Summary
Due to differences across species, the mechanisms of cell fate decisions determined in mice cannot be readily extrapolated to humans. In this study, we developed a feeder- and xeno-free culture protocol that efficiently induced human pluripotent stem cells (iPSCs) into PLZF+/GPR125+/CD90+ spermatogonium-like cells (SLCs). These SLCs were enriched with key genes in germ cell development such as MVH, DAZL, GFRα1, NANOS3, and DMRT1. In addition, a small fraction of SLCs went through meiosis in vitro to develop into haploid cells. We further demonstrated that this chemically defined induction protocol faithfully recapitulated the features of compromised germ cell development of PSCs with NANOS3 deficiency or iPSC lines established from patients with non-obstructive azoospermia. Taken together, we established a powerful experimental platform to investigate human germ cell development and pathology related to male infertility.
Keywords: pluripotent stem cells, spermatogonia, infertility, non-obstructive azoospermia
Highlights
-
•
SLCs and haploid cells are formed from PSCs via a feeder- and xeno-free condition
-
•
In-vitro-developed SLCs are enriched with germ cell-specific genes
-
•
NANOS3 deficiency compromises SLC derivation from PSCs
-
•
iPSCs derived from NOA patients display disturbed germ cell development
In this article, Wang and colleagues established a feeder- and xeno-free system to robustly induce human pluripotent stem cells (PSCs) into spermatogonia-like cells. This chemically defined induction protocol faithfully recapitulated the features of compromised germ cell development of PSCs with NANOS3 deficiency or iPSC lines established from patients with non-obstructive azoospermia.
Introduction
Germ cell development is a complicated multiple-step biological process. In mice, primordial germ cells (PGCs), the initial germ cell population, arise from proximal epiblast at embryonic day 6.5, and subsequently migrate to the genital ridge (Ginsburg et al., 1990, Hayashi et al., 2007). Fetal male PGCs transit into gonocytes/prespermatogonia and cease dividing. After birth, prespermatogonia resume proliferation and some become spermatogonial stem cells (SSCs) (Ewen and Koopman, 2010, Hayashi et al., 2007, Saitou, 2009). Spermatocytes then start to develop from differentiating spermatogonia via meiosis in a process called spermatogenesis, and subsequently mature into haploid spermatids (Ewen and Koopman, 2010, Hayashi et al., 2007, Saitou, 2009). Many important genes for mammalian germ cell development have been extensively studied in mice. Among them, BLIMP1 acts as a key regulator in fate specification of the earliest germ cell population PGCs (Ohinata et al., 2005), whereas NANOS3+ and MVH+ cells represent the migrating PGCs and post-migrating gonocytes, respectively, in mice (Tanaka et al., 2000, Tsuda et al., 2003). Thus, null mutations of these genes lead to defects in the formation of PGCs or gonocytes before birth (Ohinata et al., 2005, Tanaka et al., 2000, Tsuda et al., 2003). In addition, PLZF, ID4, DMRT1, and GFRα1 (the receptor of GDNF) are highly expressed in post-natal SSCs, and play important roles in SSC self-renewal (Buaas et al., 2004, Costoya et al., 2004, Helsel et al., 2017, Hofmann et al., 2005, Yang and Oatley, 2014, Zhang et al., 2016). By contrast, SYCP3 (an essential member of the synaptonemal complex in meiosis), PRM1 (a protein that replaces histones in sperm DNA packaging), and ACR (ACROSIN, a major component in the acrosome of haploid spermatids), indicate the formation of meiotic cells in spermatogenesis (Florke-Gerloff et al., 1983, Lammers et al., 1994, Reeves et al., 1989). As such, these genes are well-established markers of germ cells at distinct developmental stages.
In humans, about 15% of couples suffer from male infertility with half due to male factors, but a large number of which is idiopathic (Louis et al., 2013, Oakley et al., 2008). The most commonly identified cause of non-obstructive azoospermia (NOA) so far is attributed to various deletions in long arm of Y chromosome (Yq) (Skaletsky et al., 2003, Tiepolo and Zuffardi, 1976). The top three Yq deletion intervals related to NOA were thus named as azoospermia factors (AZFs; AZFa, AZFb, and AZFc) (Skaletsky et al., 2003, Tiepolo and Zuffardi, 1976). Previously, using a human-to-mouse xenotransplantation model, Ramathal et al. (2014) demonstrated reduced formation of germ cell-like cells (GCLCs) in vivo from induced pluripotent stem cells (iPSCs) of AZF-deleted patients. However, the developmental potential and properties of these GCLCs from diseased NOA-iPSCs are yet to be fully characterized. In addition, although a number of genes (such as NANOS3) are proven crucial during murine germ cell development, their relevance to human reproduction remains to be determined mainly due to the limited access to human tissues and the lack of experimental tools.
Pluripotent stem cells (PSCs) and iPSCs possess the potential to differentiate into all lineages of cells in the body, including germ cells, thereby serving as a valuable tool to investigate regulatory mechanisms underlying germ cell development (Takahashi and Yamanaka, 2006, Thomson et al., 1998). Technical advances in the past several years make it possible to efficiently derive early germline cells from PSCs. For example, PGC-like cells (PGCLCs), can now be robustly induced from PSCs in both human and mice (Hayashi et al., 2011, Irie et al., 2015, Sasaki et al., 2015), in which BLIMP1 plays a conserved and essential role (Aramaki et al., 2013, Irie et al., 2015, Sasaki et al., 2015). In addition, functional spermatids have been generated from murine PGCLCs upon co-culture with neonatal testicular somatic cells and subsequent exposure to morphogens and sex hormones (Zhou et al., 2016). However, in vitro recapitulation of post-natal spermatogenesis, during which SSCs and haploid spermatids develop, remains to be a fundamental challenge in human biology.
Differentiation of PSCs into germ cells are commonly induced using undefined medium supplemented with fetal bovine serum (FBS), which contains unknown quantities of growth factors. The efficiency of germ cell derivation across laboratories is often inconsistent, reflecting the difficulties in standardizing use of serum, feeder cells, and animal products from batch to batch. Easley et al. (2012) recently described a protocol to robustly induce spermatogonium-like cells (SLCs) from human PSCs using defined SSC culture medium, yet still relying on STO feeders and animal products such as BSA. In addition, it remains to be determined whether these in-vitro-derived SLCs bear similarity in gene expression patterns with the in-vivo-developed spermatogonia by global transcription profiling. Further, it is of great interest to investigate whether this protocol can be utilized to recapitulate the human germ cell development under various pathological conditions. In this study, we developed a feeder-, serum-, and animal product-free culture condition, and successfully induced a high percentage of SLCs from human PSCs. Genome-wide expression analyses demonstrated a significant upregulation of germ cell-specific genes in SLCs derived from PSCs compared with in-vivo-isolated human CD90+ SSCs. In addition, we demonstrated that this protocol could be utilized in vitro to evaluate the developmental potential of SLCs from PSCs with depletion of germ cell-specific gene NANOS3, and from iPSC lines derived from NOA patients.
