Summary
We identified osteoclast defects in craniometaphyseal dysplasia (CMD) using an easy-to-use protocol for differentiating osteoclasts from human induced pluripotent stem cells (hiPSCs). CMD is a rare genetic bone disorder, characterized by life-long progressive thickening of craniofacial bones and abnormal shape of long bones. hiPSCs from CMD patients with an in-frame deletion of Phe377 or Ser375 in ANKH are more refractory to in vitro osteoclast differentiation than control hiPSCs. To exclude differentiation effects due to genetic variability, we generated isogenic hiPSCs, which have identical genetic background except for the ANKH mutation. Isogenic hiPSCs with ANKH mutations formed fewer osteoclasts, resorbed less bone, expressed lower levels of osteoclast marker genes, and showed decreased protein levels of ANKH and vacuolar proton pump v-ATP6v0d2. This proof-of-concept study demonstrates that efficient and reproducible differentiation of isogenic hiPSCs into osteoclasts is possible and a promising tool for investigating mechanisms of CMD or other osteoclast-related disorders.
Keywords: hiPSC differentiated osteoclasts, craniometaphyseal dysplasia, rare genetic bone disorder, osteoclastogenesis
Highlights
-
•
An easy-to-use protocol for differentiating osteoclasts (OCs) from hiPSCs
-
•
The use of isogenic hiPSCs to study OC defects in craniometaphyseal dysplasia (CMD)
-
•
Isogenic hiPSCs with CMD mutations in ANKH have defective osteoclastogenesis
-
•
hiPSC-OCs from CMD patients show decreased levels of ANKH and vacuolar proton pump
In this article, Chen and colleagues generated isogenic hiPSCs with or without ANKH mutations identified in patients with craniometaphyseal dysplasia (CMD). By differentiating hiPSC into osteoclasts, they demonstrate that mutations in ANKH lead to decreased osteoclast formation, marker gene expression, and bone resorption. Furthermore, hiPSC osteoclasts from CMD patients have decreased protein levels of ANKH and the vacuolar proton pump ATP6v0d2.
Introduction
Many rare genetic bone disorders result in life-long debilitating symptoms with little or no cures available. Diseases with abnormal bone homeostasis can result from dysfunctional osteoblasts, osteoclasts, or a disrupted interaction between osteoblasts and osteoclasts. Pathogenic mechanisms of rare genetic bone disorders are largely unknown, in part due to the inaccessibility of human skeletal samples and lack of animal models. Studying rare bone diseases can help to better understand normal physiological processes in bone homeostasis and complex disease patterns in common bone disorders such as osteoporosis.
Various somatic cell types have been reprogrammed into hiPSCs, which are capable of indefinite self-renewal and which theoretically can be differentiated into any cell type. While in vitro and in vivo methods for differentiating hiPSCs into osteoblasts are more advanced (Kang et al., 2016, Kanke et al., 2014, Kuhn et al., 2014, Ochiai-Shino et al., 2014), there are very few publications addressing differentiation into osteoclasts (Choi et al., 2009, Grigoriadis et al., 2010). Current hiPSC-osteoclast differentiation protocols require co-culture systems or numerous cytokines for extended periods of time (Choi et al., 2009, Grigoriadis et al., 2010). The application of hiPSC-based approaches in osteoclast-related disorders has been limited due to difficulties in differentiating hiPSCs into osteoclasts.
Here, we present a simple and reproducible method for differentiating hiPSCs into osteoclasts and apply this tool to examine osteoclast defects in craniometaphyseal dysplasia (CMD) where impaired osteoclastogenesis is a major contributor as shown in a mouse model expressing a Phe377del mutation in the progressive ankyloses gene Ank (Chen et al., 2011). CMD is characterized by progressive thickening of craniofacial bones, which can lead to blindness, deafness, facial palsy, severe headaches, and abnormal shape of long bones. Treatment of CMD is limited to risky surgeries to decompress obstructed foramina to relieve symptoms. Mutations for the autosomal dominant form of CMD have been identified in the ANKH gene and are mostly one amino acid deletions or insertions that cluster in the C terminus (Nurnberg et al., 2001, Reichenberger et al., 2001). We have used Sendai virus vectors encoding OCT3/4, SOX2, KLF4, c-MYC to generate hiPSCs from peripheral blood of healthy donors and CMD patients (Chen et al., 2013). The resulting integration-free hiPSCs are pluripotent, have normal karyotype, are capable of differentiating into cells of the three-germ layers in vivo and in vitro and are negative for transgene expression after an average of 10–13 passages (Chen et al., 2013). Here, we show that isogenic hiPSCs with CMD-causing ANKH mutation are more refractory to osteoclast formation in vitro and propose that the isogenic hiPSC approach has great potential for modeling genetic bone diseases with osteoclast defects.
