Summary
Various mesenchymal cell types have been identified as critical components of the hematopoietic stem/progenitor cell (HSPC) niche. Although several groups have described the generation of mesenchyme from human pluripotent stem cells (hPSCs), the capacity of such cells to support hematopoiesis has not been reported. Here, we demonstrate that distinct mesenchymal subpopulations co-emerge from mesoderm during hPSC differentiation. Despite co-expression of common mesenchymal markers (CD73, CD105, CD90, and PDGFRβ), a subset of cells defined as CD146hiCD73hi expressed genes associated with the HSPC niche and supported the maintenance of functional HSPCs ex vivo, while CD146loCD73lo cells supported differentiation. Stromal support of HSPCs was contact dependent and mediated in part through high JAG1 expression and low WNT signaling. Molecular profiling revealed significant transcriptional similarity between hPSC-derived CD146++ and primary human CD146++ perivascular cells. The derivation of functionally diverse types of mesenchyme from hPSCs opens potential avenues to model the HSPC niche and develop PSC-based therapies.
Keywords: pluripotent stem cell, mesenchyme, hematopoietic stem cell niche, pericyte biology, directed differentiation, mesoderm
Highlights
-
•
hPSCs generated functionally and transcriptionally distinct mesenchymal populations
-
•
CD146hiCD73hi mesenchyme expressed HSC niche genes and supported human HSPCs ex vivo
-
•
CD146loCD73lo mesenchyme drove HSPC differentiation
-
•
CD146hiCD73hi mesenchyme shared a transcriptome profile with human pericytes
Crooks and colleagues demonstrated a previously underappreciated functional and molecular heterogeneity in mesenchyme generated from human pluripotent stem cells. Two mesenchymal subsets were distinguished by the reciprocal expression of CD146, CD73, and CD140a. CD146hiCD73hi mesenchyme supported self-renewing hematopoietic stem and progenitor cells (HSPCs), expressed markers of the HSPC niche, and shared a similar molecular signature with primary human adult pericytes.
Introduction
Maintenance of bona fide self-renewing hematopoietic stem and progenitor cells (HSPCs) ex vivo remains challenging in part because of our limited ability to recapitulate the human HSPC niche in culture. Intensive research efforts have begun to uncover the cellular and molecular constituents of the niche that regulate self-renewal and differentiation of HSPCs. Through the use of knockout and transgenic mice, several cell populations have been described in terms of their spatial relationship to the bone and blood vessels of the bone marrow, and their differential expression of various markers and bioactive molecules (Ding et al., 2012, Itkin et al., 2016, Kobayashi et al., 2010, Kunisaki et al., 2013).
High expression of melanoma-associated cell adhesion molecule (CD146) identifies human pericytes, a cell type that ensheaths capillaries, venules, arterioles, and sinusoids (Crisan et al., 2008) and can establish a heterotopic hematopoietic stem cell (HSC) niche when transplanted into immunodeficient mice (Sacchetti et al., 2007). Unlike CD146− mesenchyme, monolayers of CD146++ cells isolated from primary tissue (adult adipose tissue and fetal bone marrow) can support human HSPCs co-cultured for at least 2 weeks in the absence of exogenous cytokines (Corselli et al., 2013).
We and others have shown that mesenchymal cells can be differentiated from human pluripotent stem cells (hPSCs) (Chin et al., 2016, Ferrell et al., 2014, Hoffman and Calvi, 2014, Vodyanik et al., 2010). These previous studies identified mesenchyme as a single population defined mostly by expression of CD73 and/or CD105 and absence of hematopoietic and endothelial markers. We now report that the mesenchyme generated from hPSCs is functionally and transcriptionally heterogeneous. Our studies identified a distinct subpopulation of hPSC-derived mesenchyme that expressed high levels of CD146 and CD73 and low levels of PDGFRα (CD140a) and that was capable of supporting clonogenic, engraftable, and self-renewing human HSPCs ex vivo without exogenous cytokines. In contrast CD146loCD73lo mesenchyme showed significantly less capacity to support HSPCs. Transcriptome analysis revealed that the CD146hiCD73hi cells expressed significantly higher levels than CD146loCD73lo cells of perivascular markers and niche factors known to have critical roles in HSC maintenance. HSPC support was dependent in part on cell-cell interactions and Notch signaling through stromal expression of JAG1, whereas differentiation was promoted by WNT signaling. Closer transcriptional analysis, combining data from mesenchyme generated from hPSCs and human primary tissue, revealed that dominant pathways shared by the CD146++ populations were those related to vascular development, cell adhesion, and motility. Our data suggest that hPSC-derived mesoderm can generate mesenchymal cells phenotypically, functionally, and molecularly, similar to previously identified primary pericytes that contribute to the human HSPC niche.
