Abstract
Mass spectrometry (MS) has found numerous applications in life sciences. It has high accuracy, sensitivity and wide dynamic range in addition to medium- to high-throughput capabilities. These features make MS a superior platform for analysis of various biomolecules including proteins, lipids, nucleic acids and carbohydrates. Until recently, MS was applied for protein detection and characterization. During the last decade, however, MS has successfully been used for molecular diagnostics of microbial and viral infections with the most notable applications being identification of pathogens, genomic sequencing, mutation detection, DNA methylation analysis, tracking of transmissions, and characterization of genetic heterogeneity. These new developments vastly expand the MS application from experimental research to public health and clinical fields. Matching of molecular techniques with specific requirements of the major MS platforms has produced powerful technologies for molecular diagnostics, which will further benefit from coupling with computational tools for extracting clinical information from MS-derived data.
Keywords: DNA sequencing, infectious disease, mass spectrometry, molecular diagnostics, molecular surveillance, viral infection
Using mass spectrometry (MS) for the molecular diagnosis of infectious diseases is no longer novel [1]. Work on testing human metabolites and comparing the biochemical profiles of healthy and diseased individuals to detect disease was already underway in the 1970s [2]. Interest towards the use of MS re-emerged when further advancements in the field took hold, in particular the introduction of MALDI [3] and ESI [4]. These gentle ionization technologies allowed for maintaining the integrity of nucleic acid (NA) oligomers and protein molecules with large molecular mass [3], or as laureate John Fenn said in his Nobel lecture, “the electrospray wings for molecular elephants” opened new opportunities for applying MS technology. The many uses of MS – for classification and identification of bacteria, DNA analysis, screening and diagnostic research, multiplex genotyping, sequencing, genomics research, hospital infection control and quality control testing – have already been reviewed [5–8].
MS for detecting analytes as biomarkers was previously not readily appreciated, especially for NA identification. That particular use did not merit a mention in a thorough review of the state of the molecular diagnostics of infectious diseases published not long ago and which included all basic technologies and procedures used in clinical laboratories performing molecular diagnostics [9]. In proteomic studies, however, MS quickly gained recognition as a ‘gold standard’ tool for the identification and analysis of individual proteins. Currently, however, MS is being applied far beyond its use for protein characterization to include other types of analytes, including NAs [5].
MS detection of diagnostic markers is rapid, taking only milliseconds to seconds. The preparation of the analyte is highly amenable to automation, which additionally broadens the field of possible applications. MS is easily adaptable to different scales of detection and applicable for low-, medium- and high-throughput clinical screening and diagnostic testing [10]. Sub-picomolar amounts of the analyte can be detected without fluorescence or radioactive isotope labeling or using antibodies or hybridization probes. It can be used to detect the material of interest directly [11] or after attaching an intermediate detector molecule, thus conferring high sensitivity to detection [12]. Direct detection significantly simplifies the MS diagnostic applications [10,13]. As a detection system for NAs, MS can be easily coupled with any widely available DNA amplification technique. These qualities, combined with the capability of multiplexing – that is, testing for different analytes at the same time – make MS particularly suitable for rapid molecular diagnostics in the clinical setting. MS is uniquely suited for supporting multiple diverse applications. The throughput and multiplexing capacity of MS help reduce the cost of testing.
MS principles & platforms as used for the detection of NAs
All MS applications are based on direct measurement of two intrinsic properties of the bioanalyte: molecular mass and charge. The mass spectrometer consists of three functional units. The first unit, the ion source, is used to ionize the analyte and transfer it to the gas phase. The second unit is the mass analyzer that serves to separate the ions by their mass-to-charge ratio (m/z), which in turn defines their TOF. It can have various configurations that use a vacuum chamber and a static or dynamic magnetic/electric field to separate the ions. The third unit is a detector device, an electron multiplier or fast oscilloscope, which detects the ions.
There is a great variety of ionization methods using atmospheric pressure (spray), chemicals, electrons, heat, ions, atom bombardment and so on. Most ionization techniques that require chemical or electrical ionization are too energy rich and frequently result in unpredictable decomposition of the NA analyte. Softer ionization approaches that use electrospray device or matrix carrier and laser ion source, such as ESI [4] and MALDI [3], have solved this degradation problem and account for the majority of currently used ionization techniques. Additional issues arising when working with NAs, such as depurination or generation of salt adducts, have been resolved by utilizing improved reagents and clean-up methods and by the discovery that ribonucleotides are more stable than deoxyribonucleotides [14].
ESI-MS
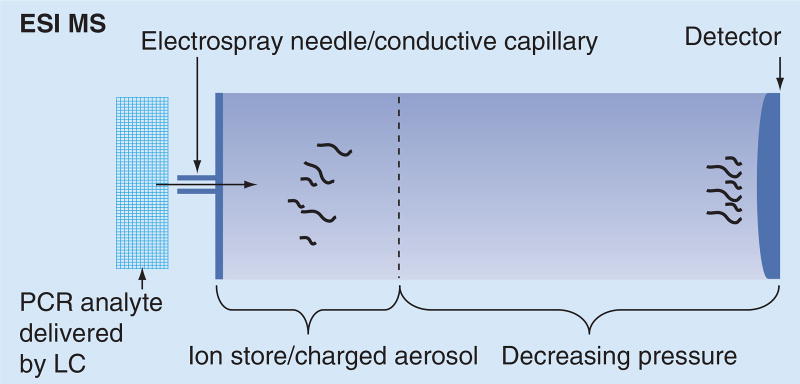
ESI-MS is a soft technique where the analyte is dissolved in organic solvent like methanol or acetonitrile, sometimes in combination with acetic acid, and then injected in a conductive capillary where high voltage is applied. This results in the emission of aerosols of charged droplets of the sample. The aerosol travels through sections of decreasing pressure, which leads to formation of gas-phase ions of the analyte. ESI is typically coupled with an automated autosampler in a well-plate format, injector and liquid chromatography, which controls the flow rate of sample delivery and buffers the zones between samples. The rate of sample detection is several minutes per sample. The analyzer/detector can be FTICR, which detects all generated ions simultaneously and differentiates them by their rotational frequency, which is then transformed into a specific mass reading or a micro-TOF. The FTICR detection has great accuracy and high resolution [15]. A schematic representation of the ESI-MS is shown in Figure 1.
Figure 1. Basic configuration of an ESI mass spectrometer for use in nucleic acid analysis.
LC: Liquid chromatography; MS: Mass spectrometry.
After the revolutionary development of this soft ionization technique [4], in a short time span the process was utilized for the analysis of PCR-amplified DNA products [16] and the study of noncovalent molecular complexes, peptides, glycans and so on. Technological improvements in DNA fragment preparation led to the introduction by Ibis Biosciences of a novel platform T5000 for rapid identification of pathogens [17]. The process has medium- to high-throughput capability, and applies ESI-MS. In essence, it combines the accuracy and sensitivity of multilocus sequence typing with the speed, throughput and accuracy of MS.
MALDI-TOF MS
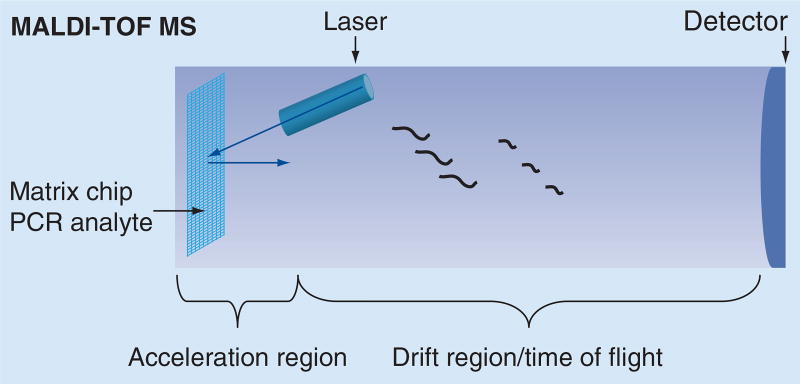
There are two major classes of matrix, ‘hot’ and ‘cool,’ for desorbing laser energy that can be used in MALDI-TOF MS. The hot matrices have higher desorption/ionization energy, which results in excessive fragmentation of the analytes, and thus are more suitable for detection of small RNA molecules. Cool matrices are used for larger oligonucleotides and cause little fragmentation. Commonly used matrices for RNA are a mix of 2,3,4- and 2,4,6-trihydroxyacetophenone, and matrices for DNA are α-cyano-4-hydroxycinnamic, 2,5-dihydroxybenzoic or 3-hydroxypicolinic acid [7]. The matrix is crystallized on a solid inert surface. The sample is then imbedded in the matrix by dispensing in nanoquantities and desorbed and ionized by short (3–4 ns) UV-laser or IR-laser pulse. The ions are accelerated by an electric field to the same kinetic energy and sent into a field-free drift flight vacuum tube where ions of different m/z values are separated from each other. The velocity of the ions depends only on the m/z – that is, heavier molecules travel at lower speed and thus reach the detector later. The detection of analytes from a single target spot can be accomplished in several seconds. For analysis of NAs, the TOF needed for the ionized analyte to reach the detector is recorded and used to derive the analyte mass. This method has resolution reaching 1 Da that allows for the discrimination of single nucleotides. A schematic representation of MALDI-TOF MS is shown in Figure 2.