Results
Differentiation of hiPSCs into Mature and Functional Osteoclasts
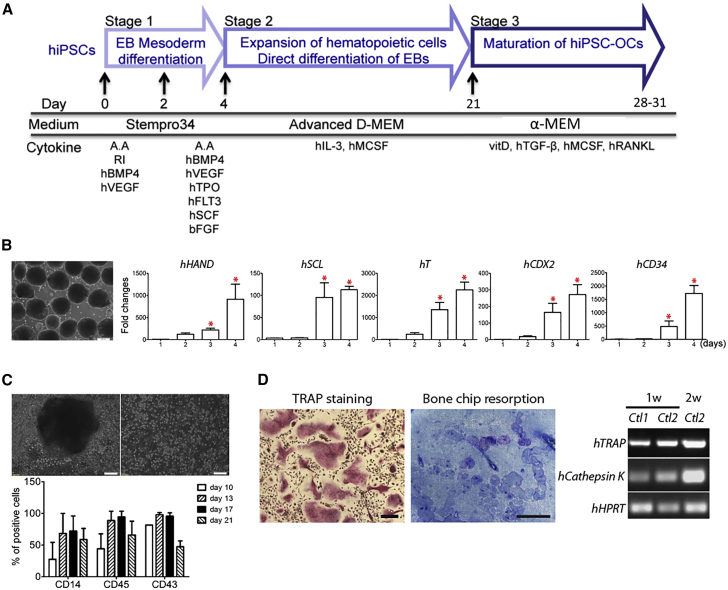
hiPSC lines used in this study were summarized in Table 1. We first used hiPSCs from healthy control individuals to optimize the osteoclast differentiation protocol by direct differentiation through embryoid bodies (EBs). This three-stage protocol consists of EB mesoderm differentiation, expansion of myelomonocytic cells, and maturation of hiPSC osteoclasts (Figure 1A).
Table 1.
hiPSC Lines with or without CMD Mutations in ANKH
| Sample # | ANKH Mutation | Age | Gender |
|---|---|---|---|
| CMD1 | Phe377del (exon 9) | 10 | male |
| CMD2 | Ser375del (exon 9) | 10 | female |
| CMD3 | Phe377del (exon 9) | 45 | female |
| Isogenic CMD hiPSCs | Phe377del (exon 9) | 33 | female |
| Control1 | none | 30 | female |
| Control2 (clone 2) | none | 33 | female |
| Control2 (clone 11) | none | 33 | female |
| Isogenic control hiPSCs | none | 10 | female |
Figure 1.
Differentiating Healthy Control hiPSCs into Osteoclasts
(A) Schematic protocol of differentiating hiPSCs into osteoclasts.
(B) Embryoid body (EB) formation and mesoderm gene expression. EBs cultured for 4 days (left panel). Scale bar, 200 μm. Expression of mesoderm marker genes in EBs cultured for 1, 2, 3, and 4 days by qPCR. ∗p < 0.05 by one-way ANOVA. Data presented are means ± SD.
(C) Myelomonocytic cell expansion. Single cells released from EBs into suspension (top panel). Scale bar, 100 μm. Percentage of cells positive for hematopoietic cell surface markers CD14, CD43, and CD45 in cells released from 10, 13, 17, and 21 day adherent EBs by flow cytometry. Data presented are means ± SD.
(D) TRAP+ osteoclasts differentiated from hiPSCs (left panel), resorption pits on bone chips (middle panel), and expression of OC marker genes, TRAP and CTSK by RT-PCR (right panel). HPRT served as internal control. Ctl1, control1; Ctl2, control2; 1w, 2w, 1 or 2 weeks in stage 3. Scale bar, 100 μm (left panel) and 200 μm (middle panel). Three independent experiments (three technical replicates per experiment) for each hiPSC line. Data were pooled from four wild-type hiPSC lines (hiPSCs from two healthy subjects, two hiPSC clones of each individual donor).
EB size can affect the differentiation efficiency of stem cells (Moon et al., 2014, Ng et al., 2005). In order to achieve better consistency in differentiation efficiency, we generated relatively uniformly sized EBs by culturing Accutase-dissociated hiPSCs on Nunclon Sphera microplates, resulting in an average of 150–250 EBs from two wells of 80% confluent hiPSCs on six-well plates (Figure 1B; see also Figures S1A and S1B). We next determined that culturing EBs in Stempro-34 medium supplemented with hBMP4 and hVEGF for 2 days followed by hBMP4, hVEGF, hSCF, hTPO, hFLT3, and hbFGF for another 2 days is most efficient for inducing the expression of mesoderm marker genes (see also Figures S1C and S1D; Table S1). Levels of HAND1 (Heart and neural crest derivatives expressed 1), SCL (TAL-1, Stem cell leukemia), T (brachyury), CDX2 (Caudal type homeobox 2), and CD34 mRNA were significantly increased in EBs cultured for 4 days, suggesting efficient mesoderm differentiation (Figure 1B).
