Abstract
Objectives
To evaluate associations between aromatase inhibitor (AI) treatment and let-7 family microRNA expression in endometriosis.
Design
In vitro study using Ishikawa cells and human endometrial stromal cells (HESC) obtained from patients with endometriosis
Setting
University research center.
Patients
Women undergoing laparoscopic surgery for endometriosis
Interventions
HESCs and Ishikawa cells treated with various letrozol concentrations and transfected with a mimic of let-7 subtypes of interest
Main Outcome Measures
microRNAs let7a-f and aromatase expression were evaluated. Migration potential after transfection with a let-7f mimic were analyzed.
Results
After letrozole treatment for 48 hours, all let-7 subtypes showed a trend toward increased expression in a dose dependent manner in Ishikawa cells, and significant differences were found in let-7b and let-7f between controls and the 20 μmol/L treated groups. Further, let-7f showed significant differences between control and 1.0 μmol/L treatment group, a typical therapeutic level, in HESCs. Transfection of a let-7f mimic decreased aromatase expression in both Ishikawa cells and HESC, and led to a significant decrease in number of migrating cells in both cell types.
Conclusions
AI treatment significantly increased expression of let-7f in Ishikawa cells and HESCs from patient with endometriosis; increased lef-7f expression effectively reduced the migration of endometrial cells. Modulation of miRNAs involved in the pathogenesis of endometriosis may have therapeutic potential for endometriosis.
Keywords: Aromatase inhibitor, endometriosis, let-7f, microRNA
Introduction
Endometriosis, defined as the proliferation of endometrial tissue outside the uterine cavity, is one of the most common gynecologic disorders affecting in approximately 10% of all reproductive-aged women, and 20–50% of women with chronic pelvic pain and/or infertility (1). Despite the high prevalence of the disease, optimal treatment of the disease still remains challenging due to the complexity of the pathogenesis and diversity of symptoms. Based on the molecular association of increased aromatase P450 expressions and estradiol production in endometriosis, aromatase inhibitors (AIs) have been suggested as an appealing concept for this estrogen-dependent chronic inflammatory disease (2, 3). Notably, several studies have demonstrated that the combination of AIs with other conventional therapy can be used to control endometriosis-associated pain and pain recurrence in premenopausal women, particularly those with pain due to rectovaginal endometriosis refractory to other medical or surgical treatment (2, 3). However, the molecular mechanisms of AI on endometriosis have not been fully clarified.
Recently, it has been shown that AI treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting aromatase gene (CYP19A1)(4). MicroRNAs (miRNAs) are a family of endogenous, small (approximately 22 nucleotides in length), noncoding, functional RNAs which regulate gene expression either by translational repression or degradation of messenger RNA transcripts after targeting the 3′ UTR. They play a pivotal role in the development and cellular homeostasis through their control of diverse biological processes and increasing evidence suggests that aberrant miRNA expression is associated with various human diseases including endometriosis (5–8). Among several miRNAs that are dysregulated in endometriosis, an association of miRNA let-7 and endometriosis has been suggested recently. In some women with severe endometriosis, a polymorphism of a let-7 miRNA binding site in the KRAS 3′ UTR leads to abnormal endometrial cell KRAS expression and that endometrial and circulating let-7 family members were expressed at lower levels in patients with endometriosis (7, 9). Based on these previous findings, we hypothesized a possible involvement of let-7 in endometriosis and in response to AI treatment. Therefore, the purpose of this study was to evaluate associations between aromatase inhibitor treatment and let-7 miRNAs expression in endometriosis and to elucidate whether modulation of miRNAs involved in the pathogenesis of endometriosis may have therapeutic potential for endometriosis.
Materials and Methods
Subjects and sample collection
The samples were collected from six patients who underwent surgery for diagnosis or treatment of endometriosis. The diagnosis of endometriosis was confirmed by histology in all women with the disease. The extent of endometriosis was determined using the American Society of Reproductive Medicine (ASRM) revised classification (10) and endometrial biopsies were obtained from women with moderate-to-severe disease (Stages III and IV) using a Pipelle catheter (Cooper Surgical, Trumbull, CT). The menstrual cycle stage was recorded for each patient as either proliferative phase (menstrual phase and until 14 days before the next menses) or secretory phase (1–13 days before the next menses) and 3 samples from each menstrual cycle were obtained. Exclusion criteria included use of hormonal medications or supplements within the 3 months before surgery, pregnancy, cancer, endometrial pathologies including polyps and submucosal/intramural fibroids. Written informed consent was obtained from all participants and approval was obtained from the Yale University School of Medicine Human Investigations Committee.
Culture of primary endometrial stromal cell and Ishikawa cell lines
Endometrium was finely minced and cells were dispersed by incubation in Hanks’ balanced salt solution containing HEPES (25mm), 1% penicillin/streptomycin, collagenase (1 mg/ml, 15 U/mg), and deoxyribonuclease (0.1 mg/ml, 1500 U/mg) for 60 min at 37°C with agitation and pipetting. The cells were pelleted; washed; suspended in Ham’s F12:DMEM (1:1) containing 10% fetal bovine serum, 1%penicillin/streptomycin, and 1% amphotericin B; passed through a 40-m cell strainer (Falcon, Franklin Lakes, NJ, USA); and plated into 75 cm2 Falcon tissue culture flasks (BD Biosciences, Franklin Lakes, NJ). Cultured HESC at 3–5 passages were used for further analysis. Ishikawa cells were maintained in MEM (Invitrogen, Carlsbad, CA) containing 2.0 mM l-glutamine and Earl’s salts, supplemented with 10% fetal bovine serum (FBS), 1% sodium pyruvate, and 1% penicillin/streptomycin.
