To the editor
Biomedical data are being produced at an unprecedented rate owing to the falling cost of experiments and wider access to genomics, transcriptomics, proteomics and metabolomics platforms1,2. As a result, public deposition of omics data is on the increase. This presents new challenges, including finding ways to store, organize and access different types of biomedical data present on different platforms. We present the Omics Discovery Index (OmicsDI - http://www.omicsdi.org), an open source platform that enables access, discovery and dissemination of omics datasets.
In 2016, a group of researchers, publishers and research funders published the first guidelines to make data Findable, Accessible, Interoperable and Re-usable (FAIR - https://www.force11.org/group/fairgroup/fairprinciples)3. The FAIR principles put specific emphasis on enhancing the ability of machines to automatically find and use the data, in addition to supporting its reuse. Challenges facing combined analyses of different datasets include achieving a common representation for datasets and their associated metadata, and the lack of protocols and tools that enable data exchange across multiple repositories. With respect to the first principle (‘Findable’), most of the available resources for the scientific community nowadays are either field specific, that is genomics, proteomics or metabolomics experimental datasets; or organism-specific, but including datasets from different omics technologies, e.g. the “Saccharomyces Genome Database” (SGD). Finding a dataset can be frustrated by the need to search individual repositories and read numerous publications. The development of consortia integrating resources e.g. ProteomeXchange and MetabolomeXchange has helped to improve findability. Nature1 and Nature Biotechnology4 have highlighted the need for dataset integration frameworks to increase findability of data. And, in the context of the European ELIXIR (https://www.elixir-europe.org/) and USA Big Data to Knowledge (BD2K)5 trans-NIH initiative, it is clear that a dedicated platform, search engines and services enabling the aggregation of omics datasets, to resources such as PubMed6 or Europe PubMed Central (EuroPMC)7, is required.
The Omics Discovery Index (OmicsDI - http://www.omicsdi.org) is an open source platform that can be used to access, discover and disseminate omics datasets. OmicsDI can integrate proteomics, genomics, metabolomics, and transcriptomics datasets (Fig. 1). To date, eleven repositories have agreed on a common metadata structure framework and exchange format, and have contributed to OmicsDI (Supplementary Notes 1–3), including proteomics databases (the PRoteomics IDEntifications (PRIDE) database, PeptideAtlas, the Mass spectrometry Interactive Virtual Environment (MassIVE) and the Global Proteome Machine Database (GPMDB)); metabolomics databases (MetaboLights, the Global Natural Products Social Molecular Networking project (GNPS), MetabolomeExpress, and the Metabolomics Workbench), the major European Genome-Phenome Archive (EGA) and transcriptomics databases (ArrayExpress and Expression Atlas). OmicsDI stores biological and technical metadata from these public datasets using an efficient indexing system (Fig. 1b) which can integrate different biological entities including genes, transcripts, proteins, metabolites and the corresponding publications from PubMed.
Figure 1.
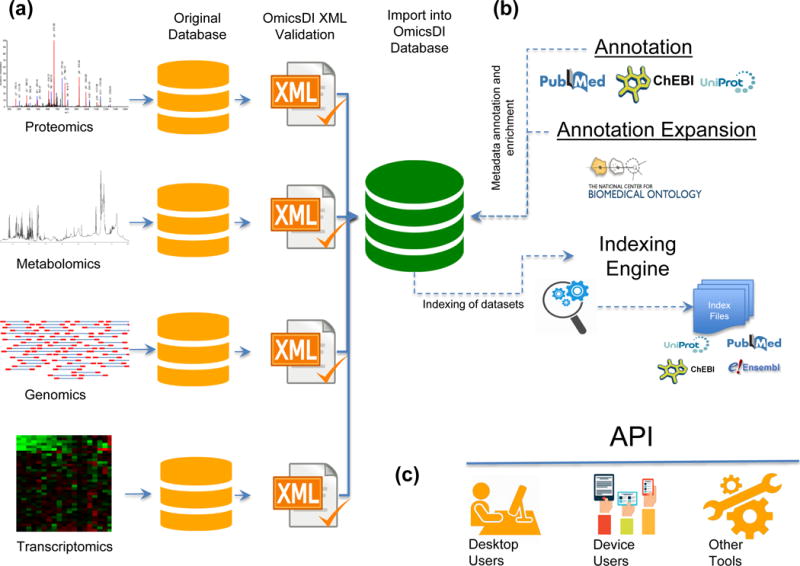
Omics Discovery Index: data standardization, annotation, index and presentation. (a) The datasets stored in public repositories are converted to a common data representation including all metadata and biological entities. The OmicsDI XML files are validated using the OmicsDI XML validator. (b) The OmicsDI XML files are then annotated using public services and databases like UniProt, ChEBI, and PubMed, and the metadata is enriched using the Annotator service. The EBI search engine generates the indexes including other related resources such as PubMed, UniProt, Ensembl and ChEBI. (c) Different clients can use the OmicsDI API to retrieve data from the resource including the web interface and the ddiR package.
