Abstract
Curcumin is a potential agent for both the prevention and treatment of cancers. Curcumin treatment alone, or in combination with piperine, limits breast stem cell self-renewal while remaining non-toxic to normal differentiated cells. We paired fluorescence activated cell sorting with RNA sequencing to characterize the genome-wide changes induced specifically in normal breast stem cells following treatment with these compounds. We generated genome-wide maps of the transcriptional changes that occur in epithelial-like (ALDH+) and mesenchymal-like (ALDH−/CD44+/CD24−) normal breast stem/progenitor cells following treatment with curcumin and piperine. We show that curcumin targets both stem cell populations by down-regulating expression of breast stem cell genes including ALDH1A3, CD49f, PROM1, and TP63. We also identified novel genes and pathways targeted by curcumin, including downregulation of SCD. Transient siRNA knockdown of SCD in MCF10A cells significantly inhibited mammosphere formation and the mean proportion of CD44+/CD24− cells, suggesting that SCD is a regulator of breast stemness and a target of curcumin in breast stem cells. These findings extend previous reports of curcumin targeting stem cells, here in two phenotypically distinct stem/progenitor populations isolated from normal human breast tissue. We identified novel mechanisms by which curcumin and piperine target breast stem cell self-renewal, such as by targeting lipid metabolism, providing a mechanistic link between curcumin treatment and stem cell self renewal. These results elucidate the mechanisms by which curcumin may act as a cancer preventive compound and provide novel targets for cancer prevention and treatment.
INTRODUCTION
The large number of newly diagnosed cases and deaths from breast cancer are indicative of the necessity for the development of novel strategies for the prevention of these diseases. While there has been a significant reduction in the number of deaths due to breast cancer since 1990, breast cancer remains the second most deadly cancer in U.S. women, with an estimated 39,840 deaths in 2010 [1]. The current strategies for breast cancer prevention are associated with toxicity or short and long term risks from surgery [2] or antiestrogen therapy [3, 4]. Antiestrogen therapy has also been shown to be effective only at preventing estrogen receptor positive (ER+) disease [5]. There is a need, therefore, for the identification and development of cancer prevention strategies that are non-toxic and prevent both ER+ and ER− disease.
Curcumin is a dietary polyphenol derived from the rhizomes of turmeric (curcuma longa), which has been widely used in traditional Indian and Chinese medicine for treatment of a range of diseases, including inflammatory conditions, diabetes, and rheumatoid arthritis [6]. Preclinical models implicate curcumin as an agent for both cancer prevention and treatment. A major issue with the use of curcumin clinically is the limited bioavailability following ingestion. A number of strategies to increase curcumin’s bioavailability have been tested, with the use of piperine as an adjuvant treatment showing up a 20-fold increase [7]. Recently, we used the mammosphere model, a primary breast tissue culture method that enriches for stem and early progenitor cells [8], to show that piperine and curcumin combined limit breast stem cell self-renewal while remaining non-toxic to normal differentiated cells [9].
Since breast tumors potentially arise from, and are sustained by, a population of progenitor or stem-like cells that harbor dysregulated self-renewal capacity, characterizing the effects of cancer preventive compounds specifically in stem and progenitor cells [10, 11] is essential. Emerging evidence suggests that normal breast, as well as breast cancer, stem and progenitor cells exist in two different states, epithelial-like and mesenchymal-like [12, 13]. Epithelial-like stem cells express aldehyde dehydrogenases (ALDH+). Mesenchymal-like stem cells are characterized by CD44+/CD24− surface expression [13]. How these distinct populations of breast stem/progenitor cells in normal tissue respond to cancer preventive agents is not well understood.