Results
Derivation of SLCs from Human PSCs under a Feeder-, Serum-, and Xeno-free Condition
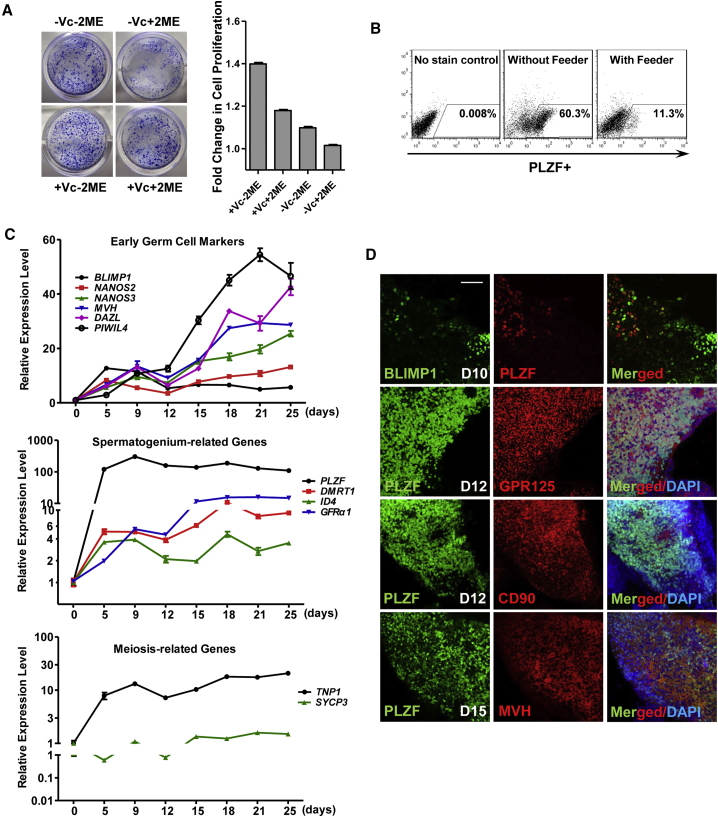
In the protocol Easley et al. (2012) described, STO feeders and SSC culture medium with 18 different components including 2-mecaptoethanol were utilized. However, with this protocol, we found a large portion of PSCs went through apoptosis during induction (Figure S1A), which may partially explain the substantially varied derivation efficiency across different iPSC cell lines in our hands. As 2-mecaptoethanol was shown to affect cell proliferation (Chen et al., 2011), whereas vitamin C may promote survival of SSCs and expression of germline genes (Blaschke et al., 2013, Wang et al., 2014), we optimized the culture medium by removal of 2-mercaptoethanol and inclusion of vitamin C. Using crystal violent staining and an MTT assay to indicate cell survival, as well as apoptosis analyses to monitor cell death, we demonstrated that a healthy differentiation culture was easily maintained across different PSC lines with more viable cells and fewer cell deaths in the optimized protocol (Figures 1A and S1A–S1C). More importantly, we were able to efficiently induce PLZF+ SLCs in the optimized medium on gelatin without any feeder cells (Figure 1B). To minimize the use of animal products in future clinical therapy, and to reduce induction variability, we further replaced BSA with 0.2%–3% xeno-free serum replacement without animal products, and obtained the comparable percentage of PLZF+ SLCs with the ones using BSA (Figure S1D, top panel).
Figure 1.
Feeder-, Xeno-, and Serum-free Induction of SLCs from hESCs
(A) Crystal violent staining of SLC colonies derived from hESCs (H1) under different conditions. Vc, vitamin C; 2ME, 2-mercaptoethanol. Cell numbers were measured by MTT assay and shown on the right. Relative absorbance of samples at 490–650 nm was calculated. Data are represented as mean of fold changes (compared with the one without Vc and 2ME) ± 1 SEM from three independent experiments.
(B) Flow cytometry analyses of PLZF+ cells on differentiated H1 hESCs under different conditions.
(C) Real-time RT-PCR analyses of germ cell-specific genes at various time points along H1 hESC differentiation. Data are represented as mean ± 1 SEM from three independent experiments.
(D) IF assays of germ cell-specific genes performed at various time points along H1 hESC differentiation. Day 10, D10; Scale bar, 50 μm.
See also Figure S1.
We next characterized the gene expression profiles along the time course of SLC induction. We found that the expression of BLIMP1, the early PGC marker, peaked at days 5–9 post-induction, followed by increased expression of key regulators that indicate formation of post-migrating gonocytes, including MVH, DAZL, NANOS2, NANOS3, and PIWIL4 between days 12 and 25 (Figure 1C, top panel). The levels of spermatogonial markers such as PLZF, DMRT1, ID4, and GFRα1 were also significantly elevated during induction (Figure 1C, middle panel). Interestingly, TNP1, the gene indicating formation of haploid cells was increased as well (Figure 1C, bottom panel), suggesting the emergence of meiotic spermatocytes and putative spermatids. Consistent with upregulated transcript levels of these germline genes, immunofluorescence (IF) assays revealed BLIMP1+ cells at day 10 post-induction (Figure 1D). These BLIMP1+ were not co-expressed with PLZF, suggesting that PLZF+ cells are not PGCs. In contrast, the germ cell-specific proteins MVH and DAZL were readily detected from the differentiated population on day 15 post-induction (Figure S1E). MVH and markers of spermatogonia (e.g., GPR125 and CD90) were largely co-stained with PLZF from days 12 to 15 during PSC differentiation (Figure 1D), indicating these PLZF+ cells are SLCs.
Putative Haploid Cells Developed from SLCs
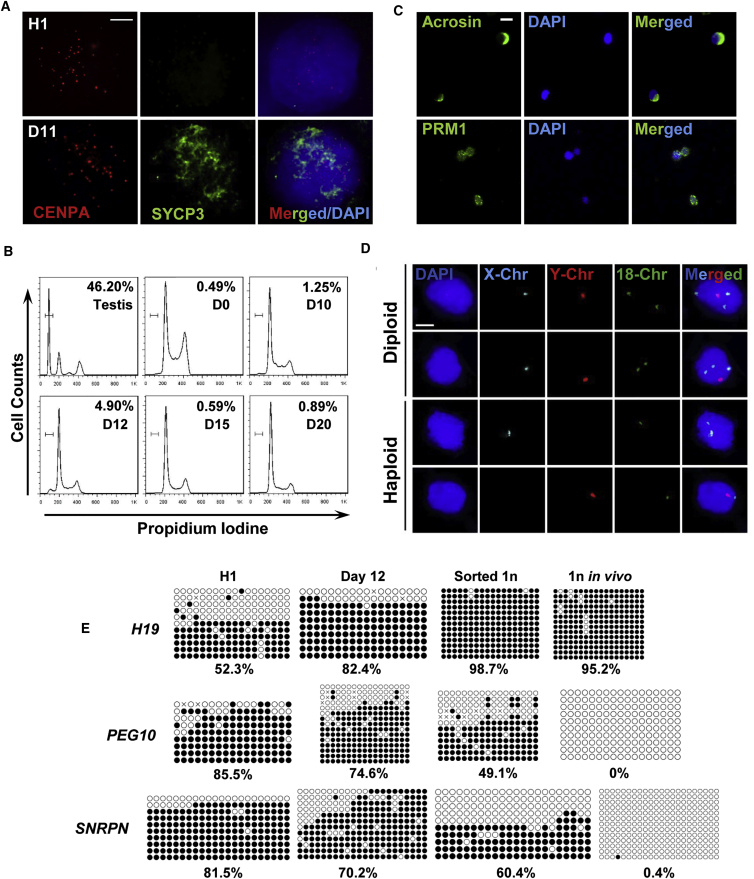
We next examined if these SLCs could go through meiosis to generate haploid spermatids. While it is difficult to obtain definitive evidence for proper segregation of homologous chromosomes, we performed DNA spreading assays using antibodies against SYCP3 and CENPA (a component of centromere) as markers of meiosis. A very small fraction of the differentiated cells (<1%) exhibited positive staining of SYCP3, but not any of their parental H1 human embryonic stem cells (hESCs) (Figure 2A). Further, the flow cytometry assay detected less than 5% haploid cells among the differentiated cells around day 12 post-induction from ESCs (Figures 2B and S1D, bottom panel). These haploid cells were collected and subjected to IF staining. Consistent with the data described above, some of these sorted haploid cells were stained positive for ACR, a unique marker for acrosome with perinuclear localization in spermatids, and as well as for PRM1, the protein protamine replacing histone in spermatids (Figure 2C). DNA content of haploid population was further analyzed via fluorescence in situ hybridization (FISH). We clearly observed that these sorted haploid cells were stained exclusively with either X or Y sex Chr, and one copy of Chr 18, or Chr 7; by contrast, the diploid cells isolated from same culture harbored both X and Y chromosomes, as well as two copies of Chr 18 or Chr 7 in the same nuclei (Figures 2D and S2C), thus confirming these sorted cells with true haploid property. In summary, our data suggest that a small proportion of SLCs induced under this feeder-free condition may have the potential to go through meiosis and form haploid cells.
