Summary
Undifferentiated spermatogonia comprise a pool of stem cells and progenitor cells that show heterogeneous expression of markers, including the cell surface receptor GFRα1. Technical challenges in isolation of GFRα1+ versus GFRα1– undifferentiated spermatogonia have precluded the comparative molecular characterization of these subpopulations and their functional evaluation as stem cells. Here, we develop a method to purify these subpopulations by fluorescence-activated cell sorting and show that GFRα1+ and GFRα1– undifferentiated spermatogonia both demonstrate elevated transplantation activity, while differing principally in receptor tyrosine kinase signaling and cell cycle. We identify the cell surface molecule melanocyte cell adhesion molecule (MCAM) as differentially expressed in these populations and show that antibodies to MCAM allow isolation of highly enriched populations of GFRα1+ and GFRα1– spermatogonia from adult, wild-type mice. In germ cell culture, GFRα1– cells upregulate MCAM expression in response to glial cell line-derived neurotrophic factor (GDNF)/fibroblast growth factor (FGF) stimulation. In transplanted hosts, GFRα1– spermatogonia yield GFRα1+ spermatogonia and restore spermatogenesis, albeit at lower rates than their GFRα1+ counterparts. Together, these data provide support for a model of a stem cell pool in which the GFRα1+ and GFRα1– cells are closely related but show key cell-intrinsic differences and can interconvert between the two states based, in part, on access to niche factors.
Keywords: spermatogonial stem cells, germ cells, telomerase, germ line, stem cells, niche, transplantation, RNA-seq, FACS
Highlights
-
•
A new method to purify GFRα1+ and GFRα1– undifferentiated spermatogonia by FACS
-
•
GFRα1+ and GFRα1– cells differ in receptor tyrosine kinase signaling
-
•
Differential surface MCAM expression can distinguish GFRα1+ and GFRα1– cells
-
•
GFRα1– cells have a third of the transplantation activity of GFRα1+ cells
In this article, Garbuzov and colleagues devise a new strategy for isolating pure populations of GFRα1+ and GFRα1– undifferentiated spermatogonia from adult testis of TertTomato reporter mice based on expression of telomerase and GFRα1. Transcriptional profiling showed a remarkable similarity between GFRα1+ and GFRα1– cells, and both populations showed elevated stem cell activity by transplantation.
Introduction
In tissues that require continuous renewal during life, stem cells fuel the generation of differentiated progeny. The ability to identify pathways important for stem cell function and to distinguish between populations with self-renewal capacity or commitment requires a robust method for isolating defined populations as well as a means for testing their stem cell potential via transplantation. In the mammalian testis, spermatogonial stem cells (SSCs) are the mitotic cells that maintain the germline by undergoing both self-renewal and differentiation, eventually yielding haploid sperm (Spradling et al., 2011). The exact identity and nature of the SSC pool remains incompletely understood.
SSCs, and all cells with transplantation potential in the testis, reside in a population of “undifferentiated type A” spermatogonia (A-undiff), named originally based on their undifferentiated morphology (Huckins, 1971). These rare cells on the basement membrane of seminiferous tubules are found as single cells, pairs, and chains of 4 to 16 cells (termed Asingle, Apair, and Aaligned4-16, respectively), as incomplete cell division in this compartment results in elongating cell syncytia. “Undifferentiated” spermatogonia mature to become “differentiated” spermatogonia, which are marked by expression of the cell surface receptor Kit (Schrans-Stassen et al., 1999). During each cycle of spermatogenesis, the vast majority of spermatogonia migrate luminally to enter meiosis. Based on histological observations, it was proposed that the SSC pool is comprised only of the Asingle cells, and that division into Apair represents commitment to a transiently amplifying progenitor (de Rooij, 1973, Huckins, 1971, Oakberg, 1971).
Recent studies have identified a number of genes that are expressed on a subset of Asingle cells, including Bmi1, Pax7, and Id4 (Aloisio et al., 2014, Helsel et al., 2017, Komai et al., 2014). In support of the Asingle model, transplantation of ID4-GFPBright spermatogonia from juvenile testis achieved a high transplantation efficiency (Helsel et al., 2017). However, whether all ID4+ cells function as SSCs in the adult or whether ID4 marks the entire population of SSCs is unclear.
Short-chain undifferentiated spermatogonia tend to express GFRα1, the cell surface receptor for the key self-renewal factor glial cell line-derived neurotrophic factor (GDNF) (Meng et al., 2000). Lineage tracing using GFRα1– CreER knockin mice revealed that GFRα1+ cells can give rise to long-term labeling of the germ cell compartment, indicating that SSCs reside within the GFRα1+ population (Hara et al., 2014, Nakagawa et al., 2007). Only a subset of undifferentiated spermatogonia express GFRα1. Seventy percent of undifferentiated spermatogonia do not express GFRα1, including 10%–30% of Asingle and 25%–50% of Apair (Gassei and Orwig, 2013, Grasso et al., 2012, Nakagawa et al., 2010), and the functional properties of these cell types are largely unexplored. The behavior of GFRα1– undifferentiated spermatogonia has been inferred by analyzing Neurogenin3-positive (NGN3+) cells, whose expression imperfectly marks the GFRα1– state. Analysis of NGN3-CreER knockin mice showed that NGN3+ cells can give rise to long-term labeling in a small subset of tracing events homeostatically, and to a greater degree after injury (Nakagawa et al., 2007, Nakagawa et al., 2010). However, approximately 10% of NGN3+ cells are also GFRα1+, so whether self-renewal potential is found outside of the GFRα1+ compartment remains unknown. Alternative approaches are required to understand the properties of GFRα1– spermatogonia.
Transplantation is a rigorous assay for stem cell potential and has been used extensively to quantify functional SSCs (Brinster and Zimmermann, 1994). Previous work has revealed that the SSC pool may reside within spermatogonia expressing Thy1, Itga6, Itgb1, Cdh1, Id4, and Pax7, among others (Aloisio et al., 2014, Helsel et al., 2017, Kubota et al., 2003, Phillips et al., 2010, Shinohara et al., 1999, Tokuda et al., 2007). Although GFRα1– expressing spermatogonia are thought to be among the most primitive cells in the SSC differentiation hierarchy, attempts to transplant this population did not show enrichment for SSC-repopulating activity (Buageaw et al., 2005, Grisanti et al., 2009).
We previously discovered that PLZF+ undifferentiated spermatogonia are characterized by high levels of telomerase, the enzyme that synthesizes telomere DNA repeats at chromosome ends (Pech et al., 2015). By generating TertTomato/+ reporter knockin mice, we identified a gradient of Tert transcription in the testis and used it to isolate undifferentiated spermatogonia. We also found that telomere dysfunction in Tert−/− mice induced depletion of the PLZF+ A-undiff pool over time, providing a cellular mechanism to explain the established infertility phenotype in telomerase knockout mouse strains (Lee et al., 1998, Pech et al., 2015). In this study, we develop methods to isolate highly purified populations of GFRα1–positive and GFRα1–negative undifferentiated spermatogonia from the testes of adult TertTomato/+ reporter mice and from wild-type mice. We leverage these techniques to define transcriptome-wide features and functional differences between these two cell populations that define the SSC pool.