The goal of this study was to comprehensively characterize the effects of these compounds specifically in these two populations of breast stem cells. By pairing fluorescence activated cell sorting (FACS) with low-input, high-throughput RNA sequencing (RNA-seq), we generated genome-wide maps of the transcriptional changes that occur in epithelial-like and mesenchymal-like normal breast stem cells following treatment with curcumin and piperine. Our results confirm that these compounds target breast stem cell self-renewal in both stem cell populations by down-regulating expression of breast stemness genes. Additionally, we identify novel genes and pathways targeted by curcumin relevant to breast stem cells, including genes involved in lipid metabolism. These results elucidate the mechanisms by which curcumin and piperine target breast stem cells and provide insight into genes and pathways involved in stem cell regulation in the normal human breast.
MATERIALS AND METHODS
Materials
Curcumin (98% pure) and piperine (BioPerine; 95% pure piperine) were donated by Sabinsa Corporation (Piscataway, NJ). Curcumin and piperine were diluted in DMSO to form a stock solution for dissolution in cell culture media. MCF7 and MCF10A cells were obtained from American Type Culture Collection (ATCC). SUM149 cells were obtained from Asterand.
Human Normal Breast Tissue Dissociation
Normal (non-pathogenic) breast tissue was isolated from women undergoing voluntary reduction mammoplasty at the University of Michigan hospital. The study protocol was approved by the University of Michigan Institutional Review Board. Breast tissue was mechanically and enzymatically digested as previously described [8, 9].
Mammosphere Formation
Single cells were plated in ultralow attachment plates (Corning) at a density of 100,000 viable cells/mL for primary cells and a density of 20,000 viable cells/mL for SUM149, MCF10A and MCF7 cells. Primary mammospheres formed for 7–10 days in serum-free mammary epithelial basal medium (MEBM) supplemented with 1 ug/mL hydrocortisone, 50ug/mL insulin, 20 ng/mL EGF, B27, 20 ug/mL gentamycin, and 1x antibiotic-antimycotic in the presence of either curcumin, piperine, or vehicle (DMSO) control. Each experiment was performed in triplicate and sphere number quantified manually. Previous work established that 5uM curcumin, individually or in co-treatment with 5uM piperine, was sufficient to inhibit primary and secondary mammosphere formation [9]. To characterize interindividual variation in response to curcumin and piperine, we treated cells isolated from 13 mammoplasty reduction patients and quantified primary sphere formation.
Flow Cytometry and Curcumin Treated Sorted Cell Populations
Primary breast cells from 3 individuals were sorted on a MoFlo Astrios flow cytometer. Cells were first stained for a hematopoetic, fibroblast, and endothelial cell lineage depletion cocktail that consisted of biotinylated antibodies targeted against CD45, HLA-DR, CD14, CD31, CD41, CD19, CD235a, CD56, CD3, CD16, and CD140b (all from eBioscience, except for CD140b (Biolegend) and CD41 (Acris)). Next, cells were stained with Alexafluor750-streptavidin, Alexafluor750 LIVE/DEAD Fixable Dead Cell Stain (Invitrogen), CD24-Brilliant Violet 421 (Biolegend), CD44-APC (BD), and Aldefluor (Stem Cell Technology). Single color and isotype controls were included for compensation and gating purposes. Aldefluor-positive gating was based on DEAB (negative) controls. Viability of cells post sorting was confirmed via trypan blue exclusion. Sorted cell populations were plated in mammosphere formation conditions and treated with either 5 µM curcumin, 5 µM piperine, both 5 µM curcumin and 5 µM piperine, or vehicle control. After 24 hours, total RNA was isolated from each treated cell population using the RNEasy Micro Kit (Qiagen) with on column DNase treatment.
High Throughput RNA Sequencing
RNA concentration and quality was determined using a Nanodrop (Thermo) and Bioanalyzer (Agilent). Due to the low level of input RNA due to a small number of cells following FACS sorting and treatment, we depleted ribosomal RNAs with Ribominus (Life) and prepared sequencing libraries utilizing the SMARTer Stranded RNA-Seq kit (Clontech) following the manufacturer’s recommended protocol. Libraries were multiplexed (4 per lane) and sequenced using paired end 50 cycle reads on a HiSeq 2500 (Illumina) at the University of Michigan DNA Sequencing Core Facility. Due to the high redundancy in base pairs introduced at the beginning of each read by the SMARTer library preparation kit, one lane of PhiX control was included per flowcell.