Figure 2.
Human SLCs Undergo Meiosis and Form Putative Haploid Cells
(A) Meiosis DNA spreading assays were performed with antibodies against CENPA and SYCP3 on H1 hESCs and their derivative SLCs at day 11 post-differentiation. Scale bar, 5 μm.
(B) Flow cytometry analyses of percentage of haploid cells formed from H1 ESCs at various time points along differentiation. Mouse testes were used as control.
(C) IF of key proteins that participated in meiosis, including ACR and PRM1, on sorted haploid cells differentiated from H1 ESCs at day 12. Scale bar, 10 μm.
(D) FISH analyses of sex chromosomes and Chr 18 on diploid cells and haploid cells sorted from the same differentiation culture at day 12. Scale bar, 5 μm.
(E) Bisulfate sequencing on imprinting loci of parental H1 ESCs, SLCs, sorted haploid cells at day 12 post-differentiation, and spermatids collected from biopsied human testes. White circles, unmethylated CpG; black circles, methylated CpGs.
See also Figure S2.
One unique feature of a germ cell is its characteristic changes in imprinting status during development: DNA methylation at imprinted loci is removed at the PGC stage and re-established in spermatogenesis (Bird, 2002, Monk et al., 1987). With bisulfate sequencing, we demonstrated that the paternal imprinted loci of H19 were highly methylated in SLCs and in sorted haploid cells, as one would expect for in vivo collected spermatids (Figure 2E). Similarly, the DNA methylation levels of maternal imprinted PEG10 and SNRPN regions were decreased compared with those of undifferentiated parental H1 ESCs. However, these maternal imprinted loci were not completely demethylated as the in vivo haploid control were (Figure 2E). Therefore, these in-vitro-derived haploid cells may need to be further reprogrammed to fully mirror their in vivo counterparts.
Genome-wide Analyses of In-Vitro-Derived SLCs and Haploid Cells from PSCs
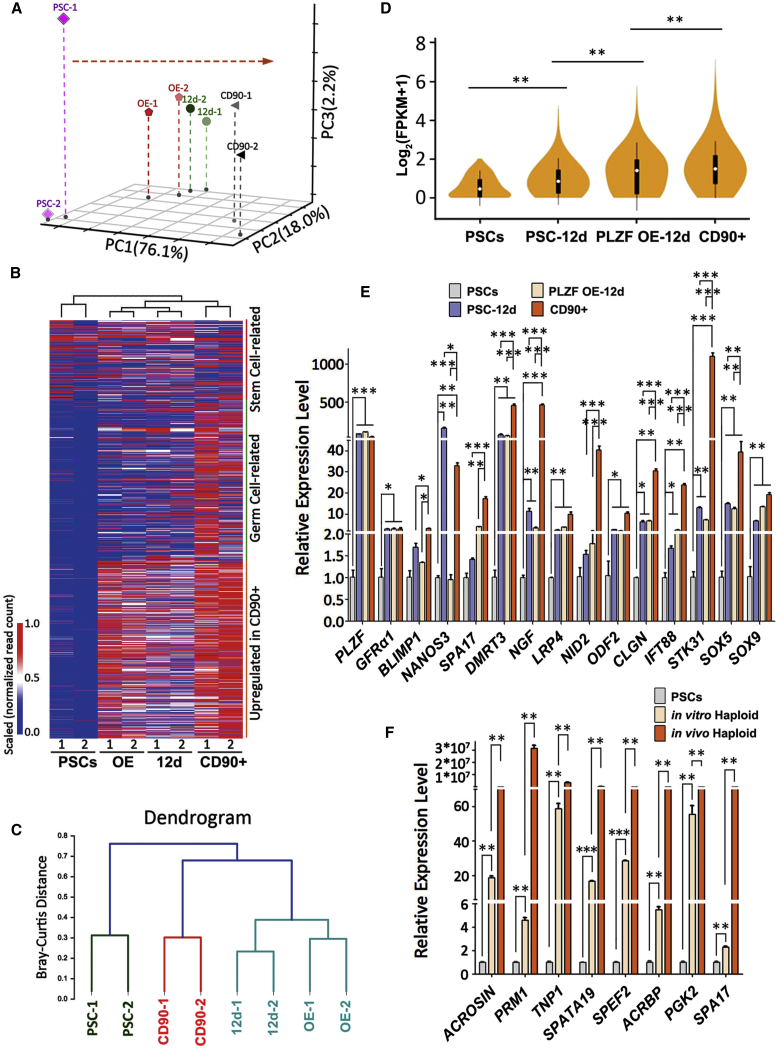
To determine if SLCs derived from PSCs have similar gene expression patterns as their in vivo counterparts at genome-wide level, CD90+ cells enriched for spermatogonia were isolated from biopsied human testes of two obstructive azoospermia (OA) patients, and their transcript profiles were compared with those of in-vitro-induced SLCs via RNA sequencing. We also included PSCs with enforced PLZF expression in this assay to explore the potential role of PLZF during germ cell derivation. We did not observe much significant alteration of SCL formation or gene expression profiles upon PLZF overexpression (Table S1), suggesting that PLZF itself is not enough to drive PSCs to become SLCs. Principal-component analysis showed a directional and progressive transition of cellular properties of SLCs toward in vivo CD90+ spermatogonia (Figure 3A; Table S1).
Figure 3.
Gene Expression Analyses on SLCs and Sorted Haploid Cells Derived from iPSCs
(A) Principal-component analysis (PCA) analyses on PSCs, SLCs from day 12 of PSC differentiation (12d) with or without PLZF overexpression (OE), and CD90+ spermatogonia sorted from biopsied testes of OA patients.
(B) Dendrogram hierarchy clustering performed on PSCs, SLCs, and CD90+ cells sorted from biopsied testes of OA patients.
(C) Heatmap on 1,815 transcripts enriched with germ cell-specific factors and genes highly expressed in CD90+ cells were analyzed on PSCs, SLCs, and CD90+ cells collected from biopsied samples.
(D) Violin map of transcript expression of germ cell-specific genes were analyzed on PSCs, SLCs, and sorted CD90+ cells. ∗∗p < 0.01.
(E) Real-time RT-PCR analyses of germ cell-specific genes in PSCs or SLCs from day 12 of PSC differentiation with and without PLZF overexpression, compared with in-vivo-isolated CD90+ spermatogonia.
(F) Real-time RT-PCR analyses of genes involved in meiosis on PSCs, or haploid cells sorted from differentiated PSCs, compared with in-vivo-formed round spermatids isolated from two OA patients. (E and F) Data are represented as mean ± 1 SEM from four independent experiments.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S3.
We next investigated whether those SLCs were enriched with germ cell-related genes. After interrogating 29 online gene ontology (GO) terms from EMBL_EBI, we pulled out 761 transcripts with purported roles in germ cell development and low expression (with fragments per kilobase of transcript per million mapped reads [FPKM] < 0.23) in undifferentiated PSCs. In addition, we included 350 transcripts that were related to stem cells from GO terms and 926 transcripts that were highly expressed in the CD90+ cells from this study (Table S3). Therefore, in total the levels of 1,815 transcripts (1,101 unique genes) were examined on both in-vitro-derived SLCs and in vivo CD90+ cells (Figure S3B; Table S3). Both cluster and heatmap analyses revealed that the expression patterns of germline genes in PLZF+ cells closely resembled those of CD90+ spermatogonia, but not their parental PSCs (Figures 3B and 3C; Tables S2 and S3). Upon excluding the 350 transcripts that were expressed in PSCs, violin map analyses clearly demonstrated an increased expression pattern of germ cell-specific transcripts in SLCs and PLZF-overexpressed cells toward CD90+ spermatogonia (Figure 3D). The key genes enriched in SLCs included PLZF, GFRα1, ID4, NANOS3, DMRT1/3, and PIWIL4, etc. (Figure S3C), all of which are important for germ cell development (Buaas et al., 2004, Carmell et al., 2007, Costoya et al., 2004, Helsel et al., 2017, Looijenga et al., 2006, Tsuda et al., 2003). We confirmed the high expression of these genes with independent real-time RT-PCR assays (Figures 3E, S3D, and S3E).
