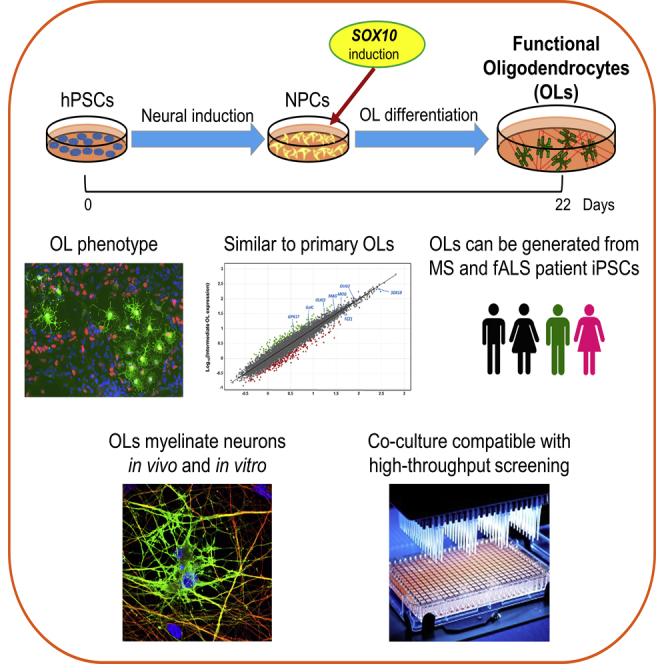
Summary
Scarce access to primary samples and lack of efficient protocols to generate oligodendrocytes (OLs) from human pluripotent stem cells (hPSCs) are hampering our understanding of OL biology and the development of novel therapies. Here, we demonstrate that overexpression of the transcription factor SOX10 is sufficient to generate surface antigen O4-positive (O4+) and myelin basic protein-positive OLs from hPSCs in only 22 days, including from patients with multiple sclerosis or amyotrophic lateral sclerosis. The SOX10-induced O4+ population resembles primary human OLs at the transcriptome level and can myelinate neurons in vivo. Using in vitro OL-neuron co-cultures, myelination of neurons by OLs can also be demonstrated, which can be adapted to a high-throughput screening format to test the response of pro-myelinating drugs. In conclusion, we provide an approach to generate OLs in a very rapid and efficient manner, which can be used for disease modeling, drug discovery efforts, and potentially for therapeutic OL transplantation.
Keywords: oligodendrocyte, induced pluripotent stem cells (iPSCs), multiple sclerosis, amyotrophic lateral sclerosis, myelination, disease modeling, drug screening
Graphical Abstract
Highlights
-
•
SOX10 is sufficient to generate myelinating human OLs from hPSCs in only 22 days
-
•
SOX10-induced OLs resemble primary human OLs at the transcriptome level
-
•
The methodology allows efficient generation of OLs from MS and ALS patients
-
•
OL-neuron co-cultures respond to myelinating drugs in a high-throughput setting
In this article, García-León JA and colleagues demonstrate the generation of functional oligodendrocytes (OLs) from human pluripotent stem cells in a rapid and efficient manner by the single overexpression of SOX10. Generated OLs resemble primary OLs at the transcriptome level and can myelinate neurons both in vivo and in vitro. Neuron-OL co-cultures, adapted to high-throughput screening formats, responded to drugs affecting myelination.
Introduction
Oligodendrocytes (OLs) are the central nervous system (CNS) glial cells responsible for axonal myelination. However, the complete roles of OLs are still only partially understood and vary depending on the CNS region wherein they reside (Marques et al., 2016). Myelination in the CNS is essential for proper signal conduction along neuronal axons and for maintaining brain homeostasis. Defects in myelin production and/or maintenance are the predominant pathological feature of several diseases, including leukodystrophies and multiple sclerosis (MS) (Franklin et al., 2012). In addition to myelination, OLs have a role in trophic and metabolic support of neurons, fueling oxidative phosphorylation in the mitochondria of axons. This trophic support is disrupted in amyotrophic lateral sclerosis (ALS), and defects in OL function contribute to ALS onset and progression (Lee et al., 2012b).
Lack of insight in human OL biology is in large part a consequence of the limited access to human OLs and difficulties in maintaining these cells in vitro. Therefore, having access to human OLs would represent a major step forward in studies aimed at understanding mechanisms that are deregulated in diseases with OL involvement. Moreover, this would allow to test and study if and how candidate drugs affect the process of OL myelination and remyelination (Mei et al., 2014, Lariosa-Willingham et al., 2016), and/or the trophic support provided by OLs.
With the isolation of human embryonic stem cells (hESCs) and the development of human induced pluripotent stem cell (hiPSC) technology, a number of protocols have been developed to generate OLs from hPSCs (Nistor et al., 2005, Hu et al., 2009, Wang et al., 2013, Douvaras et al., 2014), recapitulating in vitro the molecular signals and events that occur during in vivo OL development, leading to myelinating OLs (Wang et al., 2013, Douvaras et al., 2014). Despite recent optimizations (Douvaras and Fossati, 2015), these protocols remain inefficient and variable in terms of OL yield and, importantly, require very long differentiation times (>100 days to generate myelin basic protein (MBP)-positive OLs). These issues have precluded the use of patient-specific iPSC-derived OLs to elucidate human OL biology and disease, and use such cells as platform for drug screening.
Here, we describe that, by the overexpression of the single transcription factor (TF) SOX10 in hPSC-derived neural precursors (NPCs), it is possible to generate surface antigen O4 (O4)-positive and MBP+ OLs within only ∼20 days from the PSC stage. The transcriptome of hPSC-derived O4+ cells resembles that of primary intermediate OLs. Similar OL production in terms of efficiency and time course was obtained from patients with MS or familial ALS (fALS) compared with healthy donors. Finally, grafting into homozygous shiverer (Shi−/–) mouse brain slices and co-culture with hPSC-derived neurons confirmed the myelination capability of SOX10-induced OLs in in vivo and in vitro contexts. All hPSC-derived OL-neuron co-cultures were also adapted to high-throughput screening (HTS) formats allowing demonstration of enhanced myelin production by different compounds.
Results
Selection of TFs Involved in OL Specification
To define which TFs could promote efficient OL differentiation from hPSCs, we selected 16 TFs known to function in OL specification and/or maturation: ASCL1, AXIN2, MYRF, MYT1, OLIG1, OLIG2, NKX2-2, NKX6-1, NKX6-2, SOX2, SOX8, SOX9, SOX10, ST18, ZEB2, and ZNF536 (Cahoy et al., 2008, Pozniak et al., 2010, Weng et al., 2012, Najm et al., 2013, Yang et al., 2013). The coding regions of these genes were individually cloned in the FUW lentiviral doxycycline-inducible expression vector. As reported (Carey et al., 2009), we demonstrated efficient overexpression of each TF in an inducible manner in our experimental settings (Figures S1A and S1B).
Initial Screening
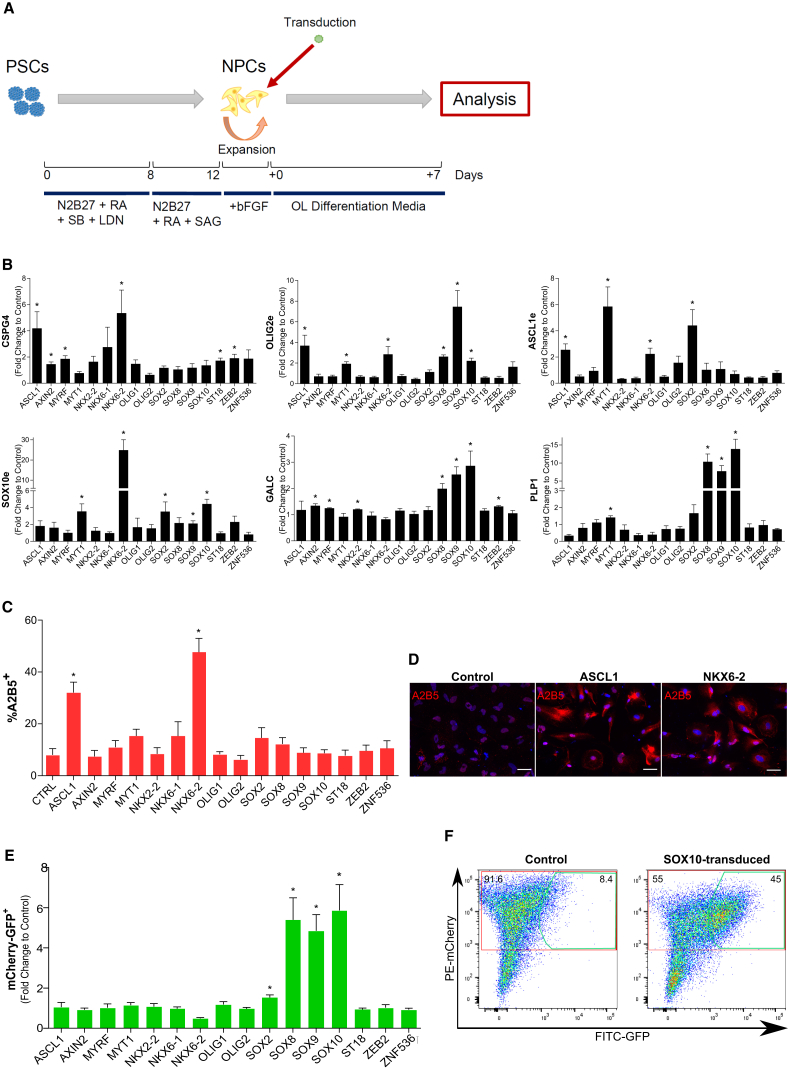
An initial screen of the 16 selected TFs was performed to identify TFs that induced early, intermediate, and late OL fate. NPCs were generated from hPSCs by dual SMAD inhibition in the presence of retinoic acid (RA) and Sonic hedgehog (SHH) agonist (Chambers et al., 2009). More than 95% of the day 12 PSC progeny stained positive for NPC markers (SOX2 and NESTIN) and most of them (81.8% ± 2.7%) expressed the ventral progenitor marker HOXB4 (Figure S1C). NPCs were further expanded using basic fibroblast growth factor (bFGF), and then transduced with each of the 16 individual TFs and cultured in OL differentiation medium (Figure 1A).