RNA-Seq Data Analysis
Computational analysis was performed using the Flux high-performance computer cluster hosted by Advanced Research Computing (ARC) at the University of Michigan. Raw sequencing read quality was assessed utilizing FastQC. Sequencing reads were concatenated by sample and read in pair using SeqTK. The first three nucleotides of the first read in each read pair were trimmed, as recommended by Clontech, using Prinseq 0.20.3. A splice junction aware build of the human genome (GRCh37) was built using the genomeGenerate function from STAR 2.3.0 [14]. Read pairs were aligned to the genome using STAR, using the options “outFilterMultimapNmax 10” and “sjdbScore 2”. The aligned reads were assigned to genomic features (GRCh37 genes) using HTSeq-count, with the set mode “union”. We conducted differential expression testing on the assigned read counts per gene utilizing edgeR [15]. Separate analysis were conducted for each stem/progenitor cell type (luminal and basal), adjusting for study subject as a covariate using glmLRT. To reduce the dispersion of the dataset due to lowly expressed genes, genes with a mean aligned read count less than five across all samples were excluded from analysis. Normalized counts per million were estimated utilizing the “cpm” function in edgeR [15]. Genes were considered differentially expressed between conditions at a false discovery rate adjusted p-value < 0.05 [16]. To compare how genes identified as differentially expressed between the ALDH+ and ALDH−/CD44+/CD24− overlap with previously reported breast stem and progenitor cell gene expression signatures, we compared log fold changes in expression in our data for genes identified as uniquely upregulated (logFC >1) in CD49f+/EpCAM-(“mammary stem”) and CD49f+/EpCAM+ (“luminal progenitor”) cell populations [17]. A total of 752 (out of 943) genes from the CD49f+/EpCAM− expression signature and 277 (out of 359) genes from the CD49f+/EpCAM+ expression signature were expressed at detectable levels in our samples and included in the analysis.
Pathway analyses
Differentially expressed pathways were identified utilizing iPathwayGuide (Advaita). A directional analysis was conducted on all genes by including p-value of the differential expression test between groups as an effect size measure and log2 fold difference in expression as a measure of effect direction. Biological pathways were considered differentially expressed at a p-value <0.05.
RNA Expression Validation
To validate two of the most differentially expressed genes identified by RNA-seq, SCD and HMOX1, normal breast cells from three different individuals were plated in anchorage independent conditions for 24 hours with 5uM curcumin. RNA was extracted as described above, and SCD and HMOX1 expression were measured by quantitative real-time PCR, normalizing SCD gene expression to the geometric mean expression of GAPD and ACTB. The qPCR primers utilized were: SCD FWD:TTCCCGACGTGGCTTTTTCT, SCD RVS:AGCCAGGTTTGTAGTACCTCC; HMOX1 FWD: ACTGCGTTCCTGCTCAACATCC, HMOX1 RVS: GGCTCTGGTCCTTGGTGTCATG; ACTB FWD:GGCACCCAGCACAATGAAG, ACTB RVS:CCGATCCACACGGAGTACTTG; GAPD FWD: CGACAGTCAGCCGCATCTT, GAPD RVS: GTTAAAAGCAGCCCTGGTGAC.