Results
Purification of GFRα1+ and GFRα1– Undifferentiated Spermatogonia from Adult TertTomato/+ Reporter Mice
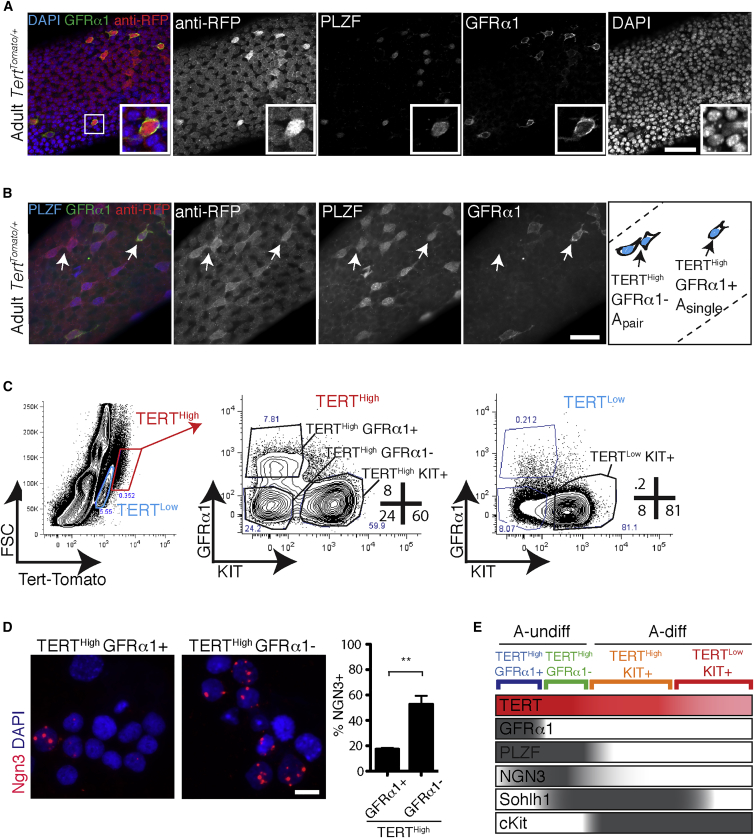
Using a Tert-Tomato transcriptional reporter of telomerase activity, we previously showed that TERTHigh KIT– population represents a pure population of PLZF+ undifferentiated spermatogonia (Pech et al., 2015). To determine the relationship between telomerase expression and GFRα1 expression, we performed whole-mount microscopy on adult seminiferous tubules. Triple-staining for PLZF, GFRα1 and Tomato revealed that effectively all GFRα1+ cells express Tert-Tomato (99.3% ± 0.5%; Figure 1A). Tert-Tomato expression was similarly homogeneous (99.8% ± 0.1%) within GFRα1– PLZF+ undifferentiated spermatogonia, which included both long and short chains of cells (Figure 1B). Thus, both GFRα1+ and GFRα1– undifferentiated spermatogonia are characterized by high Tert promoter activity.
Figure 1.
High Telomerase Expression Enables the Purification and Characterization of GFRα1+ and GFRα1– Undifferentiated Spermatogonia
(A) Whole-mount analysis of adult seminiferous tubules immunostained for GFRα1, PLZF, and anti-RFP in TertTomato/+ seminiferous tubules. A total of 99.3% ± 0.5% of GFRα1+ PLZF+ cells were Tert-Tomato+ (N = 370 cells; N = 4 mice); 99.8% ± 0.1% GFRα1– PLZF+ cells were Tert-Tomato+ (N = 1900 cells; N = 6 mice). Scale bar, 50 μm.
(B) Whole-mount analysis of adult seminiferous tubules immunostained for GFRα1, PLZF, and anti-RFP in TertTomato/+ seminiferous tubules. White arrows point to TERTHigh GFRα1− A-paired (left arrow) and TERTHigh GFRα1− A-single (right arrow) spermatogonia. Scale bar, 50 μm.
(C) Flow cytometry measurement of GFRα1 and KIT expression in TERTHigh cells. Panels are representative of at least six independent FACS runs.
(D) In situ hybridization for NGN3 mRNA on FACS-sorted cells of the indicated immunophenotypes. Percentage of NGN3+ cells was quantified. Mean and SEM are shown. Scale bar, 25 μm. N = 5–6 mice; at least 2,000 cells counted per condition. ∗∗p = 0.012.
(E) Interpretation of identities of various sorted cell types, based on whole-mount, cytospin, immunophenotype, and neonatal time course data.
To develop a method for the isolation of GFRα1+ and GFRα1– undifferentiated spermatogonia using fluorescence-activated cell sorting (FACS), dissociated adult testes were stained with antibodies against GFRα1 and KIT. We previously found that the TERTHigh population comprised approximately 50% KIT– cells, representing undifferentiated spermatogonia, and 50% KIT+ cells, representing early differentiating spermatogonia. TERTHigh KIT– cells transplanted efficiently, whereas TERTHigh KIT+ cells failed to transplant (Pech et al., 2015). FACS of whole testes showed a subpopulation of TERTHigh KIT– cells were GFRα1+, and that the GFRα1+ cells represented approximately one-third of the TERTHigh KIT– population, consistent with whole-mount analysis. GFRα1+ cells were absent in all other populations, including TERTLow cells and TERTHigh KIT+ (Figure 1C). To further assess the identity of the purified TERTHigh KIT– GFRα1+ and GFRα1– populations, we isolated them by FACS and employed RNA in situ hybridization (ISH) to assay for NGN3 mRNA, whose expression has been used as a surrogate marker for GFRα1– spermatogonia. RNA ISH on sorted TERTHigh KIT– GFRα1+ and GFRα1– populations showed that 17% ± 1% of GFRα1+ cells express Ngn3, whereas 53% ± 7% of GFRα1– cells express Ngn3 (p = 0.012, t test) (Figure 1D). These results indicate that Ngn3 is differentially expressed between the TERTHigh KIT– GFRα1+ and GFRα1– populations of undifferentiated spermatogonia, but that Ngn3 expression alone is insufficient to discriminate the GFRα1+ and GFRα1– populations.
These results provide strong evidence for the successful isolation of GFRα1+ and GFRα1– undifferentiated spermatogonia, based on intrinsic Tert promoter strength and cell surface phenotypes (summary in Figure 1E). Employing the Tert-Tomato reporter was essential for successful purification of these populations. Sorting based on GFRα1 expression alone did not allow isolation of a pure population of GFRα1+ cells, nor did it allow the discrimination of GFRα1– undifferentiated spermatogonia due to background staining for GFRα1 in the meiotic cells and spermatids (Figure S1A). Enrichment for A-undiff cells using the Tert reporter allowed detection of distinct GFRα1+ and GFRα1– A-undiff populations, enabling subsequent molecular and functional studies.
Isolation and Transcriptional Profiling of Four Distinct Spermatogonial Populations
To identify the differences between the GFRα1+ and GFRα1– undifferentiated spermatogonia we performed RNA sequencing (RNA-seq) to identify the transcriptional features of each population. From four independent TertTomato/+ mice, we sequenced the transcriptomes of FACS-purified TERTHigh GFRα1+ KIT– cells and TERTHigh GFRα1– KIT– cells. To put the transcriptional profiles in the context of spermatogenesis and differentiation, we also sequenced the population of TERTHigh KIT+ cells and TERTLow KIT+ cells (Pech et al., 2015), which represent early and late differentiating spermatogonia, respectively (Pech et al., 2015) (Figure S1). In addition, we isolated TERTHigh Oct4-GFP double-positive spermatogonia from postnatal day 6 (P6) juveniles. Spermatogonia isolated from juvenile testes are enriched for stem cell activity and have previously been used to gain insights about adult SSCs (Helsel et al., 2017, Kanatsu-Shinohara et al., 2011).
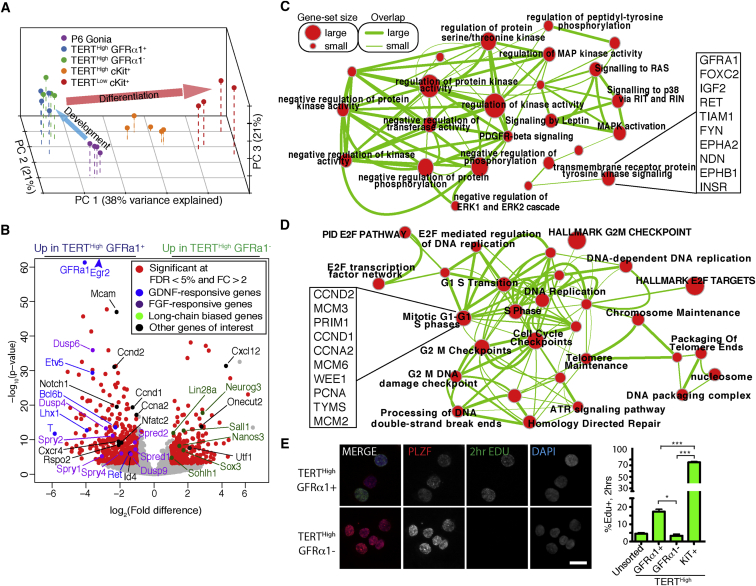
By principal-component analysis (PCA) and unsupervised hierarchical clustering, biological replicates from the same populations clustered together, confirming our ability to isolate pure populations with discrete identities (Figures 2A and S2A). PCA showed three principal components that explain a large proportion of the variance between the populations. The four adult populations lined up along axis PC1 in an order that recapitulated differentiation: the TERTHigh GFRα1+ KIT– cells and TERTHigh GFRα1– KIT– populations were the leftmost populations, followed by TERTHigh KIT+ cells further right, and with TERTLow KIT+ cells as the rightmost population (Figure 2A). By PCA, the GFRα1+ and GFRα1– spermatogonia cluster together, and neither population is closer to the more differentiated TERTHigh KIT+ cells. Our PCA analysis also showed that the P6 spermatogonia were significantly different from adult populations as they were separated from the rest of the samples along the PC2 axis (Figure 2A). We conclude that the PC1 axis captured the gene expression changes associated with differentiation, while the PC2 axis reflected changes associated with postnatal maturation. These data highlight the relatedness of the TERTHigh GFRα1+ and TERTHigh GFRα1– populations in the undifferentiated spermatogonia compartment.