Results
Heterogeneity of Embryonic Mesoderm-Derived Mesenchymal Cells
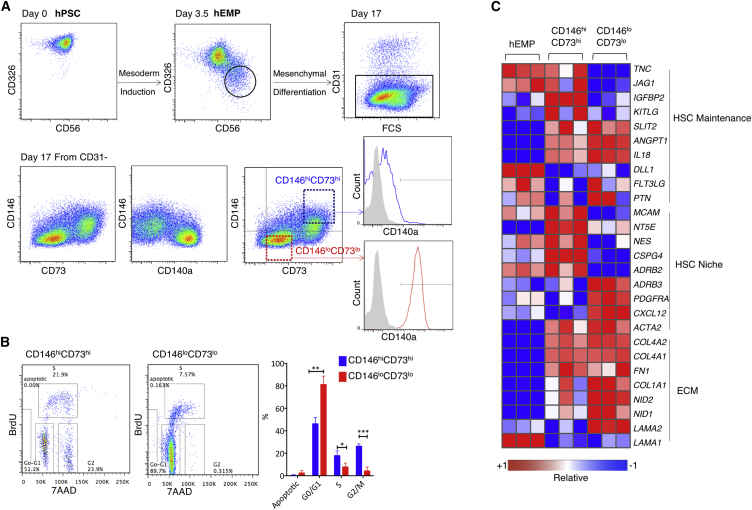
We have previously characterized a human embryonic mesoderm progenitor (hEMP) population derived from hPSCs that marks the onset of mesoderm commitment and has the potential to generate a broad range of mesodermal derivatives, including mesenchyme, endothelium, and bone, three lineages that play a crucial role in the hematopoietic niche (Chin et al., 2016, Hoffman and Calvi, 2014). hEMPs were isolated at day 3.5 of mesoderm differentiation from H1 embryonic stem cells (Evseenko et al., 2010) (Figure 1A), and re-cultured using conditions that favor mesenchymal differentiation. After a further 14 days, cultures contained a mixture of CD31+CD45− endothelial cells and CD31−CD45− mesenchymal cells. The mesenchymal cells consisted of at least two populations that could be discriminated based on expression of CD146, CD73, and CD140a (PDGFRα) (Figure 1A). High co-expression of CD146 and CD73 identified a largely CD140a− population, whereas CD146lo cells expressed intermediate levels of CD73 and high levels of CD140a. This inverse expression pattern between CD146 and CD140a was consistent with mesenchyme derived from primary human lipoaspirates (Figure S1A). Despite the differential expression of CD146, CD73, and CD140a, both hPSC-derived mesenchymal subsets expressed classic mesenchymal markers CD90, CD105, CD44, and PDGFRβ (Figure S1B). Mesenchymal differentiation from two other hPSC lines, UCLA3 and UCLA6, yielded similar populations to the H1 line, with an inverse relationship of CD146 and CD73 to CD140a expression (Figures S2A and S2B). Although few CD146hiCD73hi cells were present early in differentiation (days 4–5), the inverse expression pattern of CD146 and CD73 to CD140a was present throughout mesenchymal differentiation (Figure S2C).
Figure 1.
Immunophenotypic Fractionation of hPSC-Derived Mesenchyme Reveals Transcriptionally Distinct Populations
(A) hEMPs were generated from hPSC and isolated at day 3.5 by FACS for further differentiation in mesenchymal conditions. After 2 weeks, two distinct populations were identified within the CD31− (non-endothelial) cells, which were also CD45− (non-hematopoietic, not shown). CD146loCD73lo cells (red) were CD140a+, and CD146hiCD73hi cells (blue) were CD140lo or CD140a− (representative data using H1 cells of n = 16 experiments).
(B) Cell-cycle analysis of hEMP-derived mesenchymal differentiation cultures at week 2, pulsed with bromodeoxyuridine (BrdU) for 40 min, and gated on CD31− mesenchyme, as shown in Figure 1A, as either “CD146hiCD73hi” or “CD146loCD73lo.” n = 3 biological replicates. The y axis shows the percentage of each mesenchymal phenotype in each stage of cell cycle, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(C) Selected genes from RNA-seq profiling of hEMP, and hEMP-derived CD146hiCD73hi and CD146loCD73lo mesenchyme (as in Figure 1A). Relative expression shown for each gene using its min/max moderate expression estimates as reference. n = 3 biological replicates.
Error bars represent SEM. See also Figures S1 and S2.
To better understand whether the phenotypes above identified distinct mesenchymal cell types, we focused our studies on cells that represented the extremes of these phenotypes: a CD146hiCD73hi population and a CD146loCD73lo population (Figure 1A). Bromodeoxyuridine pulsing of hEMP-derived mesenchymal cultures showed that a significantly higher frequency of CD146hiCD73hi cells were cycling relative to CD146loCD73lo cells (Figure 1B) (p ≤ 0.05).
RNA sequencing (RNA-seq) profiling of the two mesenchymal populations demonstrated that 916 genes were differentially expressed (558 up- and 358 downregulated in CD146hiCD73hi relative to CD146loCD73lo cells; false discovery rate [FDR] < 0.01) (Figure 1C). Across all hESC lines tested, CD146hiCD73hi mesenchyme expressed significantly higher levels of pericyte markers CSPG4 (which encodes NG2), NES (NESTIN), LEPR (leptin receptor), and the β2-adrenergic receptor ADRB2 (Crisan et al., 2008, Hoffman and Calvi, 2014, Kunisaki et al., 2013), and HSC regulatory genes, KITLG, IGFBP2, and TNC (Ding et al., 2012, Huynh et al., 2008, Nakamura-Ishizu et al., 2012, Zhang et al., 2008) (Figures 1C, S1C, S2A, and S2B). In contrast, the CD146loCD73lo mesenchyme expressed significantly higher levels of the niche factors CXCL12 and ADRB3 (Greenbaum et al., 2013, Méndez-Ferrer et al., 2008) (Figure 1C). Expression of extracellular matrix genes was upregulated after the hEMP stage in both mesenchymal phenotypes (Figure 1C).
Mesenchymal Support of HSPCs Ex Vivo
The ability of hEMP-derived CD146hiCD73hi and CD146loCD73lo cells to support HSPCs ex vivo was assessed by co-culturing cord blood (CB)-derived CD34+ cells on monolayers generated from each mesenchymal population. To determine the specific effect of each stromal subset, cultures were performed in basal medium with a low concentration of serum (5%) and without addition of cytokines.