siRNA Knockdown of SCD
We utilized siRNA directed against SCD in MCF10A cells to characterize the effects of SCD downregulation on breast stem cell regulation. MCF10A cells were transfected with the ON-TARGETplus SCD siRNA SMARTpool, at concentrations ranging from 10–50 nM, using the DharmaFECT-1 transfection reagent (Dharmacon). MCF10A cells were also transfected with the ON-TARGETplus Non-targeting siRNA pool (Dharmacon) and ON-TARGETplus GAPD Control siRNA as negative and positive, respectively, transfection controls. SCD knockdown at the RNA level was confirmed by quantitative real-time PCR as described above. Changes in SCD protein expression were quantified by western blot after 2, 4, and 7 days, following an initial treatment with SCD siRNA for 24 hours, utilizing a primary anti-SCD antibody (abcam ab19862), used at 1:1,000 and a Primary Anti-beta-Actin HRP antibody (Sigma Aldrich A3854) used at 1:25,000. Western blot results were analyzed using Li-Cor Image Studio to determine relative intensity of SCD1 bands and beta-actin bands. Relative SCD intensity was compared between the knockdown and control biological replicates (n=3) at each time point. Effects on cellular proliferation were quantified by the MTT Cell Proliferation assay kit (ATCC) following the manufacturer’s recommended protocol. Changes in mammosphere formation and CD44+/CD24− cell proportions between SCD knockdown and control cells were analyzed as described above. For mammosphere formation experiments, cells were exposed to the siRNA transfection reagents for 24 hours in attachment conditions before being plated in mammosphere formation conditions. Each transfection experiment was performed in triplicate.
Palmitoleic Acid and Curcumin Co-Treatment
To quantify whether the effects of curcumin on primary mammosphere formation are mediated through SCD downregulation, we co-treated primary normal breast cells with both curcumin and palmitoleic acid, a major monounsaturated fatty acid substrate synthesized by SCD. Palmitoleic acid (Cayman) was suspended in ethanol and conjugated to fatty acid free BSA as previously described [18] to generate a stock solution. Normal breast cells from 3 individuals were cultured in mammosphere formation conditions, as described above, in the presence of 5 or 10 µM curcumin with or without supplementation with 50 µM palmitoleic acid. Proportion of mammospheres formed was compared relative to vehicle control treated cells.
Statistical Analysis
Mammosphere formation, cellular proliferation, protein expression, and CD44+/CD24− stem cell proportions were compared between treatment groups by 2-sided t-test. Differences in RNA expression, measured by qPCR, between SCD siRNA knockdown and control cells were determined by the 2(-Delta Delta C(T)) method [19]. Statistical significance for these experiments was set at p<0.05. All statistical analyses were conducted in R 3.0.2 [20].
RESULTS
Curcumin and piperine inhibit mammosphere formation
To confirm and extend previous findings of the inhibitory effect of curcumin on mammosphere formation [9], we exposed MCF7 cells, SUM149 cells, and primary human breast cells to curcumin and piperine in vitro. Curcumin inhibited mammosphere formation in both MCF7 and SUM149 in a dose dependent manner (Figure 1A and B), as well as qualitatively decreasing mammosphere size (representative images, Figure 1C). Treatment with curcumin and piperine in combination was found to decrease primary mammosphere formation at a greater rate than curcumin treatment alone. In cell lines assayed here, and in previous studies [9], 5 µM curcumin treatment was found to significantly inhibit mammosphere formation without inducing acute cytotoxicity, as assayed by trypan blue exclusion. In primary breast cells isolated from voluntary mammoplasty patients (n=13), treatment with 5 µM curcumin, 5 µM piperine, and both agents inhibited primary mammosphere formation (Figure 1D).
Figure 1.