Figure 1.
Screening of the 16 Selected TFs to Evaluate Their Influence on OL Specification
(A) Scheme of the strategy followed for NPC generation, expansion and transduction with lentiviral vectors.
(B) Gene expression changes of early (CSPG4 and OLIG2e), intermediate (ASCL1e and SOX10e), and late (GALC and PLP1) OPC markers measured by qRT-PCR, expressed as fold change relative to control (NPCs transduced with eGFP).
(C) Staining for A2B5 in the different transduced cell progeny on day +7.
(D) Representative images of the A2B5 stainings of vector control and ASCL1 and NKX6-2 transduced cells. Hoechst 33258 (blue) was used as nuclear marker.
(E) Fold change in the expression of the MCS5-SOX10 reporter as a result of the overexpression of the different TFs relative to control (cells transduced with empty vectors) after 7 days in OL differentiation media.
(F) Example of the expression of the MCS5-SOX10 reporter (GFP-FITC) within mCherry+ cells in vector control and SOX10-transduced progeny.
Data represented as mean ± SEM of N = 3–4 independent experiments. ∗p < 0.05. Scale bars: 50 μm.
To identify the TFs that enhanced OL lineage differentiation, we performed qRT-PCRs for early (OLIG2 and CSPG4), intermediate (ASCL1 and SOX10), and late (GALC and PLP1) OL lineage markers 7 days after transduction (Figure 1B). Overexpression of ASCL1, NKX6-2, or MYT1 induced a significant increase in endogenous (e) transcripts for OLIG2e, CSPG4, and ASCL1e, while expression of the more mature OL genes, GALC and PLP1, was significantly induced by overexpression of SOX8, SOX9, or SOX10. We also performed immunostaining for A2B5, a marker for intermediate oligodendrocyte precursor cells (OPCs) (Figures 1C and 1D). In the absence of TF overexpression, we detected 8.02% ± 2.46% A2B5+ cells, consistent with the fact that differentiation was induced for only 7 days. By contrast, and in line with the qRT-PCR data, 32.05% ± 4.04% and 47.63% ± 5.33% A2B5+ cells were identified following overexpression of ASCL1 and NKX6-2, respectively.
We also transduced the NPCs with a vector containing the MCS5-SOX10 enhancer region, which is an efficient and specific reporter for human OL lineage cells (Pol et al., 2013). Following co-transduction of NPCs with the individual TFs combined with the MCS5-SOX10 vector, expression of GFP (activity of the reporter) was evaluated by fluorescence-activated cell sorting (FACS) 7 days later. As the MCS5-SOX10 vector also contained a constitutive mCherry cassette, the fraction of GFP+ cells within the mCherry+ population was quantified. A 5- to 6-fold increase in eGFP+/mCherry+ cells was identified in NPCs transduced with either SOX8, SOX9 or SOX10, in line with the increased expression of mature OL markers (Figures 1E and 1F).
Thus, overexpression of ASCL1, NKX6-2, and MYT1 induced early-intermediate OL lineage transcripts and proteins, while overexpression of SOX8, SOX9, and SOX10 activated the MCS5-SOX10 enhancer-based reporter and induced expression of late OL genes (GALC and PLP1). The effect of these six TFs was then further analyzed.
Screening with the Six Selected TFs
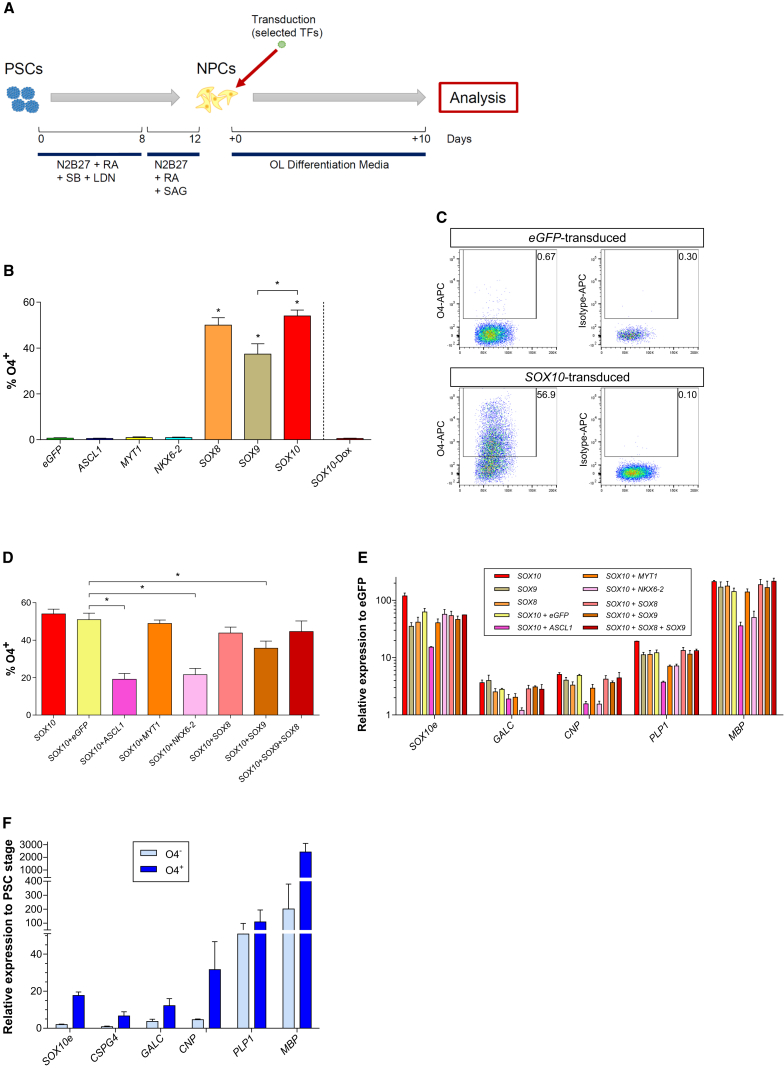
We used anti-O4 antibody staining (Sommer and Schachner, 1981) to further define which of the six TFs caused differentiation to mid- and late-stage OL lineage cells. Day 12 NPCs, without bFGF expansion, were transduced with the six TFs individually to enable assessment of the shortest period required to generate OLs from hPSCs, and to avoid possible lineage skewing due to bFGF-based NPC expansion (Furusho et al., 2015). OL differentiation was assessed on day 10 after transduction (22 days from undifferentiated hPSCs) (Figure 2A).
Figure 2.
Impact of the Six Selected TFs on OPC/OL Specification Alone or in Combination
(A) Scheme of the strategy followed for NPC generation and direct transduction for a total period of 22 days from the PSC stage.
(B) Evaluation of the expression of O4 by FACS in NPCs transduced with the selected six TFs alone after 10 days in OL differentiation media.
(C) Representative dot plots of O4 expression and isotype control by FACS of cells transduced with either SOX10 or eGFP.
(D) Percentages of O4 expression when NPCs were transduced for 10 days with different combinations of the TFs.
(E) Gene expression levels of OPC/OL markers 10 days following transduction with single or combinations of the six TFs selected.
(F) Gene expression levels for OPC/OL markers in FACS sorted O4+ and O4– cells 10 days after SOX10 induction. Expression levels normalized to GAPDH. SOX10e refers to its endogenous expression.
Data represented as mean ± SEM of N = 3–5 independent experiments. ∗p < 0.05.
Less than 1% of NPCs transduced with an eGFP control vector were O4+. Transduction of NPCs with ASCL1, NKX6-2, or MYT1 did not increase the fraction of O4+ cells, consistent with the finding that these TFs induced immature/intermediate OPC lineage. However, 50.02% ± 3.21%, 37.35% ± 4.51%, and 54.05% ± 2.52% of NPCs transduced with SOX8, SOX9 or SOX10 were O4+, respectively (Figure 2B). We next tested if combined overexpression of SOX10 with any of the other five TFs would further enhance the proportion of O4+ cells. However, no further increase in O4+ cells was seen with any TF combination over SOX10 alone (Figure 2D).
