Goda et al. provide molecular evidence that body temperature rhythm (BTR) is regulated distinctly from locomotor activity rhythms and show that diuretic hormone 31 receptor/calcitonin receptor is an ancient specific mediator of BTR during the active phase in organisms ranging from ectotherms to endotherms.
Keywords: DH31R, Calcr, calcitonin receptor, circadian rhythm, body temperature rhythm, temperature preference rhythm, thermoregulation
Abstract
Daily body temperature rhythm (BTR) is essential for maintaining homeostasis. BTR is regulated separately from locomotor activity rhythms, but its molecular basis is largely unknown. While mammals internally regulate BTR, ectotherms, including Drosophila, exhibit temperature preference rhythm (TPR) behavior to regulate BTR. Here, we demonstrate that the diuretic hormone 31 receptor (DH31R) mediates TPR during the active phase in Drosophila. DH31R is expressed in clock cells, and its ligand, DH31, acts on clock cells to regulate TPR during the active phase. Surprisingly, the mouse homolog of DH31R, calcitonin receptor (Calcr), is expressed in the suprachiasmatic nucleus (SCN) and mediates body temperature fluctuations during the active phase in mice. Importantly, DH31R and Calcr are not required for coordinating locomotor activity rhythms. Our results represent the first molecular evidence that BTR is regulated distinctly from locomotor activity rhythms and show that DH31R/Calcr is an ancient specific mediator of BTR during the active phase in organisms ranging from ectotherms to endotherms.
The body temperature rhythm (BTR) is one of the most conspicuous outputs of the circadian clock (Aschoff 1983; Krauchi 2002; Weinert 2010) and is crucial for maintaining homeostasis in metabolism and sleep as well as entraining the peripheral clock in mammals (Refinetti and Menaker 1992; Gilbert et al. 2004; Krauchi 2007a,b; Buhr et al. 2010; Morf and Schibler 2013). In humans, body temperature increases during wakefulness and decreases during sleep (Duffy et al. 1998). As daily variations in BTR are robust and parallel fluctuations in locomotor activity rhythms, BTR is widely used to monitor circadian rhythms in mammals. The molecular mechanisms that regulate BTR remain largely uncharacterized, although a study in which subsets of neurons in the brains of rats were surgically ablated suggests that locomotor activity rhythms and BTR are controlled by different output pathways that originate from the suprachiasmatic nucleus (SCN) (Saper et al. 2005). In humans, body temperature fluctuates even when locomotor activity is restricted (Smith 1969; Gander et al. 1986), and BTR and locomotor activity rhythms can be experimentally dissociated, a phenomenon known as spontaneous internal desynchronization (Lavie 2001). These accumulating data suggest that BTR is likely controlled separately from locomotor activity rhythms. However, no molecular evidence supporting this possibility has been reported. Therefore, there is a critical need to identify genes that regulate BTR.
While mammals and birds internally generate heat to regulate BTR, some ectotherms, such as Drosophila, rely on behavioral strategies to regulate their daily body temperature changes (Stevenson 1985b; Refinetti and Menaker 1992; Ellis et al. 2007; Hamada et al. 2008; Dillon et al. 2009). We previously demonstrated that Drosophila exhibit a temperature preference rhythm (TPR), in which the preferred temperature increases during the day and decreases at the transition from day to night; i.e., night onset (Kaneko et al. 2012). As flies are ectotherms and their body temperature is therefore close to that of the ambient environment (Stevenson 1985a,b), the Drosophila TPR produces a daily rhythm in body temperature through the selection of a preferred temperature. Importantly, our previous data suggest that TPR is regulated separately from locomotor activity rhythms, as is the case for mammalian BTR (Kaneko et al. 2012). Therefore, the Drosophila TPR resembles mammalian BTR. Given that the molecular mechanisms underlying locomotor activity rhythms and sleep are well conserved from Drosophila to mammals (Sehgal and Mignot 2011; Dubowy and Sehgal 2017), we used Drosophila to identify the genes that regulate BTR.
To identify the mechanisms that underlie TPR, we focused on the secretin receptor family of G-protein-coupled receptors (GPCRs), which play important conserved roles in not only circadian rhythms and sleep modulation (Taghert and Nitabach 2012; Bedont and Blackshaw 2015; Kunst et al. 2015) but also hypothalamus-mediated processes in mammals (McCoy et al. 2013; Wellman et al. 2015; Tan et al. 2016). One member of the secretin receptor family of GPCRs, the pigment-dispersing factor receptor (PDFR), is critical for the synchronization of the circadian clock in pacemaker cells and is required for robust circadian behavioral output in Drosophila (Taghert and Nitabach 2012). Importantly, PDFR is a functional homolog of vasoactive intestinal peptide (VIP) receptor 2 (Vipr2) in mammals. Although we initially expected that PDFR would be the major regulator of TPR, Pdfr mutation causes a partially abnormal TPR phenotype only at night onset (Zeitgeber time 10 [ZT10]–ZT15) (Goda et al. 2016). To better understand TPR, we therefore investigated the role of another secretin family GPCR, diuretic hormone 31 receptor (DH31R), given that this protein shares a ligand with PDFR; i.e., DH31 (Johnson et al. 2005; Mertens et al. 2005; Shafer et al. 2008).
Here, we determined that DH31R mediates TPR during the daytime (active phase for flies) but does not mediate locomotor activity rhythms. Surprisingly, we also found that the mouse homolog of DH31R, calcitonin receptor (Calcr), mediates BTR during the night (active phase for mice). Calcr is a member of the secretin family of GPCRs and is known to participate in calcium homeostasis in osteoclasts (Masi and Brandi 2007). Since Calcr is not involved in locomotor activity rhythmicity (Doi et al. 2016), these findings provide the first molecular evidence that BTR is regulated separately from locomotor activity rhythms. Although the mechanisms underlying thermoregulation in Drosophila and mammals are completely different, our data identify the calcitonin receptors DH31R and Calcr as fundamental ancient mediators for daily BTR in both flies and mice.
Results
DH31R mediates TPR
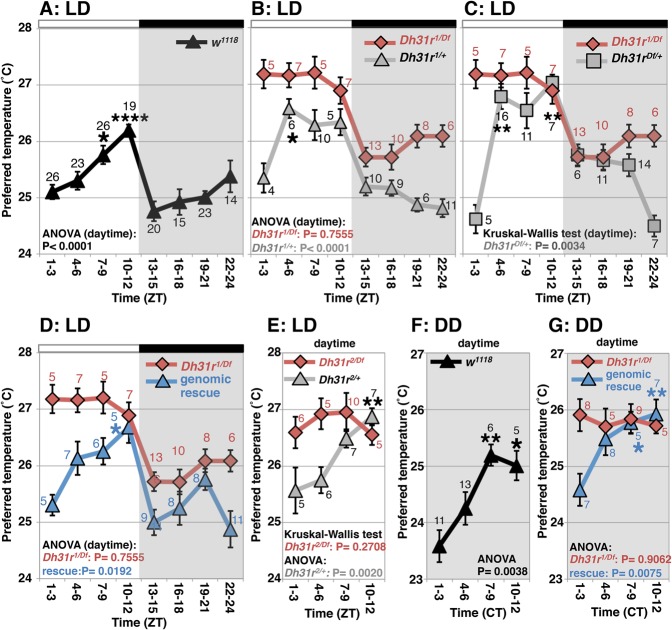
We demonstrated previously that flies exhibit a TPR (Kaneko et al. 2012). In w1118 (control) flies, the preferred temperature increased during the daytime (ZT1–ZT12) and decreased at night onset (ZT10–ZT15) (Fig. 1A). Due to the reasons mentioned above, we focused on flies mutant for Dh31r [Dh31rf05546/Df(2R)BSC273, referred to here as Dh31r1/Df] (Supplemental Fig. S1A). In Dh31r1/Df flies, the mRNA levels of Dh31r in the head were 38% of those levels observed in w1118 flies (Supplemental Fig. S1B). We found that Dh31r1/Df flies preferred a constant temperature of ∼27°C during the daytime (ZT1–ZT12; ANOVA: Dh31r1/Df, P = 0.7555) and displayed TPR profiles different from those of w1118 flies (Fig. 1B,C, red). However, Dh31r1/Df flies exhibited a normal decrease in the preferred temperature at night onset (ZT10–ZT15) (Fig. 1A–C). Heterozygous flies (Dh31r1/+ or Dh31rDf/+), which were used as controls, showed a normal TPR (Fig. 1B,C, gray). These data suggest that Dh31r1/Df flies exhibit an abnormal daytime TPR but a normal night-onset TPR.
Figure 1.
Dh31r mediates daytime TPR. TPR in Dh31r mutants and controls under 12-h light:12-h dark (LD) cycles (A–E) and constant darkness (DD) conditions (F,G). (A) TPR in w1118 flies. (B) Comparison of TPR between the Dh31r mutant Dh31r1/Df (red line) and the heterozygous control Dh31r1/+ (gray line). (C) Comparison of TPR between Dh31r1/Df (red line) and the heterozygous control Dh31rDf/+ (gray line). (D) Comparison of TPR between Dh31r1/Df (red line) and the genomic rescue mutant (rescue [Dh31r], Dh31r1/Df) (blue line). (E) Comparison of TPR between the Dh31r mutant Dh31r2/Df (red line) and its control, Dh31r2/+ (gray line). (F) TPR in w1118 flies in DD. (G) Comparison of TPR between Dh31r1/Df (red line) and the genomic rescue mutant (blue line) in DD. (ZT0) Lights on; (ZT12) lights off; (CT) circadian time; (CT0–CT12) subjective day; (CT13–CT24) subjective night. The daytime shown is from ZT1–ZT3 to ZT10–ZT12. The numbers represent the number of assays. The results of one-way ANOVA or the Kruskal-Wallis test for the data obtained during the daytime are shown. (****) P < 0.0001; (**) P < 0.01; (*) P < 0.05, the Tukey-Kramer test or Kruskal-Wallis test compared with ZT1–ZT3 (Supplemental Table S1).
To confirm that the Dh31r mutation caused the observed abnormal TPR, we used another mutant, Dh31rf06589/Df(2R)BSC273 (Dh31r2/Df) (Supplemental Fig. S1A), in which the mRNA levels of Dh31r in the head were 40% of that in w1118 flies (Supplemental Fig. S1B). We found that Dh31r2/Df flies consistently preferred a higher temperature during the daytime, while control (Dh31r2/+) flies exhibited a normal daytime TPR (Fig. 1E). This result is consistent with those obtained for the Dh31r1/Df flies (Fig. 1B,C). Furthermore, we created a genomic rescue fly line by inserting the Dh31r minigene in the Dh31r1/Df background. We observed that the genomic rescue flies (rescue [Dh31r], Dh31r1/Df) restored the daytime TPR (ANOVA: rescue, P = 0.0192) (Fig. 1D, blue). Thus, we concluded that Dh31r is involved in regulating the daytime TPR.
To assess whether the defect in the daytime TPR is endogenously entrained, we examined the TPR of Dh31r1/Df flies maintained under constant darkness (DD) (Fig. 1F,G). While w1118 flies exhibited time-dependent changes in preferred temperature during the subjective daytime (Fig. 1F), Dh31r1/Df flies did not show such fluctuations during the subjective daytime in DD and consistently preferred a temperature of ∼26°C in DD (ANOVA: P = 0.9062) (Fig. 1G, red). Furthermore, the genomic rescue (rescue [Dh31r], Dh31r1/Df) flies displayed restored TPR during the subjective daytime in DD (ANOVA: rescue, P = 0.0075) (Fig. 1G, blue). Notably, we showed previously that light positively affects temperature preference, a phenomenon referred to as light-dependent temperature preference (LDTP) (Head et al. 2015); here, the flies preferred a higher temperature in light than in dark conditions. Thus, we concluded that DH31R mediates the daytime TPR and that its regulation is driven by the endogenous clock. As the disruption of the daytime TPR (ZT1–ZT12) is a robust phenotype of Dh31r mutants, we subsequently focused on the daytime TPR to examine the role of the DH31R.
