Abstract
Shiga toxin-producing Escherichia coli (STEC) strains are the main cause of bacillary dysentery, although STEC strains generally induce milder disease symptoms compared to Shigella species. This study aimed to determine the virulence of STEC using the nematode Caenorhabditis elegans as a model host. Worm killing, fertility and bacterial colonisation assays were performed to examine the potential difference in the virulence of STEC strains compared to that of the control E. coli OP50 strains on which worms were fed. A statistically significant difference in the survival rates of C. elegans was observed in that the STEC strains caused death in 8–10 days and the E. coli OP50 strains caused death in 15 days. STEC strains severely reduced the fertility of the worms. The intestinal load of bacteria in the adult stage nematodes harbouring the E. coli OP50 strains was found to be 3.5 log CFU mL-1. In contrast, the STEC strains E15, E18 and E22 harboured 4.1, 4.2 and 4.7 log CFU ml−1 per nematode, respectively. The heat-killed STEC strains significantly increased the longevity of the worms compared to the non-heated STEC strains. In addition, PCR-based genomic profiling of shiga toxin genes, viz., stx1 and stx2, identified in selected STEC strains revealed that these toxins may be associated with the virulence of the STEC strains. This study demonstrated that C. elegans is an effective model to examine and compare the pathogenicity and virulence variation of STEC strains to that of E. coli OP50 strains.
Introduction
Enteropathogenic Escherichia coli (EPEC) causes life-threatening infections in humans as a consequence of the production of shiga-like toxins. Shiga toxin-producing Escherichia coli (STEC) strains such as O157:H7 and non-O157 that consists of 6 serogroups, including O104, O111, O121, O145, O103 and O126, cause severe diarrhoea and haemorrhagic colitis (HC), and they can also lead to life-threatening diseases like haemolytic uremic syndrome (HUS)[1]. STEC is a pathogenic form of E. coli that causes dysentery similar to Shigella but with minor symptoms [2,3]. STEC is recognized as a diverse group of pathogens that closely resembles Shigella as it shows high similarity in specific pathogenic characteristics and certain metabolic traits [3–7]. An outbreak of diarrhoea in Germany caused by entero-aggregative STEC was associated with a high percentage of patients developing HUS. In early May 2011, this led to 782 cases of HUS (29 deaths) and 3128 non-HUS cases (17 deaths), making it the largest outbreak of HUS in the world [8]. Remarkably, the outbreak strain was serotyped as a novel O104:H4, which was not reported previously and has been associated with very few HUS cases [9]. Furthermore, there was an increase in illnesses due to non-O157 STEC strains caused by serogroups O26, O45, O103, O111, O121, and O145 as well as outbreaks attributed to STEC O26:H11, O111:H8, and O121:H19 [10]. Therefore, it is necessary that the toxic mechanisms of these bacteria be investigated further.
It is generally accepted that the presence of Shigella strains arose multiple times from several independent ancestral E. coli strains and that these strains are more appropriately classified as a group of pathogenic E. coli [11–14]. Enteroinvasive E. coli (EIEC), on the other hand, is thought to have evolved later than Shigella from different ancestral strains of E. coli [15]. In addition, further research must be done to characterize the virulent effects of STEC, which will prevent the appearance new evolved strains. Several animal models have been proposed for studying enterohemorrhagic E. coli (EHEC) infections [16–18]. However, it has been difficult to identify certain bacterial virulence factors due to the unavailability of a suitable model system to study the disease mechanism as well as numerous ethical considerations. Lack of a suitable animal model system currently hinders the development of an in vivo study of a STEC virulent strain based on systematic methods [19]. As an alternative to existing mammalian pathogenesis models, researchers have applied techniques in the study of human-pathogen interactions using Caenorhabditis elegans as a simple but suitable model system [20].
C. elegans is a free-living nematode and is ubiquitous in the soil environment. It is easy to culture and can be kept in a frozen state for a long duration during the hibernating stage. The steps are considered relatively simple, and it has the advantage of allowing observation of the progression from the cell to the fertilized worm. The primary food source of the nematodes in the soil environment mainly depends on bacteria, which can affect growth development, fecundity and survival [21]. The virulence mechanisms elucidated in C. elegans infected by select pathogens were found to be very similar to those in the human host, including colonisation with biofilm formation on the worms, fatal infection of the intestine, and killing by toxins [22]. Additionally, a study has been previously conducted in C. elegans on host-pathogen interactions that led to the study of the change in behavioural mechanisms. It also facilitated in distinguishing virulent and a virulent strains [23]. Therefore, C. elegans has been established as a suitable model system for the study of virulence and behaviour mechanisms of pathogenic bacteria. The intoxication and paralysis of C. elegans by strains of the EHEC serotype O157:H7 have also been documented [24–25].
Previous studies were performed on the toxicity and virulence mechanism of pathogenic strains in the nematode [26]. Studies have been conducted on the fecundity and lifespan enhancement with C. elegans as a suitable model due to its simple and short life cycle [27]. Recent studies have used the C. elegans nematode as an infection host for STEC[28–30]. C. elegans has been extensively used as a suitable, simple and reliable model to study host–pathogen interactions and identify virulence mechanisms [31–36]. In this study, we tested whether the C. elegans model can be used to distinguish virulence between toxin-producing E. coli (STEC) like O157:H7 strains and non-O157 strains, consisting of the 4 serogroups O104, O111, O121 and O145, by quantifying nematode survival rates. Further experiments demonstrated the toxic roles using the heat-killed bacterial assay, the chemotaxis assay, PCR primers for the identification of stx1 & stx2 toxin genes and the bacterial colonisation-based fertility assay as the assessing parameters (S1 Fig).
Results
Population density of STEC strains affects the rate of developmental progression
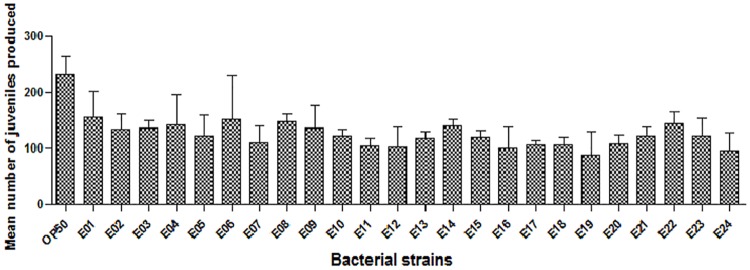
To determine whether STEC strains infect and kill C. elegans in this study concerning pathogenic and virulence mechanisms, we fed C. elegans 24 different STEC strains, which were compared to C. elegans that were given the E. coli OP50 strain as a sole food source. Single virgin hermaphrodite C. elegans were fed each STEC strain and analysed for the growth development and offspring production rate. Nematodes feeding on pathogenic E. coli (O157 and non-O157 strains) were severely infected compared to those on E. coli OP50 plates. After 4 days of exposure, the mean number of juveniles (young stage) that the C. elegans produced on the 24 STEC strains was significantly lower compared to that of the control (E. coli OP50) (Fig 1). All STEC strains reduced the reproductive capability by 30–70% compared to that of the E. coli OP50 strain. In contrast, there was no difference detected among the STEC serotypes O104, O111, O121, O145 and O157. This provides evidence that all of the 24 STEC strains used in the study decrease the size of young adult nematodes when compared to standard E. coli OP50 as the food source.
