Summary
Despite intensive efforts to optimize the process, reprogramming differentiated cells to induced pluripotent stem cells (iPSCs) remains inefficient. The most common combination of transcription factors employed comprises OCT4, KLF4, SOX2, and MYC (OKSM). If MYC is omitted (OKS), reprogramming efficiency is reduced further. Cells must overcome several obstacles to reach the pluripotent state, one of which is apoptosis. To directly determine how extensively apoptosis limits reprogramming, we exploited mouse embryonic fibroblasts (MEFs) lacking the two essential mediators of apoptosis, BAK and BAX. Our results show that reprogramming is enhanced in MEFs deficient in BAK and BAX, but only when MYC is part of the reprogramming cocktail. Thus, the propensity for Myc overexpression to elicit apoptosis creates a significant roadblock to reprogramming under OKSM conditions. Our results suggest that blocking apoptosis during reprogramming may enhance the derivation of iPSCs for research and therapeutic purposes.
Keywords: mouse embryonic fibroblasts: reprogramming, induced pluripotent stem cells, mitochondrial apoptosis, BAK, BAX, MYC, p53
Highlights
-
•
We generated MEFS lacking two essential mediators of apoptosis, BAK and BAX
-
•
Loss of BAK and BAX significantly enhances MEF reprogramming in the presence of MYC
-
•
Thus, mitochondrial apoptosis limits reprogramming of MEFs in the presence of MYC
-
•
The integrity of the genome is not reduced in reprogrammed MEFs lacking BAK and BAX
In this article, Heath, van Delft, and colleagues show that mitochondrial apoptosis limits OKSM-mediated reprogramming of MEFs. Not only is reprogramming of MEFs lacking the two essential mediators of mitochondrial apoptosis, BAK and BAX, significantly enhanced in the presence of MYC, but reprogramming in these conditions does not compromise genome integrity.
Introduction
Reprogramming somatic cells to a pluripotent state can be achieved through ectopic expression of pluripotency transcription factors (Takahashi and Yamanaka, 2006). The resulting induced pluripotent stem cells (iPSCs) hold immense promise as tools for research and regenerative therapy. To harness their full potential, studies have scrutinized the molecular mechanisms underpinning reprogramming, and sought to identify factors able to improve this inherently inefficient process (Esteban et al., 2010, Onder et al., 2012). One well-characterized means of enhancing reprogramming is to inhibit the activity of TP53/TRP53 (hereafter, p53) (Hong et al., 2009, Kawamura et al., 2009, Marion et al., 2009, Utikal et al., 2009). The improved reprogramming efficiency of p53-deficient cells has been attributed to the ability of p53 to trigger senescence and apoptosis. While senescence has been confirmed as a major reprogramming barrier (Banito et al., 2009, Kawamura et al., 2009), the significance of mitochondrial apoptosis is less clear.
Mitochondrial apoptosis is a conserved intrinsic cell death program controlled by interactions between diverse members of the BCL2 protein family, of which there are three functional classes: the pro-survival BCL2-like proteins, the pro-apoptotic BH3-only proteins, and the apoptosis effector proteins, BAK and BAX (Figure S1). In healthy cells, the BCL2-like survival proteins suppress mitochondrial apoptosis by restraining BAK and BAX. However, in response to various forms of cellular stress, including cytokine deprivation, DNA damage, and oncogene activation, BH3-only proteins are triggered to initiate apoptosis both by inhibiting BCL2-like proteins and by directly activating BAK and BAX. Once activated, BAK and BAX oligomerize on the outer mitochondrial membrane to drive its rupture (Czabotar et al., 2013). The ensuing mitochondrial outer membrane permeabilization (MOMP) results in the efflux of pro-apoptogenic factors, including cytochrome c, to the cytosol. At this point, the cell is irreversibly committed to death, and a cascade of proteolytic caspases is activated that brings about the ordered demolition of cellular components.
Deciphering the role of apoptosis in cellular processes can be complicated by the fact that each of the three classes of BCL2 family proteins comprises multiple family members, and that each of these proteins has a certain propensity to interact with and regulate the others (Chen et al., 2005, Czabotar et al., 2014, Willis et al., 2007). As such, there is a large degree of functional overlap and redundancy within the family. Importantly for our studies, however, BAK and BAX are the only two BCL2 family members able to drive MOMP, and in their combined absence, the mitochondrial apoptosis pathway is completely abolished (Lindsten et al., 2000).
