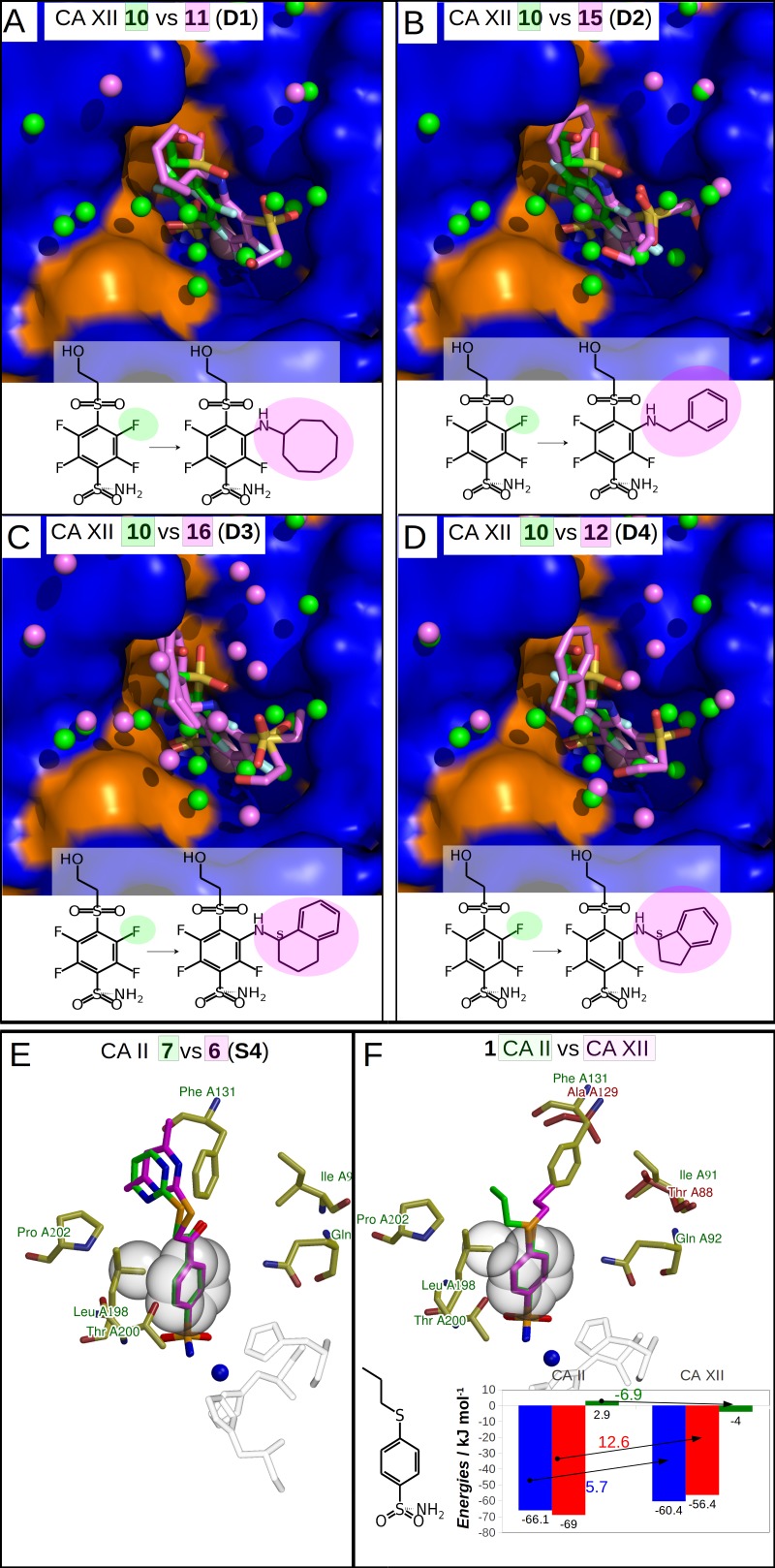
Figure 4. Dissimilar binders.
(A–D) “Dissimilar binders”: matched pairs of crystal structures. Inhibitors bound to CAs are presented in the same orientation. The structural differences of the inhibitors in pairs are shaded by green or pink. The ligands in crystal structures are colored accordingly. Water molecules found in crystal structures are shown as green and pink spheres. Zn2+ ion is shown as a magenta sphere. The protein surface of CA active site is colored orange for hydrophobic residues (Val, Ile, Leu, Phe, Met, Ala, Gly, and Pro) and blue for the residues with charged and polar side chains (Arg, Asp, Asn, Glu, Gln, His, Lys, Ser, Thr, Tyr, Trp, and Cys). (A) Compounds 10 (green, PDB ID 5MSB) and 11 (pink, PDB ID 4Q0L) are bound to CA XII (pair D1). (B) Compounds 10 (green, PDB ID 5MSB) and 15 (pink, PDB ID 4QJW) bound to CA XII (pair D2). (C) Compounds 10 (green, PDB ID 5MSB) and 16 (pink, PDB ID 5LLP) bound to CA XII (pair D3). (D) Compounds 10 (green, PDB ID 5MSB) and 12 (pink, PDB ID 5LLO) bound to CA XII (pair D4). (E) Compounds 7 (green, PDB ID 3SBI) and 6 (pink, PDB ID 3SBH) bound to CA II (colored yellow). (F) Comparison of the compound 1 position bound to CA II (1 is green, CA II is colored yellow, PDB ID 5LLG) and CA XII (1 is pink, CA XII side chains are red, PDB ID 4WW8) and corresponding binding thermodynamics. The histogram in the insert shows the comparison of binding thermodynamics: ΔG is blue, ΔH—red and −TΔS—green. The histidine side chains holding the active site Zn2+ (blue sphere) ion are transparent in (E) and (F). The Leu198 conservative between isoforms CA II and XII and benzene rings of compounds are shown in CPK representation which marked the specific aliphatic-aromatic interaction.