Ostrem et al. 2013); (B) stapled peptides (example shown is SAH-SOS1); (C) macrocyclic peptides (example shown is cyclorasin 9A5); (D) monoclonal antibodies; (E) intrabodies (the VH and VL regions of an antibody linked by a polypeptide or disulfide bond); (F) affibodies; and (G) monobodies (the tenth fibronectin type III domain of human fibronectin).
PEPTIDE-BASED INHIBITORS
Linear Peptides
The first peptidyl Ras inhibitors were discovered by Clark et al. (1996), who compared the sequences of several Ras effector proteins and identified a consensus Ras-binding sequence shared among a subset of the Ras effectors. They showed that peptides containing this sequence from Raf-1 (Table 1, compound 1) and NF1-GAP (Table 1, compound 2) blocked NF1-GAP stimulation of Ras GTPase activity and Ras-mediated activation of mitogen-activated protein kinases, with IC50 values of ∼45 µm. Similarly, Barnard et al. (1998) systematically tested short peptides corresponding to different regions of H-Ras and Raf-1 for in vitro inhibition of the Ras-Raf association and found that two of the Raf-1 peptides, E94CCAVFR100 (Table 1, compound 3) and C95CAVFRL101 (Table 1, compound 4), were significant inhibitors of Ras-Raf binding. Peptide C95CAVFRL101 had an IC50 value of 7 µm and also inhibited Ras-RalGDS binding. Xu and Luo (2002) screened 3.5 × 107 peptide aptamers and isolated two peptides (Table 1, compounds 5 and 6) that blocked the Ras–Raf interaction in vitro with IC50 values of 100 and 500 nm, respectively. Impressively, the peptides bound selectively to RasG12V over wild-type Ras and, when expressed in COS cells, inhibited Raf kinase activation in cell culture. More recently, Gareiss et al. (2010) identified a moderately potent dodecapeptide ligand (Table 1, compound 7) that binds to both H-Ras and K-Ras. However, the peptide did not alter the intrinsic GTPase activity of Ras or compete for Ras binding with Raf and showed no cellular activity.
Table 1.
A list of peptide-based Ras inhibitors and their inhibition mechanisms
Stapled Peptides
Stapled peptides have emerged as a powerful tool for disrupting protein–protein interactions by mimicking the secondary structure of crucial binding motifs (Walensky and Bird 2014). The preformed α-helical structure minimizes the entropic penalty associated with ligand–protein interaction, greatly improves the proteolytic stability of the peptides, and in some cases increases the membrane permeability of the peptides. Based on the observation that the guanine nucleotide exchange factor SOS binds to Ras primarily via an α-helical motif, Patgiri et al. (2011) designed a hydrogen bond surrogate (HBS) helical peptide (Table 1, compound 8), a special variant of stapled peptides, which had 56% helical content and bound to Ras-GDP with moderate affinity (KD = 158 µM). 1H-15N heteronuclear single quantum coherence (HSQC) nuclear magnetic resonance (NMR) studies indicated that the peptide interacts with the switch I/II region on Ras, overlapping with the SOS-binding site (Fig. 1B). Remarkably, the HBS peptide was cell-permeable and reduced epidermal growth factor (EGF)-stimulated Ras activation and downstream signaling events in HeLa cells. More recently, Leshchiner et al. (2015) designed a hydrocarbon-stapled α-helical peptide with much greater Ras-binding affinity, SAH-SOS1 (KD <200 nm) (Table 1, compound 9 and Fig. 2B), also corresponding to the Ras-binding helical motif in SOS. SAH-SOS1 was insensitive to the nucleotide-bound states of Ras. NMR and docking studies showed that SAH-SOS1 interacts with the SOS-binding site on Ras and inhibits nucleotide association of K-Ras. Importantly, SAH-SOS1 down-regulated AKT/MEK/ERK phosphorylation and reduced the viability of different K-Ras mutant cancer cells with IC50 values of 5–15 µm. Furthermore, SAH-SOS1 reduced the AKT and ERK phosphorylation levels in Drosophila tissues.
Macrocyclic Peptides
Because Ras-effector interactions are not mediated by α-helical motifs, rational design of stapled peptides against the effector-binding site is not an option. As described above, linear peptide ligands against the effector-binding site have been discovered, but they are generally weak binders. In addition, linear peptides face other challenges, including proteolytic degradation and the lack of membrane permeability. Meanwhile, macrocyclic peptides have emerged as an effective modality for inhibition of protein–protein interactions over the past decade (Dougherty et al. 2017). With molecular mass generally in the range of 500–2000, macrocyclic peptides are 3–5 times larger than conventional small-molecule drugs and possess binding surfaces similar in size to that of PPI interfaces. They also have a balanced conformational flexibility/rigidity that is conducive to recognizing the large flat protein surfaces. In addition, macrocyclic peptides have greatly increased proteolytic stability, especially when unnatural amino acids (e.g., d-amino acids) are incorporated into their structures.