Letrozole treatment
Letrozole was obtained from Ivy Fine Chemicals (Cherry Hill, NJ). Endometrial stromal cells from patients with endometriosis and Ishikawa cells were harvested from the culture flask using Trypsin/EDTA 0.05% and were plated in six-well plates in 200μℓ of media as follows; Ham’s F12:DMEM (1:1) containing 10% fetal bovine serum, 1%penicillin/streptomycin, and 1% amphotericin B for HESC and MEM (Invitrogen, Carlsbad, CA) containing 2.0 mM l-glutamine and Earl’s salts, supplemented with 10% fetal bovine serum (FBS), 1% sodium pyruvate, and 1% penicillin/streptomycin for Ishikawa cell lines, at 37 °C in 95% air and 5% CO2 in a humidified environment. At 80% confluency, letrozole was added at concentrations of 0.0 μmol/L, 0.1 μmol/L, 1.0 μmol/L, 10 μmol/L or 20 μmol/L. Three wells for each sample were used, and all culture dishes were incubated in the presence of 5% CO2 at 37°C for another 48 hours as the half-life of letrozole is about 48 hours.
Cell Transfection
Cells were cultured to 80% to 90% confluence after being seeded into 6-well plates and were transfected with hsa-mir-7f mimic, a chemically synthesized, double-stranded RNA which mimics mature endogenous miRNA after transfection into cells, or hsa-mir-negative control (Ambion, Inc., Austin, TX) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions at a final concentration of 50 nM. The transfected cells were harvested at 48 hours after transfection.
RNA extraction and Quantitative real-time PCR
To assess aromatase mRNA levels, total RNA was extracted from cultured cells using the Qiagen RNeasy isolation kit (Qiagen). All samples were treated with RNase-free DNase (Ambion, Austin, TX) to remove the possibility of genomic DNA contamination. RNA samples were analysed by a NanoDrop ND 2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). mRNA was reverse-transcribed into cDNA using the iScript cDNA Synthesis Kit at 46°C for 40 min in a reaction mixture of 20ml (Bio-Rad Laboratories, Hercules, CA). The resultant cDNA mixtures were stored at −20°C. Gene transcripts were amplified by real-time PCR using the Bio-Rad iCycler iQ system (Bio-Rad Laboratories). Real-time PCR was performed using the iQ SYBR Green supermix kit (Bio-Rad Laboratories). Reaction mixture included cDNA template (1 μg), forward and reverse primers, ribonuclease-free water and the iQSYBR Green supermix, for a final reaction volume of 25 μℓ. The thermal cycling conditions were initiated by uracil-N-glycosylase activation at 50 C for 2 min and initial denaturation at 95 °C for 10 min, and then 40 cycles at 95 °C for 15 sec, and annealing at 60 °C for 20 sec. Melting analysis was performed by heating the reaction mixture from 74 °C to 99 °Cat a rate of 0.2 °C/sec. Threshold cycle (Ct) and melting curves were acquired by using the quantitation and melting curve program of the Bio-Rad iCycler iQ system (Bio-Rad Laboratories). Only data with clear and single melting peaks were taken for further data analysis. Each reaction was performed in triplicate. The mRNA level of each sample for aromatase was normalized to GAPDH expression. Relative mRNA level was presented using the formula 2−ΔCT.
To assess miRNA expression levels, RNA was extracted from cultured cells using the miRVana™ RNA Isolation Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s specifications, and eluted with 50 μℓ of nuclease-free water. The yield of RNA was determined using a NanoDrop ND-2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). For miRNA detection, we employed the Poly (A) RT-PCR method using Invitrogen NCode miRNA First-Strand cDNA Synthesis MIRC-50 kit (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instructions. Total RNA (100 ng) from each sample was reverse transcribed and miRNAs were quantified using the iQ SYBR Green supermix kit (Bio-Rad Laboratories) with the specific forward primers to let-7a–f, and the universal reverse primer complementary to the anchor primer. Reaction mixture included 2.5 μℓ of cDNA, 12.5 μℓ of iQSYBR Green Supermix, 1.0 μℓ of forward primer, 1 μℓ of Universal qPCR Primer, and 8 μℓ RNase-free water for a final reaction volume of 25 μℓ. The thermal cycling conditions were initiated by uracil-N glycosylase activation at 50 °C for 2 min and initial denaturation at 95 °C for 10 min, and then 40 cycles at 95°C for 15 sec, and annealing at 60°C for 20 sec. Threshold cycle (Ct) and melting curves were acquired by using the quantitation and melting curve program of the Bio-Rad iCycleriQ system (Bio-Rad Laboratories). Anchor RT primer was used as the template for negative control and U6 small nuclear RNA was used as a control to determine relative miRNA expression (11, 12). Relative mRNA level was determined using the formula 2−ΔCT.
Primers for let-7a–f, aromatase, GAPDH and the U6 genes were obtained from the W. M. Keck Oligonucleotide Synthesis Facility (Yale University): let-7a forward, TGAGGTAGTAGGTTGTATAGTT; let-7b forward, TGAGGTAGTAGGTTGTGTGGTT; let-7c forward, TGAGGTAGTAGGTTGTATGGTT; let-7d forward, AGAGGTAGTAGGTTGCATAGTT; let-7e forward, TGAGGTAGGAGGTTGTATAGTT; let-7f forward, TGAGGTAGTAGATTGTATAGTT; U6 forward, CTCGCTTCGGCAGCACA, reverse, AACGCTTCACGAATTTGCGT; aromatase forward, TGGAATTATGAGGGCACATCC; aromatase reverse, AGGAATCTGCCGTGGGAGA; GAPDH forward, GAAGGTGAAGGTCGGAGTC; GAPDH reverse, GAAGATGGTGATGGGATTTC.