Figure 2. Basic configuration of a MALDI mass spectrometer for use in nucleic acid analysis.
MS: Mass spectrometry.
Both ESI-MS and MALDI-TOF MS, given the appropriate analyzer, are sensitive to small variations in the primary structure of NAs and are capable of discriminating between even single nucleotide differences. This capacity of MALDI-TOF MS was significantly improved with the development of better matrices; methods to eliminate adduct-forming agents and novel biochemistries [18]. The predominant current use of MALDI-TOF MS is in the fields of genotyping/SNP typing, methylation analysis, quantitative gene expression analysis and resequencing [19].
Pathogen detection & identification
The presence of certain phenotypic, biochemical or genetic characteristics has been used routinely in most clinical laboratories for identification and differentiation of microbes. Molecular diagnostics are based on the specific recovery and detection of certain genomic fragments. NA characterization techniques such as plasmid profiling, fragment length polymorphisms and PCR-based systems significantly improved the sensitivity, specificity and throughput of detection of microbial pathogens directly from clinical samples. Application of these techniques, however, is limited mostly to the detection of pathogens that are difficult to culture in vitro.
Sequence analysis of amplified microbial DNA allows for even more accurate characterization of pathogens. Molecular identification of bacteria and viruses is based on the significant sequence specificity of NAs in different organisms. The first applications of MS to molecular microbiological diagnostics were in detection of PCR products generated from genomic regions of pathogens. The pathogen specificity can be detected using the entire PCR fragment or site-specific cleavage products of the fragment. Many sensitive pathogen identification assays require amplification or a particular labeling of the signature molecule. Visualization or detection of the NA generally involves size separation or detection of the labeled probe. One of the first successful applications of both MALDITOF and ESI approaches were to characterize PCR amplicons of the cystic fibrosis transmembrane conductance regulator gene [16]. A similar assay principle was applied shortly after to the successful identification of immobilized PCR products of hepatitis B virus (HBV) obtained from serum, demonstrating that identification by MS is a fast and reliable method for pathogen detection [20].
Although the application to pathogen detection was particularly successful, the mere use of MS as a detection system in place of, for example, gel electrophoresis did not resonate with many researchers, mainly because of the seeming complexity of MS. It also coincided with the introduction of the real-time PCR, attractive with its ease of detection and quantification of PCR products [21], which took away for a while from the development of the MS applications to molecular diagnostics. However, MS not only surpasses electrophoresis in rapidity and sensitivity as a detection system, but allows for precise sizing of the molecule of interest and also provides information on nucleotide composition and charge. The temporarily neglected potential of MS was revived with the expanding need for development of higher throughput DNA analysis techniques and discovery of novel MS applications to molecular diagnostics beyond simple detection [22]. Yet another advantage of the MS platforms in general is that they allow for the simultaneous detection of multiple analytes. Initially, MS methods were applied to the simultaneous detection of two bacterial pathogens [23], but further along came the multiplexed detection of human herpesviruses [24], human papillomaviruses (HPVs) [25] and variants of HBV [26].
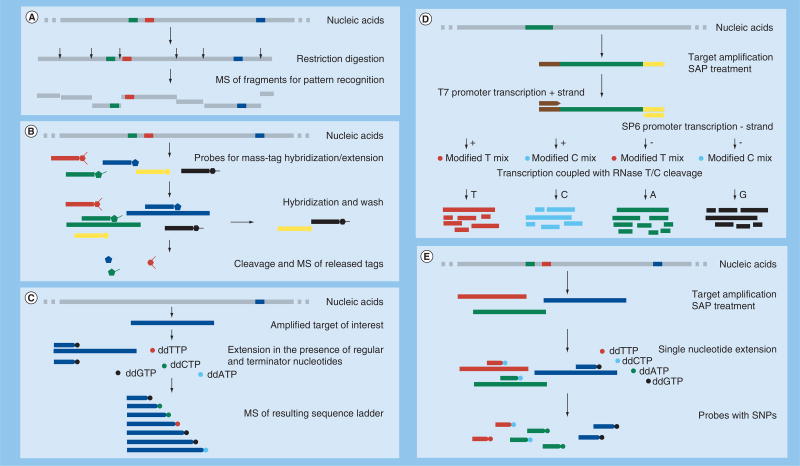
Restriction fragment length polymorphism (RFLP) has also been a tool of choice for organism identification and detection of polymorphisms. Coupling RFLP, illustrated in Figure 3A, to MS produced especially efficient technology. Replacement of gel electrophoresis with MS for the separation of restriction fragments generated by RFLP was recently implemented for the detection of avian influenza viruses [27,28], genotyping of hepatitis C virus (HCV) [29] and detection of drug-resistant HBV variants [30]. The application of MS to analyzing the products of robust and sensitive NA technologies such as real-time PCR, RFLP or uracil DNA glycosylation improved identification of pathogens not merely because of the high-throughput capability of MS, but also because a new layer of accurate molecular information could be acquired.
Figure 3. Biochemical processes for comprehensive molecular analysis of specific gene targets as applied to analysis by mass spectrometry.
(A) Restriction fragment length polymorphism – the complexity of large target molecules is reduced by enzymatic cleavage at predetermined sites, the resulting small molecules are analyzed by MS and the obtained mass patterns are used to infer organism identification. (B) Mass-tag multiplexing – each target is detected by a probe tagged by a cleavable small molecule, unused probes are removed/washed, the tags of the used probes are cleaved and used for detection by MS; detected tags indicate presence of the target of interest. (C) Generation of sequence ladder in the presence of one specific primer, regular nucleotides, dideoxynucleotides and polymerase, resulting in random termination of the fragments at any and all positions, allowing the discrimination of two fragments by one added nucleotide. (D) Sequencing by transcription with T6 and SP6 polymerases, coupled to RNase A single base-specific cleavage. The process queries both strands of each amplicon of interest. The resulting mass fingerprint contains information about all four nucleotides and is used to derive the fragment’s sequence, genotype and heterogeneity and to discover new mutations by in silico pattern comparison to a comprehensive reference set. (E) SNP identification by multiplex PCR. Each product is then queried by a specific probe designed immediately upstream from a SNP of interest and then extended in the presence of ddNTP mix. The resulting extended probes present a mass signature that identifies the SNP sequence. Depending on the assay design (e.g., T5000) the multiplex product could be used directly for MS with the resulting mass patterns used for organism identification.
MS: Mass spectrometry; SAP: Shrimp alkaline phosphatase.