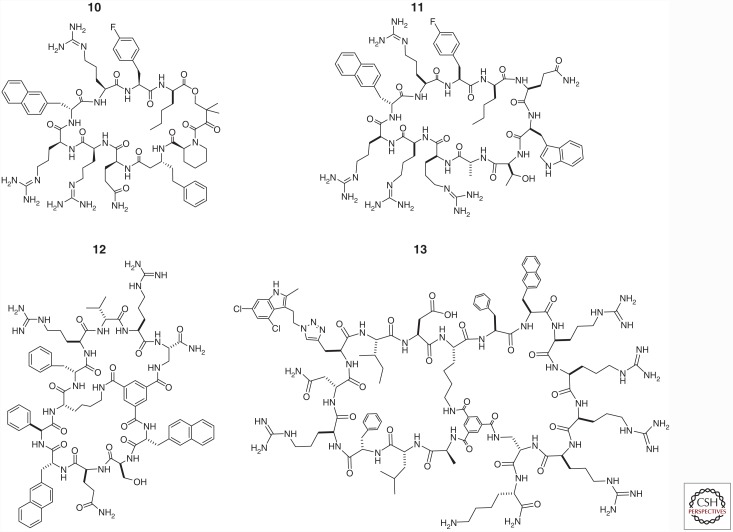
In an attempt to generalize the mode of action by rapamycin, Wu et al. (2013) constructed a one-bead–two-compound (OBTC) cyclic peptide library containing a fixed FKBP-binding motif fused with a randomized peptide sequence made of 25 different natural and unnatural amino acid building blocks. Screening of the library against K-RasG12V identified a hit peptide, “compound 12” (Table 1, compound 10 and Fig. 3), which inhibited the Ras–Raf RBD interaction with an IC50 value of 0.5 µm, in the absence of FKBP. As expected from its large size and hydrophilic structure, compound 10 was impermeable to the cell membrane and had no cellular activity. Interestingly, the structure of compound 10 contained a pentapeptide motif, Arg-Arg-nal-Arg-Fpa (where Fpa is l-4-fluorophenylalanine and nal is d-β-naphthylalanine), which bears a similarity to a family of cyclic cell–penetrating peptides (CPPs) (e.g., cyclo (Phe-Nal-Arg-Arg-Arg-Arg-Gln), where Nal is l-β-naphthylalanine) (Qian et al. 2013, 2016). The investigators subsequently constructed a second-generation library, in which the CPP-like motif was retained, while the remaining structure was replaced with a randomized peptide sequence of 0–5 amino acids (Upadhyaya et al. 2015). Screening of the second-generation library against K-RasG12V identified two hits that blocked the Ras-Raf association in vitro, were cell-permeable, and showed modest antiproliferative activity against cancer cells. One of the hits, cyclorasin 9A, was further optimized through medicinal chemistry efforts to produce an improved compound, cyclorasin 9A5 (Table 1, compound 11 and Fig. 3), which orthosterically blocked the Ras–Raf interaction in vitro (IC50 = 120 nm), was readily cell-permeable, and showed a sixfold selectivity for Ras-GTP over Ras-GDP. NMR studies suggested that cyclorasin 9A5 binds to an extended area between the switch I and II loops, which overlaps with the effector-binding site (Fig. 1B). When tested against Ras mutant lung cancer cells, cyclorasin 9A5 dose dependently inhibited the Ras–Raf interaction and abolished MEK, ERK, and AKT phosphorylation at low μm concentrations. It showed moderately potent antiproliferative activity against a panel of lung cancer cell lines (e.g., EC50 ∼3 µm against H1299 cells) and induced apoptosis of both wild-type and mutant Ras cancer cells at low micromolar concentrations, consistent with its ability to simultaneously block both the RAF/MEK/ERK and the PI3K/AKT/mTOR pathways. Transfection of H1299 cells with a constitutively active form of AKT rendered the cancer cells substantially less sensitive to cyclorasin 9A5, providing further support that the observed anticancer activity was caused by specific inhibition of Ras signaling by the cyclic peptide.
Figure 3.
Structures of macrocyclic peptidyl Ras inhibitors.
In a separate study, Upadhyaya et al. (2014) synthesized a highly structurally constrained bicyclic peptide library in the OBTC format and screened the library against K-RasG12V. They discovered two different classes of Ras ligands, with one class of ligands (e.g., cyclorasin B3) apparently bound at or near the effector-binding site and blocked the Ras-Raf association (KD = 2.6 µm). Cyclorasin B3 (Table 1, compound 12 and Fig. 3) competed with monocyclic peptide 10 and Raf-1 RBD for binding to K-RasG12V. Interestingly, the second class of Ras ligands (e.g., cyclorasin B2) apparently bound to a yet-unidentified site, different from the effector-binding site (KD = 0.49 µm), as it did not inhibit the Ras–Raf interaction. As expected, both classes of cyclic peptides were impermeable to the cell membrane and had no significant activity during cellular assays. Efforts to improve both the potency and cell-permeability of the library hits are currently ongoing in the authors’ laboratory.