Thus far, attempts to resolve the role, if any, of mitochondrial apoptosis during reprogramming have primarily focused on upstream regulators, including BCL2, the archetypal pro-survival BCL2 family member, and PUMA, a p53-regulated BH3-only protein (Kawamura et al., 2009, Lake et al., 2012, Li et al., 2013). These studies have produced conflicting data, with two studies reporting significant enhancement of reprogramming efficiency when apoptosis was blocked, either through BCL2 overexpression or PUMA depletion (Kawamura et al., 2009, Li et al., 2013), and another reporting that PUMA depletion promoted reprogramming under OKS but not OKSM conditions (Lake et al., 2012).
To directly evaluate the influence of apoptosis on the efficiency of reprogramming, we used mouse embryonic fibroblasts (MEFs) lacking both BAK and BAX. Such cells provide a definitive model system in which to study cellular processes in the absence of mitochondrial apoptosis (Lindsten et al., 2000). Here, we report that Bak−/−;Bax−/− MEFs undergo reprogramming with markedly increased efficiency compared with their wild-type (WT) counterparts in OKSM, but not OKS conditions. Remarkably, they traverse this process without incurring significantly increased genomic instability. Our data conclusively demonstrate that mitochondrial apoptosis imposes a strong barrier to OKSM-mediated iPSC induction, and that this roadblock is MYC-dependent. While the overall efficiency of iPSC derivation is markedly improved with ectopic Myc expression, the heightened sensitivity to apoptosis curtails its full benefit in reprogramming.
Results and Discussion
Mitochondrial Apoptosis: A Barrier to iPSC Derivation by OKSM Expression
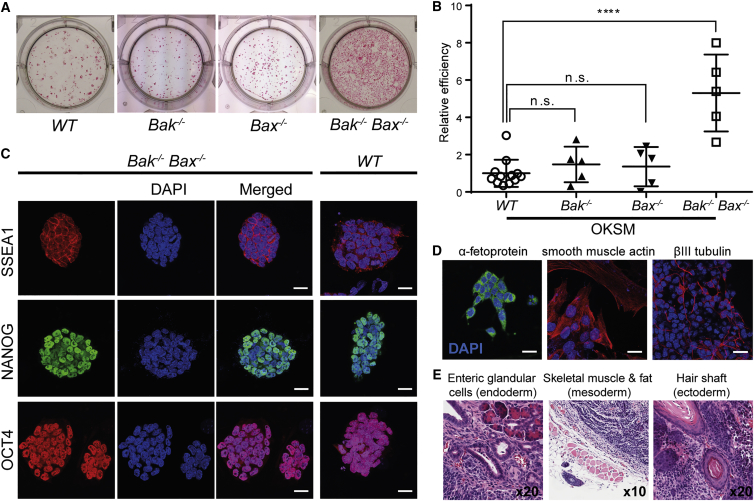
To examine the role of mitochondrial apoptosis in reprogramming, WT and Bak−/−;Bax−/− MEFs were transduced with lentiviral OKSM factors. After 18 days of reprogramming, Bak−/−;Bax−/− MEFs yielded alkaline phosphatase (AP)-positive colonies (Figure 1A) that were morphologically indistinguishable from those derived from WT MEFs (Figure S2A). Moreover, they displayed markedly improved reprogramming efficiency compared with WT MEFs, as measured by AP activity (Figure 1B) and verified by immunocytochemical staining of the pluripotency marker, NANOG (Figure S2B). Meanwhile, deficiency in either BAK or BAX alone did not significantly affect the yield of AP-positive colonies (Figures 1A and 1B). This is consistent with their functional redundancy in mediating MOMP, and proves that the increased reprogramming efficiency of Bak−/−;Bax−/− MEFs is attributable to attenuation of mitochondrial apoptosis.
Figure 1.
Enhanced Production of iPSCs in the Absence of Apoptosis Yields Fully Pluripotent Cells
(A) Alkaline phosphatase (AP) staining of WT, Bak−/−, Bax−/−, and Bak−/–;Bax−/– MEFs at 18 days post transduction with OKSM reveals enhanced reprogramming efficiency in Bak−/–;Bax−/– cells.
