Abstract
HIV-1 envelope spike (Env) is a type I membrane protein that mediates viral entry. Recent studies showed that the transmembrane domain (TMD) of the Env forms a trimer in lipid bilayer and that disruption of the TMD could significantly alter the antigenic properties of the Env. The TMD structure has several peculiar features that remain difficult to explain. One is the presence of an arginine R696 (three in the trimer) in the middle of the TM helix. Additionally, the N- and C-terminal halves of the TM helix form trimeric cores of opposite nature (hydrophobic for the N half and hydrophilic for the C half). Here we determined the membrane partition and solvent accessibility of the TMD in bicelles that mimic a lipid bilayer. Solvent paramagnetic relaxation enhancement analysis showed that the R696 is indeed positioned close to the center of the bilayer, but, surprisingly, can exchange rapidly with water as indicated by hydrogen-deuterium exchange measurements. The solvent accessibility of R696 is likely mediated by the hydrophilic core, which also showed fast water exchange. In contrast, the N-terminal hydrophobic core showed extremely slow solvent exchange, suggesting the trimer formed by this region is extraordinarily stable. Our data explain how R696 is accommodated in the middle of the membrane while reporting the overall stability of the Env TMD trimer in lipid bilayer.
Graphical Abstract
The HIV envelope glycoproteins (Env) are the sole antigens on the virion surface relevant for vaccine design. Recently, the NMR structure of the trimeric transmembrane domain (TMD) of the HIV-1 Env was determined. The structure revealed a highly unusual feature that three conserved arginines (one per protomer) are in the middle of the transmembrane helices, suggesting the unfavorable placement of three charges in the middle of the membrane. Using a combination of ideal bicelles, solvent paramagnetic relaxation enhancement analysis, and hydrogen-deuterium exchange measurement, we provide direct evidences that the controversial arginines can access water from the bulk solvent via the C-terminal hydrophilic core of the TMD, thus allowing them to hydrate despite being in the middle of a lipid bilayer. We also found that the N-terminal hydrophobic core of the TMD forms an extremely stable trimer, perhaps for maintaining the yet unknown conformation of the MPER. Our study provides an explanation for how the membrane-embedded arginines are tolerated in the Env TMD while reporting the overall stability of the trimeric membrane anchor of the HIV-1 Env in lipid bilayer.
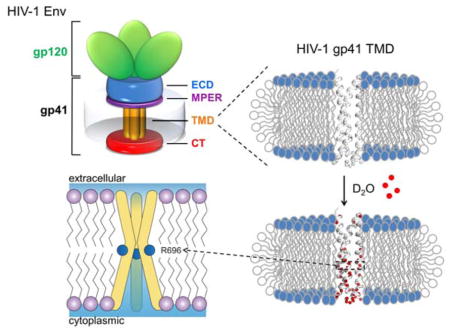
The HIV-1 envelope spike (Env) is a glycoprotein used by the HIV-1 virus to target and infiltrate specific host cells1–2. The mature Env spikes [trimeric (gp160)3, cleaved to (gp120/gp41)3] are the sole antigens on the virion surface3–4. The gp120 and gp41 are the receptor recognition and membrane fusion proteins, respectively. Conformational changes in gp120 when triggered by binding to receptor (CD4) and co-receptor (e.g., CCR5 or CXCR4) lead to a cascade of refolding events in gp41, and ultimately to membrane fusion2, 5–7.
A plethora of structures of the ectodomain (ECD) of the gp120/gp41 complex have been determined5–15, but relatively little is known about the transmembrane (TM) and membrane proximal regions of gp41 due to challenges of preserving the native-like folding of these regions in membrane mimetic media. Mutagenesis studies showed that the transmembrane domain (TMD) of gp41 is not merely a membrane anchor, but plays critical roles in membrane fusion and viral infectivity16–20. Moreover, recent studies showed that truncations in the cytoplasmic tail of gp41 could alter the antigenic surface of the Env ECD on the opposite side of the membrane21, suggesting that the ECD, the TMD, and the membrane proximal regions are conformationally coupled, and thus the conformational stability of the TMD is an important consideration for immunogen design in B-cell based HIV-1 vaccine development.