Western blot analysis
Cultured cells were lysed in Cell Lysis Buffer (Cell Signaling Technologies, Boston, MA) and centrifuged at 12,000 rpm for 2 min. The protein content was quantified by the BCA assay method by using a protein assay kit (Bio-Rad Laboratories, Richmond, VA.). Twenty micrograms of total protein were mixed 1:1 with Laemmli sample buffer (Bio-Rad Laboratories, Inc.) and heated at 95 C for 5 min. Proteins were resolved under reducing conditions on 4–10% gradient sodium dodecyl sulfate-polyacrylamide gel in 3 [Nmorpholino] propanesulfonic acid buffer system (Invitrogen) and transferred to a nitrocellulose membrane by using a Transblot apparatus (Bio-Rad) at 100 V for 2 h at 4 °C. Membranes were blocked at room temperature for 1 h with 5% BSA Tris-buffered saline [10 mM Tris-HCl (pH 7.4), 0.5 M NaCl] plus Tween 20 [0.2% (vol/vol)] (TBST). After three washes for 5 min each with TBST, membranes were incubated overnight at 4 °C with primary antibody. The rabbit polyclonal anti-aromatase antibody (ab35604, Abcam, Cambridge, MA) was used at 1:500 dilution. After this incubation, membranes were washed three times as before and then incubated at room temperature for 1 h with the appropriate horseradish peroxidase-conjugated antibody at 1:10000 dilution. After three washes for 5 min each with TBST, proteins were detected using enhanced chemiluminescence (PerkinElmer,Waltham, MA). GAPDH antibody (14C10; Cell Signaling, Beverly, MA) was used as internal loading control at 1:5000 dilution following the same procedure explained above. The appropriate horseradish peroxidase-conjugated antibody was used at 1:2000.
Migration assay
The migration assay for transfected cultured cells was carried out using 8-μm pore size polycarbonate membrane (Millipore, USA) with the 24-well plates. Freshly trypsinized and washed cells were suspended in serum free media and 200 μl of cells (5 × 104 cells)/well were placed in the top chamber of each insert. 600 μl of medium containing 10% FBS was added into the lower chambers. After incubating for 72 hr at 37°C in a 5% CO2 humidified incubator, cells were fixed and stained with hematoxylin. The cells in the inner chamber were removed with a cotton swab and the cells attached to the bottom side of the membrane were counted and imaged under an inverted microscope (Olympus Corp. Tokyo, Japan) at ×200 magnification over ten random fields in each well.
Treatment in vitro with Estradiol (E2) and letrozole
After starvation for 12 h, cells were seeded into 6-well plates in DMEM-F12 (HESCs) or MEM medium (Ishikawa cells) without phenol red, containing 10% charcoal/dextran-stripped FBS and 1% penicillin/streptomycin. When cells were grown to 80% confluence, cells were treated with E2 (Sigma-Aldrich) at doses of 10−5–10−10 M or vehicle for 48 h. To examine the effect of letrozole on let-7f expression in the presence of E2, cells were treated simultaneously with letrozole at concentrations as determined above and demonstrated in figure 1 (20.0 μmol/L for Ishikawa cells, and 1.0 μmol for HESCs) and RNA was extracted as previously described.
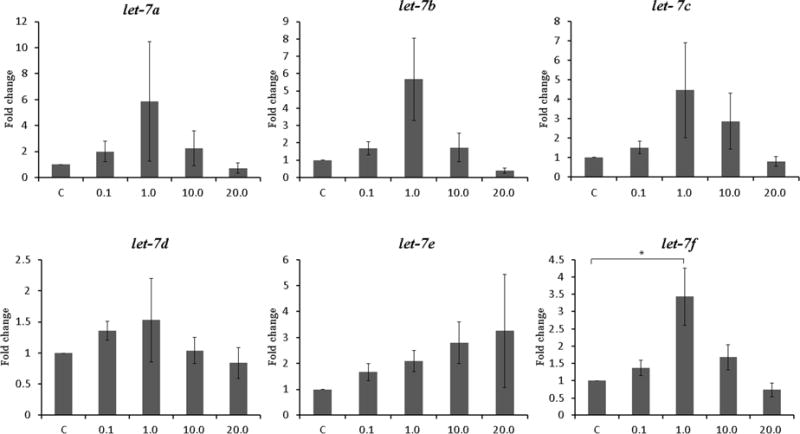
Figure 1. Expression of let-7 family after treatment with letrozole for 48 hours. The concentration of letrozole is shown on the X axis in μmol/L. Data are expressed as mean ± S.E.M. *P < 0.05, **P<0.01.