(B) Quantification of AP-positive colonies generated by each genotype relative to WT. Values shown are mean ± SD. ∗∗∗∗p < 0.0001, n ≥ 5 independent experiments, each with an independently derived MEF line. n.s., not significant.
(C) Expression of the pluripotency markers SSEA1, NANOG, and OCT4 in independent Bak−/−, Bax−/− iPSC colonies passaged at least five times. During this time, the colonies maintained a characteristic iPSC morphology (see also Figure S2).
(D) Bak−/−;Bax−/− embryoid bodies were produced from isolated iPSCs by the hanging drop method. These were cultured on adhesive surfaces for 2 weeks prior to staining for differentiation markers representing the three germ layers: α1 fetoprotein (endoderm), α smooth muscle actin (mesoderm), and βIII tubulin (ectoderm).
(E) Hematoxylin and eosin-stained sections of Bak−/−;Bax−/− iPSC-derived teratomas reveals differentiated tissues representative of all three germ layers.
Scale bars, 20 μm.
To further assess the pluripotency of colonies generated from Bak−/−;Bax−/− MEFs, we cultured them over at least five passages to confirm their self-renewal capacity, during which time they maintained a shiny, rounded morphology and expression of the pluripotency markers SSEA1, NANOG, and OCT4 (Figure 1C). These observations are consistent with those reported for embryonic stem cells deficient in BAK and BAX (Wang et al., 2015).
Next, to assess the differentiation capacity of Bak−/−;Bax−/− iPSCs, we generated embryoid bodies by the hanging drop method. Cells from three independent lines spontaneously differentiated into cells representative of all three germ layers (Figure 1D). Finally, we investigated the in vivo differentiation capacity of the Bak−/−;Bax−/− iPSC lines by injecting them subcutaneously into mice. The resulting teratomas contained tissues derived from all three germ layers (Figure 1E).
Elevated Caspase-3/7 Activity during OKSM Reprogramming Suggests MYC-Stimulated Apoptosis Forms a Significant Barrier to Reaching Pluripotency
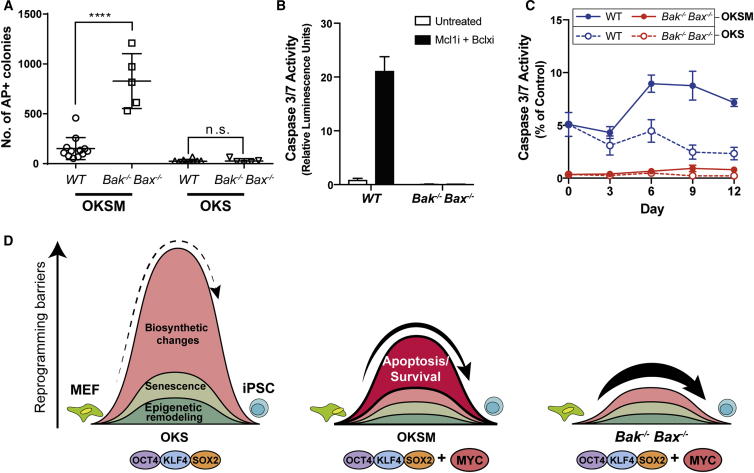
Having observed that Bak−/−;Bax−/− MEFs reprogrammed more efficiently upon the expression of OKSM factors, we next tested whether this would also hold true when MEFs were reprogrammed without MYC, given its propensity to promote apoptosis. When MEFs were reprogrammed with only OKS, there was a striking decrease in iPSC formation compared with MEFs reprogrammed with OKSM (Figure 2A), consistent with previous reports (Nakagawa et al., 2008). However, there was no significant difference in reprogramming efficiencies between Bak−/−;Bax−/− and WT cultures (Figure 2A); this suggests that in the absence of ectopic MYC expression, apoptosis does not significantly limit reprogramming. Conversely, we hypothesize that ectopic MYC expression drives elevated levels of apoptosis that in turn limits the efficiency of OKSM reprogramming.
Figure 2.
Apoptosis Is a Myc-Induced Reprogramming Roadblock
(A) Bak−/−;Bax−/− MEFs transduced with OKSM produced significantly more AP-positive colonies compared with WT MEFs. In OKS conditions, WT and Bak−/−;Bax−/− MEFs produced significantly fewer colonies, and there was no significant difference (n.s.) between them.