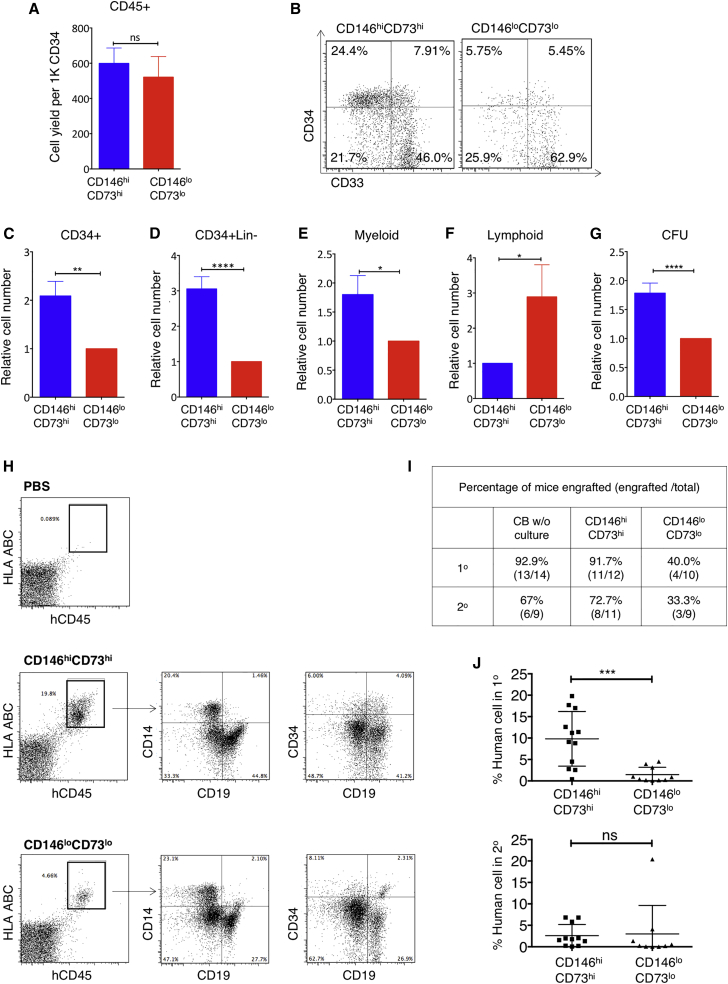
Although total hematopoietic (CD45+ cells) output was similar with both mesenchymal populations (Figure 2A), co-cultures with CD146hiCD73hi cells retained a significantly higher frequency and number of total CD34+ cells than co-culture with CD146loCD73lo mesenchymal cells (Figures 2B and 2C) (p < 0.01 and p < 0.001, respectively, n = 16). In particular, support of the more primitive CD34+Lin− subset was consistently seen in the CD146hiCD73hi co-cultures (Figure 2D). Myeloid output was higher in CD146hiCD73hi co-cultures, whereas more B lymphoid cells were generated on CD146loCD73lo mesenchyme (Figures 2E and 2F). Significantly more clonogenic cells (colony-forming unit [CFU]-C) were maintained in CD146hiCD73hi co-cultures than CD146loCD73lo co-cultures (p < 0.001, n = 16; Figure 2G). Of note, multiple other hPSC-derived phenotypic subsets were screened based on their CD146 and CD140a expression; irrespective of CD140a expression, those subsets with high levels of CD146 best supported HSPCs (Figure S3A).
Figure 2.
CD146hiCD73hi Mesenchyme Supports Ex Vivo Maintenance of Clonogenic, Engraftable, and Self-Renewing HSPCs
CD146hiCD73hi and CD146loCD73lo mesenchymal cells generated from H1-derived hEMP were isolated and co-cultured as monolayers with CB CD34+ cells in 5% serum without added cytokines. After 2 weeks, hematopoietic cells were assayed (A–G) in vitro and (H–J) in vivo.
(A) Total number of live (DAPI–) CD45+ cells recovered from co-cultures, shown as cell yield normalized to 1,000 input CD34+ cells.
(B) Representative FACS analysis of CD45+ gated cells from co-cultures.
(C–G) Cell yield of CD34+, CD34+ Lin−, CD14+ myeloid, CD10+/CD19+ B lymphoid cells, and CFU (after re-plating) recovered from 2-week CD146hiCD73hi co-cultures, normalized to CD146loCD73lo co-cultures.
(A and C–G) n = 16 independent experiments, each in duplicate. Shown are mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001; unpaired two-tailed Student's t test.
(H) Flow cytometry showing gating for the detection of human cells (hCD45+HLA ABC+) and multi-lineage reconstitution after primary transplant. PBS control with no cells injected.
(I) Proportion of mice with engraftment, i.e., detection of >1% human cells after primary transplant (1°) with 1 × 105 CD45+ cells obtained either from fresh CB CD34+ cells (CB w/o culture), or 2 weeks after co-culture on CD146hiCD73hi or CD146loCD73lo.
(J) Engraftment (percent of human cells) of mice 6 weeks after primary and secondary transplants (n = 5 independent experiments, total 9–14 mice per group; ∗∗∗p <.0001; ns, not significant).
Error bars represent SEM. See also Figure S3.
When Lin−CD34+ CD38− cells, a more primitive HSC-enriched population were studied, the co-culture system was modified by adding exogenous growth factors to allow sufficient cell growth for analysis. In the presence of cytokines, both mesenchymal populations were able to support CD34+ cells for 2–4 weeks. However, only the CD146hiCD73hi mesenchyme could maintain CD34+CD38−CD90+CD45RA−, phenotypic long-term HSCs (n = 4, p < 0.05), and CD34+CD33− cells (Figure S3B), demonstrating again the functional differences between the two mesenchymal populations in HSPC support.