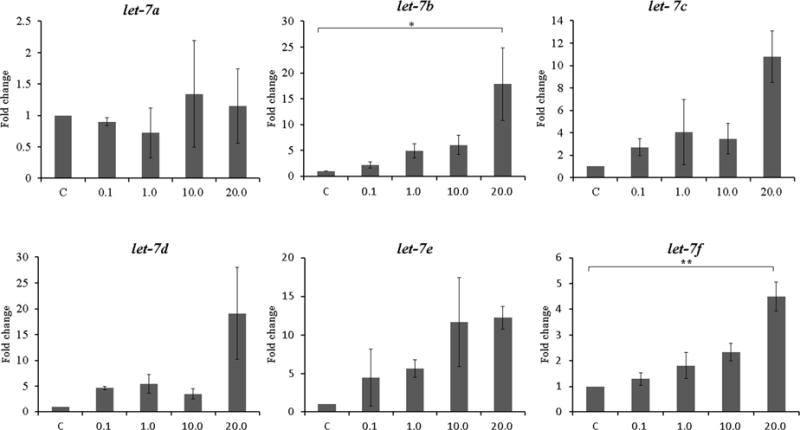
A. Ishikawa cell lines (N=3)
B. Endometrial stromal cells from subjects with endometriosis (N=6)
Statistical Analysis
Data were presented as mean ± SEM. All of the data from in vitro experiments were normally distributed as assessed by Kolmogorov-Smirnov test or Shapiro-Wilk test and were examined with one-way ANOVA, followed by Tukey test for multiple comparisons where appropriate. Student’s t-test was used to determine the significance of difference in aromatase mRNA expressions after transfection and number of migrating cells per field in migration assay. SPSS 16.0 (SPSS Inc, Chicago, IL, USA) was used for statistical analysis and a P<0.05 was considered statistically significant.
Results
Letrozole treatment increases expression of let-7 in Ishikawa cell lines and endometrial stromal cells from patients with endometriosis
To evaluate the association between aromatase inhibitor treatment and the expression of let-7, we treated Ishikawa cell lines and endometrial stromal cells from patients with endometriosis with several concentrations of letrozole for 48 hours. The effects of aromatase inhibitor treatment on let-7 expression in Ishikawa cells and endometrial stromal cells are shown in Figure 1. In Ishikawa cell, let-7 subtypes showed a trend toward increased expression in a dose dependent manner, and significant differences were noted between the control and the 20.0 μmol/L treated group in let-7b (17.84-fold increase, P=0.034) and let-7f (4.49-fold increase, P=0.001) (Figure 1A). In endometrial stromal cells from patients with endometriosis, significant differences between the control and letrozole treated group were noted in let-7f expression. Let-7f expression was significantly increased compared to the control at letrozole concentration of 1.0 μmol/L, a typical therapeutic level (3.43-fold increase, P=0.026). Since significant expression changes in response to letrozole treatment in both Ishikawa cell lines and endometrial stromal cells were found in let-7f, further analysis were carried out using this let-7 subtype.
Let-7f transfection decreases aromatase expression in Ishikawa cells and endometrial stromal cells
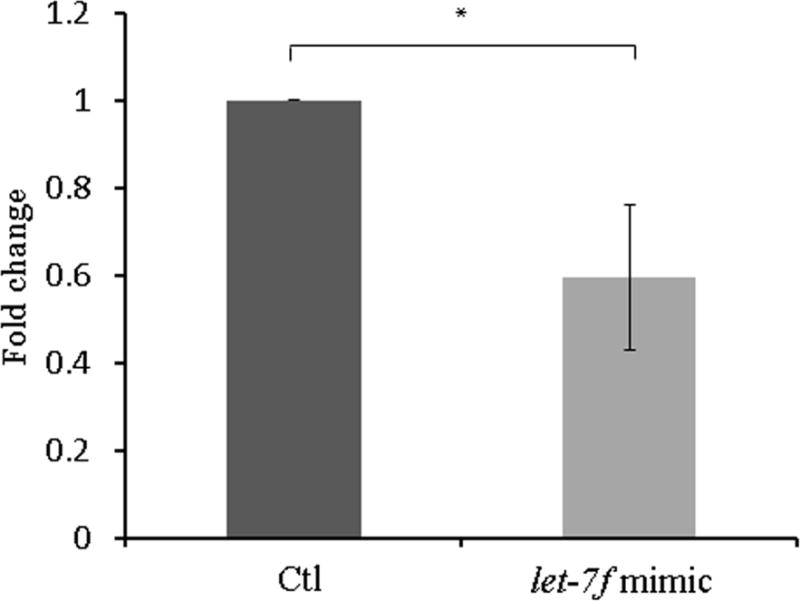
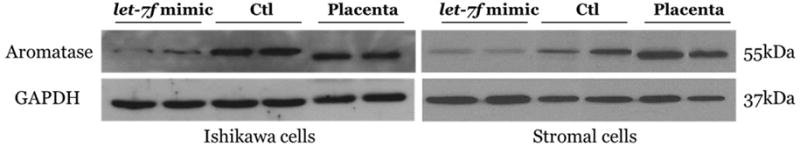
To evaluate transfection efficacy, let-7f expression was measured 48 hours after transfection with a let-7f mimic in Ishikawa cell lines and endometrial stromal cells. Treatment with the let-7f mimic resulted in an approximately 100-fold increase in let-7f expression compared with has-mir-negative control treatment (data not shown). Since let-7f has been shown to target the aromatase gene (4), we evaluated aromatase expression in Ishikawa cell lines and endometrial stromal cells from the patients with endometriosis after let-7f mimic transfection. After 48 hours of let-7f mimic treatment, aromatase mRNA expression was significantly decreased in both Ishikawa cell lines and endometrial stromal cells (0.36-fold increase, P=0.002 and 0.61-fold increase, P=0.025, respectively) (Figure 2A, B). Western blot analysis showed aromatase protein expression (molecular mass 55 kDa) and corresponding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) loading control (molecular mass 42 kDa) and confirmed decreased aromatase protein expressions by let-7f mimic treatment (Figure 2C).