Dh31r mutants maintain normal molecular oscillations in the brain
Given that the Dh31r1/Df mutant showed an abnormal TPR, we sought to determine whether molecular clock oscillations are dampened in the Dh31r1/Df mutant. To this end, we performed immunostaining using antibodies against the proteins encoded by the circadian clock genes TIMELESS (TIM) and VRILLE (VRI) (Allada and Chung 2010). There are ∼150 central pacemaker cells in the fly brain, and these cells have functions similar to those of mammalian SCN neurons. The pacemaker cells can be divided into groups of lateral neurons (s-LNv, l-LNv, and LNd) and dorsal neurons (DN1, DN2, and DN3) based on their locations in the brain (Allada and Chung 2010). We observed a robust rhythmic oscillation of TIM and VRI expression in LNvs, DN1s, DN2s, and DN3s in both Dh31r1/Df flies and w1118 flies (Supplemental Fig. S2), indicating that the rhythmic oscillations of the molecular clock are intact in the Dh31r mutant. These data therefore suggest that DH31R regulates the clock output but not the clock molecular machinery.
DH31R is expressed in clock cells and nonclock peptidergic cells
To examine where DH31R is expressed in the brain, we generated an anti-DH31R antibody. DH31R is not expressed in the mushroom body (MB) (data not shown), but bright signals were detected when DH31R was ectopically expressed in the MB using OK107-Gal4 (a MB driver) (Supplemental Fig. S3A). This result suggests that the anti-DH31R antibody specifically recognized the ectopically expressed DH31R.
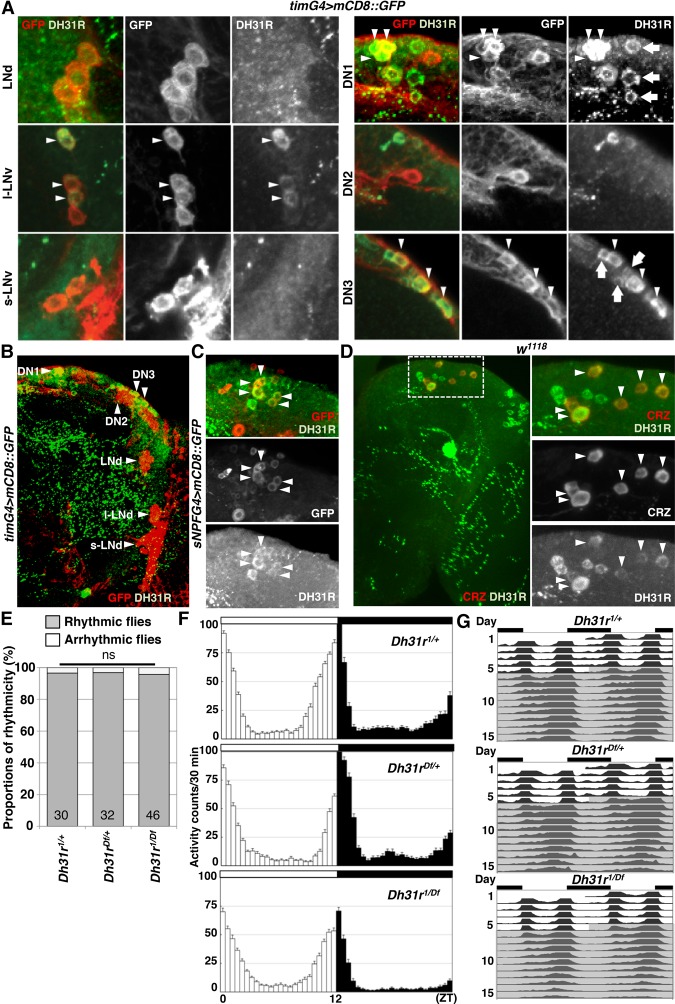
First, we sought to identify where DH31R is expressed in the brain. The results of previous studies using microarray analysis, in vivo physiological analyses, and RNA sequencing (RNA-seq) analysis (Shafer et al. 2008; Kula-Eversole et al. 2010; Abruzzi et al. 2017) suggest that DH31R is expressed in LNvs. We therefore performed immunostaining with an anti-DH31R antibody using tim-Gal4 (a clock cell driver) flies (Fig. 2A,B). We found that DH31R was strongly expressed in DN1s and DN3s but absent in s-LNvs, LNds, and DN2s (Fig. 2A; Supplemental Fig. S3B,C). Although DH31R signals were detected in l-LNvs, these signals were relatively weak (Fig. 2A; Supplemental Fig. S3B,D). Notably, the intensity of the DH31R signals in l-LNvs was variable, and only 24 of 30 brain hemispheres (80%) showed positive DH31R signals in the l-LNv (11 brain hemispheres showed strong signals, and 13 brain hemispheres showed weak signals).
Figure 2.
DH31R is expressed in clock cells but is not required for locomotor activity rhythms. (A,B) DH31R antibody staining using a timG4>mCD8::GFP (tim-Gal4/+; UAS-mCD8::GFP/+) fly brain. The clock cells are labeled by GFP (red), and DH31R signals are shown in green. The arrowheads indicate GFP-expressing clock cells that overlap with cells containing DH31R signals. The arrows indicate DH31R-expressing, GFP-negative cells. (C) DH31R antibody staining using sNPFG4>mCD8::GFP (sNPF-Gal4/+; UAS-mCD8::GFP/+) flies. The sNPF-Gal4-positive cells are labeled by GFP (red), and the DH31R signals are shown in green. The arrowheads indicate GFP-expressing cells that overlap with cells expressing DH31R. (D) DH31R and CRZ antibody staining in w1118 flies. The DH31R signals are shown in green, and the CRZ signals are shown in red. The arrowheads indicate CRZ-expressing cells that overlap with cells expressing DH31R. (E) Comparison of percentages of rhythmic (gray bar) and arrhythmic (white bar) flies between Dh31r mutant (Dh31r1/Df) and control (Dh31r1/+ and Dh31rDf/+) flies. The proportions of rhythmic and arrhythmic flies in DD over 10 d were compared using χ2 analysis. The numbers in the bar graph represent the number of flies. (ns) No significance. (F) The averaged activity profiles over 4 d (day 2–5) in LD conditions for each genotype (Dh31r1/+, Dh31rDf/+, and Dh31r1/Df). The white and black columns indicate the mean activity levels in 30 min during the daytime and nighttime, respectively. The white (daytime) and black (nighttime) bars above the profiles represent the lighting conditions. The number of flies in each genotype is the same as in E. (G) Double-plotted averaged actogram of rhythmic flies over 5 d in LD and 10 d in DD for each genotype (Dh31r1/+, Dh31rDf/+, and Dh31r1/Df). Only the rhythmic flies in E were used.
A previous study with a different anti-DH31R antibody showed that DH31R is expressed in corazonin (CRZ)-expressing cells located in the dorsal region of the brain (Johnson et al. 2005). Therefore, we also confirmed that the DH31R and CRZ signals in the cells in the dorsal brain region overlapped (Fig. 2D; Supplemental Fig. S3E). As CRZ is known to be expressed in short neuropeptide F (sNPF)-expressing cells (Supplemental Fig. S3F; Nassel et al. 2008), we also performed immunostaining with sNPF-Gal4 flies. We found that DH31R is expressed in sNPF-Gal4-expressing cells in the dorsal region of the brain (Fig. 2C). Taken together, our data indicate that DH31R is expressed in a subset of clock neurons (DN1s, DN3s, and l-LNvs) and in other nonclock peptidergic neurons (sNPF- and CRZ-expressing neurons) located in the dorsal regions of the brain (Supplemental Fig. S3G).
To examine the residual DH31R expression levels in Dh31r1/Df flies, we compared the DH31R signals between w1118 and Dh31r1/Df flies (Supplemental Fig. S4A,B). Because we observed stronger DH31R expression in clock cells at the dorsal brain area, we focused on the DN1 and DN3 areas. We found that the DH31R signals in Dh31r1/Df flies in the DN1 and DN3 areas were 56% and 59% of the respective levels in w1118 flies (Supplemental Fig. S4A,B). Thus, the data suggest that DH31R expression is lower in the DN1 and DN3 areas in Dh31r1/Df mutants compared with w1118 flies.
DH31R does not coordinate locomotor activity rhythms
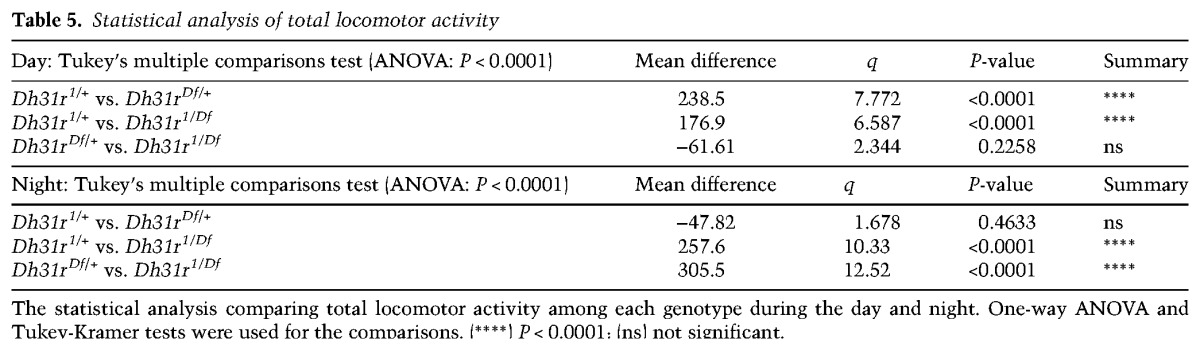
As DH31R belongs to the same group of class II GPCRs as PDFR (Kunst et al. 2015) and is expressed in the subset of clock neurons that is important for locomotor activity rhythms, we sought to determine whether DH31R is also involved in generating locomotor activity rhythms. We found that Dh31r1/Df mutants still maintained robust free-running rhythmicity (96% rhythmic; power = 939.6 ± 79.9) (Fig. 2E–G; Table 1). The mutant animals showed a period (24.0 h ± 0.1 h) very similar to that of controls (Dh31r1/+ and Dh31rDf/+; 23.9 h ± 0.0 h and 23.8 h ± 0.1 h, respectively) (Table 1) and maintained their normal morning anticipation (Fig. 2F; Tables 2, 3). Notably, the total activity level of Dh31r1/Df was slightly lower than that of controls (Dh31r1/+ and Dh31rDf/+) during the night (Fig. 2F; Tables 4, 5). Additionally, the knockdown of Dh31r expression in all neurons using a pan-neuronal Gal4 driver (elav-Gal4>UAS-Dcr2; Dh31r-RNAi1) and its controls (elav-Gal4/+; UAS-Dcr2/+ and UAS-Dh31r-RNAi1/+) resulted in normal locomotor activity rhythms (Tables 1–3). These results suggest that DH31R does not regulate the rhythmicity, period, or morning anticipation of locomotor activity rhythms.
Table 1.