This was confirmed by studies testing OPC/OL marker transcripts in cells transduced with SOX8, SOX9, or SOX10 alone, or SOX10 in combination with the other five TFs (Figure 2E). SOX10e, GALC, CNP, PLP1, and MBP expression was induced 5- to >100-fold following transduction with SOX10 alone. Transduction with either SOX8 or SOX9 induced similar, albeit somewhat lower levels of these transcripts. Combinations of SOX10 with any of the other TFs did not further enhance marker expression.
We next FACS sorted O4+ and O4– subpopulations 10 days after transduction with SOX10. The O4+ fraction was highly enriched for cells expressing OPC/OL marker transcripts in comparison with the O4– fraction (Figure 2F), confirming that expression of O4 is specific for intermediate and late OL lineage cells. Thus, overexpression of the TF SOX10 alone is sufficient to induce differentiation of NPCs toward the OL lineage. Subsequent studies were designed to further characterize the SOX10-induced cells.
SOX10-Induced Cells Express Typical Markers of OLs
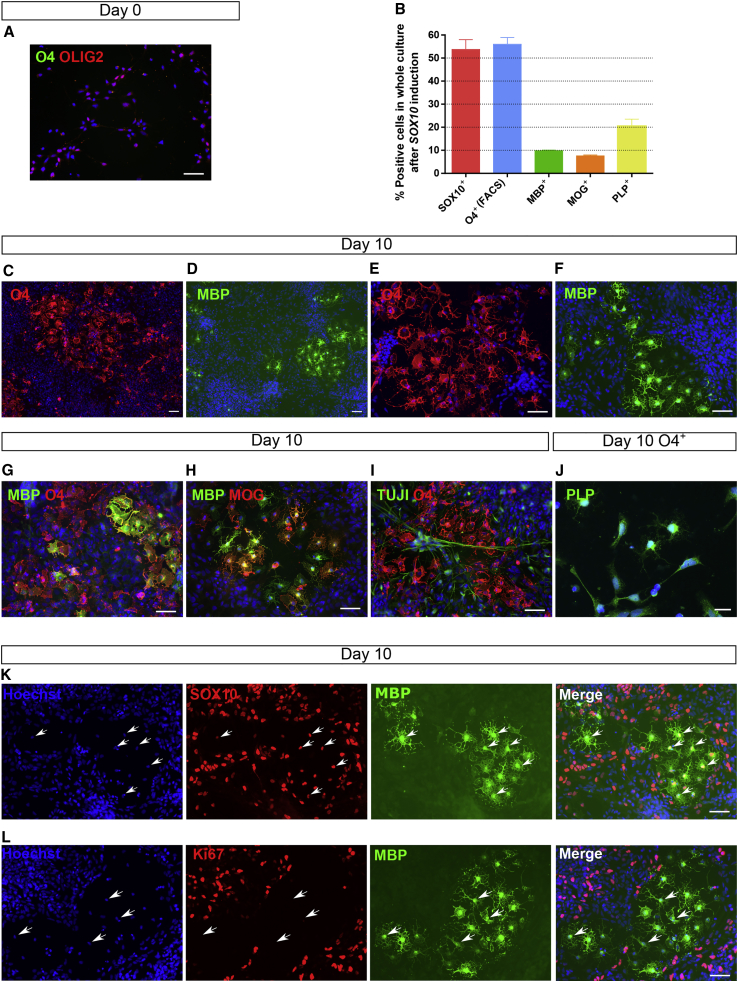
We next tested if SOX10-induced progeny expresses, in addition to O4, other typical OL markers. Immunostaining on day 10 SOX10-induced cells (22 days from hPSC stage; without prior O4+ enrichment), demonstrated that day 12 NPCs stained positive for OLIG2 but not O4 (Figure 3A), while SOX10-transduced cells 10 days after induction were negative for OLIG2 (not shown). Approximately 50%–60% of day 22 NPC progeny stained positive for O4 (in line with the FACS data; Figures 3B, 3C, and 3E) and O1 (Figure S2B), and that 97.15% ± 8.19% SOX10+ cells co-expressed O4. In addition, 21.48% ± 2.09% SOX10+ cells also stained positive for MBP (Figures 3D, 3F, and 3G), with 71.67% ± 2.43% of these cells co-expressing MOG (Figure 3H). PLP expression was found as well within the SOX10-induced cells (20.35% ± 3.19%), and remained expressed in most (94.60% ± 2.57%) of the O4+-purified cells (Figure 3J). These myelin protein-expressing cells displayed a more mature OL morphology, with extended membrane sheaths and highly branched processes (Figures 3C–3J).
Figure 3.
Phenotypic Characterization of SOX10-Induced OLs by Immunocytochemistry
(A) Untransduced NPCs express the early OPC marker OLIG2 but not O4.
(B) Quantification of OPC/OL marker expression in the whole culture after 10 days of SOX10 induction.
(C–H) Representative images of the cultures showing expression of the OL markers O4, MBP, and MOG. (I) TUJI+ neurons are present in a small proportion.
(J) Expression of the proteolipid protein (PLP) in O4+ cells isolated on day 10 following SOX10 induction.
(K) The expression of SOX10 was found in all MBP+ OLs (exemplified with white arrows).
(L) None of the MBP+ OLs expressed the Ki67 marker (exemplified with white arrows), and hence present a postmitotic phenotype.
Representative images of N = 3–5 independent differentiations. Hoechst 33258 (blue) was used as nuclear marker. Scale bars: 50 μm; 20 μm for (J).
Aside from O4+ cells, we also found rare TUJI+ neurons (<5%; Figure 3I), but no GFAP+ astrocytes in the culture. We also assessed if Schwann cells (SCs) were present in the culture, by staining for the SC-specific peripheral myelin protein 22 (PMP22; Figures S2C and S2D). SOX10-induced progeny did not contain PMP22+ cells, indicating that only OLs and not peripheral SCs were generated.
To further prove that generation of O4+/MBP+ cells was due to SOX10 overexpression, we co-stained SOX10-induced progeny with SOX10 and MBP. All MBP+ cells co-expressed SOX10, demonstrating that SOX10 expression was required for the generation of MBP+ OLs (Figure 3K). Furthermore, no O4 or MBP expression was observed in eGFP-transduced NPCs after 10 days of induction (Figures S2E and S2F).
The yield of O4+ cells on day 22 was approximately 240% of the day 12 NPCs, and 24,000% of the day 0 PSCs. This high yield, also from NPCs, reflects the presence of proliferative OPCs in the culture that give rise not only to mature OLs but also to other OPCs. In fact, Ki67+ cells were present throughout differentiation (Figure 3L), with 24.22% ± 1.20% of Ki67+ cells present in the whole culture on day 10 after SOX10 induction. However, all MBP+ cells were Ki67− (Figure 3L), and hence post-mitotic, consistent with the acquisition of a mature OL phenotype.
SOX10-Induced Cells Resemble Primary OLs at the Transcriptome Level
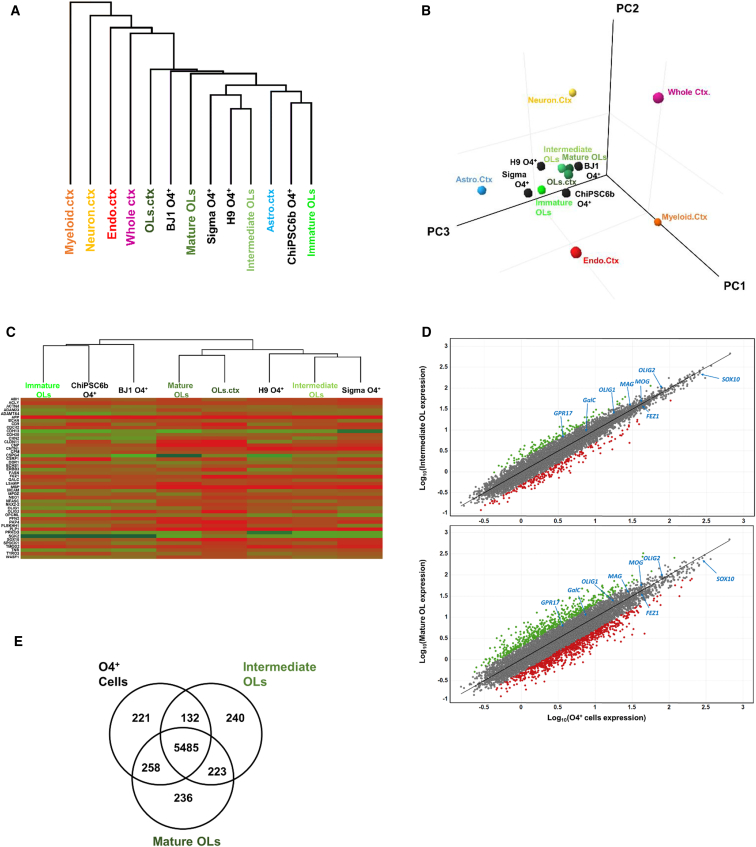
To assess if SOX10-induced OLs resembled primary OLs, we performed RNA-sequencing (RNA-seq) on purified O4+ cells derived from four different hPSC lines: the hESC H9 and the hiPSC ChiPSC6b, Sigma-iPSC0028, and BJ1 (healthy donor-derived) lines. The transcriptome of the O4+ cells was combined with published transcriptome data from different brain cells (GalC+ OLs, CD90+ neurons, CD45+ myeloid cells, HepaCAM+ astrocytes, BSL-1+ endothelial cells, and the whole cortex) (Zhang et al., 2016, Abiraman et al., 2015).
