Summary
Spermatogenesis requires retinoic acid (RA) induction of the undifferentiated to differentiating transition in transit amplifying (TA) progenitor spermatogonia, whereas continuity of the spermatogenic lineage relies on the RA response being suppressed in spermatogonial stem cells (SSCs). Here, we discovered that, in mouse testes, both spermatogonial populations possess intrinsic RA-response machinery and exhibit hallmarks of the differentiating transition following direct exposure to RA, including loss of SSC regenerative capacity. We determined that SSCs are only resistant to RA-driven differentiation when situated in the normal topological organization of the testis. Furthermore, we show that the soma is instrumental in “priming” TA progenitors for RA-induced differentiation through elevated RA receptor expression. Collectively, these findings indicate that SSCs and TA progenitor spermatogonia inhabit disparate niche microenvironments within seminiferous tubules that are critical for mediating extrinsic cues that drive fate decisions.
Keywords: retinoic acid, RAR gamma, spermatogonial stem cells, ID4, GFRA1
Highlights
-
•
Contrary to previous dogma, SSCs do express RARγ, as well as other RAR/RXR variants
-
•
Following direct exposure, SSCs exhibit an RA signaling response
-
•
SSCs are protected from RA by the niche microenvironment in the testis
-
•
Signals from the soma prime progenitors for RA-driven differentiation
Lord et al. have demonstrated that, contrary to previous assumptions, spermatogonial stem cells do express a functional complement of retinoic acid and retinoid X receptors (RARs/RXRs) and rely on protection from an undisturbed niche microenvironment to prevent loss of the spermatogenic reservoir to RA-induced differentiation.
Introduction
Gametes are the eternal link between generations and are key to the continuation and diversity of animal populations. In mammalian males, continual spermatogenesis is required for fertility, generating millions of genetically unique spermatozoa daily. Both the robustness and continuity of this process is reliant on a small pool of spermatogonial stem cells (SSCs) that are rare in number, estimated to comprise 0.01%–1% of the total testis cell population depending on the species (Tegelenbosch and de Rooij, 1993). In mice, the SSC pool is thought to exist as a subset of the Asingle spermatogonial population closely associated with the basement membrane of the seminiferous tubules (reviewed by Lord and Oatley, 2017). These Asingle SSCs undergo mitotic proliferation in order to achieve self-renewal, or alternatively, to produce transit amplifying (TA) progenitor cells that no longer possess stem cell capacity and undergo clonal expansion, remaining connected by intercellular bridges to produce chains ranging from 2 (Apaired) to 16 cells (Aaligned). A round of spermatogenesis is initiated when a pulse of retinoic acid (RA) induces a majority of the TA progenitor spermatogonia to transition from the undifferentiated type A to differentiating type A1 state. In support of the Asingle model (Huckins, 1971, Oakberg, 1971), and more recently the “revised Asingle model” (de Rooij, 2017, Lord and Oatley, 2017), the propensity for spermatogonia to respond to the RA signal is directly proportional to chain length, with ∼88% of the undifferentiated spermatogonial population undergoing the A to A1 transition in response to RA, including 63% of Apaired, 95% of Aaligned4, and 100% of Aaligned8-16, while none of the Asingle spermatogonia make the differentiating transition. Importantly, SSCs must be resistant to the RA signal or the foundation for continuity of the spermatogenic lineage will be lost (Ikami et al., 2015, Tegelenbosch and de Rooij, 1993).
While it has long been established that RA signaling is absolutely required for spermatogenesis (van Pelt and de Rooij, 1990, Wolbach and Howe, 1925), with vitamin A-deficient models experiencing infertility by consequence of a loss of the differentiating spermatogonial pool (Li et al., 2011), the mechanisms by which SSCs resist the differentiating signal that influences closely situated progenitors remains unresolved. A recent study by Ikami et al. (2015) proposed that responsiveness or resistance to RA signaling in spermatogonia is dictated by “on” or “off” expression of the retinoic acid receptor (RAR) variant γ. In that study, the “SSC” pool was considered to be all cells expressing the glial cell line-derived neurotrophic factor (GDNF) family receptor a1 (GFRA1), while the TA progenitor pool was identified as the Neurogenin3 (NEUROG3) expressing population of spermatogonia. Based on immunohistochemical analyses, the GFRA1+ population was described as having little to no RARγ expression, while RARγ expression in the NEUROG3+ progenitors was high. Although these findings imply a simple hierarchical classification of spermatogonia as RA-resistant SSCs and RA-responsive progenitors, it is at odds with other findings, and does not necessarily fit with known dynamics of the spermatogonial-soma interactions in mammalian testes. First, studies by Gely-Pernot et al. (2012) demonstrated that conditional ablation of Rarγ within the spermatogonial population specifically is insufficient to block the differentiating transition, that expression of RARγ is evident in a portion of the GFRA1+ spermatogonial population, and that induction of classical RA-responsive genes Stra8 (stimulated by retinoic acid 8) and Kit (KIT proto-oncogene receptor tyrosine kinase) can be achieved via both the RARγ and RARα variants in spermatogonia. Indeed, widespread impediment of spermatogonial responsiveness to RA signaling was only observed when expression of both Rarα and Rarγ were ablated conditionally in spermatogonia (Gely-Pernot et al., 2012). Second, several previous studies have demonstrated that GFRA1+ and NEUROG3+ spermatogonial populations are not mutually exclusive as SSCs and TA progenitors, respectively (Buageaw et al., 2005, Ebata et al., 2005, Grisanti et al., 2009, Yoshida et al., 2004) (expression profiles of all markers discussed/utilized in this study are depicted in Figure S1). Third, both Rarγ and Rarα gene expression is detectable in Inhibitor of DNA binding 4 (ID4)-eGFP+ and ID4-eGFP– spermatogonial populations that are highly enriched for SSCs and progenitors, respectively (Chan et al., 2014). Lastly, the hierarchical model proposed by Ikami et al. eliminates an influence from the soma in modulating the RA response in spermatogonia, which is at odds with the well-established dynamic role that multiple testis somatic cell populations play in the biosynthesis and clearance of RA (Hogarth et al., 2015, Kent et al., 2015, Tong et al., 2013).
Here, we aimed to further clarify the modes by which RA signaling is modulated in spermatogonial subtypes to preserve the SSC pool and therefore continuity of the spermatogenic lineage. Using the Id4-eGfp transgenic mouse model in which the ID4-eGFP+ cells are SSCs and ID4-eGFP– cells are mostly progenitors (Chan et al., 2014, Helsel et al., 2017b), RARα and RARγ expression was detected at similar levels in both populations of spermatogonia from testes and primary cultures. In addition, we found expression of both RARα and RARγ in GFRA1+ spermatogonia. Using primary cultures of undifferentiated spermatogonia that consist of SSCs and progenitors, we found that direct exposure to RA elicits hallmark responses of the differentiating transition in both populations and functional transplantation analyses confirmed depletion of the SSC pool. Lastly, we found that intricate niche microenvironments created by the soma in the testis work to simultaneously protect the SSCs from exogenous RA, while priming progenitors to be highly responsive to the RA pulse. Taken together, these findings support an adapted model for modulation of RA responsiveness in mammalian testes for which the soma is key to preservation of the SSC pool during successive rounds of spermatogenesis.
