Abstract
Group A rotaviruses (RVAs) are the leading cause of severe gastroenteritis and eventually death among infants and young children worldwide, and disease prevention and management through vaccination is a public health priority. In August 2009, Rotarix™ was introduced in the South African Expanded Programme on Immunisation. As a result, substantial reductions in RVA disease burden have been reported among children younger than 5 years old. Rotavirus strain surveillance post-vaccination is crucial to, inter alia, monitor and study the evolution of vaccine escape strains. Here, full-genome sequence data for the 11 gene segments from 11 South African G1P[8] rotavirus strains were generated, including 5 strains collected from non-vaccinated children during the 2004–2009 rotavirus seasons and 6 strains collected from vaccinated children during the 2010 rotavirus season. These data were analyzed to gain insights into the overall genetic makeup and evolution of South African G1P[8] rotavirus strains and to compare their genetic backbones with those of common human Wa-like RVAs from other countries, as well as with the Rotarix™ and RotaTeq™ G1P[8] vaccine components. All 11 South African G1P[8] strains revealed a complete Wa-like genotype constellation of G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1. On the basis of sequence similarities, the South African G1P[8] strains (with the exception of strain RVA/Human-wt/ZAF/1262/2004/G1P[8]) were closely related to each other (96–100% identity in all gene segments). Comparison to the Rotarix™ and RotaTeq™ G1P[8] vaccine components revealed a moderate nucleotide identity of 89–96% and 93–95%, respectively. The results indicated that none of the gene segments of these 11 South African G1P[8] strains were vaccine-derived. This study illustrates that large-scale next generation sequencing will provide crucial information on the influence of the vaccination program on evolution of rotavirus strains. This is the first report to describe full genomic analyses of G1P[8] RVA strains collected from both non-vaccinated and vaccinated children in South Africa.
Keywords: South Africa, rotavirus G1P[8], diarrhea stool, children, genomic analyses
INTRODUCTION
Group A rotavirus (RVA) infection is a global public health concern and an important cause of pediatric hospitalization due to severe diarrhea [Parashar et al., 2009]. RVA is the main etiologic agent of acute gastroenteritis in infants and young children under the age of 5 years worldwide [Estes and Kapikian, 2007] and an estimated 453,000 children aged <5 years die from rotavirus diarrhea each year, with >85% of these deaths occurring in low-income countries of Africa and Asia [Parashar et al., 2009; Tate et al., 2012]. In sub-Saharan Africa alone, rotavirus-associated gastroenteritis is responsible for an estimated 308,579 deaths annually [Sanchez-Padilla et al., 2009]. In South Africa, 1 in 62 children are hospitalized during their first 2 years of life, and 323 die annually due to rotavirus diarrhea [Sanchez-Padilla et al., 2009; Seheri et al., 2010].
Rotaviruses belong to the family Reoviridae and possess a segmented double-stranded RNA (dsRNA) genome composed of 11 segments encoding six nonstructural proteins (NSP1–NSP6) and six structural proteins (VP1–4, VP6, and VP7). The two outer capsid proteins, VP7 and VP4, elicit strain-specific neutralizing antibody responses and are used to classify RVA strains into G (glycoprotein) and P (protease-sensitive) genotypes, respectively [Estes and Kapikian, 2007]. An extended classification system for RVA strains based on all 11 gene segments was developed by the international Rotavirus Classification Working Group. This system defines the following genotypes: Gx-P[x]-Ix-Rx-Cx-Mx-Ax-Nx-Tx-Ex-Hx, based on nucleotide identity cut-off values for the genome segments encoding for VP7, VP4, VP6, VP1, VP2, VP3, NSP1, NSP2, NSP3, NSP4, and NSP5, respectively. Presently, 27 G, 37 P, 17 I, 9 R, 9 C, 8 M, 18 A, 10 N, 12 T, 15 E, and 11 H genotypes have been described [Matthijnssens et al., 2011; Guo et al., 2012; Papp et al., 2012; Trojnar et al., 2013; Jere et al., 2014]. Globally, the majority of human RVA strains possess either the Wa-like genotype constellation (I1-R1-C1-M1-A1-N1-T1-E1-H1) or the DS-1-like genotype constellation (I2-R2-C2-M2-A2-N2-T2-E2-H2) [Matthijnssens et al., 2008a; Matthijnssens and Van Ranst, 2012].
In humans, at least 5 common rotavirus strains (G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8]) circulate worldwide, with G1P[8] strains constituting the majority of human RVA infections [Gentsch et al., 2005; Banyai et al., 2012]. The VP7 and VP4 proteins, which are the primary target for vaccine development, contain multiple antigenic epitopes that can induce the production of neutralizing antibodies. Antigenic changes in these proteins may impact the efficacy of RVA vaccines [Bucardo et al., 2007; Maranhao et al., 2012; Zeller et al., 2012]. The VP7 gene encodes 326 amino acids and carries 9 variable regions (VR-1 to VR-9), with four of them comprising the antigenic epitopes A, B, C, and F corresponding to VR-5, VR-7, VR-8, and VR-9, respectively [Dyall-Smith et al., 1986; Green et al., 1989; Kirkwood et al., 1993]. Activation of the VP4 protein (776 amino acids) requires the proteolytic cleavage into VP8* and VP5*, which contain four (8–1 to 8–4) and five (5–1 to 5–5) antigenic epitopes, respectively [Estes and Kapikian, 2007].
Rotavirus is able to evolve rapidly via genetic drift, genomic rearrangement, duplications, and deletions of gene sequences, as well as through the zoonotic transmission of strains. The segmented genome also facilitates genetic shift (reassortment) between two different virus strains co-infecting the same cell [Kirkwood, 2010]. Complete genomic analyses of common human RVA strains from different countries are needed to obtain conclusive data on their overall genetic makeup and evolutionary patterns [Matthijnssens et al., 2008b].
Although G1P[8] is a predominant genotype globally [Gentsch et al., 2005; Santos and Hoshino, 2005; Banyai et al., 2012], only few full genomes of recent human G1P[8] RVA strains from China [Shintani et al., 2012], Bangladesh [Rahman et al., 2010], India [Arora and Chitambar, 2011; Ghosh et al., 2011], and the USA [Banyai et al., 2011] have been analyzed so far. Based on limited whole-genome-based studies of common human RVAs, it has been hypothesized that a stable Wa-like genetic backbone might be circulating in the majority of the recent Wa-like common human RVAs, such as G1P[8], facilitating the proliferation of these strains worldwide [Rahman et al., 2010; Ghosh et al., 2011]. However, complete genomic analyses of common human Wa-like RVAs from different countries across the globe are required to substantiate this hypothesis. Also, countries like South Africa that are conducting post-vaccine introduction rotavirus surveillance must be aware that vaccine strains can be associated with acute gastroenteritis and that these vaccine-derived strains may be present in surveillance samples. Methods for detecting RotaTeq™ [Donato et al., 2012] and Rotarix™ [Rose et al., 2010] vaccine strains have been described. Furthermore, it is important to recognize that Rotarix™ detection will potentially be more difficult than RotaTeq™ detection since Rotarix™ is derived from a human rotavirus strain and exhibits high sequence similarities to the wild-type G1P[8] rotavirus strains. However, advanced sequencing methods, such as next-generation sequencing, may be used to differentiate Rotarix™ from wild-type rotaviruses.
Currently, two live oral vaccines; Rotarix™ developed by GlaxoSmithKline Biologicals (Rixensart, Belgium) and RotaTeq™ by Merck (Blue Bell, PA, USA) have been licensed in more than 100 countries and are being introduced into routine immunization programs in the United States and other countries in Latin America, Europe, Africa and Asia. As a result of RVA vaccine implementation, substantial reductions in RVA disease burden have been reported [Glass et al., 2006; Patel et al., 2009; Curns et al., 2010; Tate et al., 2012]. RotaTeq™ possesses genes encoding the human rotavirus serotypes G1–G4 and P1A[8] in a bovine rotavirus background. The reassortants are expected to stimulate both homotypic responses and serotype-specific protection to these common rotavirus serotypes [Glass et al., 2006; Vesikari et al., 2007; Matthijnssens et al., 2010]. Rotarix™ is a monovalent vaccine derived from a G1P1A[8] human strain [Ward and Bernstein, 2009] aimed at inducing both homotypic and heterotypic immune responses after two doses and cross protecting against different serotypes. It has been shown to be effective in the prevention of severe rotavirus gastroenteritis caused by G1P[8], G3P[8], G4P[8], and G9P[8] [Ruiz-Palacios et al., 2006]. The nucleotide sequences of VP7 and VP4 of Rotarix™ and RotaTeq™ have been published [Glass et al., 2006; Vesikari et al., 2007; Matthijnssens et al., 2010; Zeller et al., 2012]. Although the efficacy of both vaccines is high in developed countries, it is aberrantly lower in developing countries [Sanchez-Padilla et al., 2009; Madhi et al., 2012; Seheri et al., 2012; Steele et al., 2012; Tate et al., 2012]. As a result of the high mortality associated with rotavirus infection, disease prevention and management in Africa through vaccination is a public health priority [Neuzil et al., 2010]. To date, up to nine countries in Africa namely; South Africa, Zambia, Ghana, Rwanda, Botswana, Malawi, Sudan, Morocco, and Tanzania, have recently introduced RVA vaccination into their immunization programs [Armah et al., 2010; Benhafid et al., 2012; Cunliffe et al., 2012; Seheri et al., 2012].
In South Africa, the monovalent G1P[8] Rotarix™ was introduced in the Expanded Program on Immunization in August 2009, and vaccine coverage was estimated at 67% in 2010. Also, a significant reduction in morbidity rates among children younger than 5 years old and a dramatic delay in the rotavirus season by 8 weeks was observed when comparing pre- and post-vaccination data [Seheri et al., 2012]. Rotavirus strain G1P[8] has been identified as one of the major causes of childhood diarrhea among South African children [Sanchez-Padilla et al., 2009]. However, to date, there are no reports on the full genomic analyses of G1P[8] RVA strains from South Africa. Thus, the complete genomes of 11 human G1P[8] RVA strains collected from both non-vaccinated and vaccinated children were analyzed here to gain insights into the overall genetic makeup and evolution of the recent South African G1P[8] strains detected from non-vaccinated and vaccinated children and to compare their genetic backbones with those of common human Wa-like RVAs from other countries, as well as with Rotarix™ and RotaTeq™ G1P[8] vaccine components.
MATERIALS AND METHODS
Ethical Approval
The Ethical approval to conduct this study was obtained from the Medunsa Research Committee with the research number: MR58/2003, MP46/2005, and MREC/P/17/2012:PG. Informed consent was obtained from the parent or guardian before a stool sample was taken.
Study Site and Samples
The samples were collected at the Dr George Mukhari Academic Hospital and Brits Oukasie Clinic between 2004 and 2010 from 11 South African black children less than 3 years of age presenting and/or hospitalized for dehydrating diarrhea as part of a rotavirus burden of disease surveillance studies conducted at the University of Limpopo’s Medunsa campus (MR58/2003, MP46/2005, and MREC/P/17/2012:PG). The history of illness, treatment provided, and vaccination status were determined. Stool samples were collected from each child; these included five samples from non-vaccinated children collected during the 2004–2009 rotavirus seasons and six samples from vaccinated children collected during the 2010 rotavirus season. The mean age of the five non-vaccinated children was 14.6 months (range: 5–34 months; three males and two females), while the mean age of the six vaccinated children was 12.7 months (range: 3–23 months; four males and two females).
Two doses of Rotarix™ were given at 6 and 14 weeks of age along with oral polio vaccine (OPV) (given only at 6 weeks), diphtheria, tetanus, acellular pertussis, inactivated polio vaccine and Haemophilus influenza type b combined (DTaP-IPV/Hib), hepatitis B vaccine and pneumococcal conjugated vaccine, as part of the South African Expanded Program on Immunization. The exception was a 3 months old baby (DPRU 2061) who received only one dose of the rotavirus vaccine at 6 weeks.
The stool samples were screened for RVA using the ProSpecT Rotavirus Microplate kit (Thermo Fisher Scientific, Basingstone, England) according to the manufacturers’ intructions and stored at +4°C for molecular characterization studies.
