Abstract
Spherical nucleic acids (SNAs) can be potent sequence-specific stimulators of antigen presenting cells (APCs). When loaded with peptide antigens, they can be used to activate the immune system to train T-cells to specifically kill cancer cells. Herein, the role of peptide chemical conjugation to the DNA, which is used to load SNAs with antigens via hybridization, is explored in the context of APC activation. Importantly, though the antigen chemistry does not impede TLR-9 regulated APC activation, it significantly augments the downstream T-cell response in terms of both activation and proliferation. A comparison of three linker types, (1) noncleavable, (2) cleavable but nontraceless, and (3) traceless, reveals up to an 8-fold improvement in T-cell proliferation when the traceless linker is used. This work underscores the critical importance of the choice of conjugation chemistry in vaccine development.
Subtle changes in the chemical architecture of nanoparticle constructs can significantly influence biological function, including biodistribution properties,1–3 drug release,4–6 and cellular internalization.7–10 To rationally design nanoparticles with desired properties, researchers should focus on characterizing the attributes that can be systematically changed and where structure–function relationships can begin to be defined. For example, SNA architectures, synthesized by arranging linear oligonucleotides on the surfaces of nanoparticle templates, have shown promise as probes in diagnostics11 and as therapeutic lead compounds in medicine.12 In the latter category, their ability to enter cells via endosomal pathways and agonize or antagonize toll-like receptors makes them highly promising immunomodulatory agents.13 In the case of cancer vaccines, SNAs can also be used to carry antigens that provide selective training of the immune system through T-cell activation and proliferation. From a chemistry perspective, this creates both a challenge and an opportunity. Out of the many ways of combining components required for T-cell activation and proliferation, which ones are best, and do they result in significant differences in efficacy?
The way antigen molecules are incorporated in synthetic vaccines could impact not only quantities of antigen delivered to APCs but also the processing and chemical structure of the antigen. Indeed, for small molecule and peptide delivery, activity can be highly dependent on the type of conjugation chemistry employed.14–16 When designing the next generation of vaccines, such as immunostimulatory SNAs, it is imperative to understand the impact of the conjugation chemistry used to attach the antigen to the DNA that loads the antigen on the SNA construct. Specifically, because chemical modifications can influence peptide antigenicity, it may be important to devise general strategies that can be used with a wide array of peptides, to deliver pristine antigens with no chemical appendages.
Herein we report the use of three linkage types, a disulfide reduction-activated traceless linker, a disulfide reduction-activated cleavable linker, and a noncleavable linker (Figure 1A,B), for attaching a human melanoma-specific antigenic peptide, gp100 (KVPRNQDWL), to SNAs. The study is designed to probe the importance, or lack thereof, of generating pristine antigens for immune activation. The gp100 antigen was chosen as a model system because of its clinical relevance to human diseases and high potential for translation.17
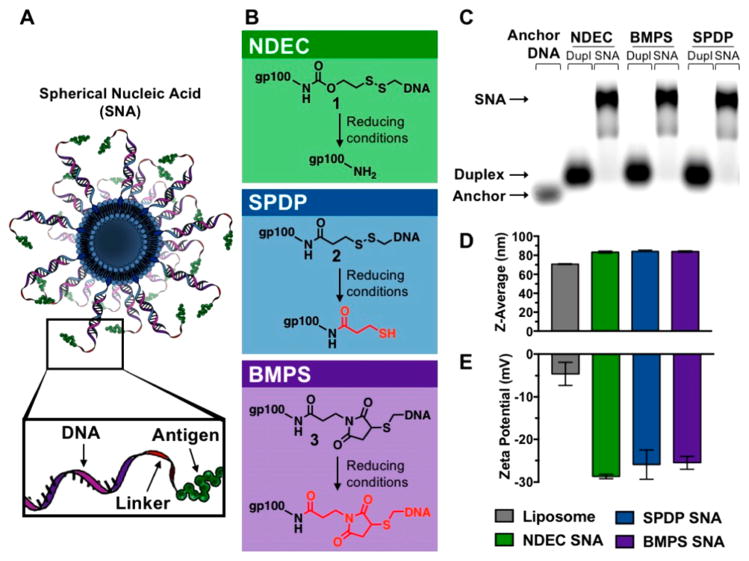
Figure 1.