Fig 1. Fecundity of STEC strains in C. elegans compared to that of the E. coli OP50 strain.
Brood sizes of C. elegans were examined at 20°C. n>10 in each case.
Effect of STEC strains on C. elegans as a killing factor
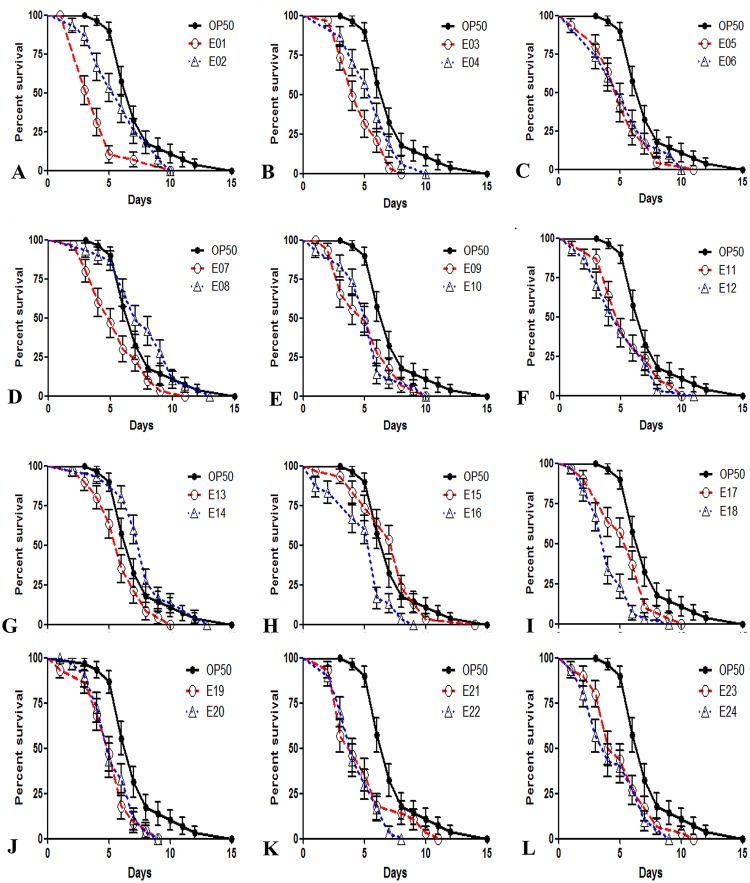
In order to determine the suitability of C. elegans as an animal model for studying STEC pathogenesis in vivo, the EHEC serotypes O104, O111, O121, O145 and O157 were tested for their ability to kill C. elegans. Nematodes were grown on a lawn of STEC strains, which were cultured on NGM plates at 20°C, and their growth development was observed for changes in body size, darkness of the intestine, and pharyngeal (mouth cavity muscles) activity based on pumping. The experiment revealed spontaneous movement in nematodes that were grown on a lawn of the avirulent E. coli OP50 strain. The nematodes grown on STEC strains were severely infected compared to those grown on control E. coli OP50 plates after 4 days, as evidenced by their pale appearance, the dilated intestinal lumen and the absence of pharyngeal pumping. As shown in Fig 2, each STEC serotype displayed a robust virulent phenotype that significantly reduced the lifespan of the nematode compared to that of the E. coliOP50 control. Nematodes feeding on E. coli O104:H12 B471 1.2673 (E1) plates displayed a median lifespan (LD50) of 4.24±0.43 days compared to the control median lifespan of 7.37±0.43 days for nematodes feeding on E. coli OP50 plates (Fig 2a). Nematodes feeding on E. coli O111:NM B473 96–3166 (E3) and E. coli O121:NM B483 9918 (E12) plates exhibited median lifespans of 4.75±0.29 days and 5.03±0.44 days, respectively, compared to that for nematodes feeding on the E. coli OP50 strain(Fig 2b and 2c). Also, the nematodes feeding on STEC O145:NM B489 BCL73 (E18) and E. coli O157:H7 B493 C7927(E22) plates appeared to have median lifespans of 4.17±0.31 days and 4.53±0.3 days, respectively, compared to that for nematodes feeding on the E. coli OP50 strain (Fig 2d and 2e). While almost all target bacteria reduced the lifespan of worms, E. coli O145:NM B485 83–75 (E14) and E. coli O145:NM B486 14728(E15) increased the survival rate of the worms. This result indicates that most STEC strains used in the study significantly reduce the longevity (lifespan) of C. elegans wild type N2.
Fig 2. Life span assay for C. elegans infected with STEC strains compared to that for C. elegans infected with the control E. coli OP50 strain.
C. elegans grown on STEC plates were significantly short-lived compared to those grown on E. coli OP50 plates. The E. coli OP50 strain was compared to 24 different STEC strains (A-L).A) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E1 and E2 strains. C. elegans grown on both E1 andE2 exhibited a decreased life span at the end of 10 days, but C. elegans grown on E. coli OP50 survived for 15 days. B) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E3 and E4 strains. C. elegans grown on both E3 and E4 exhibited a decreased life span, but when compared to C. elegans grown on E. coli OP50, C. elegans grown on E3 survived for 8 days, and C. elegans grown on E4 survived for 10 days. C) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E5 and E6 strains. C. elegans grown on both E5 and E6 exhibited a decreased life span at the end of 10 days, but C. elegans grown on E. coli OP50 survived for 15 days. D) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E7 and E8 strains. C. elegans grown on both E7 and E8 exhibited a decreased life span at the end of 10 days, but C. elegans grown on E. coli OP50 survived for 15 days. E) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E9 and E10 strains. C. elegans grown on both E9 and E10exhibited a decreased life span at the end of 10 days, but C. elegans grown on E. coli OP50 survived for 15 days. F) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E11 and E12 strains. C. elegans grown on both E11 and E12 showed a decreased life span at the end of 10 days, but C. elegans grown on E. coli OP50 survived for 15 days. G) Longevity of C. elegans grown on the E. coli OP50 strain compared that of C. elegans grown on the E13 and E14 strains. C. elegans grown on both E13 and E14 exhibited a decreased life span at the end of 10 days, but C. elegans grown on E. coli OP50 survived for 15 days. H) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E15 and E16 strains. C. elegans grown on both E15 and E16 exhibited a decreased life span at the end of 9 days, but C. elegans grown on E. coli OP50 survived for 15 days. I) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E17 and E18 strains. C. elegans grown on E17 exhibited a decreased life span at the end of 10 days, and C. elegans grown on E18 surprisingly survived for 6 days, but C. elegans grown on E. coli OP50 survived for 15 days. J) Longevity of C. elegans grown on the E. coli OP50 strain compared that of C. elegans grown on the E19 and E20 strains. C. elegans grown on both E19 and E20 exhibited a decreased life span at the end of 9 days, but C. elegans grown on E. coliOP50 survived for 15 days. K) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E21 and E22 strains. C. elegans grown on E21 showed a decreased life span at the end of 10 days, and in contrast, C. elegans grown on E22 survived for 7 days, but C. elegans grown on E. coli OP50 survived for 15 days. L) Longevity of C. elegans grown on the E. coli OP50 strain compared to that of C. elegans grown on the E23 and E24 strains. C. elegans grown on E23 showed a decreased life span at the end of 11 days, and in contrast, C. elegans grown on E24 survived for 9 days, but C. elegans grown on E. coli OP50 survived for 15 days.
