Abstract
Homologous recombination is a major driver of bacterial speciation. Genetic divergence and host association are important factors influencing homologous recombination. Here, we study these factors for Campylobacter fetus, which shows a distinct intraspecific host dichotomy. Campylobacter fetus subspecies fetus (Cff) and venerealis are associated with mammals, whereas C. fetus subsp. testudinum (Cft) is associated with reptiles. Recombination between these genetically divergent C. fetus lineages is extremely rare. Previously it was impossible to show whether this barrier to recombination was determined by the differential host preferences, by the genetic divergence between both lineages or by other factors influencing recombination, such as restriction-modification, CRISPR/Cas, and transformation systems. Fortuitously, a distinct C. fetus lineage (ST69) was found, which was highly related to mammal-associated C. fetus, yet isolated from a chelonian. The whole genome sequences of two C. fetus ST69 isolates were compared with those of mammal- and reptile-associated C. fetus strains for phylogenetic and recombination analysis. In total, 5.1–5.5% of the core genome of both ST69 isolates showed signs of recombination. Of the predicted recombination regions, 80.4% were most closely related to Cft, 14.3% to Cff, and 5.6% to C. iguaniorum. Recombination from C. fetus ST69 to Cft was also detected, but to a lesser extent and only in chelonian-associated Cft strains. This study shows that despite substantial genetic divergence no absolute barrier to homologous recombination exists between two distinct C. fetus lineages when occurring in the same host type, which provides valuable insights in bacterial speciation and evolution.
Keywords: Campylobacter fetus, homologous recombination, speciation, host association, reptile, whole genome sequencing
Introduction
There are several underlying processes driving bacterial speciation. One of these processes is homologous recombination, in which genetic material is exchanged between two identical or similar molecules of DNA. Here, the effect of homologous recombination on speciation is studied in Campylobacter fetus, which shows several distinct genetically divergent host-associated lineages.
Campylobacter fetus is recognized as an important veterinary and occasional human pathogen (van Bergen et al. 2008; Wagenaar et al. 2014). Three C. fetus subspecies are currently recognized: C. fetus subspecies fetus (Cff) and venerealis (Cfv), which are closely related and occur in mammals, primarily ungulates, and C. fetus subsp. testudinum (Cft), which is genetically divergent from Cff and Cfv, and primarily occurs in reptiles (Gilbert et al. 2014). These subspecies show a strict host dichotomy: Cff and Cfv have never been isolated from reptiles, whereas Cft has never been isolated from mammals, excluding occasional human cases, in which contact with reptiles is suspected (Patrick et al. 2013). The current situation shows two major dominating coherent lineages of C. fetus in mammals (Cff and Cfv) and reptiles (Cft). However, during independent studies assessing the Campylobacter diversity in reptiles, a genetically distinct C. fetus lineage, comprising multilocus sequence types 43 and 69, was obtained from captive-held red-footed tortoises (Chelonoidis carbonaria) in Taiwan and the Netherlands, adding up to the genetic diversity of C. fetus (Wang et al. 2013; Gilbert et al. 2014). Surprisingly, phylogenetic analysis based on 16S rRNA and multilocus sequence typing (MLST) showed that this lineage was highly related to mammal-associated Cff and Cfv, although it was isolated from reptiles (Wang et al. 2013).
Noteworthy, recombination was virtually absent between mammal-associated Cff/Cfv and reptile-associated Cft, which suggested allopatric speciation within C. fetus (Gilbert, Miller, Yee, Zomer, et al. 2016). A barrier to recombination was apparent, but it remained to be shown whether this was caused by the host dichotomy (mammal- or reptile-associated) or by intrinsic factors which inhibit recombination between these lineages. The C. fetus lineage closely related to mammal-associated Cff and Cfv, yet isolated from reptiles, might play a pivotal role in our understanding of these processes.
Based on whole genome comparison, we explore the homologous recombination between two genetically divergent bacterial lineages, which both occur naturally in a reptilian host, and provide novel insights in bacterial speciation and evolution.