Figure 2.
RNA-Seq Reveals That GFRα1+ Spermatogonia Are Defined by a Transcriptional Signature of Active GDNF and FGF Signaling
(A) Principal-component analysis (PCA) of transcriptomes from five isolated spermatogonial populations from adult and postnatal day 6 (P6) juvenile.
(B) Volcano plot of expression profiles comparing TERTHigh GFRα1+ KIT– to TERTHigh GFRα1– KIT– cells.
(C) MAP/ERK/protein phosphorylation cluster generated by Cytoscape Enrichment Map of gene set enrichment analysis (GSEA) results for TERTHigh GFRα1+.
(D) Cell-cycle/proliferation cluster generated by Cytoscape Enrichment Map of GSEA results for TERTHigh GFRα1+.
(E) Indicated cell types were sorted and cytospun. A 2 hr EdU pulse was visualized using Click chemistry, and the cells were then immunostained for the undifferentiated spermatogonia marker PLZF. Scale bar, 25 μm. Percentage of EdU+ cells was quantified. Mean and SEM are shown. (N = 5 mice; N = 900–10,000 cells per condition). ∗p < 0.05; ∗∗∗p < 0.001.
GFRα1+ Spermatogonia Are Defined by a Transcriptional Signature of Active GDNF and Fibroblast Growth Factor Signaling
By differential expression analysis, we identified 578 significantly upregulated and 430 significantly downregulated genes in TERTHigh GFRα1+ versus TERTHigh GFRα1– cells (5% false discovery rate and 2-fold change cutoff; Figure 2B; Table S1). The GFRα1 receptor was one of the most differentially expressed genes, and its co-receptor Ret was also highly differentially expressed (16.9-fold change, q = 3.9 × 10−58; 2.7-fold, q = 3.2 × 10−5, respectively). Id4, a marker of Asingle cells was also shown to be enriched in the GFRα1+ population (1.4-fold, q = 4.5 × 10−5). Furthermore, many of the most significantly upregulated genes in TERTHigh GFRα1– cells were factors known to be enriched in long-chained A-undiff cells Ngn3 (7.3-fold q = 5.3 × 10−16), Nanos3 (2.8-fold, q = 3.9 × 10−7), Sohlh1 (2.1-fold q = 4.3 × 10−4), Sox3 (3.3-fold q = 5.26 × 10−5), and Lin28a (2.2-fold q = 1.6 × 10−10) (Chakraborty et al., 2014, Phillips et al., 2010, Suzuki et al., 2009, Suzuki et al., 2012) Thus, these transcriptomic studies have captured key markers of both GFRα1+ and GFRα1– undifferentiated spermatogonia.
GFRα1– GDNF signaling is required for both the in vivo maintenance of SSCs and their ex vivo culture (Kanatsu-Shinohara et al., 2003, Meng et al., 2000). Many genes originally identified as being GDNF responsive in germline stem cell culture systems (Oatley et al., 2006) were highly enriched in TERTHigh GFRα1+ cells compared with TERTHigh GFRα1– spermatogonia: T (57.3-fold, q = 2.4 × 10−10), ETV5 (12.3-fold, q = 3.4 × 10−27), EGR2 (19.8-fold, q = 4.4 × 10−99), EGR3 (5.7-fold, q = 4.7 × 10−5), BCL6B (4.8-fold, q = 4.5 × 10−12), Tspan8 (1.2-fold, q = 7.3 × 10−3), and Lhx1 (15.5-fold, q = 3.0 × 10−11) (Figure 2B). In addition to GDNF, fibroblast growth factor (FGF) signaling is required for maintenance of SSCs in vivo and in culture (Hasegawa and Saga, 2014). Sprouty and Spred families of receptor tyrosine kinase (RTK) inhibitors and the DUSP family of MAPK phosphatases are transcriptionally induced during FGF responses (Branney et al., 2009), and this family of genes was highly upregulated in TERTHigh GFRα1+ cells (Figure 2B). These findings provide evidence that TERTHigh GFRα1+ cells actively receive FGF signals in vivo. Taken together, these transcriptomic studies indicate a specific induction of GDNF- and FGF-regulated genes in GFRα1+ cells.
To understand the transcriptomes in these two populations more generally, we analyzed the differentially expressed genes by gene set enrichment analysis and visualized the results using Cytoscape Enrichment Map (Merico et al., 2010). Three major clusters of enriched functional gene sets were found in genes upregulated in GFRα1+ cells (p-value cutoff, 0.001). The first cluster involves RTK-RAS-MAPK signaling and protein phosphorylation (Figure 2C). The RAS-MAPK pathway is known to be important downstream of GDNF and FGF signaling (Hasegawa et al., 2013, He et al., 2008). Numerous gene sets related to RTK-RAS-MAPK signaling were associated with genes upregulated in GFRα1+ cells. Many core genes in this enrichment cluster are the GDNF/FGF-responsive genes highlighted in the volcano plot (Figure 2B).
The second cluster involves cell-cycle progression, E2F transcription factor targets, DNA packaging, and replication (Figure 2D). These findings suggest a difference in cell-cycle activity between the GFRα1+ and GFRα1– undifferentiated spermatogonia. To test this idea, we assayed proliferation by in vivo 5-ethynyl-2′-deoxyuridine (EdU) labeling, followed by FACS purification, cytospin, and EdU detection. We found a marked difference in cell-cycle status between the two subpopulations of undifferentiated spermatogonia: GFRα1+ cells showed an S-phase fraction of 17.4% ± 1.4%, while GFRα1– cells exhibited an S-phase fractions of 3.3% ± 0.9% (p = 0.033, t test) (Figure 2E). These finding are consistent with the role for GDNF in spermatogonial self-renewal and proliferation (Meng et al., 2000, Tadokoro et al., 2002). The third, smaller cluster involved genes associated with morphogenesis and epithelium development (Figure S2B). Taken together, transcriptome and cell-cycle data suggest that GFRα1+ undifferentiated spermatogonia receive critical self-renewal and proliferation signals from their environment and that downregulating these pathways is an important characteristic of the transition to the GFRα1– state.
To understand the relationships between GFRα1+ and GFRα1– undifferentiated spermatogonia and other selections of undifferentiated spermatogonia, we compared the transcriptomes of GFRα1+ and GFRα1– cells with those of ID4-Bright cells, which show high transplantation potential, and ID4-Dim cells (Helsel et al., 2017). By differential expression analysis, the ID4-Bright and GFRa1+ undifferentiated spermatogonia were similar in expression of genes important for SSC maintenance and self-renewal, including: GFRa1, Taf4b, Zbtb16, Bcl6b, Lhx1, T, and Pou3f1, among others (Figure S3A). ID4 was enriched in ID4-Bright cells compared with GFRa1+ cells. Surprisingly, we found a number of genes associated with differentiated spermatogonia enriched in the ID4-Bright cells, including Stra8, Alcam, Sycp1, Nanos3, Dmrt1, Sox3, Sohlh1, and Kit (Figure S3A). A similar pattern was also seen in a comparison of GFRa1– cells with ID4-Bright cells (Figure S3B). PCA on the two ID4+ populations and our five populations to assess their relatedness revealed that both the ID4-Bright and the ID4-Dim cells cluster closely with the TERTHigh cells we isolated from day 6 neonates (P6) (Figure S3C). This relationship was also seen using unsupervised hierarchical clustering (Figure S3D). This similarity may occur because the ID4 populations in Helsel et al. were isolated from day 8 neonatal mice. Thus, the ID4-Bright cells from P8 mice are most similar transcriptionally to TERTHigh neonatal spermatogonia, and less similar to adult GFRα1+ and GFRα1– cells. These data highlight potential molecular differences between neonatal and adult SSC populations.
