Abstract
In several organ systems the transitional zone between different types of epithelia is a hotspot for pre-neoplastic metaplasia and malignancy1–3. However, the cell-of-origin for the metaplastic epithelium and subsequent malignancy, remains obscure1–3. In the case of Barrett’s oesophagus (BE), intestinal metaplasia occurs at the gastro-oesophageal junction, where stratified squamous epithelium transitions into simple columnar cells4. Based on different experimental models, several alternative cell types have been proposed as the source of the metaplasia, but in all cases the evidence is inconclusive and no model completely mimics BE with the presence of intestinal goblet cells5–8. Here, we describe a novel transitional columnar epithelium with distinct basal progenitor cells (p63+ KRT5+ KRT7+) in the squamous-columnar junction (SCJ) in the upper gastrointestinal tract of the mouse. We use multiple models and lineage tracing strategies to show that this unique SCJ basal cell population serves as a source of progenitors for the transitional epithelium. Moreover, upon ectopic expression of CDX2 these transitional basal progenitors differentiate into intestinal-like epithelium including goblet cells, thus reproducing Barrett’s metaplasia. A similar transitional columnar epithelium is present at the transitional zones of other mouse tissues, including the anorectal junction, and, importantly, at the gastro-oesophageal junction in the human gut. Acid reflux-induced oesophagitis and the multilayered epithelium (MLE) believed to be a precursor of BE are both characterized by the expansion of the transitional basal progenitor cells. Taken together our findings reveal the presence of a previously unidentified transitional zone in the epithelium of the upper gastrointestinal tract and provide evidence that the p63+ KRT7+ basal cells in this zone are the cell-of-origin for MLE and BE.
Barrett’s oesophagus is the precursor lesion of oesophageal adenocarcinoma which has registered an approximately 800% increase in incidence over the past four decades9. Histologically, BE is characterized by the replacement of the stratified squamous epithelium of the distal oesophagus with simple columnar epithelial cells which often express characteristics of intestinal differentiation (e.g. CDX2+, Alcian blue+)3. During disease progression, MLE composed of cells with both squamous and columnar features has been considered as a precursor for BE10. However, the cell-of-origin for MLE and BE remains controversial. Five different models have been proposed to explain BE pathogenesis (Extended Data Fig. 1), each involving different cell types. These include the transdifferentiation of oesophageal squamous epithelium5,6 or circulating bone marrow cells11, and the expansion of submucosal glandular epithelium12, gastric cardia mucosa7 or “residual embryonic cells (RECs)” located at the SCJ8. Some of these studies present inconsistent evidence between in vitro and in vivo experiments5,6,13. More importantly, none of the experimental models recapitulates the pathological changes characteristically associated with BE in humans, e.g. presence of intestinal goblet cells5–8,11. We therefore considered the possibility that other cell types function as the cell-of-origin of MLE and BE.
We previously showed that SOX2 overexpression leads to basal cell hyperplasia in the squamous epithelium of Krt5-CreER;R26CAG-loxp-stop-loxp-Sox2-IRES-EGFP (hereafter referred as Krt5-CreER; R26Sox2-GFP) mice14 (Extended Data Fig. 2a). Intriguingly, we also found a hyperplastic transitional columnar epithelium at the SCJ, which in mice is located at the fore- and hindstomach junction (Fig. 1a, d, Extended Data Fig. 2f) (n=7). This epithelium consists of basal (p63+ KRT5+), and luminal cells (KRT8+) (Fig. 1a, c). The latter, in contrast to the neighboring keratinized stratified squamous epithelium, secrete mucin [Alcian blue+ periodic acid-Schiff (PAS)+] and express the intestinal marker AGR2 (Extended Data Fig. 2b–c). Furthermore, the expanded transitional epithelium expresses the BE marker KRT715 (Fig.1b). The pathological presentation is similar to human MLE with the co-presence of squamous (p63+ KRT5+) and columnar (KRT8+ KRT7+) cells10. To determine whether basal cells serve as progenitors for the columnar epithelium, we examined the expression of GFP and SOX2 in Krt5-CreER; R26Sox2-GFP mutants. Previous studies have shown that genetic lineage tracing allows the identification of stem/progenitor cells in different tissues16,17. We found that lineage-tagged GFP+ cells are not only present in the stratified squamous epithelium14, but also in the amplified transitional epithelium (SOX2hi) (Fig.1b, Extended Data Fig. 2d). These findings support that basal cells serve as progenitors for the SCJ transitional epithelium. Notably, the adjacent columnar cells lining the cardia mucosa are GFP−, indicating that they are not derived from basal progenitor cells (Fig.1a–c, Extended Data Fig. 2e). Conversely, the transitional columnar epithelium does not express the cardia mucosal protein Claudin18 (Fig. 1c, Extended Data Fig. 2e). Consistently, the cardia mucosa (Lgr5+) does not contribute to the transitional epithelium in Lrg5-CreER; R26LacZ mice (Extended Data Fig. 2g)7. Furthermore, bile acid reflux, a strong risk factor for BE, also leads to the expansion of the transitional columnar epithelium in mice undergoing oesophageo-gastroduodenal anastomosis surgery (Extended Data Fig. 3a, c). Lineage tracing demonstrated that the expanded columnar epithelium is generated by p63+ basal progenitor cells in p63-CreER; R26tdTomato mice (Extended data Fig. 3c). Notably, metaplastic cells were not observed in the oesophagus or other part of the forestomach (Extended Data Fig. 3b). Together these findings in models driven by both genetic and environmental changes suggest that basal cells (p63+ KRT5+) in the transitional epithelium serve as progenitors for KRT7+ BE-like epithelium (Fig. 1d).
Figure 1. An expanded columnar epithelium consisting of basal and luminal cells at the esophageal squamous-columnar junction (SCJ) in Krt5-CreER;R26Sox2-GFP mutants.
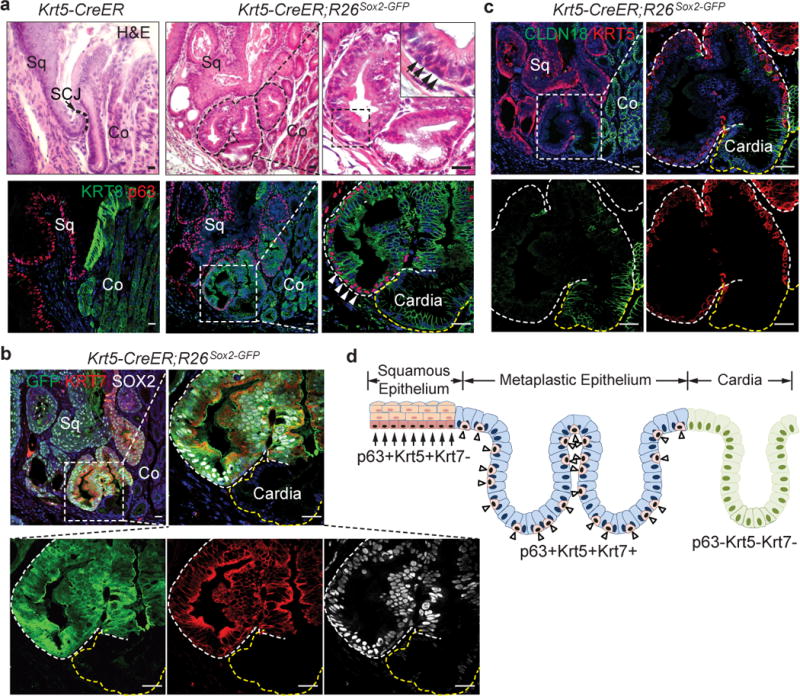
a, SOX2 overexpression leads to the formation of multilayered epithelium (MLE, dotted line) with basal (p63+, arrowheads) and luminal cells (KRT8+). n=7 per group. b, Basal progenitors at the SCJ generate the MLE (KRT7+GFP+SOX2hi). n=7. c, Basal cells in the MLE express KRT5 (white line) but not Claudin18 which labels the cardia mucosa (yellow line). n=7. d, The MLE is located between the stratified squamous epithelium and the simple columnar cardia. Note two distinct basal progenitors at the SCJ. Sq, stratified squamous; Co, columnar. Scale bar, 20μm.
These results prompted us to test whether a non-keratinized transitional columnar epithelium exists in the SCJ of normal mice. As previously described18,19, the keratinized stratified squamous epithelium consists of basal cells (p63+ KRT5+ KRT7−) and differentiated suprabasal cells (Loricrin+ Involucrin+) (Fig. 2a and Extended Data Fig. 4a). We also identified at the SCJ a narrow non-keratinized epithelial zone composed of basal cells (p63+ KRT5+ KRT7+) and luminal cells (KRT7+ Loricrin− Involucrin− Claudin 18−) (Fig. 2a and Extended Data Fig. 4a–b). Next, we asked whether the proliferative transitional basal cells (Ki67+) serve as progenitors for the luminal cells (Extended Data Fig. 4c). We first performed lineage tracing with the p63-CreER mouse line20. As expected, p63-CreER labels the squamous basal cells and their progeny (Fig. 2b). Interestingly, scattered individual basal cells in the transitional epithelium were also labeled 24hrs after a single low-dose of Tamoxifen (Fig. 2b). Two weeks after injection, a single isolated cluster of lineage-labeled cells (Tomato+ KRT7+) confined to the transitional epithelium was observed (Fig. 2b), suggesting that KRT7+ cells are generated by transitional basal cells. We then established a Krt7-CreER knockin mouse line. Notably, a single low-dose Tamoxifen injection induced recombination in the basal cells of the transitional epithelium, but not in any of the neighboring squamous epithelium (Fig. 2b). The lineage-labeled cells expand and generate the transitional epithelium (KRT7+) (Fig. 2b). Together, these lineage tracing results support the idea that the distinct basal cells (p63+ KRT7+) serve as progenitors for the SCJ transitional epithelium.
Figure 2. A unique basal progenitor population (p63+ KRT7+) maintains the transitional epithelium at the SCJ.
