Abstract
The ubiquitin proteasome system degrades the great majority of proteins in mammalian cells. Countless studies have described how ubiquitination promotes the selective degradation of different cell proteins. However, there is a small but growing literature that protein half-lives can also be regulated by post-translational modifications of the 26S proteasome. This article reviews the ability of several kinases to alter proteasome function through subunit phosphorylation. For example, PKA and DYRK2 stimulate the proteasome’s ability to degrade ubiquitinated proteins, peptides, and ATP, while one kinase, ASK1, inhibits proteasome function during apoptosis. Proteasome phosphorylation is likely to be important in regulating protein degradation because it occurs downstream of many hormones and neurotransmitters, in conditions that raise cAMP or cGMP levels, after calcium influx following synaptic depolarization, and during phases of the cell cycle. Beyond its physiological importance, pharmacological manipulation of proteasome phosphorylation has the potential to combat various diseases. Inhibitors of phosphodiesterases by activating PKA or PKG can stimulate proteasomal degradation of misfolded proteins that cause neurodegenerative or myocardial diseases and even reduce the associated pathology in mouse models. These observations are promising since in many proteotoxic diseases, aggregation-prone proteins impair proteasome function and disrupt protein homeostasis. Conversely, preventing subunit phosphorylation by DYRK2 slows cell cycle progression and tumor growth. However, further research is essential to determine how phosphorylation of different subunits by these (or other) kinases alter the properties of this complex molecular machine and thus influence protein degradation rates.
Keywords: Proteasome phosphorylation, protein degradation, ubiquitin proteasome system, protein kinase, proteasome activation, protein homeostasis
INTRODUCTION
The great majority of proteins in mammalian cells are degraded by the ubiquitin proteasome system (UPS). In this pathway, proteins are modified by the covalent attachment of multiple ubiquitin molecules via an enzymatic cascade (1). Formation of a ubiquitin chain on a protein targets it for rapid degradation by the 26S proteasome. This ATP-dependent proteolytic complex is composed of the hollow, cylindrical 20S core particle (CP), within which proteins are degraded, and at one or both ends, a 19S regulatory particle (RP) which binds the ubiquitinated substrate, disassembles the ubiquitin chain, and unfolds and translocates the polypeptide into the core particle (2–4). In the mammalian genome, there are about 650 ubiquitin ligases (E3s) and over 30 different ubiquitin conjugating enzymes (E2s), which together provide great selectivity and tight regulation to the ubiquitination step (5). Consequently, it has been widely assumed that the rate of degradation of a protein is determined solely by its rate of ubiquitination. However, there is growing evidence that many ubiquitinated proteins bind to the proteasome, are deubiquitinated and dissociate without degradation (4, 6–8). Also, it is now clear that the ability of the 26S proteasome to degrade ubiquitinated proteins is tightly regulated and can be altered by several postsynthetic mechanisms. One form of proteasome regulation that not only seems to be of major physiological importance, but also has clear therapeutic potential is subunit phosphorylation (9). This article reviews our present understanding of this mode of regulation of proteasome function and its biological significance.
The degradation of ubiquitin conjugates by the 26S proteasome involves multiple steps that are tightly coordinated and linked to ATP hydrolysis by the six AAA+ ATPase subunits (Rpt1-6), which form the base of the 19S particle (3). After a ubiquitinated protein binds reversibly to receptor subunits on the 19S particle (Rpn1, Rpn10, or Rpn13), the ubiquitin chains are hydrolyzed by three proteasome-associated deubiquitiylating enzymes (Usp14, Uch37, and Rpn11) (10). This removal of ubiquitin prevents the degradation of ubiquitin with the substrate and enables the ubiquitins to be reused in subsequent rounds of degradation. If a substrate contains a loosely-folded domain in addition to the ubiquitin chain, it can become tightly bound to the 26S complex and committed to degradation (8, 11). The protein is then translocated through the ATPase ring (Rpt1-6) and the gated pore in the 20S particle’s outer α-ring. Upon binding ATP, the ATPases’ C-terminal HbYX residues dock into intersubunit pockets in the α-ring and trigger gate opening and peptide hydrolysis (12). Translocation through the ATPases’ central channel causes unfolding of the upstream domains, enabling substrate passage through the narrow gated channel into the 20S particle and proteolytic digestion. This gate, which is formed by the N-termini of its α-subunits, helps prevent nonspecific entry and degradation of cell proteins and the escape of partially digested substrates (13). Protein hydrolysis occurs in the central chamber of the 20S particle and is catalyzed by its three peptidase sites, which have chymotrypsin-like, trypsin-like, and caspase-like specificities and are present on each of the two central β-rings (14).
The degradation of different proteins by the UPS is regulated primarily by their rates of ubiquitylation by one of the many ubiquitin ligases present in every cell. The selectivity of the E3s for certain proteins is often determined by post-translational modifications of the substrate, especially phosphorylation (15). In addition, the proteasome can be regulated by multiple mechanisms, including association with additional activating complexes, such as PA28αβ and PA200 (16), deubiquitinating enzymes, such as Usp14 (6), ubiquitin ligases, such as Ube3c/Hul5 (17, 18) or regulatory proteins such as AIRAP (19) and ZNF216 (20). Cells also can enhance their degradative capacity after proteasome inhibition by expressing additional 26S proteasomes (21, 22) and specialized subunits of the 20S core particle are present in thymus, testes, and immune tissues (23). The proteasomes in immune tissues and after IFN-γ treatment are called immunoproteasomes and degrade proteins at normal rates, but their peptidase sites are more efficient in generating peptides appropriate for antigen presentation (24).
In addition to these types of regulation, mass spectrometry studies have described hundreds of post-synthetic modifications on proteasome subunits (25, 26). A variety of intriguing reports have described alterations in proteasome function after subunit modification by O-GlcNAc (27), ADP-ribosylation (28), and ubiquitylation (17, 29). These modifications all merit further study to clarify their physiological importance. Unfortunately, the overwhelming majority of the post-translational modifications detected by mass spectrometry, especially the many examples of subunit phosphorylation, have not been experimentally investigated. Their influence, if any, on proteasome functions, remain unknown.
This article focuses on the phosphorylation of proteasome subunits by different kinases. Although the structure, enzymatic activities, and cellular functions of the 26S proteasomes have been extensively investigated (2, 4, 30), proteasome regulation by protein kinases has received relatively little attention. Nevertheless, there is considerable evidence discussed below that certain types of proteasome phosphorylation not only appear to be of major physiological importance in regulating protein turnover, but also have therapeutic potential to combat various proteotoxic diseases or cancer. Our goal in preparing this review is also to stimulate further interest in this area. Consequently, we shall also highlight a number of outstanding questions about proteasome function and regulation that remain to be explored.
cAMP via Protein Kinase A (PKA) stimulates proteasome activities
Cyclic adenosine monophosphate (cAMP) was not only the first intracellular “second messenger” to be discovered (31), but also the first intracellular signalling system implicated in proteasome regulation (32). A rise in intracellular cAMP mediates the effects of many hormones and neurotransmitters and is critical in multiple metabolic responses, especially ones involved in the mobilization of tissue energy reserves. The elucidation of the pathways for cAMP production and its signaling mechanisms have accelerated our understanding of many other cellular regulatory systems. Increasing intracellular cAMP concentration, either through stimulating its synthesis by a G protein-regulated adenylyl cyclase or inhibiting its hydrolysis by one of the cell’s eleven distinct cyclic nucleotide phosphodiesterases, leads to cAMP binding to protein kinase A (PKA) (33, 34). This association of cAMP with PKA’s two inhibitory subunits causes their dissociation and allows its two catalytic subunits to phosphorylate numerous cellular proteins, including subunits of the proteasome’s 19S regulatory complex.
When cAMP levels rise markedly in cells after treatment with forskolin, a natural product which stimulates adenylyl cyclases, or rolipram, which inhibits the major cAMP-specific phosphodiesterase, PDE4 (35), PKA’s catalytic subunits become associated with the 26S proteasome (36), and proteasome activity is enhanced in multiple ways. Both treatments were shown to increase the 26S proteasome’s peptidase activity in crude extracts of all cell types studied thus far (36). Furthermore, the 26S proteasomes purified from the treated cells were more active in the degradation of ubiquitinated proteins, the 26S proteasome’s physiological substrates, as well as short peptides that are specific substrates of each of the core particle’s three peptidase sites (37). Additionally, ATP hydrolysis by the 26S, which drives ubiquitin conjugate degradation (38), was also stimulated. These enhanced activities were clearly due to phosphorylation of the 26S because they could be reduced toward normal levels upon incubation with protein phosphatase 1 (PP1) or by treating the cells with the kinase inhibitor H89 (36).