Elevated Melanocyte Cell Adhesion Molecule Is a Cell Surface Marker of the GFRα1+ State
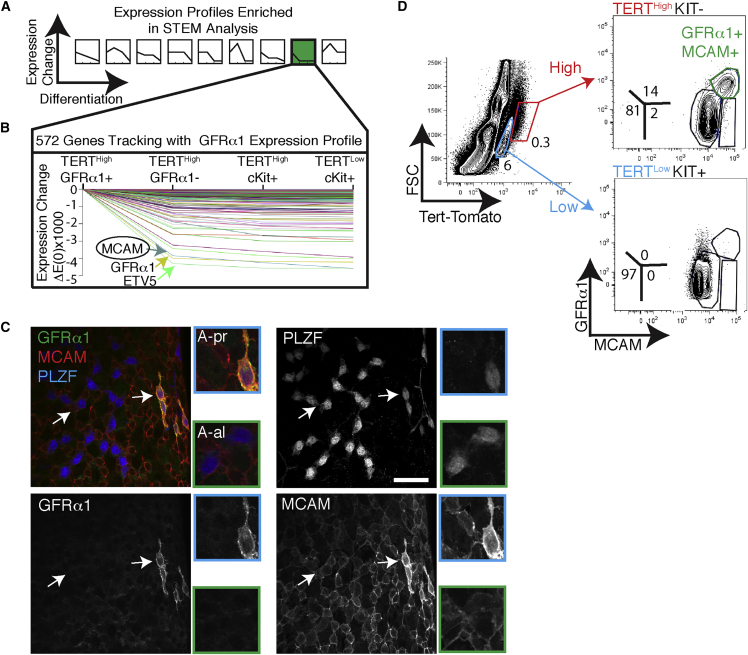
Tert reporter mice enabled efficient purification of many spermatogonial subsets, but the lack of good antibodies specific for undifferentiated spermatogonial subpopulations currently makes it difficult to isolate these cells from wild-type mice. To identify new markers, we leveraged our transcriptome datasets from four distinct subpopulations of undifferentiated and differentiated spermatogonia. We used Short Time-series Expression Miner (STEM) (Ernst and Bar-Joseph, 2006) to compare gene expression simultaneously across all four populations. STEM analysis takes an ordered collection of expression datasets, assigns each gene to a profile bin and computationally identifies statistically enriched profiles. STEM analysis of TERTHigh GFRα1+ KIT– cells, TERTHigh GFRα1– KIT– cells, TERTHigh KIT+, and TERTLow KIT+ cells identified nine enriched profiles (Figure 3A). We were specifically interested in genes that followed the expression profile of GFRα1 (Figure 3A, green pattern). STEM identified GFRα1, ETV5, and melanocyte cell adhesion molecules (MCAMs) as having highly similar expression profiles (Figure 3B and Table S2). MCAM, an immunoglobulin-superfamily surface protein shown to enrich for transplantation activity (Kanatsu-Shinohara et al., 2012), was also one of the top ten most differentially expressed genes between GFRα1+ and GFRα1– cells (Figure 2B; 4.6-fold change, q = 4.4 × 10−44). Thus, MCAM represents a candidate cell surface marker with an expression pattern similar to GFRα1. To test similarity at the protein level, we investigated MCAM expression using whole-mount immunostaining. MCAM protein was enriched in a subset of PLZF+ undifferentiated spermatogonia (Figure 3C). Elevated MCAM expression was restricted to short-chain PLZF+ cells. Co-staining revealed that these MCAMHigh cells were exclusively GFRα1+ PLZF+ spermatogonia (Figure 3C). Therefore, our analysis enabled the discovery of an independent marker, MCAM, for the GFRα1+ subset of undifferentiated spermatogonia.
Figure 3.
MCAM Is a Cell Surface Marker of the GFRα1+ State
(A) Nine patterns of gene expression changes across adult spermatogonial populations identified as statistically significant by Short Time-series Expression Miner (STEM).
(B) Details on STEM pattern no. 8, containing genes with peak expression in TERTHigh GFRα1+ cells, with diminished expression in all other cell types. Genes of interest are highlighted. The entire list of 575 genes is found in Table S2.
(C) Whole-mount analysis of tubules triple-stained for MCAM, GFRα1, and PLZF. All 76/76 GFRα1+ cells were MCAMHigh. White arrows point to cells shown in greater magnification in the panels to the right. Scale bar, 50 μm. N = 3 mice.
(D) Flow cytometry measurement of GFRα1 and MCAM expression in TERTHigh KIT– cells and TERTLow KIT+. Panels are representative of at least six independent FACS runs.
Isolation of Both GFRα1+ and GFRα1– Undifferentiated Spermatogonia Based on Differential MCAM Expression Using FACS
As MCAM is expressed on the cell surface, we tested its ability to enrich for spermatogonial populations using FACS. Staining dissociated tubules for GFRα1 and MCAM revealed a population of double-positive GFRα1+ MCAM+ cells in the TERTHigh KIT– fraction. This double-positive population was not present in the TERTLow KIT+ fraction (Figure 3D). These GFRα1+ MCAM+ cells isolated by FACS correspond to the same population seen in whole-mount immunostaining (Figure 3C).
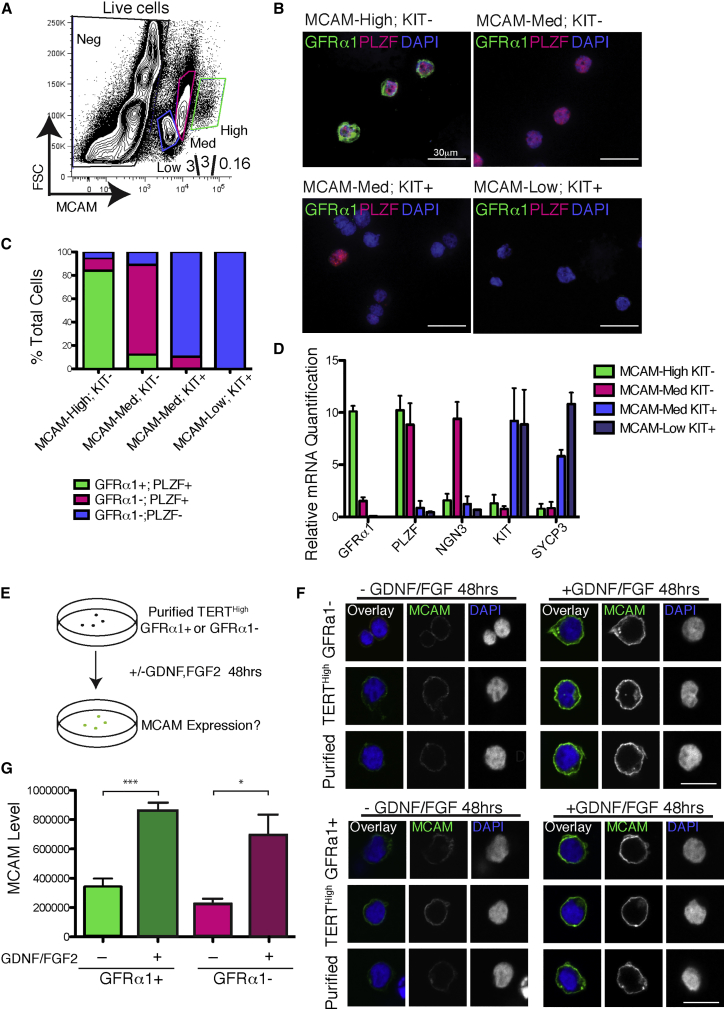
These results suggested that MCAM can be used to isolate GFRα1+ cells without relying on the Tert-Tomato reporter mouse or on GFRα1 staining. To test this hypothesis, we stained dissociated tubules from wild-type mice using antibodies against MCAM and KIT. By flow cytometry, the MCAM signal was sufficiently strong to separate three distinct populations by MCAM expression level: MCAMHigh, MCAMMed, and MCAMLow (Figure 4A). The MCAMHigh population was highly enriched for KIT– cells (85%–90% KIT–) (Figure S4A). The MCAMMed and MCAMLow populations were predominantly KIT+ cells (Figures S4B and S4C). To determine if MCAM levels along with staining for KIT allow us to isolate GFRα1+ and GFRα1– undifferentiated spermatogonia, we sorted MCAMHigh KIT– cells, MCAMMed KIT– cells, MCAMMed KIT+ cells, and MCAMLow KIT+ cells, cytospun them, and stained them for GFRα1 and PLZF (Figure 4B). The MCAMHigh KIT– cells were 84% GFRα1+ PLZF+. The MCAMMed KIT– population was 77% GFRα1– PLZF+. The MCAMMed KIT+ population was 90% GFRα1– PLZF–. And the MCAMLow KIT+ population was 100% GFRα1– PLZF– (Figure 4C). Therefore, this approach allows the isolation of highly enriched populations of A-undiff GFRα1+ spermatogonia as MCAMHigh KIT– and A-undiff GFRα1– spermatogonia as MCAMMed KIT–.