Principal-component analysis and unsupervised hierarchical clustering on the entire transcriptome identified a very distinct cluster encompassing different brain-derived OL samples, as well as the SOX10-induced O4+ cells, separated from neurons, astrocytes and other non-ectodermal-derived cells (Figures 4A and 4B). To further characterize the maturity of the generated O4+ OLs, we also included samples consisting of immature, intermediate, and mature OLs isolated from human fetal tissues (Abiraman et al., 2015). Comparison of OL-specific markers (Cahoy et al., 2008, Nielsen et al., 2006) revealed that O4+ cells derived from H9 and Sigma-iPSC0028 lines co-clustered with intermediate OLs, while ChiPSC6b and BJ1 O4+ cells clustered more closely to immature OLs (Figure 4C).
Figure 4.
Global Transcriptome Comparison of SOX10-Induced Cells with Primary Different Brain Lineages
(A) Hierarchical clustering of whole-genome expression profile of SOX10-induced O4+ cells (black), different primary OL samples (green), myeloid cells (orange), neurons (yellow), endothelial cells from cortex (red), astrocytes (light blue), and whole cortex (pink).
(B) Principal-component analysis (PCA) of different samples mentioned in (A).
(C) Hierarchical clustering and heatmap comparison of OL-specific markers among PSC-derived O4+ cells and primary OLs.
(D) Pairwise scatterplot of log10 gene expression of PSC-derived O4+ cells (N = 12) versus intermediate (N = 6) and mature (N = 3) primary OLs. Gray, genes expressed at similar levels (<2-fold difference), with blue arrows identifying key OL-specific genes; red and green dots, genes that are >2-fold higher or >2-fold less expressed in PSC-derived O4+ cells compared with primary intermediate and mature OLs.
(E) Venn diagram depicting the number of genes expressed in common or differently (>2-fold change) in SOX10-induced O4+ cells and primary intermediate and mature OLs.
Next, we compared SOX10-PSC O4+ cells (average values of all PSC-derived cells) with different primary OLs using a sequence alignment map (Figures 4D and 4E). We identified 916 (13.48%) differentially expressed genes between mature OLs and PSC-O4+ cells (fold change >2 and false discovery rate < 0.05) (699 up- and 217 downregulated genes in PSC-O4+ cells) and 240 (3.53%) differentially expressed genes between intermediate OLs and PSC-O4+ cells (all upregulated in PSC-O4+ cells) (Table S1), confirming that O4+ cells were highly similar to mature/intermediate primary OLs (Figure 4E). Over 85% (47/53) of OL-specific genes (including MAG, MOG, SOX10, and OLIG2) were expressed at comparable levels in O4+ cells and primary OLs (Figure 4D).
Lastly, we performed gene ontology (GO) analysis to identify classes of genes that were similarly expressed between O4+ hPSC-derived cells and primary OLs. When compared with primary intermediate OLs, a higher number of shared GO pathways were obtained, including those referred to CNS development as well as to OL development (Table S2). In addition, other terms were related to cytoskeleton organization and protein modifications, pathways associated to OLs (Nielsen et al., 2006). Overall, these results support the notion that SOX10-induced O4+ cells are highly comparable at the transcriptome level with primary OLs, especially intermediate OLs.
Inducible Single-Copy SOX10 Overexpression from the AAVS1 Locus Efficiently Induced OL Cell Conversion
To avoid effects of random integration of the SOX10 transgene resulting from lentiviral transduction, and also to avoid the use of this technology for OL generation, we created an hESC line wherein SOX10 was introduced in the safe harbor locus AAVS1, using recombinase-mediated cassette exchange in hPSC lines containing an FRT-flanked cassette, which contained a hygromycin-resistance/thymidine kinase selection cassette (Ordovás et al., 2015). This created 100% homogeneous hESCs containing either an SOX10 or an SOX10-eGFP cassette under a doxycycline-inducible promoter (Figures S4A and S4B).
Addition of doxycycline to hESCs induced the expression of SOX10 or SOX10-eGFP in >99% of cells (Figures S4C and S4D). Induction of SOX10 on day 0, without prior neural commitment, did not result in the generation of MBP+ OLs (not shown). When hPSCs were first fated to NPCs for 8 days, followed by addition of doxycycline and culture in OL differentiation medium, already 50% O4+ cells were found on day 4, and 89.3% ± 0.6% by day 7, which was sustained at later time points (Figure S4E). The emergence of O4+ cells was accompanied by progressively increased levels of OPC/OL markers, in both SOX10 and SOX10-eGFP transgenic lines (Figure S4F). Immunostaining further demonstrated the OL identity of the cells: MBP+ cells could be detected by day 7, and its expression increased progressively by day 10 of induction (day 18 of the overall differentiation culture; Figure S4G). No contamination with neurons was observed.
SOX10-Induced OLs Myelinate Both Shiverer Mouse and hPSC-Derived Cortical Neurons
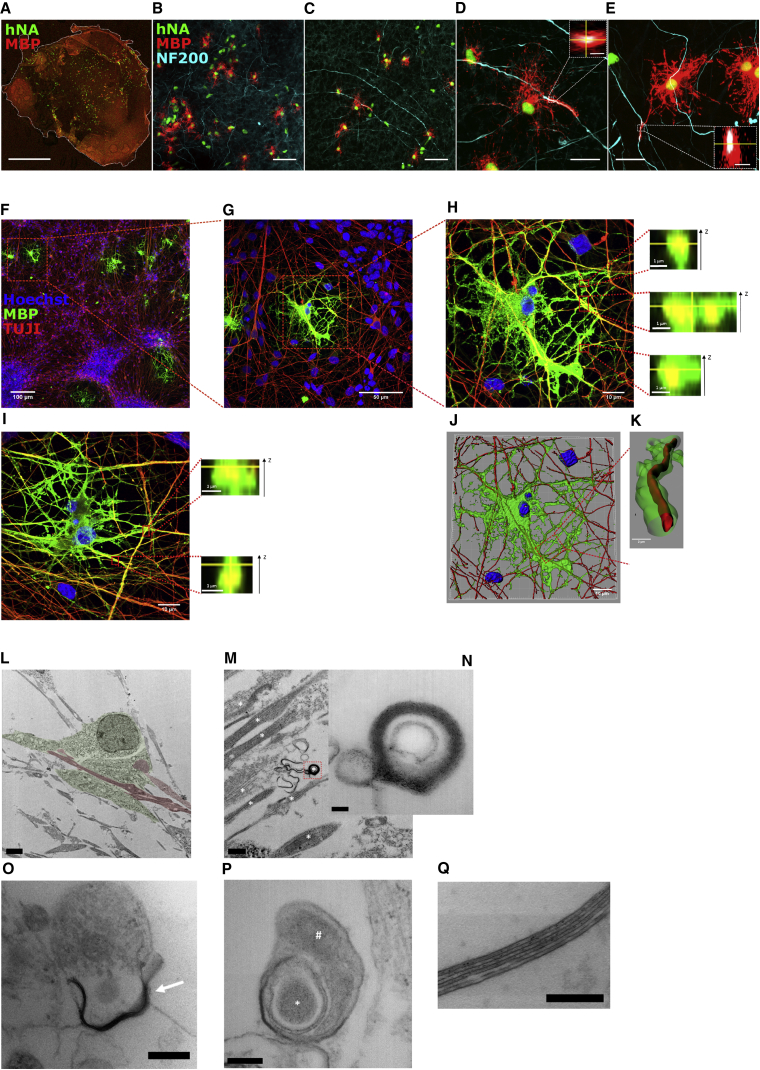
To demonstrate that SOX10-induced O4+ cells have functional characteristics of OLs, we tested if they were capable of myelinating neuronal axons. To address myelination in an in vivo context, purified O4+ cells were injected in brain slices from homozygous shiverer (shi/shi) mice (MBP deficient) and slices were analyzed 10 days later. Immunostaining demonstrated efficient engraftment and homogeneous spreading of the transplanted human hNA+ cells within the tissue, with 48.13% ± 4.15% of cells also expressing MBP (Figures 5A–5C). Moreover, MBP+ OL projections wrapping NF200+ neuronal axons could be observed 10 days after injection (Figures 5D and 5E).
Figure 5.
SOX10-Induced OLs Myelinate Both Shiverer Mouse Brain Slices and hPSC-Derived Cortical Neurons
(A–E) Confocal microscopy images of O4+ purified SOX10-induced cells injected into Shi−/− mouse brain slices 10 days after injection. (A) Whole-slice image showing homogeneous dispersion of the injected cells (human nuclear antigen, hNA+). (B and C) Overview images of slices showing grafting and maturation of the injected cells. (D and E) Detailed images showing MBP+ prolongations annealing and ensheathing NF200+ neuronal axons.