We also examined the transcript levels of genes involved in the development of ectoderm (45 transcripts), mesoderm (227 transcripts), and endoderm (109 transcripts), respectively (Table S4). Interestingly, 40%–50% of genes related to mesoderm and 36%–39% to endoderm development were upregulated in both CD90+ cells and PSC-derived SLCs, much higher than the percentage of genes (24%) involved in ectodermal lineage specification (Figure S3F). These observations thus suggest a potential correlation of germ cell development and endo-mesodermal differentiation.
We further sorted PSC-derived haploid cells, and compared their expression patterns with those of in-vivo-isolated haploid spermatids from OA patients. Interestingly, the expression of 98 meiosis-indicative transcripts, including PRM1, TNP1, and SPATA19, etc., was significantly upregulated in haploid cells from both sources (Figures 3F and S3G; Table S3), albeit at relatively lower transcript levels observed from the in-vitro-derived haploid population compared with the ones developed in vivo. Taken together, our results demonstrate that the in-vitro-derived SLCs and haploid cells from PSCs bear high similarity with their in vivo counterparts in the expression of key genes during germ cell development.
NANOS3 Deficiency Compromises the Formation of SLCs from PSCs
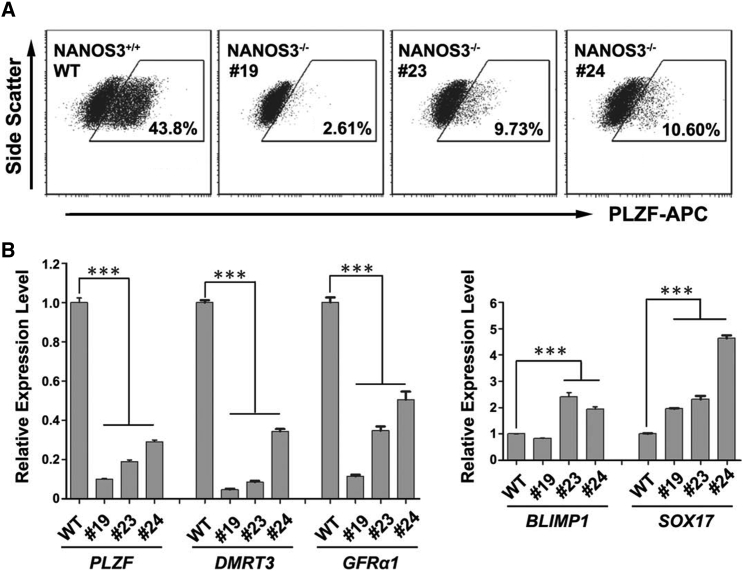
NANOS3 is a conserved germ cell-specific gene, and has been used as a marker for human PGCLCs (Irie et al., 2015, Tsuda et al., 2003). We found that NANOS3 was highly expressed during PSC differentiation toward SLCs (Figure 1C), and thereby indicates a role of NANOS3 in SLC derivation. To investigate whether our differentiation system can reflect altered germ cell development, and to delineate the function of NANOS3 in this process, we created several NANOS3 knockout H1 ESC lines via CAS9/CRISPR techniques (Figures S4A and S4B) (Ran et al., 2013). We compared three cell lines with large deletions in the first exon of NANOS3 gene against a control line without any mutation (Figures S4A and S4B). Marked reduction of NANOS3 expression in these mutated lines was confirmed by real-time RT-PCR assays (Figure S4C). In addition, compared with wild-type control PSCs, all three lines with NANOS3 deletion displayed significantly fewer PLZF+ cells during differentiation (Figure 4A). Consistently, the transcript levels of PLZF, DMRT3, and GFRα1 were dramatically reduced in NANOS3-deficient lines (Figure 4B). Interestingly, expression levels of two genes (i.e., BLIMP1 and SOX17) in PGC formation were largely upregulated (Figure 4B), suggesting a possible accumulation of early germ cell progenitors due to blockage of SLC formation. Taken together, these data prove that our in vitro differentiation system is able to recapitulate disturbed germ cell formation upon loss of function of key germline genes such as NANOS3.
Figure 4.
NANOS3 Deficiency Affects Formation of SLCs from PSCs
(A) Flow cytometry analyses of PLZF+ cells on wild-type H1 ESCs or ESCs with NANOS3 null mutations at day 13 post-differentiation.
(B) Real-time RT-PCR analyses on wild-type H1 ESCs or ESCs with NANOS3 deletions at day 13 post-differentiation. Data are represented as mean ± 1 SEM from three independent experiments. ∗∗∗p < 0.001.
See also Figure S4.
Establishing iPSC Lines from NOA Patients
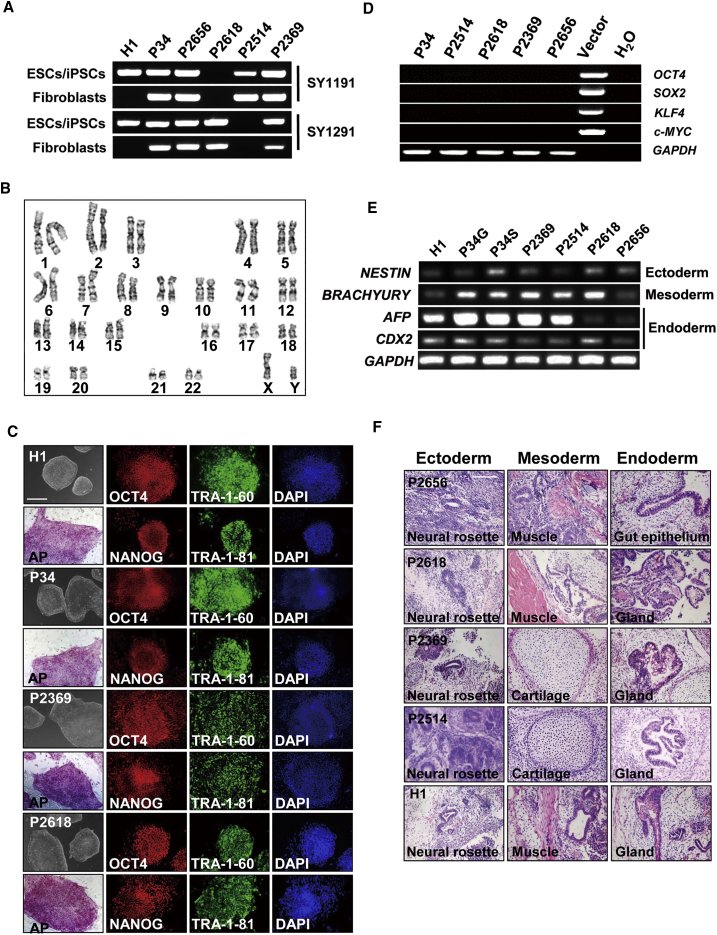
To further evaluate whether our protocol can be used to model pathological germ cell development in vitro, we obtained six iPSC lines from four NOA patients and two male controls. Of these, five lines were derived from skin biopsy samples with classical retroviral infection, and one (i.e., P24) from peripheral blood cells using electroporation with non-integrated episomal plasmids to introduce core pluripotency factors (Wen et al., 2016). Among four patients, two (P2369 and P2656) were diagnosed as NOA with Sertoli cell-only syndrome, whereas two (P2514 and P2618) had microdeletions at the AZFc locus in both parental fibroblasts and established iPSC lines, as detected by PCR-based analyses of Yq genomic markers (Yq sequence-tagged sites) (Figures 5A and S5A). We predicated that these microdeletions would lead to reduced DNA copies of multiple coding genes in the AZFc region according to the genomic map (Figure S5B). Further qPCR analyses indeed demonstrated fewer copies of BPY2, DAZ, and CDY1 in fibroblasts and iPSC lines from patients P2514 and P2618 (Figure S5C). In addition, these iPSC lines exhibited the same DNA fingerprinting of short tandem repeats as their source somatic cells (Figure S5D). Collectively, our data demonstrate that these iPSC lines faithfully reflect the genotypes of patients and control donors.