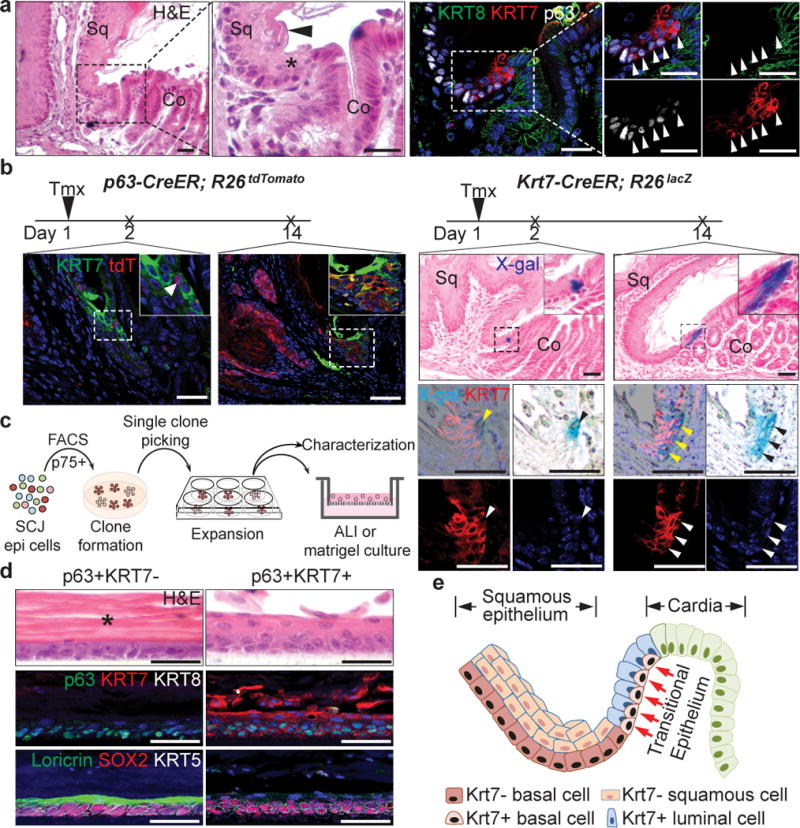
a, The transitional epithelium containing basal progenitors (p63+ KRT7+ KRT8−, arrowheads) is located between the stratified squamous and simple columnar cardia epithelium. n=11. b, The transitional basal cells serve as progenitors for the transitional epithelium as shown by lineage tracing with the p63-CreER and Krt7-CreER alleles. n=3. c, Schematic depicts culture of the transitional basal progenitor cells. d, Distinct epithelium reconstituted by p63+KRT7− and p63+KRT7+ basal progenitors in air-liquid interface (ALI) culture. Note the thick keratin layer (star) and Loricrin+ cells are not present in the epithelium reconstituted by the transitional basal progenitors. n=5 per group. e, Diagram shows the transitional basal progenitor cells (p63+ KRT7+) and luminal cells (KRT7+) in the transitional epithelium. Scale bar, 20 μm.
Next, we tested whether the transitional basal cells self-renew and differentiate to reconstitute the transitional epithelium using both 2D and 3D clonal culture. We used the nerve growth factor receptor (p75) to purify basal progenitor cells20,21. p75+ basal cells in the SCJ include two populations, squamous (p63+ KRT7−) and transitional (p63+ KRT7+) basal cells (Extended Data Fig. 4d, e). Consistently, FACS-sorted single basal cells expanded to form two types of colony, p63+ KRT7− and p63+ KRT7+ (Fig.2c, Extended Data Fig. 4f). They can be maintained for six passages in the presence of Wnt-3A, R-Spondin1, Noggin and Fgf10, growth factors that were successfully used to maintain squamous basal cells and BE organoids (n=5)14,22,23. The two distinct basal progenitor populations did not interconvert when reseeded at single-cell densities even after five passages. Moreover, two types of epithelium (keratinized Vs non-keratinized) were generated by the two basal cell populations in both air-liquid interface (ALI) culture and 3D organoid (Fig. 2d, Extended Data Fig. 4g). Notably, the p63+ KRT7+ transitional basal cells only generate a non-keratinized transitional epithelium (KRT7+ Loricrin−), while the p63+ KRT7− squamous basal cells consistently only produce a keratinized squamous epithelium (Loricrin+) (Fig. 2d, Extended Data Fig. 4g). Together these findings demonstrate that two distinct basal progenitor populations exist in the upper GI tract where they are individually responsible for maintaining the keratinized stratified squamous and the non-keratinized transitional epithelium (Fig. 2e). It is noteworthy that a transitional epithelium composed of p63+ KRT7+ basal and KRT7+ luminal cells is also present in several other tissues, including the rectum-anus and cervix SCJs (Extended Data Fig. 4h–i).
Wang et al. previously reported that RECs, a quiescent epithelial population (p63− KRT7+) persist on the upper luminal surface of the gastric epithelium and serve as the cell-of-origin for BE8 (Extended Data Fig. 1e). RECs expanded following the loss of basal cells in adult Krt14-CreER;R26-DTA mice upon Tamoxifen injection8. According to this study RECs (p63− KRT7+) and basal cells (p63+ KRT7−) are two unrelated populations in development. The authors proposed that during development p63+ KRT7− basal cells expand from the oesophagus to the forestomach and gradually replace the initial columnar cells (p63− KRT7+) except at the SCJ where they remain and become quiescent RECs8 (Extended Data Fig. 5a). The study further proposed that p63 deletion blocks the downward expansion of basal cells, and consequently the forestomach is lined by p63− KRT7+ RECs. By contrast, we found that KRT7 and p63 are co-expressed in both stratified squamous and SCJ transitional epithelium at embryonic (E) day 11.5, 12.5, 14.5, and 16.5 (Extended Data Fig. 5b, d). While basal cells in the squamous epithelium of the oesophagus and forestomach lose the expression of KRT7 at E18.5, basal cells in the SCJ transitional epithelium remain KRT7+ (Extended Data Fig. 5c–d). Lineage tracing results demonstrate that the columnar epithelium lining the p63−/− oesophagus, forestomach and SCJ is indeed derived from p63 promoter active basal progenitors in p63CreER/CreER;R26LacZ (p63 null) embryos (Extended Data Fig. 5e–h). Consistent with previous findings24, the oesophageal epithelium in E18.5 mutants remains simple columnar, similar to the normal epithelium lining the E11.5 oesophagus and forestomach25 (Extended Data Fig. 5i).
BE is characterized by the increased expression of CDX226, a transcription factor critical for intestinal development. Ectopic CDX2 expression promotes the differentiation of gastric secretory epithelium into intestinal-like cells (e.g. goblet cells)27. Ectopic CDX2 expression, however, fails to promote intestinal differentiation of the squamous epithelium of the oesophagus6. To test whether CDX2 expression drives intestinal differentiation of the transitional basal progenitor cells, we first generated Krt5-rtTA;otet-CDX2-T2A-mCherry mice (Extended Data Fig. 6a). Ectopic CDX2 expression initially promoted the expansion of the transitional epithelium (p63+ KRT7+) one week after doxycycline exposure (Extended Data Fig. 6b). At four weeks, the metaplastic cells (KRT8+) began to express AGR2 accompanied by a reduction in the levels of p63 (Extended Data Fig. 6c). At eight weeks, while p63+ cells were still present in some parts of the metaplastic epithelium reminiscence of MLE, BE epithelium (p63−KRT7+ KRT8+) was also observed in the SCJ (Extended Data Fig. 6d). Intestinal metaplasia became more prominent at 13 weeks after CDX2 expression (Fig. 3a), and the Barrett-like domains contain numerous goblet cells (MUC2+ TFF3+) with characteristic mucous secreting vesicles (Alcian blue+ PAS+) (Fig. 3b, d, Extended Data Fig. 7a–b). Notably, a mixture of MLE (p63+ KRT5+ KRT8+ KRT7+) and BE (p63− KRT5− KRT8+ KRT7+) was readily detected in the expanded SCJ (Fig. 3c, Extended Data Fig. 7c). Furthermore, the transcript levels of multiple intestinal genes including Villin1, Agr2, Muc2, Muc4 and TFF3 increased in the metaplastic epithelium isolated by Laser-Capture Microdissection (Extended Data Fig. 7d). By contrast, the levels of p63 and KRT5 were significantly downregulated (Extended Data Fig. 7d). No metaplastic changes were observed in the SCJ of otet-CDX2 mice (Fig. 3b, Extended Data Fig. 7e), and CDX2 did not induce intestinal differentiation of squamous basal progenitor cells in the stratified epithelium of the oesophagus and forestomach (Extended Data Fig. 7f). Metaplasia persisted and expanded even after withdrawal of doxycycline for 11 weeks (Extended Data Fig. 8a–c). Importantly, similar pathological changes were observed in the SCJ of Krt7-CreER, R26rtTA; otet-CDX2 mice following CDX2 expression (Fig. 3e–f, Extended Data Fig. 9a–c). As expected, MLE or Barrett’s metaplasia did not develop in the neighboring squamous epithelium (Fig. 3e–f). Consistently, intestinal metaplasia only occurred in p63+ KRT7+ but not p63+ KRT7− basal cells upon in vitro CDX2 overexpression (Extended Data Fig. 10). Take together our results support the idea that the transitional basal progenitor cells maintaining the SCJ transitional epithelium generate BE under experimental conditions.
Figure 3. CDX2 overexpression promotes intestinal metaplasia of the transitional basal progenitor cells at the SCJ.