(B) WT and Bak−/−;Bax−/− MEFs were treated for 6 hr with S63845 and A-1331852 to inhibit the pro-survival factors, MCL1 and BCLx, respectively. At this point, all WT cells exhibited high caspase-3/7 activity. In contrast, Bak−/−;Bax−/− MEFs exhibited negligible caspase-3/7 activity, even in the presence of S63845 and A-1331852.
(C) Caspase-3/7 assays were conducted every 3 days during the first 12 days of reprogramming in OKSM and OKS conditions. WT MEFs transduced with OKSM factors exhibited significantly higher caspase-3/7 activity compared with WT MEFs transduced with OKS. Meanwhile, Bak−/−;Bax−/− MEFs exhibited very low caspase-3/7 activity throughout the experiment, independently of whether they were transduced with OKSM or OKS.
(D) Proposed model: exogenous MYC enhances reprogramming by inducing biosynthetic changes favorable to reprogramming, including increased transcription, translation, and a switch from oxidative phosphorylation to a glycolytic mode of energy production. However, exogenous MYC also raises an apoptosis barrier, which is abolished in Bak−/−;Bax−/− cells.
Values are mean ± SD, ∗∗∗∗p < 0.0001, n ≥ 5 independent experiments, each with an independently derived MEF line.
To determine whether apoptosis was indeed elevated during OKSM relative to OKS reprogramming, we measured caspase-3/7 activity over the first 12 days of reprogramming using the Caspase-Glo 3/7 Assay System (Promega). To establish a reference for these assays, we induced apoptosis in 100% of MEFs by treating them with small molecule inhibitors of the pro-survival proteins, MCL1 (S63845; Kotschy et al., 2016) and BCLXL for 6 hr (A-1331,852; Leverson et al., 2015). As expected, robust caspase-3/7 activity was elicited in WT but not Bak−/−;Bax−/− MEFs (Figure 2B). WT MEF cultures reprogrammed with OKSM exhibited elevated caspase-3/7 activity that peaked at approximately 10% of the level observed in the apoptotic reference sample (Figure 2C). Furthermore, caspase-3/7 activity was consistently higher in WT MEFs reprogrammed with OKSM compared with OKS throughout the duration of the experiment (Figure 2C). In contrast, we detected very low caspase-3/7 activity in reprogramming Bak−/−;Bax−/− MEFs (Figures 2B and 2C).
These data are consistent with prior findings that ectopic MYC expression sensitizes cells to apoptosis (McMahon, 2014, Pelengaris et al., 2002) and lead us to conclude that mitochondrial apoptosis constitutes a major barrier to reprogramming when MYC is included as one of the pluripotency factors (Figure 2D). Thus, while MYC enhances overall reprogramming efficiency through reinforcing cell proliferation and growth, this is mitigated in part by an increase in BAK/BAX-mediated apoptosis.
p53-Mediated Apoptosis Is Not a Major Barrier to OKS Reprogramming
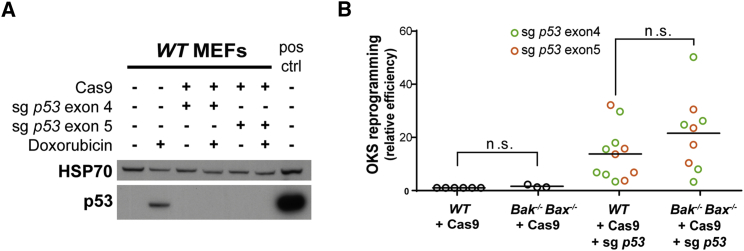
Previous studies have established p53 as a barrier to both OKSM and OKS-mediated reprogramming. While p53 drives a variety of cellular responses, it is primarily thought to hinder iPSC derivation by provoking senescence and apoptosis. Based on our observations that apoptosis was not a major barrier to OKS-mediated reprogramming (Figure 2A), we reasoned that deficiency of BAK and BAX would add little further advantage to p53-deficient cultures in OKS conditions. To test this, we introduced p53 mutations in primary MEFs using CRISPR/Cas9 targeting. Western blots confirmed that single-guide RNAs (sgRNAs) targeting either exon 4 or exon 5 of p53 were highly effective at ablating p53 expression (Figure 3A; Aubrey et al., 2015). When reprogrammed with OKS, WT and Bak−/−;Bax−/− MEFs expressing Cas9 together with sgRNA targeting p53 both formed AP-positive iPSCs more efficiently than controls expressing only Cas9 (Figure 3B). However, we observed no significant difference between the reprogramming capacity of WT or Bak−/−;Bax−/− MEFs in the absence of p53 (Figure 3B). This confirms that when mitochondrial apoptosis is blocked in OKS conditions, other p53-driven responses, such as senescence, are likely to pose the primary reprogramming roadblocks.