Figure 5.
Establishing iPSCs from NOA Patients
(A) PCR detection of Y-Chr microdeletion with sequence-tagged site in control hESCs (H1), derived iPSC lines, and their parental fibroblasts.
(B) A representative karyotyping image from iPSC line P2514 derived from NOA patients.
(C) Morphology, AP staining, and IF analyses of pluripotency factors (including OCT4, NANOG, TRA-1-60, and TRA-1-81) on established iPSC lines and control hESCs (H1). Scale bar, 100 μm.
(D) Exogenous expression of transgenes used in the derivation of iPSC lines.
(E) RT-PCR analyses of key genes in three germ layer development on embryoid bodies differentiated from control hESCs (H1) and established iPSC lines.
(F) Histology analyses on teratoma sections from control hESCs (H1) and iPSC lines. Scale bar, 50 μm.
See also Figures S5 and S6.
These iPSC cells are morphologically similar to hESCs. They proliferated every 5–7 days per passage and displayed normal karyotypes (22, XY) as human cells (Figures 5B and S5E). They exhibited high alkaline phosphatase activities and expressed pluripotency markers, including TRA-1-81, TRA-1-60, OCT4, and NANOG, similar to the existing hESC line H1 at both transcript and protein levels (Figures 5C, S6A, and S6B). No exogenous gene expression was detected in these iPSCs at the passages that we used for differentiation (Figure 5D).
During spontaneous differentiation of these iPSC lines via embryoid body formation, we observed expression of key genes in three-germ-layer development at levels that were comparable with differentiated H1 ESCs under the same conditions (Figures 5E and S6C). All iPSC lines formed teratomas within 2 months post-injection into nude mice, and derivatives from three germ layers were clearly observed from tumor sections (Figure 5F), suggesting that these iPSCs possess normal differentiation potential as ESCs. Taken together, these data confirm that we have successfully established iPSC lines from NOA patients and controls.
Modeling Germ Cell Development from NOA-iPSC Lines
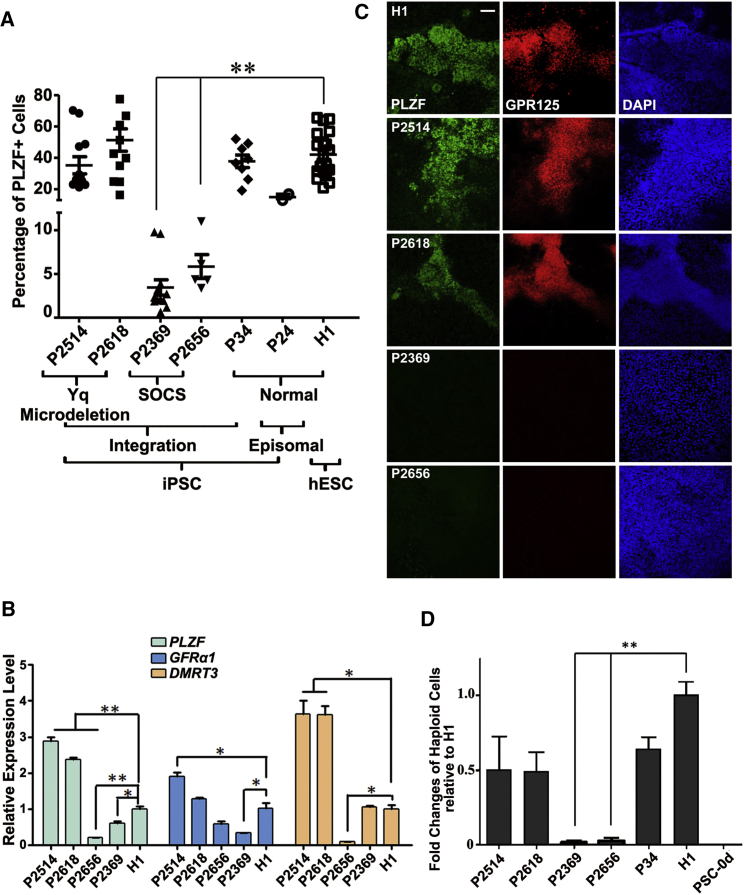
We next investigated the potential of NOA-iPSCs in germ cell development in vitro. Interestingly, we found a significantly reduced percentage of PLZF+ cells upon induction in two iPSC lines (P2369 and P2656) from NOA patients with Sertoli cell-only syndrome (Figure 6A), albeit that their differentiation ability into three germ layers remained normal. Consistently, their transcript levels of PLZF, GFRα1, or DMRT3 were also significantly decreased (Figure 6B). No PLZF+/GPR125+ cells at day 12 post-differentiation in these two lines were detected by IF (Figure 6C). In addition, very few haploid spermatids were developed from these two differentiated NOA-iPSC lines (Figure 6D). By contrast, the other two NOA-iPSC lines with AZFc microdeletion displayed a relatively normal amount of PLZF+ SLCs and slightly fewer haploid cells, compared with normal PSC lines (Figure 6D), in agreement with their relatively mild symptoms of blocked spermatogenesis in clinical diagnosis. In summary, these data demonstrate that our protocol provides a feasible tool to evaluate both physiological and pathological processes of germ cell developments in vitro.
Figure 6.
Evaluating Germ Cell Developmental Potential from NOA-iPSCs
(A) Percentage of PLZF+ cells analyzed by flow cytometry on differentiated NOA-iPSCs, control iPSCs, and H1 ESCs, at days 12–15 post-induction. At least three iPSC clones from each NOA patient and P34 control were independently examined. Each dot in the graph represents one independent differentiation experiment.
(B) Real-time RT-PCR analyses of spermatogenesis-related genes in SLCs from differentiated NOA-iPSCs and control H1 ESCs.
(C) IF assays of spermatogonium-related genes performed at day 12 post-differentiation of NOA-iPSCs and control H1 ESCs. Scale bar, 50 μm.
(D) Flow cytometry analyses of putative haploid spermatids from differentiated NOA-iPSCs at day 12 compared with wild-type hESCs (H1) or control iPSCs (P34). iPS 0d: undifferentiated iPSC line P34. (B and D) Data are represented as mean ± 1 SEM from more than six independent experiments.
∗p < 0.05; ∗∗p < 0.01.
Discussion
Development of in vitro differentiation models of PSCs into germ cells will greatly facilitate research on human reproduction. Despite recent advances in early PGC derivation (Hayashi et al., 2011, Irie et al., 2015, Sasaki et al., 2015), formation of late spermatogonia from PSCs and in vitro recapitulation of human male infertility remain as major challenges. In this study, we circumvented the problem of cell death in the Easley et al. (2012) protocol with medium optimization by removing 2-mercaptoethanol and adding vitamin C. We successfully developed a standardized and reproducible feeder- and xeno-free protocol that enables efficient differentiation of human PSCs toward SLCs. Using this system, we revealed that NANOS3 deficiency compromised SLC derivation from human PSCs. In addition, we demonstrated dramatically decreased formation of SLCs and haploid cells upon differentiation of NOA-iPSC lines from patients with Sertoli cell-only syndrome. These findings prove our system to be a powerful tool to investigate molecular pathways that govern germ cell development and to understand pathological causes of human male infertility.
In our model, in-vitro-derived SLCs displayed key features of spermatogonia, with upregulation of germline genes and specific epigenetic imprinting patterns, albeit that their global transcript dynamics were somewhat different from in-vivo-isolated CD90+ cells. This may be the result of an incomplete induction of PSCs to germ cell lineages; alternatively, this can also be explained in part by the heterogeneity of differentiated cell populations. To date, few specific surface markers have been defined to identify prospermatogonia or spermatogonia at distinct developmental stages in humans. Although PLZF and CD90 are also expressed on somatic cells, these PLZF+ SLCs largely co-stained with the germ cell-specific marker MVH, suggesting their germline lineages. These PLZF+ SLCs likely represent a heterogeneous spermatogonial population enriched from the process of PSC differentiation. In addition, these SLCs exhibited significantly increased expression in key genes for PGC development (i.e., BLIMP1, MVH, DAZL, NANOS2, and NANOS3) and spermatogonial formation (i.e., PLZF, ID4, GFRα1, and DMRT1). Proteins for germ cell-specific genes MVH and DAZL were readily detected in the differentiated cell population on day 15 post-PSC induction. As one of the most important criteria to define germ cell lineages of derived spermatogonia is to examine whether they contribute to spermatogenesis in vivo, future investigation is required to determine whether these SLCs can reconstitute testes without endogenous germ cells.