In order to facilitate participation in OmicsDI by repositories and the future integration of other omics fields (e.g. interactomics) we have developed a set of data integration guidelines and metadata requirements. The level of annotation required is flexible and many repositories only provide a subset of the metadata included in our guidelines. Data with varying amounts of annotation can be made ‘Accessible’ in OmicsDI by using a flexible metadata schema that classifies datasets as either mandatory, recommended or additional. A flexible exchange system based on the OmicsDI XML format and application programming interfaces (APIs) has been developed. Each repository needs to generate these file formats to join the OmicsDI platform. In order to facilitate integration, a stand-alone open source Java tool has been developed (‘OmicsDI XML validator’). It allows the detection of metadata related format errors as well as inconsistencies in the dataset representation (Supplementary Note 4).
Different repositories use their own data models, metadata representation and identifiers, e.g. controlled vocabularies (CVs) and ontologies. To address any interoperability problems that arise, OmicsDI includes a metadata normalization and annotation expansion step for every dataset that is integrated (Fig. 1b). These harmonization steps standardize experimental and technical metadata, the identifiers for the biological entities, and the references to external resources. For example, for any publication named using the Digital Object Identifier (DOI) or a citation, the matching PubMed identifier is inserted during harmonization (Supplementary Notes 5–6). If the name of an organism is provided using free-text the annotation step during harmonization converts it to an NCBI taxonomy identifier. Different datasets can include different terms for the same concept within the same context8 e.g. a protein can also be referred to as a gene product. To overcome this type of problem, an ontology-based annotation expansion step is applied using the ontology tool ‘Annotator’9, and every relevant phrase in the metadata (title, description, sample and protocols) and the corresponding publication (title and abstract) is enriched with the relevant synonyms, ontology and CV terms.
OmicsDI is a lightweight discovery tool that comprises more than 81,116 omics datasets (December 2016) from eleven different repositories and includes four omics types (transcriptomics: 67,361; proteomics: 6,281; genomics: 8,093; and metabolomics: 847). The number of datasets from human, model organisms and non-model organisms (excluding human) is uniformly distributed among repositories and omics types (Fig. 2a), highlighting the diversity of datasets. To the best of our knowledge, OmicsDI is the first resource that integrates datasets from different omics fields and databases into one framework and web interface.
Figure 2.
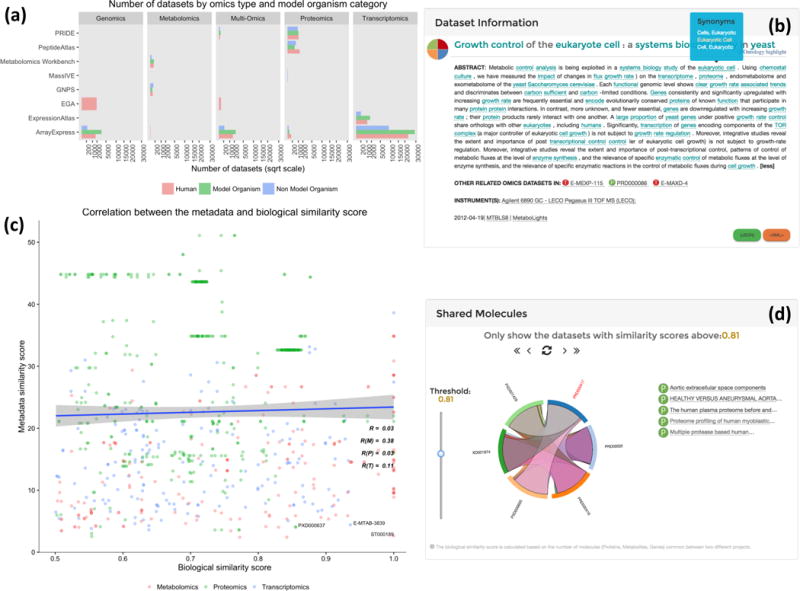
Distributions of OmicsDI datasets. (a) Distribution of datasets per omics type and organism category including model organisms, non-model organisms (excluding human) and human. (b) The dataset view showing the other related omics datasets, including the ontology highlighting option to extract the most relevant terms in the metadata. (c) Pearson-correlation plot between the metadata similarity score and the biological similarity score, across transcriptomics (T), proteomics (P) and metabolomics (M) datasets. (d) The shared molecules box shows all datasets with a biological similarity score of more than 0.5, with a slider allowing a user to increase the cutoff value (here set to 0.81).