For stage 2, EBs were transferred to gelatin-coated plates and cultured with hM-CSF and hIL-3. We collected floating monocytes released from adherent EBs after 10, 13, 17, and 21 days for fluorescence-activated cell sorting analysis. The expansion of myelomonocytic populations was assessed by expression analysis of CD14, CD43, and CD45. At day 17, CD14, CD43, and CD45 levels of hiPSC monocytes consistently expressed highest with less variability than at day 13 and dropped at day 21 (Figure 1C; see also Figure S2). Cells collected at day 17 of stage 2 culture were processed into stage 3 by supplementing with vitamin D, hTGFβ, hM-CSF, and hRANKL to generate mature osteoclasts, which stained positive for TRAP, resorbed bone chips, and expressed osteoclast marker genes TRAP (Tartrate-resistant acid phosphatase type 5; APC5) and CATHEPSIN K (CTSK) (Figure 1D).
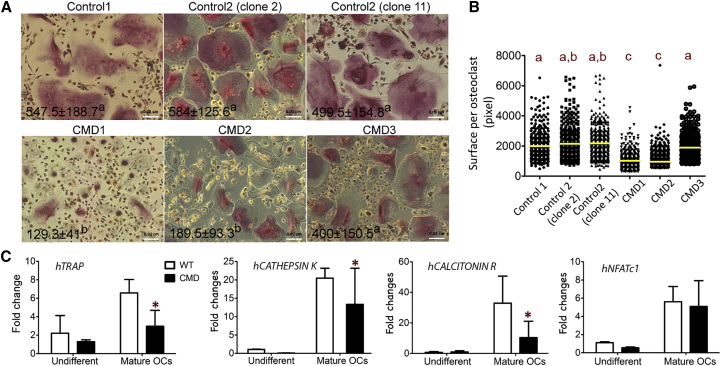
CMD hiPSCs Are More Refractory to Osteoclast Differentiation In Vitro
We next applied our osteoclast differentiation protocol to hiPSCs from three CMD patients (CMD1, CMD2, CMD3). Analysis of osteoclastogenesis progression was performed at day 10 of stage 3. CMD1 and CMD2 hiPSCs formed less TRAP+ multinucleated cells (nuclei ≥3), whereas the number of osteoclasts from CMD3 was statistically insignificant from healthy controls (Figure 2A). Smaller-sized osteoclasts is a characteristic feature observed in a CMD mouse model and in peripheral blood cultures of CMD patients (Chen et al., 2009, Chen et al., 2011). Consistently, hiPSC-derived osteoclasts from patients CMD1 and CMD2 were also reduced in size (Figure 2B). CMD hiPSCs (combined data from CMD1, 2, and 3) showed significantly reduced expression of osteoclast marker genes TRAP, CTSK, and CALCITONIN RECEPTOR, while NFATc1 (Nuclear factor of activated T-cells 1) did not significantly differ between control and CMD patient samples (Figure 2C). All osteoclast markers significantly increased in mature osteoclasts compared with undifferentiated osteoclasts from healthy donors and CMD patients (Figure 2C). We noticed a large variability in osteoclast numbers and marker gene expression within CMD patients (see also Figure S3). These differences may result from the variability in age, sex, or genetic background. To minimize these variations, we decided to generate isogenic hiPSCs and performed further analyses, including bone resorption assays, in these cells.
Figure 2.
Osteoclasts Differentiated from Healthy Control and CMD hiPSCs
(A) Comparison of control and CMD osteoclasts derived from hiPSCs. Numbers of TRAP+ multinucleated cells (nuclei ≥3) are shown (mean ± SD) in the left corners of the images. Different letters (a, b) indicate statistically significant difference (p < 0.05) by one-way ANOVA. Scale bar, 100 μm.
(B) Decreased sizes of osteoclasts derived from CMD1 and CMD2 hiPSCs. Yellow bars indicate the mean value of each group (n ≥ 400 osteoclasts measured per group). Different letters (a, b, c) indicate statistically significant difference (p < 0.05) by one-way ANOVA.
(C) Decreased expression of TRAP, CATHEPSIN K, CALCITONIN RECEPTOR mRNA message in osteoclasts differentiated from CMD hiPSCs shown by qPCR. ∗p < 0.05 by two-way ANOVA. Data presented are means ± SD. Three independent experiments (technical replicates) were performed for each hiPSC line.