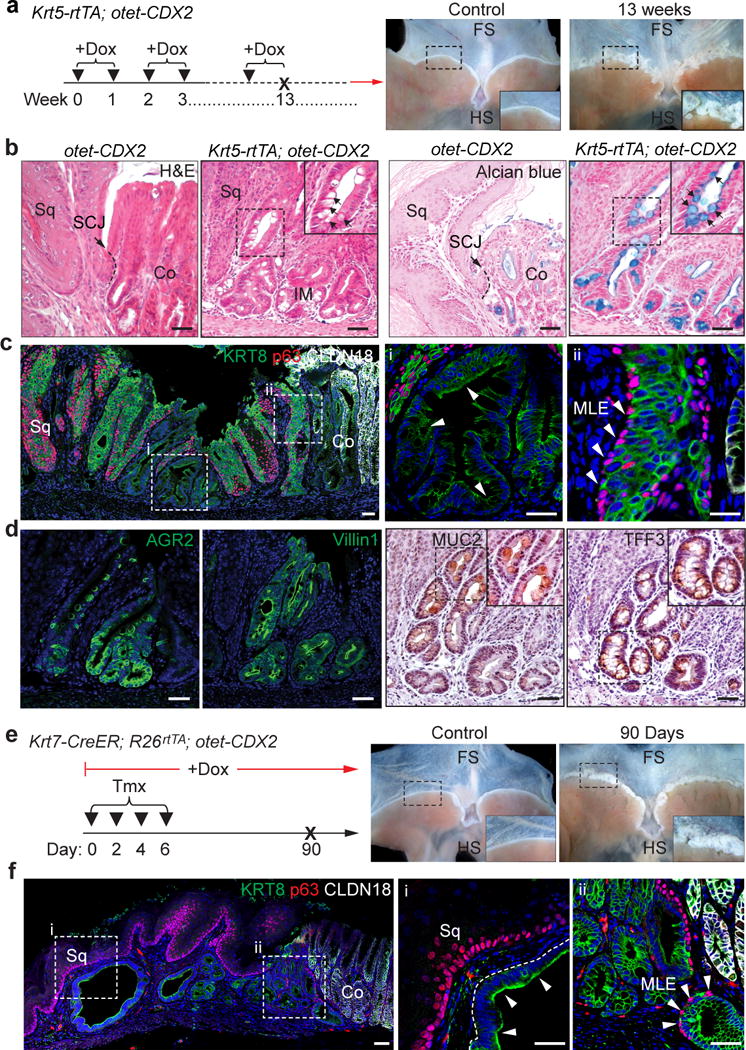
a, CDX2 overexpression in basal cells leads to metaplasia (insert) in the SCJ of Krt5-rtTA; otet-CDX2-T2A-mCherry mice. n=5 per group. b-c, CDX2 overexpression leads to intestinal metaplasia with goblet cells (black arrows) and the loss of p63 protein in some metaplastic cells (white arrows), while the MLE contains p63+ and KRT8+ cells. n=5 per group. d, Metaplastic cells express the intestinal proteins Villin1 and AGR2, and the goblet cell markers MUC2 and TFF3. n=5 per group. e, CDX2 overexpression in KRT7+ cells leads to metaplasia (insert) at the SCJ of Krt7-CreER; R26rtTA; otet-CDX2-T2A-mCherry mice. n=4 per group. f, CDX2 overexpression leads to the loss of p63 protein in the metaplastic epithelium (arrows). Note the presence of MLE with underlying p63+ basal cells and KRT8+ luminal cells. n=4 per group. Scale bar, 50 μm.
Next, we asked whether the transitional epithelium is also present in the human SCJ which unlike in the mouse is located at the gastro-oesophageal junction (Fig. 4a). Significantly, a similar columnar epithelium composed of basal (p63+ KRT5+ KRT7+) and luminal cells (p63−KRT7+) was observed (Fig.4a–c). Flow cytometric analysis indicated the presence of two distinct basal progenitor populations (p63+ KRT7− Vs p63+ KRT7+) (Extended Data Fig. 11a). The two basal cell populations sorted by FACS can be passaged for four times in the presence of Wnt3A/Noggin/R-Spondin/Fgf10. More importantly, p63+ KRT7+ but not p63+ KRT7− basal cells generate intestinal-like epithelial cells in organoid culture upon CDX2 overexpression (Fig. 4d, Extended Data Fig. 11b–d). The transitional epithelium with underlying basal cells is dramatically expanded in patients with long-term bile acid reflux (Fig. 4e and Extended Data Fig. 11e). The expansion of the transitional epithelium became more prominent in biopsies with mixture of MLE and BE (Fig. 4f and Extended Data Fig. 11f–g). The expression of p63 and KRT5 was then lost in BE only samples (n=12), while Barrett’s epithelium gained the expression of CDX2, Villin 1 and AGR2 (Extended Data Fig. 11h).
Figure 4. The transitional epithelium is also present in the human SCJ and amplified during BE pathogenesis.
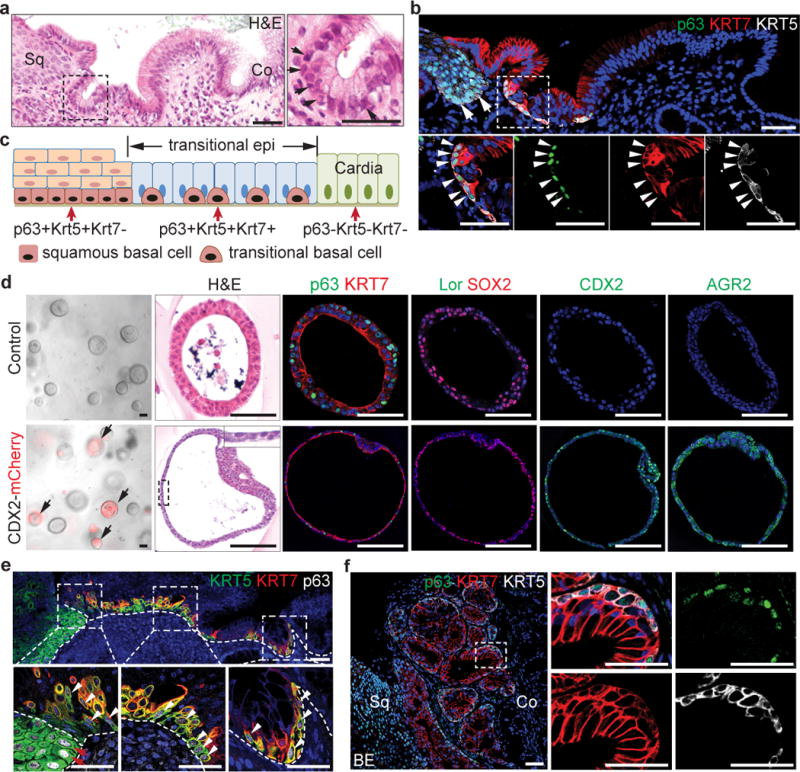
a, The transitional epithelium consists of basal progenitors (arrows) and luminal cells. n=5. b, Basal progenitor cells in the transitional epithelium express p63, KRT5 and KRT7 (arrowheads). Note that basal cells of the stratified squamous (sq) cells are KRT7− (arrows). n=5. c, Schematic shows two types of basal progenitor cells in the human SCJ. d, CDX2 overexpression promotes intestinal metaplasia of p63+KRT7+ cells in 3D organoid culture. n=6 per group. e, The transitional epithelium with underlying basal cells is dramatically expanded in patients with gastro-oesophageal acid reflux. Dotted lines indicate the basement membrane. n=3. f, The transitional epithelium with basal cells is amplified in BE mixed with MLE. n=5. Me, mesenchyme. Scale bar, 50 μm.
In summary, we have identified a novel transitional columnar epithelium at the SCJ where BE and its precursor MLE exclusively occur. We have used lineage tracing, in vitro 2D and organoid culture to support the idea that a distinct basal cell population (p63+ KRT5+ KRT7+) maintains the transitional columnar epithelium at homeostasis. We further show that these transitional basal progenitor cells differentiate into intestinal cells, including goblet cells, following CDX2 expression. Notably, a similar transitional epithelium with underlying p63+ KRT7+ basal cells is present in the human gastro-oesophageal junction, and expansion of the transitional basal progenitors is associated with BE pathogenesis. Moreover, a similar transitional epithelium exists in at least two other tissue junctions throughout the body. In the future, characterization of the progenitor cells for these different transitional epithelia and how they respond to pathologic conditions such as acid reflux, hormonal modulation, virus infection and chemical agents will likely yield critical information related to the etiology of tissue specific metaplasia and tumorigenesis.
Methods
Mice
Krt5-CreER, Krt5-rtTA, R26loxp-stop-loxp-Sox2-IRES-GFP, Lgr5-CreER mice were previously described17,20,28,29. R26tdTomato, R26LacZ, R26rtTA mice were purchased from the Jackson Laboratory. p63-CreER mice were kindly provided by Dr. Jianmin Xu of Baylor College of Medicine30. The otet-CDX2-T2A-mCherry transgenic line was generated by pronuclear injection of linearized DNA. The resulting transgenic mice are maintained on a C57BL/6 and 129SvEv mixed background. Two individual lines were generated and exhibit a consistent expression pattern. The Krt7-CreER knockin line was generated by replacing CreER cassette at the ATG of the Krt7 gene and maintained on a C57BL/6 and 129SvEv mixed background. To lineage trace Lgr5+ cells 2mg per 20g bodyweight Tamoxifen was injected into Lgr5-CreER; R26LacZ mice or controls. To lineage trace p63+ and KRT7+ cells, p63-CreER; R26tdTomato and Krt7-CreER; R26LacZ mice were injected with low-dose of Tamoxifen (0.2mg per 20g body weight) intraperitoneally. To overexpress CDX2, Krt5-rtTA; otet-CDX2-T2A-mCherry mice (6–8 weeks old) were fed with doxycycline drinking water (2 mg/ml) every other week and Krt7-CreER; R26rtTA; otet-CDX2-T2A-mCherry mice were administrated with 4 doses of tamoxifen, followed with doxycycline drinking water. Samples were collected for analysis at various timepoints as indicated in the text. Otet-CDX2-T2A-mCherry mice fed with doxycycline water were included as controls. All mice were maintained in Columbia University’s animal facility according to institutional guidelines. All mouse experiments were conducted in accordance with procedures approved by the Institutional Animal Care and Use Committee at Columbia University. All relevant ethical regulations were followed.
Bile Acid Reflux Model
To generate bile acid reflux mouse models, oesophageo-gastroduodenal anastomosis was performed by Dr. Xiaoxin Chen as previously described31. Briefly, eight-week-old p63-CreER; R26tdTomato mice injected with 4 doses of tamoxifen were administered anesthetics pre-mixed ketamine (80mg/kg) and xylazine (12 mg/kg) in normal saline by intraperitoneal injection. Oesophageo-gastroduodenal anastomosis was performed through an upper midline incision. Two 0.5 cm incisions were made on the oesophagus and the duodenum on the anti-mesenteric border, and then were anastomosed together with accurate mucosal to mucosal opposition. Samples were collected for analysis 18 weeks after surgery.