Heat-treated STECin the C. elegans killing assay
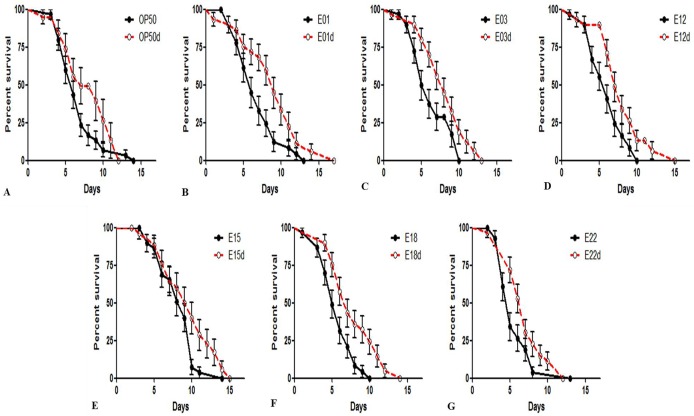
In order to determine the difference between heat-killed bacteria and live bacteria, we determined the lifespan of C. elegans fed heat-killed STEC strains (pellet). The effect of heat-killed STEC (80°C, 10min) strains in N2nematodes was quantitatively assessed at 20°C. It was determined that all of the heat-treated STEC strains significantly increased the survival rate of C. elegans compared to that of the non-heat-killed STEC strains. With respect to the control E. coli OP50 strain, these results revealed that the N2 worms fed heat-killed STEC (E1, 3, 12, 15, 18, 22) bacterial strains and the control E. coli OP50 strain had significantly longer lifespans (Fig 3). In contrast, a shorter lifespan was observed for C. elegans that were fed the non-heat-treated (live) E. coli bacterial strains. As shown in Fig 3a, the heat-killed E. coli OP50 strain increased the longevity of the worms (7.81±6.57 days) compared to the live E. coli OP50 strain. Likewise, the heat-treated bacteriaE1 (8.78±0.77 days), E3 (8.01±0.53 days), E12 (7.85±0.60 days), E15 (9.4 6 ± 0.68 days), E18 (7.72±0.58 days) and E22 (7.05±0.45 days) improved the survival rate of C. elegans compared to the live STEC strains E1 (6.78 ±0.51 days), E3 (6.21 ± 0.47 days), E12 (5.95 ± 0.42 days), E15 (8.20 ± 0.45 days), E18 (5.64 ± 0.38 days) and E22 (5.52 ± 0.41 days). These results suggest that heat treatment at 80°C causes the STEC strains to lose their toxin effect when compared to the live STEC strains in the C. elegans model.
Fig 3. Toxicity of heat-treated and live strains in the C. elegans model.
A) Heat-killed E. coli OP50 compared to live E. coli OP50. B) Heat-killed E1: O104 compared to live E1: O104. C) Heat-killed E3: O111 compared to live E3: O111. D) Heat-killed E12: O121 compared to live E12: O121. E) Heat-killed E15: O145 compared to live E15: O145. F) Heat-killed E18: O145 compared to live E18: O145. G) Heat-killed E22: O157 compared to live E22: O157. The survival curves of nematodes feeding on both heat-killed STEC strains and the E. coli OP50 strain were examined. The live strains were compared to the heat-killed strains. All heat-treated bacterial strains used in this assay increased the lifespan of C. elegans when compared to the control strains. This suggests that the toxic factors of STEC strains are heat-labile.
Identification of the stx1 and stx2 genes in STEC strains
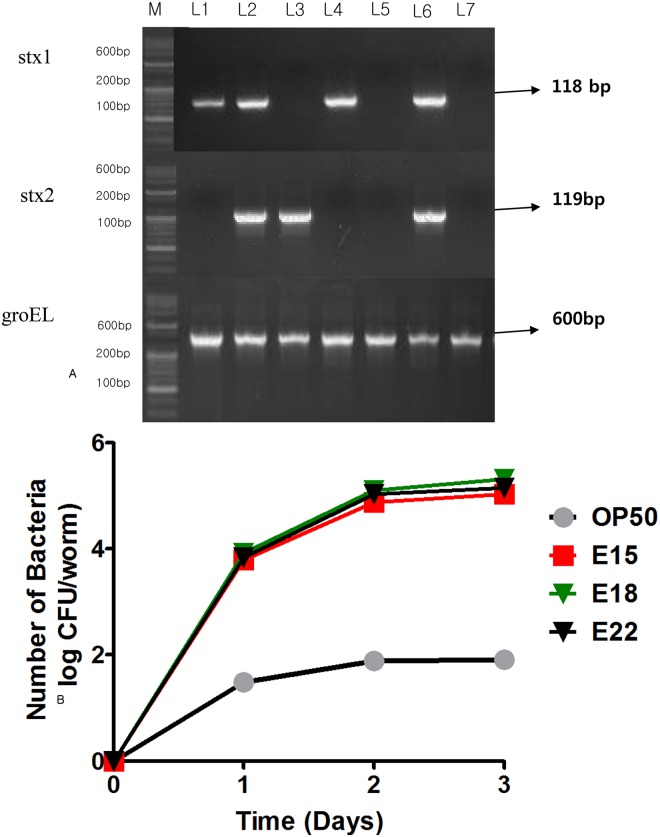
Conventional PCR was performed to confirm that STEC strains (L1—E1; L2—E3; L3—E12; L4—E15; L5—E18; L6—E22; L7 –OP50) produce stx1 and stx2 toxins. Based on our results, E1, E3 and E22 reduced the brood size and the lifespan of C. elegans. Compared to E. coli OP50, which possesses the stx1 gene, E12 and E18 lack the stx1 gene. In addition, E15, which reduced the progeny and affected the survival rate of the worms, possessed the stx1 gene (Table 1). Based on the PCR assay, the STEC toxin gene was confirmed by the detection of virulence genes (stx1 and stx2) in addition to groEL. The 3 primer pairs (Table 2) used in the assay do not interfere with each other and generate amplification products 118, 119, and 600bp in size (Fig 4a).