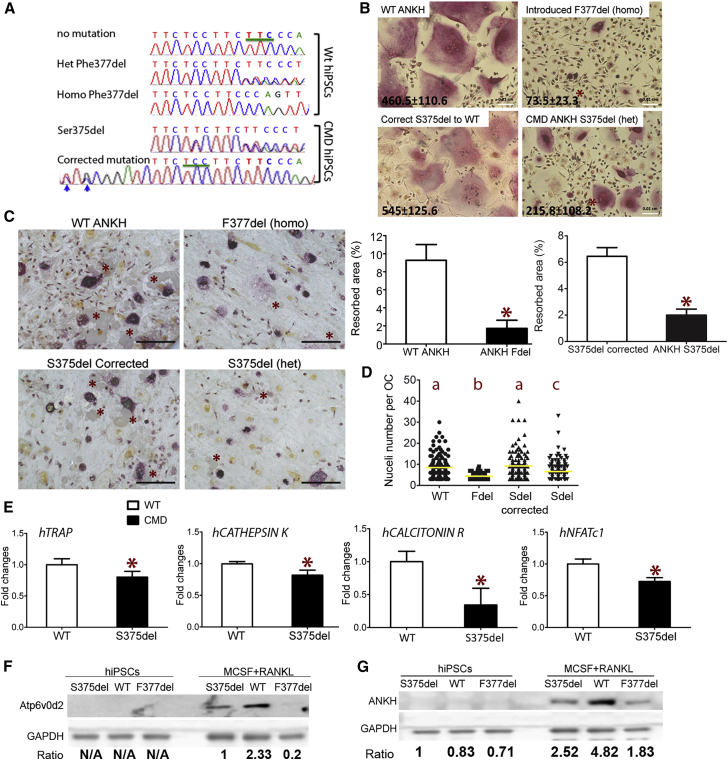
CMD Mutations in ANKH Diminish Isogenic hiPSC Differentiation into Osteoclasts
Isogenic hiPSCs have identical genetic background except for a disease mutation (genetic variant), allowing researchers to determine the impact of a specific mutation (Cong et al., 2013, Ran et al., 2013, Yoshimi et al., 2016). We introduced the ANKH Phe377del (F377del) mutation into healthy hiPSCs (healthy donor 2) and corrected a Ser375del ANKH mutation from a CMD patient (CMD2) by CRISPR/Cas9 technology. Inserted mutation or correction of ANKH was confirmed by sequencing (Figure 3A).
Figure 3.
Osteoclasts Differentiated from Isogenic hiPSC with or without CMD ANKH Mutations
(A) Electropherograms showing partial sequences of ANKH exon 9 with insertion of Phe377del into wild-type hiPSCs and correction of Ser375del ANKH in CMD hiPSCs. Blue arrows indicate silent mutations introduced to minimize recutting of a repaired site.
(B) Decreased numbers of TRAP+ multinucleated osteoclasts formed from isogenic CMD mutant hiPSCs compared with wild-type controls. Numbers of TRAP+ multinucleated cells (nuclei ≥3) are shown (mean ± SD) in the left corners of the images. ∗ indicates statistical significance compared with the parental isogenic hiPSCs by Student’s t test. Scale bar, 100 μm.
(C) Decreased resorbed area on bone chips by osteoclasts derived from isogenic CMD mutant hiPSCs compared with wild-type controls. ∗ on bone images (left panel) indicate resorption pits. Bar figures show the percentage of resorbed area (mean ± SD). Scale bar, 200 μm. ∗ indicates statistical significance compared with the parental isogenic hiPSCs by Student’s t test.
(D) Decreased nuclei numbers in CMD hiPSC-derived osteoclasts (n = 70–156 cells per group). Yellow bar indicates the mean value of each group. Different letters (a, b, c) indicate statistically significant difference (p < 0.05) by one-way ANOVA followed by Bonferroni post hoc test.
(E) Decreased expression of TRAP, CATHEPSIN K, CALCITONIN RECEPTOR, NFATc1 in osteoclasts differentiated from isogenic hiPSCs with a S375del mutation in ANKH by qPCR. ∗p < 0.05 by Student's t test. Data presented are means ± SD.
(F) A representative Atp6v0d2 (40 kDa) immunoblot of hiPSCs (left panel) and hiPSC-differentiated osteoclasts (right panel) with or without S375del or F377del ANKH mutations. GAPDH served as loading control. Samples with wild-type and a S375del mutation in ANKH are isogenic hiPSCs. The F377del cell lysate is from patient CMD1 hiPSCs and hiPSC-derived osteoclasts. Numbers below indicate ratio of Atp6v0d2 to GAPDH.