(A) Schematic design of the immunostimulatory SNA. (B) Three distinct linker chemistries were used to make the antigen-DNA conjugates 1–3: NDEC (traceless), SPDP (cleavable), and BMPS (noncleavable), respectively. (C) Cholesterol-modified cyanine-5 (Cy5)-tagged anchor DNA, conjugate and anchor duplex, and SNA were characterized using 1% agarose gel, imaged by Cy5 fluorescence. (D,E) DLS shows an increase in diameter along with a decrease in zeta potential, measured at pH 7, between the bare liposome and the SNAs. Samples for DLS were prepared without the Cy5 modification.
Immunostimulatory SNAs were synthesized using a liposomal core with TLR9-stimulatory CpG B oligonucleotides (see Table S1 for sequences), tagged with a Cy5 dye, and immobilized on the core surface through intercalation by using a cholesterol anchor on the 3′ end.18,19 Antigens were attached to the SNA as one of three gp100–DNA conjugate types, 1–3, made with DNA complementary to the CpG adjuvant. CpG anchor stands were all hybridized to the conjugates prior to their addition to liposomes, and these duplexes were added at a 75:1 ratio to liposomes. All design parameters, such as the 1:1 ratio of antigen to adjuvant, DNA and gp100 concentrations were kept constant across the SNA structures investigated, only the identity of the linker differed.
Conjugates 1–3 were synthesized by first attaching one end of the linker to a peptide amine, followed by attachment of thiolated DNA to the other. The amine residue of the antigen was used as a chemical point for conjugation because this strategy can be adapted to other antigens, all of which have at least one primary amine at their N-terminus. The three distinct linker chemistries were chosen to test general considerations for antigen attachment (Figure 1B). A commercially available noncleavable linker (N-(β-maleimidopropyloxy) succinimide ester, BMPS) was used to create conjugate 3, which has no readily cleavable bonds. A commercially available cleavable linker (succinimidyl 3-(2-pyridyldithio)propionate, SPDP) was used to prepare conjugate 2, which cleaves in the reducing environment of the cell but leaves a molecular pendant group (3-mercaptopropionamide) attached to the antigen. Finally, a traceless linker (4-nitrophenyl 2-(2-pyridyldithio)ethyl carbonate, NDEC)15,16,20–22 was incorporated to create conjugate 1 (see Figures S1–S2 for synthesis). The traceless linker incorporates a disulfide, which upon reduction, results in an intramolecular cyclization that releases the antigen in an unmodified form.
We incubated conjugates 1–3 at pH 7.4 with 10 mM glutathione and characterized the decomposition products using PAGE and MALDI-MS. These experiments confirm that, under cell-mimicking reduction conditions,23 the BMPS conjugate 3 does not release an antigen, the SPDP conjugate 2 releases an antigen that is modified with a chemical pendant, and the NDEC conjugate 1 regenerates an unmodified gp100 peptide (see Figures S3–S5). We also characterized the rate of conjugate cleavage by synthesizing 1–3 using a fluorescein labeled gp100 peptide and a quencher-containing oligonucleotide to form a FRET reporter. The fluorescence of this reporter increases upon cleavage of the linkage between the peptide and DNA. We find that conjugates 1 and 2 have similar cleavage half-lives of approximately 31 and 54 min in 20 mM GSH, respectively. Conjugate 3 did not show an increase in fluorescence (Figure S6).
SNAs synthesized with the three conjugates were characterized by agarose gel electrophoresis. A shift in electrophoretic mobility was observed between the single stranded CpG DNA, the duplex with the gp100–DNA conjugate, and the SNA (Figure 1C). Additionally, the SNAs all have indistinguishable z-average hydrodynamic diameter, of 83.7 ± 0.4 nm (PDI 0.075 ± 0.012). An increase of approximately 13 nm over the bare liposomes (Figure 1D). The ζ-potentials of the SNAs were on average –26.7 ± 1.7 mV, a decrease of approximately 20 mV compared to the bare liposomes, which can be attributed to the added negative charge carried by the DNA backbone (Figure 1E).