Many proteasome subunits and 26S-associated proteins contain the PKA target sequences and therefore can potentially be phosphorylated by PKA. Analysis of affinity-purified 26S proteasomes from the forskolin- or rolipram-treated cells by PhosTag gel electrophoresis, in which proteins migrate more slowly when phosphorylated (39), and mass spectrometry, indicated that the 19S subunit Rpn6 was phosphorylated at serine 14 by PKA. PKA had been reported to activate the proteasome via phosphorylation of the ATPase subunit Rpt6 (32), but this modification was not increased in HEK293 cells upon forskolin or rolipram treatment (36). cAMP-induced Rpn6 phosphorylation at S14 has now been observed in many cell types and tissues with a phosphorylation site-specific antibody (Lokireddy et al, submitted). Furthermore, over-expression of a phosphomimetic Rpn6 serine 14 to aspartic acid mutant in HEK293 cells increased proteasomal ATPase and peptidase activities, while the phosphodead serine 14 to alanine mutant reduced them to below control levels. Thus, Rpn6 phosphorylation is sufficient to stimulate proteasome activities (36).
In the 19S regulatory particle, Rpn6 seems ideally situated to influence multiple proteasome activities. Cryo-electron microscopy studies have shown that Rpn6 not only interacts with Rpt6, one of the six ATPase subunits, but also reaches the 20S core particles’ outer ring and interacts with α2, one of its seven α-subunits (40), whose N-termini form the gated channel for substrate entry. Raising cAMP also seemed to increase the fraction of 26S proteasomes containing two 19S complexes (36, 41), and it’s possible that this increase in doubly-capped particles contributes to the enhanced activity. Surprisingly, an increase in doubly- and singly- capped proteasomes and peptidase activity was observed in human stem cells when Rpn6 was over-expressed (42). Furthermore, when over-expressed in C. elegans, Rpn6 prolonged lifespan, especially in oxidative stress and heat shock conditions (43). Thus this subunit, even without phosphorylation, seems to serve a key regulatory role and may promote the formation or stability of the 26S proteasomes.
The stimulation of proteolysis by cAMP was evident within minutes after forskolin addition and did not require new protein synthesis (36). Raising cAMP levels and Rpn6 phosphorylation not only increased multiple activities of purified 26S proteasomes, but also enhanced the degradation of some cell proteins selectively. To ascertain whether the degradation of all cell proteins was enhanced similarly, Lokireddy and colleagues differentially radiolabeled two broad sets of cell proteins with pulses of labeled amino acids for different durations. Surprisingly, increasing intracellular cAMP levels stimulated the degradation of the short-lived (i.e. newly synthesized) fraction of cell proteins, while the breakdown of long-lived proteins, which comprise the bulk of cellular proteins, was unaffected. This forskolin-induced increase in protein breakdown occurred without a concomitant rise in the levels of ubiquitinated proteins. On the contrary, total ubiquitin conjugate levels in the cells fell rapidly after forskolin addition due to the accelerated proteasomal activities.
The short-lived proteins include various regulatory and misfolded proteins that are degraded on the order of minutes to a few hours, while the long-lived components, tend to have half-lives of many hours or days (44). Short-lived proteins are degraded exclusively by the UPS, and this is the first example in which their degradation has been found to be selectively altered. Other factors known to stimulate overall proteolysis (e.g. mTOR inhibition, serumm deprivation or overexpression of constitutively active FOXO transcription factors) only increase the breakdown of long-lived proteins (44, 45). Increasing cAMP levels also enhanced the degradation of several rapidly degraded proteins that are ubiquitinated by different enzymes, including the N-end rule system (Ub-R-GFP) (46), the UFD pathway (UbG76V-GFP) (47), and the hydrophobic degron (GFP-CL1, aka GFPu), as well as a few short-lived endogenous proteins, including the transcription factors SP1 (32), Nrf2 and c-Myc (36). By contrast, no stimulation of autophagic proteolysis in HEK293 cells was seen under these conditions. Of particular interest was the finding that raising cAMP pharmacologically, or expressing the phosphomimetic Rpn6 mutant stimulated the breakdown of several aggregation-prone mutant proteins associated with human neurodegenerative diseases, including mutant FUS and TDP43, which cause Amyotrophic Lateral Sclerosis (ALS), and a mutant Tau that causes Frontotemporal Dementia (36, 48) (see below).
Because cAMP and PKA mediate the actions of many hormones, and neurotransmitters, these findings suggested that proteasome activation occurs not only with the very high cAMP concentrations achieved pharmacologically, but also in various physiological conditions that raise cAMP. Probably the best characterized cAMP-mediated metabolic responses are the activation of glycogen breakdown and the inhibition of glycogen synthesis in liver and muscle by glucagon or epinephrine. Treatment of hepatocytes with glucagon increased Rpn6 phosphorylation at serine 14, stimulated proteasome activity, and selectively enhanced the degradation of short-lived proteins (Lokireddy et al, submitted). Epinephrine caused a similar proteasome activation in hepatocytes and hearts. Epinephrine and cAMP are critical in activating the sympathetic “Fight or Flight” response which occurs during exercise. In muscle biopsies from human volunteers after vigorous exercise (49) and in rat muscles after repetitive stimulation, the 26S proteasomes contained Rpn6 phosphorylated at serine 14 and were more active (Lokireddy et al, submitted). cAMP levels also rise in the liver and skeletal muscles after a brief fast. The 26S proteasomes purified from these mouse tissues after food-deprivation for 12–48 hours were phosphorylated on Rpn6 at serine 14 and exhibited greater activities without any increase in proteasome number. After food deprivation, this rapid enhancement of 26S activity must occur simultaneously with the increase in protein ubiquitination and autophagy caused by the fall in mTOR activity (45) and it clearly precedes the transcriptional adaptations and large increases in protein breakdown and muscle wasting in fasting (50).
Because short-lived proteins comprise a small fraction of cell mass, it is unclear how their accelerated breakdown may provide a selective advantage to the organism during exercise, fasting, or other hormonal responses that increase intracellular cAMP levels. Their degradation is unlikely to provide the organism with significant metabolic energy, in contrast to the cAMP-mediated breakdown of glycogen or lipids or the accelerated breakdown of long-lived cell proteins in starvation that provides amino acids for gluconeogenesis. The faster degradation of short-lived proteins in muscle upon exercise may promote the efficient elimination of proteins damaged by the repeated contractions or by free radicals released from mitochondria. Alternatively, the faster degradation of short-lived proteins may help cells adapt to the altered physiological conditions by rapidly destroying certain preexistent regulatory proteins. Their clearance would thus complement the concomitant stimulation of new gene expression by cAMP and CREB and together accelerate the adaptation to the new physiological demands.
Therapeutic potential of stimulating the proteasome via agents that raise cAMP
A primary function of the UPS is to eliminate misfolded or damaged proteins whose accumulation is potentially toxic (51). Pharmacological treatments that stimulate their degradation represents a potential new approach to reduce the accumulation of the misfolded proteins that cause many human diseases. The demonstration that activators of adenylyl cyclases or inhibitors of phosphodiesterases can promote the degradation of mutant proteins that cause Amyotrophic Lateral Sclerosis or Frontotemporal Dementia and other tauopathies (36, 48) raised the possibility that such agents could represent novel therapies for these and other proteotoxic disease. A number of pharmacological agents are known that raise cAMP in target tissues, and many inhibitors of PDE4 have been synthesized, including three approved for human use (35).