Free-running rhythms
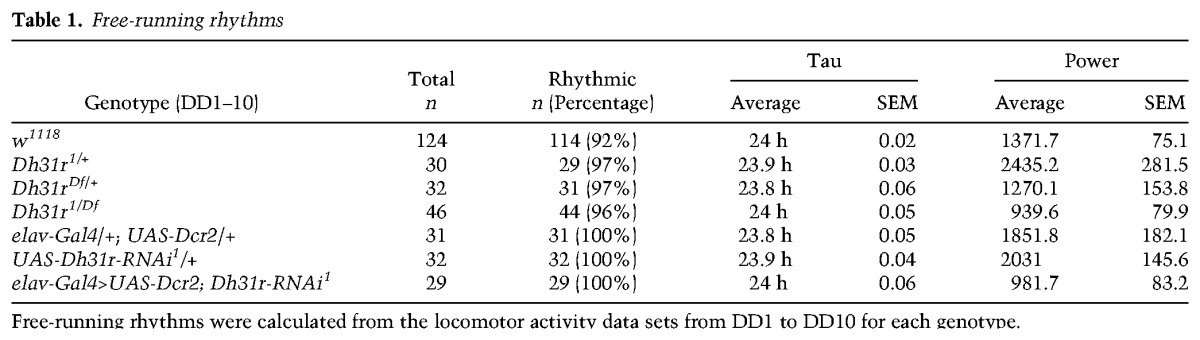
Table 2.
Morning anticipation index (AI)
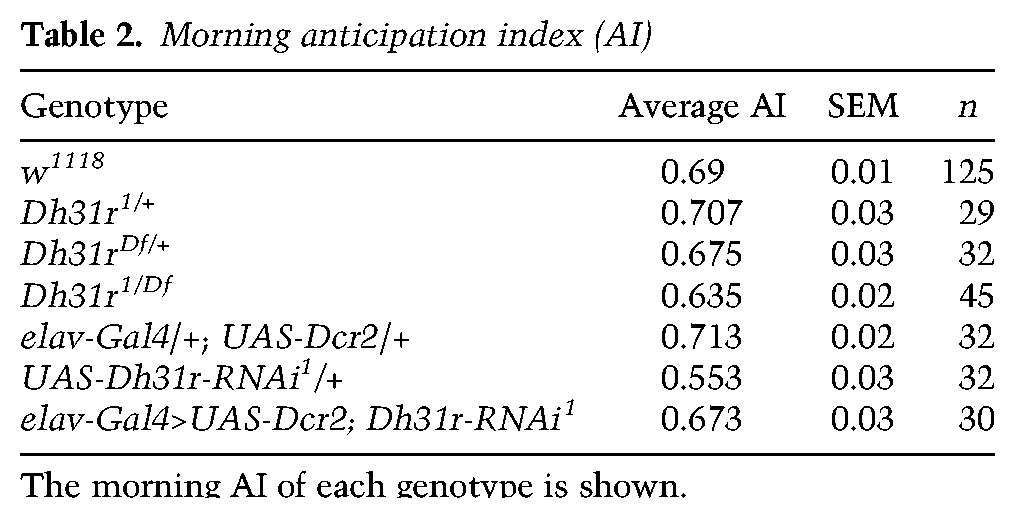
Table 3.
Statistical analysis of morning anticipation
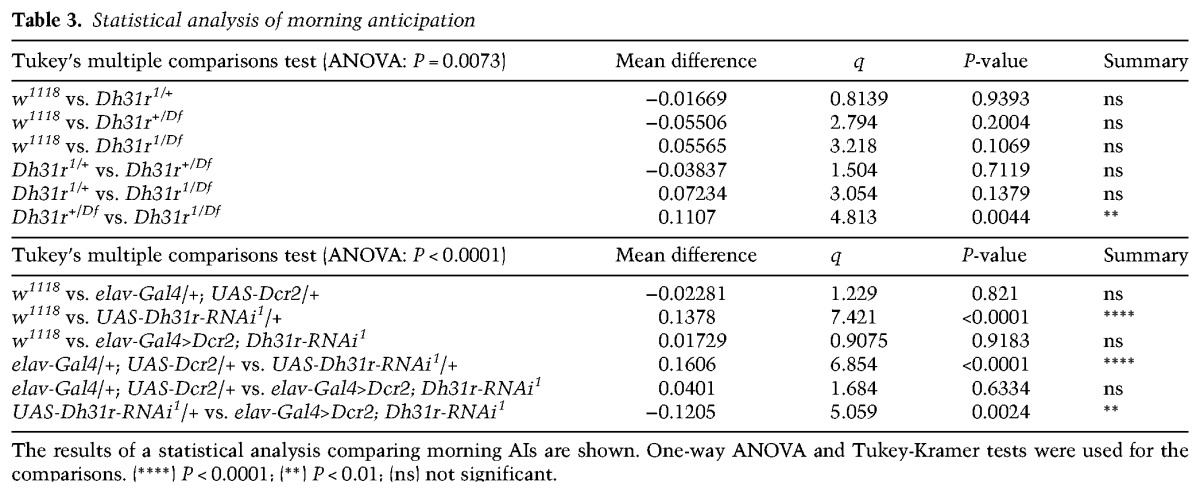
Table 4.
Total locomotor activity
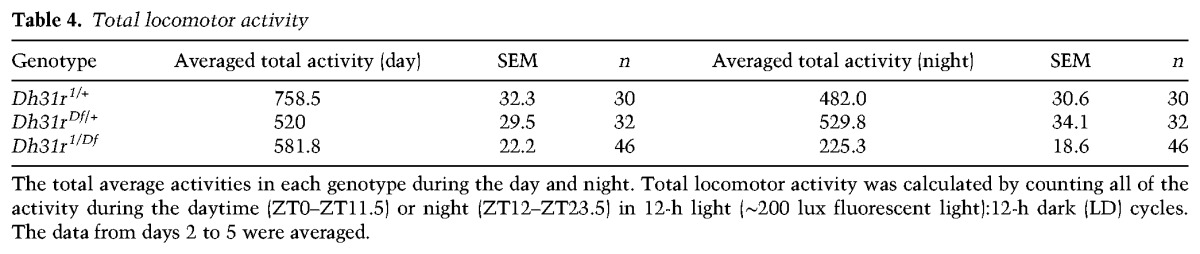
Table 5.
Statistical analysis of total locomotor activity
DH31R expression in clock cells is sufficient for the daytime TPR
To determine which DH31R-positive cells are important for the daytime TPR, we performed RNAi-mediated knockdown of Dh31r. To ensure that the phenotype was not caused by off-target effects, we used two independent RNAi lines, Dh31r-RNAi1 and Dh31r-RNAi2, which target different regions of Dh31r mRNA. When RNAi-mediated knockdown of Dh31r was performed in all neurons of Dh31r-RNAi1 and Dh31r-RNAi2 flies using the pan-neuronal driver elav-Gal4, the flies consistently preferred a higher temperature and exhibited an abnormal daytime TPR. In contrast, control flies exhibited a normal daytime TPR (Fig. 3A,B). These data suggest that reducing Dh31r expression in all neurons causes an abnormal daytime TPR. To examine whether DH31R in clock cells is important for the daytime TPR, we performed RNAi-mediated knockdown of Dh31r only in clock cells using tim-Gal4 (Fig. 3C,D). These flies maintained their daytime TPR (Fig. 3C,D, red lines), albeit they preferred an elevated temperature (∼25.5°C at ZT1–ZT3) compared with the controls (Fig. 3C, gray lines). This result suggests that reducing Dh31r expression only in clock cells does not completely disrupt the daytime TPR.
Figure 3.
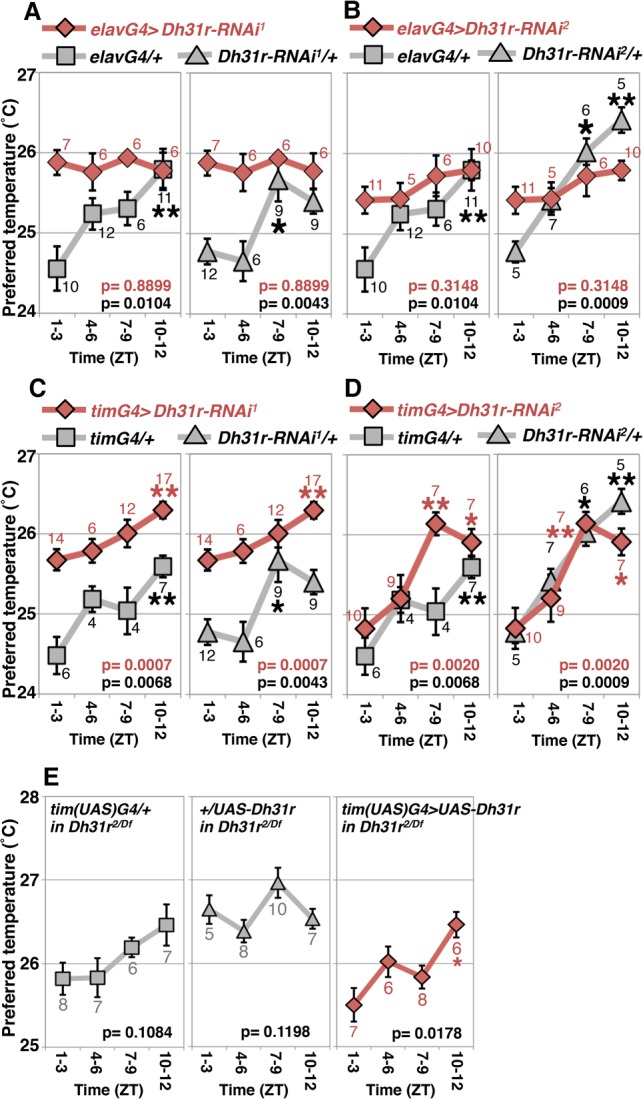
Dh31r expression in clock cells is sufficient for TPR. (A–D) Comparison of TPRs during the daytime in RNAi knockdown flies (red line) and control flies (gray line). (A,B) RNAi-mediated knockdown of Dh31r in all neurons using elavG4 (elav-Gal4, a pan-neuronal driver) with Dh31r-RNAi1 (UAS-Dh31r-RNAi1) (A) or Dh31r-RNAi2 (UAS-Dh31r-RNAi2) (B). (C,D) RNAi-mediated knockdown of Dh31r in all clock cells using timG4 (tim-Gal4, an all-clock cell driver) with Dh31r-RNAi1 (UAS-Dh31r-RNAi1) (C) or Dh31r-RNAi2 (UAS-Dh31r-RNAi2) (D). (E) Dh31r expression in all clock cells using tim(UAS)-Gal4 and UAS-Dh31r [tim(UAS)G4>UAS-Dh31r; red line] and the corresponding controls [tim(UAS)G4/+ and UAS-Dh31r/+; gray lines] in the Dh31r2/Df mutant background. The numbers represent the numbers of assays. The results of one-way ANOVA or the Kruskal-Wallis test for the data obtained during the daytime are shown. (**) P < 0.01; (*) P < 0.05, the Tukey-Kramer test or Kruskal-Wallis test compared with ZT1–ZT3 (Supplemental Table S1).
We next used the Dh31r-cDNA rescue approach to determine whether DH31R expression in clock cells is sufficient for TPR. When Dh31r was expressed in clock cells in the Dh31r2/Df background, the flies exhibited a restored daytime TPR (timG4>U-Dh31r, Dh31r2/Df) (Fig. 3E, red line), whereas UAS control in the Dh312/Df mutant flies exhibited a severely abnormal TPR (+/UAS-Dh31r, Dh31r2/Df) (Fig. 3E, gray line). The Gal4 control in the Dh312/Df mutant flies showed a small increase in daytime TPR, but this effect was not statistically significant (timG4/+, Dh31r2/Df; ANOVA: P = 0.1084) (Fig. 3E, gray lines). As a P element with UAS is inserted into the Dh31r gene locus in Dh31r2 flies, a low level of Dh31r expression may still be induced by tim-Gal4 in the absence of UAS-Dh31r. In turn, this effect might lead to a small increase in the daytime TPR. However, given that Dh31r-cDNA flies exhibited a significant increase in the daytime TPR compared with control flies (Fig. 3E, red line), these data suggest that DH31R expression in clock cells rescued the Dh31r mutant phenotype.