Nucleic Acid Extraction
The dsRNA genome was extracted from stool samples following previously described methods [Potgieter et al., 2009; Nyaga et al., 2013]. Briefly, TRI-REAGENT-LS (Molecular Research Center, Cincinnati, OH) was mixed with a 10% stool suspension at a ratio of 3:1 and incubated for 5 min at room temperature. Chloroform was added to the suspension, followed by centrifugation at 4°C for 15 min at 16,000g. The supernatant was added to a tube containing isopropanol to precipitate the RNA. The RNA was collected by centrifugation at room temperature for 30 min at 16,000g. The dsRNA pellet was then re-suspended in 95 μl elution buffer (Qiagen, Hilden, Germany). The excess ssRNA in the extract was removed by adding lithium chloride (Sigma, St. Louis, MO) at a concentration of 2 M and incubating at 4°C for 16 hr, followed by centrifugation at 16,000g for 30 min before further purification. The integrity of the dsRNA was analyzed on a 1% agarose tris-borate-ethylenediaminetetraacetic acid (TBE) gel stained with ethidium bromide.
Sequencing
Rotavirus RNA was diluted 1:30 or 1:10 based on an initial quality control RT-PCR. A 0.9 μl volume of diluted RNA was used in each of 11 Qiagen One-Step RT-PCR reactions to amplify full-length rotavirus segments using segment-specific primers (Supplementary Data—Appendix). Reverse transcription was performed for 30 min at 45°C, followed by 50 cycles of PCR (denaturation: 10 sec, 94°C; annealing: 1 min, 55°C; extension: 3 min, 68°C). RT-PCR products were verified on 1% agarose gels and excess primers and dNTPs were removed by treatment with Exonuclease I and shrimp alkaline phosphatase (37°C for 60 min, followed by incubation at 72°C for 15 min, respectively). Amplicons were quantitated using SYBR Green assay, and all 11 amplicons per genome were pooled in equimolar amounts.
Pooled rotavirus amplicons were sheared for 15 min and Ion Torrent compatible barcoded adapters were ligated to the sheared DNA using the Ion Xpress Plus Fragment Library Kit (Life Technologies, Carlsbad, CA) to create 200-base pair (bp) libraries. Barcoded libraries were pooled in equal volumes and cleaned with the Ampure XP Reagent (Agencourt Bioscience, Beverly, MA). Quantitative PCR was performed on the pooled barcoded libraries to assess their quality and to determine the template dilution factor for emulsion PCR. The pool was diluted appropriately and amplified on Ion Sphere Particles (ISPs) on the Ion One Touch instrument (Life Technologies). The amplified pool was cleaned and enriched for template-positive ISPs on the Ion One Touch ES instrument (Life Technologies). Sequencing was performed on the Ion Torrent PGM using an Ion 316 chip. In addition, the sequence-independent single-primer amplification (SISPA) method [Djikeng et al., 2008; Djikeng and Spiro, 2009] was used for each sample on pooled amplicons, and the products were sequenced on the Illumina MiSeq v2 instrument.
The sequence reads from the Ion Torrent PGM were sorted by barcode, trimmed, and de novo assembled using CLC Bio’s clc_novo_assemble program, and the resulting contigs were searched against custom full-length rotavirus A segment nucleotide databases to find the closest reference sequence for each segment. All sequence reads were then mapped to the selected reference rotavirus A segments using CLC Bio’s clc_ref_assemble_long program. At loci where both Ion Torrent and Illumina sequence data agreed on a variation (as compared to the reference sequence), the reference sequence was updated to reflect the difference. A final mapping of all next-generation sequences to the updated reference sequences was performed with CLC Bio’s clc_ref_assemble_long program.