The current differentiation protocol was optimized mainly for SLC formation, and the efficiency of meiosis is therefore not robust. Approximately 5% of haploid cells were generated using our system. However, the maternal imprinting loci PEG10 and SNRPN in these haploid cells were not completely demethylated, as one would expect for their in vivo counterparts. In addition, the expressions of genes involved in meiosis were not upregulated to the level as sorted spermatids from testes. Therefore, further reprogramming for these in-vitro-derived haploid cells may be needed to fully mirror their in vivo counterparts. On the other hand, unlike their parental PSC lines or diploid cells collected from the same culture, these cells contained either the X- or Y-Chr and a single copy of Chr 7 or Chr 18, proving that they are true haploid cells and unlikely to come from aneuploidy caused by prolonged in vitro culture. Our data therefore suggest that a small portion of SLCs may have the capability to go through meiosis with appropriate induction; however, ultimately thorough functional evaluations are required to determine whether these in-vitro-derived haploid cells are competent spermatids to fertilize human oocytes. A recent report showed that mouse PGCLCs were able to complete meiosis upon co-culture with neonatal testicular somatic cells and sequential exposure to morphogens and sex hormones (Zhou et al., 2016). It will therefore be of great interest to explore whether these protocols for PGCLC derivation (Irie et al., 2015, Sasaki et al., 2015), spermatogonial induction (Easley et al., 2012 and our study), and functional spermatid generation (Zhou et al., 2016) can be implemented synergistically to fully recapitulate the process of human germ cell development in vitro.
Of the four NOA-iPSC lines we investigated, two were from NOA patients with clinically diagnosed Sertoli cell-only syndrome. We demonstrated that these lines displayed significantly reduced formation of SLCs and haploid cells. This is unlikely due to differentiation variability among iPSC lines as the results were confirmed with at least three independent iPSC clones from each NOA patient. Furthermore, we utilized two iPSC derivation protocols, one of which was performed on peripheral mononuclear blood cells from a male control with non-integrating plasmids (Wen et al., 2016). We obtained comparable efficiency in the SLC formation from control iPSCs using both protocols, and thus excluded potential impacts from residual exogenous genes on germ cell development that were introduced by iPSC derivation. In addition, deficiency of germ cell-specific gene NANOS3 indeed compromised SLC formation from PSCs, and thus prove that our PSC differentiation protocol is able to model the pathological germ cell development in vitro.
In the current study, relatively normal development of germ cells from two iPSC lines with AZFc microdeletion were observed. This is not surprising given that the human AZFc locus harbors multiple copies of coding genes, for example, four copies of DAZ, two of CDY1, and three of BPY2 (Skaletsky et al., 2003, Tiepolo and Zuffardi, 1976). In addition, some of these Y-Chr genes have autosome homologs. Examples include CDYL at Chr 13 as a homolog for CDY1, and DAZL at Chr 3 for DAZ (Skaletsky et al., 2003, Tiepolo and Zuffardi, 1976). Therefore, symptoms due to AZFc microdeletion are usually correlated with the specific copies of genes that are deleted, and may be compensated by their autosome homologs. Such gene amplification and redundancy during evolution not only reflect the physiological importance of these genes, but also allow for diversification and precise specification of their roles in human. However, this also makes it difficult to decipher the exact biological functions of these genes solely based on animal models. Therefore, our PSC in vitro differentiation protocol provides a much needed experimental platform to systematically investigate the unique roles of human-specific genes in germ cell development. Notably, DAZ only exists in human and primate genomes (Kee et al., 2009). Although its homolog DAZL plays an indispensable role in mouse SSC formation, consistent with the findings in this study, patients with AZFc microdeletion (or with loss of DAZ gene) often have SSCs remaining in testes despite oligospermia at relatively later adulthood (Nickkholgh et al., 2015, Tiepolo and Zuffardi, 1976). These suggest that genes (e.g., DAZ) at the AZFc region may not play eminent roles for the fate specification and self-renewal of human SSCs.
Recent studies just began to elucidate molecular players that govern the derivation of early germ cell progenitors from human PSCs. For example, Irie et al. (2015) found that activation of SOX17, a gene involved in endodermal fate specification, was essential for human PGCLC formation from naive PSCs, whereas Sasaki et al. (2015) reported that PGCLCs developed robustly from human PSCs through a stage called incipient mesoderm-like cells. The current study demonstrated that genes for mesoderm and endoderm differentiation were largely upregulated in these SLCs, supporting potential roles of meso-endodermal specifiers in human germ cell development. Although BLIMP1 and NANOS3 were both expressed in PGCLCs from mouse and human PSCs (Irie et al., 2015, Tsuda et al., 2003), the expression of BLIMP1 peaked earlier than that of NANOS3, and was largely augmented upon NANOS3 deficiency in our culture. In addition, only about 5% BLIMP1+ cells developed from PSCs, much fewer than PLZF+ SLCs (>50% in most lines). It is possible that BLIMP1+ PGCLCs only transiently appear before NANOS3+ germ cells during PSC differentiation. NANOS3 deficiency may reduce downstream SSCLC formation and in turn causes blocked PGCLC differentiation. Alternatively, it is also possible that NANOS3 but not BLIMP1 directly contributes to SLC development from PSCs. Future investigation into pathways that regulate human germ cell development will provide molecular and mechanistic insights into SSC fate specification.
Experimental Procedures
Testicular Biopsy, Skin Specimens, and Periphery Blood Sample Collection
Testicular specimens were biopsied from three OA patients and immediately placed aseptically in PBS containing 1,000 units/mL penicillin and streptomycin (Gibco) for further analyses. Skin samples were obtained from four NOA patients and one male donor, whereas 2 mL peripheral blood was collected from the other male control. The diagnosis of OA and NOA patients were confirmed by pathological examination of testes via biopsy. This study was approved by the Institutional Ethical Review Committee of Ren Ji Hospital (license number 2012-01), Shanghai Jiao Tong University School of Medicine, and all participants provided written consent.
Cell Culture and hESC/iPSC Differentiation
Participant-specific PSCs were induced according to published protocols from skin biopsy samples, with the exception of iPSC line P24, which was established from peripheral blood cells (Takahashi and Yamanaka, 2006, Wen et al., 2016). Human ESC line H1 and iPSC lines were maintained in chemically defined Essential 8 medium (STEMCell Technologies). Cells were passaged every 5–7 days and dissociated by 1 mg/mL Collagenase IV (Millipore). We started to induce PSCs differentiation into germ cells in differentiation medium when they reached 80%–90% confluence (components of differentiation medium mentioned below all come from Invitrogen/Gibco, unless otherwise noted): A-MEM, 2 mM L-glutamine, 1× Insulin-Transferrin-Selenium-X, 0.2% BSA or substituted by 0.2%–3% KnockOut SR XenoFree CTS (Gibco), 1 ng/mL recombinant human basic fibroblast growth factor (bFGF), 20 ng/mL recombinant human GDNF (Sino Biological), 0.2% chemically defined lipid concentrate, and 200 μg/mL vitamin C (Sigma). Medium was changed every day until the collection of cells for analyses at each time point. No passage of cells was performed during differentiation. About 20%–30% of cells might have gone through cell death upon differentiation before day 6, but much less apoptosis was observed afterward. PLZF+ cells emerged around day 6, and by day 12, usually more than 50% of cells were PLZF+. We collected cells for analyses at days 12–15 post-differentiation, unless otherwise noted. For comparison performed in Figure S6C, PSCs were differentiated via embryoid body formation in serum containing medium (Knockout DMEM supplied 20% FBS without bFGF, all from Gibco). PLZF overexpressing cell line was established by introducing PLZF transgene driven by EF1α promoter into H1 ESCs. For construction NANOS3 knockout PSC lines, H1 ESCs were infected with vector in which the expression of Cas9 could be induced by doxycycline. The lentiviral vector containing gRNAs specifically against NANOS3 and puromycin transgene were introduced afterward for selection of drug-resistant colonies. The region spanning the gRNA-targeted region was amplified from genomic DNA to confirm individual colonies that harbored null mutations of NANOS3 at both alleles. Primers and gRNA used in this study are listed in Table S5.