To develop a general approach to discovering cell-permeable macrocyclic inhibitors against intracellular targets, Trinh et al. (2016) designed a bicyclic peptide library in which one ring consisted of a fixed CPP motif (e.g., Phe-Nal-Arg-Arg-Arg-Arg), while the second ring featured 5.7 million different peptide sequences. To increase the probability of high-affinity binding to K-Ras, the investigators included propargylglycine as a building block for the degenerate sequence and attached 4,6-dichloro-2-methyl-3-aminoethylindole (DCAI), a previously reported weak ligand for K-Ras (Maurer et al. 2012), to the alkyne side-chain by click chemistry. It was anticipated that most of the 5.7 million peptides should be cell permeable. Indeed, screening of the library against K-RasG12V followed by medicinal chemistry gave a cell-permeable and moderately potent K-Ras inhibitor (“compound 49”) (Table 1, compound 13 and Fig. 3), which inhibited the Ras–Raf interaction with an IC50 value of 3.4 µm. Compound 13 competed with DCAI for Ras binding and a control peptide without the DCAI moiety showed 80-fold lower affinity to K-RasG12V, indicating that the DCAI moiety plays a critical role in Ras binding and the bicyclic peptide binds to an area that overlaps with the switch I/II regions and the DCAI-binding pocket (Fig. 1B). Compound 13 (but not the control peptide without DCAI) inhibited Akt and MEK phosphorylation in cancer cells in a dose-dependent manner and induced apoptosis of lung cancer cells at 10 µm concentration.
Very recently, Sakamoto et al. (2017) discovered a highly potent and K-RasG12D-selective cyclic peptide inhibitor by screening a disulfide-cyclized peptide library displayed on T7 phage against the recombinant K-RasG12D. To obtain mutant-selective inhibitors, the researchers used both positive and negative screening steps, with thorough subtraction of phage bound to wild-type K-Ras. One of the library hits, KRpep-2 (Ac-RRCPLYISYDPVCRR-NH2), showed 14-fold selectivity for K-RasG12D over wild-type K-Ras (KD values of 51 and 700 nm, respectively). Subsequent sequence optimization resulted in cyclic peptide KRpep-2d (Table 1, compound 14), which inhibited SOS-mediated nucleotide exchange activity of wild-type and G12D K-Ras with IC50 values of 42 and 1.6 nm, respectively. At 30 µm peptide concentration, KRpep-2d inhibited the phosphorylation levels of ERK1/2 and the proliferation of A427 lung cancer cells (which harbor a K-RasG12D mutation), but not A549 lung cancer cells (which contain a K-RasG12C mutation). The high peptide concentration needed to cause cellular effects (30 µm, relative to IC50 of 1.6 nm for inhibition of nucleotide exchange) was attributed to limited cellular entry and inactivation of KRpep-2d on reduction of its disulfide bond inside the mammalian cytosol.
PROTEIN-BASED INHIBITORS
Anti-Ras Antibodies
Shortly after the discovery of the ras oncogene, researchers explored the possibility of using anti-Ras antibodies as potential anticancer therapeutics. Feramisco et al. (1985) reported that microinjection of antibodies specific for a mutant K-Ras protein into NRK cells transformed by V-K-ras gene caused a transient reversion of the cells to a normal phenotype. Similarly, Kung et al. (1986) found that microinjection of a neutralizing monoclonal antibody against H-Ras (Y13-259) neutralized the transforming activity of a coinjected bacterially synthesized H-Ras protein and that the neutralization effect was blocked by coinjection of excess H-Ras protein. In addition, microinjection of Y13-259 into transformed NIH3T3 cells (obtained by DNA transfection of NIH3T3 cells with a mutant H-ras gene) reversed their transformed phenotypes. Whereas these early studies showed the validity of using anti-Ras antibodies for treatment of Ras-driven cancers, their clinical application was hampered by two technical challenges: (1) how to effectively deliver the antibodies into cancer cells, and (2) how to maintain the structural integrity of the antibodies, whose multiple disulfide bonds would be cleaved inside the reducing environment of the mammalian cytosol.
To overcome these challenges, researchers prepared antibody fragments, consisting of single-chain variable region domains (scFv) derived from Y13-259 and other anti-Ras antibodies (Fig. 2E). These antibody fragments are more stable intracellularly (because of their reduced number of disulfide bonds) and are also easier to produce recombinantly, especially in bacteria. Cochet et al. (1998) showed that intracellular expression of an scFv of Y13-259 led to the specific inhibition of the Ras signaling pathway in Xenopus laevis oocytes and NIH3T3 fibroblasts. Moreover, neutralizing Ras with the scFv specifically promoted apoptosis in vitro in human cancer cells but not in untransformed cells. As a step toward cancer gene therapy, they showed that intratumor transduction of HCT116 colon carcinoma cells with the anti-Ras scFv using an adenoviral vector elicited sustained tumor regression in nude mice. Tanaka and Rabbitts (2003) developed an intracellular antibody capture (IAC) technology to isolate antibody fragments against protein targets of interest. The antibody fragments were further engineered to generate “intrabodies” that can be stably expressed intracellularly. By screening a phage display library of 7 × 109 intrabodies against H-RasG12V, the investigators discovered a potent binder, iDab#6, which interacted selectively with mutant H-RasG12V loaded with GTPγS (KD = 6.2 nm) over wild-type H-Ras and Ras-GDP and blocked Ras-GTP from interacting with effector proteins as well as downstream signaling pathways (Tanaka et al. 2007). Mutagenesis and X-ray crystallographic studies revealed that iDab#6 bound to the switch I/II regions on Ras (Fig. 1B). Expression of iDab#6 