Table 1. Bacterial (E. coli) strains used in the study and their respective sources.
| Arbitrary | Isolatesa | ||||||
|---|---|---|---|---|---|---|---|
| ID No.b | Original ID no. | Serotype | Source(reference) | stx1 | stx2 | groEL | |
| OP50 | E. coli OP50 | - | - | + | |||
| E1 | B471 | 1.2673 | O104:H12 | Co | + | - | + |
| E2 | B472 | JB1-95 | O111:H- | H | NT | NT | NT |
| E3 | B473 | 96–3166 | O111:NM | H | + | + | + |
| E4 | B475 | TB226 | O111:NM | H | NT | NT | NT |
| E5 | B476 | 8361 | O111:H8 | H | NT | NT | NT |
| E6 | B477 | 12893 | O111:H8 | H | NT | NT | NT |
| E7 | B478 | 14895 | O111:H8 | H | NT | NT | NT |
| E8 | B479 | DA-1 | O121:NM | H | NT | NT | NT |
| E9 | B480 | 97–3068 | O121:H19 | H | NT | NT | NT |
| E10 | B481 | 03–4064 | O121:NM | H | NT | NT | NT |
| E11 | B482 | 11435 | O121:H19 | H | NT | NT | NT |
| E12 | B483 | 9918 | O121:NM | H | - | + | + |
| E13 | B484 | 10896 | O121:NM | H | NT | NT | NT |
| E14 | B485 | 83–75 | O145:NM | H | NT | NT | NT |
| E15 | B486 | 14728 | O145:NM | H | + | - | + |
| E16 | B487 | 940941 | O145:H- | H | NT | NT | NT |
| E17 | B488 | 6383 | O145:NM | H | NT | NT | NT |
| E18 | B489 | BCL73 | O145:NM | Co | - | - | + |
| E19 | B490 | 8235 | O145:NM | H | NT | NT | NT |
| E20 | B491 | 6896 | O145:NM | H | NT | NT | NT |
| E21 | B492 | sakai | O157:H7 | H | NT | NT | NT |
| E22 | B493 | C7927 | O157:H7 | H | + | + | + |
| E23 | B494 | FSIS413-95 | O157:H7 | Gb | NT | NT | NT |
| E24 | B495 | 380–94 | O157:H7 | H | NT | NT | NT |
Bacterial strains used in this study and specification of three primers based on conventional PCR assay (H- Human; Gb- ground beef; Co- cow, NT- Not Tested)
Table 2. Oligonucleotide primer sequences used in this study with respect to product size.
| Targeting genes | Primers | Primer Sequence (5’→3’) | Product Size(bp) | Annealing Temperature (°C) | Source |
|---|---|---|---|---|---|
| stx1 | STX1-F-O157 | GAAAGCGATGCAGCTATTA | 789 | 60 | Chou et al., 2013 |
| STX1-R-O157 | GGATAATTTGTTTGCAGTTG | ||||
| stx2 | STX2-F-O157 | TATTATTTAAATGGGTACTGTGC | 1073 | 60 | |
| STX2-R-O157 | ATGTGTCATCCTCATTATACTTG | ||||
| groEL | GroEL-F-O157 | CCGTAACGTAGTTCTGGATA | 1493 | 60 | |
| GroEL-R-O157 | CTAAGTCAGCTGCATCGTT |
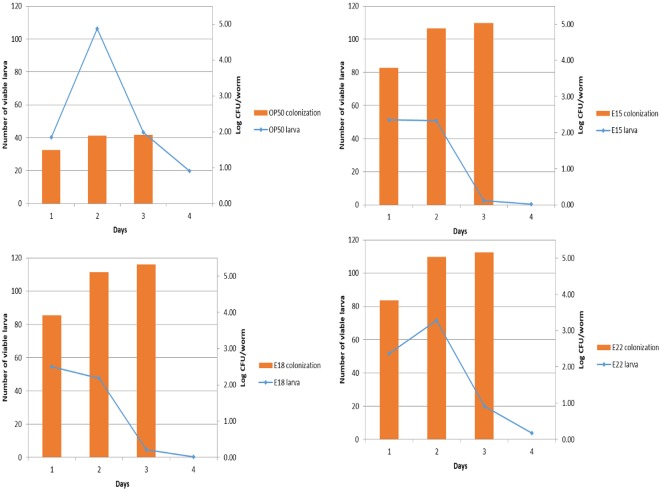
Fig 4. Conventional PCR and colonisation assay.
Conventional PCR: Based on the results of the life span assay, 6 STEC strains (L1—E1; L2—E3; L3—E12; L4—E15; L5—E18; L6—E22; L7 –OP50) out of the 24 strains were selected for further testing for the presence of toxin genes (stx1, stx2 and groEL) using 3 primer pairs (STX1-F/R-O157; STX2-F/R-O157; GroEL-F/R-O157) targeting the genes specific for toxin production(stx1, stx2 and groEL). Colonisation assay: The STEC strains colonised in the intestinal tract of L2 worms (log CFU/worm). p<0.001 was obtained for comparison with the E. coli OP50 control group by the t-test. The total numbers of worms (C. elegans) tested in each group are indicated by n.
Larval population density predetermines adult lifespan
Similar to the pathogenic colony formation in the human gastrointestinal tract, we observed the colony formation of STEC strains in the intestine of C. elegans. These results indicated that the intestines of the worms possessed a high colony number (3.34 and 4.70 log CFU/worm on each day) of E22 on the first and third days. In contrast, there were 3.23 and 2.40 log CFU/worm of E18 and E15, respectively, on the first day and subsequently 4.23 and 4.12 log CFU/worm of E18 and E15, respectively, on the third day of growth (Fig 4b). However, C. elegans was colonised with 1.92 and 3.59 log CFU/worm of the control E. coli OP50 strain on each day. Hence, the tested STEC strains (E15, E18 and E22) colonised to a greater degree in C. elegans compared to the E. coli OP50 strain. This result correlates with the reduction in the life span of C.elegans. STEC colonisation is proportional to a decrease in the longevity and fertility of the worm (Fig 5). In particular, E22 was observed to have accumulated to more than 1 log CFU/worm in all days compared to the control E. coli OP50 strain.
Fig 5. Correlation between the life span and fertility of C. elegans infected with STEC strains.
The STEC (E15, E18 and E22) strains showed an increase in colonisation, which correlates with a decrease in longevity and fertility of the worm (C. elegans). This is considered evidence of STEC pathogenicity in the C. elegans model.
Discussion
The C. elegans model has been used to determine the pathogenesis and host susceptibility to various pathogens [37]. Specifically, bacterial virulence and host susceptibility mechanisms identified in C. elegans are often well preserved. Thus, many bacterial mutants that are avirulent in C. elegans are also avirulent in other mammals [38]. Moreover, a variety of pathogens with specific modes of infection were analysed using the C. elegans model.
Previous studies have utilized C. elegans as an in vivo model to study the molecular mechanism regarding the virulence of gene-based pathogens of bacterial species. Previous studies reported on the toxic effects and pathogenicity of other enteropathogenic group (EEPC) strains with virulence factor expression and inflammatory responses in C. elegans [39]. Indole-based toxicity in C. elegans is likely to be protective in mammals because they suppress the expression of bacterial virulence genes and shiga toxin at high concentrations [40]. Moreover, our findings suggest that STEC strains possessing the toxin-producing gene [41–42] reduce the life span of C. elegans (Fig 3). Likewise, most of the Shigella toxins have been found to be exotoxins [43–44], and they belong to a small molecular protein family such as peptides and have been reported to be heat sensitive [45]. Our results highlight the utility of C. elegans-pathogen systems in conjunction with heat-treated E. coli, which showed reduced toxicity. Indeed, these methodologies may be broadly applicable in identifying the relationship between host-toxin interaction and the characterization of the toxin properties in many organisms [5]. Most food poisoning is caused by the toxicity of non-E. coli O157 strains as well as E. coli O157:H7. Although many studies have reported on the toxicity of E. coli O157:H7, there is little research on the toxicity of non-O157 strains. Therefore, we identified the virulence of pathogenic E. coli, including O157 strains and non-O157 strains, using C. elegans as a model system.