Results
Expression of RAR/RXR Isoforms in SSC and TA Progenitor Populations
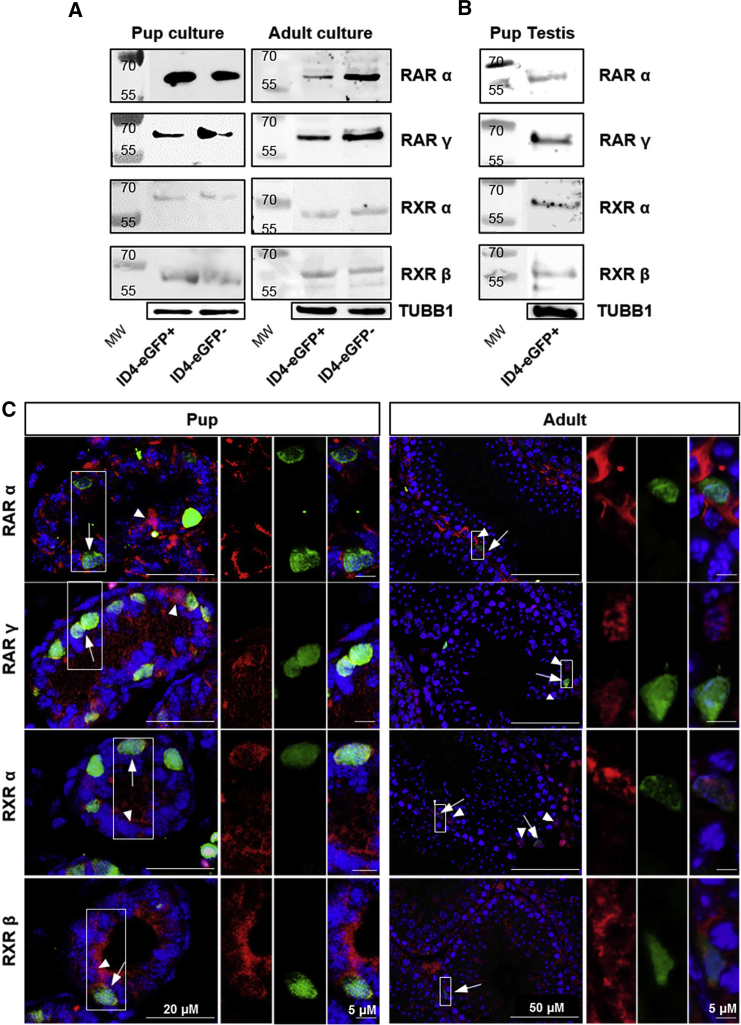
RA signaling is mediated by heterodimers of RARs and RXRs, each consisting of α, β, and γ variants. In order to compare expression profiles for these isoforms in highly enriched SSC and progenitor populations, we utilized primary cultures of undifferentiated spermatogonia generated from Id4-eGfp transgenic mice (Chan et al., 2014). In previous studies that used RNA sequencing (RNA-seq) to compare ID4-eGFP+/SSC and ID4-eGFP–/progenitor populations, we identified similar transcript levels for Rarα, Rarγ, Rxrα, and Rxrβ (Chan et al., 2014) (fragments per kilobase of transcript per million fragments mapped values provided in Figure S2B). In the current study, we validated these findings using RT-PCR (Figure S2A). Also, in agreement with other studies that explored the spermatogonial population overall (Gely-Pernot et al., 2012, Gely-Pernot et al., 2015, Ghyselinck et al., 1997), we confirmed that expression of Rarβ and Rxrγ is not detectable in cultured spermatogonia (Figure S2A). Further, using western blot analyses to assess expression of these factors at the protein level, we found expression of RARα (55 kDa), RARγ (58 kDa), RXRα (53–60 kDa), and RXRβ (57 kDa) in both ID4-eGFP+/SSC and ID4-eGFP–/progenitor populations from primary pup cultures (Figure 1A). Next, we explored whether the expression profile of RARs/RXRs in cultures derived from pups was similar in those derived from adult mice. In agreeance with data generated using the post-natal day (P) 6–8 pup cultures, expression of RARα, RARγ, RXRα, and RXRβ was detected in ID4-eGFP+/SSC and ID4-eGFP–/progenitor populations in adult cultures, as depicted by both RT-PCR and Western blot analyses (Figures S2C and 1A).
Figure 1.
Expression of RAR and RXR Variants in both SSC and Progenitor Spermatogonial Populations
(A and B) Representative images of Western blot analyses for RARα, RARγ, RXRα, and RXRβ in ID4-eGFP+ and ID4-eGFP– populations in primary cultures of spermatogonia established from P6–8 pup or adult mice (A) and ID4-eGFP+ spermatogonia isolated directly from the testes of pups (B). Tubulin (TUBB1) loading controls are included beneath each image.
(C) Representative images of immunostaining for expression of RARα, RARγ, RXRα, and RXRβ by ID4-eGFP+ and ID4-eGFP– spermatogonia in cross-sections of testes from pups and adult mice. Arrows and arrowheads indicate ID4-eGFP+ or ID4-eGFP– spermatogonia co-expressing RARα, RARγ, RXRα, or RXRβ, respectively. Boxed regions are magnified in images to the right. Negative controls are presented in Figure S2.
Considering that a period of in vitro maintenance could have altered expression profiles, we next aimed to determine whether the RAR/RXR variants are expressed by the SSC population in vivo. The outcomes of RT-PCR and Western blot analyses confirmed that RARα, RARγ, RXRα, and RXRβ are all expressed by ID4-eGFP+/SSCs isolated directly from P6–8 testes (Figures S2C and 1B). Note that examination of ID4-eGFP– spermatogonia in testes is challenging because, unlike the ID4-eGFP– population from primary cultures of undifferentiated spermatogonia, fluorescence-activated cell sorting (FACS) directly from the testis also yields somatic cells and differentiating spermatogonia that are known to express RARs and RXRs (Chan et al., 2014, Helsel et al., 2017b). Thus, we were unable to compare expression profiles between ID4-eGFP+ and ID4-eGFP– population in testes. Similarly, our ability to assess expression in ID4-eGFP+ spermatogonia from adult testes using cell sorting is limited by an ID4-eGFP fluorescence signal in spermatocytes (Chan et al., 2014). Thus, in conjunction with Western blot and RT-PCR analyses of cells isolated from cultures or pup testes directly, in situ assessment by immunofluorescence staining was used to confirm expression of RARα/γ and RXRα/β in ID4-eGFP+ spermatogonia in cross-sections of testes from both P6 and adult mice (Figure 1C, arrows). As expected, RAR/RXR expression was also identified in ID4-eGFP– spermatogonia (Figure 1C, arrowheads), throughout the nucleus and cytoplasm (Figures 1C and S3B). Images of negative control sections with normal immunoglobulin G (IgG) as the primary antibody are presented in Figure S3A. Subcellular localization of RARs was further visualized in ID4-eGFP+/SSCs and ID4-eGFP–/progenitors in immunocytochemistry experiments performed on spermatogonia taken from primary cultures (depicted in Figure S3B). Further examples of RAR expression in the testis have been provided using the undifferentiated spermatogonia marker LIN28 (Figure S1), with distinct overlay being evident between LIN28 and RAR expression (Figure S3C).
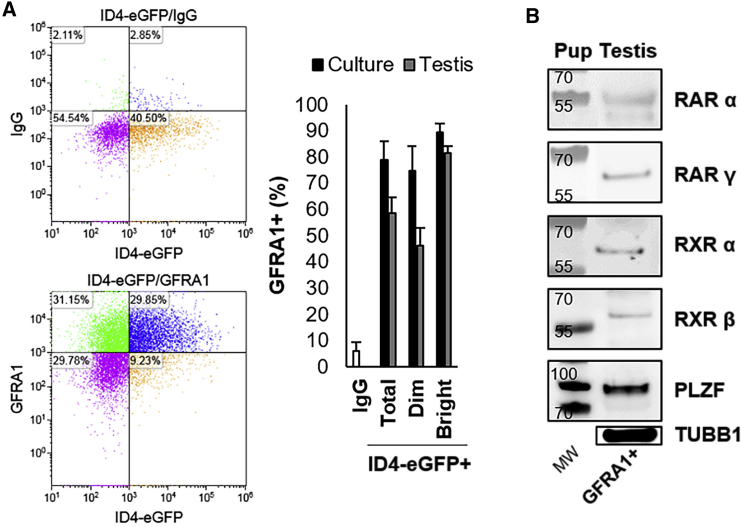
Lastly, we explored whether the expression of RAR and RXR isoforms by spermatogonia with stem cell characteristics is unique to the ID4-eGFP+ population. Based on the outcomes of immunostaining analyses, a recent study reported that RARγ is not expressed by GFRA1+ spermatogonia, which have also been considered to represent the SSC pool (Ikami et al., 2015). Here, we aimed to confirm this finding using different approaches. First, using flow cytometric analysis we found that the vast majority of ID4-eGFP+/SSCs (including both ID4-eGFPBright and ID4-eGFPDim subsets of the population) in primary cultures of spermatogonia, and directly from the pup testis, are GFRA1+, implying that the population does express RARs/RXRs (Figure 2A). Indeed, these findings are aligned with those in the literature in which ID4 and GFRA1 expression are highly analogous (Helsel et al., 2017b, Chan et al., 2014) (depicted in Figure S1). Second, using western blot analysis of FACS-isolated populations from testes of P6–8 mice we found that the GFRA1+ population expresses RARα, RARγ, RXRα, and RXRβ, similar to the ID4-eGFP+ population (Figure 2B). Taken together, these results suggest that both SSC and progenitor populations express a receptor complement for induction of RA signaling, and, thus, differential expression is unlikely to account for the resistance of SSCs to RA-induced differentiation in vivo. Importantly, it should be noted that the RARγ antibody used in this study has been subjected to stringent validation, and does not react with cells in testes of Rarγ null mice (Gely-Pernot et al., 2015).
Figure 2.
RAR/RXR Variants Are Expressed in the GFRA1+ Spermatogonial Population
(A) Quantification of the percentage of the ID4-eGFP+, ID4-eGFPBright, and ID4-eGFPDim populations that are GFRA1+ in primary cultures of undifferentiated spermatogonia and in pup testes using FACS analysis (representative images on left). Data are mean ± SEM for n = 3 different cultures or animals (biological replicates).
(B) Representative images of western blot analyses for RARα, RARγ, RXRα, and RXRβ in the GFRA1+ population isolated directly from pup testis. PLZF expression was assessed to confirm successful isolation of the undifferentiated spermatogonia population. Tubulin (TUBB1) expression was used as a loading control.