In an earlier study, we determined the NMR structure of the gp41 TMD in bicelles (made of DMPC lipid and DHPC detergent)22 using a gp41 fragment (residues 677–716) from a clade D HIV-1 isolate 92UG024.2, designated gp41HIV1D(677–716). The TMD forms a well-structured trimer, almost helical all the way from the N- to the C-terminal end. The structure shows two unusual features. One is the presence of an arginine (R696), three in the trimer, in the middle of the TM helices (Fig. S1a), implying three unbalanced charges in the hydrophobic core of the membrane. R696 is highly conserved with only lysine substitution at this position in different strains. Another feature is that the N- and C- terminal halves of the trimer are assembled differently. The region N-terminal to R696 (686–695) resembles a coiled-coil motif that forms a hydrophobic core (Fig. S1b), whereas the region C-terminal to R696 (697–712) assembles a core that is largely hydrophilic (Fig. S1c). To the best of our knowledge, there are no prior examples of this type of assembly for single-pass TM proteins.
The above features of the gp41 TMD are certainly difficult to comprehend. In particular, the three lipid-facing arginines in the middle of the TMD have no nearby acidic residues to form hydrogen bond with. In this study, we performed extensive analysis of the membrane insertion and solvent accessibility of the HIV-1 gp41 TMD in bicelles using the same protein construct, gp41HIV1D(677–716) (denoted gp41TMD for simplicity), used previously for structure determination22. Moreover, complementary dynamics information was obtained using NMR relaxation measurements. Our data provide an explanation for how the membrane-embedded arginines are tolerated in the gp41 TMD and revealed regions of the TMD with very different trimer stability.
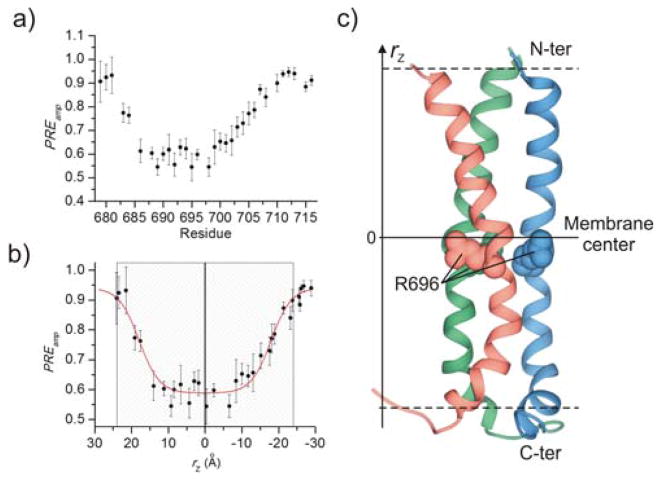
Isotropic phospholipid bicelles are versatile systems for investigating protein immersion in membrane23. We first investigated the membrane partition of the gp41 TMD to determine the true position of the R696 in the membrane using a solvent paramagnetic relaxation enhancement (PRE) method developed previously for bicelles24. This approach is based on the notion that if the bicelle is sufficiently wide, the lateral solvent PRE becomes negligible, thus allowing the use of measurable solvent PRE to probe residue-specific depth immersion of the protein in the bilayer region of the bicelle (Fig. S2a). The protein gp41TMD was reconstituted in bicelles with DMPC/DHPC molar ratio (q) of 0.5 (Fig. S2b), and titration of the water-soluble paramagnetic probe Gd-DOTA outside the bicelles provided residue-specific PRE amplitudes (PREamp) (Fig. 1a, S2c; Table S2). To determine the position of the TMD trimer relative to the bilayer center, we calculated, for each residue i, the distance (rz) along the 3-fold symmetry axis, which is parallel to the bilayer normal, from the amide proton to an arbitrary reference point based on the NMR structure of the TMD trimer. This calculation converted PREamp vs. (residue number) to PREamp vs. rz, which was then analyzed using the sigmoidal fitting method24 (Fig. S3) to position the TMD structure relative to the center of the bilayer (Fig. 1b). Moreover, the sigmoidal fit in Fig. 1b shows that the PREamp reaches the maximal value at about 24 Å away from the bilayer center on either side, indicating that the bilayer thickness around the protein is ~48 Å (with L679 and Q710 aligned with the two lipid-solvent boundaries). The membrane partition of the gp41 TMD shows that the R696 is indeed deep within the hydrophobic core of the membrane (Fig. 1C; Table S3).