(F–Q) Confocal and transmission electron microscopy (TEM) images of the co-culture between hPSC-derived neurons and SOX10-induced O4+ cells. (F) Overview of the co-culture. (G) Amplification of a representative region with mature OLs extending their processes over neuronal axons. (H and I) Magnifications of an OL showing, at different locations, the complete ensheathment of neuronal axons. (J) 3D reconstruction suggesting the wrapping of axons by an OL. (K) Detailed 3D reconstruction of a region showing complete ensheathment of an axon (red) by MBP+ prolongations (green). Nuclei counterstained with Hoechst 33258 (blue). (L) TEM image showing neuronal axons (red) surrounded by an OL (green). (M) Myelin-like structures are found in regions with axon tracts. (N) Magnification of a myelinating structure where compact myelin formation can be observed. (O) TEM example where myelination of a neuronal axon is partially complete (arrow). (P) Detailed micrograph of an OL prolongation (#) starting to form a layered myelin sheet around a neuronal structure (∗). (Q) Magnification showing the formation of a multilayer compact myelin structure.
Scale bars: 1 mm (A), 50 μm (B and C), 20 and 2 μm inserts (D and E), 2 μm (L), 0.5 μm (M), 100 nm (N), 200 nm (O), 200 nm (P), and 200 nm (Q).
We also assessed if myelination occurred in in vitro co-cultures. We generated cortical neurons from human iPSCs as described previously (Shi et al., 2012). Neuronal progenitors were replated and allowed to mature for 10–14 days. O4+ cells, isolated and purified on day 10 following SOX10 induction, were co-cultured with cortical neuronal progeny for an additional 20 days in OL myelination medium. Regions wherein TUJI+ axons were aligned with MBP+ OLs could already be seen a few days later (not shown). By day 20 of co-culture, O4+ cells extended MBP+ regions aligned with axons at multiple locations (Figures 5F–5K). Transverse sections of reconstructed confocal microscopy images demonstrated the presence of MBP+ extensions fully wrapping neuronal axons (Figures 5H and 5I). 3D reconstructions also demonstrated the presence of MBP+ sheaths surrounding TUJ1+ axonal prolongations (Figures 5J and 5K). At the ultrastructural level, cytoplasmic regions of OLs were frequently observed surrounding neuronal axons (Figures 5L and 5M) and were able to form multilayer compact myelin sheaths (Figure 5Q). Early myelination of neuronal axons was also observed (Figures 5O and 5P). These results indicate that SOX10-induced OLs matured into myelinating OLs that ensheathed and wrapped axons in both an in vivo context, as well as when co-cultured with hPSC-derived neurons in vitro.
Efficient Generation of OLs from iPSCs Derived from Patients with MS and ALS
To determine the robustness of the protocol, and to demonstrate that OLs can also be generated from iPSCs of patients with neurodegenerative diseases wherein OLs have been shown to play a causal role, we compared the generation of OLs from hPSCs from healthy donors (hESC-H9 and hiPSC ChiPSC6b, Sigma-iPSC0028, and BJ1 lines), with iPSC lines from two primary progressive MS (PPMS) patients and from two patients with a familial form of ALS (fALS) caused by mutations in the genes superoxide dismutase (SOD1A4V) or C9ORF72.
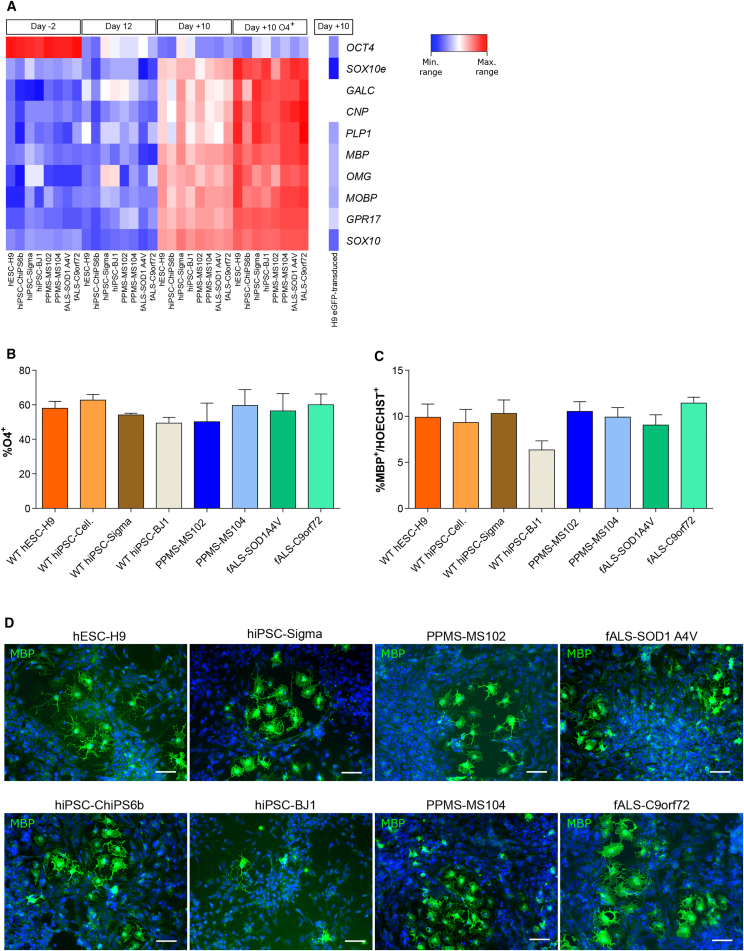
We found no substantial differences in the expression of intermediate and late OL marker transcripts among the eight cell lines analyzed at different time points (Figure 6A). In addition, no significant differences in the efficiency of generating O4+ cells were seen among the lines (50%–65% O4+ cells; Figure 6B). Finally, approximately 10% of MBP+ cells were present on day 22 in the SOX10-induced NPC progeny from all lines examined, with the exception of BJ1-derived cells, which contained only 6.34% ± 0.70% MBP+ cells (Figure 6C). In addition, MBP+ progeny from PSCs of healthy donors and from PPMS and fALS patients had similar morphology (Figure 6D).
Figure 6.
Generation of OLs from Different Lines Including MS and fALS Patients
(A) Heatmaps of qRT-PCR analysis performed at different stages of OPC/OL differentiation induced by SOX10 overexpression comparing the eight different hPSCs used: four lines derived from healthy donors (hESC-H9, hiPSCs ChiPSC6b, Sigma, and BJ1), two from primary progressive MS patients (PPMS-102 and -104), and two from patients with familial ALS with mutations in SOD1 (fALS-SOD1 A4V) and C9orf72 (fALS-C9orf72) (SOX10e = endogenous SOX10).
(B) O4 expression by FACS of the eight hPSC lines analyzed after 10 days in OL differentiation medium.
(C) Quantification of the total percentage of MBP+ cells.
(D) Representative images of MBP+ OLs obtained after 10 days from the eight hPSC lines.
Hoechst 33258 (blue) was used as nuclear marker. Scale bars: 50 μm.
Data represented as mean ± SEM of N = 3 independent experiments per line.
Thus, the SOX10-mediated differentiation protocol could generate intermediate and mature OLs with similar efficiencies also from PPMS and fALS iPSC lines only after 22 days of differentiation. This proves the robustness of the protocol irrespective of the iPSC lines used to generate O4+/MBP+ OLs.
Creation of a Myelinating Co-culture System for High-Throughput Drug Screening
Currently, no good assays are available to identify and validate drugs that can enhance myelination. The existing platforms are based mostly on primary murine OLs cultured in the absence (Mei et al., 2014, Lee et al., 2012a) or presence of neurons (Deshmukh et al., 2013, Lariosa-Willingham et al., 2016). Recently, in vitro myelination systems based on murine PSCs suitable for high-throughput screening (HTS) have been developed (Najm et al., 2015, Kerman et al., 2015). However, such a model using hPSC-derived neuronal and OL progeny has not yet been described.
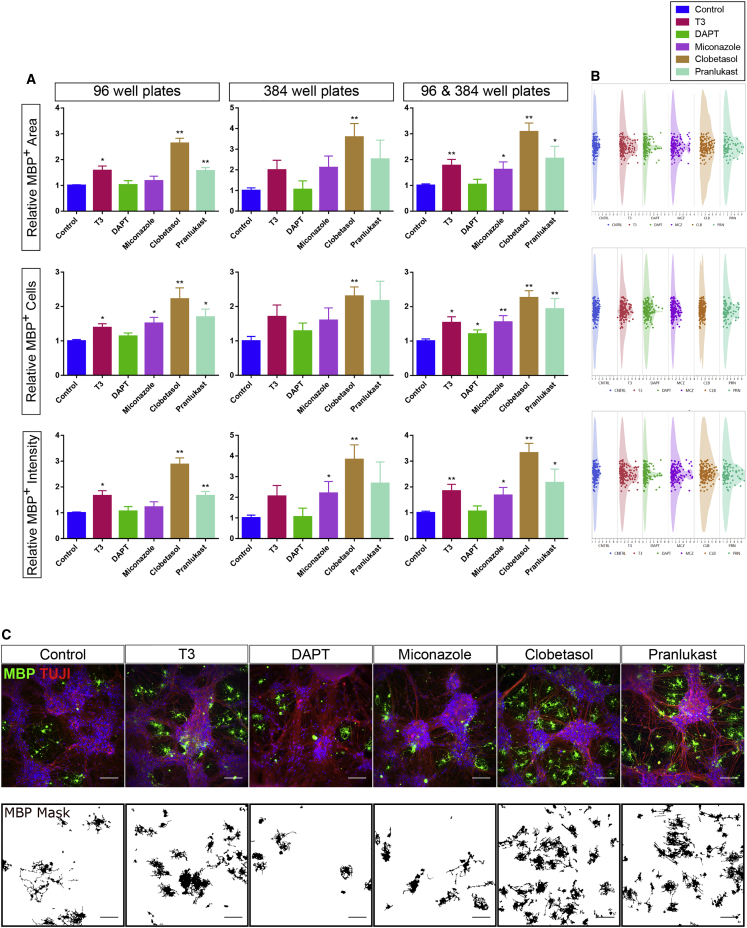
To enable HTS approaches, we adapted and optimized the cortical neuron-OL co-culture system to 96- and 384-well plate format. Following induction of SOX10 for 10 days, purified O4+ cells could be cryopreserved, with >90% cells remaining viable and functional after thawing (not shown). To test if hPSC-derived neuron-OL co-cultures allow identification of myelination-enhancing factors (e.g., triiodothyronine, T3) or different medications, SOX10-induced purified O4+ cells were cultured on top of maturing neurons for 20 days, and expression of MBP evaluated. Co-cultures were supplemented or not with two drugs identified in a murine stem cell-derived HTS to enhance MBP+ expression, miconazole and clobetasol (Najm et al., 2015). In addition, we tested the effect of the γ-secretase and Notch signaling inhibitor DAPT, reported to have a positive influence on in vitro myelination of murine OPCs (Lariosa-Willingham et al., 2016), and pranlukast, a blocker of the olig2-targeted G-protein-coupled receptor Gpr17, reported to enhance remyelination and OL survival, but not yet tested in in vitro screens (Ou et al., 2016). The readout included assessment of the total MBP+ area, number of MBP+ cells, and MBP+ intensity (Figure 7).