CD146hiCD73hi Mesenchyme Maintains Human HSPCs with Repopulating Ability and Self-Renewal Potential
To understand whether support of engraftable HSPCs was influenced by the different mesenchymal subsets, equal numbers of CD34+ cells were co-cultured for 2 weeks with CD146hiCD73hi or CD146loCD73lo cells, and then equal numbers of total hematopoietic cells from the co-cultures were transplanted into sublethally irradiated NOD/SCID/IL-2 receptorγ−/− (NSG) mice. Of note, the co-culture conditions were not designed to expand the number of CD34+ cells, as the cultures did not include growth factors (in contrast to all current ex vivo expansion protocols). Based on flow cytometry analysis of bone marrow (Figure 2H), a similar frequency of mice transplanted with hematopoietic cells co-cultured with CD146hiCD73hi cells engrafted (albeit at lower levels) compared with those transplanted with fresh (non-cultured) CD34+ cells (11/12 versus 13/14, respectively) (Figures 2I and S3C), whereas only 40% of mice transplanted with hematopoietic cells from CD146loCD73lo co-cultures demonstrated any detectable engraftment (≥1% huCD45+HLA+ cells in bone marrow) (p < 0.01) (Figure 2I). CD34+ progenitors and multi-lineage reconstitution (CD19+ lymphoid cells and CD14+ myeloid cells) were detected in the bone marrow, spleen, and peripheral blood of all mice that received hematopoietic cells from CD146hiCD73hi co-cultures (Figure 2H). Levels of engraftment were also significantly higher in animals that received CD146hiCD73hi co-cultures compared with CD146loCD73lo co-cultures (p < 0.001) (Figure 2J). Irrespective of stromal co-culture conditions, mice that demonstrated primary engraftment could engraft in secondary recipients (Figure 2J).
Together, these data indicate that at least two functionally distinct mesenchymal populations were generated from hPSC-derived mesoderm; high CD146 and CD73 expression marked a mesenchymal subpopulation capable of maintaining engraftable HSPCs ex vivo, while CD146loCD73lo mesenchyme had significantly less HSPC-supportive capacity.
HSPC Support Is Sustained through Intercellular Interactions, in Part by Notch Signaling
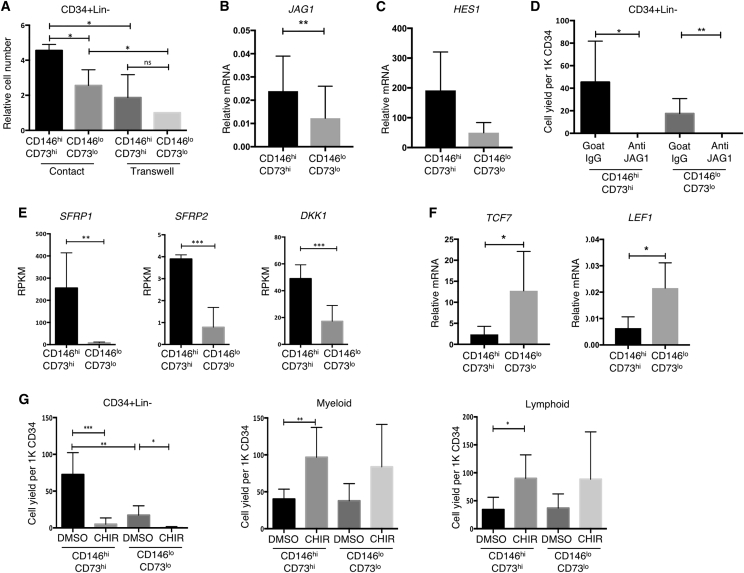
Prevention of direct contact between CD146hiCD73hi cells and CB CD34+ cells using a transwell culture system significantly reduced the number of CD34+Lin− cells maintained in culture (p < 0.05, n = 3) (Figure 3A). Output of CD146loCD73lo co-cultures was also reduced in transwells, although to a lesser degree. Thus intercellular mechanisms provide at least part of the HSPC-supportive capacity of CD146hiCD73hi mesenchyme.
Figure 3.
Notch Inhibition and Activation of Wnt Signaling in hPSC-Mesenchyme Co-cultures Leads to Loss of HSPC
(A) Shown is relative number of CD34+Lin− cells in contact and transwell co-culture normalized to yield from CD146loCD73lo transwell co-cultures (n = 3 independent experiments, mean ± SEM. ∗p < 0.05).
(B) qRT-PCR of JAG1 in CD146hiCD73hi and CD146loCD73lo mesenchyme (n = 14 biological replicates, ∗∗p < 0.01).
(C) qRT-PCR of HES1 in CD34+ cells recovered from CD146hiCD73hi and CD146loCD73lo mesenchyme after 7 days of co-culture without extra cytokines (n = 5 biological replicates, p = 0.1789).
(D) Number of CD34+ Lin− cells harvested from hPSC-mesenchyme after co-culture in the presence of human JAG1 blocking antibody or control goat IgG (∗p < 0.05, ∗∗p < 0.01, n = 3 independent experiments).
(E) Expression of WNT inhibitors SFRP1, SFRP2, and DKK1 in CD146hiCD73hi and CD146loCD73lo mesenchyme (n = 3 biological replicates, RNA-seq ∗∗FDR < 0.01, ∗∗∗FDR < 0.001).
(F) qRT-PCR of WNT target genes TCF7 and LEF1 in Lin+ cells recovered from CD146hiCD73hi and CD146loCD73lo mesenchyme after 7 days of co-culture without extra cytokines (n = 5 biological replicates, ∗p < 0.05).
(G) CD34+ Lin−, myeloid and lymphoid cell numbers after activation of Wnt signaling by addition of 3 μM CHIR99021 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant; n = 3 independent experiments).
Error bars represent SEM. See also Figure S4.
Notch signaling is a well-established cell contact-dependent paracrine factor and deletion of Jagged1 (encoded by Jag1) results in premature exhaustion of the murine HSC pool (Poulos et al., 2013). Both mesenchymal subsets expressed the Notch ligand JAG1, with significantly higher levels in CD146hiCD73hi mesenchyme (Figures 3B, S2A, and S2B); DLL4 and DLL1 were not expressed in either mesenchymal population (not shown). Expression of the Notch target HES1 tended to be higher in the CD34+ cells co-cultured with CD146hiCD73hi mesenchyme compared with CD146loCD73lo supported hematopoietic cells (p = 0.1789) (Figure 3C). Addition of a neutralizing antibody to human JAG1 caused a profound (>300-fold) decrease in the number of CD34+Lin− cells in CD146hiCD73hi co-cultures (p ≤ 0.05, n = 3) and 250-fold decrease in CD146loCD73lo co-cultures (Figure 3D). Of note, cell output was not significantly reduced when anti-JAG1 was added to co-cultures on OP9, a mouse stromal cell line, thus excluding non-specific cytotoxicity from the blocking antibody (Figure S4A).