Isolation of SCJ Basal Progenitor Cells and FACS sorting
Mouse or human SCJ regions were cut and incubated in Dispase for 30 min at room temperature. Digestion was stopped by adding DMEM medium with 10% FBS. The epithelium was detached from the mesenchyme with forceps and digested for 20min in 0.1% Trypsin. Digestion was stopped with 10% FBS-containing medium and filtered through 40μm nylon strainer. Single cell suspensions were incubated in FITC conjugated mouse anti-p75 (1:10, ab62122, Abcam) prior to sort. Cell sorting was performed using a LSRII flow cytometer (BD Biosciences). Cells were collected in DMEM with 2% BSA and cultured immediately onto rat type IV collagen–coated (BD Biosciences) tissue culture dishes (100 single cells/10cm plate). Cells were cultured in Advanced DMEM/F12 basic medium with 1X N2 (Invitrogen) and 1X B27 (Invitrogen), 1X Glutamax (Life Technologies), 1X HEPES, 1X Penicillin/Streptomycin (Gibco), 1 mM N-Acetyl-L-Cysteine (Sigma), 100 μM Gastrin (Sigma), 10 mM Nicotinamide (Sigma), 10 μM SB202190 (Sigma), 50 ng/mL Epidermal growth factor (Invitrogen), 100 ng/mL Noggin (R&D Biosystems), 100 ng/mL Wnt3A (Millipore), 100 ng/mL R-Spondin (Nuvelo), 100 ng/ml Fgf10 (R&D Biosystems), and 500 nM A8301 (Tocris)22. Individual basal cell clones were observed 7 days after seeding, and these individual clones were divided into two halves, one for immunostaining of p63 and KRT7 and the other for reseeding onto 24-well plate for ALI or organoid culture in Matrigel22. The medium was refreshed every other day. For Flow cytometric analysis, isolated single cells were fixed by Fixation/Permeabilization working solution for 30 minutes and then incubated for 1 hour at 4 degree with primary antibodies: FITC conjugated mouse anti-p75 (1:10, ab62122, Abcam), Alexa Fluor 647 conjugated mouse anti-KRT7 (1:50, sc-23876, Santa Cruz Biotechnology), rabbit anti-p63 (1:200, sc-8343, Santa Cruz Biotechnology) or control IgG followed by washing and incubation in Alexa Fluor 555 anti-rabbit antibody (1:500, Jackson ImmunoResearch Laboratories). Flow cytometry analysis was performed on BD FACS Canto cytometers.
Lentiviral Production and Transduction
CDX2-T2A-mcherry was cloned into the lentiviral expression vector pCDH. Lentiviral particles were generated by transfecting 293T cells with the lentiviral expression vectors using Fugene HD. After two days of virus production, lentiviral-containing supernatants were harvested, filtered (0.45μm), concentrated by Lenti-X concentrator (Clontech) and incubated with human basal progenitor cells in the presence of polybrene (8ng/ml) for 1 hour. Infected cells were mixed with Matrigel (growth factor-reduced, 1:1 ratio) and cultured in 24-well Transwell inserts (Falcon). Dox was added to the medium to induce CDX2 expression.
Tissue Preparation, Histology, and Immunostaining
Tissues were fixed in 4% paraformaldehyde overnight and processed as previously described13. For cryosections, organoids were fixed in 4% paraformaldehyde in PBS at 4°C overnight, placed in 30% sucrose in PBS, and embedded in OCT. The primary antibodies used for immunostaining analysis include: rabbit anti-p63 (1:200, sc-8343, Santa Cruz Biotechnology); mouse anti-p63 (1:500, MAB4135, Millipore); chicken anti–KRT8 (1:1000, ab107115, Abcam); chicken anti-GFP (1:1000, GFP-1020, Aves Labs); rabbit anti-SOX2 (1:500, WRAB-1236, Seven Hills); mouse anti-KRT7 (1:200, Ab9021, Abcam); rabbit anti-CLDN18 (1:1000, ab203563, Abcam); chicken anti-KRT5 (1:1000, 905901, Biolegend); mouse anti-Ki67 (1:200, 550609, BD Biosciences); mouse anti-Involucrin (1:1000, MS-126-P0; Thermo Fisher Scientific); rabbit anti-Loricrin (1:2000, 905101, Biolegend); rabbit anti-RFP (1:100, ab62341, Abcam); rabbit anti-CDX2 (1:500, ab76541, Abcam); rabbit anti-Villin1 (1:200, 16488-1-AP, Protein Group); rabbit anti-AGR2 (1:200, PA5-34517, Thermo Fisher Scientific); rabbit anti-Muc2 (1:200, sc15334, Santa Cruz Biotechnology); rabbit anti-TFF3 (1:200, PAB656, Cloud-Clone Corporation). Biotinylated or fluorescent secondary antibodies were used for detection and visualization. Images were obtained using Nikon SMZ1500 Inverted microscope (Nikon). Confocal images were obtained with a Zeiss LSM T-PMT confocal laser-scanning microscope (Carl Zeiss).
Alcian Blue and Periodic Acid–Schiff Staining
Alcian blue and Periodic acid–Schiff staining were performed as previously described32. In brief, for Periodic acid-Schiff staining, sections were treated with Periodic Acid Solution for 5 minutes, then stained in Schiff’s reagent (395B, Sigma) for 5 minutes and counterstained with Hematoxylin (GHS3, Sigma). For Alcian blue staining, sections were treated with 3% acetic acid solution for 3 minutes, then stained in Alcian blue (A3157, Sigma) for 5 minutes and counterstained with Nuclear Fast Red (N8002, Sigma).
X-gal Staining
The oesophagus and stomach were fixed in 4% paraformaldehyde for 30 minutes at room temperature, followed by X-gal staining overnight at 37°C. X-gal–stained samples were then dehydrated with isopropanol and embedded in paraffin for sectioning as previously described14.
Laser-Capture Microdissection (LCM) and RNA Isolation
Fresh SCJ tissues from CDX2 overexpression mice at different stages were embedded in OCT and the sections were mounted onto the RNAsezap (Ambion) treated pen-membrane Slides (Carl Zeiss). The metaplastic epithelium was cut and collected into RNAse-free tubes using a Laser Microdissection system (Carl Zeiss). Total RNA was isolated using the Arcturus PicoPure RNA Kit (Cat No.12204, Applied Biosystems). Concentration of RNA was determined using NanoDrop 2000c system (Thermo Scientific).
Reverse Transcription and Real-time PCR
RNA reverse transcription was performed using the Super-Script III First-Strand SuperMix (Invitrogen) according to the manufacturer’s instructions. cDNA was subjected to quantitative real-time PCR using the StepOnePlus Real-Time PCR Detection System (Applied Biosystems) and iTaq Universal SYBR Green Supermix (Bio-Rad). The transcript levels of genes were normalized to Actin expression. All real-time quantitative PCR experiments were performed in triplicate. PCR primers were designed using the Lasergene Core Suite (DNASTAR Inc.), and the sequences of primers are described in supplemental information (Supplemental Table 1).
Human Gastro-oesophageal Junction and Barrett’s Oesophagus
Human SCJ biopsies were provided by Drs. Julian Abrams and Timothy Wang at the Department of Medicine under Columbia University IRB approval. Slides containing human samples were provided by the Department of Pathology at the University of Rochester. The pathological changes were validated by three pathologists. Adult participants from a cohort of control subjects or subjects with acid reflux and Barrett’s oesophagus were assessed and provided under IRB approval at the University of Rochester. All participants in this study provided written informed consent. All relevant ethical regulations were followed. Basic information about patients are listed in Supplemental Table 2.
Statistics and reproducibility
Statistical analysis was done using unpaired two-tailed Student’s t-test. Results were shown mean ± s.e.m.; p values < 0.05 were considered statistically significant. The investigators were not blinded to allocation during experiments and outcome assessment in animal studies, as no statistics were performed.
Data availability statement
The authors declare that the main data supporting the findings pf this study are available within the paper and its supplementary information. Any additional data are available from the corresponding authors upon request.
Extended Data
Extended Data Figure 1. Multiple models have been proposed to explain the cell of origin for BE.
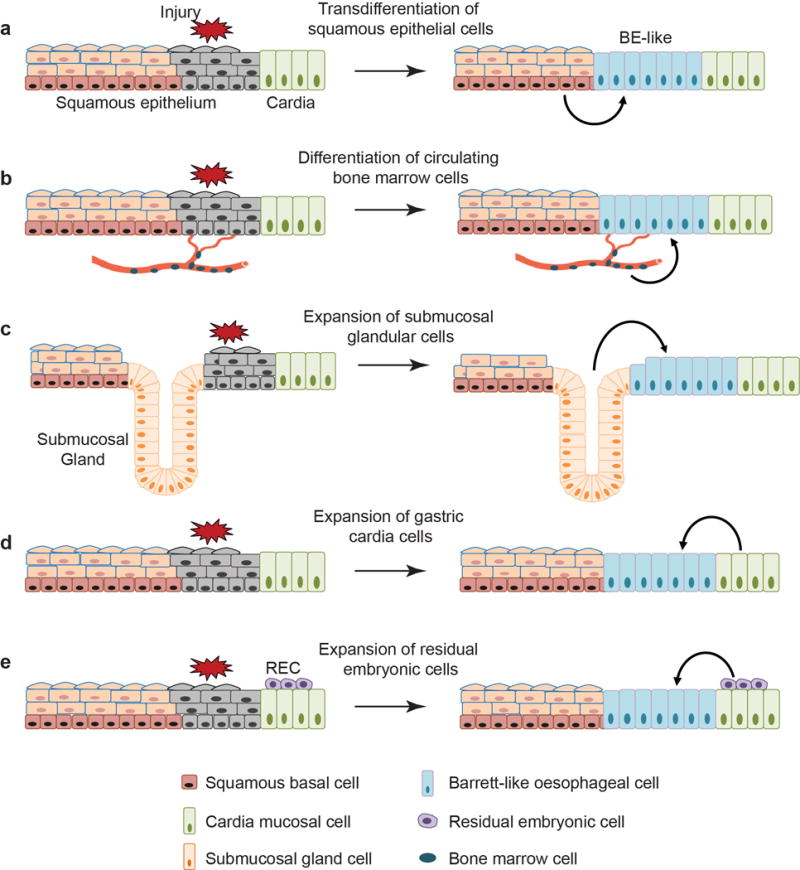
a, Transdifferentiation of the stratified squamous oesophageal epithelium into Barrett’s epithelium. b, Transdifferentiation of circulating bone marrow cells into Barrett’s epithelium. c, Expansion of the oesophageal submucosal gland leads to BE. d, Stem/progenitor cells (Lgr5+) in the cardia mucosa differentiate into BE. e, Expansion of the quiescent residual embryonic cells (RECs) at the SCJ leads to BE formation. Note that none of the studies recapitulates the pathological changes characteristically associated with BE in humans, e.g. presence of intestinal goblet cells.