Figure 7.
All hPSC-Derived OL-Neuron Co-culture Can Address Myelinating Drugs in an HTS Setting
(A) Relative MBP expression to control cultures (0.1% DMSO and no T3) of the different compounds and drugs affecting myelination: T3 (60 ng/mL), DAPT (1 μM), miconazole (1 μM), clobetasol propionate (5 μM) and pranlukast (22.8 μM). Three variables were assessed: total MBP+ area, number of MBP+ cells and total MBP+ intensity. Shown are results for co-cultures in 96-well plates, in 384-well plates, or the combination of the two.
(B) Gaussian dot plots of the average values of all single wells in the six culture conditions (N = 160 for all conditions except for pranlukast, where N = 90) for the three variables based on MBP expression.
(C) Representative images of the co-cultures assessing the effects of the different tested conditions on myelination and of the MBP+ mask identified for the analysis.
Hoechst 33258 (blue) was used as nuclear marker. Scale bars: 100 μm. Data are represented as average values per plate ± SEM of each condition of N = 7–8 independent experiments for 96-well plates, N = 6–7 for 384-well plates, and N = 12–15 for both 96- and 384-well plates. ∗p < 0.05, ∗∗p < 0.01.
MBP expression increased 1.5- to 2-fold when T3 was added compared with control (DMSO) conditions (Marta et al., 1998). Miconazole induced a similar increase, and addition of pranlukast induced a 1.5- to 2.5-fold increase in MBP expression. The highest levels of MBP expression were observed for clobetasol, reaching a 3- to 4-fold increase of MBP expression compared with control, which was significantly higher than the effect of T3 when matched-pair analysis was performed (Figure S5). By contrast, DAPT did not enhance MBP expression. Similar results were obtained in both 96- and 384-well assay formats (Figure 7A), with little variation between intra- or inter-plate replicates, and for all three MBP-staining readouts.
Thus, the fully human neuron-OL co-culture system described here can be successfully scaled across multiple formats, allowing HTS to identify and validate compounds involved in myelination. This system could be used as well to study the mechanisms underlying OL-mediated neuronal dysfunction or death.
Discussion
Our understanding of the complex OL biology and mechanisms underlying several OL diseases has been limited by the difficulties of obtaining human OLs. Since the implementation of hESCs/hiPSC technology, different methods have been described whereby OLs can be generated from hPSCs (Nistor et al., 2005, Hu et al., 2009, Wang et al., 2013, Douvaras et al., 2014), but inefficiently and, therefore, impeding the use of these cells for disease modeling or drug development.
To overcome these issues, we hypothesized that overexpression of TFs known to play a role in OL specification and/or maturation would enhance the efficiency as well as the speed with which OL could be generated from hPSCs. We selected 16 TFs known to regulate human and/or murine OL specification and/or maturation (Cahoy et al., 2008, Pozniak et al., 2010, Weng et al., 2012, Najm et al., 2013, Yang et al., 2013) and tested if their overexpression would enhance OPC/OL fating. By a combination of methods, we demonstrated that overexpression of SOX10 alone in NPCs induced ∼60% of cells expressing O4, of which 20% also co-expressed the myelin protein MBP.
The SOX genes are TFs with a characteristic high-mobility group domain, involved in developmental specification and classified into nine groups based on sequence similarity and function (Wegner, 2010). Among them, the SOXE group of TFs, composed of SOX8, SOX9 and SOX10, is involved in specification of myelinating glial cells, both SCs and OLs (Weider and Wegner, 2017). We obtained late OPC specification when either SOX8, SOX9 or SOX10 were overexpressed, consistent with the previously described role of these TFs during OL specification. Although redundant functions have been described for these three TFs due to the high sequence preservation of the functional domains, Sox10 is considered as the main Sox TF regulating OL specification and maturation, with also epigenetic functions (Weider et al., 2013, Vogl et al., 2013). As SOX10 is known to be involved in SC development (Weider et al., 2013), we also assessed if SCs were concomitantly induced in our cultures. However, we could not detect cells with typical SC markers. Thus, although SOX10 plays a role in maturation and myelin production of both cell types, the lack of SC induction in our assay is likely due to the fact that the NPC and subsequent differentiation conditions are OL specific and not supportive for SC development, in line with their differential developmental origin compared with OLs (Zawadzka et al., 2010).
The basic helix-loop-helix protein OL lineage transcription factor 2 (Olig2) is considered to be the main TF responsible for OPC specification. Most OPCs generated from NPCs in the motor neuron progenitor region of the spinal cord express Olig2, and Olig2 is believed to orchestrate OL differentiation by inducing the expression of Nkx2.2 and later that of Sox10 (Sim et al., 2011). To mimic conditions present in the motor neuron progenitor region during NPC induction from PSCs, we combined dual SMAD inhibition with the caudalizing and ventralizing factors, RA and SHH agonist, respectively. This resulted in 51.7% ± 4.6% of NPCs that expressed OLIG2 and 81.76% ± 2.67% the ventral marker HOXB4 (Franklin et al., 2012, Lee et al., 2012b). Overexpression of SOX10 alone in these NPCs resulted in the specification and maturation of OLs. However, overexpression of SOX10 in uncommitted hPSCs did not result in OPCs and differentiation (not shown), demonstrating that SOX10 alone cannot directly induce an OL fate in hPSCs, but only in already neural-committed precursors. Of note, we did not detect neural/glial antigen 2 (NG2)- or platelet-derived growth factor receptor α (PDGFRa)-positive cells following SOX10 overexpression, suggesting that SOX10 causes the direct generation of late OPCs/OLs from OLIG2+ NPCs without an intermediate OPC stage.
Recently, Ehrlich et al. (2017) published a protocol for OL generation from hPSCs also based on TF overexpression. They demonstrated that SOX10 was the only TF capable of inducing O4 expression, which was further enhanced when combined with OLIG2 and NKX6.2. They observed a similar frequency of O4+ cells and final maturation to MBP+ OLs at the end of differentiation as we describe here. However, the Ehrlich et al. protocol required 28 days to generate ∼70% O4+ cells, and an additional 7 days for efficient MBP+ OL production after NPC transduction. By contrast, our protocol requires 10 days following NPC transduction with SOX10 to generate similar levels of O4+ cells of which 20% also express MBP.
Although the reasons for the different requirement of three versus one TF to generate O4+ cells from NPC are not clear, we hypothesize that these differences might be due to differences in the initial NPC generation. Ehrlich et al. induced neural specification via embryoid bodies in the presence of SMAD inhibitors and the Wnt activator and caudalizing molecule CHIR99021, combined with the ventralizing SHH agonist Purmorphamine. By contrast, we induced NPCs by dual SMAD inhibition in adherent cultures, and in the presence of low concentrations of RA, essential for efficient generation of OLIG2+ precursors and subsequent OPC specification (Douvaras et al., 2014). The fact that we only need the single TF SOX10 may allow future studies to develop methods that will directly or indirectly target endogenous SOX10 to generate OLs that could be used clinically.
To further characterize the OL progeny, we performed RNA-seq analysis on day 22 O4+-purified OLs, and compared their transcriptome with previously described datasets from primary neural populations including immature to mature OLs (Zhang et al., 2014, Zhang et al., 2016, Abiraman et al., 2015). This demonstrated that SOX10-induced O4+-purified cells clustered with bona fide OLs and differed from other brain cells. We found slight discrepancies in the levels of OL maturity among the four lines tested, which may be the consequence of variability between cells from which O4+ cells were derived, as these cells have different developmental origins, are derived from genetically distinct individuals and/or hiPSCs may differ in their level of epigenetic reprogramming (Bilic and Izpisua Belmonte, 2012). In addition, we included in the analysis the OLs described by Ehrlich et al. (2017) (Figure S3), finding that all OLs clustered together and the OLs generated by both studies presented highly shared transcriptomes with primary OLs, indicating that TF-induced OLs resemble primary OLs at the transcriptome level.