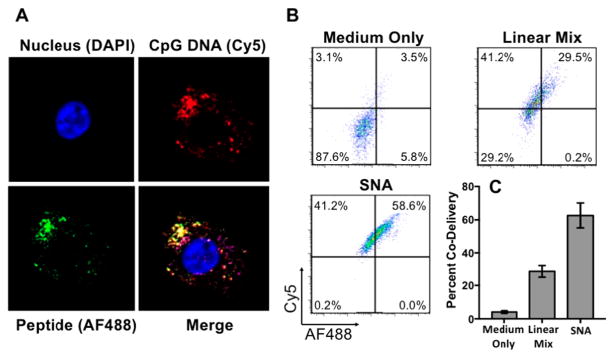
Codelivery of both adjuvant and antigen is crucial for efficient T-cell activation.24 In order to characterize the codelivery of these components, we used bone marrow-derived dendritic cells (DCs) as a model system, because they are the most effective professional APCs of the immune system.25 Confocal microscopy images show that both the AlexaFluor488 (AF488)-labeled gp100 antigen (green) and Cy5-labeled CpG adjuvant (red) have been internalized by DCs after incubation with 1-SNAs for 15 min (Figure 2A). We further quantified the codelivery of these components using flow cytometry (Figure 2B). The SNA architecture formulation resulted in a doubling of codelivery efficiency (double positive of AF488 and Cy5) compared to the linear mixture, as measured over background fluorescence control (medium only) (Figure 2C). In addition, we observed no significant effect of 1-SNA on cell viability at concentrations below 1 μM using an MTT assay (Figure S7).
Figure 2.
(A) Confocal microscopy images show gp100 antigen (AF488, green) and the CpG adjuvant (Cy5, red) inside mouse dendritic cells. (B,C) Flow cytometry measurements after 15 min incubation. Values are an average of three replicates (see Figure S8 for additional replicates).
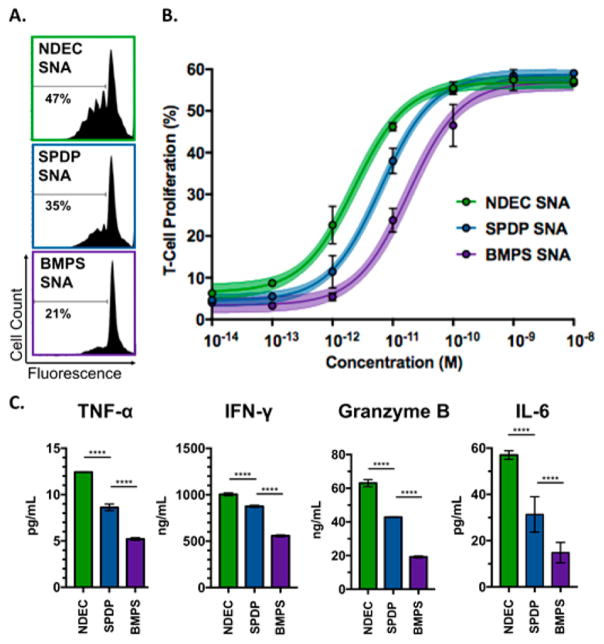
T-cell receptor transgenic CD8+ T-cells (from pmel-1 mice) specifically recognizing gp100 were used to study the efficacy of the immunostimulatory SNAs at eliciting gp100-specific CD8+ T-cell responses.26 The splenocytes from pmel-1 mice were treated with each SNA individually at different concentrations for 72 h to determine a dose–response curve (Figure 3A,B).27,28 We observed that CD8+ T-cell proliferation (measured by eFluor 450 dilution) was dependent upon linkage type, the only parameter that differs across the three SNAs. The extent of proliferation was similar across the three structures when splenocytes were treated at the highest concentration range (1–10 nM in gp100); however, at lower concentrations, the T-cell proliferation differed significantly among the three treatment groups (1–100 pM in gp100). Notably, 1-SNAs even produced detectable T-cell proliferation at 100 fM treatment whereas the two other SNAs failed to show any effect. The calculated EC50 values indicate that 1-SNA (EC50 = 2.3 pM) is approximately three times more potent than 2-SNA (EC50 = 6.4 pM), which itself is approximately three times more efficacious than 3-SNA (EC50 = 18 pM). This observation reveals the significance of antigen conjugation chemistry on the ability of SNAs to induce antigen-specific T-cell proliferation.
Figure 3.
(A) Flow cytometry data showing CD8+ T-cell proliferation following incubation of pmel-1 splenocytes with the three types of SNAs at 10 pM concentration. (B) Dose–response curve of SNA treatment on T-cell proliferation. Average and standard deviation for three replicates are shown for each point (see Figure S9 for additional replicates). The curves are three-parameter dose–response fits with a shaded 95% confidence interval of the fit. (C) Secreted TNF-α, IFN-γ, Granzyme B, and IL-6 were quantified by ELISA at a 10 pM treatment concentration, ****p < 0.0001.