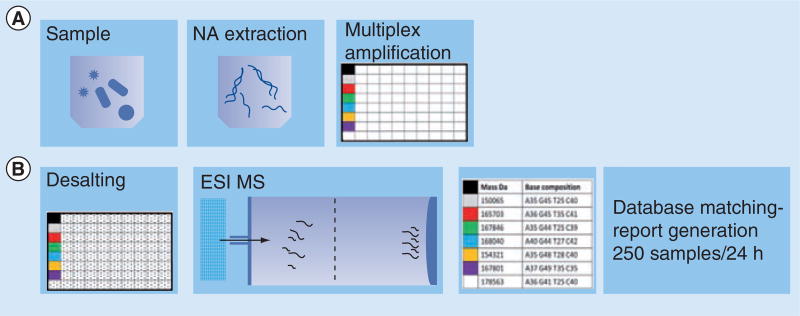
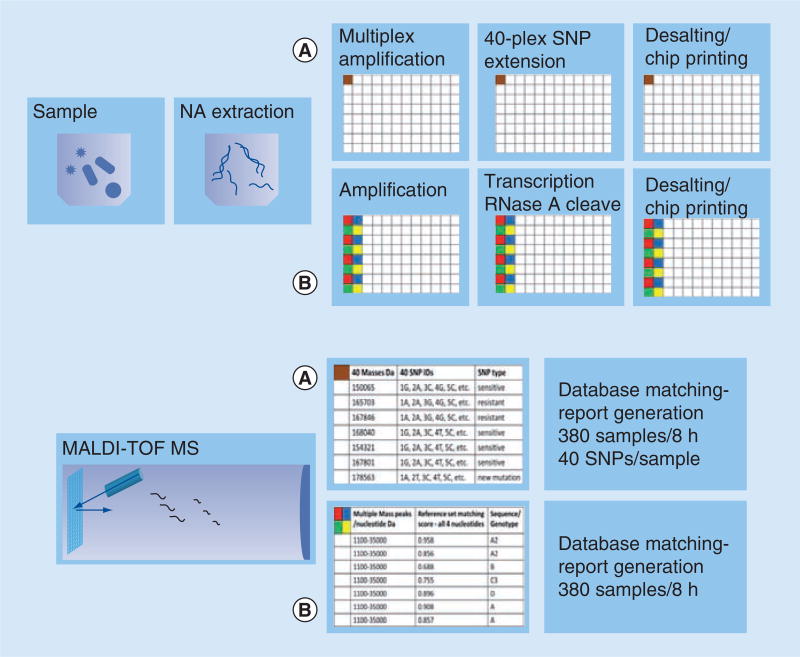
Phylogenetic analysis of the 16S rDNA-gene sequence for the purpose of microbial identification has been considered a gold standard for over 20 years. Specific and reproducible molecular identification of Mycobacterium spp. was successfully achieved by base-specific cleavage of transcripts generated from 16S rDNA amplicons and MS separation of the resulting RNA fragments [31]. The new biosensor technology, triangulation identification genetic evaluation of biological risks (TIGER) [32], on the Ibis T5000 platform was developed using ESI-MS. The molecular element of the test starts with 16S RNA as a base, and adds on the use of broad-range primers aimed at essential housekeeping genes from bacterial, fungal and protozoan pathogens or specific genomic regions of viruses. The targeted PCR amplicons are mixed with an internal calibrant amplicon of known concentration. Analysis is accomplished by matching the base composition of the detected fragments to a database of composition signatures, predetermined for every particular organism. As the different isolates of each pathogen differ slightly, the system handles the diversity by building the probability space from the composition constraints for each species. By a stepwise process of joint maximum likelihood and maximum likelihood clustering, the sample is assigned an organism ID and its abundance estimated. The algorithm involves more than one locus per pathogen. The redundancy affords as high an accuracy of pathogen identification as can be achieved by direct sequencing [15,32]. PLEX-ID is the more current version of the technology and it expands to viral detection [33]. A schematic representation of the test and the process pipeline are shown in Figure 4.
Figure 4. Process pipeline for multiplex pathogen detection and identification by nucleic acid testing on an ESI platform.
(A) Processes that are performed off the MS platform. (B) Processes that are executed on the MS platform.
MS: Mass spectrometry; NA: Nucleic acid.
TIGER found many applications including detection and identification of Acinetobacter species [34], a novel coronavirus responsible for the recent SARS outbreak [35] and alphaviruses [36]. The same platform has been applied very successfully to global surveillance of emerging influenza viruses [27,37], quantitative detection of orthopoxviruses [38] and vector-borne flaviviruses [39] and characterization of zoonotic influenza viruses prior to pandemic spread [40]. The more recent modification of the platform with focus on viral typing, PLEX-ID, is now certified for in vitro diagnostics in Europe.
Although identification of pathogens using MS is usually accomplished by direct detection of the pathogen-specific NAs, alternative approaches based on using cleavable mass-tags have been recently developed for indirect detection of pathogens [41,42]. Avoiding many limitations of the direct detection of NAs, such as the aforementioned fragmentation and salt adduct formation, the mass-tag approaches offer greater robustness and sensitivity of detection as well as ease of multiplexing. The indirect detection methods take advantage of the MS ability to detect and differentiate among fairly small molecular tags that can be cleaved from NAs to identify a specific target that may itself not be amenable to MS because of size constraints and is illustrated in Figure 3B. A recent development is based on using a photocleavable linker with tagged molecules for PCR and ELISA quantification coupled to MS [43]. However, the effective application of the mass-tag methods to the multiplex detection of pathogens is still awaiting further validation.
DNA sequencing
Nothing else showcases the capacity of MS to accurately identify single-point nucleotide differences as its application to DNA sequencing. MALDI-TOF MS has been applied to sequencing of short DNA fragments for over 25 years. MALDI-TOF MS of the products from base-specific, dideoxynucleotide chain-termination DNA-sequencing reactions catalyzed by modified T7 DNA polymerase allows reading of short stretches of sequence (45 nucleotides in length) with as little as 5 fmol material [44]. Another early success was recorded with 63-nucleotide long sequencing ladders generated on an immobilized synthetic template for which the resolved sequence included the primer. In essence, this is a solid-phase Sanger DNA sequencing followed by detection of the extended products by MALDI-TOF MS [45], as illustrated in Figure 3C. The sequencing capacity of that approach is limited to the range of possible mass-size acquisition window; that is, the range of mass sizes of DNA products that can be effectively resolved by MS. In addition, the length of sequences is restricted by the presence of primer in each extension DNA product.
Thus, coupling MS to Sanger DNA sequencing seemed to be impractical, albeit applicable, for de novo sequencing. However, it is particularly suitable for comparative DNA sequencing aiming at the detection of one or very few mutations in short genomic fragments or for the resolution of hard-to-sequence fragments that could not be separated reliably in the standard electrophoretic manner due to compression or false termination. MS is quite suitable as a fast, high-fidelity alternative to conventional sequencing procedures for which high-throughput automated screening for mutations is needed [14].
The aforementioned applications exemplified the feasibility of DNA sequencing using MS; however, they did not take advantage of the MS capacity for accurate resolution of complex molecular mixtures and, therefore, could not compete successfully with gel-based technologies. MS, however, is more sensitive to the nucleotide composition of NAs than electrophoresis as used in conventional sequencing, thus allowing for a greater resolution of short DNA or RNA fragments and providing an opportunity for development of novel MS applications for sequencing using base-specific cleavage of target DNA. For these applications, many oligonucleotide-size fragments generated by the cleavage may have the same length but different nucleotide compositions. Separation of such fragments presents a significant challenge for gel electrophoresis-based technologies, but the fine resolution afforded by MS allows for accurate identification of these fragments.
The critical improvement of the molecular technique that allowed resequencing of fragments of significant size came less than a decade ago with the introduction of the transcription-coupled base-specific RNase A cleavage [46]. The technology uses a PCR amplicon generated with gene-specific primers with one being flanked with T7- and the other with SP6-promoter regions. The amplicon is then subject to transcription by either T7 or SP6 polymerase in the presence of all four nucleotides to obtain transcripts from both DNA strands. UTP or CTP are modified to prevent cleavage by RNase A. Each reaction is carried out isothermally where transcription and cleavage occur simultaneously. The products are resolved by MALDI-TOF MS, resulting in mass patterns that are matched automatically to in silico generated cleavage patterns of an appropriate reference-sequence set. A schematic representation of the process and a process pipeline are shown in Figures 3D & 5, respectively. Although this approach is somewhat inefficient for de novo sequencing, as it requires a substantial reference database and hence great computational power, it has numerous applications for resequencing, genotyping and mutation discovery.
Figure 5. Process pipeline for multiplex single nucleotide polymorphism detection/identification and nucleic acid sequencing by testing on a MALDI-TOF platform.