Figure 4.
MCAM Levels Can be Used to Isolate Both GFRα1+ and GFRα1– Undifferentiated Spermatogonia and Are Responsive to GDNF/FGF
(A) Flow cytometry measurement of MCAM levels in whole adult testis from wild-type mice.
(B) Indicated populations were sorted from wild-type mice, cytopun, and stained for GFRα1, PLZF, and DAPI.
(C) Quantification of (B) showing fraction of GFRα1+ PLZF+, GFRα1– PLZF+, and GFRα1– PLZF– cells in each MCAM population. N = 3 mice pooled; N = 1,524 cells.
(D) qRT-PCR for indicated SSC and differentiation markers from cells sorted based on MCAM expression and KIT. Mean and SEM are shown.
(E) Experimental outline of cell culture experiments. Indicated cells populations were sorted and cultured in basal GS medium supplemented with or without 50 ng/mL GDNF and 20 ng/mL FGF2. Forty-eight hours later, anti-MCAM immunofluorescence was performed.
(F) Effect of GDNF/FGF on MCAM expression. TERTHigh GFRα1+ cells and TERTHigh GFRα1– cells were stained for MCAM and DAPI after 48 hr of culture. Scale bar, 15 μm.
(G) Quantification of (E). (N = 4 mice; N = 50 cells). Mean and SEM are shown. ∗p < 0.05; ∗∗∗p < 0.001.
To validate our data, we performed qRT-PCR for a variety of marks of undifferentiated and differentiated spermatogonia. By qPCR, MCAMHigh KIT+ cells expressed high levels of GFRα1 and PLZF mRNA. MCAMMed KIT– cells expressed 10-fold less GFRα1 mRNA than MCAMHigh KIT+ and high levels of PLZF and NGN3. MCAMMed KIT+ cells and MCAMLow KIT+ expressed high levels of KIT and SYCP3 mRNA (Figure 4D). Our findings provide a robust protocol for isolation of phenotypically defined subtypes of undifferentiated spermatogonia from adult wild-type mice. MCAM provides a marked advantage over GFRα1 staining due to an improved signal-to-noise ratio with this combination of antigen and antibody.
Our RNA-seq analysis suggests that the main difference between GFRα1+ and GFRα1– undifferentiated spermatogonia is active GDNF/FGF signaling. We wondered if MCAM were a GDNF/FGF-responsive gene. To test this hypothesis, we used FACS to isolate pure populations of GFRα1+ and GFRα1– cells and then cultured them in germline stem cell medium either with or without GDNF/FGF (Figure 4E) (Kanatsu-Shinohara et al., 2003). The high level of MCAM expression in freshly isolated SSCs was indeed dependent on exposure to GDNF/FGF: ex vivo culture of TERTHigh GFRα1+ cells in the absence of these cytokines led to a rapid downregulation of surface MCAM levels. Similarly, TERTHigh GFRα1– cells exposed to GDNF/FGF had higher MCAM levels than TERTHigh GFRα1– cells cultured in the absence of these cytokines (Figure 4F). Quantifying the changes in MCAM antibody staining showed a significant induction of MCAM expression with GDNF/FGF exposure (Figure 4G). Thus, both GFRα1+ and GFRα1– retain the ability to respond in culture to GDNF/FGF, leading to robust induction of MCAM. Taken together, GFRα1+ and GFRα1– undifferentiated spermatogonia are efficiently isolated on the basis of surface MCAM expression, and elevated MCAM protein in GFRα1+ cells likely reflects active GDNF/FGF signaling in this compartment.
Elevated Stem Cell Repopulating Activity in Both GFRα1+ and GFRα1– Undifferentiated Spermatogonia
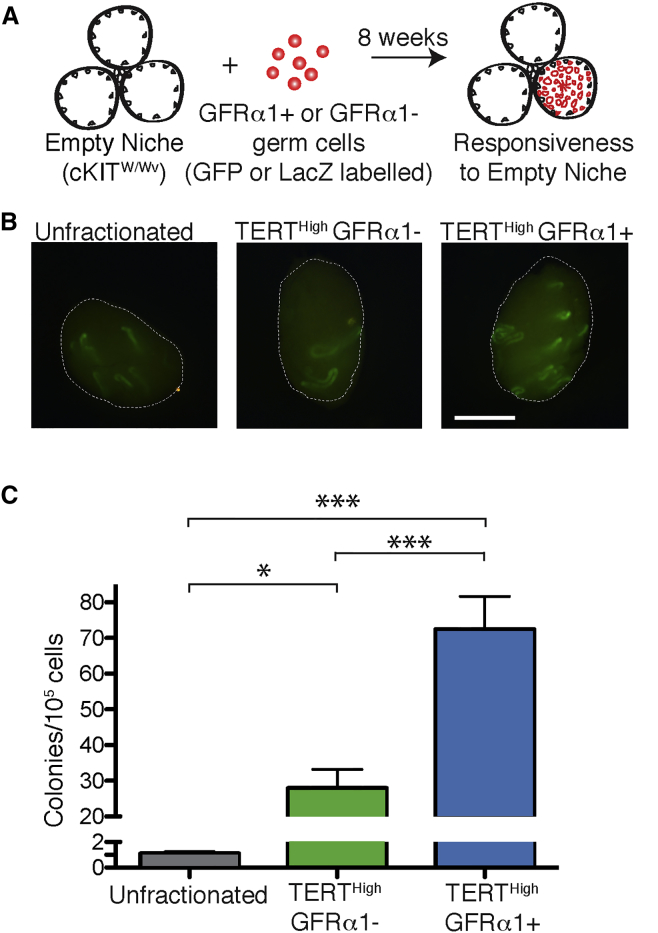
Based on the similarities between the GFRα1+ and GFRα1– cells, we hypothesized that the self-renewal capacity of GFRα1– cells may be revealed upon transplantation into an empty niche and we leveraged our ability to isolate both GFRα1+ and GFRα1– cells to compare their relative transplantation potential side-by-side. FACS-purified cells were assayed in terms of their ability to colonize the tubules of KitW/Wv mice, which lack SSCs (Brinster and Zimmermann, 1994). Donor cells were permanently marked by breeding TertTomato/+ mice to mouse strains with ubiquitous expression of either EGFP or β-galactosidase. FACS-sorted TERTHigh GFRα1+ and TERTHigh GFRα1– cells from adult donors were injected separately into the seminiferous tubules of KitW/Wv recipients via the efferent bundle (Figure 5A). Stem cell frequency in bulk germ cells was assessed by transplanting FACS-sorted live cells that were not fractionated by antigen expression (unfractionated). Two months later, the total number of colonies was counted (Figure 5B).
Figure 5.
Elevated Stem Cell Repopulating Activity in GFRα1+ and GFRα1– Undifferentiated Spermatogonia
(A) Experimental outline of transplant experiments. Tert-Tomato cells permanently labeled by ubiquitous GFP or LacZ expression were transplanted into sterile KitW/Wv recipients. Colonies were counted 2 months post-injection.
(B) Representative EGFP epifluorescence in recipient KitW/Wv mice 8 weeks after transplantation of cells shown in (A). White lines represent boundary of the testis. “Unfractionated” represents the transplantation of FACS-sorted live cells not fractionated by Tert-Tomato expression or immunophenotype. Scale bar, 2 μm.
(C) Quantification of transplant results shown in (B). Colony counts were normalized to 105 cells. Mean and SEM are shown. p Values are from two-tailed Mann-Whitney test. N = 16–18 recipient testes per condition. ∗p = 0.019 ∗∗∗p < 0.0005.
TERTHigh GFRα1+ cells transplanted at high efficiencies, achieving 65-fold enrichment for stem cell activity over unfractionated germ cells (72.5 ± 37 colonies per 100,000 cells; p < 0.0001 U test). TERTHigh GFRα1– cells also showed robust transplantation, albeit at lower frequencies than TERTHigh GFRα1+ cells (p < 0.0005 U test; GFRα1+ versus GFRα1– ). TERTHigh GFRα1– cells showed 25-fold enrichment in stem cell transplantation compared with unfractionated germ cells (28 ± 22 colonies per 100,000 cells; p = 0.0189 U test) (Figure 5C). Histological analysis confirmed that both types of A-undiff spermatogonia were capable of full reconstitution of spermatogenesis post-transplant (Figure S5B). Importantly, stringent sorting conditions led to very high purity of donor cell preparations, as confirmed by re-analysis of the sorted cells prior to transplant (Figure S5A). Thus, we find that GFRα1+ cells show high transplantation efficiency, but that GFRα1– cells also retain significant transplantation potential. Given that the pool of GFRα1– undifferentiated spermatogonia is three times as large as the GFRα1+ pool, the total number of GFRα1– stem cells is comparable with the total number of GFRα1+ stem cells in the testis.