Phylogenetic Analysis
Sequences were aligned using the MUSCLE program within MEGA version 5 software [Tamura et al., 2011]. Once aligned, the DNA Model Test program implemented in MEGA [Tamura et al., 2011] was used to identify the optimal evolutionary models that best fit the sequence datasets. Using corrected Akaike Information Criterion (AICc), the following models were found to best fit the sequence data for the indicated genes: TN93 + G + I (VP1, VP2, VP3, VP6, NSP1, NSP2, and NSP4), GTR + G + I (NSP5, VP4, and VP7), and TN93 + G (NSP3). Using these models, maximum likelihood trees were constructed using MEGA version 5 with 500 bootstrap replicates to estimate branch support. Nucleotide and amino acid distance matrixes were prepared using the p-distance algorithm in MEGA version 5 [Tamura et al., 2011].
RESULTS
The Genotype Constellation of South African G1P[8] Strains From Non-Vaccinated and Vaccinated Children
The names and characteristics of the South African G1P[8] strains analyzed in this study are presented in Table I. The accession numbers for each gene of the South African G1P[8] study strains (in bold), as well as those from the GenBank used for comparison in these analyses are in the Appendix. The nucleotide sequence analyses for all genes from the South African G1P[8] strains analyzed revealed a consensus genotype constellation of G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1, respectively, according to the proposed classification system [Matthijnssens et al., 2008b]. Phylogenetic analyses of the 11 genes established the genetic relationships of the South African strains when compared with a global collection of rotavirus genotypes.
TABLE I.
Characteristics of the South African G1P[8] From Non-Vaccinated and Vaccinated Children
| Genotypes
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VP7 | Lineage | VP4 | Lineage | VP6 | VP1 | VP2 | VP3 | NSP1 | NSP2 | NSP3 | NSP4 | NSP5 | |
| Length of gene sequenced (nucleotidea) | 1,023 | 2,328 | 1,320 | 3,267 | 2,695 | 2,530 | 1,519 | 1,003 | 1,015 | 701 | 631 | ||
| Presence of ORFb | C | C | C | C | C | C | C | C | C | C | C | ||
| Strain name | |||||||||||||
| Non-vaccinated children | |||||||||||||
| RVA/Human-wt/ZAF/1289/2007/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/2325/2009/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/2306/2009/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/1262/2004/G1P[8] | G1 | II | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/2330/2009/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| Vaccinated children | |||||||||||||
| RVA/Human-wt/ZAF/2035/2010/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/2030/2010/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/2052/2010/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/2061/2010/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/1492/2010/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
| RVA/Human-wt/ZAF/1544/2010/G1P[8] | G1 | I | P[8] | P[8]-III | I1 | R1 | C1 | M1 | A1 | N1 | T1 | E1 | H1 |
The length of the sequenced gene is given in nucleotides.
C = complete ORF.
Sequence Analyses of VP7