(G) A representative ANKH (52 kDa) immunoblot of undifferentiated hiPSCs (left panel) and hiPSC-derived osteoclasts (right panel) with or without S375del or F377del ANKH mutations. GAPDH served as loading control. Samples with wild-type and a S375del mutation are isogenic hiPSCs. The F377del cell lysate is from patient CMD1 hiPSCs and hiPSC-derived osteoclasts. Numbers below indicate the ratio of ANKH to GAPDH. Three independent experiments (technical replicates) were performed for each isogenic hiPSC line.
For this proof-of-concept study, we used two pairs of isogenic hiPSCs, hiPSCs with or without a homozygous F377del and hiPSCs with or without a heterozygous S375del mutation in ANKH. We expected that both ANKH mutations have similar effects on osteoclastogenesis and the effects of ANKH mutations are dose dependent. While differentiating into osteoclasts, the CMD mutant isogenic hiPSCs formed significantly less osteoclasts and resorbed less on bone chips than those expressing wild-type ANKH (Figures 3B and 3C). As expected, hiPSCs with the homozygous CMD mutation (F377del) showed the least osteoclast numbers (Figure 3B). Osteoclasts differentiated from isogenic hiPSCs with ANKH mutations had reduced numbers of nuclei, which is an indication of a fusion defect (Figure 3D). TRAP, CTSK, CALCR, and NFATc1 were significantly reduced in isogenic hiPSC-derived osteoclasts with a Ser375del mutation (Figure 3E).
A published study showed in a single CMD patient that the osteoclast-reactive vacuolar proton pump was not expressed but was expressed in cells derived from the patient's mother (Yamamoto et al., 1993). Vacuolar proton pump subunit D2 (v-ATPase V0 domain subunit d2, Atp6v0d2) is expressed in osteoclasts, not in osteoblasts, and plays an important role in osteoclast fusion (Lee et al., 2006). We first examined the expression of Atp6v0d2 in isogenic hiPSCs by immunoblots. Atp6v0d2 was not expressed in undifferentiated hiPSCs but was expressed in hiPSC-derived osteoclasts (Figure 3F). hiPSC-derived osteoclasts with ANKH mutations expressed lower levels of Atp6v0d2 compared with wild-type lines (Figure 3F). We next examined the expression of ANKH in hiPSCs during osteoclast differentiation. We had previously shown that Ank mRNA levels in wild-type and Ank knockin (KI) mice (AnkKI/KI) increase comparably in mature osteoclasts (Chen et al., 2011). We observed similar findings in hiPSC-differentiated osteoclasts (data not shown). ANKH protein was almost undetectable in undifferentiated hiPSCs but was significantly increased in osteoclasts differentiated from hiPSCs (Figure 3G). Interestingly, ANKH protein levels were higher in wild-type hiPSC osteoclasts compared with CMD hiPSC osteoclasts (Figure 3G). Reduced ANKH protein expression was also found in osteoblasts from bone explant cultures from a CMD patient compared with a healthy donor (see also Figure S4). These data demonstrate that isogenic hiPSC-derived bone cells can overcome obstacles of studying rare genetic bone disorders because obtaining human specimens such as bone for explant cultures is difficult, while hiPSCs can easily be obtained from peripheral blood of patients. It is yet unknown whether the reduced amount of ANKH protein is due to decreased osteoclast differentiation in CMD hiPSCs, to abnormal protein processing, or to instability of mutant ANKH. However, these data suggest that CMD pathogenesis may be partially due to decreased amounts of functional ANKH.
Discussion
The potential of hiPSCs in disease modeling or treatment is widely recognized, and methods for generating hiPSCs are well established. The main obstacle is consistency in differentiating hiPSCs into specific cell lineages. Published methods for differentiating hiPSCs into osteoclasts use (1) co-culturing hiPSCs with OP9 cells, a newborn calvarial-derived stromal cell line from mice deficient in macrophage colony stimulating factor (M-CSF) or (2) direct differentiation of hiPSC-derived EBs supplemented with multiple cytokines (Choi et al., 2009, Grigoriadis et al., 2010, Jeon et al., 2016). One of the critical steps of the co-culture method is to match proper cell densities of undifferentiated hiPSCs and OP9 cultures simultaneously. OP9 cells are commonly used to stimulate hematopoietic differentiation from embryonic stem cells and can rapidly differentiate into adipocytes especially at high cell density (Nakano et al., 1994, Wolins et al., 2006). To differentiate hiPSCs into osteoclasts, multiple cytokines are required. While 6–7 cytokines were used by Grigoriadis et al. (2010) for a minimum of 14 days to promote hematopoietic specification and myeloid expansion, we reduced medium supplementation with these many cytokines to 2 days during mesoderm differentiation. We then adapted a published protocol using M-CSF and IL-3 for myeloid differentiation (Jeon et al., 2016, Panicker et al., 2012). The advantages of our method include the following: (1) no need for co-cultures; (2) generation of uniformly sized EBs to minimize variations in differentiation efficiency; (3) rapid expansion of CD14+, CD43+, and CD45+ populations; (4) less cytokines used; and (5) sufficient numbers of osteoclasts for subsequent molecular analyses.