Activation of proteasomes should be of particular interest for therapy in neurodegenerative disorders, because of the growing evidence that proteolysis by the UPS is impaired in such diseases (52). In several disease models, the accumulation of aggregation-prone mutant proteins in cells and mouse tissues interferes with protein breakdown by the 26S proteasome. For example, infection of neurons with the toxic form of PrPSC, the agent that causes Bovine Spongiform Encephalitis and Creutzfeldt-Jakob disease, decreases, protein degradation, and 26S proteasome activity (53). Additionally, the association of aggregated PrPSC with purified proteasomes impairs its gating mechanism (54). Similarly, the overexpression in mouse brains of a mutant Tau protein (P301L) that causes human Frontotemporal Dementia leads to the accumulation of ubiquitinated proteins, phospho-tau, and the widely-studied UPS substrate UbG76V-GFP (48). Furthermore, the 26S proteasomes purified from these brains have a reduced ability to hydrolyze ubiquitinated proteins, peptides and ATP (48). Impairment of proteasome function has also been found in a mouse model of Charcot Marie Tooth 1B neuropathy, which is caused by a mutation in Myelin Protein Zero (J. VerPlank and L. Wrabetz, unpublished), and in a mouse model of desmin-related cardiomyopathy (55, 56) (see below). Thus, proteasome defects seem to represent a common mechanism in human diseases caused by accumulation of aggregation-prone proteins. Treatment of mice expressing mutant tau (P301L) with rolipram not only increased brain proteasome function but also decreased the accumulation of polyubiquitinated proteins and mutant Tau protein (48). Thus, pharmacological treatments causing phosphorylation of the proteasome by PKA are a promising approach to reduce the buildup of these toxic proteins.
Protein Kinase G (PKG) enhances proteasome activity and protein degradation
There is compelling evidence that the cGMP-dependent kinase, Protein Kinase G (PKG), can also phosphorylate and activate the 26S proteasome. PKG resembles PKA in structure and is composed of two catalytic subunits and two regulatory subunits, which in the absence of cGMP inhibit the catalytic subunits (49). Wang and colleagues have shown that overexpression of these catalytic subunits in neonatal rat ventricular myocytes results in increased proteasomal peptidase activity in crude lysates and accelerated degradation of the model UPS substrate GFPu (also termed GFP-CL1) (55). Similar changes in proteasome activity and GFPu degradation were seen when intracellular cGMP levels were increased by treating these cells with sildenafil, an inhibitor of a cGMP-specific phosphodiesterase, PDE5 (55). Sildenafil is widely used to treat erectile dysfunction and pulmonary hypertension, because it causes vasodilation by enhancing the PKG-dependent relaxation of vascular smooth muscles (35).
The finding that PKG stimulates proteasome activity in the heart also has potential therapeutic applications because there is growing evidence that impairment of the UPS contributes to the pathogenesis of several inherited cardiomyopathies (56). For example, desmin-related cardiomyopathy is caused by mutations in desmin, a muscle-specific intermediate filament protein, or by mutations in αβ-crystallin (Cryαβ), an abundant molecular chaperone that helps maintain desmin in its properly folded state (57). In the hearts and muscles of patients with this disease, and also in tissues of a transgenic mouse that express the human CryαβR120G mutation, ubiquitinated aggregates of αβ-crystallin and other proteins accumulate (55). Treatment of these mice with sildenafil increased the proteasome’s peptidase activity in the heart and reduced the accumulation of polyubiquitinated proteins and aggregated Cryαβ. Furthermore, sildenafil stimulated the degradation of mutant Cryαβ in the cultured cardiac myocytes and also reduced the cardiac hypertrophy and heart failure seen in mice expressing this mutated protein (55). Because activation of PKG affects many cellular processes and cardiovascular responses (e.g. vasodilation), it is unclear if the reduced hypertrophy resulted from the increased proteasome activity and more rapid proteolysis. Nevertheless, these findings are exciting and clearly indicate the therapeutic potential of PKG activation in certain proteotoxic diseases especially in certain cardiovascular diseases (e.g. cardiac failure) where raising cAMP could be harmful. However, it remains to be ascertained whether cAMP and cGMP promote the degradation of the distinct types of misfolded protiens. Beyond its basic biochemical interest, such information could also determine which is the best agent to use to treat specific disease.
Unfortuantely many other important mechanistic questions concerning cGMP’s effects have not been investigated. It is unclear what subunit(s) and what residue(s) are phosphorylated by PKG and necessary for proteasome activation. Ranek et al (55, 58) reported that PKG activation in cardiac cells caused an acidic shift of the 20S subunit β5 and the 19S ATPase subunit Rpt6, strongly suggesting their phosphorylation. As noted above, Rpt6 had also been reported to be phosphorylated at serine 120 by PKA (32) and CAMKIIα (59, 60) (see below). However, the residue(s) phosphorylated by PKG was not identified, nor was site-specific mutagenesis performed. In human neuroblastoma cells, raising cGMP also stimulates 26S peptidase activities, but does not cause Rpn6 S14 phosphorylation (VerPlank and Goldberg, unpublished observation). Thus, PKG and PKA activate proteasomes by distinct mechanisms, which presumably leads to distinct effects on protein turnover.
cGMP production in the heart can also be stimulated when the neurotransmitter acetylcholine binds to cardiac M2 Muscarinic receptors. Accordingly, stimulation of M2 muscarinic receptors with pilocarpine in cultured ventricular myocytes caused PKG activation and increased degradation of the GFPu and proteasome peptidase activity in the cell lysates. Conversely, blocking the M2 receptor with the antagonist methoctramine reduced PKG activation, as well as proteasome activity and proteolysis (58). Even though sympathetic stimulation of the heart by epinephrine and parasympathetic stimulation by acetylcholine have opposite effects on heart rate and cardiac output, it is intriguing that both enhance cardiac proteasome activity, apparently through phosphorylation of different 26S subunits by PKA and PKG. Although both appear to accelerate degradation of certain proteins via the UPS, it is likely that they promote the degradation of distinct sets of cell proteins in order to aid in the adaptation to the increased or decreased cardiac output.
Calcium/Calmodulin-dependent protein Kinase II (CAMKII) and neuronal function
Neuronal depolarization has been shown to alter proteasome localization in cells and proteasome activity, apparently by causing an influx of calcium (60–62). In hippocampal neurons, synaptic potentials or activation of the NMDA receptors leads to calcium entry and the translocation of 26S proteasomes from the dendrites into the spines (60, 61). These synaptic potentials also cause the movement to dendritic spines of CAMKIIα, a member of the calcium/calmodulin-dependent family of protein kinases (63). This translocation of proteasomes is dependent on CAMKIIα autophosphorylation (60), which maintains CAMKIIα in an active state after the dissipation of the calcium spike. This prolongation of CAMKIIα activity is important in the synaptic adaptations responsible for learning and memory (64), in which proteolysis also plays a role (65). Proteasome translocation decreased the level of ubiquitinated proteins in the spines (60, 61) and thus appears to lead to a localized enhancement of protein degradation.
Djakovic and colleagues (62) showed that depolarization of neurons with the GABA receptor antagonist, bicuculline, increased both proteasome peptidase activity in the lysates and the degradation in neurons of a proteasomal substrate requiring ubiquitination (GFPu) and one not requiring ubiquitination (ornithine decarboxylase). This stimulation of proteasome activity did not occur when CAMKIIα was inhibited and thus, presumably requires 26S phosphorylation by this enzyme (62). By contrast, CAMKIIα’s catalytic activity was not essential for proteasome translocation into the spines (60). Thus, although proteasome translocation and the increase in activity are both triggered by calcium influx, they must occur by distinct mechanisms.
Purified CAMKII was initially reported to phosphorylate the ATPase subunit Rpt6 (62) at serine 120 (60) on isolated 26S proteasomes and phosphorylation of Rpt6-S120 was also detected with a phospho-specific Rpt6-S120 antibody in neurons after depolarization with bicuculline (59). However, a phosphomimetic mutation of Rpt6-S120 did not enhance proteasome activities (59). Perhaps in addition to Rpt6, CAMKIIα phosphorylates another subunit, which may also be required for the stimulation of activity. Alternatively, another calcium-dependent signaling mechanism may be involved in triggering proteasome activation. In the presence of calcium, calmodulin binds to several proteasome subunits and proteasome-interacting proteins (66) and may alter 26S function allosterically without involvement of a kinase. Nevertheless, Rpt6-S120 phosphorylation is important for neuronal plasticity because expression of a phosphodead mutation diminished both synaptic strength and the outgrowth of dendritic spines that is usually induced by synaptic activity (59, 67). Both the increased synaptic strength and outgrowth of spines are also blocked by proteasome inhibitors (67–69). Furthermore, the level of p-Rpt6-S120 increased in the amygdala of rats during the formation and retrieval of fear-associated memories (70, 71) as well as in the nucleus accumbens and prefrontal cortex of mice after cocaine administration (Gentry Patrick, personal communication). Thus, Rpt6 phosphorylation seems to be important for multiple aspects of brain function, but its precise effects on proteasome activities and protein turnover are still uncertain.