As shown in Figure 1A, the G1 human and animal rotavirus strains used in this analyses segregated into seven lineages (I–VII) [Arista et al., 2006; Le et al., 2010] and the South African G1 strains from non-vaccinated and vaccinated children clustered into two such genetic lineages, I and II. The VP7 lineage I included 10 (4 from non-vaccinated and 6 from vaccinated children) South African G1 strains within sub-lineage Ic which includes strains that circulated from 1996 to 2010. Only one South African strain from this study was detected in the VP7 lineage II; this strain was collected in 2004 from a non-vaccinated child and groups together with the Rotarix™ vaccine strain and other strains that circulated globally from 1994 to 2003. Sequence similarities among South African non-vaccinated and vaccinated strains ranged between 94–100% and 100%, respectively. Comparison of the VP7 nucleotide and deduced amino acid sequences from the South African G1 strains from non-vaccinated and vaccinated children to a global collection of G1 sequences from the GenBank database, showed a close relationship to previously identified G1 strains from humans (nucleotide, 85–98% and amino acid, 91–98%) in lineages I–VI. However, when compared to animal G1 strains in lineage VII, there was a moderate relationship of nucleotide and amino acid identities in the range of 84–86% and 92–93%, respectively. In addition, comparison with the Rotarix™ VP7 gene (RVA/Vaccine/USA/Rotarix™-RIX4414/1988/G1P1A[8]) showed they share moderately high identities (nt, 93–94% and aa, 95–96%), while the RotaTeq™ G1 strain (RVA/Vaccine/USA/RotaTeq™-WI79-9/1992/G1P75) shares slightly lower similarities (nt, 91–93% and aa, 93–95%). The strain RVA/Human-wt/ZAF/1262/2004/G1P[8] detected from a non-vaccinated child in 2004 is more distantly related to the rest of the South African G1 strains (nt, 93–94% and aa, 94%), but is closely related to the G1 strains in lineage II (including the Rotarix™ vaccine G1 strain) sharing nt and aa identities in the range of 97–98%. In contrast, the South African G1 strains share a lower nt (73%) and aa (74%) relationship to non-G1 strains, such as the G2 strain (DS-1 used as an outgroup in this analyses).
Fig. 1.
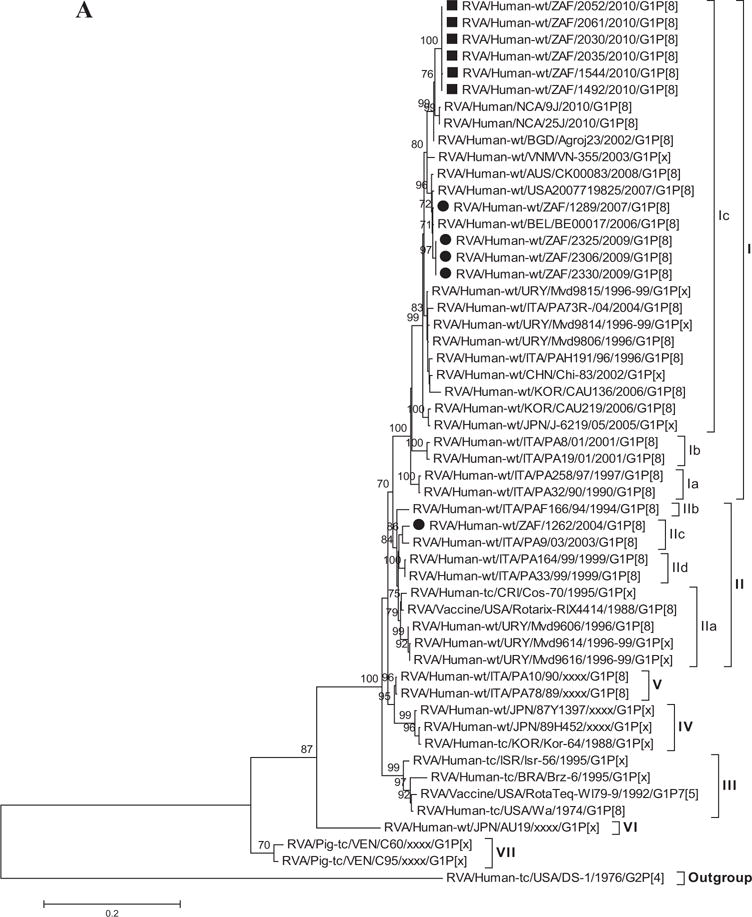
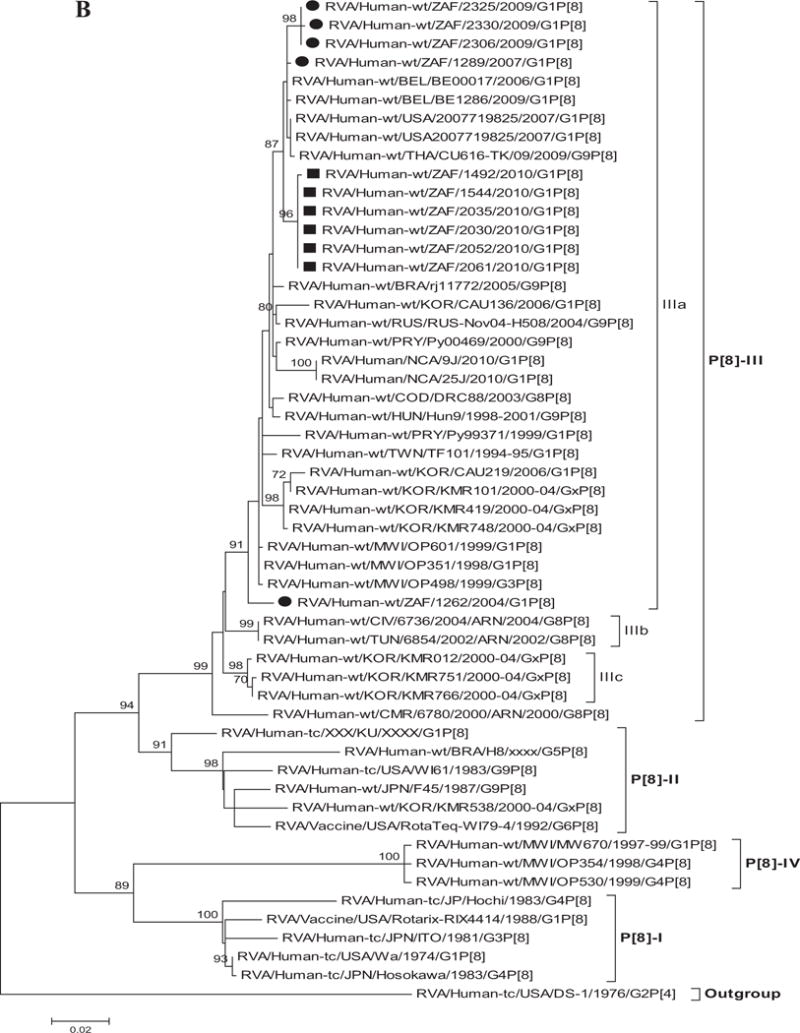
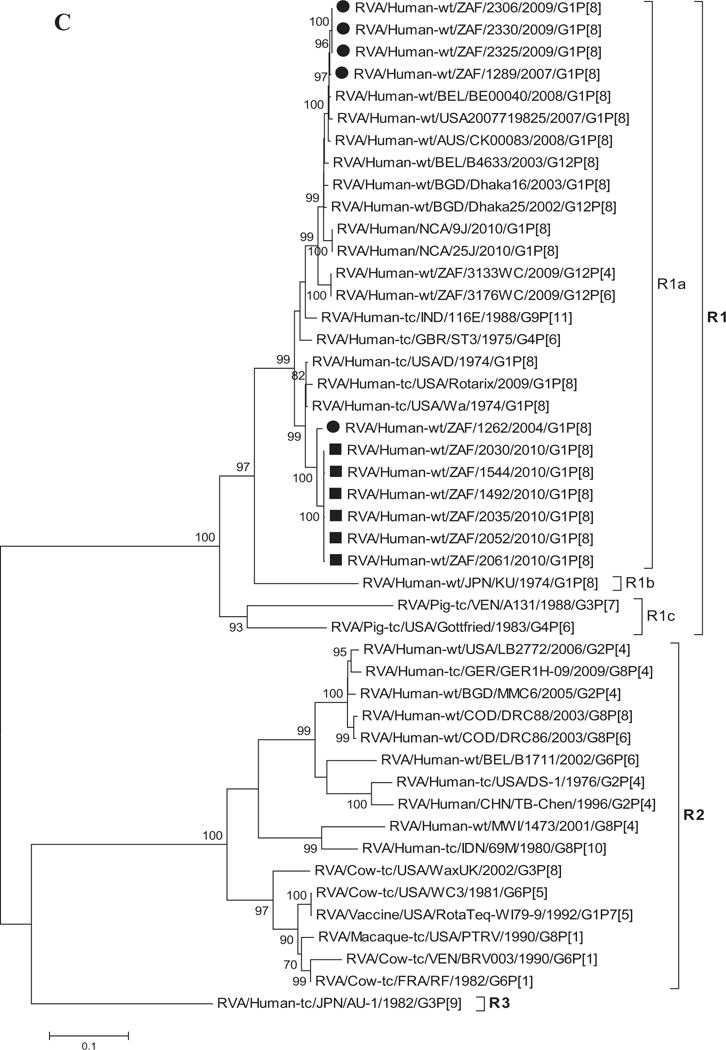
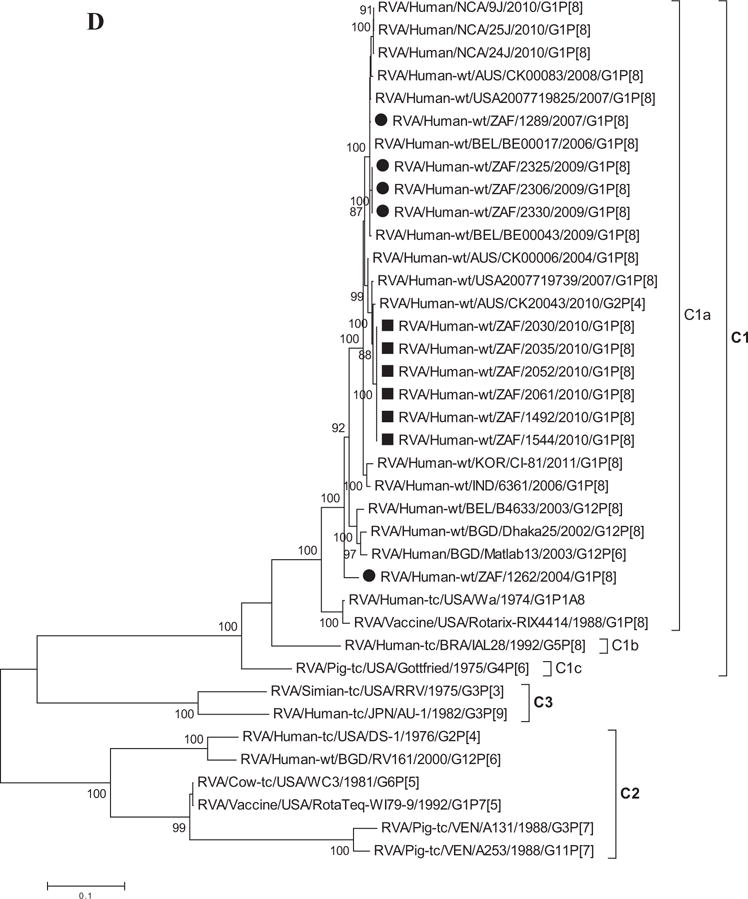
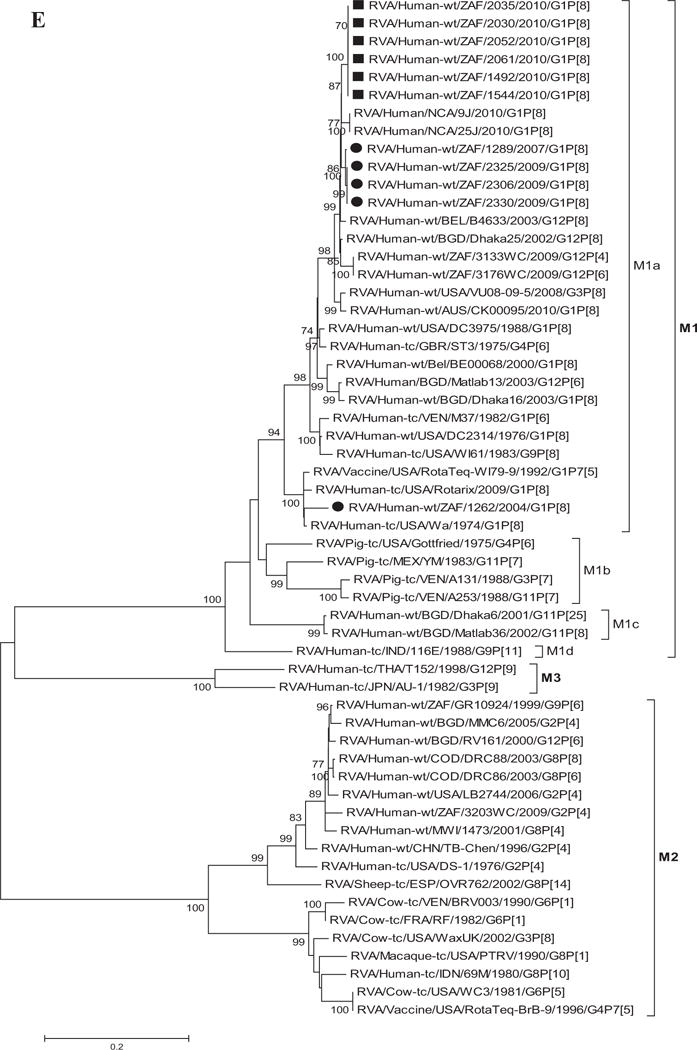
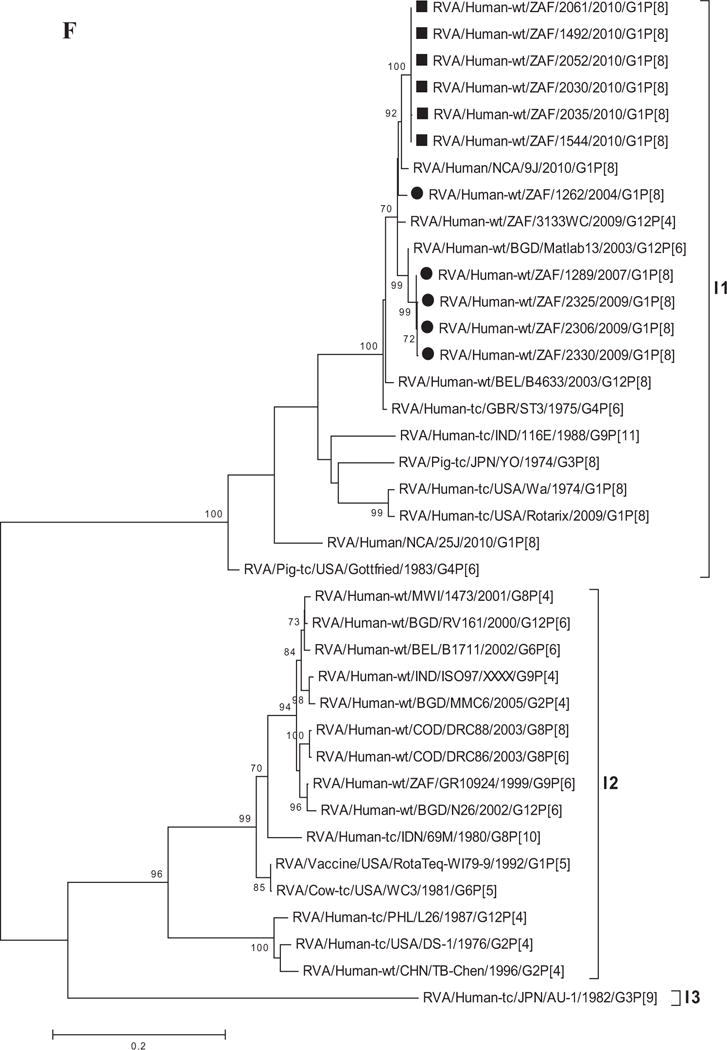
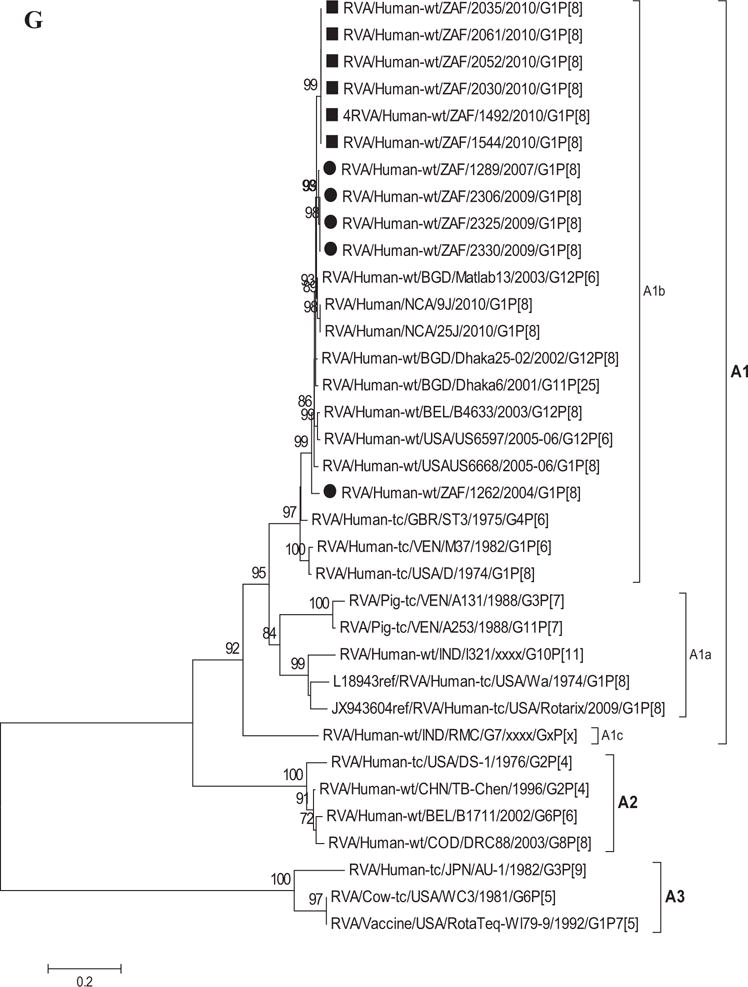
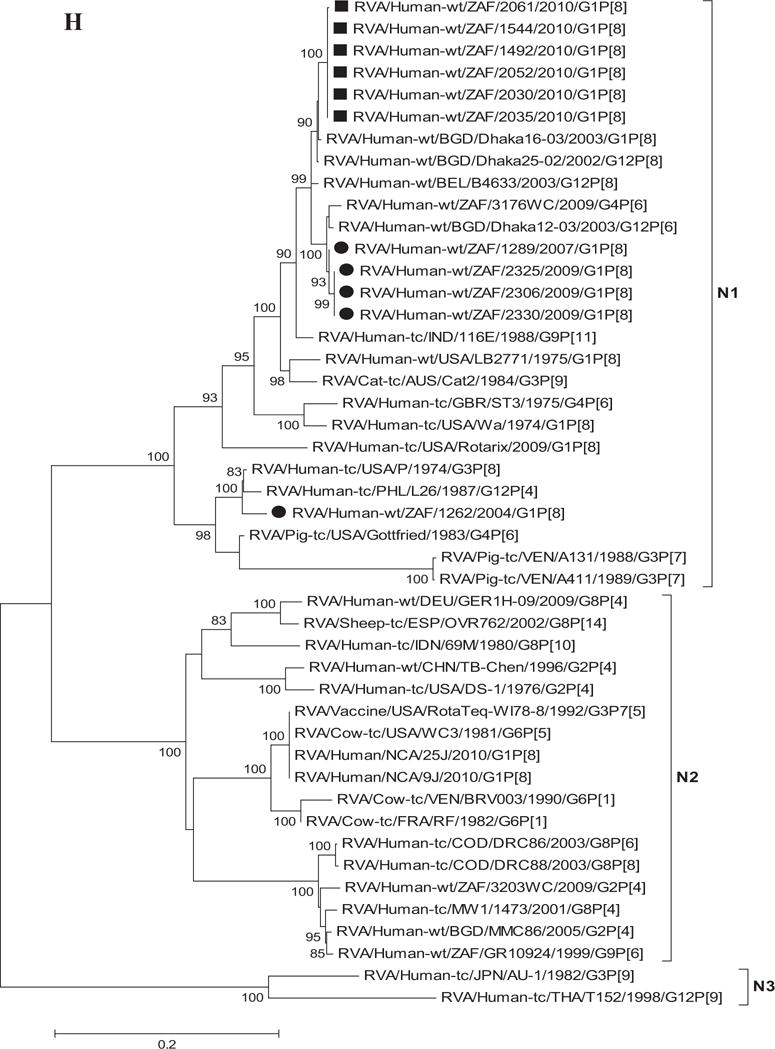
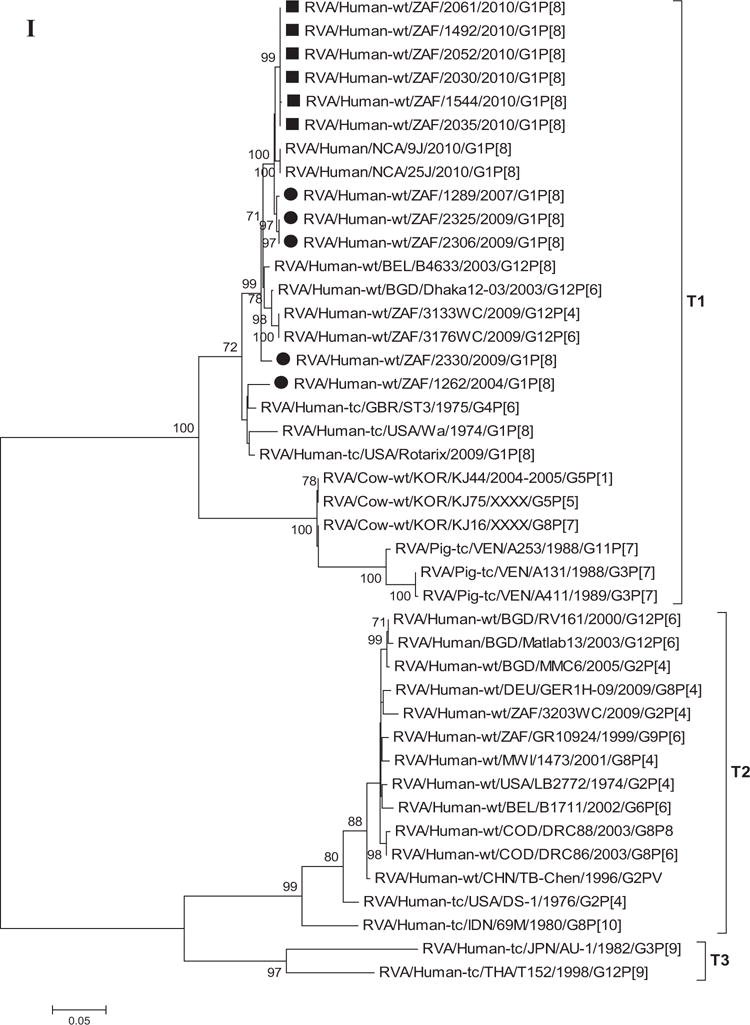
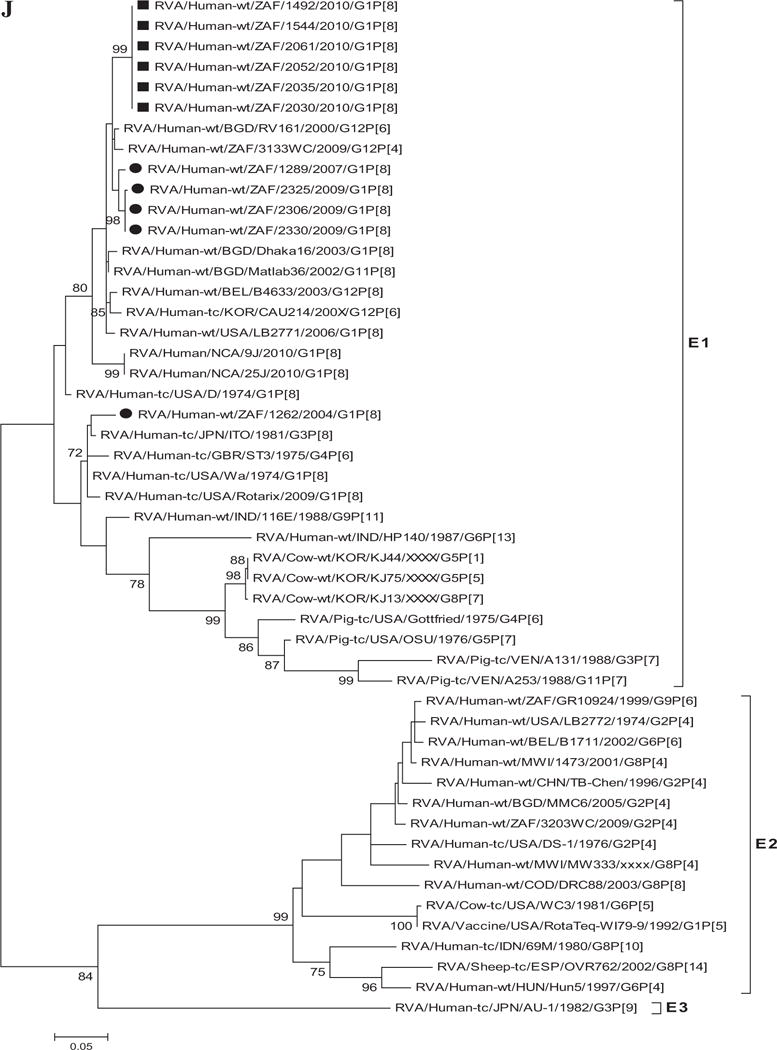
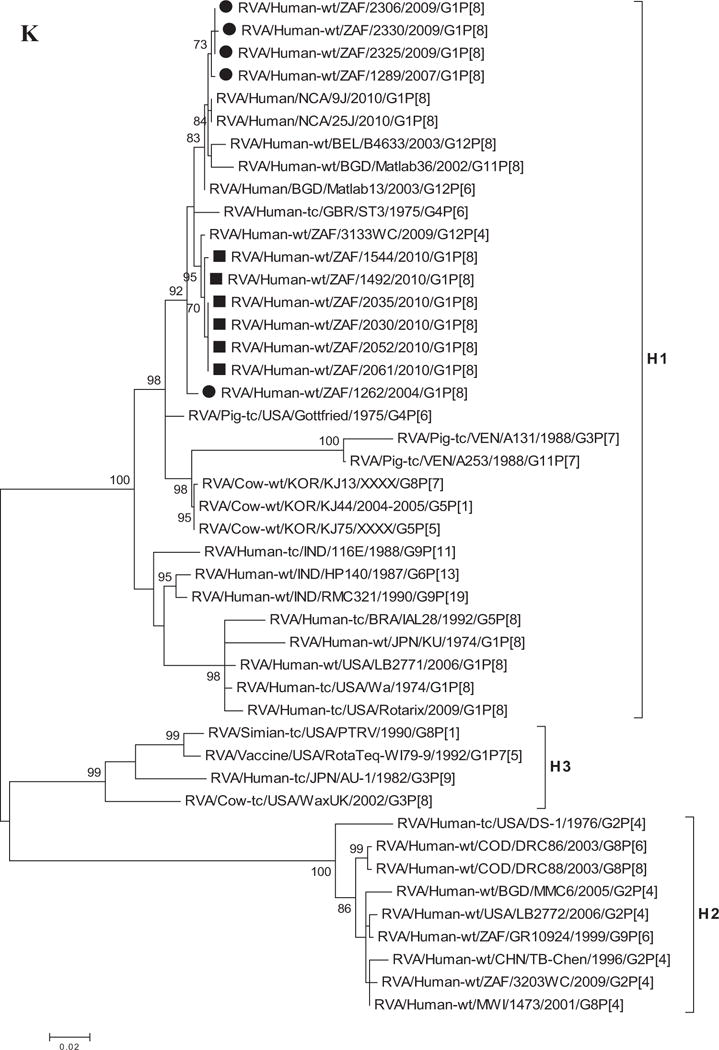
A–K: Maximum likelihood phylogenetic trees built in MEGA version 5 with bootstrap statistics as support show the genetic relationships of nucleotide sequences of VP7 (A), VP4 (B), VP1 (C), VP2 (D), VP3 (E), VP6 (F), NSP1 (G), NSP2 (H), NSP3 (I), NSP4 (J), and NSP5 (K) of human G1P[8] rotaviruses from non-vaccinated and vaccinated South Africa children with known human and animal rotavirus strains from GenBank database. The trees were drawn to scale. Only bootstrap values of 70% and greater are shown. The strains labeled with filled squares indicate G1P[8] from vaccinated and filled circles are from non-vaccinated children. The scale bar at the bottom of the trees indicates genetic distance.
The amino acid sequences for all the South African G1 strains in this study contained the conserved proline and cysteine residues that were described in 2002 [Ciarlet et al., 2002]. The glutamine residue (trypsin cleavage site) at position 51 was also conserved [Stirzaker et al., 1987; Jere et al., 2011]. Furthermore, only one potential N-linked glycosylation site at position 69 was present among the South African strains, Rotarix™ vaccine, RotaTeq™ G1 component and the global collection of human and animal G1 strains representing the seven previously described lineages [Arista et al., 2006; Le et al., 2010] (Fig. 2).
Fig. 2.