We used Rock inhibitor (Y-27632) to improve survival of Accutase-dissociated hiPSCs as shown previously (Watanabe et al., 2007). Efficient mesoderm differentiation is achieved by effects of several cytokines. BMP4 induces primitive streak and early hematopoietic gene expression and, together with VEGF, can increase expression of SCL and CD34 (Pick et al., 2007). SCF promotes in vitro proliferation of primitive hematopoietic progenitors (Hoffman et al., 1993). Together SCF, FLT-3, and TPO play several important roles in human hematopoiesis (Bhatia et al., 1997, Zandstra et al., 1998). bFGF stimulates cell proliferation during hematopoietic differentiation (Pick et al., 2007).
The murine CD11b−/lowB220−CD3−CD115highCD117+ population in bone marrow has the highest osteoclastogenic activity (Jacquin et al., 2006), but human osteoclast precursors are less defined. Choi et al. (2009) enriched myeloid progenitors by expanding hiPSC-derived Lin−CD34+CD43+CD45+ cells and Grigoriadis et al. (2010) used CD45+ as a progenitor marker. We evaluated the myelomonocytic cell expansion by CD14, CD43, and CD45. Although CD43+ and CD45+ are highly expanded, the CD14+ population appears to be variable between samples. CD14+ cells have been shown to form mature and functional osteoclasts and CD14− cells express osteoclastogenic factors to support CD14+ cells (Costa-Rodrigues et al., 2011, Taylor et al., 2012). In this study, we did not perform population selection by cell sorting.
The pathogenic mechanisms leading to CMD are complex. To date, the only known function of ANK/ANKH is to transport pyrophosphate (Ho et al., 2000). A KI mouse model (AnkKI/KI) expressing the human Phe377del mutation in Ank replicates many skeletal features of human CMD (Chen et al., 2009). In AnkKI/KI mice, the extracellular PPi levels were comparable with wild-type mice possibly due to compensatory effects of Enpp1 (Chen et al., 2011). Shared CMD phenotypes between Anknull/null and AnkKI/KI mice and decreased ANKH protein in CMD hiPSC-derived osteoclasts suggest that decreased functional ANKH partially contributes to CMD. However, we believe that it involves more than just loss of function of the pyrophosphate transporting activity of ANKH because Ank knockout mice do not fully replicate the CMD phenotype (Chen et al., 2009). In this study, we did not report PPi levels in hiPSC-derived osteoclasts.
Lack of osteoclast-reactive vacuolar proton pump expression was reported in one CMD patient where the ANKH mutation had not been identified (Yamamoto et al., 1993). This patient displayed severe sclerotic diaphyseal bone, which is not typical for most cases of CMD caused by ANKH mutations (Yamamoto et al., 1993). Our data showed significantly reduced expression of Atp6v0d2 in isogenic hiPSC-derived osteoclasts expressing CMD mutant ANKH compared with wild-type cells. Whether decreased Atp6v0d2 is caused by lower levels of ANKH and how decreased numbers of vacuolar proton pumps affect CMD pathogenesis needs to be investigated in a future study.
Osteoclast defects in CMD have been shown in our mouse model and in human peripheral blood cultures (Chen et al., 2011, Yamamoto et al., 1993). This study identifies osteoclast defects in CMD using patient-specific hiPSC cultures. Although human osteoclasts can be derived from peripheral blood, it is not realistic to repeatedly obtain blood or bone marrow samples from patients with rare disorders. Therefore, our protocol for differentiating hiPSCs into osteoclasts with high efficiency and reproducibility enables researchers to generate a sufficient supply of human osteoclasts. This proof-of-concept study shows that osteoclasts differentiated from hiPSCs can provide an unlimited source of cells for modeling genetic bone diseases with osteoclast defects and will allow us to investigate the underlying molecular defect as well as to test potential therapeutics.
Experimental Procedures
The animal protocol (101425-0819) was approved by the Institutional Animal Care and Use Committee (IACUC) of University of Connecticut Health, and all work was performed in an AAALAC accredited facility. All studies involving human protocols were in accordance with guidelines of the Institutional Review Board of the University of Connecticut Health (IRB protocol 09-199). Integration-free hiPSC lines from peripheral blood were generated as previously described (Dutra et al., 2013). Information on age, gender, and ANKH mutations of hiPSC lines are summarized in Table 1. We used two different clones (clone2 and clone11) from control2 hiPSC line. Three independent experiments were performed for each hiPSC line (technical triplicates). Statistical analysis was performed using Prism 5 software (GraphPad Software). A p value of less than 0.05 was considered statistically significant. Additional methods are available in Supplemental Experimental Procedures.