Figure 3.
p53-Mediated Apoptosis Is Not a Barrier to OKS Reprogramming
(A) Western blot of p53 protein in primary MEFs expressing Cas9 and sgRNAs targeting either exon 4 or 5 confirms p53 knockout in both examples. Cells were treated with doxorubicin to increase p53 protein to detectable levels. In the final lane, transformed MEFs overexpressing p53 were used as a positive control.
(B) WT and Bak−/−;Bax−/− MEFs expressing Cas9 alone or together with sgRNA targeting p53 were reprogrammed with OKS for 18 days, at which point AP-positive colonies were enumerated. The data are presented as efficiency relative to the WT-Cas9 control in each experiment. Bars represent the mean values for n ≥ 3 independent experiments, each with an independently derived MEF line. n.s., not significant.
Genomic Integrity of Bak−/−;Bax−/− iPSCs Is Comparable with that of WT iPSCs
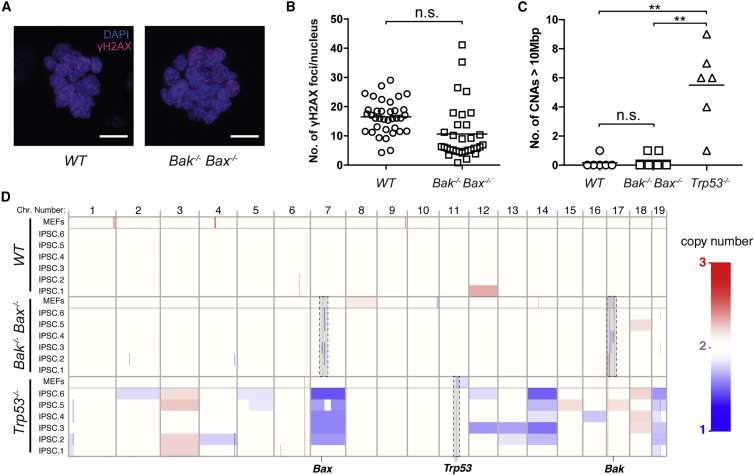
Apoptosis is sometimes regarded as a mechanism to safeguard genome integrity. A block in mitochondrial apoptosis might therefore have deleterious effects on genomic integrity during reprogramming. To investigate this, we examined DNA double-strand breaks and copy number alterations (CNAs) in several independent Bak−/−;Bax−/− iPSC lines that had been propagated for two or more passages. Using γH2AX as a marker, we quantified DNA double-strand breaks and found no significant difference between WT and Bak−/−;Bax−/− iPSC lines (Figures 4A and 4B). Furthermore, using low pass whole genome sequencing, we observed no significant difference in the number of CNAs in iPSC clones derived from Bak−/–;Bax−/– MEFs compared with those derived from WT MEFs (Figures 4C, 4D, S3A, and S3B). Although this method is not sensitive enough to detect single-nucleotide aberrations that may occur during reprogramming, these results are consistent with reports that the process of reprogramming is not inherently mutagenic (Young et al., 2012) and that embryonic stem cells lacking BAK and BAX maintain a normal karyotype (Wang et al., 2015). In contrast, iPSC clones derived from p53-deficient MEFs exhibited a significantly higher number of CNAs compared with the Bak−/–;Bax−/– and WT groups (Figures 4C, 4D, and S3C). In summary, MEFs unable to execute mitochondrial apoptosis display enhanced reprogramming capacity with OKSM without notable changes in genome stability.
Figure 4.
Enhanced Production of iPSCs in the Absence of Apoptosis Yields Cells with DNA and Genome Stability Comparable with that of WT iPSCs
Manually picked iPSC clones were cultured prior to assessment of genomic stability.
(A) Representative confocal images of iPSC colonies (passage 5) stained for γH2AX, which marks DNA double-strand breaks. Scale bars, 20 μm.