Both DH31 and PDF are involved in regulating the daytime TPR
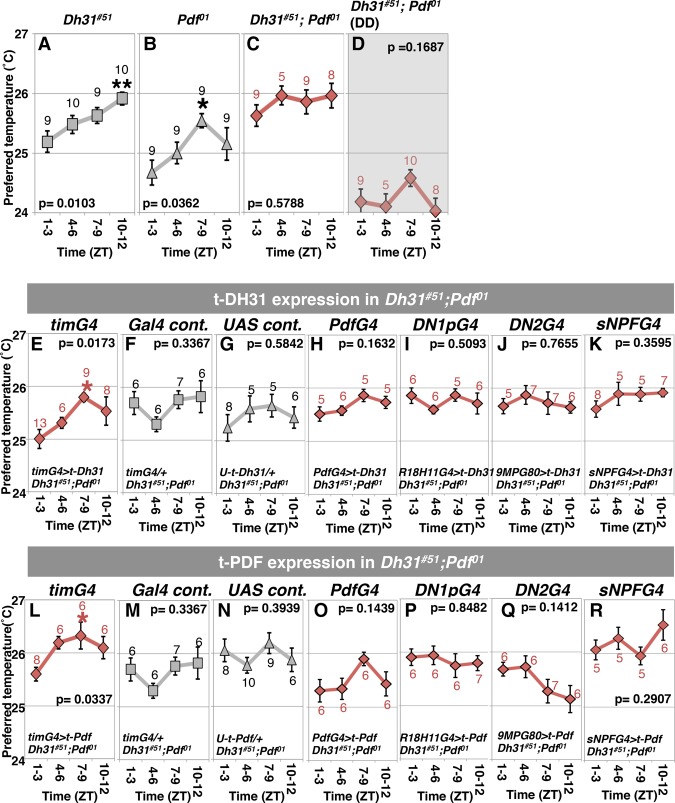
Because DH31R is required for the daytime TPR, we speculated that the DH31R ligand DH31 is also important for the daytime TPR. However, we found previously that the loss-of-function mutant Dh31#51 exhibited a normal daytime TPR (Fig. 4A; Goda et al. 2016). Similarly, PDF-null flies (Pdf01) also exhibited a daytime TPR, with advanced peaks at ZT7–ZT9 (Fig. 4B; Goda et al. 2016). Therefore, flies lacking either DH31 or PDF still exhibit a daytime TPR.
Figure 4.
The neuropeptide DH31 acts on clock cells to mediate daytime TPR. (A–D) TPRs during the daytime. (A) Dh31#51 in LD. (B) Pdf01 in LD. (C) Dh31#51; Pdf01 in LD. (D) Dh31#51; Pdf01 in DD. For Dh31#51 and Pdf01, the same data that are presented in Figure 3, A and B (Goda et al. 2016), are shown. (E–K) TPRs during the daytime in Dh31#51; Pdf01 flies with t-Dh31 expression. (E) All clock cells (tim-Gal4>UAS-t-Dh31). (F) tim-Gal4 control (tim-Gal4/+). (G) UAS-t-Dh31 control (UAS-t-Dh31/+). (H) t-Dh31 in LNvs (Pdf-Gal4>UAS-t-Dh31). (I) t-Dh31 in DN1ps (R18H11-Gal4>UAS-t-Dh31). (J) t-Dh31 in DN2s (Clk9M-Gal4; Pdf-Gal80>UAS-t-Dh31). (K) t-Dh31 in sNPF-expressing cells (sNPF-Gal4>UAS-t-Dh31). (L–R) TPRs during the daytime in Dh31#51; Pdf01 flies with t-Pdf expression. (L) All clock cells (tim-Gal4>UAS-t-Pdf). (M) tim-Gal4 control (tim-Gal4/+). (N) UAS-t-Pdf control (UAS-t-Pdf/+). (O) t-Pdf in LNvs (Pdf-Gal4>UAS-t-Pdf). (P) t-Pdf in DN1ps (R18H11-Gal4>UAS-t-Pdf). (Q) t-Pdf in DN2s (Clk9M-Gal4; PdfGal80>UAS-t-Pdf). (R) t-Pdf in sNPF-expressing cells (sNPF-Gal4>UAS-t-Pdf). The numbers represent the numbers of assays. The results of one-way ANOVA or the Kruskal-Wallis test for the data obtained during the daytime are shown. (**) P < 0.01; (*) P < 0.05, the Tukey-Kramer test or Kruskal-Wallis test compared with ZT1–ZT3 (Supplemental Table S1).
Given that DH31 can activate not only DH31R but also PDFR (Mertens et al. 2005; Shafer et al. 2008; Choi et al. 2009) and that PDF is an important neuropeptide responsible for orchestrating the activity of pacemaker neurons (Nitabach and Taghert 2008; Yao and Shafer 2014), we hypothesized that DH31 and PDF compensate for each other in mediating the daytime TPR. To examine this possibility, we generated Dh31#51; Pdf01 double-mutant flies. These flies consistently selected a daytime temperature of ∼26°C and lost rhythmic changes in temperature preference during the daytime in 12-h light:12-h dark (LD) (Fig. 4C), phenocopying the Dh31r1/Df mutant (Fig. 1B,C).
We subsequently tested TPR under DD conditions (Fig. 4D) and found that Dh31#51; Pdf01 flies lost the rhythmic daytime TPR and consistently preferred a temperature of ∼24°C, which was lower than their preferred temperature under LD conditions due to the LDTP phenotype (Fig. 4D). These data suggest that both DH31 and PDF mediate the daytime TPR via the endogenous clock.
Membrane-tethered DH31 (t-DH31) expression in clock cells restores the daytime TPR
To assess whether DH31 acts on clock cells to mediate the daytime TPR, we used t-Dh31 transgenic flies in the Dh31 and Pdf double-mutant background (Fig. 4E–K; Choi et al. 2009). The membrane-tethered peptide has both linker and anchor peptides that couple with the cell membrane, resulting in cell-autonomous binding and activation of its receptors on specific cells (Choi et al. 2009). We found that t-DH31 expression in clock cells using tim-Gal4 restored a rhythmic TPR during the daytime, with advanced peaks at ZT7–ZT9 (Fig. 4E). However, control flies still showed an abnormally flattened daytime TPR (Fig. 4F,G). These data suggest that DH31 acts on clock cells to mediate the daytime TPR.
To test the possibility that a specific subset of clock neurons is important for TPR, we expressed t-DH31 in either DN1s (R18H11-Gal4) or LNvs (Pdf-Gal4) in the Dh31#51; Pdf01 double-mutant. However, these flies still showed an abnormal daytime TPR and did not display restored daytime TPR (Fig. 4H,I). Furthermore, because we showed previously that DH31 acts on DN2s to regulate night-onset TPR (ZT10–ZT15) (Goda et al. 2016), we also expressed t-DH31 in DN2s using Clk9M-Gal4; Pdf-Gal80. However, these flies also did not display a restored daytime TPR (Fig. 4J). Moreover, as DH31R was expressed in sNPF-Gal4 cells (Fig. 2C), we expressed t-DH31 in nonclock neuropeptide cells (sNPF-Gal4 cells). However, these flies did not exhibit a restored daytime TPR (Fig. 4K). Therefore, t-DH31 expression can rescue the Dh31#51; Pdf01 double-mutant phenotype only when it is expressed in all clock neurons by tim-Gal4.
t-PDF expression in the clock cells also restores the daytime TPR
As flies in which t-DH31 expression was induced in clock neurons exhibited only a phase-advanced TPR, these flies did not exhibit a full recovery of the Dh31 and Pdf double-mutant phenotype (Fig. 4), raising the possibility that PDF also plays a role in mediating the daytime TPR. To address this possibility, we asked whether t-PDF expression in clock cells could also restore the daytime TPR. When t-PDF was expressed in clock cells using tim-Gal4, the flies showed an increased daytime TPR with advanced peaks at ZT7–ZT9 (Fig. 4L). This phenotype was very similar to the flies expressing t-DH31 in clock cells in the Dh31#51; Pdf01 double mutant (Fig. 4E). On the other hand, t-PDF expression in LNvs, DN1s, DN2s, or sNPF-Gal4 cells did not restore the daytime TPR (Fig. 4O–R), indicating that t-PDF expression can rescue the Dh31#51; Pdf01 double-mutant phenotype only when expressed in all clock neurons. Thus, either t-DH31 or t-PDF expression in all clock neurons partially rescues the Dh31#51; Pdf01 double mutant.
Mouse Calcr is expressed in the SCN shell
The Drosophila TPR, similar to the mammalian BTR, is regulated separately from locomotor activity rhythms (Kaneko et al. 2012) and follows a time-of-day-dependent pattern similar to that of the human BTR (Duffy et al. 1998). Therefore, the Drosophila TPR may share mechanistic features with the mammalian BTR. As many molecular mechanisms underlying locomotor activity rhythms and sleep are well conserved from Drosophila to mammals (Sehgal and Mignot 2011; Dubowy and Sehgal 2017), we sought to determine whether mouse homologs of DH31R mediate BTR in mice.
The closest known homologs of Drosophila Dh31r in mammals are Calcr and Calcr-like receptor (Calcrl) (Table 6; Supplemental Fig. S5; Kunst et al. 2015). The amino acid sequence similarities between DH31R and Calcr and between DH31R and Calcrl are 67.9% and 67.4%, respectively, in the seven-transmembrane domain and 74.6% and 81.0%, respectively, in the N-terminal region. We and others reported previously that Calcr is highly expressed in the SCN in rats and mice (Nakamoto et al. 2000; Becskei et al. 2004; Doi et al. 2016), but Calcrl is not expressed in the SCN (data not shown). Importantly, these expression patterns are reproducibly observed in the Allen Brain Atlas (Lein et al. 2007). We therefore further characterized Calcr expression in the SCN by in situ hybridization and immunohistochemistry (Fig. 5).
Table 6.
Sequence homology between the Drosophila DH31R and the human secretin family GPCRs
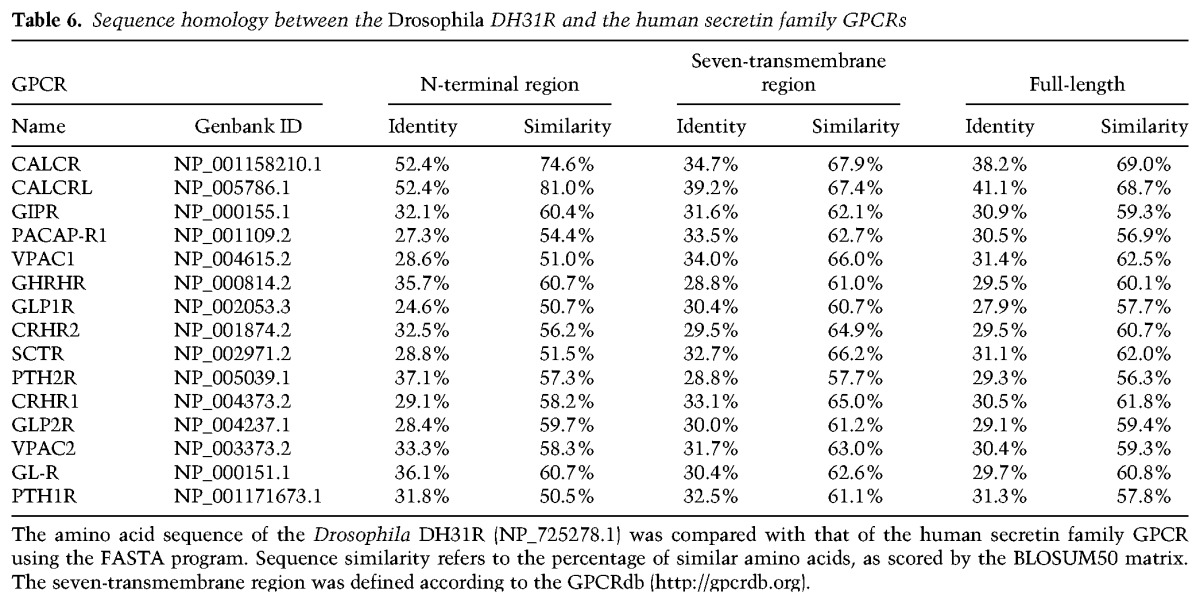
Figure 5.