Figure 2. Aromatase mRNA expression and western blot analysis after treatment with the let-7f mimic and has-mir-negative control for 48 hours. Data are expressed as mean ± S.E.M. *P < 0.05, **P<0.01.
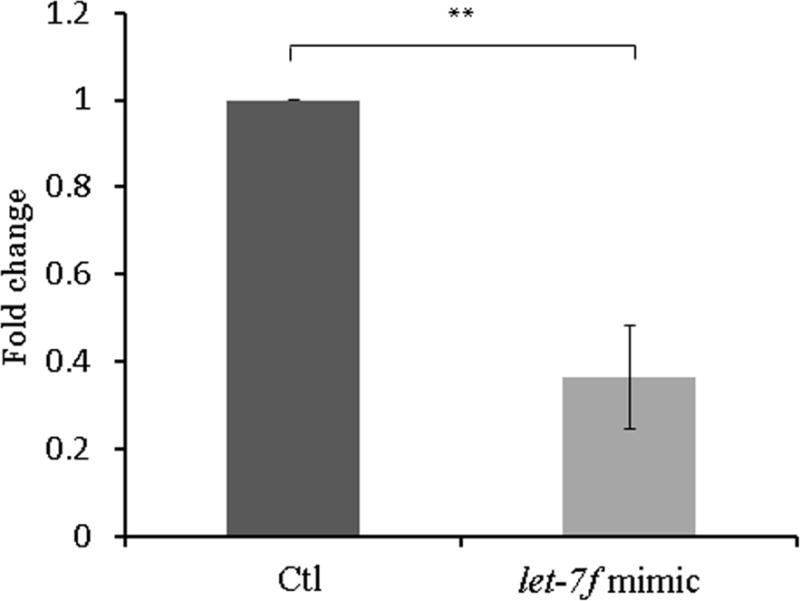
A. Ishikawa cell lines (N=4)
B. Endometrial stromal cells from subjects with endometriosis (N=4)
C. Western blot analysis of aromatase expressions treated with let-7f mimic or has-mir-negative control for 48 hours.
Let-7f decreases cell migration in Ishikawa cell lines and endometrial stromal cells
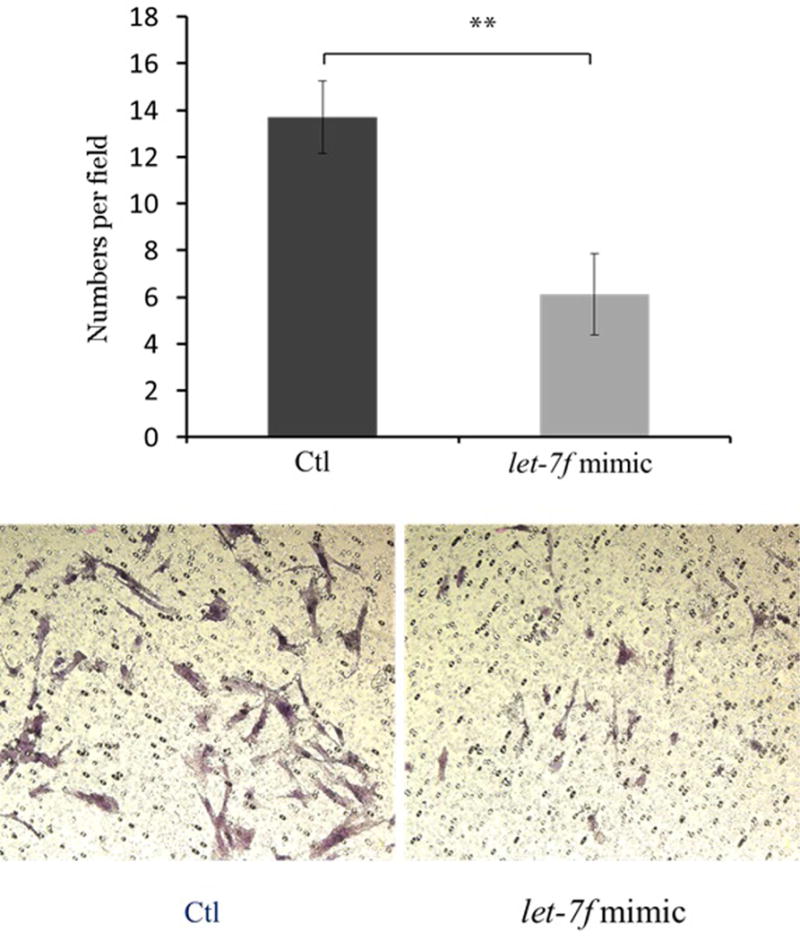
To determine the effects of let-7f on cell migration in endometriosis, Ishikawa cell lines and endometrial stromal cells from the patients with endometriosis were transfected with the let-7f mimic. Cell migration was evaluated 72 hours after transfection using Millicell cell culture inserts. Migrating cell numbers were significantly decreased after let-7f treatment of Ishikawa cells compared to the control (4.3 ± 0.9 vs. 7.5 ± 0.7, P=0.032) (Figure 3A). In endometrial stromal cells from women with endometriosis, the effects on cell migration by let-7f treatment was even more pronounced, showing a significant decrease in number of migrating cells after 72 hours of let-7f transfection (6.1 ± 1.74 vs. 13.7 ± 1.56, P=0.009) (Figure 3B).
Figure 3. Effects on let-7f on cell migration in Ishikawa cell lines and endometrial stromal cells from patients with endometriosis using Millicell cell culture insert system (top) and representative fields of migration cells on the membrane; magnification of ×200 (bottom). Data are expressed as mean ± S.E.M. *P < 0.05, **P<0.01.