Extended Data Figure 2. Expansion of the columnar epithelium at the SCJ in Krt5-CreER;R26Sox2-GFP mutants.
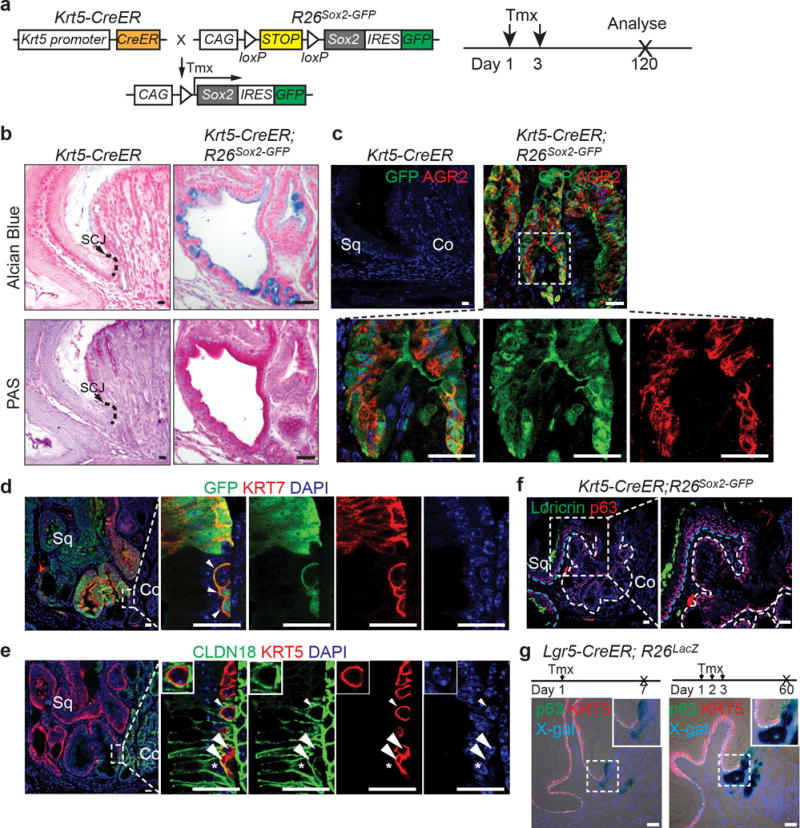
a, Schematic depicts generation of Krt5-CreER;R26Sox2-GFP (SOX2 overexpression) mutants. b, The columnar epithelium secretes mucin as indicated by Alcian blue and periodic acid-Schiff staining. n=7 per group. c, The mucin secreting cells (AGR2+) are derived from KRT5+ basal progenitor cells as verified by the lineage tracing tag GFP. n=7. d, High magnification picture of Figure 1b to show that expanded GFP+KRT7+ basal progenitor cells invade underneath of the cardia mucosa upon SOX2 overexpression. n=7. e, High magnification picture of Figure 1c to show that expanded basal cells (KRT5+) invade and intercalate with the cardia mucosal epithelium (CLDN18+) upon SOX2 overexpression. Note KRT5+ cells (arrow and arrowheads) do not express CLDN18. Conversely, CLDN18+ cell does not express KRT5 (star). n=7. f, The columnar epithelium does not express the squamous cell marker Loricrin. n=7. The white and blue dotted lines indicate the amplified columnar epithelium and the stratified squamous epithelium, respectively. g, Co-staining of X-gal with KRT5 and p63 indicates Lgr5+ cardia progenitor cells do not contribute to KRT5+ or p63+ basal cells in Lgr5-CreER; R26lacZ mice with both short-term tracing and long-term tracing. n=3 per group. Scale bar, 20 μm.
Extended Data Figure 3. Basal progenitor cells contribute to columnar metaplasia following anastomosis surgery-induced bile acid reflux.
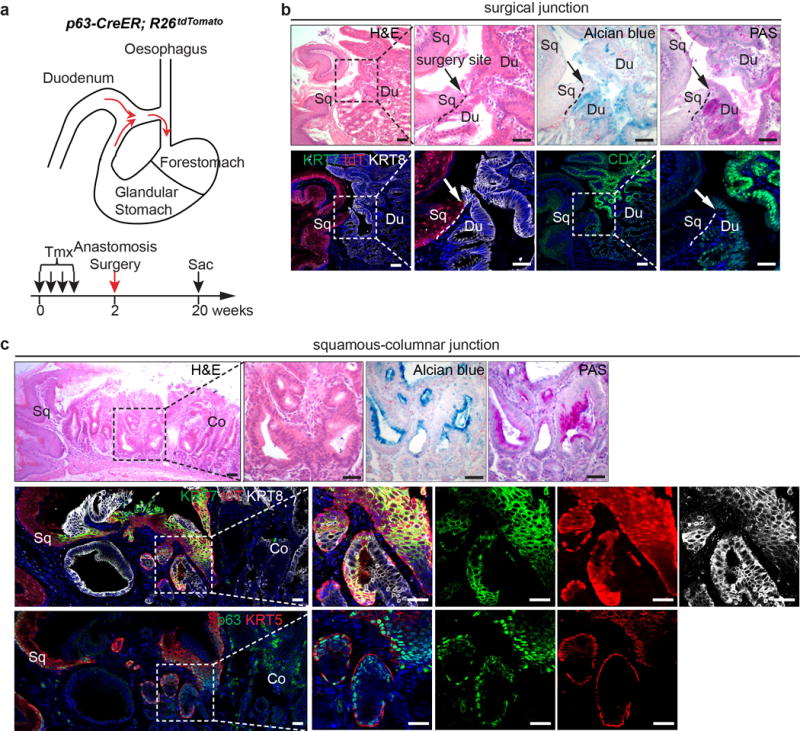
a, Schematic depicts the use of oesophageal-duodenal anastomosis and lineage tracing in p63-CreER;R26tdTomato mice. b, Metaplasia does not occur at the distal oesophagus where the anastomosis surgery site is located (arrow). Note that Alcian blue, PAS and CDX2 label the intestinal but not the squamous oesophageal epithelium. n=5. c, Lineage-labeled transitional epithelial cells (tdT+) expand and resemble the multilayered epithelium, expressing the columnar markers (KRT7, KRT8) and basal cell markers (KRT5 and p63). The expanded epithelium secretes mucin as indicated by Alcian blue and PAS staining. n=5. Scale bar, 20 μm.
Extended Data Figure 4. The transitional columnar epithelium is present at the SCJ of normal mice.
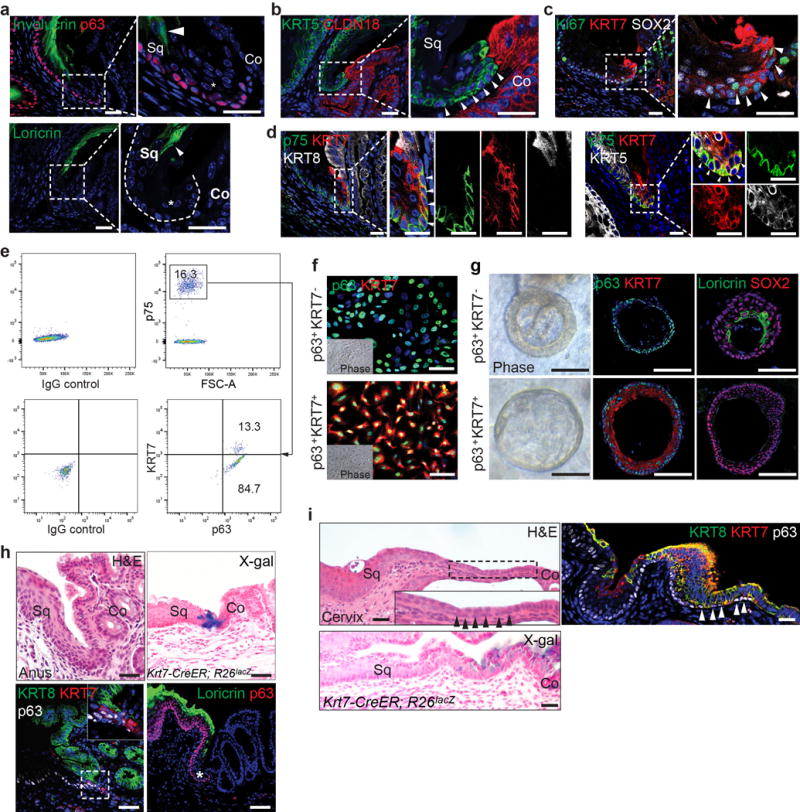
a, The transitional epithelium (star) does not express Involucrin and Loricrin, markers labeling the stratified squamous epithelium. n=11. b, Basal cells of the transitional epithelium express KRT5 but not the cardia epithelial marker CLDN18. n=11. c, The KRT7+ transitional basal cells are highly proliferative (Ki67+, arrowheads). n=3. d, The transitional basal cells of at the SCJ express p75, KRT7 and KRT5, but not KRT8. Note that p75 and KRT5 are also expressed in the neighboring squamous basal cells. n=5. e, FACS analysis reveals p75+ basal cells include two subpopulations, squamous basal cells (p63+ KRT7−) and transitional basal cells (p63+ KRT7+). n=3 independent experiments. f, A representative culture of p63+KRT7− and p63+KRT7+ basal progenitor cells. Note that p63+ KRT7+ transitional basal cells in the colony are loose unlike the cobblestone characteristic of squamous basal cell colony (p63+ KRT7−). n=5 per group. g, p63+KRT7− and p63+KRT7+ basal progenitor cells generate keratinized (Loricrin+, KRT7−) and non-keratinized (Loricrin−, KRT7+) epithelium in organoid, respectively. n=5 per group. h, i, The transitional epithelium is also present at the SCJ of the anus (h) and cervix (i). Lineage trancing with the Krt7-CreER mouse line confirmed KRT7+ cells serve as progenitors for the transitional epithelium. n=3 per group. Abbreviation: Sq, stratified squamous epithelium; Co, columnar epithelium. Scale bar, 20 μm.
Extended Data Figure 5. Loss of p63 prevents the stratification of KRT7+ columnar epithelium during embryonic development.