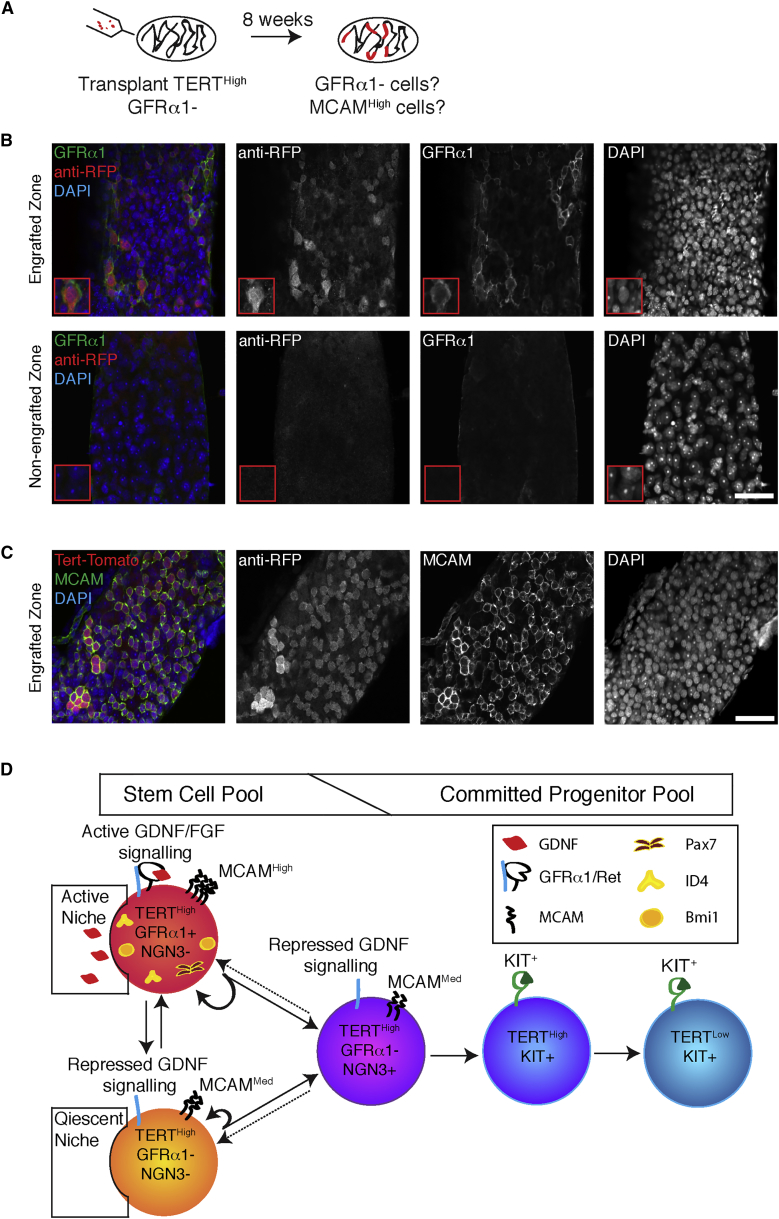
GFRα1– A-Undiff Cells Regenerate the GFRα1+ State after Transplantation
The considerable stem cell potential in GFRα1– spermatogonia prompted us to explore the features of GFRα1– stem cell self-renewal. To address whether spermatogenesis from GFRα1– cells entailed the regeneration of GFRα1+ cells, whole-mount analysis of GFRα1 expression was performed on recipient testes 2 months post-transplantation of TERTHigh GFRα1– cells (Figure 6A). These GFRα1– cells robustly gave rise to colonies containing TERTHigh GFRα1+ cells (Figure 6B). The colonies also contained TERTHigh MCAMHigh cells (Figure 6C). Our data show the ability, in vivo, of GFRα1– cells to respond to niche signals and convert to the GFRα1+ state. These data highlight the functional similarity between the GFRα1+ and the GFRα1– undifferentiated spermatogonia as revealed by the transplantation assay.
Figure 6.
In Vivo Conversion of GFRα1– Undifferentiated Spermatogonia to GFRα1+ Undifferentiated Spermatogonia
(A) Experimental outline of transplant experiments. GFRα1– Tert-Tomato cells permanently labeled by ubiquitous GFP or LacZ expression were transplanted into sterile KitW/Wv recipients. Tubules were stained for MCAM and GFRα1 2 months post transplant.
(B) GFRα1 expression in colonies arising from transplanted TERTHigh GFRα1– cells. Tert-Tomato used as a marker for the donor cells. Staining results are compared with regions of testis that were not colonized. Scale bar, 50 μm.
(C) MCAM expression in colonies arising from transplanted TERTHigh GFRα1– cells. Tert-Tomato used as a marker for the donor cells. Scale bar, 50 μm.
(D) Model for a flexible hierarchy of adult spermatogonia. Cell surface features of different spermatogonial subtypes are highlighted. GFRα1– spermatogonia represent a poised state, competent to either differentiate or convert to GFRα1+ spermatogonia in a context-dependent fashion.
Discussion
A Heterogeneous Adult Germline Stem Cell Pool: GFRα1+ and GFRα1– Undifferentiated Spermatogonia Are Closely Related
We leveraged differences in Tert promoter strength, together with cell surface marker expression, to isolate pure populations of phenotypically defined spermatogonia subsets from adult testis. Our approach enables the isolation of both GFRα1+ and GFRα1– undifferentiated spermatogonia for functional and molecular analysis. These two populations of A-undiff cells show overall relatedness with key differences in signaling pathways. Both populations showed elevated frequencies of transplantation compared with unfractionated spermatogonia, with GFRα1+ cells transplanting approximately 2.6-fold more efficiently than GFRα1– cells. Most data have converged on the idea that SSCs are restricted to the short-chain population of A-undiff cells. Consistent with this idea, short-chain A-undiff cells are present throughout the spermatogenic cycle, whereas longer-chain A-undiff cells are ultimately depleted from the population through differentiation. Our data show that SSCs are highly enriched in the GFRα1+ fraction. These results are consistent with those of other laboratories using reporter mice for GFRα1+ and other markers within this short-chain population, including ID4, BMI1, and PAX7 (Aloisio et al., 2014, Helsel et al., 2017, Komai et al., 2014). It has been argued that these latter markers of subpopulations of short-chain A-undiff cells represent the true stem cells, but direct molecular and functional comparisons of GFRα1+ cells bearing ID4, BMI1, or PAX7, with their GFRα1+ counterparts without expression of ID4, BMI1, or PAX7, have been lacking. Such direct comparisons will be required to understand functional heterogeneity within the GFRα1+ population. Our studies enabling the isolation of GFRα1+ cells using either GFRα1+ antibodies in conjunction with TertTomato/+ mice, or MCAM antibodies in wild-type mice, may allow these ideas to be tested directly. In addition, approximately 20% of short-chain A-undiff cells are GFRα1– in steady state, and these cells have not been isolated or characterized for stem cell activity.
Our findings indicate that GFRα1– cells exhibit a surprising capacity for transplantation. SSCs in this fraction may reside in the short-chain GFRα1– fraction or in the elongating chains of PLZF+ A-undiff cells, or both these populations. If residing within the short-chain GFRα1– fraction, these cells may be in equilibrium with GFRα1+ cells, or may have unique characteristics that have not yet been revealed. If the SSCs defined here in the GFRα1– fraction reside in the elongating A-undiff population, the residual stem cell activity may reflect that some or many of these cells have not yet committed to differentiate. Our results showing elevated SSC activity in the TERTHigh GFRα1– fraction suggests that many cells in this population have not yet fully committed. The ability of these cells to successfully transplant is also consistent with the likelihood that many GFRα1– cells are fated to differentiate to A-aligned spermatogonia during the spermatogenic cycle, as transplantation tests the ability of cells to function as stem cells. These distinctions are important to define the cellular and molecular mechanisms of self-renewal in the mammalian testis. We note that although the transplantation activity was 2.6 times lower in GFRα1– undifferentiated spermatogonia compared with GFRa1+ undifferentiated spermatogonia, GFRα1– cells are more abundant than their GFRα1+ counterparts, making the total number of potential stem cells comparable in each population. These data provide support for a model in which the GFRα1+ and GFRα1– cells together comprise a stem cell pool, and that some GFRα1– cells can convert to the GFRα1+ state based on exposure to niche factors (Figure 6D).