To further evaluate the impact of conjugation chemistry on T-cell activation, we quantified the release of IFN-γ, TNF-α, granzyme-B, and IL-6 for all three SNAs using ELISA at a 10 pM SNA treatment concentration (Figure 3C). Consistent with results of T-cell proliferation, we show that T-cells treated with the traceless 1-SNAs secrete higher levels of the cytokine activation markers IFN-γ and IL-6, compared to the 2-SNA and 3-SNA groups. This shows that traceless NDEC conjugation chemistry leads to higher T-cell activation. Granzyme B and TNF-α secretion, which results from 1-SNA treatment, is also higher than all other groups, indicating the increased potential of T-cell-mediated killing of tumor cells.
Optimum T-cell activation and proliferation depend on MHC-antigen-TCR binding as well as the activation state of the APCs. The observed differences in SNA efficacy could be due to different levels of APC activation. Therefore, we compared the activation levels of DCs across the SNA types by quantifying the expression of the costimulatory markers, CD40 and CD80 (Figure S10). All SNA types caused upregulation in the expression of the two receptors compared to a medium only control. No difference in APC activation among the three SNA types was observed, indicating that the activation of DCs, caused by the interaction of CpG oligonucleotides with TLR receptors in the endosomes, is likely independent of the linkage chemistry used to form the gp100–DNA conjugates.
Taken together, these data show that the choice of linker chemistry used to conjugate an antigen, gp100, to the immunostimulatory SNA has a significant impact on potency and has implications for vaccine development. Importantly, these findings support our hypothesis that the chemistry used to conjugate the antigen to an SNA cannot be chosen based simply on synthetic convenience, but instead the choice should be made by considering its impact on the immunogenicity of the delivered antigen. This knowledge underscores the impact of conjugation chemistry on immunostimulatory nanotherapeutic constructs and will inform the design of future vaccines, beyond those based upon the SNA architecture.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award U54CA199091. S.W. was supported by a fellowship associated with the Chemistry of Life Processes Predoctoral Training Program T32GM105538 at Northwestern University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors also acknowledge support from the Vannevar Bush Faculty Fellowship program sponsored by the Basic Research Office of the Assistant Secretary of Defense for Research and Engineering and funded by the Office of Naval Research grant N00014-15-1-0043. The project described was also supported by the IDP Sherman Fairchild Foundation through the Robert H. Lurie Comprehensive Cancer Center, and the Prostate Cancer Foundation and Movember Foundation under award 17CHAL08
Footnotes
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b12579.
Experimental details, oligonucleotide sequences, and characterization data (PDF)
References
- 1.Zhang G, Yang Z, Lu W, Zhang R, Huang Q, Tian M, Li L, Liang D, Li C. Biomaterials. 2009;30(10):1928–36. doi: 10.1016/j.biomaterials.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu H, Moynihan KD, Zheng Y, Szeto GL, Li AV, Huang B, Van Egeren DS, Park C, Irvine DJ. Nature. 2014;507(7493):519–22. doi: 10.1038/nature12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Townson JL, Lin YS, Agola JO, Carnes EC, Leong HS, Lewis JD, Haynes CL, Brinker CJ. J Am Chem Soc. 2013;135(43):16030–3. doi: 10.1021/ja4082414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HK, Thompson DH, Jang HS, Chung YJ, Van den Bossche J. ACS Appl Mater Interfaces. 2013;5(12):5648–58. doi: 10.1021/am400977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunn SS, Tian S, Blake S, Wang J, Galloway AL, Murphy A, Pohlhaus PD, Rolland JP, Napier ME, DeSimone JM. J Am Chem Soc. 2012;134(17):7423–30. doi: 10.1021/ja300174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang RH, Hu CM, Luk BT, Gao W, Copp JA, Tai Y, O’Connor DE, Zhang L. Nano Lett. 2014;14(4):2181–8. doi: 10.1021/nl500618u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perrault SD, Walkey C, Jennings T, Fischer HC, Chan WC. Nano Lett. 2009;9(5):1909–15. doi: 10.1021/nl900031y. [DOI] [PubMed] [Google Scholar]