Thus, although expression of JAG1 was significantly higher in CD146hiCD73hi stroma, HSPC support was dependent on JAG1-mediated activation of Notch signaling in both types of hPSC-mesenchymal co-cultures.
Wnt Inhibitors Are Highly Expressed in CD146hiCD73hi Stroma
Across the entire RNA-seq dataset, the most highly differentially expressed gene between CD146hiCD73hi and CD146loCD73lo cells was the Wnt antagonist Secreted Frizzled-Related Protein 1 (SFRP1) (33-fold difference, FDR < 0.01) (Figure 3E). Expression of other Wnt inhibitors was also significantly higher in CD146hiCD73hi mesenchyme including SFRP2 (FDR < 0.001) and DKK1 (FDR < 0.001) (Figure 3E). Expression of Wnt downstream targets TCF7 and LEF1 was significantly higher in hematopoietic cells supported by CD146loCD73lo co-cultures compared with those by CD146hiCD73hi mesenchyme (n = 5, p < 0.05) (Figure 3F).
Although neither addition of soluble SFRP1 or inhibition of SFRP1 had any detectable effect on HSPCs in either CD146hiCD73hi or CD146loCD73lo co-cultures (Figure S4B), addition of a Wnt agonist, the GSK-3 inhibitor CHIR99021 (CHIR), promoted rapid differentiation into both myeloid and lymphoid lineages, and loss of CD34+Lin− cells in both CD146hiCD73hi and CD146loCD73lo co-cultures (Figure 3G). This latter observation is consistent with previous findings that canonical Wnt signaling regulates hematopoiesis in a dosage-dependent fashion, with high Wnt activation detrimental to HSC self-renewal leading to depletion of the LT-HSC pool (Famili et al., 2016, Luis et al., 2011). It is possible that functional redundancy among Wnt inhibitors obscures the impact of manipulating expression of single factors such as SFRP1 on HSPC support during co-culture.
hPSC-Derived CD146hiCD73hi Mesenchyme Shares a Coordinated Gene Expression Profile that Resembles that of Primary Human Pericytes
Given the complexity of signals produced in the HSC niche, it seems likely that no single factor or mechanism mediates the differential support of hematopoiesis from different mesenchymal subpopulations. As molecular characterization and functional studies of hPSC-derived CD146hiCD73hi and CD146loCD73lo mesenchyme showed strong parallels to populations from primary human tissue, we performed comparative transcriptome analysis of each phenotypic population derived from hPSCs with those from primary adipose tissue (freshly isolated or after culture).
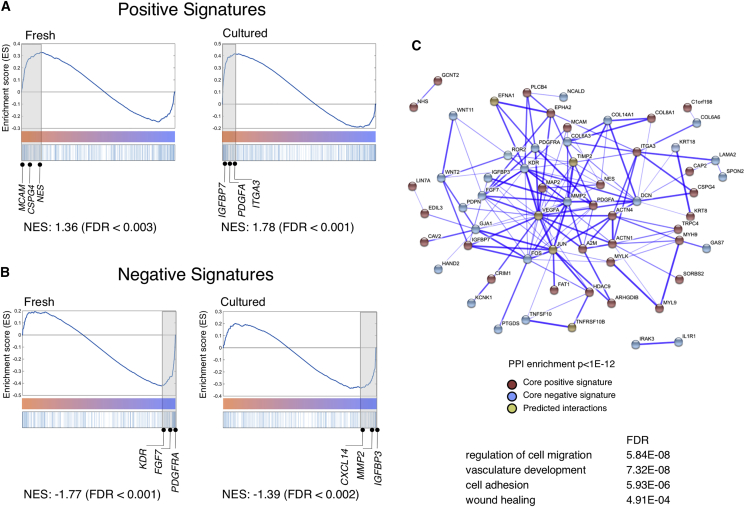
Principal-component analysis showed that the populations clustered mostly based on source rather than immunophenotype (data not shown). Similar observations that tissue-imprinted genes constitute major variances in unsupervised analysis have been reported in other comparative studies (Charbord et al., 2014). To uncover shared functional gene sets across stromal lines with HSPC-supportive capacity, we performed gene set enrichment analysis (Subramanian et al., 2005) as follows. The most up- and downregulated genes in hPSC-CD146hiCD73hi cells (compared with hPSC-CD146loCD73lo cells) were defined as positive and negative signatures, respectively (FDR < 0.01 and at least 2-fold difference). These signatures of hPSC-derived CD146hiCD73hi cells were compared with the global expression profiles of primary CD146+ pericytes, ranked by differential expression to primary CD146− cells. Significant enrichment was found between the positive signatures of hPSC-derived CD146hiCD73hi cells with genes upregulated in adult pericytes (Figure 4A, enrichment score of 1.36 and 1.78 in fresh and cultured pericytes respectively). Similarly, the negative signatures of hPSC-CD146hiCD73hi cells were significantly enriched in genes highly downregulated in primary pericytes (Figure 4B, enrichment score of −1.77 and −1.39). Thus hPSC- and adipose-derived CD146hiCD73hi cells exhibited similar transcriptional signatures.
Figure 4.
hPSC-Derived CD146hiCD73hi Mesenchyme Shares a Molecular Signature with Primary Human Adult Pericytes
(A and B) Gene set enrichment analysis of hPSC-derived mesenchyme and lipoaspirate-derived populations (both freshly sorted and cultured). (A) Positive and (B) negative signatures were defined, respectively, as up- and downregulated genes in hPSC-derived CD146hiCD73hi compared with hPSC-derived CD146loCD73lo cells. The x axis represents genes ranked from positive to negative fold change in lipoaspirate-CD146+ compared with lipoaspirate-CD146− cells. Genes in the core enrichment set are highlighted (shaded gray boxes) and selected genes in this core enrichment set are shown.