Activation of RA Signaling in SSC and Progenitor Populations
Considering that both SSC and progenitor spermatogonial populations in cultures and testes express RA receptors, we next aimed to assess whether this is sufficient to elicit a signaling response in both populations, and whether small fluctuations in RAR/RXR expression between SSC and progenitor subsets seen in Figure 1A translate to a differential RA response. To test this with direct exposure to RA, primary cultures of undifferentiated spermatogonia established from either pup (P6–8) or adult Id4-eGfp transgenic mice were treated with 0.5 μM all-trans RA or vehicle (DMSO) for 16 hr as described previously (Yang et al., 2013), followed by examination of the expression level of the RA-responsive genes Stra8 and Kit across ID4-eGFP+/SSC and ID4-eGFP–/progenitor populations. In these experiments, we reasoned that, if an intrinsic mechanism acts to block the RA response in SSCs, suppressed or reduced expression of RA-responsive genes would occur in the ID4-eGFP+ population compared with the ID4-eGFP– progenitors. Alternatively, if the RA response is blocked/prevented in SSCs by action of extrinsic factors in the niche, we would expect that direct exposure to RA in vitro would stimulate an equivalent STRA8/KIT response to that seen in the ID4-eGFP–/progenitor pool.
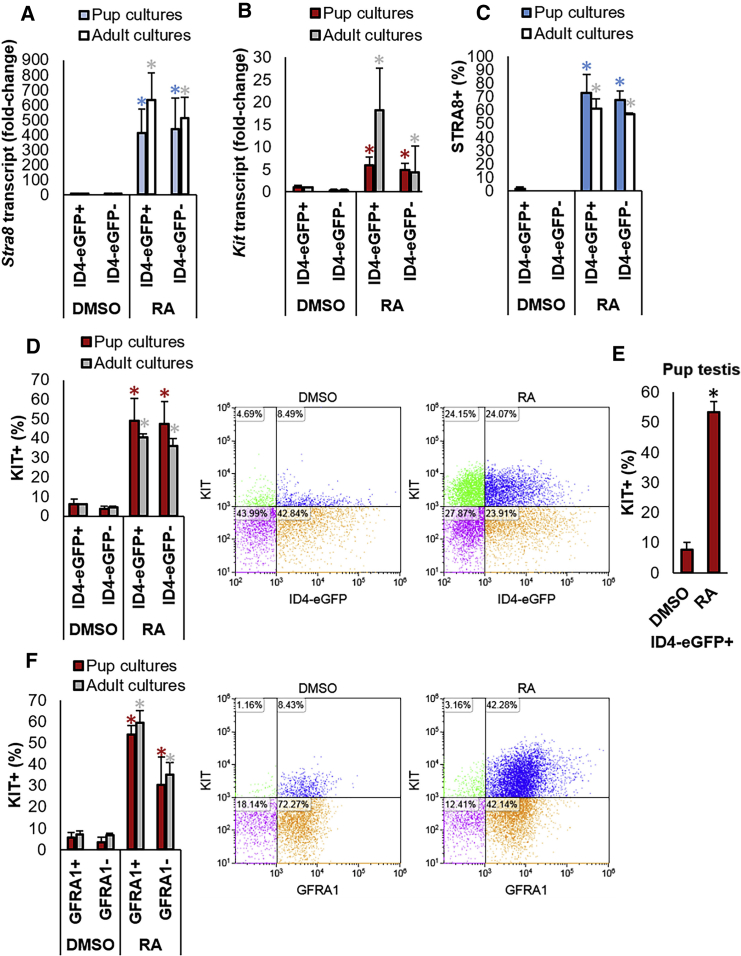
Using qRT-PCR analysis, we measured a significant (p < 0.01) induction of both Stra8 and Kit gene expression in ID4-eGFP+ and ID4-eGFP– populations of pup and adult cultures (n = 4 for each age) treated with RA compared with DMSO (Figures 3A and 3B). Furthermore, no difference in the level of induction between the two populations was measured for either of the parameters examined. These findings were further supported via analysis of STRA8 and KIT expression at the protein level. Based on immunocytochemical staining, 73% (±13.5%) and 61.3% (±7.0%) of the ID4-eGFP+ population in pup and adult cultures, respectively, were found to be STRA8+ at 16 hr post-RA treatment, which was not significantly different (p > 0.05) compared with the 67.5% (±7.1%) and 57.0% (±1.0%) of the ID4-eGFP– populations in pup and adult cultures, respectively (Figures 3C and S4A). The percentage of cells that were STRA8+ in all cultures treated with DMSO was negligible. Using flow cytometric analysis, we measured a significant (p < 0.05) increase in KIT+ cells for both ID4-eGFP+ and ID4-eGFP– populations treated with RA compared with DMSO (Figure 3D). In cultures derived from pups (n = 5), 49.0% (±11.6%) of ID4-eGFP+ and 47.5% (±11.4%) of ID4-eGFP– cells were KIT+ following RA exposure representing an 8- and 12-fold increase compared with DMSO treatment, respectively. For cultures derived from adult mice, 40.7% (±1.7%) and 36.1% (±3.8%) of cells were found to be KIT+ after RA treatment, representing a 7- and 8-fold increase compared with DMSO treatment, respectively. As with STRA8 expression, no significant differences (p > 0.05) in the percentage of KIT+ cells were detected between RA-treated ID4-eGFP+ and ID4-eGFP– populations for both pup and adult cultures. Next, we examined whether the RA response in the SSC population was an artifact associated with long-term in vitro culture. To achieve this, the SSC-enriched ID4-eGFP+ population was isolated from testes of P6–8 mice and subjected to an identical treatment regimen. Similar to cultured cells, 53.5% (±3.6%) of the ID4-eGFP+ cells became KIT+ following RA exposure, compared with only 7.6% (±2.6%) of the population following DMSO treatment (Figure 3E). Lastly, we explored whether these results are unique to the ID4-eGFP+ population by examining GFRA1+ cells, which are often considered to also represent the SSC pool and have been reported to be refractory to RA exposure (Ikami et al., 2015). Using flow cytometric analysis for KIT+ cells, we found that 54.0% (±5.6%) and 59.7% (±4.4%) of GFRA1+ cells were KIT+ after RA exposure in pup (n = 3) and adult (n = 3) cultures, respectively, which was significantly (p < 0.01) increased compared with the 5.6% (±1.7%) and 7.0% (±2.4%) of KIT+ cells in DMSO-treated cultures, respectively (Figure 3F).
Figure 3.
Responsiveness of SSC and Progenitor Spermatogonia to Direct RA Exposure In Vitro
(A and B) qRT-PCR analysis for relative Stra8 (A) and Kit (B) gene expression in ID4-eGFP+/SSC and ID4-eGFP–/progenitor populations of primary undifferentiated spermatogonial cultures established from P6–8 pups (blue or red bars, respectively) or adult (white or gray bars, respectively) mice treated with DMSO (control) or all-trans retinoic acid (RA). Both genes are hallmark responders of the RA signaling response in spermatogonia. Data are mean ± SEM for n = 3 different cultures (biological replicates). ∗Significantly different at p < 0.05 from DMSO control.
(C and D) Quantification of the percentage of ID4-eGFP+/SSC and ID4-eGFP–/progenitor cells in primary undifferentiated spermatogonial cultures established from pup or adult mice expressing STRA8 (C) or KIT (D) protein following DMSO (control) or RA treatment. Data were generated by immunocytochemical staining (STRA8+ cells, representative images provided in Figure S3) or flow cytometric analysis (KIT+ cells, representative histograms provided in D) of single-cell suspensions and are presented as mean ± SEM for n = 3 different cultures (biological replicates). ∗Significantly different at p < 0.05 from DMSO control.
(E) Quantification of the percentage of KIT+ cells in the ID4-eGFP+ population isolated from pup testes and treated with DMSO (control) or RA. Data are mean ± SEM for cell from n = 3 different animals. ∗Significantly different at p < 0.05 from DMSO control.
(F) Quantification of the percentage of KIT+/GFRA1+ cells in primary cultures of undifferentiated spermatogonia established from pup or adult mice following treatment with DMSO (control) or RA. Data were generated from flow cytometric analysis (representative scatterplots are presented) and are mean ± SEM for n = 3 different cultures (biological replicates). ∗Significantly different at p < 0.05.
Collectively, these findings confirm that not only are RARs and RXRs expressed in SSC and TA progenitor spermatogonial subsets, but the signaling pathway is functionally intact. Further, these results suggest that the resistance of SSCs to RA-induced differentiation in vivo is reliant on these cells being situated within a protective niche, and that intrinsically expressed factors are not sufficient for blocking the RA response. Notably, the trends are not unique to the dose of RA utilized because an equivalent KIT response occurs in ID4-eGFP+ and ID4-eGFP– populations at a range of RA concentrations; from 0.5 μM to 0.5 fM, with a stepwise reduction in the percentage of KIT+ cells with each reduction in RA concentration (Figure S5).