The top panel of A and B processes are performed off the MS platform, and then followed by acquisition executed on the MS platform. (A) Process line provides multiplexed SNP detection and identification. (B) Process line provides sequencing information. The estimated samples/hour refers to the capacity of the MS.
MS: Mass spectrometry; NA: Nucleic acid.
A recent comprehensive review of the available DNA sequencing strategies covered six different approaches to sequencing, namely: dideoxynucleotide terminators, cyclic array, sequencing-by- hybridization, micro-electrophoresis, MS and nanopore sequencing [47]. It acknowledged the role of MS as a fine tool to address specific problems that cannot be resolved by other methods, especially in the area of direct RNA sequencing, resequencing and methylation analysis [48], but rightly argued that MS may not outcompete the next-generation sequencing (NGS) approaches such as cyclic array (Roche, Illumina, Helicos, Applied Biosystems) and nanopore sequencing (Agilent, Oxford Nanopore Technologies, Noblegen) for de novo high-throughput applications.
Methylation
Methylation is a central epigenetic process, key to understanding pathogenesis and carcinogenesis; it has particular importance for gene regulation and is strongly implicated in the development of cancer and the response to infectious agents [49]. There is an accepted relationship between DNA methylation, chromatin structure and human malignancies [50]. It has been proposed that the host uses chromatin methylation as a defense against DNA viruses, because most CpG sites in the host genomes are methylated so as to regulate gene expression and facilitate the recognition of pathogen-associated DNA [51]. Methylation has been identified as a host defense mechanism in the event of infection with HBV, and it has been demonstrated that the methylation of viral DNA leads to downregulation of the HBV gene expression [52].
The first assay for the analysis and precise quantification of methylation of CpG positions in simplex and multiplex reactions was based on MALDI-TOF MS [53]. The methylation assays are based on the same principle as the aforementioned resequencing assay; however, they require sodium bisulfate treatment of the DNA of interest prior to other enzymatic procedures. The treatment preferentially deaminates cytosine residues to uracil, leading to change of sequence. Comparison of treated and untreated samples that are then cleaved by RNase A reveals the number and location of methylated sites.
Pathogen genotyping
Genotyping of infectious agents is needed to study the agent’s epidemiology, to detect transmission and to make clinical decisions on the appropriate course of treatment [54]. Certain viral genotypes have been associated with development of more aggressive acute or chronic disease or development of cancer. Different strategies for strain discrimination like hybridization, ligation, cleavage and primer extension (most popular) have been combined with MS to detect genotypes [7].
A large body of data has been presented showing the reliability of discriminating sequences differing at a single nucleotide position, so permitting rapid characterization of both cultured and uncultured bacteria [55]. Genotype identification has been achieved by MS separation of DNA fragments generated from 16S PCR products by base-specific DNA degradation with uracil DNA glycosylation in combination with alkaline treatment. Strategies based on the base-specific degradation of DNA and RNA molecules were proven to be very sensitive to minor variation in nucleotide sequence among genetically related organisms [10,46,55]. Specific and reproducible typing of Mycobacterium spp. has been achieved successfully by coupling such molecular approaches to MS analysis; for example, highly multiplexed spoligotyping [56] and multilocus sequence typing [57].
Examples of MALDI-TOF MS for viral typing include an efficient primer extension assay based on the 5´-untranslated region developed for the identification of the 11 major genotypes and over 70 subtypes of HCV [58]. An alternative HCV genotyping assay is based on restriction fragment mass polymorphism (RFMP) analysis of the 5´-untranslated region. This assay has the practical advantage of being capable of identifying mixed viral genotypes present at concentrations as low as 0.5% and accurately determining their relative abundance without the need for genetic cloning [59]. An improved RFMP-based MALDI-TOF MS assay mediated by the use of artificially introduced type IIS restriction enzyme sites has been developed for high-resolution genotyping of HPV. The assay was shown to successfully identify at least 74 different HPV genotypes [60].
The MassARRAY® was developed specifically for assessing genetic polymorphisms and can be used for the analysis of SNPs, insertions, deletions and repeats [61]. It uses detection of mass differences between a specific probe and single nucleotide extension products of that probe; the unique mass of the extended oligonucleotide is identified by MALDI-TOF MS (Figure 3E) and the process pipeline shown in Figure 5. The MassARRAY assay is able to reliably distinguish 0.1–0.01% differences between masses of short oligonucleotides in a detection window of 3800–12,000 Da. The assay detects minority alleles that present at only 2% in DNA mixture. It is readily amenable to multiplexing and suitable for high-throughput genotyping. A sequence-based high-throughput method for the MassARRAY platform has been developed for the detection of 14 oncogenic HPV genotypes in multiplex PCR products. This approach was shown to outperform the reverse dot-blot hybridization in sensitivity, cost and turnaround time [62]. A successful assay for typing influenza viruses was developed using the ESI-MS T5000 platform [37].
Viruses present a particular challenge to typing assays because their replication machinery generates a high error rate, resulting in the extensive genetic variability of the intra- and interhost viral populations. One of the first HBV genotyping assays adapted to MS was based on MALDI-TOF [63]. Recently, an improved approach to HBV genotyping was designed and implemented using the MassARRAY platform. The assay did not only detect genotypes accurately but was capable of identifying new HBV variants [64]. Genotyping was achieved by comparison of mass patterns experimentally obtained from the S gene of the tested strain to computer-simulated HBV variants with known genotype. Multiple parameters such as viral titer, genotype, heterogeneity, quality of PCR products and MS patterns were carefully evaluated in the design and testing of this assay. The quality of PCR product was found to be the only property that significantly affected the accuracy of HBV genotyping. Assay outcomes had complete concordance with the gold-standard sequence results. Although it was not capable of specifying minor HBV genotypes in mixed-genotype infection cases, the predominant genotype was never reported erroneously. Given the sufficient quality of PCR products, MS patterns with low matching score to the reference dataset indicated either the mixed-genotype infection or presence of new HBV variants. The flexible assay design allowed for a rapid adjustment to the detection of such new HBV variants by simply updating the reference dataset with these variants. Highly amenable to automation and high-throughput detection, this assay was commended for molecular surveillance of HBV infection as a low-cost alternative to the sequence-based methods [65].
Polymorphic sites & intra-host diversity
By the middle of the last decade, there was already a clear recognition that NA analysis by MS, in particular by MALDI-TOF MS, would emerge in the post-genome sequencing era as a versatile tool for viral typing, analysis of polymorphisms, sequencing, DNA methylation and RNA expression in a multiplex manner [7]. Identifying genetic diversity by means of detection of SNP instead of using a complete genome sequence scale is a very practical approach to identifying important genetic variants of pathogens (Figure 3E). MS is especially applicable to SNP analysis [61]. After the introduction of the base-specific cleavage concept [46], MS-based SNP analysis has found many fields for its application [7,66,67]. Multilocus genotyping in massively parallel tests is becoming a preferred strategy for the accurate and high-throughput human SNP detection [18,68], with MALDI-TOF MS being among the most powerful and widely used technological platforms. A major advantage of the MS application to molecular diagnostics originates from the unmatched capacity of MS to decode reliably the composition of complex mixtures of short DNA or RNA fragments, and its amenability to high-throughput analysis of such mixtures. The base-specific degradation of PCR products has been extensively used for the specific detection of pathogens, pathogen genotypes and even minor genetic variations associated, for example, with drug resistance [69]. Resolution of products of the base-specific degradation by MS dramatically improves the speed, throughput and accuracy of detection [11,70].
Many viruses generate genetic variants spontaneously or under selective pressure of, for example, antiviral therapy [71,72]. Such variations can confer drug resistance or affect virus replication capacity, resulting in therapeutic failure. Automated MALDI-TOF MS was used for detection of 60 different known mutations in the reverse transcriptase gene, precore promoter and basal core promoter of the HBV genome. MS and direct sequencing showed only 0.1% discordance in variant calls; however, MS was able to detect twice as many minor variants as direct sequencing while achieving close to full automation and more sensitive detection [26]. The RFMP-based MALDI-TOF MS approach was developed for the detection of mutations in the HBV polymerase, particularly in the YMDD motif. The assay was found to be sufficiently sensitive for the early detection of HBV-breakthrough infections in patients on therapeutic treatment with polymerase inhibitors [73]. A similar approach showed a remarkable ability in detecting as few as 100 copies of the HBV genome. It correctly identified several known viral variants, finding minority intra-host viral variants undetectable by other tested methods and estimating the relative abundance of the variants [74]. The RFMP-based MALDITOF assay has been evaluated in comparison to the commercially available INNO-LiPA HBV DR assay based on reverse hybridization line-probe technology. The MS assay provided the greater detection sensitivity for the mutations of interest and identified additional minor intra-host viral subpopulations [75], heralding its clinical applicability.