Figure 1.
Membrane partition of gp41TMD. (a) Residue-specific PREamp. (b) The PREamp vs. rz best fitted to the symmetric sigmoidal equation (Eq. S2), where rz = 0 corresponds to the bilayer center. The gray-striped box represents the estimated thickness of the bilayer. (c) The position of the gp41TMD structure relative to the center (solid line) and boundaries (dashed lines) of the lipid bilayer. rz is parallel to the bilayer normal.
Having three basic residues in the middle of the membrane suggests that the gp41 TMD trimer might be energetically unstable. We thus investigated the trimer stability by performing the hydrogen-deuterium (H-D) exchange experiment. We first prepared a sample of (15N, 2H)-labeled gp41TMD reconstituted in bicelles (q = 0.5) at pH 6.0, and a reference 2D 1H-15N TROSY-HSQC spectrum was recorded (Fig. S4). The sample was then flash frozen and lyophilized. To initiate H-D exchange, the completely dried sample was dissolved in 99.9% D2O (pD ~6.4). The exchange was monitored as the loss of NMR signal in a series of 2D 1H-15N TROSY-HSQC spectra recorded at different time points (Fig. 2a) as exchangeable amide protons were replaced by deuterium. Since the spectra at various time points were all recorded with protein in 99.9% D2O, no corrections for NMR relaxation enhancement due to the higher D2O viscosity were required.
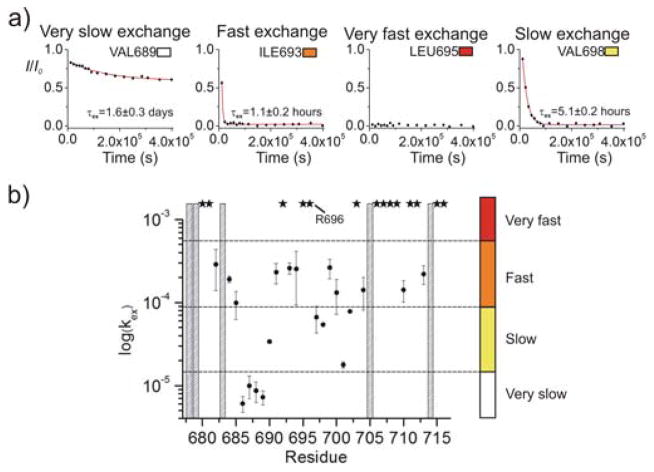
Figure 2.
Solvent accessibility of gp41TMD. (a) Signal decay over time for residues V689, I693, L695 and V698, each representing one of the four defined exchange regimes. (b) Residue-specific kex reported in logarithmic scale. Amide protons with exchange too fast to be measured are marked with stars. Residues that could not be analyzed (overlapping peaks and Pro714) are marked with gray bars. The color spectrum on the right represents the four different exchange regimes: very fast (red), fast (orange), slow (yellow), and very slow (white: very slow).