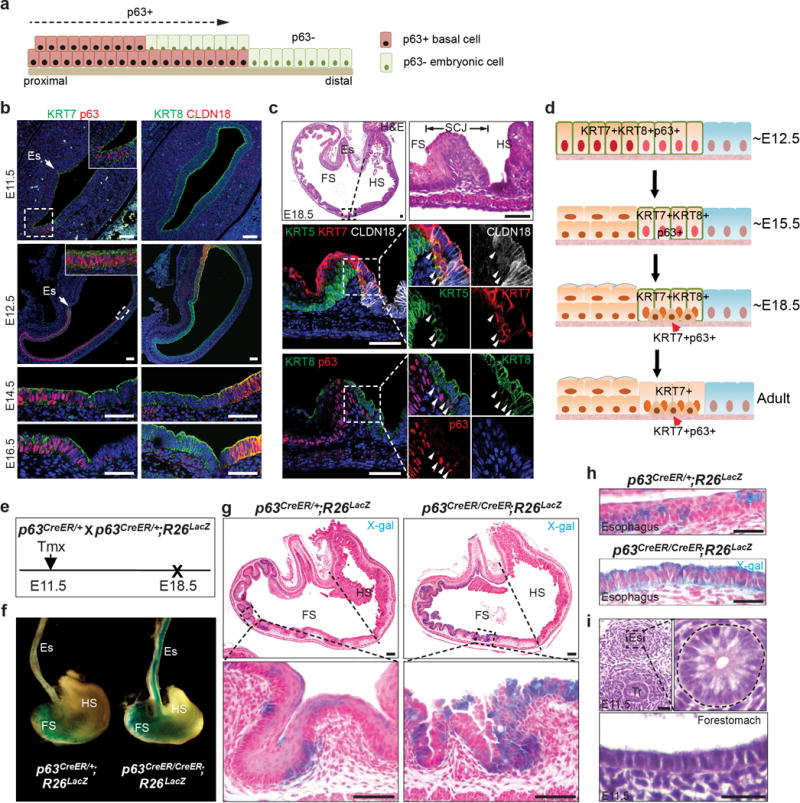
a, Diagram shows the proposed REC model of downward expansion of p63+ KRT7− basal cells and retreatment of p63−KRT7+ embryonic cells. b, The columnar epithelium lining the mouse forestomach and the SCJ expresses p63, KRT7 and KRT8 from E11.5 to E16.5. Note Claudin18 expression is limited to the hindstomach epithelium. n=5. c, Expression of KRT7 is restricted to the SCJ transitional epithelium at E18.5, and basal cells (arrowheads) express p63, KRT5, KRT7 and low levels of KRT8 but not CLDN18. n=5. d, Schematic depicts the gradual restriction of KRT7 expression to the SCJ transitional epithelium during development. Initially the simple columnar epithelium lining the forestomach and SCJ expresses both p63 and KRT7. Upon stratification the forestomach epithelium loses KRT7 expression, while basal cells at the SCJ maintain expression of both p63 and KRT7. e, Lineage tracing of epithelial progenitor cells (p63promoter active) in p63CreER/CreER; R26lacZ (p63 null) mutants. f, Whole-mount x-gal staining of the oesophagus and stomach isolated from p63CreER/CreER; R26LacZ mutants and p63CreER/+; R26LacZ controls. n=3. g, h, The simple columnar epithelium lining the forestomach (g) and oesophagus (h) of mutants is derived from basal progenitor cells (p63 promotor active) as indicated by x-gal staining. n=3 per group. i, Normal oesophagus and forestomach is lined by simple columnar epithelium at E11.5. n=3. Abbreviation: Es, oesophagus; FS, forestomach; HS, hindstomach; Tr, trachea. Scale bar, 20 μm.
Extended Data Figure 6. Ectopic CDX2 expression promotes intestinal metaplasia of the transitional epithelium at the SCJ.
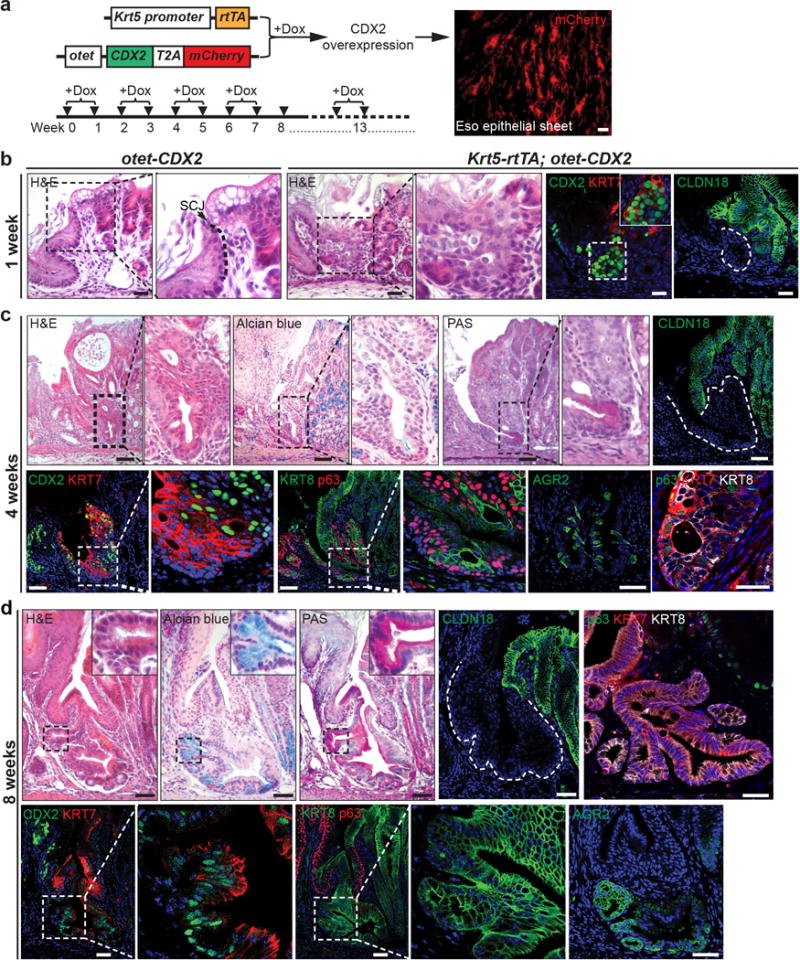
a, Targeted expression of CDX2 in basal progenitor cells of Krt5-CreER; otet-CDX2-T2A-mCherry mice. Doxycycline (Dox) water was given every other week for 3 months to induce CDX2 and mCherry expression. b, CDX2 overexpression leads to the expansion of the transitional epithelium at the SCJ at the first week of Dox treatment. n=3 per group. c, 4-week Dox treatment promotes further expansion of the transitional epithelium at the SCJ, and the epithelium presents multilayered epithelium characteristics with the co-expression of columnar markers KRT8, KRT7 and squamous marker p63. Note some basal cells start to lose p63 expression while gaining the expression of the intestinal marker AGR2. n=3 per group. d, Intestinal metaplasia is apparent following 8-week Dox treatment and some metaplastic cells lose the expression of p63. n=4 per group. Scale bar, 20 μm.
Extended Data Figure 7. Ectopic CDX2 expression promotes intestinal metaplasia of the transitional epithelium.
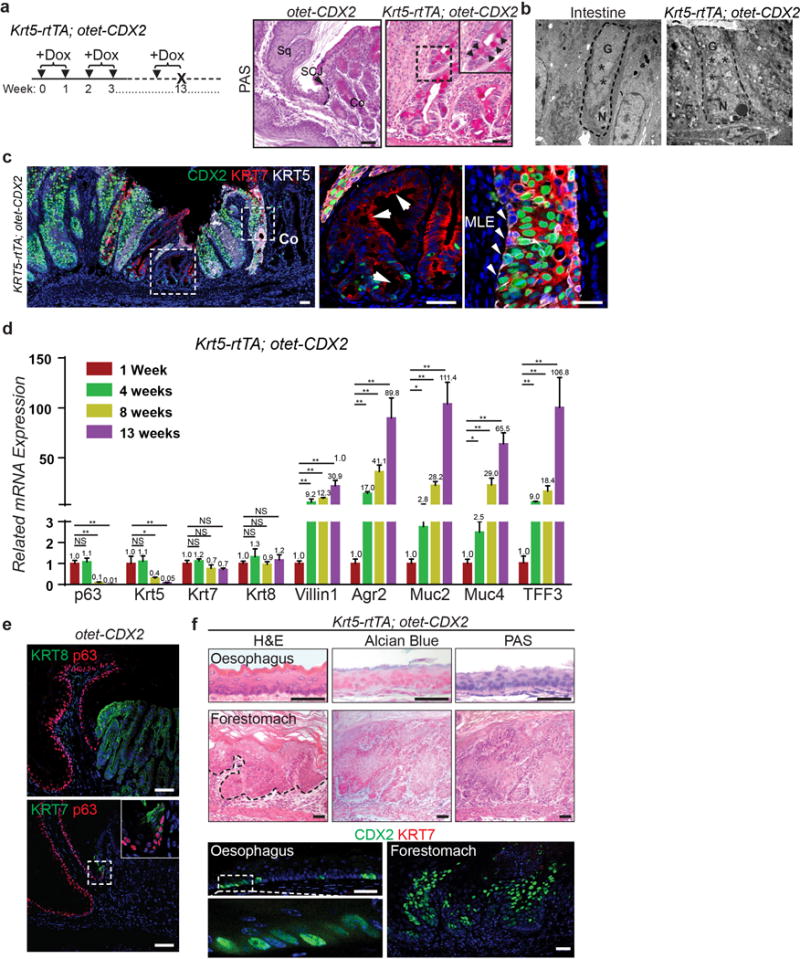
a, Intestinal metaplasia occurs in Krt5-rtTA;otet-CDX2 mutants fed with Dox-containing water for 3 months as shown by PAS staining. n=5 per group. b, Goblet cell enriched with vesicles (stars) present in the SCJ of Krt5-rtTA;otet-CDX2 mutants following 3-month Dox treatment as shown by electron microscope. n=3 per group. c, KRT7 is expressed by both BE and MLE but not the neighboring squamous epithelium. Residual CDX2 is expressed in a subpopulation of the metaplastic columnar cells although KRT5 expression is not detectable. n=5 per group. d, Reduced expression of basal cell genes (p63, Krt5) and increased expression of the intestinal genes (Villin1, Agr2, Muc2, Muc4, TFF3) during intestinalization of the SCJ following CDX2 overexpression. (ns, p>0.05; *p<0.05; **p<0.001; two-tailed Student’s t-test; n=3 independent experiments) e, Normal SCJ structure is maintained in control mice (otet-CDX2-T2A-mCherry) following 3-month Dox treatment. n=5. f, CDX2 overexpression does not promote columnar metaplasia in the oesophagus and forestomach. Squamous hyperplasia is present in the forestomach (dotted black line). Note that CDX2 expression does not induce ectopic expression of KRT7 in the stratified squamous epithelium. n=5. Abbreviation: Sq, stratified squamous epithelium; Co, columnar epithelium; G, goblet; N, nucleus. Scale bar, 20 μm.