Transcriptional Similarity, but Distinct Regulation, of GDNF and FGF Signaling in GFRα1+ and GFRα1– Spermatogonia
Transcriptional analysis of GFRα1+ and GFRα1– undifferentiated spermatogonia revealed a previously unknown similarity between the two populations, in particular when compared with transcriptomes of neonatal spermatogonia, TERTHigh KIT+ or TERTLow KIT+ spermatogonia, which each cluster separately based on PCA and unsupervised hierarchical clustering. The differentially expressed genes between GFRα1+ and GFRα1– undifferentiated spermatogonia were enriched for gene sets including cell-cycle regulation and Ras/MEK/ERK signaling downstream of GFRα1/Ret binding of GDNF. Cyclin D1, D2, and A2 were in the top of differentially expressed genes between GFRα1+ and GFRα1– cells (Figures 2B and 2D; Table S1). As cyclin D2 has been shown to be important for GS cell self-renewal and long-term culture (Lee et al., 2009), and is expressed in type A spermatogonia in vivo (Beumer et al., 2000), we speculate that cell-cycle regulation is key for the SSC population in vivo. Differences in the abundance of each population during the seminiferous cycle may also contribute to the differences in S-phase fraction measured here, as NGN3+ cells are more abundant in stages IV-VII, at which time they are not proliferating (Ikami et al., 2015).
We found that ID4-Bright cells clustered most closely with our TERTHigh neonatal spermatogonia, likely reflecting the neonatal origin of the ID4-Bright cells used for RNA-seq studies (Helsel et al., 2017). Although the neonatal ID4-Bright cells share expression of many stem cell genes with the adult GFRα1+ population, their overall transcriptomes are sufficiently different that they are most similar to other neonatal populations. These transcriptional differences may relate to expression of both SSC genes and differentiation genes within the ID4+ population. This combination of features reflects the peculiarities of the first, synchronized wave of spermatogenesis, which is faster than the adult cycle and features gonocytes that directly give rise to A2 spermatogonia (Kluin et al., 1982, van Haaster and de Rooij, 1993). The extensive RNA-seq analysis performed here highlights key differences between neonatal and adult populations of spermatogonia.
GFRα1– Spermatogonia Are Capable of Responding to GDNF/FGF Niche Signals in Culture
We found that, in culture, GFRα1– cells can respond to GDNF/FGF2 to upregulate MCAM to the levels of GFRα1+ cells. Consistent with this observation, we observed that GFRα1– cells and GFRα1+ cells share a requirement for GDNF and FGF for even short-term culture. The mechanism by which GFRα1– cells can sense GDNF/FGF is unclear, but may be due to low level receptor expression. Our molecular profiling showed a gradient of GFRα1 expression, similar to the gradient of MCAM expression (Figure 3B). Although GFRα1 mRNA is reduced by 16-fold in GFRα1– A-undiff cells compared with GFRα1+ cells (Table S1), GFRα1 mRNA remains 2.3-fold elevated in GFRα1– cells compared with TERTHigh KIT+ early differentiating spermatogonia (q = 1.39 × 10−17; Tables S2 and S3). Furthermore, FGFR1 and FGFR3 are both expressed on GFRα1+ and GFRα1– spermatogonia, and FGFR1 significantly decreases in TERTHigh KIT+ cells compared with GFRα1– cells (Table S3). Taken together, these results suggest that at least a subpopulation of GFRα1– cells retains the ability to respond to GDNF and FGF niche factors.
Isolation of Phenotypically Defined Spermatogonial Subpopulations from Adult Wild-Type Mice
Our purification of adult spermatogonia populations, together with RNA-seq, allowed us to identify MCAM as a useful cell surface marker enabling efficient purification of GFRα1+ and GFRα1– spermatogonial subtypes from adult wild-type mice. MCAM expression on spermatogonia was discovered by Kanatsu-Shinohara et al. (2012) in GS cell cultures. In vivo, MCAM expression was found on both undifferentiated and differentiating spermatogonia and CD9+ EPCAMlow MCAM+ KIT– cells were enriched for SSC activity by transplantation (Kanatsu-Shinohara et al., 2012). Subsequently, sorting for MCAM+ KIT– was used to isolate Bmi1+ undifferentiated spermatogonia (Komai et al., 2014). Our results are consistent with MCAM enriching for SSCs; however, we revealed a clear gradient of MCAM surface expression that, when coupled with KIT expression, allows isolation of nearly pure populations of GFRa1+ and GFRa1– undifferentiated spermatogonia. The ability to isolate these cells from adult wild-type mice will facilitate the study of these populations and allow for future work to define additional molecular and functional features of these cell types in steady-state spermatogenesis.
Experimental Procedures
Animals
TertTomato/+ mice were described previously (Pech et al., 2015); KitW/Wv mice were purchased (Jackson Laboratory, stock no. 100410). Experiments on adult mice were performed on males between 6 weeks and 3 months of age. All mice were treated in accordance with Association for Assessment and Accreditation of Laboratory Animal Care-approved guidelines at Stanford University.
Antibodies
The following antibodies were used for immunostaining and/or flow cytometry: MCAM-AF488 and MCAM-APC (Biolegend ME-9F1; rat monoclonal), Thy1.2-APC-Cy7 (Biolegend 3OH-12; rat monoclonal), SOHLH1 (gift of A. Rajkovic; rabbit polyclonal). Other antibodies used have been described previously (Pech et al., 2015)
Testes Dissociation and FACS Analysis
Testes were dissociated and FACS analyzed as previously described (Pech et al., 2015). For GFRα1 FACS staining, a biotinylated primary antibody was used, together with a secondary streptavidin-APC secondary (Jackson Immunoresearch) for 30 min at 4°C. All FACS experiments were performed on a single BD Aria II machine. Cells were sorted using a 100 μm nozzle in purity mode. Data were analyzed with FlowJo software (Tree Star, San Carlos, CA).
Germ Cell Transplantation
Four independent transplantations were performed. For each transplantation experiment, testes cell suspensions were prepared from two pooled adult mice and sorted as described above. A total of 16–18 recipient testes was analyzed per cell type. Donor cells were introduced into infertile KitW/Wv recipients (Jackson Laboratory) via efferent duct injection (Ogawa et al., 1997). Colonization was determined 8 weeks after injection. Colony numbers were normalized to 100,000 cells transplanted. Statistics were calculated using Prism (GraphPad Software, La Jolla, CA) using the Mann-Whitney non-parametric U test.
RNA-Seq Library Preparation
Dissociated testes cells were prepared and sorted from both testes of adult mice, as described previously (Pech et al., 2015). Four to five biological replicates were sorted per cell population. cDNA was prepared and amplified using the NuGEN Ovation V2 kit, starting from 5 to 10 ng of total RNA. cDNA was sonicated to 200 bp using a Covaris S2 machine, and 25 ng of cDNA was used to make the libraries, following standard Illumina TruSeq v2 protocols. Samples were sequenced on an Illumina HiSeq 2500 machine, with paired-end 101 bp reads.
Author Contributions
A.G. and M.F.P. carried out the majority of the experiments. A.G. performed the bioinformatics analysis. M.F.P. generated the Tert-Tomato mouse and performed whole mounts. K.H. contributed MCAM cytospin and qPCR data. M.S. performed the transplantation experiments with support from K.E.O. R.J.Z. carried out the mouse husbandry. A.G., M.F.P., and S.E.A. conceived the study, designed the experiments, and wrote the manuscript.
Acknowledgments
This work was supported by NIH grants CA197563 and AG056575. We thank members of the Artandi lab for critical comments. A.G. was funded by a National Science Foundation National Research Service Award fellowship. M.P. was supported by a National Science Foundation National Research Service Award fellowship and a T32 training grant (CA09302). We acknowledge the Stanford Neuroscience Microscopy Service (NIH grant NS069375) and the Stanford Shared FACS Facility.
Published: January 11, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and three tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.12.009.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE107694.