The robustness of the protocol was further validated by deriving OLs from two MS and two fALS patients. These MS/fALS patient-derived OLs expressed similar markers and morphology compared with healthy donor-derived cells. Among the eight lines tested, the only significant difference we found was a reduction in the proportion of MBP+ cells in SOX10-induced cells from the BJ1 line (Figure 6C). Notably, we also found a reduction in the percentage of the OLIG2+ population (51.7% versus 30.7%) after 12 days of neural induction in BJ1 cells. As hiPSCs may have skewed capacities to the differentiation toward different lineages (Bilic and Izpisua Belmonte, 2012), we hypothesize that decreased neural differentiation capacity of the BJ1 iPSCs may determine the lower frequency of mature OLs obtained at the end of the SOX10-induced protocol.
To evaluate the myelination capacity of SOX10-induced O4+ OLs, we first injected the cells into MBP−/– homozygous shiverer mouse brain slices and demonstrated not only efficient engraftment and dispersion within the slice, but also wrapping of neuronal axons by MBP+ OL prolongations in this in vivo context 10 days after injection. To further demonstrate the myelinating capabilities of generated O4+ cells in vitro, we established a fully hPSC-derived co-culture system, demonstrating that induced OLs were able to produce MBP+ extensions aligning with neuronal axons and myelin ensheathment by about 20 days following co-culture, even though SOX10 was no longer overexpressed during the last 10 days of the co-culture, suggesting that SOX10 overexpression was no longer required at this stage to support OL maturation and myelination. Myelin formation surrounding axons and the presence of compact multi-layered myelin was also observed in the co-culture system at the ultrastructural level, fully demonstrating the myelination capacity of SOX10-derived OLs.
To enable HTS for drugs that improve myelination, we successfully adapted the co-culture system to 96- and 384-well plate formats. This allowed us to demonstrate that MBP expression increased considerably when T3, a known inducer of myelin production (Marta et al., 1998), was added to the medium. Clobetasol, previously shown to enhance myelination both in vitro and in vivo (Najm et al., 2015), increased MBP expression to even higher levels than T3. MBP expression was also induced by miconazole, but to a lesser extent. Increase in MBP staining was also seen after treatment with pranlukast, a compound known to inhibit signaling via the Gpr17 receptor, and reported to enhance remyelination and OL survival in mouse (Ou et al., 2016). As this occurred already at the μM level, pranlukast might be an interesting drug candidate to be tested for (re)myelination in pathological conditions (Mogha et al., 2016). However, DAPT did not significantly increase MBP staining, in contrast with previous studies (Lariosa-Willingham et al., 2016). Differences in myelin production might be due to the fact that we tested the effect of the different compounds in OL-neuron co-cultures, while previous studies assessed myelination of OLs in the absence of neurons (Mei et al., 2014, Lee et al., 2012a, Kerman et al., 2015, Ou et al., 2016, Mogha et al., 2016).
This all-human neuron-OLs co-culture system allows to evaluate the effect of drugs on myelin expression, which should also allow modeling of pathways involved in neural diseases wherein OLs are involved, either because of defects in myelination or defects in neuronal support.
Finally, we demonstrated that induction of SOX10 expression in NPCs following a single copy insertion of SOX10 in a safe harbor locus of hPSCs (Ordovás et al., 2015) is sufficient to induce O4+ and MBP+ OL progeny. A similar methodology has recently been described by Pawlowski et al. (2017), who demonstrated that overexpression of SOX10 and OLIG2 induced cells with phenotypic features of OLs in a similar time frame as we demonstrate here, even though no functional characterization of the cells was performed. Generation of stable inducible SOX10-hPSC lines would be an invaluable tool for drug screens, as they do not suffer from random integrations and the incomplete efficiency of viral vectors, allowing a very efficient and robust generation of OLs.
In summary, we demonstrated that overerexpression of only SOX10 in hPSC-derived NPCs allows a very efficient and rapid generation of OLs from hPSCs, which can myelinate nude axons both in in vivo and in vitro contexts. The protocol is robust, as it induces with similar efficiency and speed O4+ cells from hESCs and hiPSCs of healthy donors, as well as from hiPSCs from patients with MS or fALS. A platform was also developed wherein myelination of neurons can be evaluated in an HTS format to demonstrate the effect of myelinating factors and molecules. This all-hPSC-derived neuron-OL myelinating co-culture platform has the potential to significantly improve testing and validation of candidate pro-myelinating drugs for congenital or acquired demyelinating diseases, and allow assessment of mechanisms underlying neurodegenerative diseases wherein decreased OL-mediated neuronal support is fundamental to disease onset or progression.
Experimental Procedures
Generation of NPCs and OLs from hPSCs
hPSCs were dissociated to single cells on day −2. On day 0, medium was switched to N2B27 medium supplemented with SB431542 10 μM, LDN193189 1 μM, and 100 nM RA until day 8, when SB431542 and LDN193189 were withdrawn and 1 μM Smoothened agonist added. On day 12, cells were dissociated, plated, and transduced with viral vectors. Next day, medium was changed to OL differentiation medium with the addition of 1 μg/mL doxycycline. Cells were maintained for 7–10 days.
Co-culture with Neurons and Adaptation for HTS
Cortical neurons were generated from WT hPSCs as described previously (Shi et al., 2012). Neuronal progenitors (∼50 days) were replated and allowed to mature for 10–14 days. Afterward, purified O4+ cells were seeded above the maturing neurons and cultures were maintained for 20 days in co-culture medium in the absence of T3 and AA, with the addition of doxycycline and the presence of the candidate compounds.
Animal procedures (assessment of OL myelination in brain slices derived from shiverer mice) were approved by a Dutch Ethical Committee for animal experiments.
Detailed experimental procedures can be found in the Supplemental Information.
Author Contributions
J.A.G.L. and C.M.V. designed the project and experiments. J.A.G.L. performed most of the experiments and performed analysis of the data. M.K. helped with the co-cultures and HTS experiments and analysis. R.B. helped with generation of the endogenous SOX10-expressing cell lines. D.C., J.O., and W.S.H. performed RNA-seq analysis. K.E. and P.B. helped with experiments regarding mRNA expression and immunostaining. E.W., J.C.D., and I.L. performed and analyzed the transmission electron microscopy experiments. N.G., F.d.V., B.L., and S.A.K. performed Shi−/– mouse myelination assays. B.J. and N.C. helped in the analysis of the immunostainings. J.A.G.L. and C.M.V. wrote the manuscript. C.M.V. provided scientific guidance and support. All authors read the manuscript.
Acknowledgments
We would like to acknowledge technical colleagues at SCIL (KU Leuven) for their technical support, Jonathan De Smedt for the heatmap representation of expression data, Prof. Fraser Sim (University of Buffalo, USA) for the MCS5-SOX10 reporter plasmids, Dr. Stefan Vinckier (Vesalius Research Center, VIB, KU Leuven, Belgium) for technical assistance with confocal microscopy, and Dr. John Pearson (Andalusian Center for Nanomedicine & Biotechnology, Spain) for 3D reconstruction of confocal microscopy images. J.A.G.L. has been supported by a Fellowship from the Alfonso Martín Escudero Foundation (Madrid, Spain) and M.K. by an H2020-MSCA-IF Fellowship. R.B. was funded by IWT/SB/121393 and S.A.K. by NWO-ZonMw Middelgroot (40-00506-98-10026). The work was supported by the IWT-iPSCAF grant (no. 150031) and the KUL-PF Stem Cells (no. PFO3) to C.M.V.
Published: January 11, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and seven tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.12.014.
Contributor Information
Juan Antonio García-León, Email: jgarleon@hotmail.com.
Catherine M. Verfaillie, Email: catherine.verfaillie@kuleuven.be.
Accession Numbers
The transcriptome data reported in this paper have been deposited in the Gene Expression Omnibus under GEO: GSE106984.