Extended Data Figure 8. Intestinal metaplasia is maintained even after withdrawal of Dox in Krt5-rtTA;otet-CDX2-T2A-mCherry mutants.
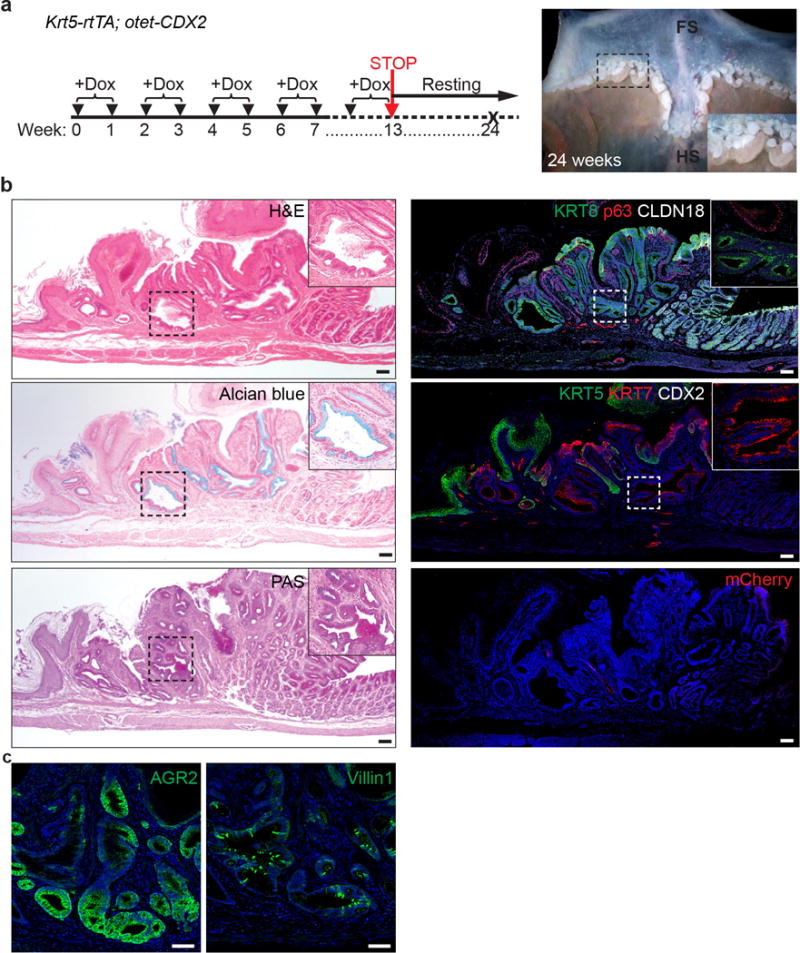
a, Severe metaplasia develops at the SCJ of mutants that have been treated with Dox for the first 13weeks and chased for another 11weeks. n=3 per group. b, The metaplastic cells remain at the SCJ even after withdrawal of Dox containing water. The metaplastic cells express KRT7 and KRT8. Note that mCherry which indicates CDX2 expression is not detected. n=3 per group. c, Metaplastic cells maintain AGR2 and Villin1 expression. n=3 per group. Scale bar, 20 μm.
Extended Data Figure 9. Ectopic CDX2 expression promotes intestinal metaplasia of the transitional epithelium in Krt7-CreER; R26rtTA; otet-CDX2-T2A-mCherry mutants.
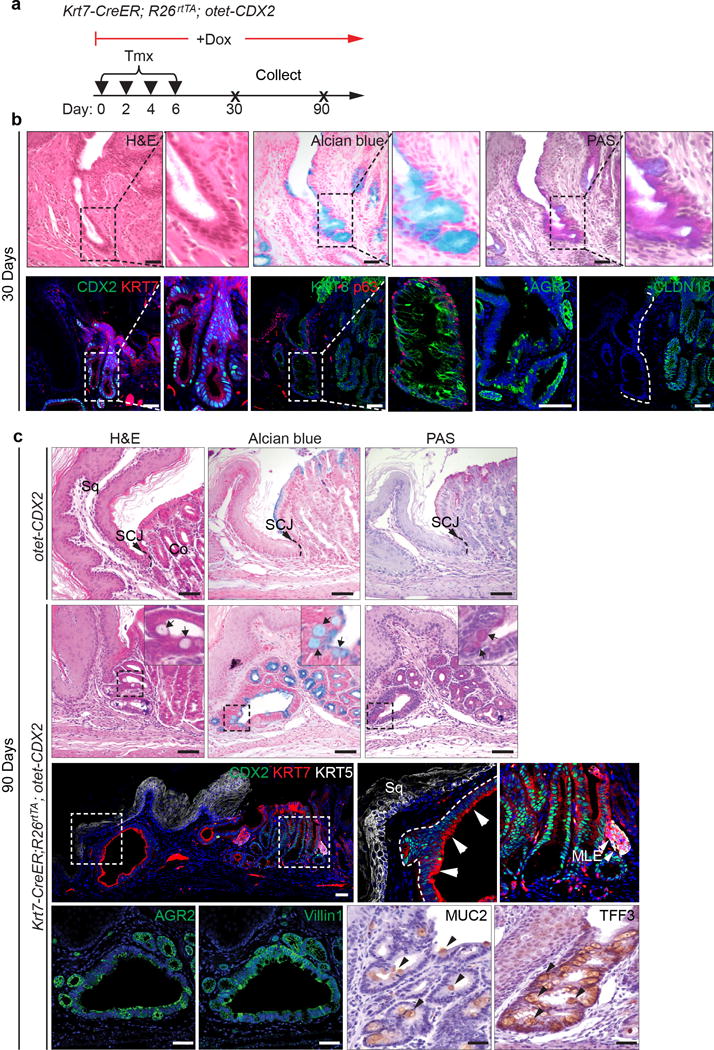
a, Schematic shows induction of CDX2 overexpression by combined treatment of Tamoxifen and Doxycycline. b, 30-day CDX2 expression drives the differentiation of the transitional basal cells into intestinal-like epithelium including goblet cells in Krt7-CreER; R26rtTA; otet-CDX2-T2A-mCherry mice. n=3 per group. c, 90-day CDX2 overexpression leads to prominent intestinal metaplasia of the transitional basal progenitor cells as indicated by intestinal markers. n=4 per group. Abbreviation: Sq, stratified squamous epithelium; Co, columnar epithelium. Scale bar, 20 μm.
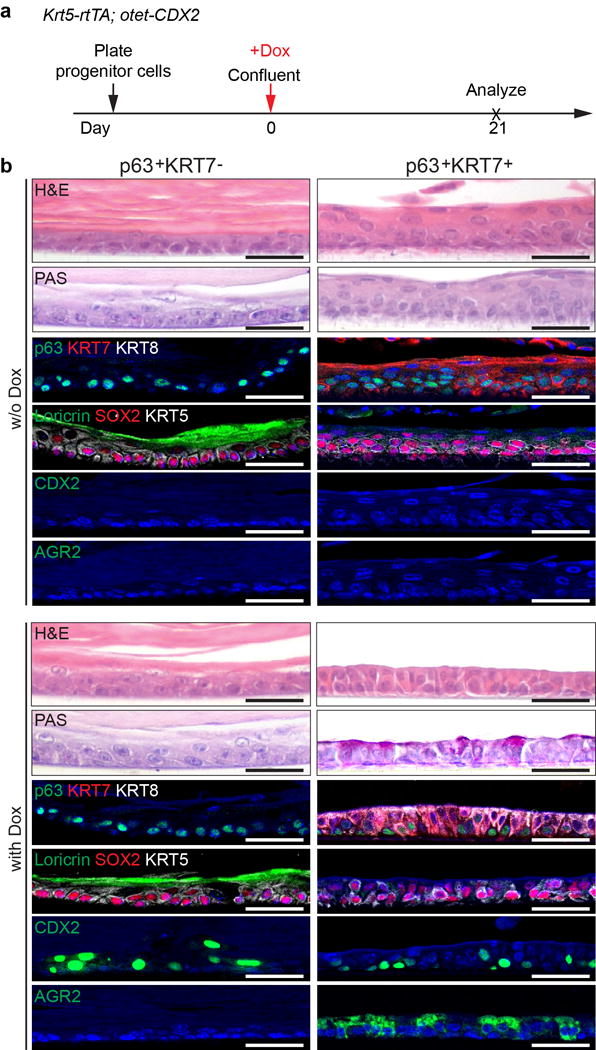
Extended Data Figure 10. Intestinal metaplasia develops in air-liquid culture of the transitional (p63+ KRT7+) but not the squamous (p63+ KRT7−) basal progenitor cells upon CDX2 expression.
a, Schematic depicts Doxycycline-induced CDX2 expression. b, Intestinal metaplasia occurs in Dox-treated transitional (p63+ KRT7+) but not squamous basal (p63+ KRT7−) progenitor cells. Note that p63+ KRT7− basal cells generate a thick keratin layer in the ALI culture, and the differentiated cells express Loricrin. n=5 per group. Scale bar, 20 μm.
Extended Data Figure 11. Different response to CDX2 overexpression in the transitional (p63+ KRT7+) and the squamous (p63+ KRT7−) basal progenitor cells in vitro. Human and mouse MLE present similar gene expression.