IF Assays
Cells were seeded and differentiated on cover slides or placed on slides by cytospin preparation, and fixed in 4% paraformaldehyde (PFA). IF was performed using the following primary antibodies: TRA-1-81 (sc-21706), TRA-1-60 (sc-21705), OCT4 (sc-9081), and PRM1 (sc-23107) from Santa Cruz Technologies; PLZF (MAB2944) from R&D; CD90 (ab133350), MVH (ab13840), and ACR (ab3983) from Abcam; GPR125 (GeneTex, GTX51219); NANOG (CST no. 3580S) and BLIMP1 (CST no. 9115S) from Cell Signaling Technology. The fluorescein-conjugated secondary antibodies (Jackson ImmunoResearch) were used at 1:200 dilution. Images were obtained with a Leica confocal microscope.
Flow Cytometry
Cells were dissociated by TrypLE, and fixed by 4% PFA for 20 min at room temperature. The fixed cells were suspended in fluorescence-activated cell sorting (FACS) buffer (PBS with 5% fetal calf serum, 0.2% Triton-100, and 0.5% Tween 20) for 25 min and centrifuged at 500 × g for 5 min before incubation with a PLZF antibody (R&D, MAB2944, 1:160 dilution) or a CD90 (BioLegend, 328109) antibody. Subsequently, the cells were washed with FACS buffer, detected with APC or PE-conjugated anti-mouse second antibody. For analyses of DNA content, dissociated cells were fixed with 0.5% PFA in PBS for 15 min, suspended in 0.1% Triton X-100 in PBS for 3 min, centrifuged at 200 × g, stained with 50 μg/mL PI (Invitrogen) and 100 ng/mL RNase (Invitrogen) at 4°C, and then analyzed with a FACS Calibur (BD Biosciences). To isolate haploid cells from in-vitro-differentiated PSCs or from patients' biopsied testes, dissociated cells were stained at 37°C for 10 min in differentiation medium containing 15 μg/mL Hoechst 33,342 (Life Technologies) and then sorted by a FACS Aria II (BD Biosciences). Apoptosis analyses were performed using FITC-Annexin V Apoptosis Detection Kit from BD Pharmingen according to the manufacturer's instructions.
FISH
FISH assays were performed according to published protocols (Easley et al., 2012). In brief, sorted cells were cytospun onto slides and fixed with Carnoy's fixative. The cytology specimens were further prepared using ZytoLight FISH-Cytology Implementation Kit according to the manufacturer's instructions (ZytoVision, Bremerhaven, Germany). Human chromosomes X, Y, and 7 were detected using ZytoLight CEN X/Y Dual Color Probe and Cen7 Probe kit (ZytoVision). Triple probes and detection kits for Chr X, Y, and 18 were purchased from Abbott Molecular. Nuclei were observed with co-staining of DAPI.
Meiotic DNA Spreading Assays
Meiotic DNA spreading assays were performed according to published protocols (Durruthy Durruthy et al., 2014, Kee et al., 2009) with minor modifications. In brief, the dissociated cells were suspended in hypo-extraction buffer (30 mM Tris, 50 mM sucrose, 17 mM Na citrate, and 5 mM EDTA, pH 8.3, Protease Inhibitor Cocktail/Santa Cruz Tech) and incubated on ice for 30 min. Following centrifugation, the cells were resuspended in 20 μL hypoextraction buffer with 60 μL 100 mM sucrose and cytospun at 180 × g for 3 min onto a slide. The cells were fixed in 4% PFA, and incubated with 0.04% photoflo (Kodak) for 5 min. Air-dried slides were blocked with 3% BSA for 30 min at room temperature and stained with SYCP3 (ab15093) and CENPA (ab13939) antibodies (both from Abcam) at 4°C overnight, and with fluorescein-conjugated secondary antibodies thereafter. Images were captured with a Leica confocal microscope.
Bisulfate Sequencing
Genomic DNA was extracted using an E.Z.N.A. MicroElute Genomic DNA Kit (Omega Bio-Tek) according to the manufacturer's protocol. About 500 ng to 1 μg of genomic DNA was bisulfate-treated followed with PCR according to published protocols (Kerjean et al., 2000, Kim et al., 2007). The PCR products were cloned into pEASY-T5 Zero Cloning Vector (TransGen Biotech), and 15 clones were sequenced for each sample. Primers are listed in Table S5.
RT-PCR, PCR, and Real-Time PCR
Total RNA from cultured cells was extracted by TRIzol and reverse-transcribed using a cDNA Synthesis Kit (TaKaRa Biotechnology). PCR and real-time PCR were performed as described previously (Zhang et al., 2014) on a Bio-Rad thermal cycler (Bio-Rad) and a Stratagene Mx3000P (Stratagene, CA, USA), respectively. Primers are listed in Table S5.
Teratoma Formation Assay
A confluent 10-cm dish containing 1–5 × 106 PSCs were digested into small clumps by collagenase IV (Gibco), resuspended with Matrigel (Gibco), and then injected subcutaneously in severe combined immunodeficient (SCID) mice, as previously described (Zhang et al., 2014). Visible tumors were formed between 5 and 8 weeks post injection and teratoma sections were examined by hematoxylin-eosin staining. All animal experimental procedures were conducted in accordance with the local Animal Welfare Act and Public Health Service Policy with approval from the Committee of Animal Experimental Ethics at East China Normal University (protocol no. m20170320).
Statistical Analysis
Data are presented as mean ± SE (SEM). All experiments were performed independently for more than three times unless otherwise stated. Statistical analyses were conducted with unpaired Student's t test using Prism Graphic software.
Other experimental methods including RNA sequencing and analyses were described in details in Supplemental Experimental Procedures.
Author Contributions
Y.Z., S.Y., D.L., and Y.W. designed the research and wrote the paper; Y.Z., S.Y., D.L., P.W., J.F., Q.M., R.K., L.S., X.G., W.C., W.D., Z.Z., and H.C. performed experiments; Z.Z., H.C., and Z.L. provided the clinically biopsied samples; Y.Z., W.Y., J.Z., and Y.W. analyzed the data.
Acknowledgments
We would like to thank Dr. Richard Mailman from Penn State University College of Medicine for his thoughtful comments on early versions of this manuscript. Thanks also go to Dr. Xiaobing Zhang at Loma Linda University for the episomal vectors in iPSC derivation. This work was supported by grants from the Ministry of Science and Technology of China (2016YFA0100300 and 2014CB964800), the National Natural Science Foundation of China (31471347 and 31771655), the Science and Technology Commission of Shanghai Municipality (16JC1404200), the National High Technology Research and Development Program (2015AA020404 and SHDC1205123), China, and the intramural research program of the National Heart Lung Blood Institute, USA (1ZICHL006058).
Published: February 1, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and five tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.001.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE108496.