OmicsDI also extends the ‘Findable’ principle by providing methods to find and link existing datasets. The annotation expansion step using synonyms enables users to find and associate datasets that cannot otherwise be found. For example, the proteomics dataset PXD002530 (http://www.ebi.ac.uk/pride/archive/projects/PXD002530) can be found in OmicsDI with the search term ‘side effects’, whereas this dataset cannot be found by searching PRIDE using that term. In PRIDE it is only possible to find the same dataset by inputting the term ‘adverse effects’ that was used in the original annotation of the dataset. By indexing the biological entities information in OmicsDI it is possible to find datasets in which the queried molecule has been reported without an exact matched term. For example, the Metabolomics Workbench dataset ST000113 can be found in OmicsDI using the metabolite name ‘Arg-[13C,15N]3’, whereas the same search will not find ST000113 in Metabolomics Workbench.
OmicsDI links datasets by two methods. First, datasets are directly linked using explicit mentions in the metadata. If the dataset is a reanalysis (e.g. PeptideAtlas dataset) of a dataset in a different member repository (e.g. PRIDE), a cross-reference in the OmicsDI XML is used to define this relation. This annotation can be provided by the original repository in the OmicsDI XML (e.g. PeptideAtlas) or can be inferred by OmicsDI during the annotation process. As of December 2016, the relations ‘Reanalyzed by’ and ‘Reanalysis of’ are already in use (Supplementary Table 1). This mechanism provides a direct link between datasets in different repositories. Second, the publication associated with a dataset can be used to link datasets that are deposited in different repositories. This enabled the linking of datasets from different databases that are however part of the same multi-omics experiment (Fig. 2b) and presents them to the user as ‘Other related omics datasets in’. As of December 2016, 4,476 datasets have been labeled by the OmicsDI annotation component as part of multi-omics experiments. While still small (5% of all OmicsDI datasets), the number of multi-omics datasets is growing (Supplementary Table 2).
OmicsDI also uses the ‘similar dataset’ concept (Supplementary Note 7). The concept of ‘Related article’ has been applied in PubMed to explore topics10. In OmicsDI, similar datasets are computed at two different levels: metadata and biological entities. Both similarity levels are estimated by comparing the weighted term vectors of each dataset using the dot (scalar) product. The distribution of the metadata similarity (Supplementary Fig. 1) and molecular similarity (Supplementary Fig. 2) are filtered depending of the distribution for each omics type. In this way OmicsDI boosts the discoverability of related datasets that use similar analytical protocols, software (Supplementary Fig. 3), or share similar biological entities (Supplementary Fig. 4). To our knowledge, this enables for the first time the association of related datasets stored in different resources. For example, for the Expression Atlas dataset E-GEOD-30999 (http://www.omicsdi.org/dataset/atlas-experiments/E-GEOD-30999), OmicsDI reports 14 related datasets. In addition, the ‘biological similarity’ score computes the number of shared biological entities among datasets without taking into account additional metadata. For example, the same dataset has five datasets with a biological similarity score above 0.5 and one dataset with a score of 0.7 (E-GEOD-41662). Of these, dataset E-GEOD-41663 is not classified as related by the metadata-based similarity, although careful reading of the associated manuscripts reveals that E-GEOD-41663 used a subset of the samples of E-GEOD-30999. This example demonstrates the value of our approach. We determined the correlation between metadata and biological similarity scores for all OmicsDI datasets (Fig. 2c). The results showed no correlation (R2 = 0.03) between both metrics across all types of omics datasets, with the highest correlation found in metabolomics approaches (R2 = 0.3). For example, the datasets PXD000637 (PRIDE), ST000189 (Metabolomics Workbench), and E-MTAB-3839 (Expression Atlas) showed a higher biological similarity score (above 0.85) and less than 5 of metadata score (Fig. 2c). These results show that both scores are orthogonal metrics supporting discovery of related datasets through complementary methods.
The OmicsDI web interface provides different views each of which focuses on a specific aspect of the data (Supplementary Notes 8–9). A metadata overview and access statistics provide a convenient entry point to browse a repository (Supplementary Fig. 5). Datasets can be searched and filtered based on annotations (e.g. species, tissue, disease), year of publication, or repository. The result of each search displays all the relevant datasets sorted using a weighted scoring function (Supplementary Fig. 6). In addition, OmicsDI provides a dataset page that includes a list of related publications and similar datasets (Fig. 2b–d). If the biological entities information is available for a given dataset, a chord diagram presents the link to related datasets with high biological similarity scores (Fig. 2d). A web service interface, including a standard RESTful API to access the data programmatically, is also provided (http://www.omicsdi.org/ws). Related libraries and packages used for OmicsDI are also available at https://github.com/BD2K-DDI. For instance, an R-package called ddiR is provided, enabling data analysis (Supplementary Note 10).