Author Contributions
R.L., Z.H., and J.K. performed molecular analysis. I.-P.C. directed the research, performed experiments, and analyzed data. I.-P.C. and E.J.R. wrote the manuscript. All authors approved the manuscript.
Acknowledgments
We appreciate help from the Stem Cell core and Flow Cytometry core at UConn Health. This work was supported by NIH/NIDCR R00 (DE021442) to I.-P.C.
Published: October 19, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.09.016.
Supplemental Information
References
- Bhatia M., Bonnet D., Kapp U., Wang J.C., Murdoch B., Dick J.E. Quantitative analysis reveals expansion of human hematopoietic repopulating cells after short-term ex vivo culture. J. Exp. Med. 1997;186:619–624. doi: 10.1084/jem.186.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.P., Fukuda K., Fusaki N., Iida A., Hasegawa M., Lichtler A., Reichenberger E.J. Induced pluripotent stem cell reprogramming by integration-free Sendai virus vectors from peripheral blood of patients with craniometaphyseal dysplasia. Cell Reprogram. 2013;15:503–513. doi: 10.1089/cell.2013.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.P., Wang C.J., Strecker S., Koczon-Jaremko B., Boskey A., Reichenberger E.J. Introduction of a Phe377del mutation in ANK creates a mouse model for craniometaphyseal dysplasia. J. Bone Miner Res. 2009;24:1206–1215. doi: 10.1359/JBMR.090218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.P., Wang L., Jiang X., Aguila H.L., Reichenberger E.J. A Phe377del mutation in ANK leads to impaired osteoblastogenesis and osteoclastogenesis in a mouse model for craniometaphyseal dysplasia (CMD) Hum. Mol. Genet. 2011;20:948–961. doi: 10.1093/hmg/ddq541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.D., Vodyanik M.A., Slukvin I.I. Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors. J. Clin. Invest. 2009;119:2818–2829. doi: 10.1172/JCI38591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Rodrigues J., Fernandes A., Fernandes M.H. Spontaneous and induced osteoclastogenic behaviour of human peripheral blood mononuclear cells and their CD14(+) and CD14(-) cell fractions. Cell Prolif. 2011;44:410–419. doi: 10.1111/j.1365-2184.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutra E.H., Chen I.P., Reichenberger E.J. Dental abnormalities in a mouse model for craniometaphyseal dysplasia. J. Dent. Res. 2013;92:173–179. doi: 10.1177/0022034512468157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriadis A.E., Kennedy M., Bozec A., Brunton F., Stenbeck G., Park I.H., Wagner E.F., Keller G.M. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010;115:2769–2776. doi: 10.1182/blood-2009-07-234690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A.M., Johnson M.D., Kingsley D.M. Role of the mouse ank gene in control of tissue calcification and arthritis. Science. 2000;289:265–270. doi: 10.1126/science.289.5477.265. [DOI] [PubMed] [Google Scholar]
- Hoffman R., Tong J., Brandt J., Traycoff C., Bruno E., McGuire B.W., Gordon M.S., McNiece I., Srour E.F. The in vitro and in vivo effects of stem cell factor on human hematopoiesis. Stem Cells. 1993;11(Suppl 2):76–82. doi: 10.1002/stem.5530110813. [DOI] [PubMed] [Google Scholar]
- Jacquin C., Gran D.E., Lee S.K., Lorenzo J.A., Aguila H.L. Identification of multiple osteoclast precursor populations in murine bone marrow. J. Bone Miner Res. 2006;21:67–77. doi: 10.1359/JBMR.051007. [DOI] [PubMed] [Google Scholar]
- Jeon O.H., Panicker L.M., Lu Q., Chae J.J., Feldman R.A., Elisseeff J.H. Human iPSC-derived osteoblasts and osteoclasts together promote bone regeneration in 3D biomaterials. Sci. Rep. 2016;6:26761. doi: 10.1038/srep26761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Shih Y.R., Nakasaki M., Kabra H., Varghese S. Small molecule-driven direct conversion of human pluripotent stem cells into functional osteoblasts. Sci. Adv. 2016;2:e1600691. doi: 10.1126/sciadv.1600691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke K., Masaki H., Saito T., Komiyama Y., Hojo H., Nakauchi H., Lichtler A.C., Takato T., Chung U.I., Ohba S. Stepwise differentiation of pluripotent stem cells into osteoblasts using four small molecules under serum-free and feeder-free conditions. Stem Cell Rep. 2014;2:751–760. doi: 10.1016/j.stemcr.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn L.T., Liu Y., Boyd N.L., Dennis J.E., Jiang X., Xin X., Charles L.F., Wang L., Aguila H.L., Rowe D.W. Developmental-like bone regeneration by human embryonic stem cell-derived mesenchymal cells. Tissue Eng. Part A. 2014;20:365–377. doi: 10.1089/ten.tea.2013.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Rho J., Jeong D., Sul J.Y., Kim T., Kim N., Kang J.S., Miyamoto T., Suda T., Lee S.K. v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 2006;12:1403–1409. doi: 10.1038/nm1514. [DOI] [PubMed] [Google Scholar]