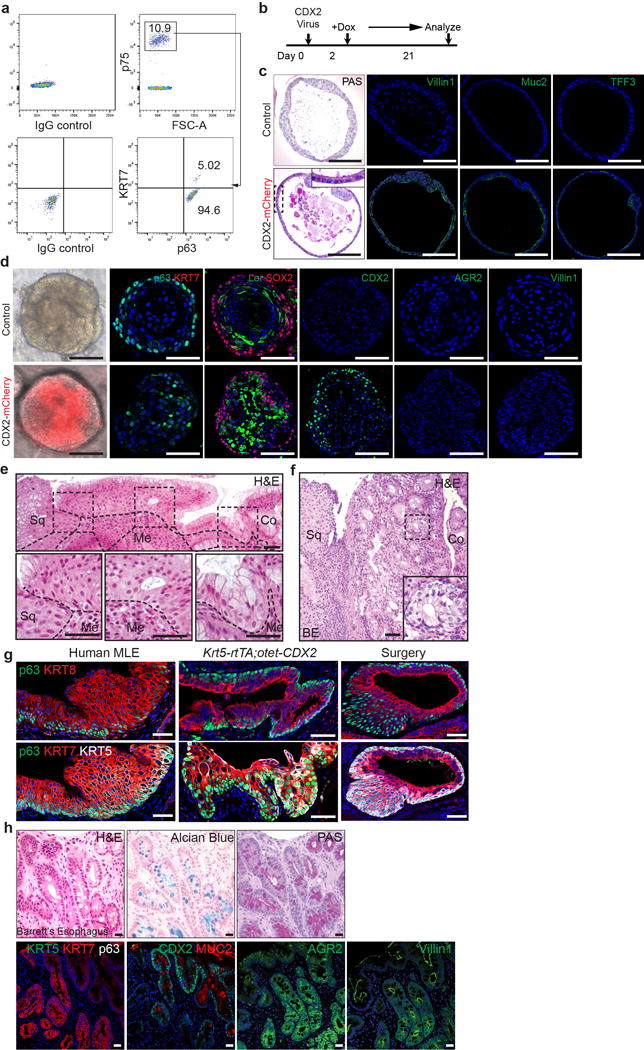
a, Two distinct basal progenitor populations (p63+ KRT7− Vs p63+ KRT7+) are present at the human SCJ as indicated by flow cytometric analysis. n=3 independent experiments. b, Schematic depicts the induction of CDX2 overexpression with Doxycycline treatment of CDX2 virus-infected human SCJ basal progenitor cells. c, CDX2 overexpression promotes intestinal metaplasia of p63+ KRT7+ cells. The metaplastic columnar cells are PAS+ and express Villin1, Muc2 and TFF3. n=6 per group. d, Ectopic CDX2 expression does not promote intestinal metaplasia of the stratified squamous epithelium in organoids formed by p63+KRT7− squamous basal cells. n=4 per group. e, The transitional epithelium with underlying basal cells is dramatically expanded in patients with long-term gastro-esophageal acid reflux. Dotted lines indicate the basement membrane. n=3. f, The transitional epithelium with basal cells is amplified in BE mixed with MLE. n=5. g, Similar phenotypic presentation of human MLE and mouse MLE developed at the SCJ following CDX2 overexpression and oesophageal-duodenal anastomosis surgery. human MLE n=10; Krt5-rtTA; otet-CDX2 mutants n=5; surgical mice n=5. h, Goblet cells in human BE is positive for Alcian blue and PAS staining. BE epithelium loses the expression of KRT5 and p63 while maintaining the expression of KRT7. Note BE gains the expression of CDX2, MUC2, AGR2 and Villin1. n=12. Scale bar, 20 μm.
Supplementary Material
Acknowledgments
We thank Dr. Brigid Hogan at Duke University and Ms. Maria R. Stupnikov at the Columbia Center for Human Development for critical reading of the manuscript. This work in Que lab is partly supported by R01DK113144, R01DK100342, R01HL132996, March of Dimes Research Grant 1-FY14-528 and the Price Family Foundation. This work is also supported by R01CA112403 and R01 CA193455 (J.X), the National Key Research and Development Program of China 2016YFA0502202 (H.W), National Natural Science Foundation of China 81728001 (J.Q), 31471121 (H.C), 81302068 and 81772994 (K.L), the Program for the Top Young Innovative Talents of Fujian Province and the International Collaborative Project of Fujian Province (project # 2017I0014, K.L).
Footnotes
Contributions
M.J. and J.Q. designed experiments, analyzed data, and wrote the manuscript. M.J. and J.Q. generated the otet-Cdx2 transgenic and Krt7-CreER knockin mouse lines. M.J. performed immunostaining, imaging, laser micro-dissection, flow cytometry, organoid culture and mouse genetics. H.L. performed quantitative PCR and immunostaining. Y.L. and J.Y. generated the CDX2 overexpression plasmid. Y.Z. performed viral work. Y.Y. generated p63 null mutants. K.L., S.L. assisted with mouse genetics. R.L., X.C., S.J. performed the acid reflux surgery. X.L., H.W., J.Z., J.S. assisted with imaging. J.X., D.L. provided the p63-CreER mouse line. Z.Z., L.Z., J.A., T.W., A. S., Q.W., H.C., X.S. provided human MLE and BE samples.
Author Information
The authors declare no competing financial interests
References
- 1.Yang EJ, et al. Microanatomy of the cervical and anorectal squamocolumnar junctions: a proposed model for anatomical differences in HPV-related cancer risk. Mod Pathol. 2015;28:994–1000. doi: 10.1038/modpathol.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirkovic J, et al. Carcinogenic HPV infection in the cervical squamo-columnar junction. J Pathol. 2015;236:265–271. doi: 10.1002/path.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spechler SJ, Souza RF. Barrett’s esophagus. N Engl J Med. 2014;371:836–845. doi: 10.1056/NEJMra1314704. [DOI] [PubMed] [Google Scholar]
- 4.McDonald SA, Lavery D, Wright NA, Jansen M. Barrett oesophagus: lessons on its origins from the lesion itself. Nat Rev Gastroenterol Hepatol. 2015;12:50–60. doi: 10.1038/nrgastro.2014.181. [DOI] [PubMed] [Google Scholar]
- 5.Milano F, et al. Bone morphogenetic protein 4 expressed in esophagitis induces a columnar phenotype in esophageal squamous cells. Gastroenterology. 2007;132:2412–2421. doi: 10.1053/j.gastro.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 6.Kong J, Crissey MA, Funakoshi S, Kreindler JL, Lynch JP. Ectopic Cdx2 expression in murine esophagus models an intermediate stage in the emergence of Barrett’s esophagus. PLoS One. 2011;6:e18280. doi: 10.1371/journal.pone.0018280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quante M, et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. 2012;21:36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, et al. Residual embryonic cells as precursors of a Barrett’s-like metaplasia. Cell. 2011;145:1023–1035. doi: 10.1016/j.cell.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaughan TL, Fitzgerald RC. Precision prevention of oesophageal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2015;12:243–248. doi: 10.1038/nrgastro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glickman JN, Chen YY, Wang HH, Antonioli DA, Odze RD. Phenotypic characteristics of a distinctive multilayered epithelium suggests that it is a precursor in the development of Barrett’s esophagus. Am J Surg Pathol. 2001;25:569–578. doi: 10.1097/00000478-200105000-00002. [DOI] [PubMed] [Google Scholar]
- 11.Sarosi G, et al. Bone marrow progenitor cells contribute to esophageal regeneration and metaplasia in a rat model of Barrett’s esophagus. Dis Esophagus. 2008;21:43–50. doi: 10.1111/j.1442-2050.2007.00744.x. doi:DES744 [pii] 10.1111/j.1442-2050.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- 12.Leedham SJ, et al. Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett’s oesophagus. Gut. 2008;57:1041–1048. doi: 10.1136/gut.2007.143339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang M, et al. BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. J Clin Invest. 2015;125:1557–1568. doi: 10.1172/JCI78850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu K, et al. Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells. Cell Stem Cell. 2013;12:304–315. doi: 10.1016/j.stem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabibi D, et al. Keratin 7 expression as an early marker of reflux-related columnar mucosa without intestinal metaplasia in the esophagus. Med Sci Monit. 2009;15:CR203–210. [PubMed] [Google Scholar]
- 16.Van Keymeulen A, Blanpain C. Tracing epithelial stem cells during development, homeostasis, and repair. J Cell Biol. 2012;197:575–584. doi: 10.1083/jcb.201201041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez P, et al. BMP signaling in the development of the mouse esophagus and forestomach. Development. 2010;137:4171–4176. doi: 10.1242/dev.056077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doupe DP, et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 2012;337:1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc Natl Acad Sci U S A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okumura T, Shimada Y, Imamura M, Yasumoto S. Neurotrophin receptor p75(NTR) characterizes human esophageal keratinocyte stem cells in vitro. Oncogene. 2003;22:4017–4026. doi: 10.1038/sj.onc.1206525. [DOI] [PubMed] [Google Scholar]
- 22.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 23.DeWard AD, Cramer J, Lagasse E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014;9:701–711. doi: 10.1016/j.celrep.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daniely Y, et al. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004;287:C171–181. doi: 10.1152/ajpcell.00226.2003. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs IJ, Ku WY, Que J. Genetic and cellular mechanisms regulating anterior foregut and esophageal development. Dev Biol. 2012;369:54–64. doi: 10.1016/j.ydbio.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips RW, Frierson HF, Jr, Moskaluk CA. Cdx2 as a marker of epithelial intestinal differentiation in the esophagus. Am J Surg Pathol. 2003;27:1442–1447. doi: 10.1097/00000478-200311000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Silberg DG, et al. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology. 2002;122:689–696. doi: 10.1053/gast.2002.31902. [DOI] [PubMed] [Google Scholar]
- 28.Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional gene expression in the epidermis of transgenic mice using the tetracycline-regulated transactivators tTA and rTA linked to the keratin 5 promoter. J Invest Dermatol. 2000;115:788–794. doi: 10.1046/j.1523-1747.2000.00144.x. [DOI] [PubMed] [Google Scholar]
- 29.Lu Y, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS One. 2010;5:e11022. doi: 10.1371/journal.pone.0011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DK, Liu Y, Liao L, Wang F, Xu J. The prostate basal cell (BC) heterogeneity and the p63-positive BC differentiation spectrum in mice. Int J Biol Sci. 2014;10:1007–1017. doi: 10.7150/ijbs.9997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao J, Liu B, Yang CS, Chen X. Gastroesophageal reflux leads to esophageal cancer in a surgical model with mice. BMC Gastroenterol. 2009;9:59. doi: 10.1186/1471-230X-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009;136:1899–1907. doi: 10.1242/dev.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the main data supporting the findings pf this study are available within the paper and its supplementary information. Any additional data are available from the corresponding authors upon request.