- 8.Li J, Wang W, He Y, Li Y, Yan EZ, Zhang K, Irvine DJ, Hammond PT. ACS Nano. 2017;11(3):2531–2544. doi: 10.1021/acsnano.6b08447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong Y, Dorkin JR, Wang W, Chang PH, Webber MJ, Tang BC, Yang J, Abutbul-Ionita I, Danino D, DeRosa F, Heartlein M, Langer R, Anderson DG. Nano Lett. 2016;16(2):842–8. doi: 10.1021/acs.nanolett.5b02428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Lin P, Perrett I, Lin J, Liao YP, Chang CH, Jiang J, Wu N, Donahue T, Wainberg Z, Nel AE, Meng H. J Clin Invest. 2017;127(5):2007–2018. doi: 10.1172/JCI92284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taton TA, Mirkin CA, Letsinger RL. Science. 2000;289(5485):1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 12.Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Science. 2006;312(5776):1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- 13.Radovic-Moreno AF, Chernyak N, Mader CC, Nallagatla S, Kang RS, Hao LL, Walker DA, Halo TL, Merkel TJ, Rische CH, Anantatmula S, Burkhart M, Mirkin CA, Gryaznov SM. Proc Natl Acad Sci U S A. 2015;112(13):3892–3897. doi: 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirosue S, Kourtis IC, van der Vlies AJ, Hubbell JA, Swartz MA. Vaccine. 2010;28(50):7897–7906. doi: 10.1016/j.vaccine.2010.09.077. [DOI] [PubMed] [Google Scholar]
- 15.Suma T, Cui JW, Mullner M, Fu SW, Tran J, Noi KF, Ju Y, Caruso F. J Am Chem Soc. 2017;139(11):4009–4018. doi: 10.1021/jacs.6b11302. [DOI] [PubMed] [Google Scholar]
- 16.Xu J, Wang JJ, Luft JC, Tian SM, Owens G, Pandya AA, Berglund P, Pohlhaus P, Maynor BW, Smith J, Hubby B, Napier ME, DeSimone JM. J Am Chem Soc. 2012;134(21):8774–8777. doi: 10.1021/ja302363r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker AB, Schreurs MW, Tafazzul G, de Boer AJ, Kawakami Y, Adema GJ, Figdor CG. Int J Cancer. 1995;62(1):97–102. doi: 10.1002/ijc.2910620118. [DOI] [PubMed] [Google Scholar]
- 18.de Titta A, Ballester M, Julier Z, Nembrini C, Jeanbart L, van der Vlies AJ, Swartz MA, Hubbell JA. Proc Natl Acad Sci U S A. 2013;110(49):19902–7. doi: 10.1073/pnas.1313152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klinman DM, Barnhart KM, Conover J. Vaccine. 1999;17(1):19–25. doi: 10.1016/s0264-410x(98)00151-0. [DOI] [PubMed] [Google Scholar]
- 20.Chen JW, Zhao MK, Feng FD, Sizovs A, Wang J. J Am Chem Soc. 2013;135(30):10938–10941. doi: 10.1021/ja405261u. [DOI] [PubMed] [Google Scholar]
- 21.Dunn SS, Tian SM, Blake S, Wang J, Galloway AL, Murphy A, Pohlhaus PD, Rolland JP, Napier ME, DeSimone JM. J Am Chem Soc. 2012;134(17):7423–7430. doi: 10.1021/ja300174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutta K, Hu D, Zhao B, Ribbe AE, Zhuang JM, Thayumanavan S. J Am Chem Soc. 2017;139(16):5676–5679. doi: 10.1021/jacs.7b01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito G, Swanson JA, Lee KD. Adv Drug Delivery Rev. 2003;55(2):199–215. doi: 10.1016/s0169-409x(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg MS. Cell. 2015;161(2):201–204. doi: 10.1016/j.cell.2015.03.037. [DOI] [PubMed] [Google Scholar]
- 25.Steinman RM, Cohn ZA. J Exp Med. 1973;137(5):1142–1162. doi: 10.1084/jem.137.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banchereau J, Steinman RM. Nature. 1998;392(6673):245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 27.Dominguez D, Ye C, Geng Z, Chen S, Fan J, Qin L, Long A, Wang L, Zhang Z, Zhang Y, Fang D, Kuzel TM, Zhang B. J Immunol. 2017;198(3):1365–1375. doi: 10.4049/jimmunol.1501399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qin L, Dominguez D, Chen S, Fan J, Long A, Zhang M, Fang D, Zhang Y, Kuzel TM, Zhang B. Oncotarget. 2016;7(38):61069–61080. doi: 10.18632/oncotarget.11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.