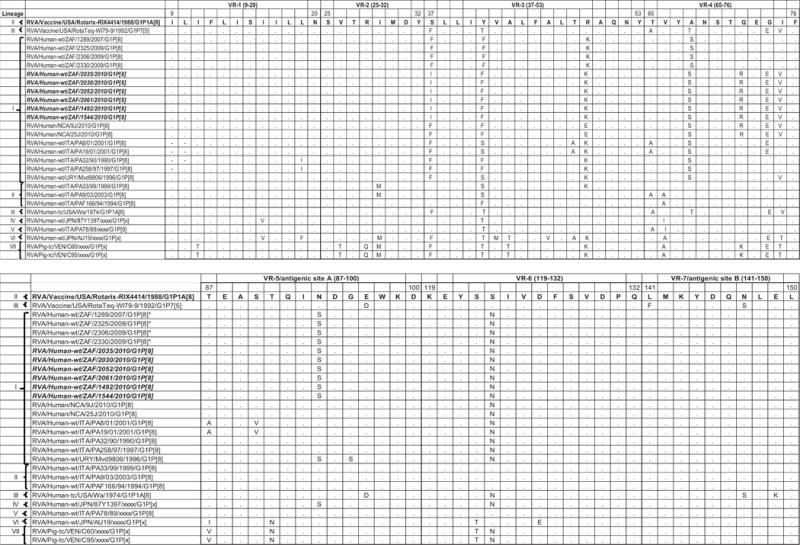
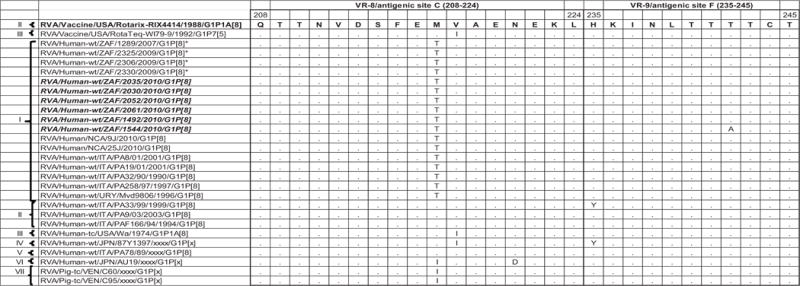
Comparison of the deduced amino acid sequence of gene segment 9 of strain South African G1P[8] from both non-vaccinated and vaccinated children to Rotarix™, G1 component of RotaTeq™ and a selection of older and contemporary G1 sequences from the GenBank. Only amino acids which differ are shown. Variable regions designated VR-1 to VR-9 are shown.
The VP7 gene sequence contains the major neutralizing sites targeted by the cytotoxic T-lymphocytes leading to the production of neutralizing antibodies by B cells [Dyall-Smith et al., 1986]. At least nine VP7 variable regions, VR-1 to VR-9 have been described [Green et al., 1989]. Six antigenic regions have been described before and are defined as A (aa 87–101), B (aa 141–150), C (aa 208–224), D (aa 291), E (aa 189), and F (aa 235–245). Antigenic regions A, B, C, and F correspond to VR-5, VR-7, VR-8, and VR-9, respectively [Dyall-Smith et al., 1986; Green et al., 1989; Kirkwood et al., 1993]. Comparison of the amino acid sequence from the Rotarix™ G1 component with sequences from the South African G1 strains revealed moderately high sequence similarities with numerous substitutions in variable regions namely: VR-3 (S37F in strains from non-vaccinated and S37I in strains from vaccinate children; Y41F and R49K in all South African G1 strains), VR-4 (A68S in strains from both non-vaccinated and vaccinated children; Q72R, G74E, and I75V in only 5 strains from vaccinated children), VR-5/antigenic site A (N94S in 10 of the South African G1 strains), VR-6 (S123N in 10 of the South African strains), and VR-8/antigenic site C (M217T in 10 of the South African G1 strains) [Dyall-Smith et al., 1986; Green et al., 1989; Kirkwood et al., 1993]. Also, comparison of the amino acid sequence from the VP7 G1 component of the RotaTeq™ vaccine with the South African G1 sequences revealed several important substitutions at VR-3 (F37I only in strains from vaccinated children; T41F in all South African G1 strains), VR-4 (T68S in strains from both non-vaccinated and vaccinated children; Q72R, E74G, and V75I only in strains from non-vaccinated children), VR-5/antigenic site A (N94S in both non-vaccinated and vaccinated children), VR-6 (S123N in both non-vaccinated and vaccinated children), and VR-8/antigenic site C (M217T in 10 of the South African G1 strains) [Dyall-Smith et al., 1986; Green et al., 1989; Kirkwood et al., 1993]. The VR-1, VR-2, VR-7/antigenic site B and VR-9/antigenic site F were highly conserved among the South African strains, Rotarix™ and RotaTeq™ (Fig. 2).
Sequence Analyses of VP4
The VP4 gene sequences of 11 South African G1P[8] strains collected from both non-vaccinated and vaccinated children were compared with representative human rotaviruses from the four established VP4 P[8] genotype lineages (P[8]-I to P[8]-IV) [Arista et al., 2006; Le et al., 2010]. Analyses of the South African P[8] strains from both non-vaccinated and vaccinated children revealed that they were closely identical with each other (97–100%). However, strains from the vaccinated children were identical in both nt and aa sequences (100%). The South African strains clustered into two distinct sub-lineages within the P[8]-III lineage of the P[8] genotype (Fig. 1B), showing 96–99% nt and 96–100% aa identities. Within this lineage are strains detected from 1995 to 2010. In addition, the South African P[8] strains were compared with non-P[8]-III lineage strains and they showed moderate relationship in nt and aa similarities in the range of 86–94% and 86–96%, respectively. Further, comparison with the Rotarix™ vaccine P[8] and RotaTeq™ vaccine P[8] genotype revealed a moderate relationship (nt, 90–91% and aa, 91–92%) and (nt, 92–93% and aa, 94%), respectively. The lowest identities were seen with non-P[8] genotype such as P[4] (nt, 86% and aa, 88–89%) which was used as an outgroup in this analyses. The VP4 spike protein is comprised of two structurally distinct regions (VP8* and VP5*) generated following trypsin activation of the virion particle [Estes and Kapikian, 2007]. The VP8* region contains four surface-exposed antigenic epitopes (8–1 to 8–4), while VP5* contains five surface exposed antigenic epitopes (5–1 to 5–5) [Zeller et al., 2012]. Sequence analyses showed that all the South African G1 strains in this study contained a conserved trypsin cleavage sites (arginine) at positions 230, 240, and 581 [Ciarlet et al., 2002; Estes and Kapikian, 2007]. A second conserved trypsin cleavage site described [Crawford et al., 2001] at residues 257 and 466 (data not shown) was observed. Also, analyses of the South African P[8] strains showed that they contained up to eight and five differences with Rotarix™ and RotaTeq™, respectively. For Rotarix™ six of the substitutions were located in VP8* epitopes 8–1 (positions E150D, N195G) and 8–3 (positions N113D, S125N, S131R, N135D) and two in VP5* epitope 5–1 (positions S383N, Y385D). The amino acid changes at positions N113D and S383N only occur between strains from South African vaccinated children and Rotarix™. On the other hand, in RotaTeq™ three of the substitutions were found in VP8* epitopes 8–1 (positions E150D and D195G) and 8–3 (position N113D) and two were in VP5* epitope 5–1 (positions R383S in non-vaccinated and R383N in vaccinated children and H385D) (Table II).
TABLE II.
Alignment of the Amino Acid Residues Corresponding to Those Defining the VP4 Neutralization Domains Designated as 8-1, 8-2, 8-3, and 8-4 in the VP8* Subunit and 5-1, 5-2, 5-3, 5-4, and 5-5 in the VP5* Subunit Between the P[8] Strain in Rotarix™ and South African RVA Strains Collected From Non-Vaccinated and Vaccinated Children
|
|
|
||||||||||||||||||||||||||||||||||||
| 8-1 |
8-2 |
8-3 |
8-4 |
5-1 |
5-3 |
5-3 |
5-4 |
5-5 |
|||||||||||||||||||||||||||||
| 100 | 146 | 148 | 150 | 188 | 190 | 192 | 193 | 194 | 195 | 196 | 180 | 183 | 113 | 114 | 115 | 116 | 125 | 131 | 132 | 133 | 135 | 87 | 88 | 89 | 383 | 385 | 387 | 392 | 393 | 397 | 439 | 440 | 433 | 458 | 428 | 305 | |
| RVA/Vaccine/USA/Rotarix-RIX4414/1988/G1P1A[8] | D | S | Q | E | S | T | N | L | N | N | I | T | A | N | P | V | D | S | S | N | D | N | N | T | N | S | Y | S | A | W | N | L | R | E | N | S | L |
| Non-vaccinated children | |||||||||||||||||||||||||||||||||||||
| RVA/Human-wt/ZAF/1289/2007/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | . | . | . | . | N | R | . | . | D | . | . | . | . | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/2325/2009/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | . | . | . | . | N | R | . | . | D | . | . | . | . | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/2306/2009/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | . | . | . | . | N | R | . | . | D | . | . | . | . | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/1262/2004/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | . | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/2330/2009/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | . | . | . | . | N | R | . | . | D | . | . | . | . | D | . | . | . | . | . | . | . | . | N | . |
| Vaccinated children | |||||||||||||||||||||||||||||||||||||
| RVA/Human-wt/ZAF/2035/2010/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | N | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/2030/2010/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | N | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/2052/2010/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | N | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/2061/2010/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | N | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/1492/2010/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | N | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/ZAF/1544/2010/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | N | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/NCA/25J/2010/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | . | D | . | . | . | . | . | . | . | . | . | . |
| RVA/Human-wt/NCA/9J/2010/G1P[8] | . | . | . | D | . | . | . | . | . | G | . | . | . | D | . | . | . | N | R | . | . | D | . | . | . | . | D | . | . | . | . | . | . | . | . | . | . |
Sequence Analyses of VP1–VP3 and VP6
Phylogenetic analyses based on VP1–VP3 and VP6 nucleotide sequences demonstrated that each gene from the South African study samples grouped into two to three separate clades with strains isolated worldwide (Fig. 1C–F) usually by year and vaccination status (i.e., 2004–2009 non-vaccinated children and 2010 vaccinated children). The collection exhibited a close genetic relationship with gene sequences of previously reported strains from GenBank belonging to the R1, C1, M1, and I1 genotypes that were isolated globally [Wyatt et al., 1983; Rahman et al., 2007; Matthijnssens et al., 2008a; Jere et al., 2011; Bucardo et al., 2012]. Nucleotide (amino acid) identities among South African strains collected from non-vaccinated children ranged from 94–100% (98–100%), 96–100% (99–100%), 90–100% (95–100%), and 97–100% (99–100%) for VP1, VP2, VP3, and VP6, respectively, while those from vaccinated children shared an absolute identity with each other. Among strains from non-vaccinated children, a complete nucleotide and amino acid similarities were shared between strains RVA/Human-wt/ZAF/1289/2007/G1P[8], RVA/Human-wt/ZAF/2325/2009/G1P[8] and RVA/Human-wt/ZAF/2330/2009/G1P[8], likewise between strains RVA/Human-wt/ZAF/2325/2009/G1P[8], RVA/Human-wt/ZAF/2306/2009/G1P[8] and RVA/Human-wt/ZAF/2330/2009/G1P[8] in their VP1–VP3 and VP6 genes. However, when the nucleotide (amino acid) homologies, of the VP1–VP3 and VP6 gene sequences of these South African strains, were compared with similar gene sequences of strains belonging to previously identified VP1–VP3 and VP6 genotypes, all were more closely related to strains in the R1, 85–99% (93–100%), C1, 88–100% (92–100%), M1, 86–99% (88–99%), and I1, 88–99% (97–100%) genotypes, respectively. These same sequences when compared to cognate gene sequences of the Rotarix™ vaccine currently being used in South Africa [Seheri et al., 2012], revealed they shared nucleotide (amino acid) homologies in the range of 95–97% (99%), 93–94% (98%), 92% (95–97%), and 89–90% (97–98%), respectively. The lowest nucleotide and amino acid similarities in the range of 75–82% and 80–93% were with strains belonging to the DS-1-like (R2, C2, M2, and I2) and AU-1-like (R3, C3, M3, and I3) genogroups.