Supplemental Information
References
- Aramaki S., Hayashi K., Kurimoto K., Ohta H., Yabuta Y., Iwanari H., Mochizuki Y., Hamakubo T., Kato Y., Shirahige K. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell. 2013;27:516–529. doi: 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blaschke K., Ebata K.T., Karimi M.M., Zepeda-Martinez J.A., Goyal P., Mahapatra S., Tam A., Laird D.J., Hirst M., Rao A. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buaas F.W., Kirsh A.L., Sharma M., McLean D.J., Morris J.L., Griswold M.D., de Rooij D.G., Braun R.E. Plzf is required in adult male germ cells for stem cell self-renewal. Nat. Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Girard A., van de Kant H.J., Bourc'his D., Bestor T.H., de Rooij D.G., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costoya J.A., Hobbs R.M., Barna M., Cattoretti G., Manova K., Sukhwani M., Orwig K.E., Wolgemuth D.J., Pandolfi P.P. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat. Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Durruthy Durruthy J., Ramathal C., Sukhwani M., Fang F., Cui J., Orwig K.E., Reijo Pera R.A. Fate of induced pluripotent stem cells following transplantation to murine seminiferous tubules. Hum. Mol. Genet. 2014;23:3071–3084. doi: 10.1093/hmg/ddu012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easley C.A.t., Phillips B.T., McGuire M.M., Barringer J.M., Valli H., Hermann B.P., Simerly C.R., Rajkovic A., Miki T., Orwig K.E. Direct differentiation of human pluripotent stem cells into haploid spermatogenic cells. Cell Rep. 2012;2:440–446. doi: 10.1016/j.celrep.2012.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewen K.A., Koopman P. Mouse germ cell development: from specification to sex determination. Mol. Cell Endocrinol. 2010;323:76–93. doi: 10.1016/j.mce.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Florke-Gerloff S., Topfer-Petersen E., Muller-Esterl W., Schill W.B., Engel W. Acrosin and the acrosome in human spermatogenesis. Hum. Genet. 1983;65:61–67. doi: 10.1007/BF00285030. [DOI] [PubMed] [Google Scholar]
- Ginsburg M., Snow M.H., McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- Hayashi K., de Sousa Lopes S.M., Surani M.A. Germ cell specification in mice. Science. 2007;316:394–396. doi: 10.1126/science.1137545. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ohta H., Kurimoto K., Aramaki S., Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Helsel A.R., Yang Q.E., Oatley M.J., Lord T., Sablitzky F., Oatley J.M. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144:624–634. doi: 10.1242/dev.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M.C., Braydich-Stolle L., Dym M. Isolation of male germ-line stem cells; influence of GDNF. Dev. Biol. 2005;279:114–124. doi: 10.1016/j.ydbio.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N., Weinberger L., Tang W.W., Kobayashi T., Viukov S., Manor Y.S., Dietmann S., Hanna J.H., Surani M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253–268. doi: 10.1016/j.cell.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K., Angeles V.T., Flores M., Nguyen H.N., Reijo Pera R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjean A., Dupont J.M., Vasseur C., Le Tessier D., Cuisset L., Paldi A., Jouannet P., Jeanpierre M. Establishment of the paternal methylation imprint of the human H19 and MEST/PEG1 genes during spermatogenesis. Hum. Mol. Genet. 2000;9:2183–2187. doi: 10.1093/hmg/9.14.2183. [DOI] [PubMed] [Google Scholar]
- Kim K.P., Thurston A., Mummery C., Ward-van Oostwaard D., Priddle H., Allegrucci C., Denning C., Young L. Gene-specific vulnerability to imprinting variability in human embryonic stem cell lines. Genome Res. 2007;17:1731–1742. doi: 10.1101/gr.6609207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers J.H., Offenberg H.H., van Aalderen M., Vink A.C., Dietrich A.J., Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell. Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looijenga L.H., Hersmus R., Gillis A.J., Pfundt R., Stoop H.J., van Gurp R.J., Veltman J., Beverloo H.B., van Drunen E., van Kessel A.G. Genomic and expression profiling of human spermatocytic seminomas: primary spermatocyte as tumorigenic precursor and DMRT1 as candidate chromosome 9 gene. Cancer Res. 2006;66:290–302. doi: 10.1158/0008-5472.CAN-05-2936. [DOI] [PubMed] [Google Scholar]
- Louis J.F., Thoma M.E., Sorensen D.N., McLain A.C., King R.B., Sundaram R., Keiding N., Buck Louis G.M. The prevalence of couple infertility in the United States from a male perspective: evidence from a nationally representative sample. Andrology. 2013;1:741–748. doi: 10.1111/j.2047-2927.2013.00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Nickkholgh B., Korver C.M., van Daalen S.K., van Pelt A.M., Repping S. AZFc deletions do not affect the function of human spermatogonia in vitro. Mol. Hum. Reprod. 2015;21:553–562. doi: 10.1093/molehr/gav022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley L., Doyle P., Maconochie N. Lifetime prevalence of infertility and infertility treatment in the UK: results from a population-based survey of reproduction. Hum. Reprod. 2008;23:447–450. doi: 10.1093/humrep/dem369. [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Payer B., O'Carroll D., Ancelin K., Ono Y., Sano M., Barton S.C., Obukhanych T., Nussenzweig M., Tarakhovsky A. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436:207–213. doi: 10.1038/nature03813. [DOI] [PubMed] [Google Scholar]
- Ramathal C., Durruthy-Durruthy J., Sukhwani M., Arakaki J.E., Turek P.J., Orwig K.E., Reijo Pera R.A. Fate of iPSCs derived from azoospermic and fertile men following xenotransplantation to murine seminiferous tubules. Cell Rep. 2014;7:1284–1297. doi: 10.1016/j.celrep.2014.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves R.H., Gearhart J.D., Hecht N.B., Yelick P., Johnson P., O'Brien S.J. Mapping of PRM1 to human chromosome 16 and tight linkage of Prm-1 and Prm-2 on mouse chromosome 16. J. Hered. 1989;80:442–446. doi: 10.1093/oxfordjournals.jhered.a110895. [DOI] [PubMed] [Google Scholar]
- Saitou M. Germ cell specification in mice. Curr. Opin. Genet. Dev. 2009;19:386–395. doi: 10.1016/j.gde.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Yokobayashi S., Nakamura T., Okamoto I., Yabuta Y., Kurimoto K., Ohta H., Moritoki Y., Iwatani C., Tsuchiya H. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell. 2015;17:178–194. doi: 10.1016/j.stem.2015.06.014. [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tanaka S.S., Toyooka Y., Akasu R., Katoh-Fukui Y., Nakahara Y., Suzuki R., Yokoyama M., Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tiepolo L., Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum. Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- Tsuda M., Sasaoka Y., Kiso M., Abe K., Haraguchi S., Kobayashi S., Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Wang J., Cao H., Xue X., Fan C., Fang F., Zhou J., Zhang Y., Zhang X. Effect of vitamin C on growth of caprine spermatogonial stem cells in vitro. Theriogenology. 2014;81:545–555. doi: 10.1016/j.theriogenology.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Wen W., Zhang J.P., Xu J., Su R.J., Neises A., Ji G.Z., Yuan W., Cheng T., Zhang X.B. Enhanced generation of integration-free iPSCs from human adult peripheral blood mononuclear cells with an optimal combination of episomal vectors. Stem Cell Reports. 2016;6:873–884. doi: 10.1016/j.stemcr.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.E., Oatley J.M. Spermatogonial stem cell functions in physiological and pathological conditions. Curr. Top. Dev. Biol. 2014;107:235–267. doi: 10.1016/B978-0-12-416022-4.00009-3. [DOI] [PubMed] [Google Scholar]
- Zhang X., Li B., Li W., Ma L., Zheng D., Li L., Yang W., Chu M., Chen W., Mailman R.B. Transcriptional repression by the BRG1-SWI/SNF complex affects the pluripotency of human embryonic stem cells. Stem Cell Reports. 2014;3:460–474. doi: 10.1016/j.stemcr.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Oatley J., Bardwell V.J., Zarkower D. DMRT1 is required for mouse spermatogonial stem cell maintenance and replenishment. PLoS Genet. 2016;12:e1006293. doi: 10.1371/journal.pgen.1006293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Wang M., Yuan Y., Wang X., Fu R., Wan H., Xie M., Liu M., Guo X., Zheng Y. Complete meiosis from embryonic stem cell-derived germ cells in vitro. Cell Stem Cell. 2016;18:330–340. doi: 10.1016/j.stem.2016.01.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.