Materials and Methods
Strains
Campylobacter fetus isolates 12S01208-4 and 12S01908-5 were obtained on February 23 and March 16, 2012, respectively, from a red-footed tortoise (Chelonoidis carbonaria) suffering from a Mycoplasma induced pneumonia. Isolates were grown on Columbia agar with 5% sheep blood (Oxoid, the Netherlands) in a microaerobic atmosphere (83.3% N2, 7.1% CO2, 3.6% H2, and 6% O2) at 37°C for 48 h. All other 61 C. fetus strains (Cff, 19 strains; Cfv, 22 strains; Cft, 20 strains) were identical to those used in previous studies (Gilbert, Miller, Yee, Zomer, et al. 2016; van der Graaf-van Bloois et al. 2016).
Whole Genome Sequencing
Sequencing of C. fetus isolates 12S01208-4 and 12S01908-5 was performed using Illumina MiSeq with 300 bp paired end reads. The reads were assembled using SPAdes 3.1.1. The average coverage was 216-354×, the number of contigs was 22-30 and the number of gaps was 23-31 for both genomes. The level of completeness (99.77%) and contamination (1.96%) was determined for both genomes based on the Campylobacter genus using CheckM 1.0.5. The whole genome sequences of C. fetus isolates 12S01208-4 and 12S01908-5 have been deposited at GenBank under accession numbers MTDX00000000 and MTDY00000000, respectively. All other C. fetus strains were sequenced as described previously (Gilbert, Miller, Yee, Zomer, et al. 2016; van der Graaf-van Bloois et al. 2016) and are present in GenBank.
Average Nucleotide Identity
As a measure of genomic relatedness the average nucleotide identity (ANI) was used (Konstantinidis and Tiedje 2005; Konstantinidis et al. 2006). Using the OrthoANIu tool (Lee et al. 2016), ANI values based on whole genome sequences were calculated for C. fetus isolates 12S01208-4 and 12S01908-5, Cff strains 04/554 and 82-40, Cft strains 03-427, 13S00388-15, 85-387, and SP3, Cfv strain 97/608, and C. iguaniorum strain 1485E.
Genome Analysis
Protein-, rRNA-, and tRNA-encoding genes were identified using Prokka (Seemann 2014). An all versus all BLAST was performed for all predicted proteins of the whole genomes (supplementary table S1, Supplementary Material online) at an E-value cutoff of 1E − 6. To determine the orthologous relationships of all proteins, protein sequences were clustered using Roary with a 75% identity cutoff (Page et al. 2015). Core genome alignment was performed using Parsnp 1.2 (Treangen et al. 2014). DNA regions present in all isolates were extracted and gaps were removed using trimAl (Capella-Gutierrez et al. 2009). Based on this core genome alignment, phylogenomic reconstruction and prediction of recombination events was performed using Gubbins (Croucher et al. 2015) with the default settings. Phylogenetic dendrograms were created using Fasttree (Price et al. 2009). A BLAST search of the predicted recombination regions of C. fetus ST69 isolate 12S01908-5 (supplementary information S1, Supplementary Material online) against the genomes of Cft strain 03-427 and Cff strain 82-40 and against the NCBI non redundant (nr) database was performed to search for particular recombination between these reptile-associated taxa and other species. The same procedure was performed for all predicted recombination regions of Cft (supplementary information S2, Supplementary Material online) and the genomes of C. fetus ST69 isolate 12S01908-5 and Cff strain 82-40. Percentage sequence identity of C. fetus ST69 recombination regions with Cft and Cff and percentage sequence identity of Cft recombination regions with C. fetus ST69 and Cff were plotted in an x–y graph using Microsoft Excel.
Multilocus Sequence Typing
The loci for the C. fetus multilocus sequence typing (MLST) scheme (van Bergen et al. 2005) were extracted from the whole genome of C. fetus isolate 12S01208-4 and submitted to the Campylobacter MLST database (www.pubmlst.org/campylobacter; last accessed March 17, 2016).