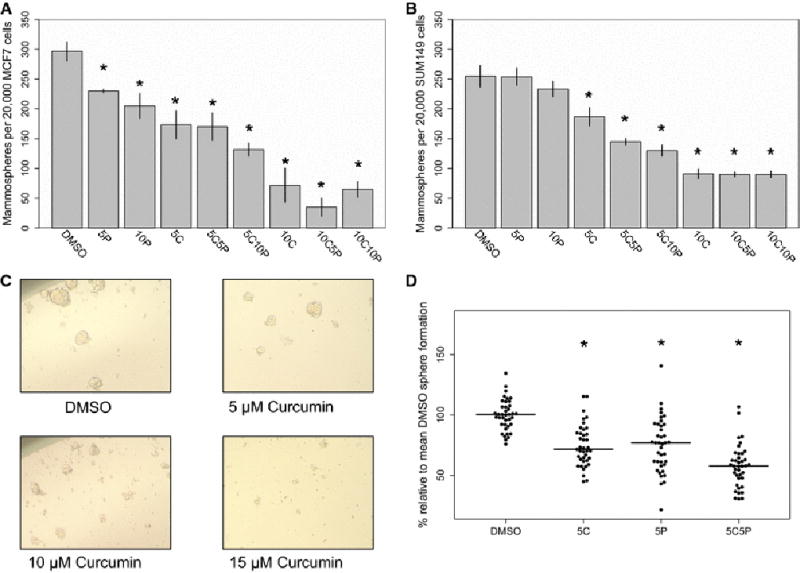
The effects of curcumin and piperine treatment on primary mammosphere number and size in cancer cell lines and normal breast cells. Curcumin and piperine were tested at multiple concentrations in cell lines (5C = 5 µM curcumin, 5P = 5 µM piperine, 5C5P = 5 µM curcumin and 5 µM piperine, for example). (A) and (B) The effects of curcumin and piperine on primary mammosphere formation in MCF7 and SUM149 cells, respectively. (C) The effects of curcumin on mammosphere size in MCF7 cells. (D) Curcumin and piperine significantly inhibited primary mammosphere formation in normal breast cells (N=13). * - p<0.05 compared to vehicle control.
Transcriptome-wide analysis of the effects of curcumin in ALDH+ and ALDH-CD44+CD24− breast cells
Differential expression in ALDH+ and ALDH−/CD44+/CD24− cells
We isolated two distinct breast cell populations enriched for stem and progenitor cells, ALDH+ (epithelial-like) and ALDH−/CD44+/CD24− (mesenchymal-like), via FACS (Figure 2A). To characterize the baseline transcriptional differences in the two populations, we compared expression between the vehicle control treated cell fractions after 24 hours in culture. Figure 2B presents a multidimensional scaling plot, with the two different cell fractions clearly clustering on the first dimension of the leading log fold change. A differential expression analysis identified 1369 genes upregulated in the ALDH−/CD44+/CD24− fraction and 1573 genes upregulated in the ALDH+ fraction (Figure 2C; Supplemental Table 1).
Figure 2.
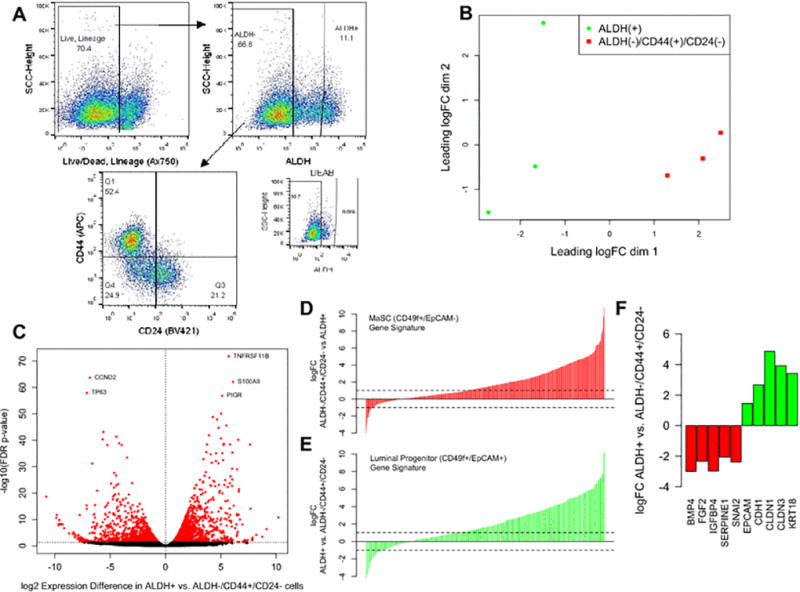
Isolation and characterization of live, lineage negative, ALDH(+) and ALDH(−)/CD44(+)/CD24(−) normal breast cells. (A) FACS gating scheme: A biotinylated lineage cocktail was used to isolate live, lineage negative cells. ALDH(+) cells were collected, with gating set based on the DEAB negative control. Finally, ALDH−/CD44+/CD24− cells (Q1) were isolated. (B) Multidimensional scaling plot of the vehicle control treated ALDH+ and ALDH−/CD44+/CD24− cells, based on the top 500 most variable genes. (C) False discovery rate (FDR) volcano plot of the log(2) ratio of gene expression between the vehicle control treated ALDH(+) and ALDH(−)CD44(+)CD24(−) cells. (D) Comparing the differences in expression in ALDH−/CD44+/CD24− and ALDH+ cells for genes uniquely expressed in mammary stem (MaSC) CD49f+/EpCAM- cells (E) Comparing the differences in expression in ALDH+ and ALDH−/CD44+/CD24− for genes uniquely expressed in luminal progenitor CD49f+/EpCAM+ cells. (F) Expression differences between ALDH+ and ALDH−/CD44+/CD24− cells for genes involved in the epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions.