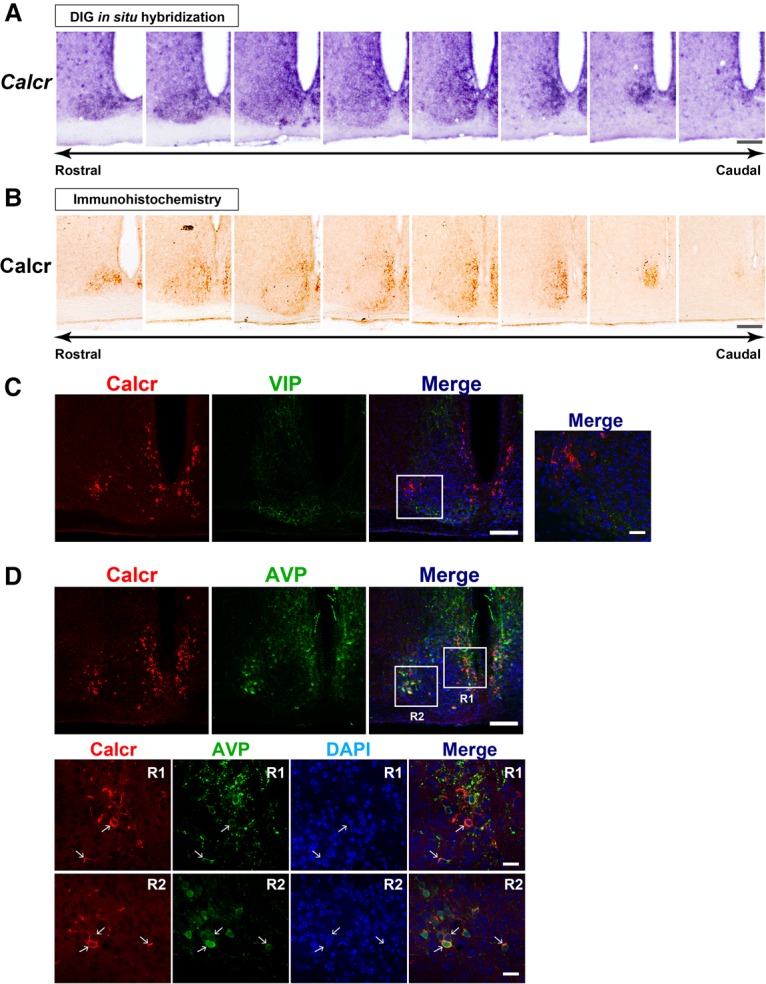
The mouse Calcr is expressed in the SCN shell. (A,B) Topographical distribution of Calcr mRNA (A) and Calcr protein (B) in serial coronal brain sections covering the entire mouse SCN in the rostral–caudal direction. Bar, 100 µm. (C) Double-label confocal immunofluorescence of Calcr and VIP in the mouse SCN. The merged image shows combined images of Calcr-based (red), VIP-based (green), and 4,6-diamino-2-phenylindole (DAPI)-based nuclear staining (blue). The boxed area is enlarged in the right panel. Bars: left, 100 µm; right, 20 µm. (D) Double-label confocal immunofluorescence of Calcr and arginine vasopressin (AVP). The boxes indicate the regions enlarged in the bottom panels. The arrows indicate cells double-immunolabeled for Calcr and AVP. Bars: top, 100 µm; bottom, 20 µm.
We found that Calcr mRNA and Calcr protein were similarly distributed from the rostral to the caudal margins of the SCN (Fig. 5A,B). Topographically, the Calcr mRNA and protein signals in SCN were detected mainly in the dorsomedial area, a region corresponding to the SCN shell. To gain additional insight into the regional distribution of Calcr in the SCN shell, double immunostaining was performed with anti-Calcr and anti-arginine vasopressin (AVP) antibodies (Fig. 5D), the latter of which was used as a marker of the SCN shell (Abrahamson and Moore 2001). We found that the distributions of AVP and Calcr partially overlapped in the SCN shell: Approximately 84% of the Calcr-positive cells expressed AVP, while ∼65% of the AVP-positive cells expressed Calcr (Fig. 5D). Double immunostaining was also performed using anti-Calcr and anti-VIP, which delineates the SCN core (Abrahamson and Moore 2001). We found that VIP and Calcr did not colocalize (Fig. 5C), suggesting that Calcr is not expressed in the VIP-ergic SCN core. Furthermore, Vipr2-expressing Calcr-immunopositive cells were observed in the SCN shell (Supplemental Fig. S6), suggesting a potential functional link between Calcr and Vipr2. Notably, this link may be similar to the situation that occurs in Drosophila, where DH31R and PDFR are expressed in the same group of clock cells (Fig. 2; Im and Taghert 2010). Taken together, these findings show that Calcr expression is localized mainly to a part of the SCN shell that partially overlaps the AVP-expressing region of the SCN.
Mouse Calcr mediates body temperature fluctuations during the night
To determine whether Calcr is involved in mediating BTR, we used Calcr knockout mice that had been backcrossed to the C57BL/6J background for >10 generations. Using both in situ hybridization and anti-Calcr immunostaining, we confirmed that Calcr mRNA and protein were absent in the SCN of Calcr knockout mice (Fig. 6A). Importantly, Calcr knockout mice showed a normal free-running period of locomotor activity rhythm as well as normal total locomotor activity. These observations are consistent with previously reported data (Doi et al. 2016) (Fig. 6B).
Figure 6.
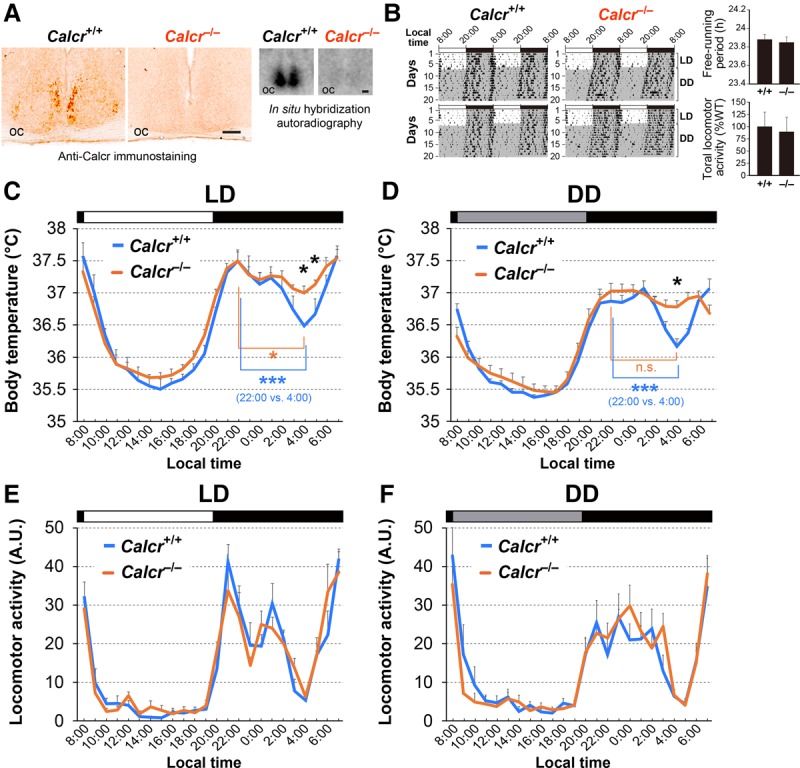
Calcr knockout mice exhibit an abnormal body temperature during the active phase. (A) Immunohistochemistry (left) and radioisotopic in situ hybridization (right) confirm the deficiency of Calcr protein and transcript levels in Calcr−/− mice in the SCN. Bar, 100 µm. (oc) Optic chiasm. (B) Double-plotted actograms of C57BL/6J-backcrossed Calcr+/+ and Calcr−/− mice over 7 d in LD and 13 d in DD. The periods of darkness are indicated by gray backgrounds. The bar graphs indicate the mean ± SEM of the free-running periods of Calcr+/+ and Calcr−/− mice and their relative total locomotor activity per day. The values were determined based on a 10-d interval taken after 3 d in DD. n = 6 (both genotypes). P = 0.521 for period; P = 0.806 for activity, unpaired t-test. (C,D) Body temperatures of Calcr+/+ and Calcr−/− mice in LD (C) and DD (D), as measured by intra-abdominally implanted thermometers. The body temperatures at each time of day for three consecutive days in LD (C) or DD (D) were averaged and smoothed once with a three-point moving average. The values shown are the mean ± SEM. n = 5 mice for each data point. (Black asterisk) P < 0.05, Calcr+/+ versus Calcr−/− (two-way ANOVA with Bonferroni post hoc test); (blue asterisks) P < 0.001, 22:00 versus 4:00 in Calcr+/+ mice; (orange asterisk) P < 0.05, 22:00 versus 4:00 in Calcr−/− mice; (n.s.) not significant (one-way ANOVA with Bonferroni post hoc test). (E,F) The temporal profiles of Calcr+/+ and Calcr−/− locomotor activity in LD (E) and DD (F). The locomotor activities of Calcr+/+ and Calcr−/− mice at each time of day on three consecutive days were averaged and plotted as the mean ± SEM. n = 5 mice per genotype.
In LD conditions, the body temperature of wild-type mice fluctuates over each 24-h period (Duffy et al. 1998). During the daytime, when the mice were primarily resting, their body temperature gradually decreased during the early phase and increased during the later phase (Fig. 6C; Shiromani et al. 2004; Nagashima et al. 2005; Gerhart-Hines et al. 2013). However, during the night, the animals’ body temperature displayed bimodal peaks in the early night and at dawn, with a deep trough late in the night (i.e., midnight trough) (Fig. 6C). The midnight trough in body temperature has been reported previously (Gerhart-Hines et al. 2013; Wolff et al. 2013), and our data confirmed that the difference between the body temperatures at the early peak (22:00) and at the trough (4:00) was statistically significant (Fig. 6C, blue asterisk, [***] P < 0.001). Furthermore, the body temperature fluctuation patterns observed over the 24-h period, including the midnight trough, were very similar in LD and DD conditions, although the overall body temperatures were slightly lower in DD than in LD (Fig. 6D). The body temperature data for individual animals in LD and DD are in Supplemental Figure S7. These data indicate that the patterns of temporal fluctuations in body temperature are controlled by the endogenous clock.
Although we found that the body temperatures of both wild-type and Calcr knockout mice fluctuated over a 24-h period, they were significantly different at midnight (Fig. 6C,D, black asterisks, [*] P < 0.05; Supplemental Fig. S7). Specifically, the body temperatures of wild-type mice showed a deep trough at midnight (Fig. 6C,D, blue asterisk, [***] P < 0.001), whereas the body temperatures of Calcr knockout mice lost the characteristic trough and remained relatively unchanged during the night in both LD (Fig. 6C) and DD (Fig. 6D) conditions (Fig. 6C,D, orange asterisk, [*] P < 0.05 for LD and n.s. for DD). These data indicate that the lack of Calcr expression causes a shallow midnight trough in body temperature, suggesting that Calcr is required for body temperature fluctuations, particularly during the night (the active phase of mice). Importantly, this finding is consistent with the findings for the TPR phenotype of the Drosophila Dh31r mutant, as we observed that the Dh31r mutation caused a flat TPR during the daytime (the active phase of flies) (Fig. 1B,C). Therefore, both Drosophila DH31R and mouse Calcr are required for body temperature fluctuation during the active phase.