in mutant Ras-transformed mouse and human cells reverted them back to the untransformed phenotype. Furthermore, retroviral delivery of a membrane-anchored iDab#6 variant into tumor cell lines prevented Ras-dependent tumorigenesis in a mouse xenograft model. Interestingly, by using a transgenic mouse model, the same investigators later found that blocking the mutant Ras-effector protein interactions by iDab#6 was effective in preventing tumor initiation and controlling tumor growth, but was not sufficient to cause tumor regression (Tanaka and Rabbitts 2010). Very recently, Yang et al. constructed a recombinant adenovirus KGHV300 that carried an anti-p21Ras intrabody and that could replicate in tumor cell lines but not in normal cell lines (Yang et al. 2016a,b; Pan et al. 2017). In KGHV300, the expression levels of the essential replication genes E1a and E1b, were controlled by the human telomerase reverse transcriptase promoter and the hypoxia response element, respectively, and the anti-p21Ras intrabody gene was controlled by the cytomegalovirus promoter. The conditional replication of KGHV300 and its antitumor efficacy were shown in several tumor cell lines in vitro and in xenograft models of human breast cancer in nude mice. The recombinant adenovirus KGHV300 provides a promising and potentially safer antitumor therapeutic for Ras-driven cancers. Finally, Shin et al. (2017) engineered a cell-permeable antibody, RT11, which enters the cytosol of mammalian cells by endocytosis, selectively binds to the Ras-GTP form of various oncogenic Ras mutants, and blocks the Ras-effector interactions. RT11 suppressed Ras signaling and exerted antiproliferative effects in a variety of Ras mutant tumor cells. When systemically administered, an RT11 variant containing a tumor-targeting moiety significantly inhibited the in vivo growth of Ras-mutant tumor xenografts in mice, but not wild-type Ras-harboring tumors.
Nonimmunoglobulin Protein Inhibitors
Compared to antibodies and their fragments, nonimmunoglobulin protein scaffolds containing no disulfide bonds are more stable under reducing conditions and easier to produce recombinantly, offering alternative modalities as macromolecular therapeutics (Löfblom et al. 2010). Affibodies are ∼6-kDa three-helix bundles containing flexible regions for sequence randomization (Fig. 2F). By screening a combinatorial affibody library displayed on bacteriophage, Grimm et al. (2010) isolated affibody molecules that bound H-Ras and Raf with 79 nm and 1.9 µm affinities, respectively. Interestingly, in a real-time biospecific interaction analysis, the two Ras targeting affibodies failed to inhibit the Ras–Raf interaction, suggesting that they bind a different site(s) from the effector protein-binding region. On the other hand, the Raf targeting affibody was able to block the Ras–Raf interaction in the same assay.
Cetin et al. (2017) developed a protein inhibitor against K-Ras using a miniprotein scaffold derived from the 10th fibronection type III domain of human fibronection (Fig. 2G). These 10-kD domains have an immunoglobulin-like fold but do not contain disulfide bonds. They contain two flexible loop regions (BC and FG) similar to the CDRH1 and CDRH3 loops at the antibody-combining site. These investigators generated a biased messenger RNA (mRNA) display library on the fibronection scaffold by incorporating the previously reported Ras-binding sequence from iDab#6 into the BC loop and a completely randomized sequence into the FG loop. Screening of the library followed by a second round of cell-based selection resulted in a hit molecule, RasIn1, which bound selectively to K-RasG12V-GTP (KD = 2.1 µm) over K-RasG12V-GDP and wild-type K-Ras and blocked the Ras–Raf interaction. Mutational analysis showed the dependence of the BC and FG loops for Ras binding. When expressed inside mammalian cells, RasIn1 colocalized with the activated RasG12V. To further increase the potency of the hit molecule, a second-generation library was constructed based on the sequence of Rasln1 and screened to give Rasln2, which showed both improved binding affinity (KD = 120 nm for K-RasG12V-GTP) and selectivity. ELISA assays suggested that Rasln2 binds to RasG12V with significantly higher affinity than the Ras-binding domain (RBD) of Raf. Similarly, Tamaskovic et al. (2016) developed designed ankyrin repeat proteins (DARPins) against Ras, which specifically blocked the Ras–RBD interactions. Breast cancer cells (which carry wild-type Ras) expressing the anti-Ras DARPins showed significantly higher susceptibility to anti-HER2 treatment (trastuzumab) than cells without RAS interference.
In addition to Ras-effector interaction blockers, proteins that allosterically inhibit Ras function have also been reported. Spencer-Smith et al. (2017) isolated a monobody molecule (NS1) from a phage display library. NS1 bound to H-Ras and K-Ras with KD values of 14 and 67 nm, respectively, regardless of the GTP/GDP-bound state, but did not bind to N-Ras. NS1 effectively inhibited growth-factor-induced signaling and oncogenic H-Ras-/K-Ras-mediated transformation. Interestingly, NS1 had no effect on SOS-mediated nucleotide exchange of H-Ras, suggesting that its binding site also differs from that of SOS. X-ray crystal structure of the NS1/H-Ras complex revealed that NS1 binds to the dimerization interface of activated H-Ras, which is located on the back side of the protein with respect to the effector- and SOS-binding sites and divergent among the different Ras isoforms (Fig. 1B). Electron microscopic analysis showed that NS1 disrupted the dimerization and nanoclustering of H-Ras and K-Ras inside cells. NS1 inhibited Ras-induced Raf activation, consistent with the notion that Ras dimerization is required for recruitment of Raf to the plasma membrane.