(C) Up- and downregulated genes common to the core sets of CD146hi from hPSC and fresh and cultured primary CD146+ cells were imported into the STRING database, which identified a network of known protein-protein interactions (enrichment p value = 0), along with a number of high-confidence (score > 0.9999) predicted interactions. Line thickness is proportional to the confidence level of each interaction. See also Table S1.
The shared core transcriptional signature obtained from both hPSC- and adipose-derived CD146++ cells (104 genes, Table S1) was imported into the STRING database to identify known and predicted protein interactions (STRING protein-protein interaction enrichment p <10 × 10−12) (Figure 4C). Of note, the dominant functional categories identified in the STRING analysis were those enriched in biological processes characteristic of pericytes, i.e., cell adhesion (FDR < 6 × 10−6), vasculature development (FDR < 7 × 10−8), regulation of cell motion (FDR < 3 × 10−8), and wound healing (FDR < 3 × 10−4), among others (Figure 4C). Therefore, the shared transcriptional signature between hPSC- and adipose-derived CD146++ cells is significantly enriched in processes involved in niche homeostasis and maintenance.
Discussion
Mesenchymal stromal/stem cells, either derived from primary tissues or differentiated from hPSC, are conventionally isolated and expanded as a population of cells loosely defined by CD73, CD105, and CD90 expression (Murray et al., 2014). Our data strongly argue that phenotypically, molecularly, and functionally distinct populations coexist within these conventional definitions of mesenchymal stroma. Previously, CD146 was shown to define mesenchymal precursors adjacent to the vasculature of multiple human organs, including bone marrow and adipose tissues (Crisan et al., 2009). Other studies have derived CD146+ pericytes from hPSCs, demonstrating the capacity of these cells to reconstruct a vascular network (Dar et al., 2012, Kusuma et al., 2013, Orlova et al., 2014). Our previous work (Corselli et al., 2013) identified immunophenotypic subsets of mesenchyme isolated from human primary tissues and found that a subpopulation of perivascular mesenchyme with high expression of CD146 possessed significantly greater HSPC supporting ability. In the current paper, we show that two phenotypically, functionally, and molecularly distinct mesenchymal populations can be faithfully recapitulated in the hPSC system. We also provide the transcriptome profiling of these mesenchymal populations, revealing the HSPC-supportive CD146hiCD73hi cells to be closely related to primary pericytes.
hPSC-derived CD146hiCD73hi mesenchyme showed high expression of NES, LEPR, and CSPG4, markers of peri-sinusoidal and peri-arteriolar cells in the murine HSC niche (Ding et al., 2012, Kunisaki et al., 2013, Méndez-Ferrer et al., 2010, Zhou et al., 2014). The relevance of these perivascular cells has recently been addressed in the context of human hematopoiesis (Corselli et al., 2013, Pinho et al., 2013). PDGFRα/CD140a, which is a long-known marker for murine mesenchymal stromal cells (MSCs), was also reported on human fetal bone marrow MSCs capable of supporting HSPC expansion (Pinho et al., 2013). In contrast, later studies using adult human bone marrow reported that MSCs with hematopoiesis-supporting ability were highly enriched within the CD140alo/− population but not in CD140a+ cells (Li et al., 2014). These conflicting results may be explained by reports of human mesenchymal stroma, including our own (Corselli et al., 2013), which suggest that CD140a expression is differentially regulated both developmentally and across tissue types. A recent report demonstrated that nestin+ mesenchyme in the murine fetal liver HSC niche expressed CD146 (Khan et al., 2016), further supporting the inclusion of CD146 in defining mesenchyme with HSC niche activity.
In the present study, both CD146hiCD73hi and CD146loCD73lo cells from hPSCs expressed known HSC niche genes, such as SLIT2 and ANGPT1. However, other niche factors, such as KITL, JAG1, and IGFBP2, were specifically upregulated in CD146hiCD73hi cells. As is likely for the components of the hematopoietic niche in vivo, we conclude the CD146hiCD73hi mesenchymal subset supports HSPCs through a combination of mechanisms that involve cell-cell contact (e.g., Notch signaling and adhesion molecules) and secreted factors (e.g., Wnt regulators and cytokines). Time course analysis for the development of CD146hiCD73hi and CD146loCD73lo cells showed that they could each be distinguished early during mesenchymal differentiation. However, clonal data are needed to understand the developmental relationships of these cell types. It should also be acknowledged that it remains unclear whether the phenotypes examined in our study represent cell types at the extremes of a functional spectrum.
The identification of distinct mesenchymal subsets generated from hPSCs provides a foundation from which developmental and functional differences can be further defined, and the application of each population for cell therapy explored. The ability to prospectively isolate human stroma with HSC-supporting capacity is particularly relevant given clinical interest in using stroma co-culture as a platform for HSC expansion (Hofmeister et al., 2007).
While this study focused on defining cellular subsets within hPSC-derived mesenchyme that would support a well-characterized HSPC population, the existence of distinct mesenchymal subsets during hPSC differentiation has broader implication for the directed differentiation of hPSCs toward definitive, functional HSCs. Further study will be needed to define the optimal hPSC-derived mesenchyme that would promote HSC specification.
Experimental Procedures
Mesoderm Differentiation from hPSCs to Generate hEMPs
The hPSC lines H1 (WiCell, Madison, WI), and UCLA3 and UCLA6 (UCLA BSCRC hESC Core) were maintained and expanded on irradiated primary mouse embryonic fibroblasts (EMD Millipore, Billerica, MA). All hPSC experiments were approved by the UCLA Embryonic Stem Cell Research Oversight (ESCRO) committee. Mesoderm commitment was induced as described previously (Evseenko et al., 2010, Hoffman and Calvi, 2014) (Supplemental Experimental Procedures). CD326−CD56+ embryonic mesoderm progenitors (hEMPs) were isolated by flow cytometry at day 3.5 (Figure 1A) and seeded onto Matrigel for mesenchymal differentiation over the next 14 days.