(B) Quantification of γH2AX foci was automated using γ-irradiated iPSC colonies as a positive control to set the threshold for fluorescence intensity. The average number of γH2AX foci in the nuclei of WT and Bak−/−;Bax−/− iPSCs is not significantly different. For this experiment, independent MEF lines derived from different embryos (n = 4 for WT; n = 3 for Bak−/−;Bax−/−) were reprogrammed and corresponding iPSC lines established. Each point represents analysis of 1 colony (15–30 nuclei), with 9–12 colonies analyzed per independently derived iPSC line. Bars represent the mean values for each genotype. n.s., not significant.
(C) Quantification of CNAs in iPSC clones (passage 2) derived from Bak−/−;Bax−/− MEFs compared with those derived from WT and p53−/− MEFs. There is no significant difference in the number of CNAs in WT and Bak;Bax DKO parental MEFs and their clonal iPSC derivatives. In contrast, iPSC clones derived from p53−/− MEFs exhibit a significantly higher number of CNAs (n = 6 iPSC clones derived from a single MEF line/genotype). Bars represent the mean values for each genotype. ∗∗p < 0.01, Mann-Whitney U test.
(D) A heatmap detailing CNAs on autosomal chromosomes (1–19, labeled at top) in MEF and clonal iPSC derivatives. Copy number is shown with a color scale, ranging from one to three copies (from blue to red). The genomic position of targeted genes is indicated along the bottom, and these loci are highlighted in the relevant samples (dashed boxes). See also Figure S3.
Conclusion
Our observations lead us to conclude that MYC-driven BAK/BAX-mediated mitochondrial apoptosis presents a significant roadblock to OKSM reprogramming. While the overall effect of forced MYC expression favors reprogramming by regulating a broad range of biosynthetic processes required for enhanced cell growth and proliferation, including transcription, translation, and energy production (Nie et al., 2012, Polo et al., 2012, Sridharan et al., 2009), its propensity to sensitize cells to mitochondrial apoptosis partly mitigates these advantages (Figure 2D). In our hands, inhibiting apoptosis in Bak−/−;Bax−/− iPSCs does not compromise their capacity to differentiate into lineages derived from all three germ layers, both in vitro and in vivo, nor have an adverse effect on genome integrity.
In conclusion, our study shows that harnessing the reprogramming-enhancing effects of MYC while selectively inhibiting its apoptotic response may be a useful strategy for efficient generation of iPSCs. We predict that studies aimed at improving our understanding of reprogramming factor stoichiometry and the specific downstream effectors of MYC will most likely make important contributions to improved iPSC derivation and application in the future.
Experimental Procedures
Mouse Embryonic Fibroblasts
Bak−/− mice (gift of Craig Thompson; Lindsten et al., 2000), Bax−/− mice (Jackson Laboratory; Knudson et al., 1995) were backcrossed to C57BL/6 mice for >10 generations, then inter-crossed to generate Bak−/−;Bax−/− double-knockout (DKO) E13.5 embryos to obtain MEFs. WT MEFs were derived from C57BL/6 mice. p53−/− MEFs (Jacks et al., 1994) were a gift of Anne Voss. All procedures performed on mice were reviewed and approved by the Animal Ethics Committee of the Walter and Eliza Hall Institute of Medical Research.
Generation of MEFs Containing p53 Mutations for OKS Reprogramming
MEFs were transduced with a lentivirus encoding doxycycline-inducible mCherry-Cas9 either alone or together with lentiviruses encoding a GFP expression marker and sgRNA targeting either exon 4 or 5 of p53 (Aubrey et al., 2015). mCherry positive or mCherry/GFP double-positive cells were then sorted on a BD FACSAria III, and treated with doxycycline (2 μg/mL) for 48 hr before reprogramming.
Generation of Lentiviruses
HEK293T cells were plated at a density of 3 × 106 cells/9 cm plate the day prior to transfection. Plasmids encoding lentiviral packaging components, the ecotropic envelope glycoprotein, and the reprogramming cassette OKSM (Sommer et al., 2009) or OKS (Chang et al., 2009) were introduced into HEK293T cells with FuGENE 6 Transfection Reagent (Promega) according to the manufacturer's instructions. The medium was replaced 24 hr post transfection. Viral supernatants were collected from cells at 48 and 72 hr post transfection, filtered (0.45 μm), and added to MEFs alongside polybrene (5 μg/mL).