There have been few studies reported previously on the interaction between pathogenic E. coli and C. elegans [46]. Our results demonstrate that STEC strains infect and kill nematodes. We showed that STEC strains decrease the brood size and lifespan of C. elegans. We also demonstrated that pathogenic E. coli strains form colonies and increase in population in the intestinal tract of worms. Moreover, our genetic analysis and heat-killed bacteria killing assay results associatedstx1 and stx2 of STEC strains with its toxicity. Our results suggest that C. elegans is a suitable alternative model for studying the infection of pathogenic bacteria, including STEC strains, in hosts in vivo.
These results suggest that C. elegans is highly susceptible to infection with most of the STEC strains, and the normal morphology of the intestine in the infected animals may be affected. The fertility and lifespan assay showed that most of the STEC strains reduced the brood size and life span of C. elegans similar to that observed for C. elegans infected with E. coli. Each of the E1, E3, E12, E18 and E22STEC strain serotypes was selected for screening for STEC strain virulence in C. elegans because they were the most toxic of the serotypes. In addition, E15, which was associated with a survival rate similar to that of E. coli OP50, was used in further experiments. Due to certain limitations in the nematode model, the worm model may not predict a 100% correlation[5]. However, infection can play a major role in the ability of human pathogens to interact with a host, the mode of virulence, and the sensitivity that can be expressed in C. elegans [47–48]. As such, it is quite possible that the clinical isolates, which were non-pathogenic to C. elegans in our assay, did not express the appropriate virulence factors. Consequently, the differences in pathogenicity for some of the clinical isolates in this study may be due to differences in the infection conditions. This observation raises fascinating questions concerning how the expression of various genes differs depending on the infection conditions (Fig 2).
Based on the results obtained, it had been determined that the STEC strains are responsible for reducing the brood size and lifespan of L2 worms. This may be due to the colonisation of virulent bacteria. It was also reported that the E. coli O157 strain causes toxicity based on colony formation in the gastrointestinal tract of humans. Hence, STEC strains may colonize in the intestine of the nematodes and cause toxicity towards C. elegans. Thus, C. elegans can be used as a model system to screen virulence mechanisms of STEC strains when compared to the control E. coli OP50 strain.
The E.coli O157:H7 strain, the most common type of STEC, harboursa virulence plasmid that possesses genes such as stx1, stx2, eae, and ehxA [49–51], which are more commonly associated with severe human diseases[52–53]. It can produce two different shiga-like toxins, stx1 and stx2, which are encoded on prophages embedded in the genome. The stx1 is very similar to the shiga toxin of Shigella dysenteriae, and stx2 is genetically and immunologically distinct from stx1 [54]. Also, a recent report suggested that stx1 of EHEC is almost entirely cell-associated and causes many diseases in humans in vivo only when it is released by bacterial lysis, whereas stx2 can be released by two different mechanisms from bacterial cells into the extracellular milieu [55]. Our PCR and electrophoresis results showed that E1, E3, E15 and E22 possessed the stx1 gene, while E12 and E18 exhibited strong virulence towards C. elegans via stx2(Table 1). As previously reported, E. coli O157:H7 possessed stx1 and stx2, and the heat-treated E. coli O157:H7 strain did not cause an early death of C. elegans in the heat-killed bacteria killing assay. It has been reported that the enzyme activity of stx1 is greatly decreased at higher temperatures (Brigotti et al., 2004), whereas stx2 is heat-stable and not inactivated at higher temperatures (Rasooly and Do, 2010). As a result, all of the heat-treated STEC strains significantly increased the lifespan of C. elegans compared to control STEC strains, as suggested by a significantly longer longevity of the L2 worms fed heat-killed target bacteria and the control E. coliOP50 strain (Fig 3). The most prominent virulence genes evaluated in the present study were significant in that they were detected at a much higher rate pathogenetically (Fig 4a), and Table 1shows the adhesion-encoding genes focG, focA, and sfaD that encode a receptor with an auto-transporter protein that were typically associated with extra-intestinal infections. In addition, theywere required for synthesis of the F1C pilus and were upregulated during a urinary tract infection (UTI) caused by the E. coli strain CFT073 [5]. It is imperative to note that the genes themselves may not be directly involved in infection, but they could represent the effect of a combination of genes interlinked with the selective virulence genes of pathogenicity [21] that were not screened for during this PCR analysis. Moreover, regulatory genes required for the mRNA expression of virulence genes may be suppressive or not present, which results in the dissimilarity between the non-virulence type and pathogenicity in the C. elegans assay [46].
In conclusion, the virulence factor of the E. coli O157 strain is primarily shiga toxin 1, and the heat-killed E. coli O157 strain could not colonize in the intestine of C. elegans and cause toxicity due to an inactivated stx1. Although some non-O157 strains are believed to be similar to the O157 strain, both E12 and E18 are considered to be toxic by forming colonies in the intestinal tract of C. elegans rather than producing shiga toxins. The relatively rapid method of conventional PCR was used to identify both the stx1 and the stx2 genes and confirm the presence of STEC. Using C. elegans as an invivo model for the study of the virulence of shiga toxin-producing E. coli, our work further focused on detecting toxin genes with similar PCR profiles in patients within a short period of time and may provide the first evidence of a link between cases consistent with common-source outbreaks. The PCR analysis is a valuable diagnostic and epidemiological tool, which facilitates the analysis of STEC before it causes severe disease in humans.
Material and methods
Bacterial strains and growth media
The STEC strains used in this study are listed in Table 1. The US Food Fermentation Laboratory Culture Collection (Raleigh, NC, USA) and the Agricultural Research Service, Eastern Regional Research Center (Wyndmoor, PA, USA) provided4 STEC E. coli O157:H7 strains and 20non-O157 STEC strains, consisting of the 4 serogroups O104, O111, O121,and O145 from various sources such as cow, ground beef, and humans. The bacterial strains were stored at –80 °C in tryptic soy broth (TSB, BD Biosciences, San Jose, CA, USA) supplemented with 30% glycerol. Each culture was streaked from frozen stocks onto tryptic soy agar (TSA, BD Bioscience) and incubated at 37 °C for 24 h.
Population density-dependent egg-laying assays
Nematode and stage of development
C. elegans is studied in four stages (L1, L2, L3 and L4). The stage immediately following egg hatching is called the L1 stage. Adult worms lay their eggs, and the stage before C. elegans lays eggs is called the L4 stage. It is maintained on 35-mm diameter NGM (Nematode Growing Media) agar plates seeded with 50μL of E. coli OP50, and it is stored at 20°C. C. elegans (N2) strains were obtained from the Caenorhabditis Genetic Center (CGC; MN).