NUCLEIC ACID–BASED INHIBITORS
siRNA
RNA interference (RNAi) provides an alternative to small-molecule and antibody-based therapeutics against challenging targets such as Ras. RNAi can, in principle, be applied to reversibly silence any target gene, including oncogenic ras mutants (as opposed to wild-type ras gene). Using short hairpin RNAs (shRNAs) to deplete K-Ras in lung and pancreatic cancer cell lines harboring K-ras mutations, Singh et al. (2009) identified two classes of cancer cell lines, those that do or do not require K-Ras to maintain viability. It was found that epithelial differentiation and tumor cell viability are associated, and that epithelial-mesenchymal transformation (EMT) regulators in “K-Ras-addicted” cancers represent candidate therapeutic targets. Since then, several recent studies have shown the validity of targeting mutant K-Ras by siRNAs as therapeutics. Khvalevsky et al. (2013) showed that siRNA targeting K-Ras mutations with a local prolonged release system knocks down K-Ras expression in vitro and in vivo (mouse models), leading to an antitumor effect. Treatment of pancreatic cancer cells with siRNA against K-RasG12D resulted in a significant decrease in K-Ras levels, leading to inhibition of proliferation and EMT. In vivo, intratumorally implanted siRNA impeded the growth of human pancreatic tumor cells and prolonged mouse survival. Pecot et al. (2014) subsequently showed that systemic delivery of K-Ras siRNAs with nanoparticles was effective for treatment of lung and colon cancers in mouse models. siRNA against K-Ras has also been chemically attached to anti-epidermal growth factor receptor (EGFR) antibodies for cancer-specific delivery to K-Ras-mutated EGFR-positive cancer cells (Bäumer et al. 2015). Finally, siRNA against mutant K-Ras has been used in combination with siRNAs against other Ras signaling pathway targets (e.g., Raf and PI3K) to treat colorectal cancer in xenograft models (Yuan et al. 2014).
Other Nucleic Acid–Based Approaches
Yu et al. (2009) reported an interesting approach to treating K-Ras mutant cancers with an oligodeoxyribonucleotide-based ribonuclease, 10-23 DNAzyme. The DNAzyme consists of a 15-nucleotide catalytic domain flanked by two target-binding sequences, and is capable of cleaving specific target mRNAs at a purine-pyrimidine dinucleotide. A K-RasG12V-specific DNAzyme was designed to cleave the mRNA of K-RasG12V (GGU→GUU) at the GU dinucleotide while leaving the wild-type K-Ras mRNA untouched. Transfection of SW480 cells (which carry homozygous K-RasG12V mutation) with the DNAzyme reduced K-RasG12V at both mRNA and protein levels, but not in HEK cells (wild-type K-Ras). Although the DNAzyme alone did not inhibit proliferation of SW480 or HEK cells, pretreatment with this DNAzyme sensitized the K-RasG12V mutant cells to anticancer agents such as doxorubicin and radiation. These results suggest the potential of using allele-specific DNAzymes in combination with other cancer therapies for more effective cancer treatment.
Another interesting approach involved direct targeting of mutant K-Ras DNA using pyrrole-imidazole polyamide bearing a reactive chloromethyl moiety (KR12) (Hiraoka et al. 2015). KR12 selectively recognizes oncogenic codon 12 K-Ras mutations and alkylates adenine N3 at the target sequence, causing strand cleavage and growth suppression in human colon cancer cells with G12D or G12V mutations. In xenograft models, KR12 infusions induced significant tumor growth suppression, with low host toxicity in K-Ras-mutated but not wild-type tumors.
CONCLUDING REMARKS AND OUTLOOK
Renewed efforts on the development of direct Ras inhibitors over the past few years have resulted in a large number of small-molecule, peptide, protein, and nucleic acid–based Ras inhibitors. Although achieving high potency and selectivity for small-molecule Ras inhibitors remains challenging (Zhou et al. 2017), highly potent and selective macromolecular Ras inhibitors have already been obtained. Some of the latter compounds (e.g., macrocyclic peptides and proteins) have shown remarkable selectivity for oncogenic Ras mutants over the wild-type protein, suggesting that selective targeting of mutant Ras proteins pharmacologically is feasible. When properly delivered into cancer cells and tissues (e.g., by intratumoral injection), some of the macromolecular Ras inhibitors have shown in vivo efficacy for preventing tumor initiation/growth and in some cases causing cancer cell apoptosis/tumor regression. Nucleic acid–based therapeutics have also shown in vivo efficacy in animal models and can, in principle, selectively target mutant Ras genes.