Alteration of SSC Functionality following Direct Exposure to RA
Next, we aimed to assess whether the activation of RA signaling responses in SSCs leads to a change in function. The experimental scheme is presented in Figure 4A. Because the percentage of undifferentiated spermatogonia that are ID4-eGFP+ is reflective of SSC content, both in vitro (Chan et al., 2014) and in vivo (Helsel et al., 2017b), we first decided to measure fluctuations in the pool in response to direct RA exposure. As reported previously (Helsel et al., 2017b), fluorescent intensity in the ID4-eGFP+ population can be subdivided into quadrants of Bright, Mid, Dim, and Negative in primary cultures derived from pups, and we observed a similar profile for cultures derived from adult cells (Figure 4B). In addition, we previously demonstrated that the density of cells with regenerative capacity in vivo is greatest in the ID4-eGFPBright population (Helsel et al., 2017b). Thus, to ensure that our findings in the ID4-eGFP+ population reflect that of the ID4-eGFPBright population, we confirmed RAR/RXR expression in ID4-eGFPBright cells from primary culture using Western blot analysis (Figures 4C and S3D). It should be noted that it was not possible to compare RAR/RXR expression in ID4-eGFPBright versus ID4-eGFPDim spermatogonia using an immunohistochemistry approach as, post-fixation, distinguishing between these “levels” of ID4-eGFP fluorescence is not possible. In support of our Western blotting analysis, however, acquisition of KIT expression following RA treatment (n = 3 cultures examined) was largely equivalent in ID4-eGFP Bright, Mid, and Dim populations of primary cultures established from either pup or adult cells (Figure 4D).
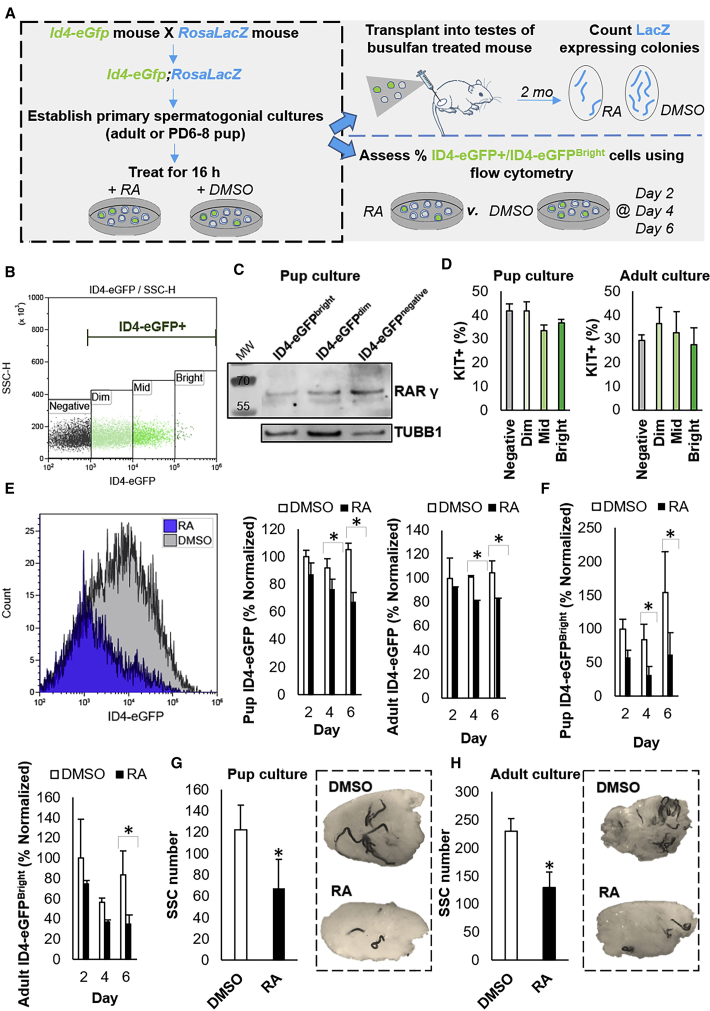
Figure 4.
Impact of Direct Exposure to RA on Regenerative Capacity of the SSC Population
(A) Experimental scheme for assessing changes in SSC content of primary undifferentiated spermatogonial cultures following direct exposure to RA. Note that all spermatogonia from the culture well (ID4-eGFP+ and ID4-eGFP–) were subjected to DMSO or RA treatment.
(B) Representative scatterplot for the ID4-eGFP+ population in primary cultures of undifferentiated spermatogonia. The population can be subdivided based on Bright, Mid, Dim, and Negative EGFP intensity and the SSC population is mostly contained in the ID4-eGFPBright subset.
(C) Representative image of western blot analysis for RARγ expression in ID4-eGFP Bright, Dim, and Negative spermatogonial pools from primary cultures. Tubulin (TUBB1) expression was used as a loading control.
(D) Quantification of KIT+ cells in the ID4-eGFP subpopulations following direct exposure to RA. Data are presented mean ± SEM for n = 3 different cultures established from 3 independent P6–8 pups or adult mice.
(E) Quantification of the percentage of cells that could be classified as ID4-eGFP+ in pup- or adult-derived primary cultures at 2–6 days after treatment with DMSO (control) or RA. Data were generated from flow cytometric analyses (representative histogram plot is presented) and are presented as mean ± SEM for n = 3 different cultures (biological replicates).
(F) Quantification of the percentage of cells in primary cultures established from pup or adult mice that were ID4-eGFPBright at 4 and 6 days after treatment with DMSO (control) or RA. Data were generated by flow cytometric analysis and are presented as mean ± SEM for n = 3 different cultures (biological replicates).
(G and H) Quantification of SSC number (donor-derived colonies of spermatogenesis/105 cells transplanted) in primary cultures established from (G) pup or (H) adult mice, transplanted 16 hr after treatment with DMSO (control) or RA. Data were generated by functional transplantation analyses (representative images of recipient testes with LacZ-stained donor-derived spermatogenic colonies are presented) and are mean ± SEM for n = 3 different cultures (biological replicates).
∗Significantly different at p < 0.05.
In order to track an impact of direct exposure to RA on functional changes of the SSC pool, we first assessed alteration of the ID4-eGFP+ population at 2, 4, and 6 days post-RA or -DMSO treatment of the entire undifferentiated spermatogonial population within the culture well. A significant (p < 0.01) reduction in the percentage of the overall ID4-eGFP+ population in primary cultures of spermatogonia established from pups and adult mice (n = 3) was measured in RA-treated cells compared with DMSO-treated cells at day 4 and 6 (Figure 4E). Likewise, the percentage of ID4-eGFPBright cells was significantly (p < 0.05) diminished following RA exposure at 4 and 6 days post-RA treatment (Figure 4F). Second, we carried out transplantation analyses to assess alteration in the regenerative stem cell content of cultures following RA treatment. Outcomes aligned with changes in the ID4-eGFPBright population (Figures 4G and 4H), in that the number of donor-derived spermatogenic colonies was significantly (p < 0.05) reduced in cultures treated with RA (67.8 ± 26.9 colonies/105 cells for pup cultures and 130.2 ± 24.7 colonies/105 cells for adult cultures) following as little as 16 hr exposure compared with DMSO-treated cultures (122.5 ± 22.7 colonies/105 cells for pup cultures and 230 ± 35.5 colonies/105 cells for adult cultures). Collectively, these data demonstrate that direct exposure to RA outside of the in vivo microenvironment elicits a response in SSCs that alters their functional state by pushing them toward differentiation.