Nematode synchronizing
The young adult worms hatched from eggs were treated with sterile water containing 0.5 M NaOH in addition to 0.5% bleaching solution (sodium hypochlorite and sodium hydroxide) for synchronization. Briefly, the assay was performed as previously described [56]. After vortexing in 2-min intervals, the worms were washed with M9 buffer via centrifugation (1200 × g for 200 sec) and resuspended, which was subsequently repeated three times. To prepare worms for all assays, approximately 3000 of the synchronized eggs were hatched in M9 buffer at 20°C overnight. The L1 larvae were subsequently transferred to NGM agar containing a lawn of E. coli OP50 and incubated at 25°C for 48 h to reach the L4 stage.
C. elegansfertility assay
L2 young adult worms were transferred to new NGM plates containing 50 μL of the target bacterial strain cultured in TSB every day until they did not produce progeny. The NGM plates containing eggs were incubated at 20°C for another 48 h, and the number of progeny was counted for each adult worm tested at the end of 3 days. The fertility of worms was monitored for wild-type L2 [57].
Lifespan assay
The longevity of C. elegans was determined by a previously described method [58]. In the experiment, 50 μL of the target bacterial strain cultured in TSB at 25°C for 24 h was spread on a 35-mm diameter NGM agar plate and incubated overnight at 37°C. Each plate was seeded with 30 C. elegans L4 stage larva or young adults grown on E. coli OP50. All plates were incubated at 25°C, and dead worms were counted every 24 h. Liveworms were transferred onto fresh NGM agar plates seeded with the target bacterial strain every 2 days. The experiment was performed in triplicate for each strain. Each plate was examined until the worms died. In order to compare the effects of the E. coli (STEC) strains, worms were grown on E. coli OP50, and the mean life span (MLS) was calculated by using the formula previously described [59].
Effect of heat on toxic compounds
Heat-killed bacteria killing assay
The assay plates were prepared as follows using the procedures outlined above for the killing assay. The STEC strains and the E. coli OP50strain were incubated overnight at 37°C in TSB medium. Using a dry bath (Vision Scientific -251D12), the bacteria were treated aseptically at 80°C for 10 min, and 50-ml aliquots were spread-plated on NGM agar as well as Eosin Methylene Blue (EMB) agar (selective media for E. coli) to identify whether the bacteria were killed by heat-treatment.
Colonisation-virulence interaction
The number of colonised worms and their bacterial load were estimated in individual progeny samples at day 3 in parallel with the number of bacteria present on the plate. Immediately following the killing assays, bacterial colonisation in the intestine of C. elegans was quantified by the spread-plating method to determine the number of live bacteria in the worm (L2). The young adult L4 stage worms were fed either STEC strains or E. coli OP50 for 3 days at 20°C. Five L2 worms were picked after 1 day and 3days. The surface bacteria of C. elegans were removed by washing the worms subsequently three times in 1ml of 1% Triton X-100, and finally,5 worms were disrupted using a pestle with 20μL of 1%Triton X-100. They were diluted to 10−1, 10−2, 10−3 and 10−4 with 1ml of M9 buffer and then plated on EMB agar. Bacterial colonies (log CFU/ml) were counted after 16 h of incubation at 37°C. Each experiment was performed on the 1st and 3rd days.
Evaluation of potential virulent genes associated with pathogenesis
Primary identification and characterization of the stx1 and stx2virulent genes in STEC responsible for toxin production were performed using conventional PCR with the primers given in Table 2. The primary PCR results for stx1, stx2, and groEL in all strains are included in the study. For the PCR, the bacterial strains were grown overnight on TS agar. One of the bacterial colonies was suspended in 100 μL of lysis buffer (50 mMKCl,10 mM Tris-HCl, pH 8.3,2.5 mM MgCl2,0.45% NP-40, 0.45% Tween 20) and 100 μLof Tris-EDTA (TE) buffer solution (pH 7.4) and boiled for 15 min at 95°C. After centrifugation at 13,000 rpm for 1 min, the supernatant was used directly in the PCR using standardized reaction conditions (denaturation at 95°C for 10 min; 35 cycles of 95°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec; and a 3-min final extension at 72°C).
Statistics
The reproducibility of the qPCR was determined using three independent experiments conducted in triplicate. The data obtained from each experiment were expressed as the mean, and the standard deviation is shown as error bars. The asterisk denotes p<0.001 compared to the E. coli OP50 control group calculated by the t-test. The total numbers of samples in each group are indicated by n (Fig 4b). Differences in the survival of C. elegans in the infection assays were determined using Graph Pad Prism version 5.00 (www.graphpad.com).
Supporting information
(PDF)
Acknowledgments
The authors would like to thank Caenorhabdtis Genetics Center (CGC), JinIl Lee, and Younghoon Kim for providing the C. elegans strains, and Mr. Park-Yong IK and Ms. Lee Hyun Ah of Kangwon National University, Central laboratory, for training and technical support for confocal analysis. This work was supported by a grant from the Ministry of Agriculture, Food and Drug Safety and Kangwon National University 14162MFDS085.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research work was carried out with the support of research grant (14162MFDS085) to DHO from Ministry of Agriculture, Food and Drug Safety and Kangwon National University. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Singh P, Sha Q, Lacher DW, Del Valle J, Mosci RE, Moore JA, et al. (2015) Characterization of enteropathogenic and Shiga toxin-producing Escherichia coli in cattle and deer in a shared agroecosystem. Frontiers in cellular and infection microbiology, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michelacci V, Tozzoli R, Caprioli A, Morabito S. (2017). Verocytotoxin-Producing Escherichia coli in the Genomic Era: From Virulotyping to Pathogenomics In Applied Genomics of Foodborne Pathogens (pp. 109–126). Springer International Publishing. [Google Scholar]
- 3.Smati M, Magistro G, Adiba S, Wieser A, Picard B, Schubert S, et al. (2017) Strain-specific impact of the high-pathogenicity island on virulence in extra-intestinal pathogenic Escherichia coli. International Journal of Medical Microbiology, 307(1), 44–56 doi: 10.1016/j.ijmm.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 4.Chou TC, Chiu HC, Kuo CJ, Wu CM, Syu WJ, Chiu WT, et al. (2013) Enterohaemorrhagic Escherichia coli O157: H7 Shiga like toxin 1 is required for full pathogenicity and activation of the p38 mitogen activated protein kinase pathway in Caenorhabditis elegans. Cellular microbiology, 15(1), 82–97. doi: 10.1111/cmi.12030 [DOI] [PubMed] [Google Scholar]