Mesenchymal Differentiation from hEMPs
To induce mesenchymal differentiation, hEMPs were seeded at 50,000 cells/well into 6-well tissue culture plates, pre-coated with Matrigel. Day 0–7 of differentiation, MesenCult Proliferation Kit, Human (STEMCELL Technologies) supplemented with SB-431542 (10 μM, R&D Systems) was used. Day 0–3, 10 μM Rock inhibitor (Y27632 hydrochloride; R&D Systems) and 10 μg/mL gentamicin (Gibco, Thermo Fisher Scientific, Waltham, MA) were also added. Day 7–14, medium was switched to EGM-2 (Lonza) supplemented with SB-431542 (10 μM). Day 14–18, cells were dissociated into single-cell suspension with Accutase (Innovative Cell Technologies, San Diego, CA), and analyzed or isolated by fluorescence-activated cell sorting (FACS) (see Supplemental Experimental Procedures).
Co-culture of Stromal Cells and CB CD34+ Cells
hEMP-derived mesenchyme (CD146hiCD73hi and CD146loCD73lo) was isolated from mesenchymal cultures after 14–18 days and re-plated in 96-well tissue culture-treated plates at 6.5 × 103 cells per well in 100 μL of EGM-2. Three days later, CB CD34+ cells (>80% purity, 1 × 105 per well) were plated on 80% confluent stroma in 200 μL of co-culture medium consisting of RPMI 1640, L-glutamine, 5% fetal bovine serum, and penicillin/streptavidin. No supplemental cytokines were added unless otherwise indicated (Lin−CD34+CD38− experiments). Cells were harvested after 2 weeks unless otherwise indicated for FACS analysis, CFU assay, and in vivo repopulating assay.
Graphical and Statistical Analysis
Graphs were generated and statistics analyzed using GraphPad Prism software. Paired parametric two-tailed t tests were used to calculate p values except in cases where normalized values were used, in which case unpaired parametric t tests were used. p < 0.05 was considered statistically significant.
Author Contributions
C.J.C. and S.L., conception and design, collection and assembly of data, data analysis and interpretation, and manuscript writing. M.C., conception and design and collection of data. D.C., RNA-seq data analysis and interpretation. Y.Z. and C.B.H., collection of data. R.H. and B.P., RNA-seq data collection. G.M.C., conception and design, data analysis and interpretation, manuscript writing, financial support, and final approval of manuscript.
Acknowledgment
We thank J. Scholes and F. Codrea at the UCLA Broad Stem Cell Research Center (BSCRC) Flow Cytometry Core for assistance with cell sorting and R. Chan for assistance with specimen processing. The research was made possible by grants from the California Institute of Regenerative Medicine (CIRM), grant numbers RB3-05217 (to G.M.C.) and TR2-01821 (to B.P.). C.J.C. and M.C. acknowledge the support of the California Institute for Regenerative Medicine Training Grant (TG2-01169), and S.L. acknowledges the support of a fellowship grant from the UCLA BSCRC Training Program. Core services were supported by the Genomics Shared Resource (GSR) in the UCLA Jonsson Comprehensive Cancer Center (NIH/NCI 5P30CA016042), the BSCRC Flow Cytometry Core, and the UCLA Center for AIDS Research (CFAR) Virology Core Lab (NIH/NIAID AI028697).
Published: January 4, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.12.005.
Accession Numbers
The accession numbers for the RNA sequence data reported in this paper are GEO: GSE77879 and GSE83443.
Supplemental Information
References
- Charbord P., Pouget C., Binder H., Dumont F., Stik G., Levy P., Allain F., Marchal C., Richter J., Uzan B. A systems biology approach for defining the molecular framework of the hematopoietic stem cell niche. Cell Stem Cell. 2014;15:376–391. doi: 10.1016/j.stem.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Chin C.J., Cooper A.R., Lill G.R., Evseenko D., Zhu Y., He C.B., Casero D., Pellegrini M., Kohn D.B., Crooks G.M. Genetic tagging during human mesoderm differentiation reveals tripotent lateral plate mesodermal progenitors. Stem Cells. 2016;34:1239–1250. doi: 10.1002/stem.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corselli M., Chin C.J., Parekh C., Sahaghian A., Wang W., Ge S., Evseenko D., Wang X., Montelatici E., Lazzari L. Perivascular support of human hematopoietic stem/progenitor cells. Blood. 2013;121:2891–2901. doi: 10.1182/blood-2012-08-451864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisan M., Chen C.W., Corselli M., Andriolo G., Lazzari L., Péault B. Perivascular multipotent progenitor cells in human organs. Ann. N. Y. Acad. Sci. 2009;1176:118–123. doi: 10.1111/j.1749-6632.2009.04967.x. [DOI] [PubMed] [Google Scholar]
- Crisan M., Yap S., Casteilla L., Chen C.W., Corselli M., Park T.S., Andriolo G., Sun B., Zheng B., Zhang L. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008;3:301–313. doi: 10.1016/j.stem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Dar A., Domev H., Ben-Yosef O., Tzukerman M., Zeevi-Levin N., Novak A., Germanguz I., Amit M., Itskovitz-Eldor J. Multipotent vasculogenic pericytes from human pluripotent stem cells promote recovery of murine ischemic limb. Circulation. 2012;125:87–99. doi: 10.1161/CIRCULATIONAHA.111.048264. [DOI] [PubMed] [Google Scholar]