Bingol and colleagues also demonstrated a localized increase in ubiquitination after calcium influx (60), which seems to lead to enhanced proteolysis and to be coordinated with the proteasome activation. When proteasome movement into the spines following NMDA receptor activation was prevented (by mutating CAMKII’s calmodulin- or NMDAR-binding sites), polyubiquitinated proteins accumulated to a greater extent than if no calcium had entered the neuron. In addition, there was a greater accumulation of polyubiquitinated proteins upon activation of NMDA receptors in the presence of a proteasome inhibitor (60). Thus, the rise in cell calcium in addition to promoting proteasome activity also stimulates ubiquitination, probably by activating E3 ligases that are dependent on calcium or calmodulin (72–74). It is noteworthy that cAMP, like calcium influx, also seemed to promote ubiquitination in addition to proteasome function (36). Thus, both seem to enhance proteolysis by the UPS through multiple, presumably linked, actions.
Dual-specificity tyrosine-regulated kinase 2 (DYRK2) and the cell cycle
In addition to these examples of proteasomal regulation by non-cell autonomous signals, there is at least one important form of 26S phosphorylation controlled through a cell-autonomous mechanism. Progression through the cell cycle depends on the phosphorylation-induced ubiquitination and proteasomal degradation of key regulatory proteins (i.e. cyclins and cyclin-dependent kinase inhibitors), which control the timing of cell cycle transitions (75). Although this crucial role of phosphorylation-dependent ubiquitination has been known for decades, the possible regulation of proteasomes during the cell cycle was not investigated until recently. Several studies have identified by mass spectrometry proteasome subunits that are phosphorylated at different stages of the cell cycle (76–78). One clear example was recently described by Guo and colleagues, who demonstrated that phosphorylation of the 19S ATPase subunit Rpt3 on threonine 25 increased in S phase and remained high through G2 and M phases (79). They therefore performed a screen of human kinases and found that DYRK2 could catalyze Rpt3 phosphorylation at T25. Moreover, the levels of DYRK2 mRNA and protein also increased during S phase and remained elevated through M phase, and thus, its expression may determine rates of Rpt3 modification.
Proteasomes purified from these phases of the cycle contained phosphorylated Rpt3, and exhibited a greater capacity to degrade short peptides and a ubiquitinated protein, as well as greater ATPase activity in the presence of a ubiquitinated protein (79). Thus, these particles showed generally similar enzymatic changes as 26S proteasomes phosphorylated by PKA on Rpn6. Prevention of this phosphorylation by a phosphodead mutation of threonine 25 to valine with CRISPR-Cas9 gene editing in human breast cancer cells prevented the stimulation of the proteasomes by DYRK2. Most importantly, these mutations slowed the degradation of long-lived cell proteins and the major cell cycle regulatory factors, p27Kip1 and p21Cip1. These cdk1 inhibitors block the transition from G1 to S phase and their accelerated degradation should thus promote cell cycle progression. As a result of the phosphodead Rpt3, cell proliferation was reduced, and apoptosis increased. Furthermore, knockdown of DYRK2 or prevention of Rpt3 phosphorylation with the phosphodead mutation reduced tumor growth in nude mice (79). This inhibition of cell proliferation in vivo strongly suggests that DYRK2 is a potential drug target for the treatment of certain cancers. In addition, DYRK2 gene amplification occurs in many cancers, and breast cancer patients with higher DYRK2 expression levels tend to have a lower survival rates (79).
While DYRK2 may regulate additional processes important for cell cycle transitions, the slow growth and cell death caused by the Rpt3 phosphodead mutation clearly demonstrate the importance of this modification in enhancing degradation of key proteins during S, G2, and M phases. To achieve a further understanding of the importance of Rpt3 phosphorylation and DYRK2’s role, it will be valuable to identify the other proteins that are ubiquitinated and degraded faster during these cell-cycle phases.
Apoptosis Signal-Regulating Kinase 1 (ASK1) and proteasome inhibition
Unlike the other examples of subunit phosphorylation discussed here, which enhance proteasome activity, phosphorylation of the 19S ATPase subunit Rpt5 by ASK1 reduces 26S activity and protein breakdown (80). ASK1 is a member of the Mitogen Activated Protein Kinase (MAPK) kinase kinase (MKK) family and is activated by several apoptotic stimuli, including death receptor ligands, lipopolysaccharides, severe oxidative and endoplasmic reticulum stress (81). After overexpression of ASK1 in HEK293 cells, proteasomal peptidase activity in cell lysates decreased, and the levels of polyubiquitinated proteins increased. Also, there was an accumulation of certain short-lived proteins that normally undergo proteasomal degradation by ubiquitin-dependent (GFPu, UbG767V-GFP) or independent (ODC-GFP) routes (80). Conversely, siRNA-mediated knockdown of endogenous ASK1 enhanced proteasomal peptidase activity and the degradation of these proteins in HEK293 cells, mouse embryonic fibroblasts, and human melanoma cells (80, 82). Thus in these cultures, endogenous ASK1 appears to cause some basal phosphorylation of the 26S proteasome that limits its activity and inhibits intracellular proteolysis. Overexpression of ASK1 also leads to its association with 19S subunits, phosphorylation of the ATPase subunit Rpt5, and reduced proteasomal ATPase activity. Moreover, 26S activity could be restored to normal levels by phosphatase treatment of the isolated particles (80). However, it remains unclear on which residue Rpt5 is modified and whether this modification by itself actually causes these changes in proteasome activity and protein turnover rates. Presumably, this ASK1-mediated decrease in proteasome function and inhibition of protein degradation contributes to the progression of apoptosis, but it is not clear why it is advantageous to decrease proteasome activity in response to apoptotic stimuli, since many steps in the apoptosis pathway require degradation of inhibitory proteins by the UPS (83).
In addition to ASK1, other kinases in the p38 MAPK signaling cascade can repress proteasome function (82, 84). Overexpression of an active mutant of p38 MAPK caused the accumulation of proteins that normally undergo rapid proteasomal degradation by ubiquitin-dependent (GFPu, UbG767V-GFP, Ub-R-GFP) or independent (ODC-GFP) routes (84). Conversely, small molecule inhibitors of p38 MAPK and siRNA-mediated knockdown of its α-isoform stimulated proteasome peptidase activity and intracellular degradation of α-synuclein (82). Furthermore, siRNA-mediated knockdown of MKK6, which like ASK1 is an upstream activator of p38 MAPK, or knockdown or chemical inhibition of MK2, a kinase that functions downstream of p38 MAPK, enhanced proteasome peptidase activity (82). Thus, although the biological significance of the regulation of proteasome function by this pathway is unclear, specific inhibitors of the p38 MAPK pathway may also be therapeutically useful to enhance proteasome activity and promote the clearance of potentially toxic proteins.
Casein Kinase II (CK2)
Multiple early studies reported that Casein Kinase II can be co-purified with the 20S proteasome and can phosphorylate the α7 subunit at serines 243 and 250 (85–87). However, recent mass spectrometric studies of the 26S proteasome have not found CK2 in affinity-purified preparations (17). This kinase may thus interact only with the free 20S particles and perhaps specifically with α-ring residues that tend to be masked in the 26S particles. CK2 is a pleiotropic kinase, whose physiological role remains incompletely understood. Unlike the other kinases discussed in this article, CK2 is constitutively active and is not known to respond to secondary messengers or phosphorylation by other kinases (88). Therefore, the phosphorylation of proteasome by CK2 is likely to occur constitutively, but may be regulated through the actions of some protein phosphatases.
The phosphorylation of serines within the C-terminal tail of mammalian α7 has been suggested to be important for the interaction between the CP and the RP (87). When mammalian α7 S243 or S250 phosphodead mutants were overexpressed in COS-7 cells, less 26S proteasomes and more 20S proteasomes were found (87). In addition, 26S proteasomes purified from yeast expressing α7 with these phosphodead mutations in its three C-terminal serines (258, 263 and 264) contained less of the large proteasome-interacting protein ECM29 (89). Several functions have been proposed for ECM29, including stabilizing 19S–20S association (90), inhibiting proteasome activity (91), and causing proteasomal dissociation during exposure to H202 (92). Though intriguing, these observations are difficult to interpret since the replacement of multiple serines in an α-subunit may greatly alter proteasome structure and interactions. Thus, the importance of this modification by CK2 and its biological consequences for proteasome function remain unclear.