OmicsDI exploits advances in metadata-based browsing to support dataset findability. The original datasets are not replicated, but are referenced. In addition to fully open datasets, life science often produces valuable datasets containing personal identifiable genetic or phenotypic data. These data are deposited in controlled-access repositories, to which access is granted after application to a data access committee (DAC). However, the metadata of controlled –access repositories is accessible, and therefore OmicsDI can integrate data from EGA (the first controlled-access repository with open, searchable metadata). The responsibility for provision of well-formatted metadata lies with the original data providers (similar to the concept of publisher data provision to PubMed). OmicsDI displays and promotes the original dataset identifiers, not only to avoid creation of another set of identifiers, but also to ensure attribution of credit to the original data providers. OmicsDI can integrate with large, broader scope efforts like the Biomedical healthCAre Data Discovery and Index Ecosystem (bioCADDIE) through shared metadata formats.
In conclusion, Omics DI provides an integrated search framework for datasets that introduces a range of modern features such as access metrics and discovery of related datasets that we now take for granted in PubMed.
Supplementary Material
Acknowledgments
This work has been supported by the US NIH BD2K grant U54 GM114833 and a National Natural Science Foundation of China grant [61501071]. AIN is supported by US National Institute of Health grant [R01-GM-094231]. YPR is supported by BBSRC ‘PROCESS’ grant [BB/K01997X/1]. MB is supported by Projects of International Cooperation and Exchanges grant [2014DFB30010]. MW is supported by an NIH grant [5P41GM103484-07]. JAV and NDT are supported by the Wellcome Trust [grant WT101477MA]. TT is supported by the BBSRC ‘ProteoGenomics’ grant [BB/L024225/1]. EWD and DSC are supported in part by grant [U24 AI117966-02S1]. SAS is supported in part by US NIH BD2K grant [1U24AI117966-01]. MW and NB were supported by NIH grant [5P41GM103484-07]. N.B. was also partially supported as an Alfred P. Sloan Fellow.
Footnotes
Author’s contributions
HH, YPR and PP developed the OmicsDI concept. YPR and MB designed and developed the web and annotation/enrichment framework. FPV, RCB and YPR developed the GPMDB reader. SS, YMP and NB developed the indexing system based on the EMBL-EBI (European Bioinformatics Institute) Search Server. YPR, EF, MS, ShS, AJC, KH, DS, JP, and MW developed the Metabolomics Workbench, Metabolome Express, MetaboLights, EGA and MassIVE readers and APIs, respectively. PZ helped to make the MetabolomeExpress schema OmicsDI-compatible. US, RP, MK contributed with the integration of ArrayExpress and Expression Atlas. NT, YPR and TT developed the PRIDE reader and contributed to the web development. EWD, DSC, RMS, NB, AIN, CS, RCJ, RL and JAV contributed to the design of the system. YPR and AB developed the ddiR package. YPR and MB, designed and implemented the biological similarity scoring system; and YPR and AB performed the data analysis. YPR, JAV and HH wrote the manuscript, with contributions from all authors.
References
- 1.Bourne PE, Lorsch JR, Green ED. Perspective: Sustaining the big-data ecosystem. Nature. 2015;527:S16–17. doi: 10.1038/527S16a. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Riverol Y, Alpi E, Wang R, Hermjakob H, Vizcaino JA. Making proteomics data accessible and reusable: current state of proteomics databases and repositories. Proteomics. 2015;15:930–949. doi: 10.1002/pmic.201400302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson MD, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prins P, et al. Toward effective software solutions for big biology. Nature biotechnology. 2015;33:686–687. doi: 10.1038/nbt.3240. [DOI] [PubMed] [Google Scholar]
- 5.Bourne PE, et al. The NIH Big Data to Knowledge (BD2K) initiative. J Am Med Inform Assoc. 2015;22:1114. doi: 10.1093/jamia/ocv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic acids research. 2016;44:D7–19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Europe PMCC. Europe PMC: a full-text literature database for the life sciences and platform for innovation. Nucleic acids research. 2015;43:D1042–1048. doi: 10.1093/nar/gku1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blake J. Bio-ontologies-fast and furious. Nature biotechnology. 2004;22:773–774. doi: 10.1038/nbt0604-773. [DOI] [PubMed] [Google Scholar]
- 9.Shah NH, et al. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC bioinformatics. 2009;10(Suppl 9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Wilbur WJ. PubMed related articles: a probabilistic topic-based model for content similarity. BMC bioinformatics. 2007;8:423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