Mitochondrial Apoptosis Assay
At 3-day intervals following transduction of MEFs with OKS or OKSM, we collected, pooled, and lysed adherent and non-adherent cells in PBS containing NP-40 (1%). Protein content was quantitated with a BCA assay, and caspase activity was measured with the Caspase-Glo 3/7 Assay System (Promega). Luminescence readings were divided by the protein content of the lysate to give a measure of caspase activity per milligram of protein, thereby correcting for the increase in cell number over the duration of the assay. To provide a positive control, we treated WT MEFs with S63845 (SYNthesis Med Chem) and A-1331852 (kind gift of Jean-Marc Garnier and Guillaume Lessene) to specifically inhibit two pro-survival Bcl2 family molecules, MCL1 and BCLx, respectively. This treatment produced synchronous apoptosis, which was morphologically evident in 100% cells after 6 hr. We designated the corresponding luminescence as the maximum expected value from the assay if all cells undergo apoptosis. Thus, at day 6, when WT-OKSM values peak at ∼10% of this value, we extrapolated this to mean that ∼10% of cells in the culture are apoptotic.
Detection of CNAs
Colonies of WT, Bak−/−;Bax−/−, and p53−/− iPSCs were picked and propagated for two passages. Genomic DNA was isolated from six clones of each genotype, plus the corresponding MEFs from which the clones were derived, using a DNeasy blood and tissue kit (Qiagen). Libraries were prepared from sheared genomic DNA (∼300 bp; 4 ng per sample) using a Truseq DNA sample preparation kit (Illumina). All 21 samples were applied to one lane of a NextSeq run, yielding ∼4 million × 81 bp paired-end reads per library. Reads were aligned to mm10 with BWA 0.7.15-r1140 (Li and Durbin, 2009) and analyzed with superFreq 0.9.21 (https://github.com/ChristofferFlensburg/superFreq) over 100 kbp bins, with increased sensitivity for CNAs with systematic variance set to 0.01. Limma-voom was used to estimate log fold change (LFC) and variance, which forms the input to a hierarchical segmentation of the genome. SuperFreq calls CNAs by comparing with a set of reference samples, which in this case comprised the WT colonies. When analyzing individual WT colonies, we compared them with a reference pool that included all other WT samples (i.e., the test sample was excluded from the reference pool). Using the superFreq calls, we counted copy number segments with an absolute LFC larger than 7% stretching over at least 10 Mbp.
Statistical Analysis
Graphs and p values were obtained with GraphPad Prism 6. Pairwise comparisons were made using Student's t test.
Author Contributions
E.J.Y.K. designed and conducted experiments, collected and assembled data, analyzed and interpreted data, wrote the paper, and approved the final manuscript. M.-L.A. designed and conducted experiments, analyzed and interpreted data, provided study materials, edited the paper, and approved the final manuscript. C.F. designed and conducted experiments, collected and assembled data, analyzed and interpreted data, edited the paper, and approved the final manuscript. I.J.M. designed experiments, provided study materials, analyzed and interpreted data, read the paper, and approved the final manuscript. F.-S.G. conducted experiments, read the paper, and approved the final manuscript. J.F. conducted experiments, read the paper, and approved the final manuscript. D.C.S.H. conceived and designed experiments, provided financial support and study materials, analyzed and interpreted data, edited the paper, and approved the final manuscript. M.F.v.D. designed and conducted experiments, provided study materials, collected and assembled data, analyzed and interpreted data, wrote the paper, and approved the final manuscript. J.K.H. conceived and designed experiments, provided financial support and study materials, analyzed and interpreted data, wrote the paper, and approved the final manuscript.
Acknowledgments
We thank Sue Mei Lim for assistance with teratoma formation assays, Siddhartha Deb for pathological analysis, Jose Polo for discussions, Marco Herold for the validated Cas9 and guide RNA constructs, and Lachlan Whitehead and Stephen Mieruszynski for image analysis. This research was supported by the Australian National Health and Medical Research Council (1043092, 1022870, 487922) and Ludwig Cancer Research, with operational infrastructure grants from the Australian Federal Government (IRISS) and the Victorian State Government (OIS).
Published: January 18, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.12.019.
Contributor Information
Mark F. van Delft, Email: vandelft@wehi.edu.au.
Joan K. Heath, Email: joan.heath@wehi.edu.au.