Our limited knowledge and future directions
The regulation of protein breakdown by proteasome phosphorylation represents a newly appreciated mode of regulation of intracellular protein degradation, but as this review emphasizes, our knowledge about the biochemical and cellular consequences of these various proteasome modifications is limited. Although the literature concerning the degradation of different proteins is now enormous, nearly all of it has focused on the specificity and control of ubiquitination rates, and until recently, there has been very little consideration of the possibility of regulation at the proteasome level (4, 9). Yet more than 300 phosphorylation sites in proteasome subunits have been detected by mass spectrometry (9), and for the vast majority of them, the responsible kinases, their frequencies, and the functional consequences, if any, are unknown. While it is likely that the great majority of these phosphorylations have no regulatory roles, a number of additional kinases are likely to influence proteasome function, since the few studies on this topic have already uncovered several well-characterized protein kinases that alter proteasome properties.
This article has focused on the several kinases that clearly influence proteasome function or seem likely to do so. However, definitive evidence of regulation of protein half-lives through 26S phosphorylation has thus far only been obtained for PKA (36) and DYRK2 (79), where phosphomimetic and phosphodead mutations have been shown to alter proteasome function. For the other kinases, it remains to be proven by mutagenesis that the phosphorylation of a specific 26S subunit actually causes the increase in enzymatic activity, and that the proteasome activation is both necessary and sufficient to cause the more rapid protein degradation in cells. Such genetic evidence is essential because these kinases may not only enhance proteasome activity, but may also stimulate ubiquitination. Many examples are known where a kinase triggers a protein’s degradation by phosphorylating it and forming a motif that is specifically recognized by a ubiquitin ligase (a “phosphodegron”) (15). Kinases may also stimulate ubiquitination by phosphorylation of a ubiquitin ligase (15) or through the phosphorylation and inhibition of a deubiquitinating enzyme.
In fact, two kinases, PKA (36) and CAMKII (60, 62), have been shown to both activate proteasomes and increase protein ubiquitination, and it seems likely that these responses are linked to favor efficient breakdown of a set of cell proteins. In such cases, ubiquitination may still specify the proteins being degraded, while proteasome activation would have more general effects to ensure rapid breakdown of the conjugated proteins. Thus, proteasome activation would seem to be an appropriate response during cellular transitions, where the proteome is being remodeled, such as in cells proceeding through the cell cycle, or in transition from the fed to fasted states, or in recovery after exercise. At such times, additional components of this pathway may also be coordinately regulated, such as shuttling factors, which deliver ubiquitin conjugates to the proteasome or the p97/VCP ATPase complex, which extracts and unfolds ubiquitinated proteins to facilitate their degradation.
Recently, major advances have been made in our knowledge of proteasome structure and catalytic activities (2, 4). Hopefully these new insights will enable us to eventually understand how the phosphorylation of specific residues can change the structure and properties of a particle containing approximately 60 subunits. Achieving this goal will be challenging because for most of the kinases discussed here, the phosphorylated subunit and the modified residue have not yet been definitively identified, and their identification has proven surprisingly difficult (82). Also, the analysis of proteasome function has often been restricted to assays in crude extracts using small fluorogenic peptide subunits, which do not necessarily measure proteasome capacity to degrade its natural substrates, ubiquitinated proteins. In only two cases, PKA and DYRK2, have the 26S proteasomes been purified and degradation of ubiquitin conjugates and ATP been studied (36, 79). Surprisingly, the phosphorylation of Rpn6 by PKA and of the ATPase subunit Rpt3 by DYRK2 seem to enhance proteasome activity similarly (i.e both increase hydrolysis of peptides, ATP, and ubiquitin conjugates), even though these two phosphorylated subunits share no structural features and differ in location, interacting subunits, and function.
The multiple proteasomal processes that are stimulated by these two kinases and inhibited by ASK1 are not independent activities. The 19S ATPases regulate gate opening into the 20S particle and thus control itsl three peptidase activities, and the rate of ubiquitin conjugate degradation is determined by the rate of ATP hydrolysis (38). Thus, the acceleration of ATPase activity by itself may account for the multiple enzymatic changes during proteasome activation. Other potentially important functional consequences of proteasome phosphorylation have not been examined for any of these kinases, such as possible effects on the three 26S-associated deubiquitinating enzymes, on binding of different types of ubiquitin conjugates.
In cells 26S proteasomes are quite heterogeneous and differ normally in their subcellular locations, content of interacting proteins, and associated regulatory complexes. Thus, kinases may phosphorylate only a fraction of cell proteasomes and alter protein turnover only in parts of the cell. Enzymes involved in cAMP, cGMP and calcium signaling and the various phosphodiesterases are not uniformaly distributed in cells (93). In fact, physiological activation of phosphorylation cascades probably occurs in only certain cellular locations. There is growing evidence that at most times, the majority of intracellular 26S particles are inactive (94, 95), and phosphorylation may serve to activate some of these latent particles, or to inactivate some proteasomes in the case of ASK1.
Although time course studies have not yet been reported, proteasome phosphorylation in most cases must be a transitory response that occurs only during certain phases of the cell cycle or following hormonal stimuli or synaptic activity. Therefore, these responses must be terminated through actions of phosphatases, whose roles in proteasome regulation have received even less attention than kinases. Nevertheless, two phosphatases, calcineurin, the calmodulin-regulated phosphatase, and UBLCP1, the ubiquitin-like domain-containing phosphatase, co-purify with 26S proteasomes (96–99). Interestingly, UBLCP1 is only found on nuclear proteasomes (97), and dephosphorylation by UBLCP1 suppressed nuclear proteasome peptidase activity (97). So, a specific role for UBLCP1 in regulation of nuclear proteasomes seems likely, but the opposing kinase(s) in the nucleus are not known.
Even though our knowledge of the underlying biology is limited, it is clear that treatments that enhance proteasome phosphorylation are a promising, new approach to combat various proteotoxic diseases. The fundamental role of the ubiquitin-proteasome pathway in protecting against the accumulation of misfolded proteins had long been recognized (51), but it had not been appreciated that physiological mechanisms exist to rapidly enhance the cell’s capacity to degrade such proteins, or that they can be manipulated pharmacologically. This exciting possibility is based on the finding that agents that raise cAMP or cGMP can stimulate the clearance of aggregation-prone, toxic proteins in cultured cells and transgenic mouse models (48, 55). Enhancing proteasome function seems like a particularly attractive approach to treat proteotoxic diseases because of the growing evidence that in such diseases, proteasome activity is impaired (probably by the protein aggregates), and that the resulting failure of protein homeostasis is deleterious and causes further accumulation of the toxic proteins (48, 53). Combatting these diseases by raising cAMP or cGMP also seems quite feasible, because many drugs that inhibit specific phosphodiesterases are known, and several are already approved by the FDA. In fact, a recent screen found ten compounds that activate proteasomes and protein breakdown, and several of them have been shown to influence cellular levels of cAMP, cGMP, or calcium (82). It is unclear at present whether activating these different kinases enhances the breakdown of the same proteins by the proteasome, and such information should help determine their potential therapeutic applications.
For other diseases, especially to treat certain cancers, blocking proteasome activation by kinase inhibition (e.g. DYRK2) or proteasome inhibition through activation of ASK1 may be a useful strategy, just as proteasome inhibitors have already greatly advanced the treatment of multiple myeloma (100). The exciting finding that preventing Rpt3 phosphorylation by DYRK2 retards growth of certain cancers (79), and that DYRK2 is induced in several cancers, provide a strong rationale for developing selective inhibitors of this enzyme. The utility and long-term consequences of such therapies are difficult to predict, since it is unclear how these various kinases affect the breakdown of cell proteins. Thus, greater knowledge about these cellular mechanisms is not only of biochemical and cell biological interest but may also have important therapeutic benefits.
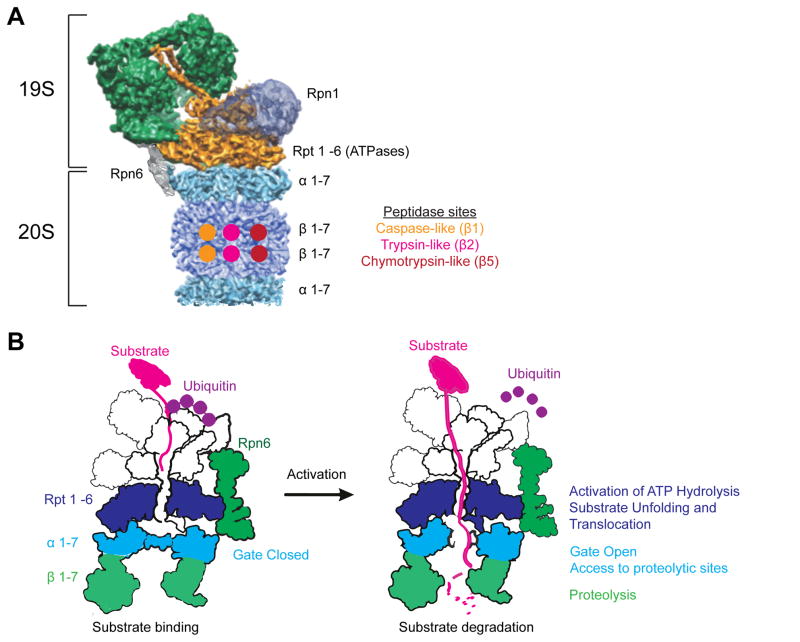
Figure 1. Structures of the 26S proteasome.