However, clinical development of these macromolecular Ras inhibitors still faces significant challenges. Because Ras is an intracellular target, any anti-Ras therapeutics must be effectively delivered into the cytosol of cancer cells. Larger molecules, such as the Ras inhibitors described in this review, violate almost every rule of Lipinski’s rule of five (Lipinski et al. 2001) and generally cannot cross the plasma membrane by passive diffusion. The currently used methods for protein and nucleic acid delivery typically require endocytic uptake of the drug/vehicle complex, resulting in their initial localization in the early endosome (Bareford and Swaan 2007). To reach a cytosolic target (e.g., Ras), the drug must escape from the endosome into the cytosol by physically crossing the endosomal membrane. Endosomal escape is poorly understood and believed to be the limiting factor for all current nonviral protein and nucleic acid delivery technologies (Varkouhi et al. 2011). Protein therapeutics additionally face the challenge of immunogenicity if administered repeatedly. Fortunately, highly active cell-penetrating peptides have recently been discovered (Qian et al. 2013, 2016) and have been applied to efficiently deliver linear and macrocyclic peptides (Lian et al. 2014; Qian et al. 2015, 2017; Upadhyaya et al. 2015), proteins (Qian et al. 2014), and nucleic acids (D Pei, unpubl.) into the cytosol of mammalian cell in vitro and in vivo. Integration of the macromolecular Ras inhibitors with the latest delivery technologies may provide macromolecular therapeutics for clinical treatment of Ras mutant cancers.
ACKNOWLEDGMENTS
The Ras-related work in the Pei group is supported by grants from the National Institutes of Health (GM062820, GM110208, and GM122459).
Footnotes
Editors: Linda VanAelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Bareford LM, Swaan PW. 2007. Endocytic mechanisms for targeted drug delivery. Adv Drug Deliv Rev 59: 748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D, Sun H, Baker L, Marshall MS. 1998. In vitro inhibition of Ras-Raf association by short peptides. Biochem Biophys Res Commun 247:176–180. [DOI] [PubMed] [Google Scholar]
- Bäumer S, Bäumer N, Appel N, Terheyden L, Fremerey J, Schelhaas S, Wardelmann E, Buchholz F, Berdel WE, Müller-Tidow C. 2015. Antibody-mediated delivery of anti-KRAS-siRNA in vivo overcomes therapy resistance in colon cancer. Clin Cancer Res 21: 1383–1394. [DOI] [PubMed] [Google Scholar]
- Berndt N, Hamilton AD, Sebti SM. 2011. Targeting protein prenylation for cancer therapy. Nat Rev Cancer 11: 775–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten CD. 2013. PI3K and MEK inhibitor combinations: examining the evidence in selected tumor types. Cancer Chemother Pharmacol 71: 1395–1409. [DOI] [PubMed] [Google Scholar]
- Castellano E, Downward J. 2011. Ras interaction with PI3K: More than just another effector pathway. Genes Cancer 2: 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E, Sheridan C, Thin MZ, Nye E, Spencer-Dene B, Diefenbacher ME, Moore C, Kumar MS, Murillo MM, Gronroos E, et al. 2013. Requirement for interaction of PI3-kinase p110α with RAS in lung tumor maintenance. Cancer Cell 24: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin M, Evenson WE, Gross GG, Jalali-Yazdi F, Krieger D, Arnold D, Takahashi TT, Roberts RW. 2017. RasIns: Genetically encoded intrabodies of activated Ras proteins. J Mol Biol 429: 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, et al. 2012. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signaling of Ras family proteins. Nat Cell Biol 14: 148–158. [DOI] [PubMed] [Google Scholar]
- Clark GJ, Drugan JK, Terrell RS, Bradham C, Der CJ, Bell RM, Campbell S. 1996. Peptides containing a consensus Ras binding sequence from Raf-1 and the GTPase activating protein NF1 inhibit Ras function. Proc Natl Acad Sci 93:1577–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochet O, Kenigsberg M, Delumeau I, Virone-Oddos A, Multon MC, Fridman WH, Schweighoffer F, Teillaud JL, Tocqué B. 1998. Intracellular expression of an antibody fragment-neutralizing p21 ras promotes tumor regression. Cancer Res 58: 1170–1176. [PubMed] [Google Scholar]
- Dougherty PG, Qian Z, Pei D. 2017. Macrocycles as protein–protein interaction inhibitors. Biochem J 474: 1109–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, et al. 2012. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149: 307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. 2008. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 14: 1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feramisco J, Clark R, Wong G, Arnheim N, Milley R, McComick F. 1985. Transient reversion of ras oncogene-induced cell transformation by antibodies specific for amino acid 12 of ras protein. Nature 314: 639–642. [DOI] [PubMed] [Google Scholar]
- Gareiss PC, Schneekloth AR, Salcius MJ, Seo SY, Crews CM. 2010. Identification and characterization of a peptidic ligand for Ras. ChemBioChem 11: 517–522. [DOI] [PubMed] [Google Scholar]