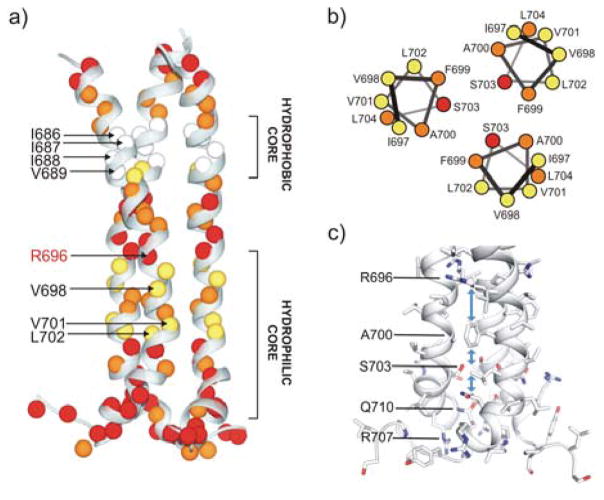
Our measurements show that H-D exchange among the TMD residues is extremely heterogeneous, e.g., the exchange time constant (τex = 1/kex) of V689 is 1.6±0.3 days whereas that of I693 is 1.1±0.2 hours (Fig. 2a; Table S4). For convenience, we define four time regimes of solvent exchange: very fast (τex < 1 hour), fast (1 ≤ τex < 3 hours), slow (3 hours ≤ τex < 1 day), and very slow (τex ≥ 1 day). The plot of exchange rate (kex) vs. (residue number) (Fig. 2b) shows that on average the N-terminal hydrophobic core has much slower H-D exchange than the C-terminal hydrophilic core (Fig. 3a). To our great surprises, the R696 amide exchanged rapidly despite being in the most buried region of the bicelles (Fig. 1c). The exchange of this residue was too fast to be measured, i.e., the peak vanished by the time of sample preparation and NMR experiment setup and acquisition (~1 hour). Since it is unlikely for water to penetrate a lipid bilayer, the fast access to D2O by the R696 ought to be mediated by the TMD structure. Incidentally, the core-facing amides of the C-terminal hydrophilic core (e.g., F699, A700 and S703) also showed fast exchange whereas the corresponding lipid-facing amides (e.g., V698, V701 and L702) showed slow exchange (Fig. 3b). These measurements strongly suggest that D2O can readily diffuse through the hydrophilic core spanning from residues R696 to R709, and that this hydrophilic core serves as a channel that mediates the fast water access by the R696. In stark contrast, just a few residues away, residues 686–689 in the N-terminal hydrophobic core exchanged very slowly (Fig. 2b; 3a), e.g., the peaks were still present even after 4.6 days (Fig. S4). The result implies that water diffusion along the trimer core stops at around R696, as it cannot pass through the hydrophobic core formed by residues 686–689. The H-D exchange measurements clearly show that the TMD of HIV-1 gp41 is very different from most of the known oligomeric TMDs in that multiple regions with very different trimer stability and water accessibility exist within the same TMD.
Figure 3.
Water accessibility of gp41TMD. (a) The four exchange regimes mapped onto the gp41TMD structure with colors defined in Fig. 2b. (b) Helical wheel representation of residues 697–704 colored according to their exchange regime. (c) Residues lining the hydrophilic core that could mediate water passage to the R696.
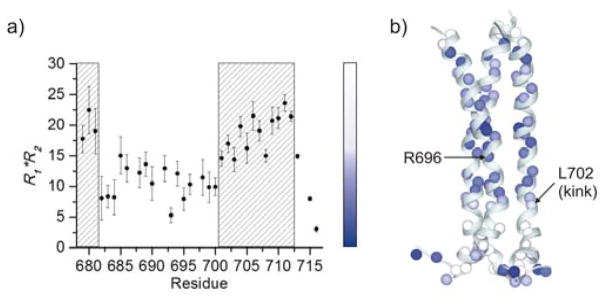
The large heterogeneity in H-D exchange could be accompanied by variations in its dynamic properties. Therefore, we studied the protein backbone dynamics by measuring 15N R1 and R2 relaxation rates (Fig. S5). As expected, both hydrophobic and hydrophilic cores are well structured (low R1 and high R2), with the protein becoming progressively more dynamic on both ends where it reaches the edges of the bilayer. While R1 and R2 are excellent probes for ns-ps time scale motions, they can also provide qualitative information on slower time scale motions (ms-μs) through their product (R1R2). It has been shown that the R1R2 product tends to be reduced by ns-ps motions but increased by ms-μs motions associated with chemical shift exchange (Rex)25. For the well-structured region of the TMD (residues 682–710), the R1R2 values of residues 682–700 are quite consistent at ~10 (Fig. 4a), indicating the lack of Rex in this region of the trimer. After A700, however, there is a sharp increase in R1R2 for residues 702–710 (Fig. 4a). We note that this sudden change of R1R2 aligns with the kink at L702 that resulted in ~30° tilt of the C-terminal segment (residues 702–710) with respect to the core region of the TM helix (residues 685–700) (Fig. 4b). The larger R1R2 of residues 702–710 suggest that the C-terminal helical segments undergo substantial conformational exchange.