The 501-nt trimmed pgm alleles of C. fetus and C. iguaniorum strain 1485E were extracted from the Campylobacter MLST database and from the genome (GenBank accession number CP009043), respectively. Alignment and phylogenetic analysis, based on the neighbor-joining method with bootstrap values using 500 repetitions, was performed using MEGA 6.05.
Results
The genomes of Campylobacter fetus isolates 12S01208-4 and 12S01908-5 are highly similar and show a high degree of synteny. In total, 34 discriminatory SNPs were identified in the core genomes of both isolates; 88.2% (30/34) were located inside recombination regions, 11.8% (4/34) were located outside recombination regions. Both isolates belong to the same multilocus sequence type, ST69. Based on the presence of LPS-biosynthesis gene wcbK, encoding a putative GDP-mannose 4, 6-dehydratase, the predicted serotype of both isolates is B or AB (Kienesberger et al. 2014; Gilbert, Miller, Yee, Zomer, et al. 2016). As in other C. fetus lineages, the S-layer encoding sap genes, considered important in C. fetus virulence (Blaser et al. 2008), are present in C. fetus ST69 isolate 12S01908-5. However, no sapCDEF genes were detected in isolate 12S01208-4. A CRISPR/Cas system is present in C. fetus ST69, including two CRISPR repeat regions (30 nt spacers; 36 and 39 repeats, respectively). However, the additional locus encoding CRISPR/Cas system-associated RAMP superfamily proteins, which is well conserved in Cff, Cfv, and Cft, was absent from both C. fetus ST69 isolates (supplementary table S1, Supplementary Material online).
Of the genes specifically present in or absent from C. fetus ST69 only, or C. fetus ST69 and either Cff/Cfv or Cft, a disproportionally high number encoded proteins related to DNA uptake and defense, such as competence, transformation system, restriction-modification system, and CRISPR/Cas system proteins (supplementary table S1, Supplementary Material online).
Notably, 60 genes were exclusively shared between C. fetus ST69 and Cft strain 13S00388-15, which was isolated from Chelonoidis denticulata, a chelonian species closely related to Chelonoidis carbonaria. Most of these genes encode hypothetical proteins, but also several phage-specific proteins and likely represent a prophage.
The tcuRABC locus, involved in catabolism of tricarballylate (a citrate analog), which has been shown present in Cft and in many other reptile-associated Campylobacter and Helicobacter taxa (Gilbert, Miller, Yee, Kik, et al. 2016; Gilbert, Miller, Yee, Zomer, et al. 2016; Gilbert et al. 2017), is absent from C. fetus ST69.
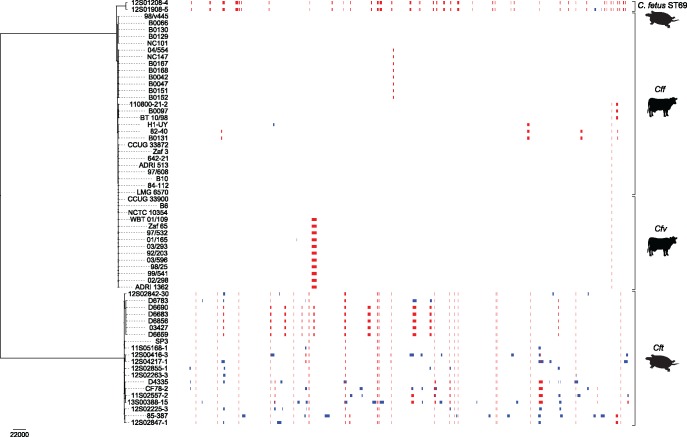
A whole genome-based phylogeny of C. fetus shows that C. fetus ST69 is most closely related to mammal-associated Cff and Cfv (fig. 1). Campylobacterfetus ST69, Cff and Cfv are highly divergent from Cft. The average nucleotide identity (ANI) was used as a measure of genomic relatedness. The ANI between both C. fetus ST69 isolates was 99.96%. Campylobacterfetus ST69 and Cff showed 98% ANI, which is well above the 95% species delineation (supplementary table S2, Supplementary Material online). The ANI between C. fetus ST69 and Cft was 92%. Although this is below the species delineation, these lineages are considered conspecific based on many shared genotypic and phenotypic characteristics, including the presence of an S-layer, as examined previously for C. fetus (Fitzgerald et al. 2014). The ANI between C. fetus ST69 and reptile-associated C. iguaniorum was 76%.