Because body temperature may be increased by locomotor activity, we speculated that higher levels of locomotor activity result in the higher body temperatures observed in Calcr knockout mice. To examine this possibility, we compared fluctuations in locomotor activity between wild-type and Calcr knockout mice in both LD (Fig. 6E) and DD (Fig. 6F) conditions; however, there was no significant difference in locomotor activity between the two groups. Therefore, we concluded that Calcr specifically mediates body temperature fluctuations during the night but does not affect locomotor activity rhythms.
Discussion
Calcitonin receptors mediate daily BTRs
Daily BTR is essential for homeostatic functions, such as metabolism and sleep (Refinetti and Menaker 1992). Several lines of evidence suggest that BTR is controlled separately from locomotor activity rhythms (Smith 1969; Gander et al. 1986; Lavie 2001; Saper et al. 2005); however, no molecular evidence supporting this model has been reported to date. Given that several mouse mutants that have defects in BTR also have defects in locomotor activity rhythms (Shiromani et al. 2004; Nagashima et al. 2005; Gerhart-Hines et al. 2013), it is crucial to find genes that specifically regulate daily BTR.
Here, we provide the first evidence that Calcr and its insect homolog, DH31R, play a role in mediating BTR during the active phase in mice and flies, respectively. We also noted several functional similarities between DH31R and Calcr with respect to the regulation of fly TPR and mouse BTR during the active phase (Supplemental Fig. S8). First, we found that DH31R and Calcr are expressed in the brain's central clock neurons in flies and mice, respectively (Figs. 2, 5). Second, although both Dh31r and Calcr mutants showed almost no fluctuations in body temperature during the active phase (daytime for flies and nighttime for mice) (Figs. 1, 6), both showed normal rhythmicity of locomotor activity (Figs. 2E–G, 6B,E,F). These observations thus revealed that calcitonin receptors play an important role in daily BTR during the active phase but not in locomotor activity in flies and mice. This molecular evidence therefore substantiates the premise that the daily profiles of body temperature and locomotor activity are controlled separately.
DH31 acts on clock cells via DH31R to mediate TPR in Drosophila
An important open question is the nature of the relationship between clock cells and DH31R in Drosophila. As (1) DH31 acts on clock cells to mediate TPR (Fig. 4) and (2) DH31R expression in clock cells rescued the Dh31r mutant phenotype (Fig. 3), DH31 likely acts on clock cells via DH31R to mediate TPR (Fig. 4). These data suggest that the DH31–DH31R interaction in clock cells is sufficient to meditate TPR.
On the other hand, our data also showed that t-PDF expression in the clock cells could partially rescue the Dh31 Pdf double-mutant phenotype (Fig. 4L), suggesting that PDF also acts on the clock cells to mediate TPR. As we determined previously that PDFR is not required for regulating the daytime TPR (Goda et al. 2016), the PDF–PDFR interaction is unlikely to play a primary role in regulating daytime TPR. Therefore, we speculate that PDF might be able to weakly activate DH31R in the intact brain, although it has been shown that PDF does not activate DH31R (Johnson et al. 2005; Choi et al. 2009). Alternatively, t-PDF overload might be artificially able to activate DH31R-neurons via PDFR or DH31R. Thus, it is of interest to further investigate the relationship between DH31, PDF, and their respective receptors in terms of the regulation of TPR.
Furthermore, we showed previously that DN2s are the main clock regulators of TPR (Kaneko et al. 2012). However, we found that DH31 and DH31R were not expressed in DN2s (Fig. 2; Supplemental Fig. S3; Goda et al. 2016) and that the oscillations of the molecular clock (VRI expression) in DN2s were intact in both Dh31r mutants (Supplemental Fig. S2) and Dh31 Pdf double mutants (T Goda, Y Umezaki, HW Seo, and FN Hamada, in prep.). These data suggest that the DH31–DH31R interaction mediates the TPR downstream from DN2s.
A possible contribution of nonclock cells to the fly TPR and mouse BTR
In Drosophila, DH31R is expressed in not only a subset of clock cells but also a population of nonclock peptidergic cells expressing sNPF and CRZ. These latter cells are located in the dorsal portion of the fly brain (Fig. 2; Supplemental Fig. S3). Although we showed that DH31R expression in clock cells restored the Dh31r mutant phenotype (Fig. 3E), RNAi-mediated knockdown of Dh31r in clock cells did not completely eliminate TPR (Fig. 3C,D), suggesting that DH31R expression by clock cells is sufficient but not necessary for TPR. Therefore, DH31R-, sNPF-, and CRZ-expressing nonclock cells likely play some roles in mediating TPR in Drosophila.
In mice, we found that Calcr is present not only in the SCN but also in regions outside the SCN, such as the subparaventricular zone (SPZ), the medial preoptic area (MPO), and the arcuate nucleus (ARC) (data not shown). These findings are consistent with those of previous studies (Nakamoto et al. 2000; Becskei et al. 2004). Although it remains unclear which Calcr-expressing neurons are responsible for mediating BTR, these Calcr-expressing non-SCN neurons may have a function similar to that of DH31R-positive nonclock cells in flies. The neurons producing neuropeptide Y (NPY), a mouse homolog of Drosophila sNPF located in the ARC, respond to an endocrine hormone known as ghrelin, which is secreted by the stomach when appetite increases (Barsh and Schwartz 2002). Gonadotropin-releasing hormone (GnRH) (Campbell et al. 2009), functionally similar to Drosophila CRZ (Tian et al. 2016), is a hormone produced in the hypothalamus and is involved in the production of the male hormone testosterone by binding to pituitary GnRH receptors. Neurons that express GnRH are located in the MPO, project to the median eminence, and secrete GnRH into the hypophyseal portal vessels, thereby regulating pituitary luteinizing hormone (LH)/follicle-stimulating hormone (FSH) release. Furthermore, the MPO is involved in thermoregulation (Morrison and Nakamura 2011), and ablation of the dorsal SPZ causes abnormal BTR but not abnormal locomotor rhythm in rats (Saper et al. 2005). Therefore, Calcr function may not be limited to clock cells, and it is possible that Calcr neurons at extra-SCN sites may contribute to mediating BTR.
The role of body temperature fluctuations during the night in mice
The mouse BTR includes two peaks, one in the early night and one at dawn, with a trough at midnight. As BTR is highly influenced by animal rest–activity states, BTR patterns temporally correlate with those of locomotor activity rhythms. Therefore, it is reasonable to speculate that the trough in the middle of the dark phase seen in wild-type mice could be due simply to the animals’ resting and may perhaps be a passive consequence of the activity burst that occurs during the early part of the night. However, given that Calcr deletion resulted in a flat body temperature throughout the night without affecting locomotor activity (Fig. 6C–F), these results strongly suggest that the midnight body temperature trough is genetically and endogenously programmed and not merely a consequence of decreased locomotor activity. Interestingly, mice deficient in the nuclear receptor Rev-erbα also show a reduced body temperature fluctuation amplitude as well as a reduced midnight trough (Gerhart-Hines et al. 2013). Notably, it appears likely that human body temperature also drops during the active phase (at approximately 3:00 pm) (Patke et al. 2017). Thus, our data raise the possibility that the midnight trough in body temperature may play a proactive role in homeostatic regulation, much like physiology and metabolism regulation; this would be an interesting subject for a future study.
Taken together, the findings of this study show that although Drosophila and mice have completely different thermoregulatory systems, they exhibit a daily BTR mediated by a conserved molecule, DH31R/Calcr. Thus, our data support the idea that circadian rhythms are evolutionarily conserved and essential for animal survival among different species. Given that studies in Drosophila have provided strong contributions to the discovery of many conserved mammalian circadian clock genes and regulatory mechanisms, we anticipate that understanding the fly TPR will provide us with novel insights into the molecular and neural mechanisms that control BTR in mammals.
Materials and methods
Fly lines and generation of transgenic flies
All of the flies were maintained under LD cycles at 25°C. In ZT, ZT0 is lights on, and ZT12 is lights off. Dh31r1and Dh31r2are P-element insertion mutants (PBac{WH}Dh31-Rf05546 and PBac{WH}Dh31-Rf06589, respectively) and were obtained from the Exelixis Collection of Harvard Medical School. Dh31rDf is a deletion mutant [Df(2R)BSC273: RRID: BDSC_23169] and was obtained from the Bloomington Drosophila Stock Center (stock no. 23169). The OK107-Gal4 (RRID: BDSC_854), elav-Gal4; UAS-Dcr2 (elavC155w1118; UAS-Dcr2) (RRID:BDSC_25750), tim-Gal4 (RRID: BDSC_7126), Pdf-Gal4 (RRID: BDSC_6900), R18H11-Gal4 (RRID: BDSC_48832), and UAS-Dcr2 (RRID: BDSC_24644) flies were obtained from the Bloomington Drosophila Stock Center. The Clk9M-Gal4; Pdf-Gal80, tim(UAS)-Gal4, UAS-t-Dh31 (t-DH31-ML:B4), and UAS-t-Pdf (t-PDF-ML:M2a) flies were kind gifts from Dr. Paul E. Hardin, Dr. Justin Blau, and Dr. Michael Nitabach, respectively. The sNPF-Gal4 (Drosophila Genomics Resource Center, 113901) flies were obtained from the Kyoto Stock Center. UAS-Dh31r-RNAi1 (Vienna Drosophila Resource Center [VDRC], 8777) and UAS-Dh31r-RNAi2 (17043R-4) flies were obtained from the VDRC and the National Institute of Genetics, Japan, respectively. All of the Gal4 driver and UAS reporter flies used in Figures 3E and 4, E–R, were generated via chromosome recombination with w; Dh31r2/Df or w; Dh31#51; Pdf01 double-mutant flies. elavc155 w; UAS-Dcr2/UAS-Dh31r-RNAi1, elavc155 w; UAS-Dcr2/+, w; UAS-Dh31r-RNAi1/+, elavc155 w; UAS-Dcr2/UAS-Dh31r-RNAi2, w; UAS-Dh31r-RNAi2/+, UAS-Dcr2 w; tim-Gal4/UAS-Dh31r-RNAi1, UAS-Dcr2 w; tim-Gal4/+, and UAS-Dcr2 w; tim-Gal4/UAS-Dh31r-RNAi2 flies were used in Figure 3, A–D. w; tim(UAS)-Gal4 Dh31r2/Dh31rDf, w; Dh31r2/UAS-Dh31r Dh31rDf, and w; tim(UAS)-Gal4 Dh31r2/UAS-Dh31r Dh31rDf flies were used in Figure 3E. All of the flies used for the behavioral experiments were on the white background.
Generation of the rescue (Dh31r), Dh31r1/Df and UAS-Dh31r fly lines
In the rescue (Dh31r), Dh31r1/Df fly line, which was obtained from P[acman] resources, the Ch321-57F06 BAC clone covers the entire Dh31r gene region, including the surrounding genes. Purified BAC DNA was injected into attP40 embryos to generate the transgenic flies (BestGene, Inc.). The rescue (Dh31r), Dh31r1/Df fly line was generated via chromosome recombination with w; Dh31r1/Df mutant flies. In the UAS-Dh31r fly line, the DNA-coding sequence of the DH31R-RB (1–430 amino acids) with a Kozak sequence and a stop codon was amplified from Dh31r-cDNA that had been isolated from Canton-S by PCR with the sense primer 5′-gaagatctACGACCATGAGCGACCAGAT-3′ and the antisense primer 5′-gctctagagTCATACCTTCTCCTCTCCTG-3′. The full-length Dh31r-cDNA was subcloned into the pUAS-stringerΔAP-attB vector (obtained from Dr. Emi Nagoshi), and the purified DNA was injected into attB embryos to generate the transgenic flies (BestGene, Inc.).