Supplemental Information
References
- Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O'Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–1432. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Banito A., Rashid S.T., Acosta J.C., Li S., Pereira C.F., Geti I., Pinho S., Silva J.C., Azuara V., Walsh M. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.W., Lai Y.S., Pawlik K.M., Liu K., Sun C.W., Li C., Schoeb T.R., Townes T.M. Polycistronic lentiviral vector for "hit and run" reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:1042–1049. doi: 10.1002/stem.39. [DOI] [PubMed] [Google Scholar]
- Chen L., Willis S.N., Wei A., Smith B.J., Fletcher J.I., Hinds M.G., Colman P.M., Day C.L., Adams J.M., Huang D.C. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Czabotar P.E., Lessene G., Strasser A., Adams J.M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- Czabotar P.E., Westphal D., Dewson G., Ma S., Hockings C., Fairlie W.D., Lee E.F., Yao S., Robin A.Y., Smith B.J. Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell. 2013;152:519–531. doi: 10.1016/j.cell.2012.12.031. [DOI] [PubMed] [Google Scholar]
- Esteban M.A., Wang T., Qin B., Yang J., Qin D., Cai J., Li W., Weng Z., Chen J., Ni S. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Hong H., Takahashi K., Ichisaka T., Aoi T., Kanagawa O., Nakagawa M., Okita K., Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Kawamura T., Suzuki J., Wang Y.V., Menendez S., Morera L.B., Raya A., Wahl G.M., Izpisua Belmonte J.C. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson C.M., Tung K.S., Tourtellotte W.G., Brown G.A., Korsmeyer S.J. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- Kotschy A., Szlavik Z., Murray J., Davidson J., Maragno A.L., Le Toumelin-Braizat G., Chanrion M., Kelly G.L., Gong J.N., Moujalled D.M. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature. 2016;538:477–482. doi: 10.1038/nature19830. [DOI] [PubMed] [Google Scholar]
- Lake B.B., Fink J., Klemetsaune L., Fu X., Jeffers J.R., Zambetti G.P., Xu Y. Context-dependent enhancement of induced pluripotent stem cell reprogramming by silencing Puma. Stem Cells. 2012;30:888–897. doi: 10.1002/stem.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leverson J.D., Phillips D.C., Mitten M.J., Boghaert E.R., Diaz D., Tahir S.K., Belmont L.D., Nimmer P., Xiao Y., Max Ma X. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci. Transl. Med. 2015;7:279ra40. doi: 10.1126/scitranslmed.aaa4642. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Feng H., Gu H., Lewis D.W., Yuan Y., Zhang L., Yu H., Zhang P., Cheng H., Miao W. The p53-PUMA axis suppresses iPSC generation. Nat. Commun. 2013;4:2174. doi: 10.1038/ncomms3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsten T., Ross A.J., King A., Zong W.X., Rathmell J.C., Shiels H.A., Ulrich E., Waymire K.G., Mahar P., Frauwirth K. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol. Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion R.M., Strati K., Li H., Murga M., Blanco R., Ortega S., Fernandez-Capetillo O., Serrano M., Blasco M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon S.B. MYC and the control of apoptosis. Cold Spring Harb. Perspect. Med. 2014;4:a014407. doi: 10.1101/cshperspect.a014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelengaris S., Khan M., Evan G. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer C.A., Stadtfeld M., Murphy G.J., Hochedlinger K., Kotton D.N., Mostoslavsky G. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells. 2009;27:543–549. doi: 10.1634/stemcells.2008-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R., Tchieu J., Mason M.J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Utikal J., Polo J.M., Stadtfeld M., Maherali N., Kulalert W., Walsh R.M., Khalil A., Rheinwald J.G., Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E.S., Reyes N.A., Melton C., Huskey N.E., Momcilovic O., Goga A., Blelloch R., Oakes S.A. Fas-activated mitochondrial apoptosis culls stalled embryonic stem cells to promote differentiation. Curr. Biol. 2015;25:3110–3118. doi: 10.1016/j.cub.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis S.N., Fletcher J.I., Kaufmann T., van Delft M.F., Chen L., Czabotar P.E., Ierino H., Lee E.F., Fairlie W.D., Bouillet P. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- Young M.A., Larson D.E., Sun C.W., George D.R., Ding L., Miller C.A., Lin L., Pawlik K.M., Chen K., Fan X. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.