Fig. 1.
—Core genome-based phylogeny for Campylobacter fetus. Identical recombination regions in two or more strains are indicated in red; unique recombination regions are indicated in blue. Campylobacter fetus ST69 is associated with reptiles (chelonians), C. fetus subsp. fetus (Cff) and venerealis (Cfv) are associated with mammals (primarily ungulates), and C. fetus subsp. testudinum (Cft) is primarily associated with reptiles.
Extensive recombination was detected in C. fetus ST69. In isolates 12S01208-4 and 12S01908-5, 5.1% and 5.5% of the gapless core genome was predicted to be recombined, respectively. In the Cft and Cff/Cfv genomes, on an average 2.9% and 0.4% of the core genome was predicted to be recombined, respectively. The ratio of base substitutions predicted to have been imported through recombination to those occurring through point mutation is 0.32 for C. fetus ST69, 0.06 for Cft, and 0.03 for Cff/Cfv, indicating that recombination is a major driver of mutation in C. fetus ST69.
A total of 56 different recombination regions were identified in the C. fetus ST69 genomes, of which two were uniquely present in isolate 12S01908-5 (fig. 1). In Cft and in Cff/Cfv, on an average 31.9 (± 5.4) and 1.5 (± 1.1) recombination regions were identified, respectively. A BLAST search against the NCBI nonredundant database showed that 80.4% (45/56) of the recombination regions in C. fetus ST69 were most closely related to Cft, 14.3% (8/56) to Cff, and 5.6% (3/56) to C. iguaniorum.
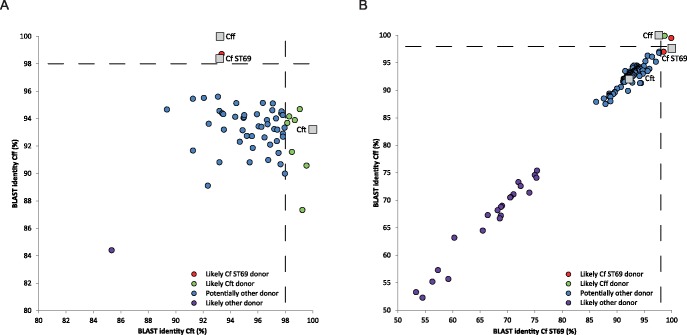
A scatter plot of the BLAST identities between the C. fetus strain 12S01908-5 recombination regions and Cff 82-40 and Cft 03-427 confirmed that most recombination regions were more homologous with Cft than with Cff (fig. 2A). On an average, the recombination regions showed 95.9% (± 2.3) homology with Cft and 93.2% (± 1.7) homology with Cff. Of the recombination regions, 98.2% (55/56) showed >87% homology with Cff and >89% homology with Cft. The homology of the recombination regions with Cft gradually increased from 89.4% to 99.5%, while this range was 87.3–98.7% for Cff. Recombination regions became scarcer with decreasing BLAST identity. Of the recombination regions, 12.5% (7/56) and 1.8% (1/56) was >98% homologous with Cft and Cff, respectively.
Fig. 2.