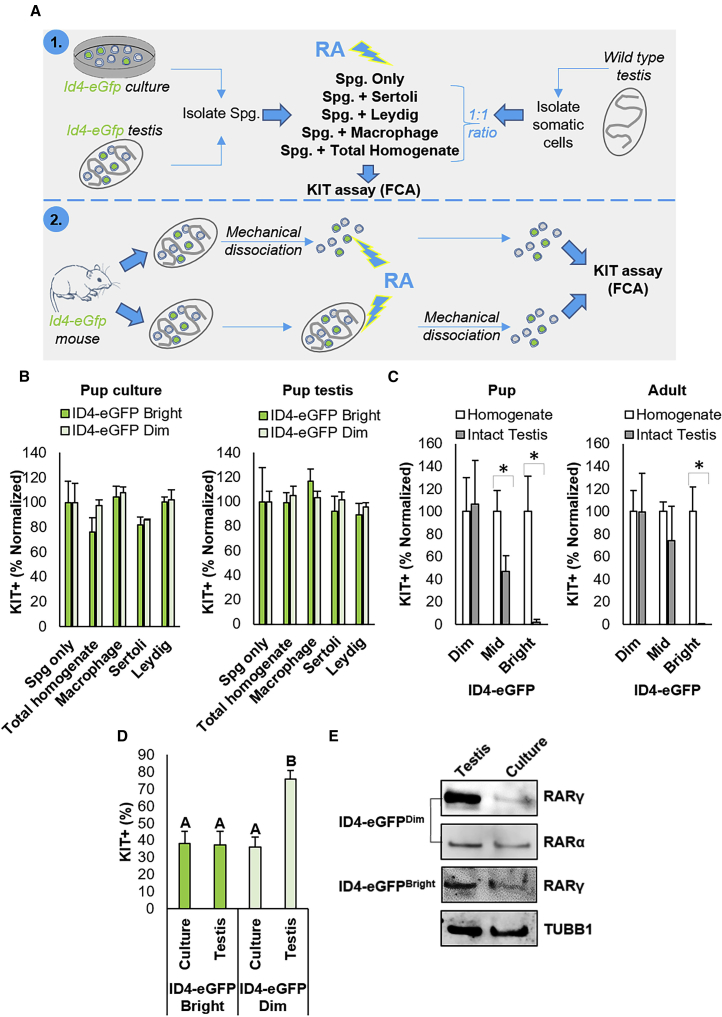
The Role of the Soma in Modulating the RA-Induced Differentiating Transition
The findings presented above strongly suggest that influences of an in vivo niche protect SSCs from the RA signal by either preventing direct exposure or by providing extrinsic signals that activate an intrinsic suppressive mechanism and that these are lacking in the culture conditions we utilized. Our experimental approach to explore this possibility is depicted in Figure 5A. First, we pre-incubated spermatogonia from culture or ID4-eGFP+ spermatogonia isolated from P6–8 testes with different testis somatic cell populations in vitro, including Leydig cells, Sertoli cells, F4/80+ testicular macrophages, or a “total testis homogenate,” in which all these cell populations were present. Combined cell types were then exposed directly to RA and assessed for KIT expression via flow cytometry as a readout for activation of the RA signaling response. Although modest fluctuations in the percentage of KIT+ cells could be identified in ID4-eGFPBright and ID4-eGFPDim populations from culture and from the testis in response to the presence of these different somatic cell populations (Figure 5B), no somatic cell population was capable of ablating the RA response in SSCs. Next, we explored whether the different spermatogonial subsets respond to RA exposure when located within a normal testicular architecture by treating testes excised from pup and adult mice in vitro (Figure 5C). Note that, in these experiments, ID4-eGFP+ spermatocytes were excluded from analyses on adult testis homogenates by preselecting for undifferentiated spermatogonia using E-cadherin antibody labeling (E-cadherin expression profile is depicted in Figure S1). In contrast to exposure as a single-cell suspension, the RA signaling response in ID4-eGFPBright cells was ablated in intact testes (Figures 5C and S4B); with a >46-fold reduction in the number of KIT+ cells compared with the ID4-eGFPBright population in single-cell suspension from the mechanically disassociated contralateral testis (p < 0.05, n = 4 for pup, n = 3 for adult, values in histogram are normalized to the testis homogenate control). The percentage of KIT+ cells was also halved in the ID4-eGFPMid population in P6–8 intact testes compared with single-cell suspensions (p < 0.05). The absence of KIT+ cells in the ID4-eGFPBright pool from RA-treated intact testes was not due to a lack of penetration of RA, as equivalent numbers of KIT+ spermatogonia were detected in the ID4-eGFPDim pool regardless of whether the testis was disassociated or not. These trends can also be appreciated in whole-mount immunofluorescence analyses of RA-treated testes in which ID4-eGFPBright/SSCs are KIT–, while the KIT+ cells are ID4-eGFPDim or ID4-eGFP– progenitors (Figure S4B).
Figure 5.
Influence of the Testicular Soma on Modulation of the RA Response in SSC and Progenitor Spermatogonia
(A) Experimental scheme for examining the influence of different testicular somatic cell populations and topographic architecture on RA responsiveness of SSC and progenitor spermatogonia using KIT flow cytometric analysis (FCA).
(B) Quantification of the percentage of KIT+ cells in the ID4-eGFP+ spermatogonial population from primary cultures and from P6–8 pup testis combined at a 1:1 ratio with Sertoli cells, Leydig cells, macrophages, or total testis homogenate (containing all somatic cell populations) and treated with RA. ID4-eGFP+ cells not combined with somatic cells (Spg only) were used as a control. Data are mean ± SEM for n = 3 different cultures or animals (biological replicates). The percentage of both ID4-eGFPBright and ID4-eGFPDim subpopulations that were KIT+ are shown normalized to the Spg only control value.
(C) Quantification of the percentage of KIT+ cells in the Bright, Mid, and Dim subsets of the ID4-eGFP+ population in disassociated single testicular cell suspensions or intact testes from pup and adult mice that were treated with RA. Data are presented as mean ± SEM for n = 3 different animals (biological replicates). ∗Significantly different at p < 0.05. Data are normalized to the digested testis control.
(D) Quantification of the percentage of KIT+ cells in ID4-eGFPBright/SSC and ID4-eGFPDim/progenitor spermatogonial populations isolated from primary undifferentiated spermatogonial cultures or testes directly and exposed to RA. Data are presented as mean ± SEM for n = 3 different cultures or animals (biological replicates). ∗Significantly different at p < 0.05.
(E) Representative images of western blot analysis for expression of RARγ and RARα in ID4-eGFPBright/SSC and ID4-eGFPDim/progenitor spermatogonial subsets isolated from primary cultures of undifferentiated spermatogonia or testes directly. Tubulin (TUBB1) was used as a loading control.
Finally, we observed that the in vivo environment is also important for priming TA progenitors to respond to the RA signal. When exposed directly to RA, the percentage of ID4-eGFPDim spermatogonia that became KIT+ was halved (76.1% [±5.0%] versus 36.3% [±5.7%]) if these spermatogonia had been subjected to long term in vitro culture, rather than treated immediately following isolation from the testis (p < 0.01) (Figure 5D). Contrastingly, when exposed directly to RA in a single-cell suspension the percentage of KIT+ cells in the ID4-eGFPBright population remained constant. To assess whether extrinsic signals in vivo “prime” the ID4-eGFPDim spermatogonia by stimulating upregulation of Rarγ expression, Western blot analysis was performed on ID4-eGFPDim and ID4-eGFPBright populations isolated directly from testes or primary cultures (Figure 5E). While RARγ levels were equivalent when comparing the two populations of ID4-eGFPBright cells, expression was markedly greater in the ID4-eGFPDim population from the testis directly compared with the ID4-eGFPDim population from primary cultures. A similar trend was not identified in comparing levels of RARα expression (Figure 5E).
Taken together, these findings demonstrate that the topological arrangement of cell types in the testis and unique architecture influence the responsiveness of SSC and TA progenitor spermatogonial populations to the RA-induced differentiating transition. Moreover, these results indicate that discrete niche microenvironments are established by cell-to-cell communication in mammalian testes to modulate extrinsic cues that influence fate decisions of stem cells and progenitors in the male germline.
Discussion
Maintenance of the SSC population in testes is critical for the preservation of male fertility. Indeed, it is from the SSC reservoir that the entirety of spermatogenesis arises in adult life. As such, it is integral that SSCs are not lost to differentiation in response to the RA signal that is released asynchronously throughout seminiferous tubules to drive the A (undifferentiated) to A1 (differentiating) transition in TA progenitor spermatogonia. At present, the mode by which SSCs and progenitor spermatogonia differentially respond to RA while co-residing in a seemingly facultative environment of the seminiferous epithelium is unresolved. Indeed, conflicting reports exist in the peer-reviewed scientific literature about whether the different subsets of spermatogonia express receptors for RA signaling. In studies of Ikami et al. (2015), this phenomenon was attributed to a lack of RARγ expression in SSCs that is gained upon the progenitor transition, thereby preventing the self-renewing SSCs from responding to the RA signal. In that study, the GFRA1+ and NEUROG3+ spermatogonia were examined in the context of the entire populations being an SSC or progenitor pool, respectively, and expression of RARγ was reported to be completely absent in GFRA1+ cells but present in NEUROG3+ cells. Although these findings make for an easy explanation for why progenitors respond to RA but SSCs do not, they are inconsistent with previous studies that have reported expression of RARγ in some GFRA1+ spermatogonia, and that a majority of these cells do indeed retain the capacity to differentiate even in the absence of spermatogonial Rarγ expression (Gely-Pernot et al., 2012).
In the current study, we utilized multiple, complimentary approaches to demonstrate expression of RARγ, as well as other RAR/RXR variants, in a spermatogonial population that is highly enriched for SSCs (i.e., ID4-eGFP+ cells) as well as the GFRA1+ population. Further, we discovered that the receptor complement is functionally intact in both SSC and progenitor subsets to elicit hallmark responses of RA signaling, and induce a loss of regenerative capacity in SSCs. So, why the discrepancy with the findings of Ikami et al. (2015)? One explanation could be that assessment of RARγ expression in the Ikami et al. study was made using immunostaining analyses, which may not be sensitive enough for detecting expression by all GFRA1+ spermatogonia. Indeed, our Western blotting results on spermatogonia isolated from the testis directly revealed that RARγ expression is higher in progenitor spermatogonia than SSCs. Thus, based on the immunostaining technique employed, detection of moderate-to-low RARγ levels within particular cellular subsets may be challenging, and more sensitive techniques are required to uncover the full spectrum of expression among all spermatogonia. In addition, we were able to detect expression of RARs by GFRA1+ spermatogonia using multiple approaches, all of which revealed expression of RARγ and RARα. Moreover, we found that direct exposure to RA elicits a hallmark signaling response in both GFRA1+ and ID4-eGFP+ spermatogonia, demonstrating that the receptor complement is functionally intact. These findings suggest that, rather than being protected from RA-induced differentiation by means of differential RARγ expression, the SSC population is hidden from the RA pulse in vivo by intricate and distinct niche microenvironments established by surrounding somatic cells in the testis.