- Ding L., Saunders T.L., Enikolopov G., Morrison S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evseenko D., Zhu Y., Schenke-Layland K., Kuo J., Latour B., Ge S., Scholes J., Dravid G., Li X., MacLellan W.R. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2010;107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famili F., Brugman M.H., Taskesen E., Naber B.E., Fodde R., Staal F.J. High levels of canonical Wnt signaling lead to loss of stemness and increased differentiation in hematopoietic stem cells. Stem Cell Reports. 2016;6:652–659. doi: 10.1016/j.stemcr.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell P.I., Hexum M.K., Kopher R.A., Lepley M.A., Gussiaas A., Kaufman D.S. Functional assessment of hematopoietic niche cells derived from human embryonic stem cells. Stem Cells Dev. 2014;23:1355–1363. doi: 10.1089/scd.2013.0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A., Hsu Y.M., Day R.B., Schuettpelz L.G., Christopher M.J., Borgerding J.N., Nagasawa T., Link D.C. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C.M., Calvi L.M. Minireview: complexity of hematopoietic stem cell regulation in the bone marrow microenvironment. Mol. Endocrinol. 2014;28:1592–1601. doi: 10.1210/me.2014-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmeister C.C., Zhang J., Knight K.L., Le P., Stiff P.J. Ex vivo expansion of umbilical cord blood stem cells for transplantation: growing knowledge from the hematopoietic niche. Bone Marrow Transpl. 2007;39:11–23. doi: 10.1038/sj.bmt.1705538. [DOI] [PubMed] [Google Scholar]
- Huynh H., Iizuka S., Kaba M., Kirak O., Zheng J., Lodish H.F., Zhang C.C. Insulin-like growth factor-binding protein 2 secreted by a tumorigenic cell line supports ex vivo expansion of mouse hematopoietic stem cells. Stem Cells. 2008;26:1628–1635. doi: 10.1634/stemcells.2008-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin T., Gur-Cohen S., Spencer J.A., Schajnovitz A., Ramasamy S.K., Kusumbe A.P., Ledergor G., Jung Y., Milo I., Poulos M.G. Distinct bone marrow blood vessels differentially regulate haematopoiesis. Nature. 2016;532:323–328. doi: 10.1038/nature17624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan J.A., Mendelson A., Kunisaki Y., Birbrair A., Kou Y., Arnal-Estapé A., Pinho S., Ciero P., Nakahara F., Ma'ayan A. Fetal liver hematopoietic stem cell niches associate with portal vessels. Science. 2016;351:176–180. doi: 10.1126/science.aad0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Butler J.M., O'Donnell R., Kobayashi M., Ding B.S., Bonner B., Chiu V.K., Nolan D.J., Shido K., Benjamin L. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nat. Cell Biol. 2010;12:1046–1056. doi: 10.1038/ncb2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki Y., Bruns I., Scheiermann C., Ahmed J., Pinho S., Zhang D., Mizoguchi T., Wei Q., Lucas D., Ito K. Arteriolar niches maintain haematopoietic stem cell quiescence. Nature. 2013;502:637–643. doi: 10.1038/nature12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusuma S., Shen Y.I., Hanjaya-Putra D., Mali P., Cheng L., Gerecht S. Self-organized vascular networks from human pluripotent stem cells in a synthetic matrix. Proc. Natl. Acad. Sci. USA. 2013;110:12601–12606. doi: 10.1073/pnas.1306562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ghazanfari R., Zacharaki D., Ditzel N., Isern J., Ekblom M., Méndez-Ferrer S., Kassem M., Scheding S. Low/negative expression of PDGFR-α identifies the candidate primary mesenchymal stromal cells in adult human bone marrow. Stem Cell Reports. 2014;3:965–974. doi: 10.1016/j.stemcr.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis T.C., Naber B.A., Roozen P.P., Brugman M.H., de Haas E.F., Ghazvini M., Fibbe W.E., van Dongen J.J., Fodde R., Staal F.J. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell. 2011;9:345–356. doi: 10.1016/j.stem.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Murray I.R., West C.C., Hardy W.R., James A.W., Park T.S., Nguyen A., Tawonsawatruk T., Lazzari L., Soo C., Péault B. Natural history of mesenchymal stem cells, from vessel walls to culture vessels. Cell Mol. Life Sci. 2014;71:1353–1374. doi: 10.1007/s00018-013-1462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Lucas D., Battista M., Frenette P.S. Haematopoietic stem cell release is regulated by circadian oscillations. Nature. 2008;452:442–447. doi: 10.1038/nature06685. [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma'ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura-Ishizu A., Okuno Y., Omatsu Y., Okabe K., Morimoto J., Uede T., Nagasawa T., Suda T., Kubota Y. Extracellular matrix protein tenascin-C is required in the bone marrow microenvironment primed for hematopoietic regeneration. Blood. 2012;119:5429–5437. doi: 10.1182/blood-2011-11-393645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova V.V., Drabsch Y., Freund C., Petrus-Reurer S., van den Hil F.E., Muenthaisong S., Dijke P.T., Mummery C.L. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler. Thromb. Vasc. Biol. 2014;34:177–186. doi: 10.1161/ATVBAHA.113.302598. [DOI] [PubMed] [Google Scholar]
- Pinho S., Lacombe J., Hanoun M., Mizoguchi T., Bruns I., Kunisaki Y., Frenette P.S. PDGFRα and CD51 mark human nestin+ sphere-forming mesenchymal stem cells capable of hematopoietic progenitor cell expansion. J. Exp. Med. 2013;210:1351–1367. doi: 10.1084/jem.20122252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulos M.G., Guo P., Kofler N.M., Pinho S., Gutkin M.C., Tikhonova A., Aifantis I., Frenette P.S., Kitajewski J., Rafii S. Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep. 2013;4:1022–1034. doi: 10.1016/j.celrep.2013.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodyanik M.A., Yu J., Zhang X., Tian S., Stewart R., Thomson J.A., Slukvin I.I. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell. 2010;7:718–729. doi: 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.C., Kaba M., Iizuka S., Huynh H., Lodish H.F. Angiopoietin-like 5 and IGFBP2 stimulate ex vivo expansion of human cord blood hematopoietic stem cells as assayed by NOD/SCID transplantation. Blood. 2008;111:3415–3423. doi: 10.1182/blood-2007-11-122119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O., Yue R., Murphy M.M., Peyer J.G., Morrison S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.