To characterize how the gene expression signatures of these two cell populations relate to previous reports of normal breast stem and progenitor cell fractions, we compared our results to previously reported gene expression signatures of mammary stem (CD49f+/EpCAM-) and luminal progenitor (CD49f+/EpCAM+) cells [17]. Relative to ALDH+ cells, ALDH−/CD44+/CD24− cells strongly overexpressed genes identified as uniquely expressed in mammary stem cells (Figure 2D). ALDH+ cells, conversely, overexpress genes identified as uniquely expressed in luminal progenitor cells (Figure 2E). These results support previous findings that ALDH+ and CD44+/CD24− isolate overlapping, but not identical, cell populations as CD49f+/EpCAM+ and CD49f/EpCAM- [13], that ALDH+ cells are enriched for a luminal progenitor-like cell population [21], and further show that these phenotypes are stable for 24 hours in culture after sorting. Additionally, epithelial-mesenchymal transition (EMT) associated genes, such as BMP4, FGF2, IGFBP4, SERPINE1, and SNAI2 were significantly upregulated in ALDH−/CD44+/CD24− cells, while conversely, epithelial-phenotype associated genes, including CDH1, EPCAM (, CLDN1, CLDN3, and KRT18 were overexpressed in the ALDH+ cells (Figure 2F). Pathway analyses identified that biological processes involved in cell adhesion, ECM-receptor interaction, focal adhesion, Hippo signaling, and steroid biosynthesis were differentially expressed between the two cell types (Table 1A).
Table 1.
The 10 most enriched KEGG biological pathways identified between (A) the vehicle control treated ALDH+ and ALDH−/CD44+/CD24− cells (B) Curcumin vs. DMSO treated ALDH+ cells (C) curcumin vs. DMSO treated ALDH−/CD44+/CD24− cells.
| (A) ALDH+ vs. ALDH−/CD44+/CD24−
| |
|---|---|
| Name | P-Value |
| Cell adhesion molecules (CAMs) | 1.42E-07 |
| Amoebiasis | 2.40E-07 |
| ECM-receptor interaction | 2.66E-07 |
| Regulation of actin cytoskeleton | 1.59E-06 |
| Tight junction | 1.95E-06 |
| Protein digestion and absorption | 5.73E-06 |
| Focal adhesion | 1.02E-05 |
| Adherens junction | 1.19E-05 |
| Hippo signaling pathway | 1.21E-05 |
| Steroid biosynthesis | 1.63E-05 |
|
| |
| (B) Curcumin vs. DMSO treated ALDH+ cells | |
|
| |
| Biosynthesis of unsaturated fatty acids | 0.0007 |
| Steroid biosynthesis | 0.002 |
| PPAR signaling pathway | 0.003 |
| Biosynthesis of amino acids | 0.003 |
| Metabolism of xenobiotics by cytochrome P450 | 0.003 |