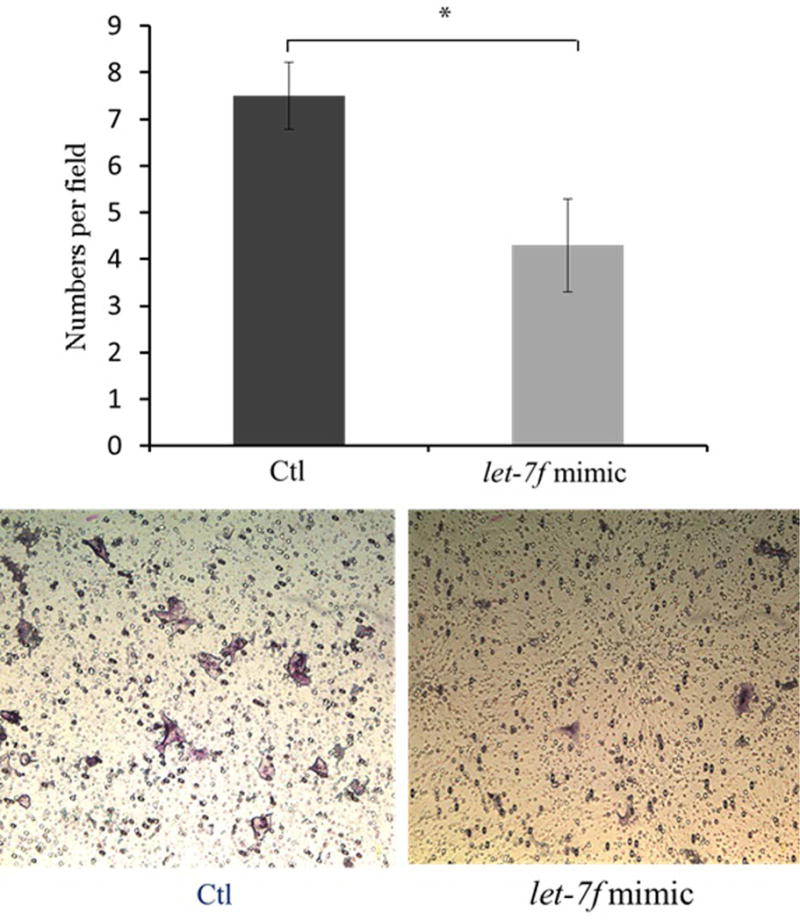
A. Ishikawa cell lines (N=5)
B. Endometrial stromal cells from subjects with endometriosis (N=5)
Estradiol fails to rescue the effect of letozole on let-7f expression
To evaluate whether the effect of letrozole on let-7f is reversed by estrogens, we treated the Ishikawa cell lines and endometrial stromal cells from women with endometriosis with several physiologic and supraphysiologic concentrations of estradiol (Figure 4). In Ishikawa cell lines and endometrial stromal cells with endometriosis, there were no significant changes in let-7f expression between the letrozole treatment group and the letrozole and estradiol treated groups, indicating that the effect of letrozole on let-7f expression is not reversed by estrogens at even greater than physiological concentrations.
Figure 4. Let-7f expression after treatment with letrozole and estradiol for 48 hours. Data are expressed as mean ± S.E.M. *P < 0.05. LZ: letrozole, E: estradiol.
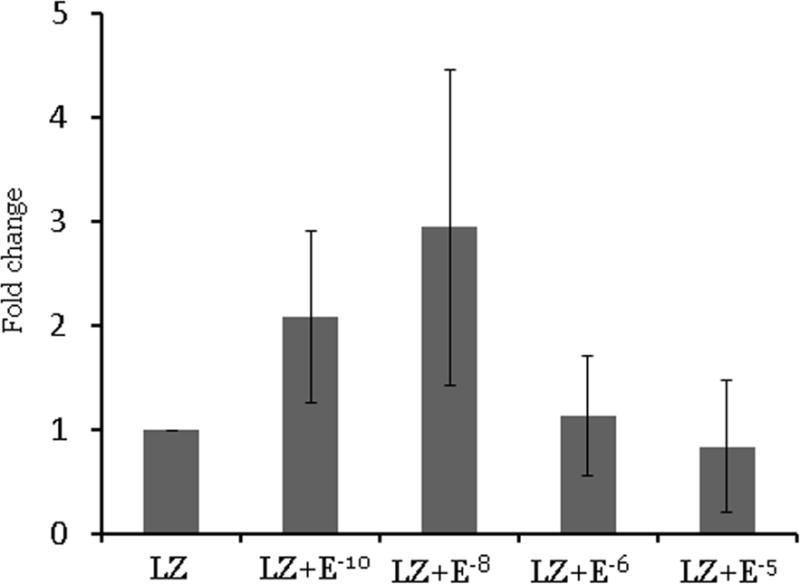
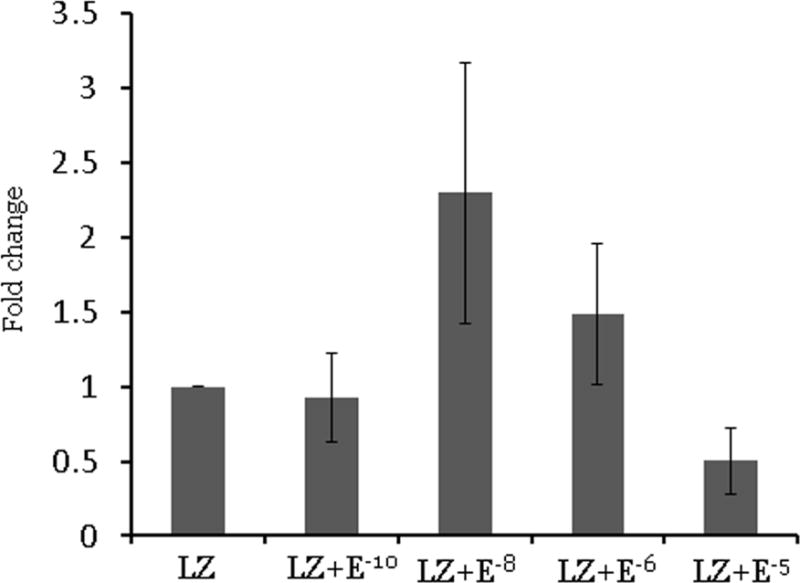
A. Ishikawa cell lines (N=3)
B. Endometrial stromal cells from subjects with endometriosis (N=3)
Discussion
In the present study we evaluated expression of the let-7 microRNA family in women with endometriosis after AI treatment; our results showed that let-7f expression was upregulated in both Ishikawa cells and endometrial stromal cells from women with endometriosis. Upregulation of lef-7f expression effectively reduced the migration potential of endometrial cell in vitro, indicating that AI may regulate let-7f expression and let-7f induced cell migration in endometriosis. After let-7f transfection, aromatase expression in these cells was down-regulated. Let7f both decreased cell migration and decreased estrogen production through inhibition of aromatase expression. Aromatase inhibitors such as letrozol may have a duel mechanism in endometriosis inhibition mediated by let7f microRNAs.
To our knowledge, this is the first study to evaluate possible association between AI and miRNAs in endometriosis. Aromatase p450 is the enzyme responsible for ovarian estrogen biosynthesis and a body of evidence indicate that aromatase activity in eutopic endometrium and ectopic endometriotic lesions play a pivotal role in the development of endometriosis (13–15). Therefore, use of AI in endometriosis has been suggested as rational approach to inhibit estrogen synthesis and to provide effective treatment for this disease entity. However, there is insufficient data concerning the detailed molecular mechanisms of AI in endometriosis. Since dysregulation of miRNAs has been recently reported to play a key role in various therapeutic responses (16), and there is a growing body of evidence indicating possible involvement of miRNAs in the pathogenesis of endometriosis (5–9), we hypothesized that AI treatment may involve alteration of miRNAs in endometriosis. Potential involvement of miRNAs in the treatment of AI has been suggested in breast cancer cell lines. Mi-128b was up-regulated in letrozole-resistant breast cancer cell lines, and alterations in miRNA profile have been reported in breast cancer specimen treated by tamoxifene combined with exemestane (17, 18).