DH31R antibody
The DNA-coding sequence of the DH31R N-terminal region (1–138 amino acids) with a stop codon was amplified from Dh31r-cDNA that had been isolated from Canton-S by PCR with the sense primer 5′-cgggatccATGAGCGACCAGATTGGCAA-3′ and the antisense primer 5′-cccgtcgacTCACCAGTTGAGGTCTTCGAGAT-3′. The N terminus of the Dh31r-cDNA was subcloned into the pGEX-6P-2 vector (GE Healthcare Life Sciences) and transformed into BL21 cells to produce GST-tagged N-terminal DH31R by IPTG induction. GST-tagged N-DH31R was purified from the cell lysates using a GST column. The molecular weight of the purified protein was ∼40 kDa, as confirmed by SDS gel electrophoresis. The purified protein was injected into rats to produce antibodies (Covance).
Immunohistochemistry of fly brains
Immunostaining was performed as described previously (Hamada et al. 2008; Tang et al. 2013) with the following modifications: 10% normal goat serum in PBST (PBS plus 0.5% Triton X-100) was used for blocking and for incubating the brain tissues with the anti-DH31R antibody, and brain fixation was performed using 4% paraformaldehyde and 7% picric acid for 30 min at room temperature prior to immunostaining with anti-DH31R. The following antibodies at the indicated dilutions were used for this experiment: rat anti-DH31R (1:200), rat anti-TIM (1:200), donkey anti-rat Cy5 (1:200; Jackson ImmunoResearch Laboratories, Inc.), rabbit anti-CRZ (1:200), donkey anti-rabbit Cy3 (1:200; Jackson ImmunoResearch Laboratories, Inc.), guinea pig anti-VRI (1:200), donkey anti-guinea pig Alexa 647 (1:200; Jackson ImmunoResearch Laboratories, Inc.), mouse anti-PDF (1:200; Hybridoma Bank), and donkey anti-mouse Cy3 (1:200; Jackson ImmunoResearch Laboratories, Inc.). The mounted brain sections were scanned using a Zeiss LSM5 Pascal confocal microscope. The images shown are digitally projected z-stacks. For anti-DH31R antibody signal quantification (Supplemental Fig. S4), ImageJ software was used to quantify the intensity of the anti-DH31R immunostaining signal in DN1 or DN3 in each brain hemisphere. After background subtraction, the average intensity of 10 brain hemispheres was calculated using Microsoft Excel.
Temperature preference behavioral assay and data analysis
Temperature preference behavior was assessed for 30 min in the light during the daytime and in the dark during the nighttime in an environmental room maintained at 25°C/65%–70% RH. The flies used for the behavioral assay were never reused. The behavioral apparatus and the conditions used are described in detail in Goda et al. (2014). It is worth noting that the preferred temperature was occasionally variable. Our recent data suggest that light exposure (Head et al. 2015) and feeding state (Umezaki et al. 2018) strongly affect preferred temperatures but not TPR.
The method used to calculate the mean preferred temperature has been described previously (Kaneko et al. 2012; Goda et al. 2014). After the 30-min behavioral assay, the number of flies that were completely on the apparatus was counted. The flies that were partially or completely on the walls of the apparatus cover were not counted or included in the data analysis. The percentage of flies within each 1°C temperature interval on the apparatus was calculated by dividing the number of flies within each 1°C interval by the total number of flies on the apparatus. The location of each 1°C interval was determined by measuring the temperature at six different points on both the top and the bottom of the apparatus. The data points were plotted as the percentages of flies within each 1°C temperature interval. The weighted mean preferred temperature was calculated by summing the products of the percentage of flies within a 1°C temperature interval and the corresponding temperature (i.e., the fractional number of flies × 18.5°C + fractional number of flies × 19.5°C + ………fractional number of flies × 32.5°C). We tested the temperature preference behavior of the flies at least five times in each time segment (ZT1–ZT3, ZT4–ZT6, ZT7–ZT9, ZT10–ZT12, ZT13–ZT15, ZT16–ZT18, ZT19–ZT21, and ZT22–ZT24). If the SEM of the averaged preferred temperature was not <0.3 after five trials, we performed additional trials until the SEM was <0.3. At least 40 experiments were necessary to obtain a full curve for a 24-h period. In each time segment, the weighted mean preferred temperatures from each trial were averaged, and the SEM was calculated.
Behavioral analysis of locomotor activity
Locomotor activity assays and data analysis were performed as described previously (Kaneko et al. 2012; Umezaki et al. 2012; Goda et al. 2016). The flies were reared under LD cycles at 25°C. Male flies (1–5 d old) were used in the locomotor activity experiments. A Drosophila activity monitoring (DAM) system (http://www.trikinetics.com) was placed in an incubator (Sanyo Scientific, MIR-154). Lights in the incubator (15-W cool white fluorescent lamps [Toshiba, FL15D]) were connected to an electric timer, and the light intensity was ∼800 lux. Locomotor activity was monitored in LD cycles for 5 d and in DD conditions for >11 d at 25°C. The data were analyzed with Actogram J software (Schmid et al. 2011). Free-running periods and power values were calculated using a χ2 periodogram (Sokolove and Bushell 1978; Umezaki et al. 2012), and flies having a power value <100 were defined as arrhythmic (Goda et al. 2016). Only the rhythmic flies were used, and these data were averaged to generate a double-plotted actogram. Morning anticipation index (AI) values were calculated as described previously (Harrisingh et al. 2007; Sheeba et al. 2010; Seluzicki et al. 2014) Briefly, AI = (total activity 3 h before lights on)/(total activity 6 h before lights on). All of the flies’ AIs over days 2–5 of the LD cycles were averaged in each genotype. AIs for different genotypes were compared using Tukey's multiple comparisons test. Total locomotor activity was calculated by counting all of the activity during daytime (ZT0–ZT11.5) or night (ZT12–ZT23.5) in LD cycles. These data from days 2–5 were averaged. Total locomotor activity in each genotype was compared by using one-way ANOVA and Tukey's multiple comparisons test.
Real-time PCR
Total RNA was isolated from the heads of 30 flies using TRIzol (Qiagen). After DNase I treatment, cDNA was generated using SuperScript III (Invitrogen). Real-time PCR was performed using SYBR Advantage qPCR mix on a StepOnePlus real-time PCR system (Applied Biosystems).
Control primers for ef1b were F (GTCATCGAGGACGACAAGGT) and R (CTTGTTGAAGGCAGCAAT), and primers for Dh31r were 5F (TACATCCTTACGCCCTTTCGTCCT) and RT-L-2R (GGCAACGCACAGACCTTGAAATGA).
Mouse strain
Mutant Calcr mice (Calcrtm1Dgen) with a mixed genetic background involving 129P2/OlaHsd × C57BL/6J were obtained from the Mutant Mouse Regional Resource Center at the University of North Carolina (https://www.mmrrc.org) and backcrossed to C57BL/6J for 10 generations. The Calcr+/− mice were then intercrossed to produce homozygous null and wild-type progenies by in vitro fertilization. All of the animal experiments were performed according to protocols approved by the Animal Care and Experimentation Committee of Kyoto University.
Measurements of mouse body temperature and locomotor activity
Single caged male littermate mice (aged 6 wk) were housed individually in light-tight ventilated closets in a temperature- and humidity-controlled facility. The animals were entrained on a LD cycle for at least 2 wk before the experiments. The core body temperatures of Calcr+/+ and Calcr−/− mice (8 wk of age) were measured using a Thermochron iButton DS1921H (Maxim Integrated), which was inserted into the peritoneal cavity as described previously (Yamaguchi et al. 2013). Following a week of convalescence, the mice were maintained in either LD or DD. The body temperatures of the animals were measured every 20 min, and three measurements obtained in each nonoverlapping 60-min interval were then averaged to obtain a single value for each clock hour. To extract circadian trends, we averaged the data obtained at each time of day on three consecutive days and smoothed the data once using a three-point moving average. Locomotor activity was simultaneously detected with passive (pyroelectric) infrared sensors (Omron, FA-05 F5B), and the data were analyzed with ClockLab software (Actimetrics) developed on MatLab (Mathworks), as described elsewhere (Yamaguchi et al. 2013).
Mouse histology
In situ hybridization was performed with free-floating brain sections (30 mm thick) using either a [33P]-labeled or a digoxigenin-labeled cRNA probe specific for mouse Calcr (nucleotides 670–1168; NM_007588), as described previously (Doi et al. 2016). Free-floating immunohistochemistry was also performed using 5 µg/mL anti-Calcr antibody (Abcam, ab11042), as described elsewhere (Doi et al. 2016), and immunoreactivity was visualized with a peroxidase-based Vectorstain Elite ABC kit (Vector Laboratories) with diaminobenzidine serving as the chromogen. For dual-label immunofluorescence, the free-floating sections were stained with 5 µg/mL anti-Calcr (rabbit polyclonal; Abcam, ab11042) along with either 4 µg/mL anti-Vipr2 (chicken polyclonal) (Doi et al. 2016), 1:1000 anti-VIP (guinea pig polyclonal; Abnova, PAB16648), or 0.2 µg/mL anti-AVP-associated neurophysin II (goat polyclonal; Santa Cruz Biotechnology, sc-27093) antibodies. Immunoreactivity was visualized using Alexa 594-conjugated anti-rabbit IgG (1:1000; Life Technologies) and Alexa 488-conjugated anti-chicken, anti-guinea pig, or anti-goat IgG (1:1000; Life Technologies) antibodies. The nuclei were visualized by staining with 4,6-diamino-2-phenylindole (DAPI).
Supplementary Material
Acknowledgments
We are grateful to Dr. Paul E. Hardin for providing the anti-VRI antibody and the Clk9M-Gal4; PdfGal80 flies, Dr. Justin Blau for providing the tim(UAS)-Gal4 flies, Dr. Michael Nitabach for providing the UAS-t-Dh31 and UAS-t-Pdf flies, Dr. Emi Nagoshi for providing the pUAS-stringerΔAP-attB vector, and Dr. Jan Veenstra for providing the CRZ antibody. We also thank the Bloomington Drosophila Fly Stock Center, the Kyoto Stock Center, Harvard Medical School, P[acman] resources, and the Vienna Drosophila Resource Center for providing the other fly lines. We thank Dr. Richard Lang, Dr. John Hogenesch, Dr. Christian I. Hong, Dr. Emi Nagoshi, Dr. Haruna Kaneko, Dr. Yosuke Ichijima, Dr. Tomotake Kanki, Dr. Masashi Yukawa, Dr. Tianyu Wang, and the Hamada and Okamura laboratory members for their comments and advice regarding the manuscript. This research was supported by a Trustee Grant and RIP funding from Cincinnati Children's Hospital, Japan Science and Technology (JST)/Precursory Research for Embryonic Science and Technology (PRESTO), the March of Dimes, and National Institutes of Health grant R01 GM107582 to F.N.H., and grants from JST/Core Research for Evolutional Science and Technology (CREST) (JPMJCR14W3), the Project for Elucidating and Controlling Mechanisms of Aging and Longevity from Japan Agency for Medical Research and Development (AMED), and the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15H01843, 15H05933, and 17H01524).