Mechanisms of viral persistence continue to be the subject of investigation. An important aspect of chronic HCV infection is the quasispecies nature of viral populations, which has been particularly well documented in the hypervariable region 1 (HVR1) of the E2 glycoprotein. Recent studies showed that characterization of the quasispecies diversity at the amino acid level could help to predict the outcome of HCV infection. The accurate characterization of HCV quasispecies requires cloning of PCR products, followed by sequencing of many clones. A method based on in vitro translation of PCR amplicons, followed by MS analysis of the resulting peptide mix was developed for characterization of HCV diversity in infected hosts [76]. The assay was shown to detect the weekly HVR1 changes in the HCV-infected chimpanzee, which coincided with emergence of neutralizing antibodies [77]. Using both a MS-based method and the conventional method of cloning and sequencing, weekly changes of the HVR1 quasispecies in the HCV-infected chimpanzee could be followed and detected with great sensitivity.
Analysis of genetic heterogeneity of viral populations was found to be especially important for evaluation of safety of live-attenuated vaccines, since the presence of even small quantities of mutants or revertants may be associated with incomplete or unstable attenuation of viral strains. However, assessment of the presence of such mutants in a viral population is laborious. MS provides a rapid and accurate platform for such assessment. Recently, DNA MassARRAY was used for evaluation of genetic variation in live-attenuated mumps virus vaccine [78]. A strong correlation with estimates of variation made using restriction enzyme cleavage analysis of PCR fragments indicated the utility of MALDI-TOF MS for routine quality control of live viral vaccines, assessment of their genetic stability and quantitative monitoring of genetic variations.
Genetic relatedness & transmission detection
Transmission is a fundamental viral property, essential for dissemination of infection and disease. As such, it is key to surveillance of infectious diseases. Molecular detection of transmissions can be a complex task. It involves not only identification of the virus, but assessment of the genetic association among its variants. The major assumption of the genome-based detection of transmissions is that the genetic composition of the viral strain that has been passed from one patient to another remains approximately ‘identical’. Detection of transmission, especially when it pertains to viruses, can often present a challenge, in particular when the agent concerned causes chronic disease – that is, the agent has the opportunity to accumulate multiple mutations.
Genetic heterogeneity is a hallmark of many viruses. HCV, for example, exists as multiple variants or quasispecies in each infected individual. Consensus sequencing of the HCV HVR1 or NS5A gene, or also NS5b, is commonly applied to determine genetic relatedness among HCV strains and to identify HCV transmission [79]. However, consensus sequence cannot adequately represent the entire HCV population present in the host, particularly in chronically infected patients where the viral genetic heterogeneity can be extensive.
Accurate identification of HCV strains involved in transmission can be achieved by matching the genetic compositions of viral populations sampled from the infected hosts. Since HVR1 is one of the most variable regions of the HCV genome, analysis of intra-host HVR1 variants is frequently used for identification and tracking of HCV transmission. Such analysis involves: separation of individual HVR1 variants either by genetic cloning of PCR amplicons or by PCR cloning using end point limiting-dilution of cDNA and sequencing of these variants [80]. NGS technologies couple the separation of genetic variants with sequencing, thus significantly simplifying the assessment of viral heterogeneity. However, the high rate of sequencing errors generated per DNA read and problematic representation of intra-host viral heterogeneity are potential hindrances to adopting NGS approaches to the detection of transmissions.
Recently, MALDI-TOF MS was explored for identification of HCV transmission [81]. MS profiles (MSPs) generated by base-specific cleavage of RNA transcripts derived from PCR fragments were used as source of information about the nucleotide sequence and structure of the intra-host viral population. The assay detected patterns of short RNA fragments or k-mers. It was found that the k-mer structure of the MS data closely reflects the heterogeneity of viral populations. Since the genetic distances estimated using MSPs are affected by sequence heterogeneity and composition of intra-host HCV populations [82], MSPs were shown to be highly applicable to evaluating phylogenetic relationships among HCV strains. The analyses revealed that there is a fundamental similarity between MSP- and sequence-based distances, making them suitable for the genetic detection of transmissions. The separation between MSP distances among genetically related and unrelated cases is clear and permitted discrimination of the HCV variants involved in the outbreaks. Such detection of transmission does not require complete assessment of phylogenetic relationships among the HCV variants and, therefore, can be achieved using a threshold and visualized using simple linkage graphs. The MSP-based detection of HCV transmissions matched the accuracy of sequence-based methods, paving the way for the application of MS to molecular surveillance of viral hepatitis [65].
Expert commentary
The fourth Annual Next Generations Diagnostics Summit held in Washington, DC, USA, in August 2012 included a session on molecular diagnostics for infectious disease, dedicated to emerging and novel technologies. The summit emphasized the important potential role of MS for the clinic, recognized that the technological discoveries that enabled the finding of multiple biomarkers have yet to be translated to the clinical practice and encouraged clinicians and scientists to adopt technologies that identify not only pathogens alone but can also reveal resistance determinants present in the pathogens.
During the last decade, MS has emerged as a rapid, cost-effective and highly reproducible technique with multiple versatile applications to molecular diagnostics. Many studies demonstrated efficient applications of MS to the detection of viral pathogens with ESI-MS being especially successful in these applications [32]. Although there are competing technologies like microchip and microarray in the field, MS stands on its own. The major advantages of MS over other competing technologies are most evident when related to investigating parameters of viral infections that require assessment of genetic heterogeneity; for example, detection of mutations, prediction of outcome of therapeutic treatment, and detection of transmissions and genetic relatedness among viral variants. Although there are other technologies such as NGS that can assess the same parameters efficiently, MS is unprecedented in its simplicity in providing dense and accurate genetic information directly associated with the variability parameters of viral infections.
When properly matched with molecular technologies compatible with the mass detection range – for example, base-specific cleavage of DNA or RNA molecules – MS provides an unparalleled capacity for cost-effective and high-throughput detection of genetic markers crucial for molecular surveillance of viral infections and disease as well as for patient management, with MALDI-TOF MS being especially useful in generating information-dense data on the genetic composition of intrahost viral populations. It appears that degrading long NA molecules into short k-mers produces MS patterns that accurately reflect not only the primary structure but also the diversity and frequency of intra-host viral variants, as represented by PCR fragments amplified from viral genomes [46]. Viral genetic heterogeneity has long been found to be associated with outcomes of infection or therapy treatment. Currently, genetic heterogeneity is assessed using NGS, genetic cloning or limited dilution followed by sequencing. Although DNA sequencing using MS is not as efficient as NGS, analysis of MS patterns directly without conversion into sequences seemed to provide adequate estimates of genetic composition of viral populations and can be used to measure genetic relatedness among viral strains and detect viral transmissions [81,82]. However, MS-generated data have structure, which is significantly different from sequences, and, therefore, it requires different computational and mathematical approaches for extraction of information relevant to physicians for patient management and public health practitioners for implementation and development of surveillance and prevention activities.
Five-year view
Application of MS to molecular diagnostics has become a very dynamic area of research with significant implications for medicine and public health. The most advanced developments have been generated by specific matching of molecular techniques with two major MS platforms: ESI and MALDI-TOF. MS as a detection system is most suited for the identification of complex genetic markers without invoking sequences. It can be envisioned that the future development of MS-based molecular diagnostics will be linked to novel methods of extracting clinically and epidemiologically relevant information such as disease severity, drug resistance, vaccine escape and transmission from the genetic markers using specifically designed computational and mathematical models [83,84]. An important aspect of the MS technology will be the potential of its application to the rapid detection of microbes causing hospital infection [85]. If coupled with appropriate rapid and sensitive technologies for the diagnosis of preventable infections, MS can also have impact in the areas of quality control of sterile blood products and food safety [86]. Integration of molecular and computational approaches with MS should produce diagnostic assays for broad, routine application in public health and clinical practice.