Stem cells of most, if not all, tissues are thought to reside in a niche microenvironment that is comprised of contributions from support cells. In mammalian testes, the SSC-soma interaction is thought to constitute a niche unit that dictates fate decisions of self-renewal or formation of progenitor spermatogonia that no longer inhabit the niche (de Rooij, 2017, Oatley and Brinster, 2012). However, recent studies have proposed that discrete niche microenvironments for SSCs do not exist, rather all spermatogonia inhabit a facultative or open environment within the seminiferous epithelium (Hara et al., 2014). Findings in the current study suggest that the arrangement of cell types within the testis provides an intricate microenvironment for SSCs to protect them from RA-induced differentiation. Indeed, while exogenous RA exposure does not induce KIT expression in the ID4-eGFPBright SSC population in the architecturally undisturbed testis, a vast majority of the population becomes KIT+ in the contralateral testis that has been mechanically disassociated into a single-cell suspension before RA exposure. Contrastingly, the ID4-eGFPDim spermatogonia exhibit a robust KIT response in both experimental situations. Thus, the simple presence of the testicular somatic cells is not sufficient to provide a protective environment in which SSCs are shielded from RA-induced differentiation, but rather it is the interconnected microenvironment that they create which provides this protection. Also, the fact that ID4-eGFPDim spermatogonia respond to RA exposure both within the intact testis and as a dispersed single-cell suspension, whereas the ID4-eGFPBright spermatogonia do not, suggests that the populations exist in different microenvironments that receive disparate extrinsic signals from the surrounding somatic support cells. These findings are supported by those recently published by Agrimson et al. (2017), in which live mice were injected with RA, and the effects on the SSC population in the undisturbed testis was assessed. In that study, SSCs in the undisturbed testes of pre-pubertal mice remained protected from differentiation in the presence of excess RA, as demonstrated by spermatogonial transplantation analyses. While stem cell content was unchanged, the percentage of STRA8+ cells in the ID4-eGFP+ population was significantly elevated. Thus, although Agrimson et al. did not compare STRA8 staining in ID4-eGFPBright versus ID4-eGFPDim cells, we can predict that this elevated STRA8+ abundance would be attributed to a response in the ID4-eGFPDim population, as reflected in the current study.
A prominent unanswered question is what are the external signals within the SSC-niche unit microenvironment in vivo that curtail the RA response? The most obvious candidates are the CYP26 enzymes that are potent degraders of RA. Both Sertoli cells and germ cells are known to express the Cyp26a1 and Cyp26b1 variants, and simultaneous knock down of Cyp26b1 in Sertoli and germ cells results in a severe subfertility phenotype (Hogarth et al., 2015). Indeed, 96% of tubules are abnormal in these CYP26B1-deficient mice, and a vast majority of tubules possess obvious vacuolization or a complete loss of germ cells by P15 (Hogarth et al., 2015). In addition, simultaneous knock down of Cyp26b1 and Cyp26a1 in Sertoli cells specifically results in overabundant and precocious STRA8 expression in the spermatogonial population (Hogarth et al., 2015). Together, these findings suggest that when the capacity for CYP26B1/A1 production in the testis is impaired, the SSC population may no longer be protected from RA-induced differentiation, and, as a consequence, the capacity for continuation of spermatogenesis is lost. Despite this, how production of these enzymes by Sertoli cells could act to degrade RA selectively within microenvironments surrounding SSC populations remains elusive. Certainly, crosstalk between germ cells and Sertoli cells is known to exist to influence gene expression. For example, spermatogonia have been shown to produce ligands such as JAG1 that activate NOTCH signaling in Sertoli cells (Garcia et al., 2017), modulating Sertoli expression of both Gdnf and Cyp26b1 (Garcia et al., 2013, Garcia et al., 2017). However, whether differential spermatogonia-somatic cell interactions exist in SSCs versus progenitors, and whether these interactions relate directly to the RA response in vivo will need to be defined by future investigation. Promisingly, however, mining of our previously published RNA-seq database (Helsel et al., 2017b) revealed that expression of the NOTCH ligands Dll1 and Dll3, as well as Jag1, is elevated in ID4-eGFPDim cells compared with ID4-eGFPBright cells. It should be noted that although low levels of Cyp26a1 and Cyp26b1 expression have been detected in ID4-eGFPBright/SSC and ID4-eGFPDim/progenitor populations specifically by our previous RNA-seq analyses (Helsel et al., 2017b), differential expression does not appear to exist between these cell types (p > 0.1), supporting findings in the current study that demonstrate a lack of intrinsic protective mechanisms against RA in the SSC population. Also, in line with these findings, knockdown Cyp26 expression in germ cells alone does not impair fertility (Hogarth et al., 2015).
In addition to the influence of the in vivo microenvironment provided by normal testicular architecture on maintenance of the SSC pool, we also discovered an important effect on “priming” progenitor spermatogonia for maximal response to RA. Specifically, we found that ID4-eGFPDim/progenitor spermatogonia isolated directly from the testis possess markedly higher levels of RARγ expression compared with the contemporary population isolated from primary culture. This elevated level of RARγ expression apparently increases the sensitivity of these cells to exogenous RA exposure, as demonstrated by a doubling in the percentage of KIT+ cells. This result marries with previous data from Ikami et al. (2015) that showed induction of ectopic expression of RARγ in GFRA1+ spermatogonia resulted in increased differentiation rates following physiological pulses of RA in all but a subset of spermatogonia (presumably the true SSCs). In the current study, RARγ levels were unchanged in SSC populations regardless of origin, isolated from the testis directly or from primary cultures of spermatogonia. These findings suggest that extrinsic signals in the testis act on the progenitor spermatogonia specifically to upregulate expression of RARγ, again supporting the notion for discrete microenvironments within the seminiferous tubules that dictate differential function of SSCs and progenitors. These experiments also highlight insufficiencies in current in vitro culture conditions in terms of maintaining progenitors in their true biological state.
In conclusion, we have provided empirical evidence that the current dogma surrounding the RA response in SSCs versus TA progenitor spermatogonia should be revisited. Specifically, we have demonstrated expression of RARα, RARγ, RXRα, and RXRβ, as well as their functionality in the SSC pool following direct RA exposure; suggesting that a lack of RARγ expression is unlikely to account for the resistance to RA-induced differentiation that SSCs exhibit in vivo. Instead, we have shown that the SSC population only remains protected from RA when the intricate architecture of the of the SSC niche microenvironment remains undisturbed; a situation that cannot be recapitulated by combining SSCs with different testicular somatic cell populations as a single-cell suspension in vitro, even in the presence of the entire complement that is present in the intact testis. Collectively, our findings depict a previously underappreciated role for testis architecture and microenvironments in modulating RA-induced differentiation in spermatogonial populations and provide compelling evidence for intricate niche microenvironments in the mammalian testis that influence fate decisions of SSC and progenitor spermatogonia. Understanding of these systems not only has the potential to be influential on our perception of stem cell niche environments in other tissue types, but may facilitate advances in our knowledge of underlying causes of male infertility, and potentially the development of novel male contraceptive agents based on RA responsiveness.
Experimental Procedures
Animals
All animal procedures were approved by the Washington State Institutional Animal Care and Use Committee. The Id4-eGfp transgenic mouse line was described previously (Chan et al., 2014). Id4-eGfp mice were crossed with Rosa26-LacZ mice (Jackson Laboratories, stock number 112073) to generate donors for the establishment of spermatogonial culture that are suitable for transplantation analyses. Recipients for spermatogonial transplantation were F1 hybrids of C57BL/6J (Jackson Laboratories, stock number 000664) and 129S1/SvlmJ (Jackson Laboratories, stock number 112073). F1 hybrids were treated with busulfan (Sigma-Aldrich, MO, USA, B2635) to eliminate endogenous spermatogenesis prior to transplantation, as described previously (Oatley and Brinster, 2006).
Cell Culture
Primary cultures of undifferentiated spermatogonia were generated from double-transgenic Id4-eGFP;Rosa26-LacZ hybrid mice that express the LacZ transgene within all germ cells, but express the EGFP transgene only in ID4+ SSCs (Chan et al., 2014, Helsel et al., 2017b). Primary cultures of spermatogonia were established from the magnetic-activated cell sorting (MACS)-sorted THY1+ fraction of testis homogenates of P6–8 or adult mice (∼2–4 months of age) as described previously (Helsel et al., 2017a, Kaucher et al., 2012, Oatley and Brinster, 2006). Where cultures were used for experimental replicates, each culture was derived from a different mouse, and was established independently. In these cultures, >90% of SSCs are captured in the ID4-eGFP+ population, while the ID4-eGFP– cells are progenitor spermatogonia that have lost the capacity for self-renewal, but maintain expression of undifferentiated markers such as Plzf, Utf1, Lin28, and Pou5f1, as well as germ cell markers such as Dazl and Ddx4 (Chan et al., 2014). Spermatogonial cultures were maintained on mitotically inactivated SIM mouse embryo-derived thioguanine- and ouabain-resistant feeder monolayers (STOs) in mouse serum-free medium (mSFM) supplemented with the growth factors GDNF (20 ng/mL; Peprotech, NJ, USA) and fibroblast growth factor 2 (1 ng/mL; Peprotech). While mSFM used for P6–8 cultures was constituted exactly as described previously (Kaucher et al., 2012, Kubota et al., 2004a, Kubota et al., 2004b), conditions for adult cultures were slightly modified in that the media was devoid of fatty acids (Helsel et al., 2017a). Also, while P6–8 cultures were kept within humidified incubators at 37°C in 5% CO2 in air, adult cultures were maintained in glycolysis-optimized conditions (Helsel et al., 2017a). Culture media was replaced every second day, and passaging onto fresh feeders was performed every 6–8 days.