Author contributions: F.N.H., M.D., T.G., and H.O. designed the research. T.G. performed the TPR behavioral experiments and created the anti-DH31R antibody and Dh31r genomic rescue fly lines. T.G., M.L.C., and V.H.N. performed Drosophila brain immunostaining. T.G. and Y.U. performed the Drosophila locomotor activity assays. M.D. and H.O. generated the C57BL/6J-backcrossed Calcr knockout mouse, and H.S. and M.D. performed the mouse BTR and locomotor activity assays. I.M. and M.D. performed mouse brain immunostaining. F.N.H., M.D., T.G., and H.O. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.307884.117.
References
- Abrahamson EE, Moore RY. 2001. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res 916: 172–191. [DOI] [PubMed] [Google Scholar]
- Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, Shafer O, Rosbash M. 2017. RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet 13: e1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R, Chung BY. 2010. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol 72: 605–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschoff J. 1983. Circadian control of body temperature. J Therm Biol 8: 143–147. [Google Scholar]
- Barsh GS, Schwartz MW. 2002. Genetic approaches to studying energy balance: perception and integration. Nat Rev Genet 3: 589–600. [DOI] [PubMed] [Google Scholar]
- Becskei C, Riediger T, Zund D, Wookey P, Lutz TA. 2004. Immunohistochemical mapping of calcitonin receptors in the adult rat brain. Brain Res 1030: 221–233. [DOI] [PubMed] [Google Scholar]
- Bedont JL, Blackshaw S. 2015. Constructing the suprachiasmatic nucleus: a watchmaker's perspective on the central clockworks. Front Syst Neurosci 9: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhr ED, Yoo SH, Takahashi JS. 2010. Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330: 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Gaidamaka G, Han SK, Herbison AE. 2009. Dendro–dendritic bundling and shared synapses between gonadotropin-releasing hormone neurons. Proc Natl Acad Sci 106: 10835–10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Fortin JP, McCarthy E, Oksman L, Kopin AS, Nitabach MN. 2009. Cellular dissection of circadian peptide signals with genetically encoded membrane-tethered ligands. Curr Biol 19: 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon ME, Wang G, Garrity PA, Huey RB. 2009. Review: thermal preference in Drosophila. J Therm Biol 34: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Murai I, Kunisue S, Setsu G, Uchio N, Tanaka R, Kobayashi S, Shimatani H, Hayashi H, Chao HW, et al. 2016. Gpr176 is a Gz-linked orphan G-protein-coupled receptor that sets the pace of circadian behaviour. Nat Commun 7: 10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubowy C, Sehgal A. 2017. Circadian rhythms and sleep in Drosophila melanogaster. Genetics 205: 1373–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy JF, Dijk DJ, Klerman EB, Czeisler CA. 1998. Later endogenous circadian temperature nadir relative to an earlier wake time in older people. Am J Physiol 275: R1478–R1487. [DOI] [PubMed] [Google Scholar]
- Ellis DJ, Firth BT, Belan I. 2007. Circadian rhythms of locomotor activity and temperature selection in sleepy lizards, Tiliqua rugosa. J Comp Physiol A Neuroethol Sens Neural Behav Physiol 193: 695–701. [DOI] [PubMed] [Google Scholar]
- Gander PH, Connell LJ, Graeber RC. 1986. Masking of the circadian rhythms of heart rate and core temperature by the rest-activity cycle in man. J Biol Rhythms 1: 119–135. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, Bugge A, Hou C, Ferrara C, Seale P, et al. 2013. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature 503: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SS, van den Heuvel CJ, Ferguson SA, Dawson D. 2004. Thermoregulation as a sleep signalling system. Sleep Med Rev 8: 81–93. [DOI] [PubMed] [Google Scholar]
- Goda T, Leslie JR, Hamada FN. 2014. Design and analysis of temperature preference behavior and its circadian rhythm in Drosophila. J Vis Exp 83: e51097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda T, Tang X, Umezaki Y, Chu ML, Hamada FN. 2016. Drosophila DH31 neuropeptide and PDF receptor regulate night-onset temperature preference. J Neurosci 36: 11739–11754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. 2008. An internal thermal sensor controlling temperature preference in Drosophila. Nature 454: 217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN. 2007. Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J Neurosci 27: 12489–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head LM, Tang X, Hayley SE, Goda T, Umezaki Y, Chang EC, Leslie JR, Fujiwara M, Garrity PA, Hamada FN. 2015. The influence of light on temperature preference in Drosophila. Curr Biol 25: 1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Taghert PH. 2010. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. J Comp Neurol 518: 1925–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Shafer OT, Trigg JS, Park J, Schooley DA, Dow JA, Taghert PH. 2005. A novel diuretic hormone receptor in Drosophila: evidence for conservation of CGRP signaling. J Exp Biol 208: 1239–1246. [DOI] [PubMed] [Google Scholar]
- Kaneko H, Head LM, Ling J, Tang X, Liu Y, Hardin PE, Emery P, Hamada FN. 2012. Circadian rhythm of temperature preference and its neural control in Drosophila. Curr Biol 22: 1851–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauchi K. 2002. How is the circadian rhythm of core body temperature regulated? Clin Auton Res 12: 147–149. [DOI] [PubMed] [Google Scholar]
- Krauchi K. 2007a. The human sleep-wake cycle reconsidered from a thermoregulatory point of view. Physiol Behav 90: 236–245. [DOI] [PubMed] [Google Scholar]
- Krauchi K. 2007b. The thermophysiological cascade leading to sleep initiation in relation to phase of entrainment. Sleep Med Rev 11: 439–451. [DOI] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M. 2010. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci 107: 13497–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst M, Tso MC, Ghosh DD, Herzog ED, Nitabach MN. 2015. Rhythmic control of activity and sleep by class B1 GPCRs. Crit Rev Biochem Mol Biol 50: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavie P. 2001. Sleep-wake as a biological rhythm. Annu Rev Psychol 52: 277–303. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176. [DOI] [PubMed] [Google Scholar]
- Masi L, Brandi ML. 2007. Calcitonin and calcitonin receptors. Clin Cases Miner Bone Metab 4: 117–122. [PMC free article] [PubMed] [Google Scholar]
- McCoy ES, Taylor-Blake B, Street SE, Pribisko AL, Zheng J, Zylka MJ. 2013. Peptidergic CGRPα primary sensory neurons encode heat and itch and tonically suppress sensitivity to cold. Neuron 78: 138–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. 2005. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron 48: 213–219. [DOI] [PubMed] [Google Scholar]
- Morf J, Schibler U. 2013. Body temperature cycles: gatekeepers of circadian clocks. Cell Cycle 12: 539–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SF, Nakamura K. 2011. Central neural pathways for thermoregulation. Front Biosci 16: 74–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima K, Matsue K, Konishi M, Iidaka C, Miyazaki K, Ishida N, Kanosue K. 2005. The involvement of Cry1 and Cry2 genes in the regulation of the circadian body temperature rhythm in mice. Am J Physiol Regul Integr Comp Physiol 288: R329–R335. [DOI] [PubMed] [Google Scholar]
- Nakamoto H, Soeda Y, Takami S, Minami M, Satoh M. 2000. Localization of calcitonin receptor mRNA in the mouse brain: coexistence with serotonin transporter mRNA. Brain Res Mol Brain Res 76: 93–102. [DOI] [PubMed] [Google Scholar]
- Nassel DR, Enell LE, Santos JG, Wegener C, Johard HA. 2008. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci 9: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. 2008. Organization of the Drosophila circadian control circuit. Curr Biol 18: R84–R93. [DOI] [PubMed] [Google Scholar]
- Patke A, Murphy PJ, Onat OE, Krieger AC, Ozcelik T, Campbell SS, Young MW. 2017. Mutation of the human circadian clock gene CRY1 in familial delayed sleep phase disorder. Cell 169: 203–215.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refinetti R, Menaker M. 1992. The circadian rhythm of body temperature. Physiol Behav 51: 613–637. [DOI] [PubMed] [Google Scholar]
- Saper CB, Lu J, Chou TC, Gooley J. 2005. The hypothalamic integrator for circadian rhythms. Trends Neurosci 28: 152–157. [DOI] [PubMed] [Google Scholar]
- Schmid B, Helfrich-Forster C, Yoshii T. 2011. A new ImageJ plug-in ‘ActogramJ’ for chronobiological analyses. J Biol Rhythms 26: 464–467. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Mignot E. 2011. Genetics of sleep and sleep disorders. Cell 146: 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seluzicki A, Flourakis M, Kula-Eversole E, Zhang L, Kilman V, Allada R. 2014. Dual PDF signaling pathways reset clocks via TIMELESS and acutely excite target neurons to control circadian behavior. PLoS Biol 12: e1001810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. 2008. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron 58: 223–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeba V, Fogle KJ, Holmes TC. 2010. Persistence of morning anticipation behavior and high amplitude morning startle response following functional loss of small ventral lateral neurons in Drosophila. PLoS One 5: e11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiromani PJ, Xu M, Winston EM, Shiromani SN, Gerashchenko D, Weaver DR. 2004. Sleep rhythmicity and homeostasis in mice with targeted disruption of mPeriod genes. Am J Physiol Regul Integr Comp Physiol 287: R47–R57. [DOI] [PubMed] [Google Scholar]
- Smith RE. 1969. Circadian variations in human thermoregulatory responses. J Appl Physiol 26: 554–560. [DOI] [PubMed] [Google Scholar]
- Sokolove PG, Bushell WN. 1978. The chi square periodogram: its utility for analysis of circadian rhythms. J Theor Biol 72: 131–160. [DOI] [PubMed] [Google Scholar]
- Stevenson RD. 1985a. Body size and limits to the daily range of body temperature in terrestrial ectotherms. Am Nat 125: 102–117. [Google Scholar]
- Stevenson RD. 1985b. The relative importance of behavioral and physiological adjustments controlling body temperature in terrestrial ectotherms. Am Nat 126: 362–386. [Google Scholar]
- Taghert PH, Nitabach MN. 2012. Peptide neuromodulation in invertebrate model systems. Neuron 76: 82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CL, Cooke EK, Leib DE, Lin YC, Daly GE, Zimmerman CA, Knight ZA. 2016. Warm-sensitive neurons that control body temperature. Cell 167: 47–59 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Platt MD, Lagnese CM, Leslie JR, Hamada FN. 2013. Temperature integration at the AC thermosensory neurons in Drosophila. J Neurosci 33: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Zandawala M, Beets I, Baytemur E, Slade SE, Scrivens JH, Elphick MR. 2016. Urbilaterian origin of paralogous GnRH and corazonin neuropeptide signalling pathways. Sci Rep 6: 28788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezaki Y, Yoshii T, Kawaguchi T, Helfrich-Forster C, Tomioka K. 2012. Pigment-dispersing factor is involved in age-dependent rhythm changes in Drosophila melanogaster. J Biol Rhythms 27: 423–432. [DOI] [PubMed] [Google Scholar]
- Umezaki Y, Hayley SE, Chu ML, Seo HW, Shah P, Hamada FN. 2018. Feeding-state-dependent modulation of temperature preference requires insulin signaling in Drosophila warm-sensing neurons. Curr Biol (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert D. 2010. Circadian temperature variation and ageing. Ageing Res Rev 9: 51–60. [DOI] [PubMed] [Google Scholar]
- Wellman LL, Yang L, Sanford LD. 2015. Effects of corticotropin releasing factor (CRF) on sleep and temperature following predictable controllable and uncontrollable stress in mice. Front Neurosci 9: 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G, Duncan MJ, Esser KA. 2013. Chronic phase advance alters circadian physiological rhythms and peripheral molecular clocks. J Appl Physiol 115: 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Suzuki T, Mizoro Y, Kori H, Okada K, Chen Y, Fustin JM, Yamazaki F, Mizuguchi N, Zhang J, et al. 2013. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342: 85–90. [DOI] [PubMed] [Google Scholar]
- Yao Z, Shafer OT. 2014. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343: 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.