Key issues.
During the last decade, mass spectrometry (MS) has emerged as a rapid, cost-effective and highly reproducible technique with multiple versatile applications to molecular diagnostics.
MALDI and ESI are two major platforms for application of MS to molecular diagnostics.
ESI-MS is especially efficient in detection of viral pathogens.
MALDI-TOF is most efficient in assessment of viral and clinical factors associated with genetic heterogeneity of viral pathogens; for example, detection of specific mutations, prediction of outcome of therapeutic treatment, detection of transmissions, identification of genotypes and assessment of fine genetic relatedness among viral variants.
MS is unprecedented in its efficacy of providing dense and accurate genetic information directly associated with clinical parameters of viral infections, thus successfully competing with such powerful technologies as next-generation sequencing.
The structure of the MS-generated data is significantly different from sequences and presents one of the major challenges to application of MS to molecular diagnostics.
Future developments in the MS-based molecular diagnostics of viral infections are contingent on the successful matching between MS and molecular technologies, and application of computational approaches to identification of complex genetic markers from MS patterns directly.
Integration of molecular and computational approaches with MS should produce diagnostic assays for broad, routine application in public health and clinical practice.
Footnotes
Financial & competing interests disclosure
The CDC has a Cooperative Research and Development Agreement with Sequenom Inc. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Abbott R, Anbar M, Faden H, et al. Diagnosis of viral infections by multicomponent mass spectrometric analysis. Clin. Chem. 1980;26(10):1443–1449. [PubMed] [Google Scholar]
- 2.Anbar M, Dyer RL, Scolnick ME. Diagnosis of infectious hepatitis by multicomponent analysis with use of field ionization mass spectrometry. Clin. Chem. 1976;22(9):1503–1509. [PubMed] [Google Scholar]
- 3.Karas M, Hillenkamp F. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 1988;60(20):2299–2301. doi: 10.1021/ac00171a028. [DOI] [PubMed] [Google Scholar]
- 4.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246(4926):64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 5.Pusch W, Kostrzewa M. Application of MALDI-TOF mass spectrometry in screening and diagnostic research. Curr. Pharm. Des. 2005;11(20):2577–2591. doi: 10.2174/1381612054546932. [DOI] [PubMed] [Google Scholar]
- 6.Drake RR, Boggs SR, Drake SK. Pathogen identification using mass spectrometry in the clinical microbiology laboratory. J. Mass Spectrom. 2011;46(12):1223–1232. doi: 10.1002/jms.2008. [DOI] [PubMed] [Google Scholar]
- 7••.Meyer K, Ueland PM. Use of matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for multiplex genotyping. Adv. Clin. Chem. 2011;53:1–29. doi: 10.1016/b978-0-12-385855-9.00001-1. MALDI-TOF mass spectrometry (MS) for large-scale multiplexed assays for fine mapping and allele discrimination. [DOI] [PubMed] [Google Scholar]
- 8••.Tost J, Gut IG. DNA analysis by mass spectrometry-past, present and future. J. Mass Spectrom. 2006;41(8):981–995. doi: 10.1002/jms.1096. Thorough review of the capabilities and potential of MS. [DOI] [PubMed] [Google Scholar]
- 9.Sakallah SA. Molecular diagnostics of infectious diseases: state of the technology. Biotechnol. Annu. Rev. 2000;6:141–161. doi: 10.1016/s1387-2656(00)06021-x. [DOI] [PubMed] [Google Scholar]
- 10.Schweickert B, Moter A, Lefmann M, Göbel UB. Let them fly or light them up: matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometry and fluorescence in situ hybridization (FISH) APMIS. 2004;112(11–12):856–885. doi: 10.1111/j.1600-0463.2004.apm11211-1210.x. [DOI] [PubMed] [Google Scholar]
- 11.Ho YP, Reddy PM. Identification of pathogens by mass spectrometry. Clin. Chem. 2010;56(4):525–536. doi: 10.1373/clinchem.2009.138867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dominguez SR, Briese T, Palacios G, et al. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J. Clin. Virol. 2008;43(2):219–222. doi: 10.1016/j.jcv.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Ecker DJ, Sampath R, Blyn LB, et al. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl Acad. Sci. USA. 2005;102(22):8012–8017. doi: 10.1073/pnas.0409920102. Advanced application of ESI MS in respiratory tract epidemiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurinke C, Oeth P, van den Boom D. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol. Biotechnol. 2004;26(2):147–164. doi: 10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- 15.Hofstadler SA, Sannes-Lowery KA, Hannis JC. Analysis of nucleic acids by FTICR MS. Mass Spectrom. Rev. 2005;24(2):265–285. doi: 10.1002/mas.20016. [DOI] [PubMed] [Google Scholar]
- 16.Doktycz MJ, Hurst GB, Habibi-Goudarzi S, et al. Analysis of polymerase chain reaction-amplified DNA products by mass spectrometry using matrix-assisted laser desorption and electrospray: current status. Anal. Biochem. 1995;230(2):205–214. doi: 10.1006/abio.1995.1465. [DOI] [PubMed] [Google Scholar]
- 17.Ecker DJ, Sampath R, Massire C, et al. Ibis T5000: a universal biosensor approach for microbiology. Nat. Rev. Microbiol. 2008;6(7):553–558. doi: 10.1038/nrmicro1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurinke C, van den Boom D, Cantor CR, Köster H. The use of MassARRAY technology for high throughput genotyping. Adv. Biochem. Eng. Biotechnol. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- 19.Cullinan A, Cantor C. Sequenom, Inc. Pharmacogenomics. 2008;9(9):1211–1215. doi: 10.2217/14622416.9.9.1211. [DOI] [PubMed] [Google Scholar]
- 20.Jurinke C, Zöllner B, Feucht HH, et al. Detection of hepatitis B virus DNA in serum samples via nested PCR and MALDI-TOF mass spectrometry. Genet. Anal. 1996;13(3):67–71. doi: 10.1016/1050-3862(95)00151-4. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (NY) 1992;10(4):413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- 22.Monforte JA, Becker CH. High-throughput DNA analysis by time-of-flight mass spectrometry. Nat. Med. 1997;3(3):360–362. doi: 10.1038/nm0397-360. [DOI] [PubMed] [Google Scholar]
- 23.Rees JC, Voorhees KJ. Simultaneous detection of two bacterial pathogens using bacteriophage amplification coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2005;19(19):2757–2761. doi: 10.1002/rcm.2107. [DOI] [PubMed] [Google Scholar]
- 24.Sjöholm MI, Dillner J, Carlson J. Multiplex detection of human herpesviruses from archival specimens by using matrix-assisted laser desorption ionizationtime of flight mass spectrometry. J. Clin. Microbiol. 2008;46(2):540–545. doi: 10.1128/JCM.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang H, Yang K, Khafagi A, et al. Sensitive detection of human papillomavirus in cervical, head/neck, and schistosomiasis-associated bladder malignancies. Proc. Natl Acad. Sci. USA. 2005;102(21):7683–7688. doi: 10.1073/pnas.0406904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luan J, Yuan J, Li X, et al. Multiplex detection of 60 hepatitis B virus variants by maldi-tof mass spectrometry. Clin. Chem. 2009;55(8):1503–1509. doi: 10.1373/clinchem.2009.124859. [DOI] [PubMed] [Google Scholar]
- 27.Deyde VM, Sampath R, Gubareva LV. RT-PCR/electrospray ionization mass spectrometry approach in detection and characterization of influenza viruses. Expert Rev. Mol. Diagn. 2011;11(1):41–52. doi: 10.1586/erm.10.107. [DOI] [PubMed] [Google Scholar]