Sequences Analyses of NSP1–NSP5
Phylogenetic analyses of NSP1–NSP5 nucleotide sequences revealed that for each gene, the South African strains from both non-vaccinated and vaccinated children grouped in small separate sub-clusters of genotypes A1, N1, T1, E1, and H1, respectively, together with other strains from around the world (Fig. 1H–K). Nucleotide (amino acid) identity values among South African strains from non-vaccinated children ranged from 96–100% (95–100%), 90–100% (95–100%), 95–100% (96–100%), 92–100% (94–100%), and 99–100% (99–100%) for NSP1, NSP2, NSP3, NSP4, and NSP5, respectively. On the other hand, strains from vaccinated children shared an almost complete identity (≥99%) with each other. Absolute nucleotide and amino acid identities were shared among strains RVA/Human-wt/ZAF/1289/2007/G1P[8], RVA/Human-wt/ZAF/2325/2009/G1P[8], and RVA/Human-wt/ZAF/2330/2009/G1P[8] and also between strains RVA/Human-wt/ZAF/2325/2009/G1P[8], RVA/Human-wt/ZAF/2306/2009/G1P[8], and RVA/Human-wt/ZAF/2330/2009/G1P[8] (NSP1 and NSP2 genes); RVA/Human-wt/ZAF/2325/2009/G1P[8] and RVA/Human-wt/ZAF/2306/2009/G1P[8] (NSP3 gene), and RVA/Human-wt/ZAF/2325/2009/G1P[8], RVA/Human-wt/ZAF/2306/2009/G1P[8], and RVA/Human-wt/ZAF/2330/2009/G1P[8] (NSP4 and NSP5 genes). However, when the nucleotide and amino acid similarities of the NSP1–NSP5 gene sequences, of the South African strains, were compared with similar gene sequences of strains belonging to already identified NSP1–NSP5 genotypes, all of them were more closely related to strains in the A1, N1, T1, E1, and H1 genotypes, in that order. Within each of these genotypes, the South African strains from both non-vaccinated and vaccinated children shared maximum nucleotide (amino acid) identity of 80–99% (80–99%), 80–99% (88–100%), 82–98% (85–100%), 81–98% (92–99%), and 90–99% (90–100%), respectively. In addition, comparison with the cognate gene sequences of Rotarix™ vaccine strain indicated 84% (83%) for NSP1, 89–91% (96–97%) for NSP2, 96% (97–98%) for NSP3, 92–97% (94–97%) for NSP4, and 93% (94%) for NSP5 genes. Repeatedly, the lowest nucleotide and amino acid similarities in the range of 63–88% and 59–91% were with strains belonging in DS-1-like (A2, N2, T2, and H2) and AU-1-like (A3, N3, T3, and H3) genogroups.
DISCUSSION
Globally, there are five common RVA genotypes namely: G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8] that are responsible for approximately 88% of all rotavirus infections in children less than 5 years of age. Genotype G1P[8] is the most common circulating strain accounting for 50–65% of all strains in developed and in some developing countries [Banyai et al., 2012]. In Africa, G1P[8] accounts for approximately 21% of all circulating RVA strains [Steele and Ivanoff, 2003; Todd et al., 2010; Banyai et al., 2012]. In 2009, the monovalent (G1P[8]) Rotarix™ vaccine was introduced in the South African Expanded Program on Immunization and in less than 2 years, a significant reduction in rotavirus-associated hospitalization midst children younger than 5 years old was observed [Seheri et al., 2012], with G1P[8] being the most common strain in circulation among South African children [Sanchez-Padilla et al., 2009]. A few previous reports from South Africa showed G1 genotype associated with either VP4 P[6] or P[8] genotypes as the predominant strain in circulation [Steele et al., 2003; Potgieter et al., 2010]. In this study, we described the full genome analyses of eleven G1P[8] strains collected from both non-vaccinated and vaccinated South African children ≤5 years of age.
Using nucleotide sequence identities and phylogenetic analyses of the complete coding sequences of the VP7–VP4–VP6–VP1–VP2–VP3–NSP1–NSP2–NSP3–NSP4–NSP5 genes of the five South African RVA strains reported here from non-vaccinated children and six strains from vaccinated children were assigned to the G1-P[8]-I1-R1-C1-M1-A1-N1-T1-E1-H1 genotypes, respectively (Fig. 1A–K). Therefore, these 11 South African G1P[8] RVA strains exhibited a typical Wa-like genotype constellation.
The emergence of new lineages or sub-lineages of G1 strains is a possible explanation for the continuous circulation of G1 rotavirus in a given geographic area as shown in previous studies in Hungary and Italy [Arista et al., 2006; Banyai et al., 2009]. Sequencing of the VP7 gene from the G1 genotype reveals temporal variation and antigenic change among strains [Diwakarla and Palombo, 1999]. VP7 gene analyses of G1 strains has conveyed significant genetic diversity and produced at least seven global lineages among the G1 strains collected from different geographic locations [Arista et al., 2006; Banyai et al., 2009; Le et al., 2010; Bucardo et al., 2012]. In this present study, phylogenetic analyses revealed close similarities (99–100%) between the South African strains from both non-vaccinated and vaccinated children. With the exception of strain RVA/Human-wt/ZAF/1262/2004/G1P[8] which clustered in G1 lineage II with the G1 of the Rotarix™ vaccine and other G1 strains collected between 1994 and 2003, the remaining ten South African G1 strains collected between 2007 and 2010 clustered in G1 lineage I. It suggests that this is a G1 strain that circulated earlier and has been replaced with recent G1 strains in lineage I. Within this lineage I, the strains from non-vaccinated and vaccinated children clustered separately. In another study from Finland, the circulation of two G1 sub-lineages (G1-I and G1-II) were reported for a period of 20 years before and after the introduction of both the Rotarix™ and the RotaTeq™ vaccines, which suggests that there was little or no variations in the deduced VP7 amino acid sequences of these strains even after the introduction of the vaccines [Hemming and Vesikari, 2013]. However, the South African G1 strains shared amino acid identities in the range of 95–96% and 93–95% to Rotarix™ and G1 component of RotaTeq™, respectively, indicating that these strains were not vaccine derived. The formation of two lineages, in a small number of collections of South African G1 strains provides evidence of genetic variation over the years. In addition, different VP7 genetic lineages were also observed between South African G1 strains from non-vaccinated and vaccinated children and the prototype G1P[8] strain (Wa-like) detected in 1974 in the USA and classified to VP7 lineage III [Jin et al., 1996; Arista et al., 2006; Le et al., 2010; Bucardo et al., 2012].
By multiple alignment, the deduced amino acid sequences of the VP7 of the South African G1P[8] strains from non-vaccinated and vaccinated children exhibited 13, 7–14, and 21 and 13, 16, and 21 mismatches in both antigenic and non-antigenic regions with those of the prototype G1P[8] strain Wa-like, VP7 of Rotarix™ and G1 component of RotaTeq™, respectively. However, when amino acid analyses of only VP7 antigenic/variable regions were considered, it showed that the South African G1P[8] strains were highly conserved in VR-1, VR-2, VR-7/antigenic site B and VR-9/antigenic site F, while multiple amino acids changes were observed in five of the other nine variable regions [Dyall-Smith et al., 1986; Green et al., 1989; Kirkwood et al., 1993]. Amino acid substitutions in antigenic regions A/VR-5 (aa 87–101), B/VR-7 (aa 141–150), and C/VR-8 (aa 208–224), especially at positions 94, 96, 147, 148, 190, 208, 211, 213, and 217, with or without glycosylation changes, are known to alter the antigenicity of viruses and enhance host immunity [Ahmed et al., 2007; Trinh et al., 2007]. Therefore, the substitutions detected at positions 94 and 217 may be important in antigenic drift in the South African rotavirus G1 strains, thereby escaping host immunity [Maranhao et al., 2012]. Also, the fact that these substitutions were observed among the G1 strains from vaccinated children could suggest the possibility of a positive selection over time and might be related to some kind of advantages regarding viral fitness, although this could not be conclusively determined in this study as selection pressure on these substitutions was not investigated.
Although VP7 is the surface protein represented in most current vaccines, VP4 which is also a surface protein is also very important in inducing protective immunity [Offit et al., 1986; Ward et al., 1993]. Previous phylogenetic data of the VP4 gene provides evidence of diversity within P[8] genotypes; four distinct lineages have been described [Arista et al., 2006; Le et al., 2010; Cho et al., 2013]. The South African G1P[8] strains though separated in three groups were all clustered in P[8]-III lineage together with strains collected globally.
Deduced amino acid residues defining the RVA VP4 epitopes have been identified by neutralization escape mutants and identifying surface exposed amino acid residues that show intergenotypic variability among prevalent human P-genotypes [Dormitzer et al., 2004; McDonald et al., 2009]. With the exception of a single residue at position N113D in the VP8* portion of all vaccinated strains, these amino acid residues were conserved among the South African G1P[8] strains (Table II). Alignment of the amino acid residues, defining the VP4 neutralization domains, revealed eight and five mismatches in the VP8* (antigenic epitopes 8–1 and 8–3) and VP5* (antigenic epitope 5–1) antigenic epitopes between the VP4 component of the South African G1P[8] strains and that of Rotarix™ and RotaTeq™ (RotaTeq™ data not shown) (Table II). With the exception of conservation at residue 190, the mutation pattern in the VP8* antigenic epitopes were similar to those reported previously [Zeller et al., 2012; Mouna et al., 2013]. Among the other genes, the VP1-3, VP6, and NSP1-NSP5 genes of all South African strains were closely related to those of several Wa-like common human RVA strains, such as G1, G3, G9, and/or G12, detected in the late 1990s and 2000s from different countries.
Overall, full genomic analyses of South African G1P[8] strains collected from both non-vaccinated and vaccinated children revealed a stable Wa-like genetic backbone that might be circulating in most of the current Wa-like common human RVAs, such as the G1P[8], G3P[8], G4P[8], and G9P[8] strains, worldwide. It has been hypothesized that RVAs with this genetic backbone have the ability to propagate extremely well in the human host, as evidenced by the detection of large numbers of Wa-like human RVA strains across the globe [Matthijnssens et al., 2008a; Rahman et al., 2010; Ghosh and Kobayashi, 2011]. Comparison of the full genomes of the South African G1P[8] strains from these two groups revealed a close genetic relationship among these RVAs, suggesting that identical G1P[8] strains with limited sequence variability might be circulating among children in South Africa. In 2002, the predominance of G1-I P[8]-I combination was reported, however in the current study the combination detected was G1-1 P[8]-III with the exception of the G1P[8] strain from 2004 which had a G1-IP[8]-II combination [Maunula and Von Bonsdorff, 2002]. Although the present study provided important insights into the origin and overall genetic makeup of the circulating human G1P[8] RVA strains in South Africa, it was limited to five strains from non-vaccinated and six from vaccinated children collected from one location. Full genomic analyses of additional RVA strains from different and varying geographical regions in South Africa will provide a better understanding of the evolutionary dynamics of the RVA strains in South Africa post-vaccine introduction.
The currently licensed RVA vaccines, Rotarix™ and RotaTeq™, have been found to be effective against the commonly detected human RVA strains, including G8 and G12 strains [Steele et al., 2012] resulting in substantial declines in rotavirus and/or diarrhea related hospitalization in many countries [Lopman et al., 2012]. Comparisons of the amino acid residues for the VP7 and VP4 antigenic domains showed several mismatches between the South African G1P[8] strains and the G1 and P[8] strains contained in Rotarix™ (which is currently being used in South Africa); however, a sharp reduction of severe gastroenteritis and hospitalization amongst South African children ≤5 years of age has been described [Seheri et al., 2012]. Therefore, to monitor the implications of these changes on the efficacy of these vaccines, molecular epidemiologic surveillance programs post-vaccine introduction must be encouraged. There is evidence that gene products other than VP7 and VP4 influence the immune response in the host following RVA vaccination [Matthijnssens et al., 2009]. Vaccine-induced immunological pressure may cause changes in these genes that are disadvantageous to the efficacy of the present RVA vaccines [Matthijnssens et al., 2009]. Consequently, large-scale complete genome-based studies on common human RVA strains from various countries are essential to identify these vaccine-induced changes in the RVA genome. This report therefore describes the first full genomic analyses of G1P[8] RVA strains collected from both non-vaccinated and vaccinated children in South Africa.
Supplementary Material
Acknowledgments
We thank Lorens Maake who assisted MMN with sample preparation for whole genome sequencing and also all the staff and students of MRC/Diarrhoeal Pathogens Research Unit together with those of the Virology group at the JCVI (especially, A. Akopov, N. Fedorova, and S. Shrivastava for their technical assistance and in-depth discussions). In addition, we thank Dr. Sunando Roy, Rotavirus Surveillance Team at CDC, for expert advice on the bioinformatics aspect of this work.