| Glycine, serine and threonine metabolism | 0.004 |
| Gastric acid secretion | 0.005 |
| Steroid hormone biosynthesis | 0.008 |
| Protein processing in endoplasmic reticulum | 0.008 |
| Measles | 0.009 |
|
| |
| (C) Curcumin vs. DMSO treated ALDH−/CD44+/CD24− cells | |
|
| |
| Antigen processing and presentation | 0.004 |
| Transcriptional misregulation in cancer | 0.01 |
| Arachidonic acid metabolism | 0.01 |
| Carbohydrate digestion and absorption | 0.01 |
| Estrogen signaling pathway | 0.01 |
| Legionellosis | 0.01 |
| Measles | 0.02 |
| Valine, leucine and isoleucine degradation | 0.02 |
| Pathways in cancer | 0.02 |
| Vitamin digestion and absorption | 0.03 |
Transcriptomic changesinduced by curcumin
To characterize the effects of curcumin in ALDH+ cells, we compared the transcriptional profiles of the sorted ALDH+ cells cultured for 24 hours with curcumin or DMSO. Unlike when comparing the expression profiles of the ALDH+ and ALDH−/CD44+/CD24− cells, there was no clear separation by treatment using MDS for 2 of the 3 samples tested (Supplemental Figure 1A). One hundred and ninety genes were differentially expressed with curcumin, with 97 genes upregulated and 93 genes downregulated (Figure 3A; Supplemental Table 2). The most significantly upregulated genes with curcumin treatment included HMOX1, SRXN1, HSPA1A, HSPA7, HSPA6, HSPA1B, and UBB, while KRT15, KRT6A, and SCD were downregulated.).
Figure 3.
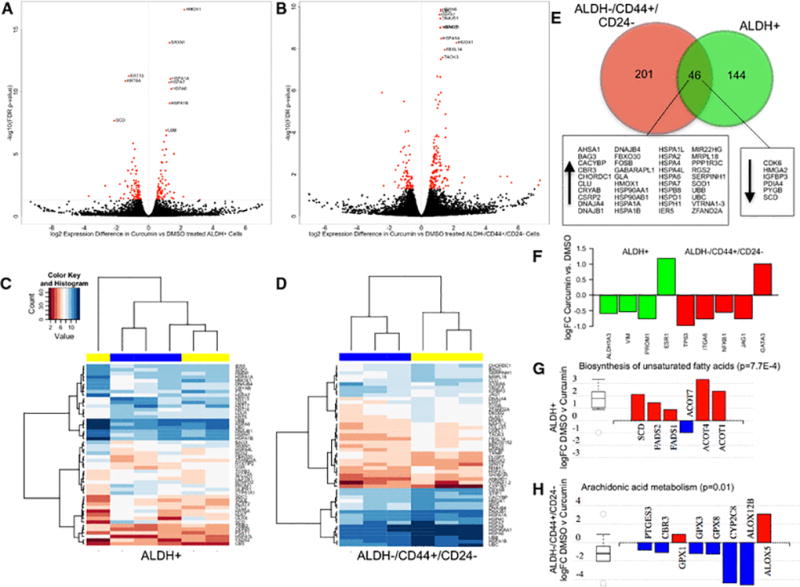
Genome-wide expression differences induced by curcumin treatment in ALDH+ and ALDH−/CD44+/CD24− breast cells. (A) and (B) FDR volcano plots of the log(2) ratio of gene expression between the 5 µM curcumin and DMSO treated ALDH+ and ALDH+/CD44+/CD24− cells. (C) and (D) Unsupervised hierarchical clustering of the log-transformed counts per million of the top 50 most differentially expressed genes in the curcumin (yellow) vs. DMSO (blue) treated ALDH+ and ALDH−/CD44+/CD24− cells. (E) Genes identified as commonly differentially expressed in both cellular fractions with curcumin treatment. (F) Expression differences of breast stem cell and differentiation related genes with curcumin treatment. (G) Expression differences in DMSO vs. curcumin treated ALDH+ cells for genes involved in biosynthesis of unsaturated fatty acids, the most significantly enriched pathway. (H) Expression differences in DMSO vs. curcumin treated ALDH−/CD44+/CD24− cells for genes involved in arachidonic acid metabolism.