- 28.Michael K, Harder TC, Mettenleiter TC, Karger A. Diagnosis and strain differentiation of avian influenza viruses by restriction fragment mass analysis. J. Virol. Methods. 2009;158(1–2):63–69. doi: 10.1016/j.jviromet.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Ilina EN, Malakhova MV, Generozov EV, Nikolaev EN, Govorun VM. Matrix-assisted laser desorption ionization-time of flight (mass spectrometry) for hepatitis C virus genotyping. J. Clin. Microbiol. 2005;43(6):2810–2815. doi: 10.1128/JCM.43.6.2810-2815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han KH, Hong SP, Choi SH, et al. Comparison of multiplex restriction fragment mass polymorphism and sequencing analyses for detecting entecavir resistance in chronic hepatitis B. Antivir. Ther. (Lond.) 2011;16(1):77–87. doi: 10.3851/IMP1702. [DOI] [PubMed] [Google Scholar]
- 31.Lefmann M, Honisch C, Böcker S, et al. Novel mass spectrometry-based tool for genotypic identification of mycobacteria. J. Clin. Microbiol. 2004;42(1):339–346. doi: 10.1128/JCM.42.1.339-346.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Hofstadler SA, Sampath R, Blyn LB, et al. TIGER: the universal biosensor. Int. J. Mass Spectrom. 2005;242(1):23–41. Thorough description of the triangulation strategy for identification of microorganisms by ESI. [Google Scholar]
- 33.Cordey S, Thomas Y, Suter P, Kaiser L. Pilot evaluation of RT-PCR/electrospray ionization mass spectrometry (PLEX-ID/ Flu assay) on influenza-positive specimens. Open Virol. J. 2012;6:64–67. doi: 10.2174/1874357901206010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ecker JA, Massire C, Hall TA, et al. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 2006;44(8):2921–2932. doi: 10.1128/JCM.00619-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampath R, Hofstadler SA, Blyn LB, et al. Rapid identification of emerging pathogens: Coronavirus. Emerging Infect. Dis. 2005;11(3):373–379. doi: 10.3201/eid1103.040629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eshoo MW, Whitehouse CA, Zoll ST, et al. Direct broad-range detection of α viruses in mosquito extracts. Virology. 2007;368(2):286–295. doi: 10.1016/j.virol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 37.Sampath R, Russell KL, Massire C, et al. Global surveillance of emerging Influenza virus genotypes by mass spectrometry. PLoS ONE. 2007;2(5):e489. doi: 10.1371/journal.pone.0000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grant RJ, Baldwin CD, Nalca A, et al. Application of the Ibis-T5000 pan-orthopoxvirus assay to quantitatively detect monkeypox viral loads in clinical specimens from macaques experimentally infected with aerosolized monkeypox virus. Am. J. Trop. Med. Hyg. 2010;82(2):318–323. doi: 10.4269/ajtmh.2010.09-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant-Klein RJ, Baldwin CD, Turell MJ, et al. Rapid identification of vector-borne flaviviruses by mass spectrometry. Mol. Cell. Probes. 2010;24(4):219–228. doi: 10.1016/j.mcp.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 40.Metzgar D, Baynes D, Myers CA, et al. Initial identification and characterization of an emerging zoonotic influenza virus prior to pandemic spread. J. Clin. Microbiol. 2010;48(11):4228–4234. doi: 10.1128/JCM.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofstadler SA, Sannes-Lowery KA, Crooke ST, et al. Multiplexed screening of neutral mass-tagged RNA targets against ligand libraries with electrospray ionization FTICR MS: a paradigm for high-throughput affinity screening. Anal. Chem. 1999;71(16):3436–3440. doi: 10.1021/ac990262n. [DOI] [PubMed] [Google Scholar]
- 42.Thompson A, Prescott M, Chelebi N, Smith J, Brown T, Schmidt G. Electrospray ionisation-cleavable tandem nucleic acid mass tag-peptide nucleic acid conjugates: synthesis and applications to quantitative genomic analysis using electrospray ionisation-MS/MS. Nucleic Acids Res. 2007;35(4):e28. doi: 10.1093/nar/gkl1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stauber J, El Ayed M, Wisztorski M, Day R, Fournier I, Salzet M. Polymerase chain reaction and immunoassay–matrix assisted laser desorption mass spectrometry using tag-mass technology: new tools to break down quantification limits and multiplexes. Anal. Chem. 2009;81(22):9512–9521. doi: 10.1021/ac901416s. [DOI] [PubMed] [Google Scholar]
- 44.Shaler TA, Tan Y, Wickham JN, Wu KJ, Becker CH. Analysis of enzymatic DNA sequencing reactions by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 1995;9(10):942–947. doi: 10.1002/rcm.1290091015. [DOI] [PubMed] [Google Scholar]
- 45.Köster H, Tang K, Fu DJ, et al. A strategy for rapid and efficient DNA sequencing by mass spectrometry. Nat. Biotechnol. 1996;14(9):1123–1128. doi: 10.1038/nbt0996-1123. [DOI] [PubMed] [Google Scholar]
- 46••.Stanssens P, Zabeau M, Meersseman G, et al. High-throughput MALDI-TOF discovery of genomic sequence polymorphisms. Genome Res. 2004;14(1):126–133. doi: 10.1101/gr.1692304. Introducing the base-specific cleavage of a target sequence as a comparative sequencing strategy for MALDI-TOF MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shendure JA, Porreca GJ, Church GM. Overview of DNA sequencing strategies. Curr. Protoc. Mol. Biol. 2008;Chapter 7(Unit 7.1) doi: 10.1002/0471142727.mb0701s81. [DOI] [PubMed] [Google Scholar]
- 48.Ragoussis J, Elvidge GP, Kaur K, Colella S. Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry in genomics research. PLoS Genet. 2006;2(7):e100. doi: 10.1371/journal.pgen.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng YB, Nagae G, Midorikawa Y, et al. Identification of genes preferentially methylated in hepatitis C virus-related hepatocellular carcinoma. Cancer Sci. 2010;101(6):1501–1510. doi: 10.1111/j.1349-7006.2010.01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Ehrich M, Nelson MR, Stanssens P, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl Acad. Sci. USA. 2005;102(44):15785–15790. doi: 10.1073/pnas.0507816102. High-throughput DNA methylation analysis by MALDI-TOF MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoelzer K, Shackelton LA, Parrish CR. Presence and role of cytosine methylation in DNA viruses of animals. Nucleic Acids Res. 2008;36(9):2825–2837. doi: 10.1093/nar/gkn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vivekanandan P, Daniel HD, Kannangai R, Martinez-Murillo F, Torbenson M. Hepatitis B virus replication induces methylation of both host and viral DNA. J. Virol. 2010;84(9):4321–4329. doi: 10.1128/JVI.02280-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tost J, Schatz P, Schuster M, Berlin K, Gut IG. Analysis and accurate quantification of CpG methylation by MALDI mass spectrometry. Nucleic Acids Res. 2003;31(9):e50. doi: 10.1093/nar/gng050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lok AS, Seeff LB, Morgan TR, et al. HALT-C Trial Group. Incidence of hepatocellular carcinoma and associated risk factors in hepatitis C-related advanced liver disease. Gastroenterology. 2009;136(1):138–148. doi: 10.1053/j.gastro.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Wintzingerode F, Böcker S, Schlötelburg C, et al. Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: a tool for rapid bacterial identification. Proc. Natl Acad. Sci. USA. 2002;99(10):7039–7044. doi: 10.1073/pnas.102165899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honisch C, Mosko M, Arnold C, Gharbia SE, Diel R, Niemann S. Replacing reverse line blot hybridization spoligotyping of the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 2010;48(5):1520–1526. doi: 10.1128/JCM.02299-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Honisch C, Chen Y, Mortimer C, et al. Automated comparative sequence analysis by base-specific cleavage and mass spectrometry for nucleic acid-based microbial typing. Proc. Natl Acad. Sci. USA. 2007;104(25):10649–10654. doi: 10.1073/pnas.0704152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Il’ina EN, Malakhova MV, Generozov EV, Govorun VM, Archakov AI, Pokrovskii VI. Using the mass spectrometry analysis for hepatitis C virus typing. Biomed. Khim. 2005;51(1):41–47. [PubMed] [Google Scholar]