—Scatter plot based on the BLAST-based nucleotide identities of all recombination regions identified in Campylobacter fetus ST69 (Cf ST69) isolate 12S01908-5 (A) and all C. fetus subsp. testudinum (Cft) strains (B). A BLAST search was performed for each recombination region and the genomes of Cft strain 03-427 and C. fetus subsp. fetus (Cff) strain 82-40 (A) and Cff strain 82-40 and Cf ST69 isolate 12S01908-5 (B). The nucleotide identities of the recombination regions are plotted as circles; the nucleotide identities of the reference genomes are plotted as squares. The dashed line shows the ≥98% nucleotide identity. x- and y-axis range 80–100% (A) or 50–100% (B).
Two unique recombination regions were identified only in C. fetus ST69 isolate 12S01908-5 (indicated in blue in fig. 1). One of those regions was 99.8% (3,801/3,808 nt) homologous with C. fetus ST69 isolate 12S01208-4. All nucleotide substitutions were synonymous and did not alter the amino acid sequence of the proteins affected. The other recombined sequence was 100% (114/114 nt) homologous with reptile-associated C. iguaniorum strains 1485E and 2463 D. Due to this recombination, the start site of the gene encoding threonyl-tRNA synthetase and the amino acid composition of the first part of the translated protein were altered, resulting in a shorter open reading frame.
In Cft, recombination regions which likely originated from C. fetus ST69 were observed as well, but to a much lesser extent than from Cft to C. fetus ST69. Of the recombination regions extracted from Cft, 83.1% (108/130) showed >87% homology with Cff and C. fetus ST69. Two well-separated clusters of recombination regions which showed 52.3–75.4% and 87.5–99.9% homology with Cff and C. fetus ST69 were identified (fig. 2B). In total, only 2.3% (3/130) of the recombination regions likely originated from C. fetus ST69 (>98% homology). These regions were detected in Cft strains 13S00388-15 and 85-387, isolated from the chelonian species Chelonoidis denticulata and Terrapene carolina, respectively. Notably, one recombination region in Cft strain 85-387, showing high homology with C. fetus ST69, contained phosphoglucosamine mutase encoding pgm (glmM), an essential housekeeping gene which is part of the C. fetus MLST scheme. A dendrogram based on all C. fetus pgm alleles present in the pubMLST database (www.pubmlst.org/campylobacter; last accessed March 17, 2016) confirmed that pgm from Cft strain 85-387 was identical to pgm from C. fetus ST69, indicating recent recombination from the C. fetus ST69 lineage to Cft strain 85-387 (supplementary fig. S1, Supplementary Material online).
Discussion
This study shows that homologous recombination between divergent bacterial lineages occurs despite substantial genetic distance when present in the same host. As shown previously, Cff/Cfv and Cft show no or very few recombination between each other (Gilbert, Miller, Yee, Zomer, et al. 2016). In contrast, no absolute barrier to recombination exists between divergent C. fetus lineages when occurring in the same host type, as shown in this study. Apparently, a species-level divergence of 8% between both lineages is no barrier to homologous recombination. Notably, most of the recombination regions detected in both C. fetus ST69 and Cft showed highest homology with C. fetus, implicating that recombination is occurring most frequently within C. fetus or a closely related Campylobacter lineage. Of the C. fetus ST69 core genome, 5.1–5.5% showed signs of recombination and the majority of the recombination regions in C. fetus ST69 were most closely related to Cft, indicating that this lineage is the most important DNA donor.
The majority of the recombination regions in C. fetus ST69 were most closely related to Cft, yet showed relative low nucleotide identity (<98%). These more divergent recombination regions likely represent ancient recombination events, followed by divergent evolution of those recombination regions based on point mutations in both donor and recipient. The gradual increase from 89.4% to 99.5% observed in the BLAST sequence identities of the C. fetus ST69 recombination regions with Cft suggests that this has been a long-term and continuous process which may still be ongoing. Indeed, parts of similar bacterial genomes can maintain the ability to recombine over long timespans before genetic isolation between two lineages is complete (Retchless and Lawrence 2007). With sets of niche-specific genes being maintained in populations that freely recombine at other loci, different parts of the genome may be genetically isolated at different times, suggesting temporal fragmentation of speciation.