A.) The 26S proteasome is composed of the 20S core particle and a 19S regulatory particle attached to one or both ends. The 20S has seven α subunits (light blue), whose N-termini form a gate that prevents nonspecific protein degradation, and seven β subunits (dark blue), three of which contain proteolytic active sites with distinct substrate preference (indicated). The 19S has six ATPase subunits (Rpt 1-6) (orange), which unfold and translocate substrates in an ATP-dependent manner into the 20S for proteolysis. Three of these ATPase subunits (Rpt 3, 5, and 6) are reported to be phosphorylated by kinases discussed here. Rpn6 is phosphorylated by PKA and is unusual because it interacts with subunits of both the 19S ATPase ring and 20S α ring.
B.) A ubiquitinated substrate is bound by one of the ubiquitin receptor subunits. If the substrate has an unstructured region, it becomes tightly bound by the proteasome, committed to degradation, and the 19S ATPase subunits (Rpt 1 - 6) are activated. Degradation does not occur every time a ubiquitinated substrate binds the proteasome because deubiqutination can occur prior to the commitment step, resulting in substrate release. If committed to degradation, the substrate is translocated in an ATP-dependent manner into the 20S where it is hydrolyzed to small peptides by the three proteolytic sites. It is currently unknown how phosphorylation of a subunit either speeds up or slows down this degradation process.
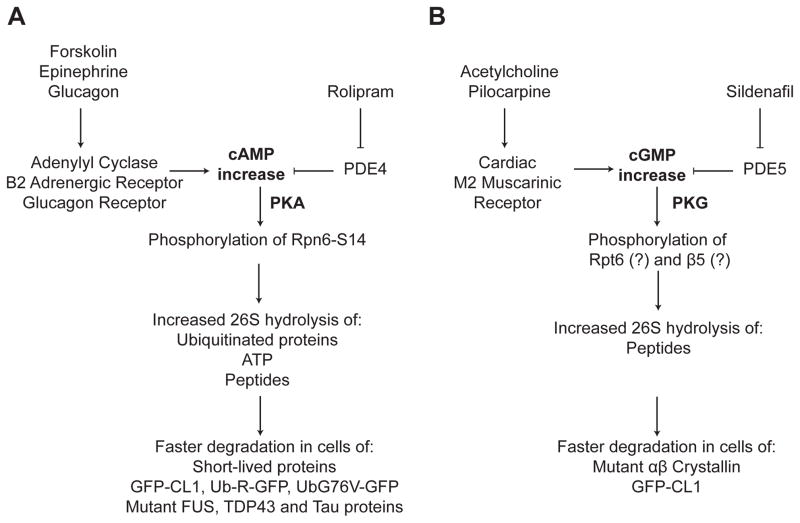
Figure 2. Our present understanding of proteasome activation by cAMP and cGMP.
A.) Hormones and compounds that raise intracellular cAMP cause PKA to phosphorylate the proteasome on 19S subunit Rpn6. The phosphorylated proteasome is more active and degrades faster short-lived proteins in all examined cells, including defined UPS model substrates and mutant proteins that cause neurodegenerative diseases.
B.) PDE5 inhibitors and cardiac M2 muscarinic receptor agonists raise intracellular cGMP and activate PKG in hearts and cultured primary cardiomyocytes. PKG phosphorylates proteasome subunits and stimulates the degradation of a mutant αβCrystallin, which causes desmin-related cardiomyopathy, and the defined UPS substrate GFP-CL1.
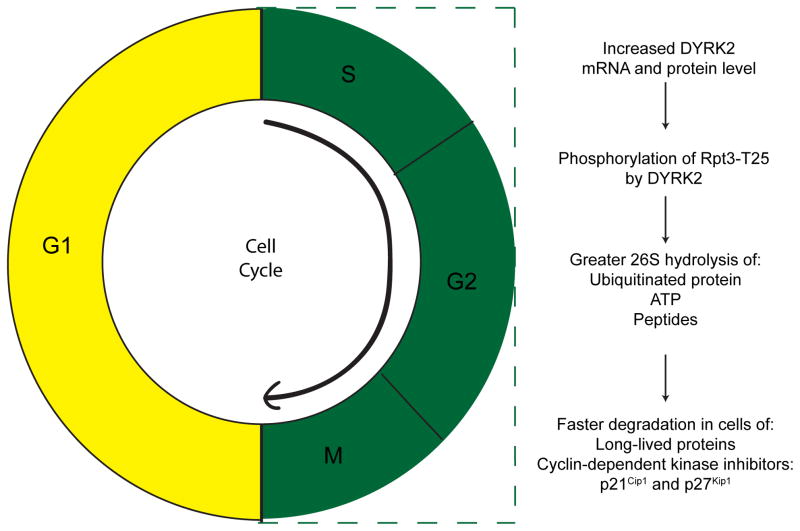
Figure 3.
Phosphorylation by DYRK2 of the proteasome 19S subunit Rpt3 during S, M, and G2 phases of the cell cycle. These phosphorylated proteasomes are more active and degrade faster long-lived proteins as well as two cyclin-dependent kinase inhibitors that inhibit the transition from G1 to S phase: p21Cip1 and p27Kip1.
Table 1.
Summary of kinases that phosphorylate 26S proteasome subunits.
| Kinase | Signaling molecule | Physiological stimuli | Phosphorylated Subunit | Effect on activity | Pharmacological stimuli |
|---|---|---|---|---|---|
| PKA | cAMP | Epinephrine Glucagon | Rpn6* | Stimulatory | PDE4 inhibitors |
| PKG | cGMP | Acetylcholine (on cardiomyocytes) | Rpt6 β5 | Stimulatory | PDE5 inhibitors |
| CAMKIIα | Calcium influx | Synaptic depolarization | Rpt6 | Stimulatory | ? |
| DYRK2 | ? | S → M phase in cell cycle | Rpt3* | Stimulatory | ? |
| ASK1 | ? | Apoptosis | Rpt5 | Inhibitory | ? |
| CK2 | ? | ? | α7 | ? | ? |
effects on proteasome activity have been confirmed by site-specific mutagenesis.