- 5.Merkx-Jacques A, Coors A, Brousseau R, Masson L, Mazza A, Tien YC, et al. (2013) Evaluating the pathogenic potential of environmental Escherichia coli by using the Caenorhabditis elegans infection model. Applied and environmental microbiology, 79(7), 2435–2445. doi: 10.1128/AEM.03501-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bommarius B, Anyanful A, Izrayelit Y, Bhatt S, Cartwright E, Wang W,et al. (2013) A family of indoles regulate virulence and Shiga toxin production in pathogenic E. coli. PLoS One, 8(1), e54456 doi: 10.1371/journal.pone.0054456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun EJ, Lee SH, Kim S, Kim SH, Kim KH, (2017) Global profiling of metabolic response of Caenorhabditis elegans against Escherichia coli O157: H7. Process Biochemistry, 53, 36–43. [Google Scholar]
- 8.Cheung MK, Li L, Nong W, Kwan HS, (2011) 2011 German Escherichia coli O104: H4 outbreak: whole-genome phylogeny without alignment. BMC research notes, 4(1), 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bae WK, Lee YK, Cho MS, Ma SK, Kim SW, Kim NH, et al. (2006) A case of hemolytic uremic syndrome caused by Escherichia coli O104: H4. Yonsei medical journal, 47(3), 437–439. doi: 10.3349/ymj.2006.47.3.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor EV, Nguyen TA, Machesky KD, Koch E, Sotir MJ, Bohm SR, et al. (2013) Multistate Outbreak of Escherichia coli O145 Infections Associated with Romaine Lettuce Consumption, 2010‡ §. Journal of food protection, 76(6), 939–944. doi: 10.4315/0362-028X.JFP-12-503 [DOI] [PubMed] [Google Scholar]
- 11.Welch RA, Burland V, Plunkett G, Redford P, Roesch P, Rasko D, et al. (2002) Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proceedings of the National Academy of Sciences, 99(26), 17020–17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paauw A, Jonker D, Roeselers G, Heng JM, Mars-Groenendijk RH, Trip H, et al. (2015) Rapid and reliable discrimination between Shigella species and Escherichia coli using MALDI-TOF mass spectrometry. International Journal of Medical Microbiology, 305(4), 446–452. [DOI] [PubMed] [Google Scholar]
- 13.Njamkepo E, Fawal N, Tran-Dien A, Hawkey J, Strockbine N, Jenkins C, et al. (2016) Global phylogeography and evolutionary history of Shigelladysenteriae type 1. Nature microbiology, 1, 16027 doi: 10.1038/nmicrobiol.2016.27 [DOI] [PubMed] [Google Scholar]
- 14.Dunne KA, Chaudhuri RR, Rossiter AE, Beriotto I, Browning DF, Squire D, et al. (2017) Sequencing a piece of history: complete genome sequence of the original Escherichia coli strain. Microbial Genomics, 3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lan R, Alles MC, Donohoe K, Martinez MB, Reeves PR (2004) Molecular evolutionary relationships of enteroinvasive Escherichia coli and Shigella spp. Infection and immunity, 72(9), 5080–5088. doi: 10.1128/IAI.72.9.5080-5088.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panda A, Tatarov I, Melton-Celsa AR, Kolappaswamy K, Kriel EH, Petkov D, et al. (2010) Escherichia coli O157: H7 infection in Dutch belted and New Zealand white rabbits. Comparative medicine, 60(1), 31–37. [PMC free article] [PubMed] [Google Scholar]
- 17.Golan L, Gonen E, Yagel S, Rosenshine I, Shpigel NY (2011) Enterohemorrhagic Escherichia coli induce attaching and effacing lesions and hemorrhagic colitis in human and bovine intestinal xenograft models. Disease models & mechanisms, 4(1), 86–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohawk KL, O’Brien AD (2011) Mouse models of Escherichia coli O157: H7 infection and shiga toxin injection. BioMed Research International, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franz E, Delaquis P, Morabito S, Beutin L, Gobius K, Rasko DA, et al. (2014) Exploiting the explosion of information associated with whole genome sequencing to tackle Shiga toxin-producing Escherichia coli (STEC) in global food production systems. International journal of food microbiology, 187, 57–72. doi: 10.1016/j.ijfoodmicro.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 20.Sarabhai S (2014) Molecular mechanism of terminaliachebula RETZ mediated quenching of Pseudomonas aeruginosa PAO1 virulence and use of Caenorhabditis elegans as the non mammalian model host system
- 21.Diaz SA, Viney M (2015) The evolution of plasticity of dauer larva developmental arrest in the nematode Caenorhabditis elegans. Ecology and evolution, 5(6), 1343–1353. doi: 10.1002/ece3.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abada EAE, Osman MES, Bandyopadhyay J (2007) Use of Caenorhabditis elegans as a Genetic Model for Bacterial Pathogenesis Studies. Korean Journal of Genetics, 29(3), 275. [Google Scholar]
- 23.Zhang Y, Lu H, Bargmann CI (2005) Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature, 438(7065), 179 doi: 10.1038/nature04216 [DOI] [PubMed] [Google Scholar]
- 24.Anyanful A, DolanLivengood JM, Lewis T, Sheth S, DeZalia MN, Sherman MA, et al. (2005) Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Molecular microbiology, 57(4), 988–1007. doi: 10.1111/j.1365-2958.2005.04739.x [DOI] [PubMed] [Google Scholar]
- 25.Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM (2008) A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nature genetics, 40(2), 181–188. doi: 10.1038/ng.2007.70 [DOI] [PubMed] [Google Scholar]
- 26.Huang X, Tian B, Niu Q, Yang J, Zhang L, Zhang K (2005) An extracellular protease from Brevibacilluslatero sporus G4 without parasporal crystals can serve as a pathogenic factor in infection of nematodes. Research in Microbiology, 156(5–6), 719–727. doi: 10.1016/j.resmic.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 27.So S, Tokumaru T, Miyahara K, Ohshima Y (2011) Control of lifespan by food bacteria, nutrient limitation and pathogenicity of food in C. elegans. Mechanisms of ageing and development, 132(4), 210–212. doi: 10.1016/j.mad.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 28.George DT, Behm CA, Hall DH, Mathesius U, Rug M, Nguyen KC, et al. (2014) Shigellaflexneri Infection in Caenorhabditis elegans: cytopathological examination and identification of host responses. PloS one, 9(9), e106085 doi: 10.1371/journal.pone.0106085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fung CC, Octavia S, Mooney AM, Lan R (2015) Virulence variations in Shigella and enteroinvasive Escherichia coli using the Caenorhabditis elegans model. FEMS microbiology letters, 362(3), 1–5. [DOI] [PubMed] [Google Scholar]
- 30.Rivas M, Chinen I, Guth BE (2016) Enterohemorrhagic (Shiga toxin-producing) Escherichia coli In Escherichia coli in the Americas (pp. 97–123). Springer International Publishing. [Google Scholar]
- 31.Utari PD, Quax WJ (2013) Caenorhabditis elegans reveals novel Pseudomonas aeruginosa virulence mechanism. Trends in microbiology, 21(7), 315–316. doi: 10.1016/j.tim.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 32.Kulshreshtha G, Borza T, Rathgeber B, Stratton GS, Thomas NA, Critchley A, et al. (2016) Red seaweeds Sarcodiothecagaudichaudii and Chondruscrispus down regulate virulence factors of Salmonella Enteritidis and induce immune responses in Caenorhabditis elegans. Frontiers in microbiology, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez-Diener I, Zamorano L, López-Causapé C, Cabot G, Mulet X, Peña C, et al. (2017) Interplay among resistance profiles, high-risk clones and virulence in the Caenorhabditis elegans Pseudomonas aeruginosa infection model. Antimicrobial Agents and Chemotherapy, AAC-01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neuhaus K, Lamparter MC, Zölch B, Landstorfer R, Simon S, Spanier B, et al. (2017) Probiotic Enterococcus faecalisSymbioflor® down regulates virulence genes of EHEC in vitro and decrease pathogenicity in a Caenorhabditis elegans model. Archives of microbiology, 199(2), 203–213. doi: 10.1007/s00203-016-1291-8 [DOI] [PubMed] [Google Scholar]