For treatment of cultured spermatogonia with all-trans RA (0.5 μM) or DMSO (vehicle), the spermatogonial clumps were separated from feeders by gentle pipetting and single-cell suspension generated by trypsin-EDTA digestion followed by exposure for 16 hr (Yang et al., 2013).
Flow Cytometry
For flow cytometric analysis, single-cell suspensions from the testis or from spermatogonial cultures were generated by trypsin-EDTA digestion as described previously (Chan et al., 2014) and analyzed using an Attune NxT Flow Cytometer (Thermo Scientific, MA, USA). For FACS, single-cell suspensions were fractionated using a Sony SH800 machine. Identification and gating of the ID4-eGFP+ and ID4-eGFP– populations from cultures and pup testes was as described previously (Chan et al., 2014). For assessment of ID4-eGFP+ populations from adult mouse testes, undifferentiated spermatogonia were labeled with a fluorescein-conjugated E-cadherin antibody by incubation at a 1:100 dilution (gated off PE/Cy7 isotype control). For KIT labeling to compare DMSO- and RA-treated spermatogonia, cells were incubated in a fluorescein-conjugated antibody at 1:200 dilution. Gating was selected based on an untreated population of undifferentiated spermatogonia from culture that do not express KIT (Yang et al., 2013). A control population in which primary antibody was omitted was also utilized. For GFRA1 labeling, cells were incubated with an unconjugated primary antibody at 1:100 dilution followed by incubation with a fluorescein-conjugated secondary antibody at 1:500 dilution. Gating was selected based on a control population in which primary antibody was omitted. All antibodies utilized for flow cytometry are listed in Table S1. All incubations for conjugated primary antibodies were conducted in DPBSS (Dulbecco's PBS with 0.1% fetal bovine serum, 10 mM HEPES, 10 mM sodium pyruvate, 1 mg/mL glucose, and 1 mg/mL penicillin/streptomycin) for 20 min on ice. GFRA1 primary antibody incubation was in mSFM overnight at 37°C, and secondary incubation was in mSFM for 2 hr at 37°C.
RT-PCR and qRT-PCR
RNA extraction of isolated spermatogonial cells was conducted using TRIzol reagent (Invitrogen, CA, USA) as per the manufacturer's instructions. Extracted RNA was treated with DNase I, and RNA concentration and purity were determined using a NanoDrop 2000 spectrophotometer (Thermo Scientific) prior to reverse transcription using oligo d(T) priming and Superscript III reverse transcriptase (Invitrogen). Integrity of resulting cDNA was confirmed via PCR using primers specific to the housekeeping gene Gapdh. Where stated, qRT-PCR was conducted using validated TaqMan probes and an ABI 7500 fast sequence detection system (Applied Biosystems, NY, USA). Transcript abundance was normalized to the constitutively expressed ribosomal protein S2 (Rps2), and calculated using the 2-ΔΔCt formula, as described previously (Yang et al., 2013). Sequences of all primers are provided in Table S2.
Western Blotting
Protein lysates from spermatogonia were collected using RIPA buffer (Thermo Scientific) supplemented with protease inhibitor (Thermo Scientific) and phosphatase inhibitor (Sigma-Aldrich) cocktails. Samples were resolved in 4%–12% Bis-Tris NuPAGE gels using an XCell SureLock Mini-Cell electrophoresis system (Invitrogen) followed by transfer to nitrocellulose membranes. Prior to addition of primary antibodies, nitrocellulose membranes were blocked in either 5% BSA in Tris-buffered saline (TBS) containing 0.1% Tween (TBST), or 5% skim milk in TBST for 1 hr at room temperature. Primary antibodies are listed in Table S1. All primary antibodies were diluted 1:2,000 in either 5% BSA/TBST (RARα and RXRβ) or 5% skim milk/TBST (RARγ and RXRα), and blots were incubated overnight at 4°C with gentle rocking. On the next day, blots were washed with TBST prior to a 1 hr incubation in secondary antibody (1:2,000 goat anti-rabbit IgG horseradish peroxidase, Santa Cruz Biotechnology) at room temperature. Blots were again washed, then developed using ECL prime chemiluminescence detection reagent (GE Healthcare, PA, USA) and visualization on a LAS 4000 imager (GE Healthcare).
Immunofluorescence Analyses
For immunofluorescent staining, testes were fixed in Bouin's solution and embedded in paraffin followed by cross-sectioning (5 μM thickness). After de-paraffinization, antigen retrieval was conducted by boiling slides in citraconic anhydride buffer (pH 7.4, 0.5% citraconic anhydride [Sigma-Aldrich] in ultrapure water) for 20 min. Following this, sections were blocked for non-specific antibody staining using 10% donkey serum in PBS for 1 hr at room temperature. The primary/secondary antibodies and dilutions were as described in Table S1. Sections were incubated in primary antibody or control IgG overnight at 4°C, followed by washing in PBS and incubation with fluorescein-conjugated secondary antibody for 2 hr at room temperature. Cross-sections were again washed in PBS, and mounted in ProLong Gold antifade reagent containing DAPI (Invitrogen).
For immunofluorescence analyses of isolated spermatogonial cell suspensions, cells were adhered to poly-lysine-coated cover slips and fixed in 4% paraformaldehyde for 10 min before permeabilization in ice-cold methanol. Non-specific antibody binding was blocked using 10% serum in 3% BSA/PBS, followed by overnight incubation with primary antibody at 4°C. On the next day, coverslips were washed in PBS, followed by incubation with secondary antibodies for 1 hr at room temperature.
Immunostained cross-sections and single cells were visualized using an IX 51 model inverted microscope (Olympus) and digital images were captured using a DP71 digital microscope camera and CellSense software (Olympus, Tokyo, Japan).
For whole-mount imaging, testes were de-tunicated, and seminiferous tubules gently teased apart on a glass slide. Tubules were placed under a coverslip in mounting media (VectaMount AQ, Vector laboratories, CA, USA) and imaged on a Leica TCS SP5 II confocal microscope.
Spermatogonial Transplantation
The relative SSC content of primary spermatogonial cultures was assayed via germ cell transplantation as described previously (Helsel and Oatley, 2017). In brief, an equal number of cells from primary cultures of undifferentiated spermatogonia established from Id4-eGfp;Rosa26LacZ mice were treated with 0.5 μM RA (Sigma-Aldrich) or DMSO (vehicle control) for 16 hr. Single-cell suspensions of the entire culture well (i.e., containing both ID4-EGFP+ and ID4-eGFP– cells) were then collected, washed by centrifugation and then suspended in mSFM at a concentration of 1 × 106 cells/mL for transplantation. For each recipient mouse testis, 10 μL of cell suspension (equivalent to 10,000 cells) was microinjected into seminiferous tubules via the efferent duct. Assessment of donor-derived spermatogenesis was conducted 2 months post-transplantation by X-Gal staining for LacZ-expressing colonies. Colony numbers were quantified using an SZ51 dissecting microscope used to derive relative SSC contents as described previously (Chan et al., 2014, Oatley and Brinster, 2006).
Isolation of Testis Somatic Cell Populations and Recombination with Spermatogonial Subsets
Enriched populations of Sertoli and Leydig cells were isolated from the one testis of several C57BL/6J adult mice (>6 weeks of age) using methodologies described by Chang et al. (2011). Alongside these cell preparations, the contralateral testes were used to generate a heterogeneous “total testis homogenate,” and also for isolation of testicular macrophage populations. To generate a single-cell suspension from the contralateral testis, the tunica albuginea was removed and automated manual dissociation was conducted using a gentleMACS machine (Miltenyi Biotech, Bergisch Gladbach, Germany). Testicular macrophages were isolated using MACS anti-F4/80 microbeads (Miltenyi Biotech), as per the manufacturer's instructions. For experiments where somatic cell populations were recombined with spermatogonial populations, each population was present at a 1:1 ratio. A 1:1 ratio was selected following preliminary experiments in which additional ratios were examined but produced identical outcomes (data not shown). It should be noted that the “total testis homogenate” provided a control in which normal spermatogonia to somatic cell ratios were maintained, allowing further confirmation that the trends being identified were biologically relevant.
In experiments where RA treatment was conducted using intact testes, the tunica albuginea was removed and the testis placed in 1 mL mSFM with growth factors and 0.5 μM RA. The contralateral testis was dissociated using gentleMACS and the total homogenate was also re-suspended in 1 mL of the mSFM/RA solution. For these experiments, RA incubation was for 4 hr to limit the impact of degeneration of the testicular tissue that would occur with a longer incubation period.
Statistical Analyses
All experiments were replicated at least three times with different biological samples. Data are presented as the mean ± SEM for the replicates. Differences between means of treatment groups was determined statistically using the one-way ANOVA or t test function of GraphPad Prism 6 software (La Jolla, CA, USA). Multiple comparison analysis was carried out using the Tukey's post-hoc test function of GraphPad. A value of p < 0.05 was considered to be statistically significant.