Abbreviations
- PKA
Protein Kinase A
- cAMP
cyclic adenosine monophosphate
- PKG
Protein Kinase G
- cGMP
cyclic guanosine monophosphate
- DYRK2
Dual-specificity tyrosine-regulated kinase 2
- ASK1
Apoptosis Signal-Regulating Kinase 1
- UPS
Ubiquitin Proteasome System
- CK2
Casein kinase 2
- CAMK2
Calcium/Calmodulin-dependent protein kinase 2
- PDE
phosphodiesterase
- NMDA
N-Methyl-D-aspartic acid
- ATP
adenosine triphosphate
- GABA
ϒ-Aminobutyric acid
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annual review of biochemistry. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Finley D, Chen X, Walters KJ. Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends in biochemical sciences. 2016;41(1):77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matyskiela ME, Martin A. Design principles of a universal protein degradation machine. Journal of molecular biology. 2013;425(2):199–213. doi: 10.1016/j.jmb.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins GA, Goldberg AL. The Logic of the 26S Proteasome. Cell. 2017;169(5):792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickart CM. Mechanisms underlying ubiquitination. Annual review of biochemistry. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 6.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee BH, Lu Y, Prado MA, Shi Y, Tian G, Sun S, et al. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature. 2016;532(7599):398–401. doi: 10.1038/nature17433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Matouschek A. Recognition of Client Proteins by the Proteasome. Annual review of biophysics. 2017;46:149–73. doi: 10.1146/annurev-biophys-070816-033719. [DOI] [PubMed] [Google Scholar]
- 9.Guo X, Huang X, Chen MJ. Reversible phosphorylation of the 26S proteasome. Protein & cell. 2017;8(4):255–72. doi: 10.1007/s13238-017-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Molecular & cellular proteomics : MCP. 2011;10(5):R110.003871. doi: 10.1074/mcp.R110.003871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peth A, Uchiki T, Goldberg AL. ATP-dependent steps in the binding of ubiquitin conjugates to the 26S proteasome that commit to degradation. Molecular cell. 2010;40(4):671–81. doi: 10.1016/j.molcel.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Molecular cell. 2007;27(5):731–44. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, et al. A gated channel into the proteasome core particle. Nature structural biology. 2000;7(11):1062–7. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 14.Gallastegui N, Groll M. The 26S proteasome: assembly and function of a destructive machine. Trends in biochemical sciences. 2010;35(11):634–42. doi: 10.1016/j.tibs.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Filipcik P, Curry JR, Mace PD. When Worlds Collide-Mechanisms at the Interface between Phosphorylation and Ubiquitination. Journal of molecular biology. 2017;429(8):1097–113. doi: 10.1016/j.jmb.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Stadtmueller BM, Hill CP. Proteasome activators. Molecular cell. 2011;41(1):8–19. doi: 10.1016/j.molcel.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Besche HC, Sha Z, Kukushkin NV, Peth A, Hock EM, Kim W, et al. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. The EMBO journal. 2014;33(10):1159–76. doi: 10.1002/embj.201386906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127(7):1401–13. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 19.Stanhill A, Haynes CM, Zhang Y, Min G, Steele MC, Kalinina J, et al. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Molecular cell. 2006;23(6):875–85. doi: 10.1016/j.molcel.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 20.Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. The EMBO journal. 2006;25(3):554–64. doi: 10.1038/sj.emboj.7600945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Molecular cell. 2010;38(1):17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sha Z, Goldberg AL. Proteasome-mediated processing of Nrf1 is essential for coordinate induction of all proteasome subunits and p97. Current biology : CB. 2014;24(14):1573–83. doi: 10.1016/j.cub.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kniepert A, Groettrup M. The unique functions of tissue-specific proteasomes. Trends in biochemical sciences. 2014;39(1):17–24. doi: 10.1016/j.tibs.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Basler M, Kirk CJ, Groettrup M. The immunoproteasome in antigen processing and other immunological functions. Current opinion in immunology. 2013;25(1):74–80. doi: 10.1016/j.coi.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Scruggs SB, Zong NC, Wang D, Stefani E, Ping P. Post-translational modification of cardiac proteasomes: functional delineation enabled by proteomics. American journal of physiology Heart and circulatory physiology. 2012;303(1):H9–18. doi: 10.1152/ajpheart.00189.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Z, Scruggs SB, Gilda JE, Ping P, Gomes AV. Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond. Journal of molecular and cellular cardiology. 2014;71:32–42. doi: 10.1016/j.yjmcc.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell. 2003;115(6):715–25. doi: 10.1016/s0092-8674(03)00974-7. [DOI] [PubMed] [Google Scholar]
- 28.Cho-Park PF, Steller H. Proteasome regulation by ADP-ribosylation. Cell. 2013;153(3):614–27. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall RS, McLoughlin F, Vierstra RD. Autophagic Turnover of Inactive 26S Proteasomes in Yeast Is Directed by the Ubiquitin Receptor Cue5 and the Hsp42 Chaperone. Cell reports. 2016;16(6):1717–32. doi: 10.1016/j.celrep.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 30.Budenholzer L, Cheng CL, Li Y, Hochstrasser M. Proteasome Structure and Assembly. Journal of molecular biology. 2017 doi: 10.1016/j.jmb.2017.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutherland EW. Studies on the mechanism of hormone action. Science (New York, NY) 1972;177(4047):401–8. doi: 10.1126/science.177.4047.401. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, Hu Y, Huang P, Toleman CA, Paterson AJ, Kudlow JE. Proteasome function is regulated by cyclic AMP-dependent protein kinase through phosphorylation of Rpt6. The Journal of biological chemistry. 2007;282(31):22460–71. doi: 10.1074/jbc.M702439200. [DOI] [PubMed] [Google Scholar]
- 33.Krebs EG. Nobel Lecture. Protein phosphorylation and cellular regulation I. Bioscience reports. 1993;13(3):127–42. doi: 10.1007/BF01149958. [DOI] [PubMed] [Google Scholar]
- 34.Fischer EH. Cellular regulation by protein phosphorylation. Biochemical and biophysical research communications. 2013;430(2):865–7. doi: 10.1016/j.bbrc.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 35.Maurice DH, Ke H, Ahmad F, Wang Y, Chung J, Manganiello VC. Advances in targeting cyclic nucleotide phosphodiesterases. Nature reviews Drug discovery. 2014;13(4):290–314. doi: 10.1038/nrd4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lokireddy S, Kukushkin NV, Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(52):E7176–85. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods in enzymology. 2005;398:364–78. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- 38.Peth A, Nathan JA, Goldberg AL. The ATP costs and time required to degrade ubiquitinated proteins by the 26 S proteasome. The Journal of biological chemistry. 2013;288(40):29215–22. doi: 10.1074/jbc.M113.482570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinoshita E, Kinoshita-Kikuta E, Koike T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nature protocols. 2009;4(10):1513–21. doi: 10.1038/nprot.2009.154. [DOI] [PubMed] [Google Scholar]
- 40.Pathare GR, Nagy I, Bohn S, Unverdorben P, Hubert A, Korner R, et al. The proteasomal subunit Rpn6 is a molecular clamp holding the core and regulatory subcomplexes together. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(1):149–54. doi: 10.1073/pnas.1117648108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asai M, Tsukamoto O, Minamino T, Asanuma H, Fujita M, Asano Y, et al. PKA rapidly enhances proteasome assembly and activity in in vivo canine hearts. Journal of molecular and cellular cardiology. 2009;46(4):452–62. doi: 10.1016/j.yjmcc.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 42.Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489(7415):304–8. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489(7415):263–8. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 44.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell metabolism. 2007;6(6):472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Zhao J, Zhai B, Gygi SP, Goldberg AL. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(52):15790–7. doi: 10.1073/pnas.1521919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein science : a publication of the Protein Society. 2011;20(8):1298–345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nature biotechnology. 2000;18(5):538–43. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 48.Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, et al. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nature medicine. 2016;22(1):46–53. doi: 10.1038/nm.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffman NJ, Parker BL, Chaudhuri R, Fisher-Wellman KH, Kleinert M, Humphrey SJ, et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell metabolism. 2015;22(5):922–35. doi: 10.1016/j.cmet.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen S, Nathan JA, Goldberg AL. Muscle wasting in disease: molecular mechanisms and promising therapies. Nature reviews Drug discovery. 2015;14(1):58–74. doi: 10.1038/nrd4467. [DOI] [PubMed] [Google Scholar]