Despite the high genetic similarity between both C. fetus ST69 isolates, small-scale differences were observed in recombination regions and gene content. As the number of discriminatory SNPs outside the recombination regions was low in the core genomes of both isolates (n = 4), these can be considered recent events. Furthermore, as 88.2% of the discriminatory SNPs in the core genomes of both isolates could be attributed to recombination, this can be considered the main driver of the short-scale divergence in C. fetus ST69. The differences in gene content could predominantly be attributed to the sap genes. The absence of sapCDEF genes in C. fetus ST69 isolate 12S01208-4 has been observed in other C. fetus strains as well and may represent a spontaneous mutation occurring in vitro (Dworkin et al. 1995; Gilbert, Miller, Yee, Zomer, et al. 2016).
Recombination also occurred in essential housekeeping genes, such as pgm in Cft strain 85-387. As pgm is part of the C. fetus MLST scheme, recombination can distort the inferences made based on MLST (Dingle et al. 2010). In addition, the observation that Cft strain 85-387, isolated in 1984, has an identical pgm allele as C. fetus ST69 isolates 12S01208-4 and 12S01908-5, isolated in 2012, indicates that both lineages occur together for at least 28 years and that no mutation has occurred in this allele during that period.
Reciprocal recombination between C. fetus ST69 and Cft strains from chelonian hosts suggests that recombination between both lineages primarily occurs in chelonian hosts. In addition, the presence of an identical prophage in C. fetus ST69 and Cft strain 13S00388-15, which was isolated from Chelonoidis denticulata, further affirms that both C. fetus lineages occur in the same niche.
Interestingly, only little recombination was observed between C. fetus ST69 and reptile-associated C. iguaniorum, which showed a prevalence of 33.3% (5/15) in Chelonoidis carbonaria and Chelonoidis denticulata and was isolated from the same animal and the same samples as both C. fetus ST69 isolates (Gilbert et al. 2014). Recombination between C. fetus ST69 and C. iguaniorum does occur, but the genetic divergence between both species may lead to an altered protein (function), and likely reduced fitness in most cases, making it less likely that these recombination regions will be fixed in the genome over time.
Cft and C. fetus ST69 diverged in isolation of each other, either in space or due to intrinsic barriers to recombination. Upon contact in a shared reptilian host, recombination between both lineages occurred. Recombination between C. fetus ST69 and Cft appears to be bidirectional. However, the number and size of recombination regions was larger from Cft to C. fetus ST69 than vice versa. This asymmetry could be the result of numerical dominance of Cft over C. fetus ST69 in the reptilian intestine, which is supported by the prevalence rates (Gilbert et al. 2014), and which has been suggested previously for C. coli showing introgression of C. jejuni DNA (Sheppard et al. 2008).
Intrinsic factors explaining the high recombination frequency in C. fetus ST69 cannot be excluded, as genes encoding transformation and restriction-modification system proteins were disproportionally highly distributed among the genes specifically shared with either Cft or Cff/Cfv, or which were specifically present in C. fetus ST69 only. In addition, the specific absence of the additional CRISPR/Cas system-associated RAMP superfamily proteins, which have been shown well conserved in Cff, Cfv, and Cft (Gilbert, Miller, Yee, Zomer, et al. 2016), from C. fetus ST69 might favor the import of exogenous DNA and enable a higher recombination frequency.