Figure 4.
Presence of ms-μs dynamics in gp41TMD. (a) Residue-specific R1R2. The regions in the striped box are characterized by greater Rex than the rest of the TMD. (b) Mapping R1R2 onto the gp41TMD structure according to the color spectrum in (a).
We have shown that although the gp41 TMD appeared visually as a simple TM helix trimer, it comprises multiple regions with very different assembly stability and water accessibility. The diverse properties of the TMD afford an explanation for how the conserved basic residue at position 696 (R/K) can be accommodated in the middle of a lipid bilayer while preserving the integrity of the trimer complex.
Due to the unusual position of the R696, the membrane partition of the gp41 TMD has been controversial20, 22, 26–27. Our solvent PRE analysis now confirms that R696 resides in the middle of the membrane (Fig. 1c). Since the large Gd-DOTA molecule can penetrate neither the lipid bilayer nor the protein core, it is an unambiguous probe for the solvent exposure and depth immersion of the TMD in bicelles. Notably, the PRE-sensitive regions in Fig. 1a, residues 681–686 and 702–710, show remarkable correlation between the slope of PREamp vs. (residue number) and the helical segment orientation relative to the 3-fold axis (or bilayer normal). For example, the slope of PREamp for residues 702–710 is smaller than that of 681–686, and this is consistent with the larger tilt of the 702–710 segment caused by the kink at L702 (Fig. S6).
The solvent PRE, however, provided no information about the water accessibility of the protein, which we probed using the H-D exchange experiment. We found, unexpectedly, that the R696, although in the middle of the bilayer, showed fast H-D exchange (too fast to be measured), indicating that water from the bulk solvent can rapidly access the R696. This is consistent with a previous observation that the R696 sidechain Hε showed a water cross-peak in a NOESY spectrum22 (Fig. S7), though it was not understood how water could possibly reach the middle of the lipid bilayer. The H-D exchange data for the C-terminal hydrophilic core (residues 697–709) provided the missing piece of the puzzle. That is, the pore-lining amides of this region all showed reasonably fast exchange (τex < 3 hours), suggesting that water can easily diffuse through the hydrophilic core to reach the R696. In contrast, the N-terminal hydrophobic core showed very slow H-D exchange (τex > 1 day), which prevents the TMD from becoming a water channel. The R969 is thus strategically located at the interface between the two regions with completely different water accessibilities. To date, the functional role of the R696 remains unknown, but different studies have suggested that it is important for efficient membrane fusion17, 19. Our data revealed properties of the gp41 TMD that enables the membrane-embedded basic residue to access water and provided a rationale for the relatively high content of polar residues in the C-terminal core of the TMD.
The very slow H-D exchange of the N-terminal hydrophobic core (residues 686–689) (τex > 1 day) suggests that this trimeric region of the gp41 TMD essentially does not dissociate, as even transient dissociation would result in the loss of the amide protons that cannot be recovered. The extreme stability could ensure that the gp41 remains trimeric in specific stages of the fusion process28–29; it could also play a role in stabilizing the prefusion state of the trimeric HIV-1 Env.
Our R1R2 measurements suggest that C-terminal residues 702–710 experience conformational exchange in ms-μs timescale (Fig. 4a). This helical segment is significantly tilted relative to the core region of the TM helix, resulting from a kink at L702. It is thus not surprising that this segment is more dynamic than the TM core. The ms-μs motion of this segment could have been caused by the absence of the gp41 cytoplasmic region, which would immediately follow our construct in the native protein. We also note that the proposed water passage through the C-terminal hydrophilic core could also benefit from conformational breathing of this region of the TMD trimer.
In conclusion, our NMR measurements provided direct evidence that the membrane-embedded R696 of HIV-1 gp41 TMD can access water from the bulk solvent via the C-terminal hydrophilic core, thus allowing the arginine to hydrate despite being in the middle of a lipid bilayer. The coexistence of the N-terminal hydrophobic and C-terminal hydrophilic cores is consistent with water permeation through only the C-terminal half of the TMD. This peculiar feature of the gp41 TMD should also apply to HIV-2 and SIV based on structural homology27. Although the function of the highly conserved R696 remains elusive, our results show that the bipolar nature of the gp41 TMD allows the natural placement of the arginine in an otherwise unfavorable lipid environment.