Supplemental Information
References
- Abiraman K., Pol S.U., O'Bara M.A., Chen G.D., Khaku Z.M., Wang J., Thorn D., Vedia B.H., Ekwegbalu E.C., Li J.X. Anti-muscarinic adjunct therapy accelerates functional human oligodendrocyte repair. J. Neurosci. 2015;35:3676–3688. doi: 10.1523/JNEUROSCI.3510-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilic J., Izpisua Belmonte J.C. Concise review: induced pluripotent stem cells versus embryonic stem cells: close enough or yet too far apart? Stem Cells. 2012;30:33–41. doi: 10.1002/stem.700. [DOI] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function (2008) J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey B.W., Markoulaki S., Hanna J., Saha K., Gao Q., Mitalipova M., Jaenisch R. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc. Natl. Acad. Sci. USA. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh V.A., Tardif V., Lyssiotis C.A., Green C.C., Kerman B., Kim H.J., Padmanabhan K., Swoboda J.G., Ahmad I., Kondo T. A regenerative approach to the treatment of multiple sclerosis. Nature. 2013;502:327–332. doi: 10.1038/nature12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P., Fossati V. Generation and isolation of oligodendrocyte progenitor cells from human pluripotent stem cells. Nat. Protoc. 2015;10:1143–1154. doi: 10.1038/nprot.2015.075. [DOI] [PubMed] [Google Scholar]
- Douvaras P., Wang J., Zimmer M., Hanchuk S., O'Bara M.A., Sadiq S., Sim F.J., Goldman J., Fossati V. Efficient generation of myelinating oligodendrocytes from primary progressive multiple sclerosis patients by induced pluripotent stem cells. Stem Cell Reports. 2014;3:250–259. doi: 10.1016/j.stemcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Mozafari S., Glatza M., Starost L., Velychko S., Hallmann A.L., Cui Q.L., Schambach A., Kim K.P., Bachelin C. Rapid and efficient generation of oligodendrocytes from human induced pluripotent stem cells using transcription factors. Proc. Natl. Acad. Sci. USA. 2017;114:E2243–E2252. doi: 10.1073/pnas.1614412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin R.J., ffrench-Constant C., Edgar J.M., Smith K.J. Neuroprotection and repair in multiple sclerosis. Nat. Rev. Neurol. 2012;8:624–634. doi: 10.1038/nrneurol.2012.200. [DOI] [PubMed] [Google Scholar]
- Furusho M., Roulois A.J., Franklin R.J., Bansal R. Fibroblast growth factor signaling in oligodendrocyte-lineage cells facilitates recovery of chronically demyelinated lesions but is redundant in acute lesions. Glia. 2015;63:1714–1728. doi: 10.1002/glia.22838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B.Y., Du Z.W., Zhang S.C. Differentiation of human oligodendrocytes from pluripotent stem cells. Nat. Protoc. 2009;4:1614–1622. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman B.E., Kim H.J., Padmanabhan K., Mei A., Georges S., Joens M.S., Fitzpatrick J.A., Jappelli R., Chandross K.J., August P., Gage F.H. In vitro myelin formation using embryonic stem cells. Development. 2015;142:2213–2225. doi: 10.1242/dev.116517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariosa-Willingham K.D., Rosler E.S., Tung J.S., Dugas J.C., Collins T.L., Leonoudakis D. Development of a central nervous system axonal myelination assay for high throughput screening. BMC Neurosci. 2016;17:16. doi: 10.1186/s12868-016-0250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Leach M.K., Redmond S.A., Chong S.Y., Mellon S.H., Tuck S.J., Feng Z.Q., Corey J.M., Chan J.R. A culture system to study oligodendrocyte myelination processes using engineered nanofibers. Nat. Methods. 2012;9:917–922. doi: 10.1038/nmeth.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Morrison B.M., Li Y., Lengacher S., Farah M.H., Hoffman P.N., Liu Y., Tsingalia A., Jin L., Zhang P.W. Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature. 2012;487:443–448. doi: 10.1038/nature11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marta C.B., Adamo A.M., Soto E.F., Pasquini J.M. Sustained neonatal hyperthyroidism in the rat affects myelination in the central nervous system. J. Neurosci. Res. 1998;53:251–259. doi: 10.1002/(SICI)1097-4547(19980715)53:2<251::AID-JNR14>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Marques S., Zeisel A., Codeluppi S., van Bruggen D., Mendanha Falcão A., Xiao L., Li H., Häring M., Hochgerner H., Romanov R.A. Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science. 2016;352:1326–1329. doi: 10.1126/science.aaf6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F., Fancy S.P., Shen Y.A., Niu J., Zhao C., Presley B., Miao E., Lee S., Mayoral S.R., Redmond S.A. Micropillar arrays as a high-throughput screening platform for therapeutics in multiple sclerosis. Nat. Med. 2014;20:954–960. doi: 10.1038/nm.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogha A., D'Rozario M., Monk K.R. G protein-coupled receptors in myelinating glia. Trends Pharmacol. Sci. 2016;37:977–987. doi: 10.1016/j.tips.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm F.J., Lager A.M., Zaremba A., Wyatt K., Caprariello A.V., Factor D.C., Karl R.T., Maeda T., Miller R.H., Tesar P.J. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat. Biotechnol. 2013;31:426–433. doi: 10.1038/nbt.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm F.J., Madhavan M., Zaremba A., Shick E., Karl R.T., Factor D.C., Miller T.E., Nevin Z.S., Kantor C., Sargent A. Drug-based modulation of endogenous stem cells promotes functional remyelination in vivo. Nature. 2015;522:216–220. doi: 10.1038/nature14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J.A., Maric D., Lau P., Barker J.L., Hudson L.D. Identification of a novel oligodendrocyte cell adhesion protein using gene expression profiling. J. Neurosci. 2006;26:9881–9891. doi: 10.1523/JNEUROSCI.2246-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistor G.I., Totoiu M.O., Haque N., Carpenter M.K., Keirstead H.S. Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation. Glia. 2005;49:385–396. doi: 10.1002/glia.20127. [DOI] [PubMed] [Google Scholar]
- Ordovás L., Boon R., Pistoni M., Chen Y., Wolfs E., Guo W., Sambathkumar R., Bobis-Wozowicz S., Helsen N., Vanhove J. Efficient recombinase-mediated cassette exchange in hPSCs to study the hepatocyte lineage reveals AAVS1 locus-mediated transgene inhibition. Stem Cell Reports. 2015;5:918–931. doi: 10.1016/j.stemcr.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Z., Sun Y., Lin L., You N., Liu X., Li H., Ma Y., Cao L., Han Y., Liu M. Olig2-targeted G-protein-coupled receptor Gpr17 regulates oligodendrocyte survival in response to lysolecithin-induced demyelination. J. Neurosci. 2016;36:10560–10573. doi: 10.1523/JNEUROSCI.0898-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski M., Ortmann D., Bertero A., Tavares J.M., Pedersen R.A., Vallier L., Kotter M.R.N. Inducible and deterministic forward programming of human pluripotent stem cells into neurons, skeletal myocytes, and oligodendrocytes. Stem Cell Reports. 2017;8:803–812. doi: 10.1016/j.stemcr.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol S.U., Lang J.K., O'Bara M.A., Cimato T.R., McCallion A.S., Sim F.J. Sox10-MCS5 enhancer dynamically tracks human oligodendrocyte progenitor fate. Exp. Neurol. 2013;247:694–702. doi: 10.1016/j.expneurol.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozniak C.D., Langseth A.J., Dijkgraaf G.J., Choe Y., Werb Z., Pleasure S.J. Sox10 directs neural stem cells toward the oligodendrocyte lineage by decreasing suppressor of fused expression. Proc. Natl. Acad. Sci. USA. 2010;107:21795–21800. doi: 10.1073/pnas.1016485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Smith J., Robinson H.P., Livesey F.J. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 2012;15 doi: 10.1038/nn.3041. 477–486, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim F.J., McClain C.R., Schanz S.J., Protack T.L., Windrem M.S., Goldman S.A. CD140a identifies a population of highly myelinogenic, migration-competent and efficiently engrafting human oligodendrocyte progenitor cells. Nat. Biotechnol. 2011;29:934–941. doi: 10.1038/nbt.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer I., Schachner M. Monoclonal antibodies (O1 to O4) to oligodendrocyte cell surfaces: an immunocytological study in the central nervous system. Dev. Biol. 1981;83:311–327. doi: 10.1016/0012-1606(81)90477-2. [DOI] [PubMed] [Google Scholar]
- Vogl M.R., Reiprich S., Küspert M., Kosian T., Schrewe H., Nave K.A., Wegner M. Sox10 cooperates with the mediator subunit 12 during terminal differentiation of myelinating glia. J. Neurosci. 2013;33:6679–6690. doi: 10.1523/JNEUROSCI.5178-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Bates J., Li X., Schanz S., Chandler-Militello D., Levine C., Maherali N., Studer L., Hochedlinger K., Windrem M., Goldman S.A. Human iPSC-derived oligodendrocyte progenitor cells can myelinate and rescue a mouse model of congenital hypomyelination. Cell Stem Cell. 2013;12:252–264. doi: 10.1016/j.stem.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M. All purpose Sox: the many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 2010;42:381–390. doi: 10.1016/j.biocel.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Weider M., Reiprich S., Wegner M. Sox appeal – Sox10 attracts epigenetic and transcriptional regulators in myelinating glia. Biol. Chem. 2013;394:1583–1593. doi: 10.1515/hsz-2013-0146. [DOI] [PubMed] [Google Scholar]
- Weider M., Wegner M. SoxE factors: transcriptional regulators of neural differentiation and nervous system development. Semin. Cell Dev. Biol. 2017;63:35–42. doi: 10.1016/j.semcdb.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Weng Q., Chen Y., Wang H., Xu X., Yang B., He Q., Shou W., Chen Y., Higashi Y., van den Berghe V. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron. 2012;73:713–728. doi: 10.1016/j.neuron.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Zuchero J.B., Ahlenius H., Marro S., Ng Y.H., Vierbuchen T., Hawkins J.S., Geissler R., Barres B.A., Wernig M. Generation of oligodendroglial cells by direct lineage conversion. Nat. Biotechnol. 2013;31:434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzka M., Rivers L.E., Fancy S.P., Zhao C., Tripathi R., Jamen F., Young K., Goncharevich A., Pohl H., Rizzi M. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell. 2010;6:578–590. doi: 10.1016/j.stem.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O'Keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.