Let-7 was the first human miRNA discovered and one of its major functions is to promote differentiation of cells (19). However, evidence indicates that the let-7 miRNA family members also function as tumor suppressors (20). Reduced expression of multiple let-7 members have been found to be associated with human malignancies and their restoration to normal levels has been reported to suppress cancer growth (21, 22). Interestingly, an association between decreased expression of several let-7 subtypes and endometriosis has been reported recently (10,12). Let-7b expression, associated with cyclin D1, an estrogen-regulated gene involved in cellular proliferation, was significantly lower in patients with endometriosis than controls, and several let-7 subtypes in sera and HESC from patients with endometriosis were also shown to be down-regulated (7, 9). Our results indicate that among let-7 subtypes, let-7f is associated with letrozole treatment in Ishikawa cells and in HESC. These results are in close agreement with recent findings showing that let-7f, which directly targeted the aromatase gene, was restored by AI treatment and suggested that AI may exert tumor-suppressing effects upon breast cancer cells by suppressing aromatase gene expression via restoration of let-7f (4). Interestingly, our findings indicate that the effects of AI on let-7f were optimal at lower doses of AI than required to affect other let7 family members, especially in endometrial stromal cells. The concentration of letrozole that led to a significant increase of let-7f expressions was at 1.0 μmol/L, which is similar to typical therapeutic level. In contrast, much higher concentrations were required to induce significant miRNA expression changes in Ishikawa cell lines, suggesting different dose of AI treatment may be required to restore miRNAs according to cell type. Furthermore, overexpression of let-7f induced inhibition of cell migration in both Ishikawa cell lines and endometrial stromal cells. These results closely resemble previous studies showing that restoration and overexpression of let-7f led to significant decrement in migration of breast cancer cell lines and gastric cancer cell (4, 22) and further support the possible therapeutic effects of AI through regulation of let-7f in endometriosis.
Let-7f also decreased the expression of aromatase. Functionally let-7f acts as an endogenous aromatase inhibitor. This function of let-7f may allow development of novel substitutes for aromatase inhibitors. Further, the effect of letrozol on let-7f was not lowered by estradiol administration. Estradiol treatment did not rescue expression of let-7. While estradiol is required for the progression of endometriosis, letrozole may function to suppress let-f7 by an estrogen independent mechanism. The effect of letrozole may be mediated in part through a mechanism other than inhibition of aromatase and diminished estradiol production. Future investigation will explore the possibility of a novel mechanism of letrozole action.
One potential limitations of the study is that we did not test the effects of AI on let-7f expression in endometriosis cells obtained directly from endometriotic lesions. Therefore, further comparison of the effects of AI on let-7f expression in cells obtained directly from the endometriosis lesions will provide more conclusive results.