- 59.Kim YJ, Kim SO, Chung HJ, et al. Population genotyping of hepatitis C virus by matrix-assisted laser desorption/ ionization time-of-flight mass spectrometry analysis of short DNA fragments. Clin. Chem. 2005;51(7):1123–1131. doi: 10.1373/clinchem.2004.047506. [DOI] [PubMed] [Google Scholar]
- 60.Hong SP, Shin SK, Lee EH, et al. High-resolution human papillomavirus genotyping by MALDI-TOF mass spectrometry. Nat. Protoc. 2008;3(9):1476–1484. doi: 10.1038/nprot.2008.136. [DOI] [PubMed] [Google Scholar]
- 61.Jurinke C, van den Boom D, Cantor CR, Köster H. Automated genotyping using the DNA MassArray technology. Methods Mol. Biol. 2002;187:179–192. doi: 10.1385/1-59259-273-2:179. [DOI] [PubMed] [Google Scholar]
- 62.Söderlund-Strand A, Dillner J, Carlson J. High-throughput genotyping of oncogenic human papilloma viruses with MALDITOF mass spectrometry. Clin. Chem. 2008;54(1):86–92. doi: 10.1373/clinchem.2007.092627. [DOI] [PubMed] [Google Scholar]
- 63.Malakhova MV, Ilina EN, Govorun VM, et al. Hepatitis B virus genetic typing using mass-spectrometry. Bull. Exp. Biol. Med. 2009;147(2):220–225. doi: 10.1007/s10517-009-0479-1. [DOI] [PubMed] [Google Scholar]
- 64.Ganova-Raeva L, Ramachandran S, Honisch C, Forbi JC, Zhai X, Khudyakov Y. Robust hepatitis B virus genotyping by mass spectrometry. J. Clin. Microbiol. 2010;48(11):4161–4168. doi: 10.1128/JCM.00813-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganova-Raeva LM, Dimitrova ZE, Campo DS, Khudyakov Y. Application of mass spectrometry to molecular surveillance of hepatitis B and C viral infections. Antivir. Ther. 2012;17(7 Pt B):1477–1482. doi: 10.3851/IMP2466. [DOI] [PubMed] [Google Scholar]
- 66.Jurinke C, Denissenko MF, Oeth P, Ehrich M, van den Boom D, Cantor CR. A single nucleotide polymorphism based approach for the identification and characterization of gene expression modulation using MassARRAY. Mutat. Res. 2005;573(1–2):83–95. doi: 10.1016/j.mrfmmm.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 67.Tang K, Oeth P, Kammerer S, et al. Mining disease susceptibility genes through SNP analyses and expression profiling using MALDI-TOF mass spectrometry. J. Proteome Res. 2004;3(2):218–227. doi: 10.1021/pr034080s. [DOI] [PubMed] [Google Scholar]
- 68.Pusch W, Wurmbach JH, Thiele H, Kostrzewa M. MALDI-TOF mass spectrometry-based SNP genotyping. Pharmacogenomics. 2002;3(4):537–548. doi: 10.1517/14622416.3.4.537. [DOI] [PubMed] [Google Scholar]
- 69.Woo HY, Park H, Kim BI, Jeon WK, Cho YK, Kim YJ. Comparison of mass spectrometric analysis and TRUGENE HBV genotyping for monitoring lamivudine resistance in chronic hepatitis B patients. Antivir. Ther. (Lond.) 2007;12(1):7–13. [PubMed] [Google Scholar]
- 70•.Emonet S, Shah HN, Cherkaoui A, Schrenzel J. Application and use of various mass spectrometry methods in clinical microbiology. Clin. Microbiol. Infect. 2010;16(11):1604–1613. doi: 10.1111/j.1469-0691.2010.03368.x. Various applications of MS methods in clinical microbiology. [DOI] [PubMed] [Google Scholar]
- 71.Pawlotsky JM. Genetic heterogeneity and properties of hepatitis C virus. Acta Gastroenterol. Belg. 1998;61(2):189–191. [PubMed] [Google Scholar]
- 72.Villet S, Billioud G, Pichoud C, et al. In vitro characterization of viral fitness of therapy-resistant hepatitis B variants. Gastroenterology. 2009;136(1):168–176. e2. doi: 10.1053/j.gastro.2008.09.068. [DOI] [PubMed] [Google Scholar]
- 73.Lee CH, Kim SO, Byun KS, et al. Predominance of hepatitis B virus YMDD mutants is prognostic of viral DNA breakthrough. Gastroenterology. 2006;130(4):1144–1152. doi: 10.1053/j.gastro.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 74.Hong SP, Kim NK, Hwang SG, et al. Detection of hepatitis B virus YMDD variants using mass spectrometric analysis of oligonucleotide fragments. J. Hepatol. 2004;40(5):837–844. doi: 10.1016/j.jhep.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 75.Kim HS, Han KH, Ahn SH, et al. Evaluation of methods for monitoring drug resistance in chronic hepatitis B patients during lamivudine therapy based on mass spectrometry and reverse hybridization. Antivir. Ther. (Lond.) 2005;10(3):441–449. [PubMed] [Google Scholar]
- 76.Ayers M, Siu K, Roberts E, Garvin AM, Tellier R. Characterization of hepatitis C virus quasispecies by matrix-assisted laser desorption ionization-time of flight (mass spectrometry) mutation detection. J. Clin. Microbiol. 2002;40(9):3455–3462. doi: 10.1128/JCM.40.9.3455-3462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77•.Yea C, Bukh J, Ayers M, Roberts E, Krajden M, Tellier R. Monitoring of hepatitis C virus quasispecies in chronic infection by matrix-assisted laser desorption ionization-time of flight mass spectrometry mutation detection. J. Clin. Microbiol. 2007;45(3):1053–1057. doi: 10.1128/JCM.02512-06. Use of MALDI-TOF MS for monitoring of viral variability. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amexis G, Oeth P, Abel K, et al. Quantitative mutant analysis of viral quasispecies by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl Acad. Sci. USA. 2001;98(21):12097–12102. doi: 10.1073/pnas.211423298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Widell A, Christensson B, Wiebe T, et al. Epidemiologic and molecular investigation of outbreaks of hepatitis C virus infection on a pediatric oncology service. Ann. Intern. Med. 1999;130(2):130–134. doi: 10.7326/0003-4819-130-2-199901190-00007. [DOI] [PubMed] [Google Scholar]
- 80.Nainan OV, Lu L, Gao FX, Meeks E, Robertson BH, Margolis HS. Selective transmission of hepatitis C virus genotypes and quasispecies in humans and experimentally infected chimpanzees. J. Gen. Virol. 2006;87(Pt 1):83–91. doi: 10.1099/vir.0.81268-0. [DOI] [PubMed] [Google Scholar]
- 81.Ganova-Raeva LM, Dimitrova ZE, Campo DS, et al. Detection of hepatitis C virus transmission by use of DNA mass spectrometry. J. Infect. Dis. 2012;207(6):999–1006. doi: 10.1093/infdis/jis938. [DOI] [PubMed] [Google Scholar]
- 82.Dimitrova Z, Campo DS, Ramachandran S, et al. Evaluation of viral heterogeneity using next-generation sequencing, end-point limiting-dilution and mass spectrometry. In Silico Biol. 2012;11(5):183–192. doi: 10.3233/ISB-2012-0453. [DOI] [PubMed] [Google Scholar]
- 83.Khudyakov Y. Molecular surveillance of hepatitis C. Antivir. Ther. (Lond.) 2012;17(7 Pt B):1465–1470. doi: 10.3851/IMP2476. [DOI] [PubMed] [Google Scholar]
- 84.Lara J, Khudyakov Y. Epistatic connectivity among HCV genomic sites as a genetic marker of interferon resistance. Antivir. Ther. 2012;12:1471–1475. doi: 10.3851/IMP2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Endimiani A, Hujer KM, Hujer AM, et al. Are we ready for novel detection methods to treat respiratory pathogens in hospital-acquired pneumonia? Clin. Infect. Dis. 2011;52(Suppl. 4):S373–S383. doi: 10.1093/cid/cir054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Bissonnette L, Bergeron MG. Multiparametric Technologies for the diagnosis of syndromatic infections. Clin. Microbiol. Newslett. 2012;34(20):159–168. Available clinical molecular microbiology tests for the diagnosis of infectious syndromes developed on multiparametric detection platforms. [Google Scholar]