- Grimm S, Lundberg E, Yu F, Shibasaki S, Vernet E, Skogs M, Nygren PÅ, Gräslund T. 2010. Selection and characterization of affibody molecules inhibiting the interaction between Ras and Raf in vitro. N Biotechnol 27: 766–773. [DOI] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. 2007. Binding of Ras to phosphoinositide 3-kinase p110α is required for Ras-driven tumorigenesis in mice. Cell 129: 957–968. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Haling JR, Chen H, Song K, Price S, Heald R, Hewitt JFM, Zak M, Peck A, Orr C, et al. 2013. Mechanism of MEK inhibition determines efficacy in mutant KRAS- versus BRAF- driven cancers. Nature 501: 232–236. [DOI] [PubMed] [Google Scholar]
- Hiraoka K, Inoue T, Taylor RD, Watanabe T, Koshikawa N, Yoda H, Shinohara K, Takatori A, Sugimoto H, Maru Y, et al. 2015. Inhibition of KRAS codon 12 mutants using a novel DNA-alkylating pyrrole-imidazole polyamide conjugate. Nat Commun 6: 6706. [DOI] [PubMed] [Google Scholar]
- Khvalevsky EZ, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, Shemi A, Golan T, Domb AJ, Yavin E, et al. 2013. Mutant KRas is a druggable target for pancreatic cancer. Proc Natl Acad Sci 110: 20723–20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. 2007. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov 6: 541–555. [DOI] [PubMed] [Google Scholar]
- Kung HF, Smith MR, Bekesi E, Manne V, Stacey DW. 1986. Reversal of transformed phenotype by monoclonal antibodies against Ha-ras p21 proteins. Exp Cell Res 162: 363–371. [DOI] [PubMed] [Google Scholar]
- Ledford H. 2015. The Ras renaissance. Nature 520: 278–280. [DOI] [PubMed] [Google Scholar]
- Leshchiner ES, Parkhitko A, Bird GH, Luccarelli J, Bellairs JA, Escudero S, Opoku-Nsiah K, Godes M, Perrimon N, Walensky LD. 2015. Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc Natl Acad Sci 112: 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian W, Jiang B, Qian Z, Pei D. 2014. Cell-permeable bicyclic peptide inhibitors against intracellular proteins. J Am Chem Soc 136: 9830–9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46: 3–26. [DOI] [PubMed] [Google Scholar]
- Löfblom J, Feldwisch J, Tolmachev V, Carlsson J, Ståhl S, Frejd FY. 2010. Affibody molecules: engineered proteins for therapeutic, diagnostic and biotechnological applications. FEBS Lett 584: 2670–2680. [DOI] [PubMed] [Google Scholar]
- Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, et al. 2012. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci 109: 5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. 2013. K-Ras (G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503: 548–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan XY, Liu XJ, Li J, Zhen SJ, Liu DX, Feng Q, Zhao WX, Luo Y, Zhang YL, Li HW, et al. 2017. The antitumor efficacy of anti-p21Ras scFv mediated by the dual-promoter-regulated recombinant adenovirus KGHV300. Gene Therapy 24: 40–48. [DOI] [PubMed] [Google Scholar]
- Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. 2011. An orthosteric inhibitor of the Ras-Sos interaction. Nat Chem Biol 7: 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecot CV, Wu SY, Bellister S, Filant J, Rupaimoole R, Hisamatsu T, Bhattacharya R, Maharaj A, Azam S, Rodriguez-Aguayo C, et al. 2014. Therapeutic silencing of KRAS using systemically delivered siRNAs. Mol Cancer Ther 13: 2876–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. 2010. RAF inhibitors transactivate RAF dimers and ERK signaling in cells with wild-type BRAF. Nature 464: 427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior IA, Lewis PD, Mattos C. 2012. A comprehensive survey of Ras mutations in cancer. Cancer Res 72: 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Liu T, Liu YY, Briesewitz R, Barrios AM, Jhiang SM, Pei D. 2013. Efficient delivery of cyclic peptides into mammalian cells with short sequence motifs. ACS Chem Biol 8: 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, LaRochelle JR, Jiang B, Lian W, Hard RL, Selner NG, Luechapanickhul R, Barrios AM, Pei D. 2014. Early endosomal escape of a cyclic cell-penetrating peptide allows effective cytosolic cargo delivery. Biochemistry 53: 4034–4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Xu X, Amacher JF, Madden DR, Cormet-Boyaka E, Pei D. 2015. Intracellular delivery of peptidyl ligands by reversible cyclization: Discovery of a PDZ domain inhibitor that rescues CFTR activity. Angew Chem Int Ed 54: 5874–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Martyna A, Hard RL, Wang J, Appiah-Kubi G, Coss C, Phelps MA, Rossman JS, Pei D. 2016. Discovery and mechanism of highly efficient cyclic cell-penetrating peptides. Biochemistry 55: 2601–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Rhodes CA, McCroskey LC, Wen J, Appiah-Kubi G, Wang DJ, Guttridge DC, Pei D. 2017. Enhancing the cell permeability and metabolic stability of peptidyl drugs by reversible bicyclization. Angew Chem Int Ed 56: 1525–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Kamada Y, Sameshima T, Yaguchi M, Niida A, Sasaki S, Miwa M, Ohkubo S, Sakamoto J, Kamaura M, et al. 2017. K-Ras (G12D)-selective inhibitory peptides generated by random peptide T7 phage display technology. Biochem Biophys Res Commun 484: 605–611. [DOI] [PubMed] [Google Scholar]