Grant sponsor: National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (partial support, contract number HHSN272200900007C); Grant sponsor: Medical Research Council and Poliomyelitis Research Foundation of South Africa
Footnotes
Conflicts of interest: none.
Disclaimer: The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention, the J. Craig Venter Institute, South African Medical Research Council, University of Limpopo, or Enteric and Diarrhoeal Diseases Programme, Global Health Program, Bill and Melinda Gates Foundation.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- Ahmed K, Nakagomi T, Nakagomi O. Molecular identification of a novel G1 VP7 gene carried by a human rotavirus with a super-short RNA pattern. Virus Genes. 2007;35:141–145. doi: 10.1007/s11262-007-0089-9. [DOI] [PubMed] [Google Scholar]
- Arista S, Giammanco GM, De Grazia S, Ramirez S, Lo Biundo C, Colomba C, Cascio A, Martella V. Heterogeneity and temporal dynamics of evolution of G1 human rotaviruses in a settled population. J Virol. 2006;80:10724–10733. doi: 10.1128/JVI.00340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, Binka FN, Steele AD, Laserson KF, Ansah NA, Levine MM, Lewis K, Coia ML, Attah-Poku M, Ojwando J, Rivers SB, Victor JC, Nyambane G, Hodgson A, Schodel F, Ciarlet M, Neuzil KM. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: A randomised, double-blind, placebo-controlled trial. Lancet. 2010;376:606–614. doi: 10.1016/S0140-6736(10)60889-6. [DOI] [PubMed] [Google Scholar]
- Arora R, Chitambar SD. Full genomic analysis of Indian G1P[8] rotavirus strains. Infect Genet Evol. 2011;11:504–511. doi: 10.1016/j.meegid.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Banyai K, Gentsch JR, Martella V, Bogdan A, Havasi V, Kisfali P, Szabo A, Mihaly I, Molnar P, Melegh B, Szucs G. Trends in the epidemiology of human G1P[8] rotaviruses: A hungarian study. J Infect Dis. 2009;200:S222–S227. doi: 10.1086/605052. [DOI] [PubMed] [Google Scholar]
- Banyai K, Mijatovic-Rustempasic S, Hull JJ, Esona MD, Freeman MM, Frace AM, Bowen MD, Gentsch JR. Sequencing and phylogenetic analysis of the coding region of six common rotavirus strains: Evidence for intragenogroup reassortment among co-circulating G1P[8] and G2P[4] strains from the United States. J Med Virol. 2011;83:532–539. doi: 10.1002/jmv.21977. [DOI] [PubMed] [Google Scholar]
- Banyai K, Laszlo B, Duque J, Steele AD, Nelson EA, Gentsch JR, Parashar UD. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: Insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30:A122–A130. doi: 10.1016/j.vaccine.2011.09.111. [DOI] [PubMed] [Google Scholar]
- Benhafid M, Rguig A, Trivedi T, Elqazoui M, Teleb N, Mouane N, Maltouf AF, Parashar U, Patel M, Aouad RE. Monitoring of rotavirus vaccination in Morocco: Establishing the baseline burden of rotavirus disease. Vaccine. 2012;30:6515–6520. doi: 10.1016/j.vaccine.2012.08.058. [DOI] [PubMed] [Google Scholar]
- Bucardo F, Karlsson B, Nordgren J, Paniagua M, Gonzalez A, Amador JJ, Espinoza F, Svensson L. Mutated G4P[8] rotavirus associated with a nationwide outbreak of gastroenteritis in Nicaragua in 2005. J Clin Microbiol. 2007;45:990–997. doi: 10.1128/JCM.01992-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucardo F, Rippinger CM, Svensson L, Patton JT. Vaccinederived NSP2 segment in rotaviruses from vaccinated children with gastroenteritis in Nicaragua. Infect Genet Evol. 2012;12:1282–1294. doi: 10.1016/j.meegid.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MK, Jheong WH, Lee SG, Park CJ, Jung KH, Paik SY. Full genomic analysis of a human rotavirus G1P[8] strain isolated in South Korea. J Med Virol. 2013;85:157–170. doi: 10.1002/jmv.23366. [DOI] [PubMed] [Google Scholar]
- Ciarlet M, Hyser JM, Estes MK. Sequence analysis of the VP4, VP6, VP7, and NSP4 gene products of the bovine rotavirus WC3. Virus Genes. 2002;24:107–118. doi: 10.1023/a:1014512314545. [DOI] [PubMed] [Google Scholar]
- Crawford SE, Mukherjee SK, Estes MK, Lawton JA, Shaw AL, Ramig RF, Prasad BV. Trypsin cleavage stabilizes the rotavirus VP4 spike. J Virol. 2001;75:6052–6061. doi: 10.1128/JVI.75.13.6052-6061.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunliffe NA, Witte D, Ngwira BM, Todd S, Bostock NJ, Turner AM, Chimpeni P, Victor JC, Steele AD, Bouckenooghe A, Neuzil KM. Efficacy of human rotavirus vaccine against severe gastroenteritis in Malawian children in the first two years of life: A randomized, double-blind, placebo controlled trial. Vaccine. 2012;30:A36–A43. doi: 10.1016/j.vaccine.2011.09.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curns AT, Steiner CA, Barrett M, Hunter K, Wilson E, Parashar UD. Reduction in acute gastroenteritis hospitalizations among US children after introduction of rotavirus vaccine: Analysis of hospital discharge data from 18 US states. J Infect Dis. 2010;201:1617–1624. doi: 10.1086/652403. [DOI] [PubMed] [Google Scholar]
- Diwakarla CS, Palombo EA. Genetic and antigenic variation of capsid protein VP7 of serotype G1 human rotavirus isolates. J Gen Virol. 1999;80:341–344. doi: 10.1099/0022-1317-80-2-341. [DOI] [PubMed] [Google Scholar]
- Djikeng A, Spiro D. Advancing full length genome sequencing for human RNA viral pathogens. Future Virol. 2009;4:47–53. doi: 10.2217/17460794.4.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, Spiro DJ. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato CM, Ch’ng LS, Boniface KF, Crawford NW, Buttery JP, Lyon M, Bishop RF, Kirkwood CD. Identification of strains of RotaTeq rotavirus vaccine in infants with gastroenteritis following routine vaccination. J Infect Dis. 2012;206:377–383. doi: 10.1093/infdis/jis361. [DOI] [PubMed] [Google Scholar]
- Dormitzer PR, Nason EB, Prasad BV, Harrison SC. Structural rearrangements in the membrane penetration protein of a non-enveloped virus. Nature. 2004;430:1053–1058. doi: 10.1038/nature02836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith ML, Lazdins I, Tregear GW, Holmes IH. Location of the major antigenic sites involved in rotavirus serotype-specific neutralization. Proc Natl Acad Sci USA. 1986;83:3465–3468. doi: 10.1073/pnas.83.10.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes MK, Kapikian A. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields virology. 5th. Philadelphia, PA: Kluwer/Lippincott, Williams and Wilkins; 2007. pp. 1917–1974. [Google Scholar]
- Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. Serotype diversity and reassortment between human and animal rotavirus strains: Implications for rotavirus vaccine programs. J Infect Dis. 2005;192:S146–S159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Kobayashi N. Whole-genomic analysis of rotavirus strains: Current status and future prospects. Future Microbiol. 2011;6:1049–1065. doi: 10.2217/fmb.11.90. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Paul SK, Yamamoto D, Nagashima S, Kobayashi N. Full genomic analyses of human rotavirus strains possessing the rare P[8]b VP4 subtype. Infect Genet Evol. 2011;11:1481–1486. doi: 10.1016/j.meegid.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Glass RI, Parashar UD, Bresee JS, Turcios R, Fischer TK, Widdowson MA, Jiang B, Gentsch JR. Rotavirus vaccines: Current prospects and future challenges. Lancet. 2006;368:323–332. doi: 10.1016/S0140-6736(06)68815-6. [DOI] [PubMed] [Google Scholar]
- Green KY, Hoshino Y, Ikegami N. Sequence analysis of the gene encoding the serotype-specific glycoprotein (VP7) of two new human rotavirus serotypes. Virology. 1989;168:429–433. doi: 10.1016/0042-6822(89)90289-4. [DOI] [PubMed] [Google Scholar]
- Guo D, Liu J, Lu Y, Sun Y, Yuan D, Jiang Q, Lin H, Li C, Si C, Qu L. Full genomic analysis of rabbit rotavirus G3P[14] strain N5 in China: Identification of a novel VP6 genotype. Infect Genet Evol. 2012;12:1567–1576. doi: 10.1016/j.meegid.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Hemming M, Vesikari T. Genetic diversity of G1P[8] rotavirus VP7 and VP8* antigens in Finland over a 20-year period: No evidence for selection pressure by universal mass vaccination with RotaTeq(R) vaccine. Infect Genet Evol. 2013;19:51–58. doi: 10.1016/j.meegid.2013.06.026. [DOI] [PubMed] [Google Scholar]
- Jere KC, Mlera L, O’Neill HG, Potgieter AC, Page NA, Seheri ML, van Dijk AA. Whole genome analyses of African G2, G8, G9, and G12 rotavirus strains using sequence-independent amplification and 454(R) pyrosequencing. J Med Virol. 2011;83:2018–2042. doi: 10.1002/jmv.22207. [DOI] [PubMed] [Google Scholar]
- Jere KC, Esona MD, Ali YH, Peenze I, Roy S, Bowen MD, Saeed IK, Khalafalla AI, Nyaga MM, Mphahlele J, Steele D, Seheri ML. Novel NSP1 genotype characterised in an African camel G8P[11] rotavirus strain. Infect Genet Evol. 2014;21:58–66. doi: 10.1016/j.meegid.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Jin Q, Ward RL, Knowlton DR, Gabbay YB, Linhares AC, Rappaport R, Woods PA, Glass RI, Gentsch JR. Divergence of VP7 genes of G1 rotaviruses isolated from infants vaccinated with reassortant rhesus rotaviruses. Arch Virol. 1996;141:2057–2076. doi: 10.1007/BF01718215. [DOI] [PubMed] [Google Scholar]
- Kirkwood CD. Genetic and antigenic diversity of human rotaviruses: Potential impact on vaccination programs. J Infect Dis. 2010;202:S43–S48. doi: 10.1086/653548. [DOI] [PubMed] [Google Scholar]
- Kirkwood C, Masendycz PJ, Coulson BS. Characteristics and location of cross-reactive and serotype-specific neutralization sites on VP7 of human G type 9 rotaviruses. Virology. 1993;196:79–88. doi: 10.1006/viro.1993.1456. [DOI] [PubMed] [Google Scholar]
- Le VP, Chung YC, Kim K, Chung SI, Lim I, Kim W. Genetic variation of prevalent G1P[8] human rotaviruses in South Korea. J Med Virol. 2010;82:886–896. doi: 10.1002/jmv.21653. [DOI] [PubMed] [Google Scholar]
- Lopman BA, Payne DC, Tate JE, Patel MM, Cortese MM, Parashar UD. Post-licensure experience with rotavirus vaccination in high and middle income countries; 2006 to 2011. Curr Opin Virol. 2012;2:434–442. doi: 10.1016/j.coviro.2012.05.002. [DOI] [PubMed] [Google Scholar]
- Madhi SA, Kirsten M, Louw C, Bos P, Aspinall S, Bouckenooghe A, Neuzil KM, Steele AD. Efficacy and immunogenicity of two or three dose rotavirus-vaccine regimen in South African children over two consecutive rotavirus-seasons: A randomized, double-blind, placebo-controlled trial. Vaccine. 2012;30:A44–A51. doi: 10.1016/j.vaccine.2011.08.080. [DOI] [PubMed] [Google Scholar]