Supplemental Information
References
- Aloisio G.M., Nakada Y., Saatcioglu H.D., Pena C.G., Baker M.D., Tarnawa E.D., Mukherjee J., Manjunath H., Bugde A., Sengupta A.L. PAX7 expression defines germline stem cells in the adult testis. J. Clin. Invest. 2014;124:3929–3944. doi: 10.1172/JCI75943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer T.L., Roepers-Gajadien H.L., Gademan I.S., Kal H.B., de Rooij D.G. Involvement of the D-type cyclins in germ cell proliferation and differentiation in the mouse. Biol. Reprod. 2000;63:1893–1898. doi: 10.1095/biolreprod63.6.1893. [DOI] [PubMed] [Google Scholar]
- Branney P.A., Faas L., Steane S.E., Pownall M.E., Isaacs H.V. Characterisation of the fibroblast growth factor dependent transcriptome in early development. PLoS One. 2009;4:e4951. doi: 10.1371/journal.pone.0004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster R.L., Zimmermann J.W. Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. USA. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buageaw A., Sukhwani M., Ben-Yehudah A., Ehmcke J., Rawe V.Y., Pholpramool C., Orwig K.E., Schlatt S. GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes1. Biol. Reprod. 2005;73:1011–1016. doi: 10.1095/biolreprod.105.043810. [DOI] [PubMed] [Google Scholar]
- Chakraborty P., Buaas F.W., Sharma M., Snyder E., de Rooij D.G., Braun R.E. LIN28A marks the spermatogonial progenitor population and regulates its cyclic expansion. Stem Cells. 2014;32:860–873. doi: 10.1002/stem.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij D.G. Spermatogonial stem cell renewal in the mouse. I. Normal situation. Cell Tissue Kinet. 1973;6:281–287. doi: 10.1111/j.1365-2184.1973.tb01617.x. [DOI] [PubMed] [Google Scholar]
- Ernst J., Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassei K., Orwig K.E. SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS One. 2013;8:e53976. doi: 10.1371/journal.pone.0053976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso M., Fuso A., Dovere L., de Rooij D.G., Stefanini M., Boitani C., Vicini E. Distribution of GFRA1– expressing spermatogonia in adult mouse testis. Reproduction. 2012;143:325–332. doi: 10.1530/REP-11-0385. [DOI] [PubMed] [Google Scholar]
- Grisanti L., Falciatori I., Grasso M., Dovere L., Fera S., Muciaccia B., Fuso A., Berno V., Boitani C., Stefanini M. Identification of spermatogonial stem cell subsets by morphological analysis and prospective isolation. Stem Cells. 2009;27:3043–3052. doi: 10.1002/stem.206. [DOI] [PubMed] [Google Scholar]
- Hara K., Nakagawa T., Enomoto H., Suzuki M., Yamamoto M., Simons B.D., Yoshida S. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell. 2014;14:658–672. doi: 10.1016/j.stem.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Namekawa S.H., Saga Y. MEK/ERK signaling directly and indirectly contributes to the cyclical self-renewal of spermatogonial stem cells. Stem Cells. 2013;31:2517–2527. doi: 10.1002/stem.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K., Saga Y. FGF8-FGFR1 signaling acts as a niche factor for maintaining undifferentiated spermatogonia in the mouse. Biol. Reprod. 2014;91:145. doi: 10.1095/biolreprod.114.121012. [DOI] [PubMed] [Google Scholar]
- He Z., Jiang J., Kokkinaki M., Golestaneh N., Hofmann M.-C., Dym M. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells. 2008;26:266–278. doi: 10.1634/stemcells.2007-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel A.R., Yang Q.-E., Oatley M.J., Lord T., Sablitzky F., Oatley J.M. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144:624–634. doi: 10.1242/dev.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat. Rec. 1971;169:533–557. doi: 10.1002/ar.1091690306. [DOI] [PubMed] [Google Scholar]
- Ikami K., Tokue M., Sugimoto R., Noda C., Kobayashi S., Hara K., Yoshida S. Hierarchical differentiation competence in response to retinoic acid ensures stem cell maintenance during mouse spermatogenesis. Development. 2015;142:1582–1592. doi: 10.1242/dev.118695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Morimoto H., Shinohara T. Enrichment of mouse spermatogonial stem cells by melanoma cell adhesion molecule expression. Biol. Reprod. 2012;87:139. doi: 10.1095/biolreprod.112.103861. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Ogonuki N., Inoue K., Miki H., Ogura A., Toyokuni S., Shinohara T. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Takashima S., Ishii K., Shinohara T. Dynamic changes in EPCAM expression during spermatogonial stem cell differentiation in the mouse testis. PLoS One. 2011;6:e23663. doi: 10.1371/journal.pone.0023663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluin P.M., Kramer M.F., de Rooij D.G. Spermatogenesis in the immature mouse proceeds faster than in the adult. Int. J. Androl. 1982;5:282–294. doi: 10.1111/j.1365-2605.1982.tb00257.x. [DOI] [PubMed] [Google Scholar]
- Komai Y., Tanaka T., Tokuyama Y., Yanai H., Ohe S., Omachi T., Atsumi N., Yoshida N., Kumano K., Hisha H. Bmi1 expression in long-term germ stem cells. Sci. Rep. 2014;4:6175. doi: 10.1038/srep06175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Brinster R.L. Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc. Natl. Acad. Sci. USA. 2003;100:6487–6492. doi: 10.1073/pnas.0631767100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.W., Blasco M.A., Gottlieb G.J., Horner J.W., 2nd, Greider C.W., DePinho R.A. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- Lee J., Kanatsu-Shinohara M., Morimoto H., Kazuki Y., Takashima S., Oshimura M., Toyokuni S., Shinohara T. Genetic reconstruction of mouse spermatogonial stem cell self-renewal in vitro by Ras-cyclin D2 activation. Cell Stem Cell. 2009;5:76–86. doi: 10.1016/j.stem.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Meng X., Lindahl M., Hyvonen M.E., Parvinen M., de Rooij D.G., Hess M.W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- Merico D., Isserlin R., Stueker O., Emili A., Bader G.D. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Nabeshima Y., Yoshida S. Functional identification of the actual and potential stem cell compartments in mouse spermatogenesis. Dev. Cell. 2007;12:195–206. doi: 10.1016/j.devcel.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Sharma M., Nabeshima Y.i., Braun R.E., Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328:62–67. doi: 10.1126/science.1182868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakberg E.F. Spermatogonial stem-cell renewal in the mouse. Anat. Rec. 1971;169:515–531. doi: 10.1002/ar.1091690305. [DOI] [PubMed] [Google Scholar]
- Oatley J.M., Avarbock M.R., Telaranta A.I., Fearon D.T., Brinster R.L. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. USA. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Arechaga J.M., Avarbock M.R., Brinster R.L. Transplantation of testis germinal cells into mouse seminiferous tubules. Int. J. Dev. Biol. 1997;41:111–122. [PubMed] [Google Scholar]
- Pech M.F., Garbuzov A., Hasegawa K., Sukhwani M., Zhang R.J., Benayoun B.A., Brockman S.A., Lin S., Brunet A., Orwig K.E. High telomerase is a hallmark of undifferentiated spermatogonia and is required for maintenance of male germline stem cells. Genes Dev. 2015;29:2420–2434. doi: 10.1101/gad.271783.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips B.T., Gassei K., Orwig K.E. Spermatogonial stem cell regulation and spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:1663–1678. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrans-Stassen B.H., van de Kant H.J., de Rooij D.G., van Pelt A.M. Differential expression of c-kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology. 1999;140:5894–5900. doi: 10.1210/endo.140.12.7172. [DOI] [PubMed] [Google Scholar]
- Shinohara T., Avarbock M.R., Brinster R.L. beta1– and alpha6-integrin are surface markers on mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 1999;96:5504–5509. doi: 10.1073/pnas.96.10.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A., Fuller M.T., Braun R.E., Yoshida S. Germline stem cells. Cold Spring Harb. Perspect. Biol. 2011;3:a002642. doi: 10.1101/cshperspect.a002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Ahn H.W., Chu T., Bowden W., Gassei K., Orwig K., Rajkovic A. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev. Biol. 2012;361:301–312. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Sada A., Yoshida S., Saga Y. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev. Biol. 2009;336:222–231. doi: 10.1016/j.ydbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- Tadokoro Y., Yomogida K., Ohta H., Tohda A., Nishimune Y. Homeostatic regulation of germinal stem cell proliferation by the GDNF/FSH pathway. Mech. Dev. 2002;113:29–39. doi: 10.1016/s0925-4773(02)00004-7. [DOI] [PubMed] [Google Scholar]
- Tokuda M., Kadokawa Y., Kurahashi H., Marunouchi T. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes1. Biol. Reprod. 2007;76:130–141. doi: 10.1095/biolreprod.106.053181. [DOI] [PubMed] [Google Scholar]
- van Haaster L.H., de Rooij D.G. Spermatogenesis is accelerated in the immature Djungarian and Chinese hamster and rat. Biol. Reprod. 1993;49:1229–1235. doi: 10.1095/biolreprod49.6.1229. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.