- 35.Essebe CN, Visvikis O, Fines-Guyon M, Vergne A, Cattoir V, Lecoustumier A, et al. (2017) Decrease of Staphylococcus aureus Virulence by Helcococcuskunzii in a Caenorhabditis elegans Model. Frontiers in cellular and infection microbiology, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson TA, Brown PD (2017) Association between the agr locus and the presence of virulence genes and pathogenesis in Staphylococcus aureus using a Caenorhabditis elegans model. International Journal of Infectious Diseases, 54, 72–76. doi: 10.1016/j.ijid.2016.11.411 [DOI] [PubMed] [Google Scholar]
- 37.Martin N, Singh J, Aballay A (2017) Natural genetic variation in the Caenorhabditis elegans response to Pseudomonas aeruginosa. G3: Genes, Genomes, Genetics, 7(4), 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ewbank JJ, Pujol N (2016) Local and long-range activation of innate immunity by infection and damage in C. elegans. Current opinion in immunology, 38, 1–7. doi: 10.1016/j.coi.2015.09.005 [DOI] [PubMed] [Google Scholar]
- 39.Yang W, Dierking K, Rosenstiel PC, Schulenburg H (2016) GATA transcription factor as a likely key regulator of the Caenorhabditis elegans innate immune response against gut pathogens. Zoology, 119(4), 244–253. doi: 10.1016/j.zool.2016.05.013 [DOI] [PubMed] [Google Scholar]
- 40.Lee JH, Kim YG, Kim M, Kim E, Choi H, Kim Y, et al. (2017) Indoleassociated predator–prey interactions between the nematode Caenorhabditis elegans and bacteria. Environmental microbiology, 19(5), 1776–1790. doi: 10.1111/1462-2920.13649 [DOI] [PubMed] [Google Scholar]
- 41.He Y, Sonnenwald T, Sprenger A, Hansen U, Dengjel J, BrucknerTuderman L, et al. (2014) RhoA activation by CNFy restores cell–cell adhesion in kindlin2deficient keratinocytes. The Journal of pathology, 233(3), 269–280. doi: 10.1002/path.4350 [DOI] [PubMed] [Google Scholar]
- 42.Akiyama Y, Futai H, Saito E, Ogita K, Sakae H, Fukunaga M, et al. (2017) Shiga Toxin Subtypes and Virulence Genes in Escherichia coli Isolated from Cattle. Japanese journal of infectious diseases, 70(2), 181–185 doi: 10.7883/yoken.JJID.2016.100 [DOI] [PubMed] [Google Scholar]
- 43.Greaney AJ, Leppla SH, Moayeri M. (2015) Bacterial exotoxins and the inflammasome. Frontiers in immunology, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sastalla I, Monack DM, Kubatzky KF (2016) Bacterial Exotoxins: How Bacteria Fight the Immune System. Frontiers in immunology, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Topalcengiz Z, Danyluk MD (2017) Thermal inactivation responses of acid adapted and non-adapted stationary phase Shiga toxin-producing Escherichia coli (STEC), Salmonella spp. and Listeria monocytogenes in orange juice. Food Control, 72, 73–82. [Google Scholar]
- 46.Hwang AB, Ryu EA, Artan M, Chang HW, Kabir MH, Nam HJ, et al. (2014) Feedback regulation via AMPK and HIF-1 mediates ROS-dependent longevity in Caenorhabditis elegans. Proceedings of the National Academy of Sciences, 111(42), E4458–E4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang D, Kirienko NV (2017) High-Throughput Genetic Screen Reveals that Early Attachment and Biofilm Formation Are Necessary for Full Pyoverdine Production by Pseudomonas aeruginosa. Frontiers in Microbiology, 8, 1707 doi: 10.3389/fmicb.2017.01707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar DKV, Choi SH, Washicosky KJ, Eimer WA, Tucker S, Ghofrani J, et al. (2016) Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Science translational medicine, 8(340), 340ra72–340ra72. doi: 10.1126/scitranslmed.aaf1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miko A, Rivas M, Bentancor A, Delannoy S, Fach P, Beutin L (2014) Emerging types of Shiga toxin-producing E. coli (STEC) O178 present in cattle, deer, and humans from Argentina and Germany. Frontiers in cellular and infection microbiology, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen J, Rump L, Ju W, Shao J, Zhao S, Brown E, et al. (2015) Virulence characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from food, humans and animals. Food microbiology, 50, 20–27. doi: 10.1016/j.fm.2015.02.007 [DOI] [PubMed] [Google Scholar]
- 51.Losada L, DebRoy C, Radune D, Kim M, Sanka R, Brinkac L, et al. (2016) Whole genome sequencing of diverse Shiga toxin-producing and non-producing Escherichia coli strains reveals a variety of virulence and novel antibiotic resistance plasmids. Plasmid, 83, 8–11. doi: 10.1016/j.plasmid.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 52.Fruth A, Prager R, Tietze E, Rabsch W, Flieger A (2015) Molecular epidemiological view on Shiga toxin-producing Escherichia coli causing human disease in Germany: Diversity, prevalence, and outbreaks. International Journal of Medical Microbiology, 305(7), 697–704. doi: 10.1016/j.ijmm.2015.08.020 [DOI] [PubMed] [Google Scholar]
- 53.Dallman TJ, Ashton PM, Byrne L, Perry NT, Petrovska L, Ellis R, et al. (2015) Applying phylogenomics to understand the emergence of Shiga-toxin-producing Escherichia coli O157: H7 strains causing severe human disease in the UK. Microbial Genomics, 1(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Johannes L, Römer W (2010) Shiga toxins—from cell biology to biomedical applications. Nature reviews. Microbiology, 8(2), 105 doi: 10.1038/nrmicro2279 [DOI] [PubMed] [Google Scholar]
- 55.Shimizu T, Ohta Y, Tsutsuki H, Noda M (2011) Construction of a novel bioluminescent reporter system for investigating Shiga toxin expression of enterohemorrhagic Escherichia coli. Gene, 478(1), 1–10. [DOI] [PubMed] [Google Scholar]
- 56.Stiernagle T (2006) Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, Worm Book, doi: 10.1895/wormbook.1.101.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cypser JR, Wu D, Park SK, Ishii T, Tedesco PM, Mendenhall AR,et al. (2013) Predicting longevity in C. elegans: fertility, mobility and gene expression. Mechanisms of ageing and development, 134(7), 291–297. [DOI] [PubMed] [Google Scholar]
- 58.Ikeda T, Yasui C, Hoshino K, Arikawa K, Nishikawa Y (2007) Influence of lactic acid bacteria on longevity of Caenorhabditis elegans and host defense against Salmonella entericaserovarenteritidis. Applied and environmental microbiology, 73(20), 6404–6409. doi: 10.1128/AEM.00704-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu D, Rea SL, Yashin AI, Johnson TE (2006) Visualizing hidden heterogeneity in isogenic populations of C. elegans. Experimental gerontology, 41(3), 261–270. doi: 10.1016/j.exger.2006.01.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.