- Maranhao AG, Vianez-Junior JL, Benati FJ, Bisch PM, Santos N. Polymorphism of rotavirus genotype G1 in Brazil: In silico analysis of variant strains circulating in Rio de Janeiro from 1996 to 2004. Infect Genet Evol. 2012;12:1397–1404. doi: 10.1016/j.meegid.2012.04.018. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Van Ranst M. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr Opin Virol. 2012;2:426–433. doi: 10.1016/j.coviro.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gomara M, Maes P, Patton JT, Rahman M, Van Ranst M. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol. 2008a;82:3204–3219. doi: 10.1128/JVI.02257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Banyai K, Estes MK, Gentsch JR, Iturriza-Gomara M, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Patton JT, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Desselberger U, Van Ranst M. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol. 2008b;153:1621–1629. doi: 10.1007/s00705-008-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, Bilcke J, Ciarlet M, Martella V, Banyai K, Rahman M, Zeller M, Beutels P, Van Damme P, Van Ranst M. Rotavirus disease and vaccination: Impact on genotype diversity. Future Microbiol. 2009;4:1303–1316. doi: 10.2217/fmb.09.96. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Joelsson DB, Warakomski DJ, Zhou T, Mathis PK, van Maanen MH, Ranheim TS, Ciarlet M. Molecular and biological characterization of the 5 human-bovine rotavirus (WC3)-based reassortant strains of the pentavalent rotavirus vaccine, RotaTeq. Virology. 2010;403:111–127. doi: 10.1016/j.virol.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Ciarlet M, McDonald SM, Attoui H, Banyai K, Brister JR, Buesa J, Esona MD, Estes MK, Gentsch JR, Iturriza-Gomara M, Johne R, Kirkwood CD, Martella V, Mertens PP, Nakagomi O, Parreno V, Rahman M, Ruggeri FM, Saif LJ, Santos N, Steyer A, Taniguchi K, Patton JT, Desselberger U, Van Ranst M. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG) Arch Virol. 2011;156:1397–1413. doi: 10.1007/s00705-011-1006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunula L, Von Bonsdorff CH. Frequent reassortments may explain the genetic heterogeneity of rotaviruses: Analysis of Finnish rotavirus strains. J Virol. 2002;76:11793–11800. doi: 10.1128/JVI.76.23.11793-11800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald SM, Matthijnssens J, McAllen JK, Hine E, Overton L, Wang S, Lemey P, Zeller M, Van Ranst M, Spiro DJ, Patton JT. Evolutionary dynamics of human rotaviruses: Balancing reassortment with preferred genome constellations. PLoS Pathog. 2009;5:e1000634. doi: 10.1371/journal.ppat.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouna BH, Benhamida-Rebai M, Heylen E, Zeller M, Moussa A, Kacem S, Van Ranst M, Matthijnssens J, Trabelsi A. Sequence and phylogenetic analyses of human rotavirus strains: Comparison of VP7 and VP8 antigenic epitopes between Tunisian and vaccine strains before national rotavirus vaccine introduction. Infect Genet Evol. 2013;18C:132–144. doi: 10.1016/j.meegid.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Neuzil KM, Armah GE, Parashar UD, Steele AD. Rotavirus in Africa: Shifting the focus to disease prevention. J Infect Dis. 2010;202:S1–S4. doi: 10.1086/653545. [DOI] [PubMed] [Google Scholar]
- Nyaga MM, Jere KC, Peenze I, Mlera L, van Dijk AA, Seheri ML, Mphahlele MJ. Sequence analysis of the whole genomes of five African human G9 rotavirus strains. Infect Genet Evol. 2013;16:62–77. doi: 10.1016/j.meegid.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Offit PA, Clark HF, Blavat G, Greenberg HB. Reassortant rotaviruses containing structural proteins vp3 and vp7 from different parents induce antibodies protective against each parental serotype. J Virol. 1986;60:491–496. doi: 10.1128/jvi.60.2.491-496.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp H, Al-Mutairi LZ, Chehadeh W, Farkas SL, Lengyel G, Jakab F, Martella V, Szucs G, Banyai K. Novel NSP4 genotype in a camel G10P[15] rotavirus strain. Acta Microbiol Immunol Hung. 2012;59:411–421. doi: 10.1556/AMicr.59.2012.3.11. [DOI] [PubMed] [Google Scholar]
- Parashar UD, Burton A, Lanata C, Boschi-Pinto C, Shibuya K, Steele D, Birmingham M, Glass RI. Global mortality associated with rotavirus disease among children in 2004. Infect Dis. 2009;200:S9–S15. doi: 10.1086/605025. [DOI] [PubMed] [Google Scholar]
- Patel M, Pedreira C, De Oliveira LH, Tate J, Orozco M, Mercado J, Gonzalez A, Malespin O, Amador JJ, Umana J, Balmaseda A, Perez MC, Gentsch J, Kerin T, Hull J, Mijatovic S, Andrus J, Parashar U. Association between pentavalent rotavirus vaccine and severe rotavirus diarrhea among children in Nicaragua. JAMA. 2009;301:2243–2251. doi: 10.1001/jama.2009.756. [DOI] [PubMed] [Google Scholar]
- Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA. Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol. 2009;90:1423–1432. doi: 10.1099/vir.0.009381-0. [DOI] [PubMed] [Google Scholar]
- Potgieter N, de Beer MC, Taylor MB, Steele AD. Prevalence and diversity of rotavirus strains in children with acute diarrhea from rural communities in the Limpopo Province, South Africa, from 1998 to 2000. J Infect Dis. 2010;202:S148–S155. doi: 10.1086/653561. [DOI] [PubMed] [Google Scholar]
- Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, Taniguchi K, Iturriza-Gomara M, Iftekharuddin N, Azim T, Van Ranst M. Evolutionary history and global spread of the emerging g12 human rotaviruses. J Virol. 2007;81:2382–2390. doi: 10.1128/JVI.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Matthijnssens J, Saiada F, Hassan Z, Heylen E, Azim T, Van Ranst M. Complete genomic analysis of a Bangladeshi G1P[8] rotavirus strain detected in 2003 reveals a close evolutionary relationship with contemporary human Wa-like strains. Infect Genet Evol. 2010;10:746–754. doi: 10.1016/j.meegid.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Rose TL, Miagostovich MP, Leite JP. Rotavirus A genotype G1P[8]: A novel method to distinguish wild-type strains from the Rotarix vaccine strain. Mem Inst Oswaldo Cruz. 2010;105:1068–1072. doi: 10.1590/s0074-02762010000800021. [DOI] [PubMed] [Google Scholar]
- Ruiz-Palacios GM, Perez-Schael I, Velazquez FR, Abate H, Breuer T, Clemens SC, Cheuvart B, Espinoza F, Gillard P, Innis BL, Cervantes Y, Linhares AC, Lopez P, Macias-Parra M, Ortega-Barria E, Richardson V, Rivera-Medina DM, Rivera L, Salinas B, Pavia-Ruz N, Salmeron J, Ruttimann R, Tinoco JC, Rubio P, Nunez E, Guerrero ML, Yarzabal JP, Damaso S, Tornieporth N, Saez-Llorens X, Vergara RF, Vesikari T, Bouckenooghe A, Clemens R, De Vos B, O’Ryan M, Human Rotavirus Vaccine Study Group Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N Engl J Med. 2006;354:11–22. doi: 10.1056/NEJMoa052434. [DOI] [PubMed] [Google Scholar]
- Sanchez-Padilla E, Grais RF, Guerin PJ, Steele AD, Burny ME, Luquero FJ. Burden of disease and circulating serotypes of rotavirus infection in sub-Saharan Africa: Systematic review and meta-analysis. Lancet Infect Dis. 2009;9:567–576. doi: 10.1016/S1473-3099(09)70179-3. [DOI] [PubMed] [Google Scholar]
- Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- Seheri LM, Page N, Dewar JB, Geyer A, Nemarude AL, Bos P, Esona M, Steele AD. Characterization and molecular epidemiology of rotavirus strains recovered in Northern Pretoria, South Africa during 2003–2006. J Infect Dis. 2010;202:S139–S147. doi: 10.1086/653559. [DOI] [PubMed] [Google Scholar]
- Seheri LM, Page NA, Mawela MP, Mphahlele MJ, Steele AD. Rotavirus vaccination within the South African Expanded Programme on Immunisation. Vaccine. 2012;30:C14–C20. doi: 10.1016/j.vaccine.2012.04.018. [DOI] [PubMed] [Google Scholar]
- Shintani T, Ghosh S, Wang YH, Zhou X, Zhou DJ, Kobayashi N. Whole genomic analysis of human G1P[8] rotavirus strains from different age groups in China. Viruses. 2012;4:1289–1304. doi: 10.3390/v4081289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele AD, Ivanoff B. Rotavirus strains circulating in Africa during 1996–1999: Emergence of G9 strains and P[6] strains. Vaccine. 2003;21:361–367. doi: 10.1016/s0264-410x(02)00616-3. [DOI] [PubMed] [Google Scholar]
- Steele AD, Peenze I, de Beer MC, Pager CT, Yeats J, Potgieter N, Ramsaroop U, Page NA, Mitchell JO, Geyer A, Bos P, Alexander JJ. Anticipating rotavirus vaccines: Epidemiology and surveillance of rotavirus in South Africa. Vaccine. 2003;21:354–360. doi: 10.1016/s0264-410x(02)00615-1. [DOI] [PubMed] [Google Scholar]
- Steele AD, Neuzil KM, Cunliffe NA, Madhi SA, Bos P, Ngwira B, Witte D, Todd S, Louw C, Kirsten M, Aspinall S, Van Doorn LJ, Bouckenooghe A, Suryakiran PV, Han HH. Human rotavirus vaccine Rotarix provides protection against diverse circulating rotavirus strains in African infants: A randomized controlled trial. BMC Infect Dis. 2012;12:213. doi: 10.1186/1471-2334-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirzaker SC, Whitfeld PL, Christie DL, Bellamy AR, Both GW. Processing of rotavirus glycoprotein VP7: Implications for the retention of the protein in the endoplasmic reticulum. J Cell Biol. 1987;105:2897–2903. doi: 10.1083/jcb.105.6.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEG A5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, the WHO-coordinated Global Rotavirus Surveillance Network 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect Dis. 2012;12:136–141. doi: 10.1016/S1473-3099(11)70253-5. [DOI] [PubMed] [Google Scholar]
- Todd S, Page NA, Duncan Steele A, Peenze I, Cunliffe NA. Rotavirus strain types circulating in Africa: Review of studies published during 1997–2006. J Infect Dis. 2010;202:S34–S42. doi: 10.1086/653555. [DOI] [PubMed] [Google Scholar]
- Trinh QD, Nguyen TA, Phan TG, Khamrin P, Yan H, Hoang PL, Maneekarn N, Li Y, Yagyu F, Okitsu S, Ushijima H. Sequence analysis of the VP7 gene of human rotavirus G1 isolated in Japan, China, Thailand, and Vietnam in the context of changing distribution of rotavirus G-types. J Med Virol. 2007;79:1009–1016. doi: 10.1002/jmv.20920. [DOI] [PubMed] [Google Scholar]
- Trojnar E, Sachsenroder J, Twardziok S, Reetz J, Otto PH, Johne R. Identification of an avian group A rotavirus containing a novel VP4 gene with a close relationship to those of mammalian rotaviruses. J Gen Virol. 2013;94:136–142. doi: 10.1099/vir.0.047381-0. [DOI] [PubMed] [Google Scholar]
- Vesikari T, Karvonen A, Prymula R, Schuster V, Tejedor JC, Cohen R, Meurice F, Han HH, Damaso S, Bouckenooghe A. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: Randomised, double-blind controlled study. Lancet. 2007;370:1757–1763. doi: 10.1016/S0140-6736(07)61744-9. [DOI] [PubMed] [Google Scholar]
- Ward RL, Bernstein DI. Rotarix: A rotavirus vaccine for the world. Clin Infect Dis. 2009;48:222–228. doi: 10.1086/595702. [DOI] [PubMed] [Google Scholar]
- Ward RL, McNeal MM, Sander DS, Greenberg HB, Bernstein DI. Immunodominance of the VP4 neutralization protein of rotavirus in protective natural infections of young children. J Virol. 1993;67:464–468. doi: 10.1128/jvi.67.1.464-468.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt RG, James HD, Jr, Pittman AL, Hoshino Y, Greenberg HB, Kalica AR, Flores J, Kapikian AZ. Direct isolation in cell culture of human rotaviruses and their characterization into four serotypes. J Clin Microbiol. 1983;18:310–317. doi: 10.1128/jcm.18.2.310-317.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller M, Patton JT, Heylen E, De Coster S, Ciarlet M, Van Ranst M, Matthijnssens J. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. J Clin Microbiol. 2012;50:966–976. doi: 10.1128/JCM.05590-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