Author Contributions
Conceptualization, T.L. and J.M.O.; Methodology, T.L. and J.M.O.; Investigation, T.L., M.J.O., and J.M.O.; Writing – Original Draft, T.L. and J.M.O.; Writing – Review & Editing, T.L. and J.M.O.; Funding Acquisition, J.M.O.
Acknowledgments
This research was supported by grant HD061665 awarded to J.M.O. from the NICHD. T.L. is the recipient of the Lalor Foundation Postdoctoral Fellowship. The authors wish to thank Ryan D. Anderson for his assistance in immunofluorescence experiments.
Published: February 1, 2018
Footnotes
Supplemental Information includes five figures and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.01.003.
Supplemental Information
References
- Agrimson K.S., Oatley M.J., Mitchell D., Oatley J.M., Griswold M.D., Hogarth C.A. Retinoic acid deficiency leads to an increase in spermatogonial stem number in the neonatal mouse testis, but excess retinoic acid results in no change. Dev. Biol. 2017;432:229–236. doi: 10.1016/j.ydbio.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buageaw A., Sukhwani M., Ben-Yehudah A., Ehmcke J., Rawe V.Y., Pholpramool C., Orwig K.E., Schlatt S. GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes. Biol. Reprod. 2005;73:1011–1016. doi: 10.1095/biolreprod.105.043810. [DOI] [PubMed] [Google Scholar]
- Chan F., Oatley M.J., Kaucher A.V., Yang Q.-E., Bieberich C.J., Shashikant C.S., Oatley J.M. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 2014;28:1351–1362. doi: 10.1101/gad.240465.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-F., Lee-Chang J.S., Panneerdoss S., MacLean J.A., Rao M.K. Isolation of Sertoli, Leydig, and spermatogenic cells from the mouse testis. Biotechniques. 2011;51:341–344. doi: 10.2144/000113764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij D.G. The nature and dynamics of spermatogonial stem cells. Development. 2017;144:3022–3030. doi: 10.1242/dev.146571. [DOI] [PubMed] [Google Scholar]
- Ebata K.T., Zhang X., Nagano M.C. Expression patterns of cell-surface molecules on male germ line stem cells during postnatal mouse development. Mol. Reprod. Dev. 2005;72:171–181. doi: 10.1002/mrd.20324. [DOI] [PubMed] [Google Scholar]
- Garcia T.X., DeFalco T., Capel B., Hofmann M.-C. Constitutive activation of NOTCH1 signaling in Sertoli cells causes gonocyte exit from quiescence. Dev. Biol. 2013;377:188–201. doi: 10.1016/j.ydbio.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia T.X., Parekh P., Gandhi P., Sinha K., Hofmann M. The NOTCH ligand JAG1 regulates GDNF expression in Sertoli cells. Stem Cells Dev. 2017;26:585–598. doi: 10.1089/scd.2016.0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gely-Pernot A., Raverdeau M., Célébi C., Dennefeld C., Feret B., Klopfenstein M., Yoshida S., Ghyselinck N.B., Mark M. Spermatogonia differentiation requires retinoic acid receptor γ. Endocrinology. 2012;153:438–449. doi: 10.1210/en.2011-1102. [DOI] [PubMed] [Google Scholar]
- Gely-Pernot A., Raverdeau M., Teletin M., Vernet N., Féret B., Klopfenstein M., Dennefeld C., Davidson I., Benoit G., Mark M. Retinoic acid receptors control spermatogonia cell-fate and induce expression of the SALL4A transcription factor. PLoS Genet. 2015;11:e1005501. doi: 10.1371/journal.pgen.1005501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghyselinck N.B., Dupé V., Dierich A., Messaddeq N., Garnier J.M., Rochette-Egly C., Chambon P., Mark M. Role of the retinoic acid receptor beta (RARbeta) during mouse development. Int. J. Dev. Biol. 1997;41:425–447. [PubMed] [Google Scholar]
- Grisanti L., Falciatori I., Grasso M., Dovere L., Fera S., Muciaccia B., Fuso A., Berno V., Boitani C., Stefanini M. Identification of spermatogonial stem cell subsets by morphological analysis and prospective isolation. Stem Cells. 2009;27:3043–3052. doi: 10.1002/stem.206. [DOI] [PubMed] [Google Scholar]
- Hara K., Nakagawa T., Enomoto H., Suzuki M., Yamamoto M., Simons B.D., Yoshida S. Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell. 2014;14:658–672. doi: 10.1016/j.stem.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel A.R., Oatley J.M. Transplantation as a quantitative assay to study mammalian male germline stem cells. Methods Mol. Biol. 2017;1463:155–172. doi: 10.1007/978-1-4939-4017-2_12. [DOI] [PubMed] [Google Scholar]
- Helsel A.R., Oatley M.J., Oatley J.M. Glycolysis-optimized conditions enhance maintenance of regenerative integrity in mouse spermatogonial stem cells during long-term culture. Stem Cell Rep. 2017;8:1430–1441. doi: 10.1016/j.stemcr.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel A.R., Yang Q.-E., Oatley M.J., Lord T., Sablitzky F., Oatley J.M. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144:624–634. doi: 10.1242/dev.146928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth C.A., Evans E., Onken J., Kent T., Mitchell D., Petkovich M., Griswold M.D. CYP26 enzymes are necessary within the postnatal seminiferous epithelium for normal murine spermatogenesis. Biol. Reprod. 2015;93:19. doi: 10.1095/biolreprod.115.129718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins C. The spermatogonial stem cell population in adult rats. 3. Evidence for a long-cycling population. Cell Tissue Kinet. 1971;4:335–349. doi: 10.1111/j.1365-2184.1971.tb01544.x. [DOI] [PubMed] [Google Scholar]
- Ikami K., Tokue M., Sugimoto R., Noda C., Kobayashi S., Hara K., Yoshida S. Hierarchical differentiation competence in response to retinoic acid ensures stem cell maintenance during mouse spermatogenesis. Development. 2015;142:1582–1592. doi: 10.1242/dev.118695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaucher A.V., Oatley M.J., Oatley J.M. NEUROG3 is a critical downstream effector for STAT3-regulated differentiation of mammalian stem and progenitor spermatogonia. Biol. Reprod. 2012;86:164. doi: 10.1095/biolreprod.111.097386. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent T., Arnold S.L., Fasnacht R., Rowsey R., Mitchell D., Hogarth C.A., Isoherranen N., Griswold M.D. ALDH enzyme expression is independent of the spermatogenic cycle, and their inhibition causes misregulation of murine spermatogenic processes. Biol. Reprod. 2015;94:12. doi: 10.1095/biolreprod.115.131458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Brinster R.L. Culture conditions and single growth factors affect fate determination of mouse spermatogonial stem cells. Biol. Reprod. 2004;71:722–731. doi: 10.1095/biolreprod.104.029207. [DOI] [PubMed] [Google Scholar]
- Kubota H., Avarbock M.R., Brinster R.L. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 2004;101:16489–16494. doi: 10.1073/pnas.0407063101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Palczewski K., Baehr W., Clagett-Dame M. Vitamin A deficiency results in meiotic failure and accumulation of undifferentiated spermatogonia in prepubertal mouse testis. Biol. Reprod. 2011;84:336–341. doi: 10.1095/biolreprod.110.086157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord T., Oatley J.M. A revised Asingle model to explain stem cell dynamics in the mouse male germline. Reproduction. 2017;154:R55–R64. doi: 10.1530/REP-17-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakberg E.F. Spermatogonial stem-cell renewal in the mouse. Anat. Rec. 1971;169:515–531. doi: 10.1002/ar.1091690305. [DOI] [PubMed] [Google Scholar]
- Oatley J.M., Brinster R.L. Spermatogonial stem cells. In: Irina K., Robert L., editors. Methods in Enzymology. Academic Press; 2006. pp. 259–282. [DOI] [PubMed] [Google Scholar]
- Oatley J.M., Brinster R.L. The germline stem cell niche unit in mammalian testes. Physiol. Rev. 2012;92:577–595. doi: 10.1152/physrev.00025.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegelenbosch R.A.J., de Rooij D.G. A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat. Res. 1993;290:193–200. doi: 10.1016/0027-5107(93)90159-d. [DOI] [PubMed] [Google Scholar]
- Tong M., Yang Q., Davis J.C., Griswold M.D. Retinol dehydrogenase 10 is indispensible for spermatogenesis in juvenile males. Proc. Natl. Acad. Sci. USA. 2013;110:543–548. doi: 10.1073/pnas.1214883110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pelt A.M., de Rooij D.G. Synchronization of the seminiferous epithelium after vitamin A replacement in vitamin A-deficient mice. Biol. Reprod. 1990;43:363–367. doi: 10.1095/biolreprod43.3.363. [DOI] [PubMed] [Google Scholar]
- Wolbach S.B., Howe P.R. Tissue changes following deprivation of fat-soluble a vitamin. J. Exp. Med. 1925;42:753–777. doi: 10.1084/jem.42.6.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Racicot K.E., Kaucher A.V., Oatley M.J., Oatley J.M. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–290. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., Takakura A., Ohbo K., Abe K., Wakabayashi J., Yamamoto M., Suda T., Nabeshima Y. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev. Biol. 2004;269:447–458. doi: 10.1016/j.ydbio.2004.01.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.