- Shin SM, Choi DK, Jung K, Bae J, Kim JS, Park SW, Song KH, Kim YS. 2017. Antibody targeting intracellular oncogenic Ras mutants exerts anti-tumour effects after systemic administration. Nat Commun 10.1038/ncomms15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. 2009. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell 15: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Longo DL, Chalmer BA. 2015. Improving prospects for targeting Ras. J Clin Oncol 33: 3650–3659. [DOI] [PubMed] [Google Scholar]
- Spencer-Smith R, Koide A, Zhou Y, Eguchi RR, Sha F, Gajwani P, Santana D, Gupta A, Jacobs M, Herrero-Garcia E, et al. 2017. Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol 13: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H. 2014. Small-molecule modulation of Ras signaling. Nat Chem Biol 10: 613–622. [DOI] [PubMed] [Google Scholar]
- Stephen AG, Esposito D, Bagni RK, McCormick F. 2014. Dragging ras back in the ring. Cancer Cell 25: 272–281. [DOI] [PubMed] [Google Scholar]
- Tamaskovic R, Schwill M, Nagy-Davidescu G, Jost C, Schaefer DC, Verdurmen WPR, Schaefer JV, Honegger A, Plückthun A. 2016. Intermolecular biparatopic trapping of ErbB2 prevents compensatory activation of PI3K/AKT via RAS–p110 crosstalk. Nat Commun 7: 11672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Rabbitts TH. 2003. Intrabodies based on intracellular capture frameworks that bind the RAS protein with high affinity and impair oncogenic transformation. EMBO J 22: 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Rabbitts TH. 2010. Interfering with RAS-effector protein interactions prevent RAS-dependent tumour initiation and causes stop–start control of cancer growth. Oncogene 29: 6064–6070. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Williams R, Rabbitts TH. 2007. Tumour prevention by a single antibody domain targeting the interaction of signal transduction proteins with RAS. EMBO J 6: 3250–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh TB, Upadhyaya P, Qian Z, Pei D. 2016. Discovery of a direct Ras inhibitor by screening a combinatorial library of cell-permeable bicyclic peptides. ACS Comb Sci 18: 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya P, Qian Z, Habir NA, Pei D. 2014. Direct Ras inhibitors identified from a structurally rigidified bicyclic peptide library. Tetrahedron 70: 7714–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya P, Qian Z, Selner NG, Clippinger SR, Wu Z, Briesewitz R, Pei D. 2015. Inhibition of Ras signaling by blocking Ras-effector interactions with cyclic peptides. Angew Chem Int Ed 54: 7602–7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkouhi AK, Scholte M, Storm G, Haisma HJ. 2011. Endosomal escape pathways for delivery of biologicals. J Control Release 151: 220–228. [DOI] [PubMed] [Google Scholar]
- Walensky LD, Bird GH. 2014. Hydrocarbon-stapled peptides: Principles, practice, and progress. J Med Chem 57: 6275–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Fang G, Rudolph J. 2012. Ras inhibition via direct Ras binding—Is there a path forward? Bioorg Med Chem Lett 22: 5766–5776. [DOI] [PubMed] [Google Scholar]
- Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, Bishop WR, Pai JK. 1997. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem 272: 14459–14464. [DOI] [PubMed] [Google Scholar]
- Winter-Vann AM, Baron RA, Wong W, dela Cruz J, York JD, Gooden DM, Bergo MO, Young SG, Toone EJ, Casey PJ. 2005. A small-molecule inhibitor of isoprenylcysteine carboxyl methyltransferase with antitumor activity in cancer cells. Proc Natl Acad Sci 102: 4336–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, U padhyaya P, Villalona-Calero MA, Briesewitz R, Pei D. 2013. Inhibition of Ras-effector interaction by cyclic peptides. Medchemcomm 4: 378–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CW, Luo Z. 2002. Inactivation of Ras function by allele-specific peptide aptamers. Oncogene 21: 5753–5757. [DOI] [PubMed] [Google Scholar]
- Yang JL, Liu DX, Zhen SJ, Zhou YG, Zhang DJ, Yang LY, Chen HB, Feng Q. 2016a. A novel anti-p21Ras scFv antibody reacting specifically with human tumour cell lines and primary tumour tissues. BMC Cancer 16: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Pan X, Zhao W, Hu Q, Ding F, Feng Q, Li G, Luo Y. 2016b. The antitumor efficacy of a novel adenovirus-mediated anti-p21Ras single chain fragment variable antibody on human cancers in vitro and in vivo. Int J Oncol 48: 1218–1228. [DOI] [PubMed] [Google Scholar]
- Young A, Lyons J, Miller AL, Phan VT, Alarcón IR, McCormick F. 2009. Ras signaling and therapies. Adv Cancer Res 102: 1–17. [DOI] [PubMed] [Google Scholar]
- Yu SH, Wang TH, Au LC. 2009. Specific repression of mutant K-RAS by 10-23 DNAzyme: Sensitizing cancer cell to anti-cancer therapies. Biochem Biophys Res Commun 378: 230–234. [DOI] [PubMed] [Google Scholar]
- Yuan T, Fellmann C, Lee CS, Ritchie CD, Thapar V, Lee LC, Hsu DJ, Grace DO, Carver J, Zuber J, et al. 2014. Development of siRNA payloads to target KRAS-mutant cancer. Cancer Discov 4:1182–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zhou Y, Prakash P, Gorfe AA, Hancock JF. 2017. Ras and the plasma membrane: A complicated relationship. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G, Papke B, Ismail S, Vartak N, Chandra A, Hoffmann M, Hahn SA, Triola G, Wittinghofer A, Bastiaens PI, et al. 2013. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signaling. Nature 497: 638–642. [DOI] [PubMed] [Google Scholar]