In conclusion, we have demonstrated that aromatase inhibitor treatment at a typical therapeutic level significantly increased expression of let-7f in Ishikawa cells and HESCs from patients with endometriosis, and that overexpression of lef-7f expression effectively reduced the migration potential of endometrial cells. These results suggest that alteration of miRNA expressions could be one of the molecular mechanisms involved in the therapeutic effects of AI treatment in endometriosis and that modulation of miRNAs involved in the pathogenesis of endometriosis may provide new therapeutic targets for endometriosis treatment.
Capsule.
Aromatase inhibitor treatment of cultured endometrial cells from the patients with endometriosis significantly increased expression of let-7f and effectively reduced the migration of endometrial cells in vitro.
Acknowledgments
Funding: This study was supported by National Institute of Health Grant U54 HD052668.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Statement: The authors have nothing to disclose.
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Giudice LC, Kao LC. Endometriosis. Lancet. 2004;364:1789–99. doi: 10.1016/S0140-6736(04)17403-5. [DOI] [PubMed] [Google Scholar]
- 2.Pavone ME, Bulun SE. Aromatase inhibitors for the treatment of endometriosis. Fertil Steril. 2012;98:1370–9. doi: 10.1016/j.fertnstert.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abu Hashim H. Potential role of aromatase inhibitors in the treatment of endometriosis. Int J Womens Health. 2014;6:671–80. doi: 10.2147/IJWH.S34684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shibahara Y, Miki Y, Onodera Y, Hata S, Chan MS, Yiu CC, et al. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. J Pathol. 2012;227:357–66. doi: 10.1002/path.4019. [DOI] [PubMed] [Google Scholar]
- 5.Teague EM, Print CG, Hull ML. The role of microRNAs in endometriosis and associated reproductive conditions. Hum Reprod Update. 2010;16:142–65. doi: 10.1093/humupd/dmp034. [DOI] [PubMed] [Google Scholar]
- 6.Petracco R, Grechukhina O, Popkhadze S, Massasa E, Zhou Y, Taylor HS. MicroRNA 135 regulates HOXA10 expression in endometriosis. J Clin Endocrinol Metab. 2011;96:E1925–33. doi: 10.1210/jc.2011-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho S, Mutlu L, Grechukhina O, Taylor HS. Circulating microRNAs as potential biomarkers for endometriosis. Fertil Steril. 2015;103:1252–60 e1. doi: 10.1016/j.fertnstert.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu CY, Hsieh TH, Tsai CF, Tsai HP, Chen HS, Chang Y, et al. miRNA-199a-5p regulates VEGFA in endometrial mesenchymal stem cells and contributes to the pathogenesis of endometriosis. J Pathol. 2014;232:330–43. doi: 10.1002/path.4295. [DOI] [PubMed] [Google Scholar]
- 9.Grechukhina O, Petracco R, Popkhadze S, Massasa E, Paranjape T, Chan E, et al. A polymorphism in a let-7 microRNA binding site of KRAS in women with endometriosis. EMBO Mol Med. 2012;4:206–17. doi: 10.1002/emmm.201100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.American Society for Reproductive Medicine. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril. 1997;67:817–21. doi: 10.1016/s0015-0282(97)81391-x. [DOI] [PubMed] [Google Scholar]
- 11.Kuwabara Y, Ono K, Horie T, Nishi H, Nagao K, Kinoshita M, et al. Increased microRNA-1 and microRNA-133a levels in serum of patients with cardiovascular disease indicate myocardial damage. Circ Cardiovasc Genet. 2011;4:446–54. doi: 10.1161/CIRCGENETICS.110.958975. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–8. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 13.Maia H, Jr, Haddad C, Coelho G, Casoy J. Role of inflammation and aromatase expression in the eutopic endometrium and its relationship with the development of endometriosis. Womens Health (Lond Engl) 2012;8:647–58. doi: 10.2217/whe.12.52. [DOI] [PubMed] [Google Scholar]
- 14.Bulun SE. Endometriosis. N Engl J Med. 2009;360:268–79. doi: 10.1056/NEJMra0804690. [DOI] [PubMed] [Google Scholar]
- 15.Bulun SE, Fang Z, Imir G, Gurates B, Tamura M, Yilmaz B, et al. Aromatase and endometriosis. Semin Reprod Med. 2004;22:45–50. doi: 10.1055/s-2004-823026. [DOI] [PubMed] [Google Scholar]
- 16.Blower PE, Verducci JS, Lin S, Zhou J, Chung JH, Dai Z, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Mol Cancer Ther. 2007;6:1483–91. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 17.Masri S, Liu Z, Phung S, Wang E, Yuan YC, Chen S. The role of microRNA-128a in regulating TGFbeta signaling in letrozole-resistant breast cancer cells. Breast Cancer Res Treat. 2010;124:89–99. doi: 10.1007/s10549-009-0716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masri S, Phung S, Wang X, Chen S. Molecular characterization of aromatase inhibitor-resistant, tamoxifen-resistant and LTEDaro cell lines. J Steroid Biochem Mol Biol. 2010;118:277–82. doi: 10.1016/j.jsbmb.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–16. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Jerome T, Laurie P, Louis B, Pierre C. Enjoy the Silence: The Story of let-7 MicroRNA and Cancer. Curr Genomics. 2007;8:229–33. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang S, He L, Zhao X, Miao Y, Gu Y, Guo C, et al. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS One. 2011;6:e18409. doi: 10.1371/journal.pone.0018409. [DOI] [PMC free article] [PubMed] [Google Scholar]