- 51.Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426(6968):895–9. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka K, Matsuda N. Proteostasis and neurodegeneration: the roles of proteasomal degradation and autophagy. Biochimica et biophysica acta. 2014;1843(1):197–204. doi: 10.1016/j.bbamcr.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 53.Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Molecular cell. 2007;26(2):175–88. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Deriziotis P, Andre R, Smith DM, Goold R, Kinghorn KJ, Kristiansen M, et al. Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry. The EMBO journal. 2011;30(15):3065–77. doi: 10.1038/emboj.2011.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128(4):365–76. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilda JE, Gomes AV. Proteasome dysfunction in cardiomyopathies. The Journal of physiology. 2017 doi: 10.1113/JP273607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLendon PM, Robbins J. Desmin-related cardiomyopathy: an unfolding story. American Journal of Physiology-Heart and Circulatory Physiology. 2011;301(4):H1220–H8. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ranek MJ, Kost CK, Jr, Hu C, Martin DS, Wang X. Muscarinic 2 receptors modulate cardiac proteasome function in a protein kinase G-dependent manner. Journal of molecular and cellular cardiology. 2014;69:43–51. doi: 10.1016/j.yjmcc.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Djakovic SN. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. 2012;32(15):5126–31. doi: 10.1523/JNEUROSCI.4427-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bingol B, Wang C-F, Arnott D, Cheng D, Peng J, Sheng M. Autophosphorylated CaMKIIα acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 2010;140(4):567–78. doi: 10.1016/j.cell.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 61.Bingol B, Schuman EM. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441(7097):1144–8. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 62.Djakovic SN, Schwarz LA, Barylko B, DeMartino GN, Patrick GN. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. Journal of Biological Chemistry. 2009;284(39):26655–65. doi: 10.1074/jbc.M109.021956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen K, Meyer T. Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science (New York, NY) 1999;284(5411):162–7. doi: 10.1126/science.284.5411.162. [DOI] [PubMed] [Google Scholar]
- 64.Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nature reviews Neuroscience. 2012;13(3):169–82. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lip PZ, Demasi M, Bonatto D. The role of the ubiquitin proteasome system in the memory process. Neurochemistry international. 2017;102:57–65. doi: 10.1016/j.neuint.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 66.Shen X, Valencia CA, Szostak J, Dong B, Liu R. Scanning the human proteome for calmodulin-binding proteins. Proceedings of the National Academy of Sciences. 2005;102(17):5969–74. doi: 10.1073/pnas.0407928102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, Hell JW, Patrick GN, et al. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74(6):1023–30. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fonseca R, Vabulas RM, Hartl FU, Bonhoeffer T, Nägerl UV. A balance of protein synthesis and proteasome-dependent degradation determines the maintenance of LTP. Neuron. 2006;52(2):239–45. doi: 10.1016/j.neuron.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 69.Karpova A, Mikhaylova M, Thomas U, Knöpfel T, Behnisch T. Involvement of protein synthesis and degradation in long-term potentiation of Schaffer collateral CA1 synapses. Journal of Neuroscience. 2006;26(18):4949–55. doi: 10.1523/JNEUROSCI.4573-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jarome TJ, Kwapis JL, Ruenzel WL, Helmstetter FJ. CaMKII, but not protein kinase A, regulates Rpt6 phosphorylation and proteasome activity during the formation of long-term memories. Frontiers in behavioral neuroscience. 2013;7:115. doi: 10.3389/fnbeh.2013.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jarome TJ, Ferrara NC, Kwapis JL, Helmstetter FJ. CaMKII regulates proteasome phosphorylation and activity and promotes memory destabilization following retrieval. Neurobiology of learning and memory. 2016;128:103–9. doi: 10.1016/j.nlm.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McGourty CA, Akopian D, Walsh C, Gorur A, Werner A, Schekman R, et al. Regulation of the CUL3 Ubiquitin Ligase by a Calcium-Dependent Co-adaptor. Cell. 2016;167(2):525–38. e14. doi: 10.1016/j.cell.2016.09.026. [DOI] [PubMed] [Google Scholar]
- 73.Wang J, Peng Q, Lin Q, Childress C, Carey D, Yang W. Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. Journal of Biological Chemistry. 2010;285(16):12279–88. doi: 10.1074/jbc.M109.086405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Braganza A, Li J, Zeng X, Yates NA, Dey NB, Andrews J, et al. UBE3B Is a Calmodulin-regulated, Mitochondrion-associated E3 Ubiquitin Ligase. The Journal of biological chemistry. 2017;292(6):2470–84. doi: 10.1074/jbc.M116.766824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borg NA, Dixit VM. Ubiquitin in Cell-Cycle Regulation and Dysregulation in Cancer. Annual Review of Cancer Biology. 2017;1(1) [Google Scholar]
- 76.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, et al. A quantitative atlas of mitotic phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(31):10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nagano K, Shinkawa T, Mutoh H, Kondoh O, Morimoto S, Inomata N, et al. Phosphoproteomic analysis of distinct tumor cell lines in response to nocodazole treatment. Proteomics. 2009;9(10):2861–74. doi: 10.1002/pmic.200800667. [DOI] [PubMed] [Google Scholar]
- 78.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Science signaling. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 79.Guo X, Wang X, Wang Z, Banerjee S, Yang J, Huang L, et al. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nature cell biology. 2016;18(2):202–12. doi: 10.1038/ncb3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Um JW, Im E, Park J, Oh Y, Min B, Lee HJ, et al. ASK1 negatively regulates the 26 S proteasome. The Journal of biological chemistry. 2010;285(47):36434–46. doi: 10.1074/jbc.M110.133777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nishida T, Hattori K, Watanabe K. The regulatory and signaling mechanisms of the ASK family. Advances in Biological Regulation. 2017 doi: 10.1016/j.jbior.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 82.Leestemaker Y, de Jong A, Witting KF, Penning R, Schuurman K, Rodenko B, et al. Proteasome Activation by Small Molecules. Cell chemical biology. 2017 doi: 10.1016/j.chembiol.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 83.Bader M, Steller H. Regulation of cell death by the ubiquitin-proteasome system. Current opinion in cell biology. 2009;21(6):878–84. doi: 10.1016/j.ceb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee SH, Park Y, Yoon SK, Yoon JB. Osmotic stress inhibits proteasome by p38 MAPK-dependent phosphorylation. The Journal of biological chemistry. 2010;285(53):41280–9. doi: 10.1074/jbc.M110.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Castano JG, Mahillo E, Arizti P, Arribas J. Phosphorylation of C8 and C9 subunits of the multicatalytic proteinase by casein kinase II and identification of the C8 phosphorylation sites by direct mutagenesis. Biochemistry. 1996;35(12):3782–9. doi: 10.1021/bi952540s. [DOI] [PubMed] [Google Scholar]
- 86.Ludemann R, Lerea KM, Etlinger JD. Copurification of casein kinase II with 20 S proteasomes and phosphorylation of a 30-kDa proteasome subunit. The Journal of biological chemistry. 1993;268(23):17413–7. [PubMed] [Google Scholar]
- 87.Bose S, Stratford FL, Broadfoot KI, Mason GG, Rivett AJ. Phosphorylation of 20S proteasome alpha subunit C8 (alpha7) stabilizes the 26S proteasome and plays a role in the regulation of proteasome complexes by gamma-interferon. The Biochemical journal. 2004;378(Pt 1):177–84. doi: 10.1042/BJ20031122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2003;17(3):349–68. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 89.Wani PS, Suppahia A, Capalla X, Ondracek A, Roelofs J. Phosphorylation of the C-terminal tail of proteasome subunit alpha7 is required for binding of the proteasome quality control factor Ecm29. Scientific reports. 2016;6:27873. doi: 10.1038/srep27873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, et al. Multiple associated proteins regulate proteasome structure and function. Molecular cell. 2002;10(3):495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 91.De La Mota-Peynado A, Lee SY, Pierce BM, Wani P, Singh CR, Roelofs J. The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. The Journal of biological chemistry. 2013;288(41):29467–81. doi: 10.1074/jbc.M113.491662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang X, Yen J, Kaiser P, Huang L. Regulation of the 26S proteasome complex during oxidative stress. Science signaling. 2010;3(151):ra88. doi: 10.1126/scisignal.2001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Langeberg LK, Scott JD. Signalling scaffolds and local organization of cellular behaviour. Nature reviews Molecular cell biology. 2015;16(4):232–44. doi: 10.1038/nrm3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, et al. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science (New York, NY) 2015;347(6220):439–42. doi: 10.1126/science.1261197. [DOI] [PubMed] [Google Scholar]
- 95.Tsvetkov P, Mendillo ML, Zhao J, Carette JE, Merrill PH, Cikes D, et al. Compromising the 19S proteasome complex protects cells from reduced flux through the proteasome. eLife. 2015:4. doi: 10.7554/eLife.08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sun S, Liu S, Zhang Z, Zeng W, Sun C, Tao T, et al. Phosphatase UBLCP1 controls proteasome assembly. Open biology. 2017;7(5) doi: 10.1098/rsob.170042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guo X, Engel JL, Xiao J, Tagliabracci VS, Wang X, Huang L, et al. UBLCP1 is a 26S proteasome phosphatase that regulates nuclear proteasome activity. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(46):18649–54. doi: 10.1073/pnas.1113170108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang W, Wei Q. Calcineurin stimulates the expression of inflammatory factors in RAW 264.7 cells by interacting with proteasome subunit alpha type 6. Biochemical and biophysical research communications. 2011;407(4):668–73. doi: 10.1016/j.bbrc.2011.03.071. [DOI] [PubMed] [Google Scholar]
- 99.Li N, Zhang Z, Zhang W, Wei Q. Calcineurin B subunit interacts with proteasome subunit alpha type 7 and represses hypoxia-inducible factor-1alpha activity via the proteasome pathway. Biochemical and biophysical research communications. 2011;405(3):468–72. doi: 10.1016/j.bbrc.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 100.Goldberg AL. Development of proteasome inhibitors as research tools and cancer drugs. The Journal of cell biology. 2012;199(4):583–8. doi: 10.1083/jcb.201210077. [DOI] [PMC free article] [PubMed] [Google Scholar]