Homologous recombination acts as a force of coherence between both lineages and can counteract speciation (Fraser et al. 2007). When occurring in identical host types recombination between divergent C. fetus lineages occurs. However, when occurring in different host types, that is, mammalian or reptilian, recombination is virtually absent (Gilbert, Miller, Yee, Zomer, et al. 2016). This study shows that no obvious barriers to homologous recombination exist between two genetically divergent bacterial lineages when colonizing the same host type. In this case, recombination is rather predicted by host specificity than by sequence divergence, supporting allopatric speciation based on host type.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Literature Cited
- Blaser MJ, Newell DG, Thompson SA, Zechner EL. (2008). Pathogenesis of Campylobacter fetus In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. Washington (DC: ): ASM Press; p. 401–428. [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2515:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, et al. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 433:e15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingle KE, et al. 2010. Genetic relationships among reptilian and mammalian Campylobacter fetus strains determined by multilocus sequence typing. J Clin Microbiol. 483:977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J, Tummuru MK, Blaser MJ.. 1995. A lipopolysaccharide-binding domain of the Campylobacter fetus S-layer protein resides within the conserved N terminus of a family of silent and divergent homologs. J Bacteriol. 1777:1734–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald C, et al. 2014. Campylobacter fetus subsp. testudinum subsp. nov., isolated from humans and reptiles. Int J Syst Evol Microbiol. 64(Pt 9):2944–2948. [DOI] [PubMed] [Google Scholar]
- Fraser C, Hanage WP, Spratt BG.. 2007. Recombination and the nature of bacterial speciation. Science 3155811:476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJ, Duim B, Timmerman AJ, Zomer AL, Wagenaar JA.. 2017. Whole genome-based phylogeny of reptile-associated Helicobacter indicates independent niche adaptation followed by diversification in a poikilothermic host. Sci Rep. 71:8387.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJ, et al. 2014. Occurrence, diversity, and host association of intestinal Campylobacter, Arcobacter, and Helicobacter in reptiles. PLoS One 97:e101599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJ, Miller WG, Yee E, Kik M, et al. 2016. Comparative genomics of Campylobacter iguaniorum to unravel genetic regions associated with reptilian hosts. Genome Biol Evol. 8:3022–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJ, Miller WG, Yee E, Zomer AL, et al. 2016. Comparative genomics of Campylobacter fetus from reptiles and mammals reveals divergent evolution in host-associated lineages. Genome Biol Evol. 8:2006–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienesberger S, et al. 2014. Comparative genome analysis of Campylobacter fetus subspecies revealed horizontally acquired genetic elements important for virulence and niche specificity. PLoS One 91:e85491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Ramette A, Tiedje JM.. 2006. The bacterial species definition in the genomic era. Philos Trans R Soc B Biol Sci. 3611475:1929–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM.. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 1027:2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Kim YO, Park S, Chun J.. 2016. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 662:1100–1103. [DOI] [PubMed] [Google Scholar]
- Page AJ, et al. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick ME, et al. 2013. Human infections with new subspecies of Campylobacter fetus. Emerg Infect Dis. 1910:1678–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP.. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 267:1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retchless AC, Lawrence JG.. 2007. Temporal fragmentation of speciation in bacteria. Science 3175841:1093–1096. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 3014:2068–2069. [DOI] [PubMed] [Google Scholar]
- Sheppard SK, McCarthy ND, Falush D, Maiden MC.. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 3205873:237–239. [DOI] [PubMed] [Google Scholar]
- Treangen TJ, Ondov BD, Koren S, Phillippy AM.. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 1511:524.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bergen MA, et al. 2005. Clonal nature of Campylobacter fetus as defined by multilocus sequence typing. J Clin Microbiol. 4312:5888–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bergen MAP, van Putten JP, Dingle KE, Blaser MJ, Wagenaar JA. (2008). Isolation, identification, subspecies differentiation, and typing of Campylobacter fetus In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. Washington (DC: ): ASM Press; p. 213–225. [Google Scholar]
- van der Graaf-van Bloois L, et al. 2016. Whole genome sequence analysis indicates recent diversification of mammal-associated Campylobacter fetus and implicates a genetic factor associated with H2S production. BMC Genomics 17:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar JA, et al. 2014. Campylobacter fetus infections in humans: exposure and disease. Clin Infect Dis. 5811:1579–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CM, Shia WY, Jhou YJ, Shyu CL.. 2013. Occurrence and molecular characterization of reptilian Campylobacter fetus strains isolated in Taiwan. Vet Microbiol. 164(1–2):67–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.