- Moon S.H., Ju J., Park S.J., Bae D., Chung H.M., Lee S.H. Optimizing human embryonic stem cells differentiation efficiency by screening size-tunable homogenous embryoid bodies. Biomaterials. 2014;35:5987–5997. doi: 10.1016/j.biomaterials.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Nakano T., Kodama H., Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- Ng E.S., Davis R.P., Azzola L., Stanley E.G., Elefanty A.G. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106:1601–1603. doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- Nurnberg P., Thiele H., Chandler D., Hohne W., Cunningham M.L., Ritter H., Leschik G., Uhlmann K., Mischung C., Harrop K. Heterozygous mutations in ANKH, the human ortholog of the mouse progressive ankylosis gene, result in craniometaphyseal dysplasia. Nat. Genet. 2001;28:37–41. doi: 10.1038/ng0501-37. [DOI] [PubMed] [Google Scholar]
- Ochiai-Shino H., Kato H., Sawada T., Onodera S., Saito A., Takato T., Shibahara T., Muramatsu T., Azuma T. A novel strategy for enrichment and isolation of osteoprogenitor cells from induced pluripotent stem cells based on surface marker combination. PLoS One. 2014;9:e99534. doi: 10.1371/journal.pone.0099534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker L.M., Miller D., Park T.S., Patel B., Azevedo J.L., Awad O., Masood M.A., Veenstra T.D., Goldin E., Stubblefield B.K. Induced pluripotent stem cell model recapitulates pathologic hallmarks of Gaucher disease. Proc. Natl. Acad. Sci. USA. 2012;109:18054–18059. doi: 10.1073/pnas.1207889109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick M., Azzola L., Mossman A., Stanley E.G., Elefanty A.G. Differentiation of human embryonic stem cells in serum-free medium reveals distinct roles for bone morphogenetic protein 4, vascular endothelial growth factor, stem cell factor, and fibroblast growth factor 2 in hematopoiesis. Stem Cells. 2007;25:2206–2214. doi: 10.1634/stemcells.2006-0713. [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenberger E., Tiziani V., Watanabe S., Park L., Ueki Y., Santanna C., Baur S.T., Shiang R., Grange D.K., Beighton P. Autosomal dominant craniometaphyseal dysplasia is caused by mutations in the transmembrane protein ANK. Am. J. Hum. Genet. 2001;68:1321–1326. doi: 10.1086/320612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.M., Kashima T.G., Hemingway F.K., Dongre A., Knowles H.J., Athanasou N.A. CD14- mononuclear stromal cells support (CD14+) monocyte-osteoclast differentiation in aneurysmal bone cyst. Lab Invest. 2012;92:600–605. doi: 10.1038/labinvest.2012.5. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Wolins N.E., Quaynor B.K., Skinner J.R., Tzekov A., Park C., Choi K., Bickel P.E. OP9 mouse stromal cells rapidly differentiate into adipocytes: characterization of a useful new model of adipogenesis. J. Lipid Res. 2006;47:450–460. doi: 10.1194/jlr.D500037-JLR200. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Kurihara N., Yamaoka K., Ozono K., Okada M., Yamamoto K., Matsumoto S., Michigami T., Ono J., Okada S. Bone marrow-derived osteoclast-like cells from a patient with craniometaphyseal dysplasia lack expression of osteoclast-reactive vacuolar proton pump. J. Clin. Invest. 1993;91:362–367. doi: 10.1172/JCI116194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi K., Kunihiro Y., Kaneko T., Nagahora H., Voigt B., Mashimo T. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat. Commun. 2016;7:10431. doi: 10.1038/ncomms10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandstra P.W., Conneally E., Piret J.M., Eaves C.J. Ontogeny-associated changes in the cytokine responses of primitive human haemopoietic cells. Br. J. Haematol. 1998;101:770–778. doi: 10.1046/j.1365-2141.1998.00777.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.