Supplementary Material
Acknowledgments
We thank Prof. Stephen C. Harrison and Prof. Bing Chen for insightful discussion. This study was supported by NIH grant AI127193 to J.J.C.
Footnotes
The Supporting Information is available free of charge on the ACS Publications website at DOI: xxxx.
The SI includes Tables S1–S4 and Fig. S1–S7, as well as the description of the NMR experiment setup and the protocols for the sample preparation and data analysis.
References
- 1.Harrison SC. Advances in Virus Research. 2005;64:231–259. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan DC, Kim PS. Cell. 1998;93(5):681–4. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 3.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Nature. 2003;422(6929):307–12. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 4.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Proc Natl Acad Sci U S A. 2003;100(7):4144–9. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan DC, Fass D, Berger JM, Kim PS. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 6.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 7.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, Stewart-Jones GB, Stuckey J, Bailer RT, Joyce MG, Louder MK, Tumba N, Yang Y, Zhang B, Cohen MS, Haynes BF, Mascola JR, Morris L, Munro JB, Blanchard SC, Mothes W, Connors M, Kwong PD. Nature. 2014;514(7523):455–61. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC. Nature. 2005;433(7028):834–41. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 9.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure Fold Des. 2000;8(12):1329–39. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 10.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang CC, Tang M, Zhang MY, Majeed S, Montabana E, Stanfield RL, Dimitrov DS, Korber B, Sodroski J, Wilson IA, Wyatt R, Kwong PD. Science. 2005;310(5750):1025–8. doi: 10.1126/science.1118398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan K, Liu JH, Wang JH, Shen S, Lu M. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caffrey M, Cai M, Kaufman J, Stahl SJ, Wingfield PT, Covell DG, Gronenborn AM, Clore GM. EMBO J. 1998;17:4572–4584. doi: 10.1093/emboj/17.16.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, Ward AB, Wilson IA. Science. 2013;342(6165):1477–83. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, Wilson IA, Ward AB. Science. 2013;342(6165):1484–90. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. J Virol. 1990;64(12):6314–8. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owens RJ, Burke C, Rose JK. J Virol. 1994;68(1):570–4. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shang L, Yue L, Hunter E. J Virol. 2008;82(11):5417–28. doi: 10.1128/JVI.02666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long Y, Meng F, Kondo N, Iwamoto A, Matsuda Z. Protein & cell. 2011;2(5):369–76. doi: 10.1007/s13238-011-1051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rotem E, Reuven EM, Klug YA, Shai Y. Biochemistry. 2016;55(7):1049–57. doi: 10.1021/acs.biochem.5b01307. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B. Science. 2015;349(6244):191–5. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dev J, Park D, Fu Q, Chen J, Ha HJ, Ghantous F, Herrmann T, Chang W, Liu Z, Frey G, Seaman MS, Chen B, Chou JJ. Science. 2016;353(6295):172–5. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt T, Situ AJ, Ulmer TS. J Phys Chem Lett. 2016;7(21):4420–4426. doi: 10.1021/acs.jpclett.6b02159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piai A, Fu Q, Dev J, Chou JJ. Chemistry. 2017;23(6):1361–1367. doi: 10.1002/chem.201604206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kneller JM, Lu M, Bracken C. J Am Chem Soc. 2002;124(9):1852–3. doi: 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- 26.Miyauchi K, Curran R, Matthews E, Komano J, Hoshino T, Engelman DM, Matsuda Z. Japanese journal of infectious diseases. 2006;59(2):77–84. [PubMed] [Google Scholar]
- 27.Chen B, Chou JJ. FEBS J. 2017;284(8):1171–1177. doi: 10.1111/febs.13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roche J, Louis JM, Grishaev A, Ying J, Bax A. Proc Natl Acad Sci U S A. 2014;111(9):3425–30. doi: 10.1073/pnas.1401397111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reuven EM, Dadon Y, Viard M, Manukovsky N, Blumenthal R, Shai Y. Biochemistry. 2012;51(13):2867–78. doi: 10.1021/bi201721r. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.