In ALDH−/CD44+/CD24− cells, curcumin treatment induced a similar pattern of change as in the ALDH+ cells, with 2 of the 3 samples having no clear separation when visualized utilizing MDS (Supplemental Figure 2A). In contrast to ALDH+ cells, approximately two times more genes were upregulated with curcumin (164) than downregulated (83) in ALDH−/CD44+/CD24− cells (Figure 3B; Supplemental Table 3). The most upregulated genes included HSPA6, UBC, HSPA7, DNAJB1, HSPA1B, BAG3, HSPA1A, HMOX1, FBXL14, and TAOK3. Unsupervised hierarchical clustering of the curcumin and DMSO treated ALDH+ and ALDH−/CD44+/CD24− cells based on the top 50 most differentially expressed genes identified a clear separation in the ALDH−/CD44+/CD24− cells (Figure 3D), but not in the ALDH+ cells (Figure 3C).
Between the curcumin and DMSO treated ALDH+ and ALDH−/CD44+/CD24− cells, 46 genes were differentially expressed in both cell populations (Figure 3E). All of these genes were differentially expressed in the same direction, and 40 of these genes were upregulated. We validated the overexpression of HMOX1 and downregulation of SCD following 24 hour curcumin treatment in cells from 3 additional normal breast reductions (Supplemental Table 4) Genes previously associated with breast “stemness” were also downregulated in the curcumin treated cells (Figure 3F). In the ALDH+ cell fraction, ALDH1A3, VIM, and PROM1 were downregulated with curcumin, while TP63, ITGA6 (CD49f), NFKB1, and JAG1 were downregulated in the ALDH−/CD44+/CD24− cells. Differentiation markers were also upregulated with curcumin in both cellular fractions: \ESR1 in the ALDH+ cells and the luminal transcriptional factor GATA3 up in the ALDH−/CD44+/CD24− cells, although these changes did not reach genome-wide statistical significance (p=5.4E-3,FDR=0.16 and p=6.0E-3; FDR=0.15, respectively).
In ALDH+ cells, pathway enrichment analysis revealed that curcumin treatment changed expression of genes involved in steroid biosynthesis, PPAR signaling, and protein processing (Table 1B). The pathway most enriched for differentially expressed genes with curcumin treatment in ALDH+ cells was biosynthesis of unsaturated fatty acids, with downregulation of fatty acid desaturases including SCD, FADS2, and FADS1 (Figure 3G). In ALDH−/CD44+/CD24− cells, we also observed an enrichment in genes involved in arachidonic acid metabolism, including downregulation of PTGES3 and ALOX12B (Figure 3H). Additionally, biological pathways involved in antigen processing, transcriptional misregulation, and estrogen signaling were also enriched in ALDH−/CD44+/CD24− cells (Table 1C).
Combined effects of curcumin and piperine in breast stem/progenitor cells
Despite a reduction in primary mammosphere formation observed in normal breast cells following 5 µM piperine treatment (Figure 1D), no genes were identified as differentially expressed = in the piperine treated ALDH+ or ALDH-CD44+CD24- cells. 4
Treatment of the ALDH+ and ALDH−/CD44+/CD24− cells with curcumin and piperine in tandem revealed many of the same genes to be differentially expressed as with curcumin only treatment (Supplemental Tables 5 and 6). Many of the same biological pathways were also enriched in curcumin and piperine co-treated vs. DMSO cells (data not shown). We compared expression in curcumin and piperine treated ALDH+ or ALDH-CD44+CD24- cells compared to curcumin alone. Differentially expressed genes in curcumin and piperine cotreated ALDH+ cells (Supplemental Table 7), included the ribosomal protein RPL26, ubquitin B (UBB), and the calcium homeostasis regulator CALHM2. In ALDH−/CD44+/CD24− cells, all of the differentially expressed genes in theco-treated cells compared to curcumin only were involved in heat shock response (Supplemental Table 7).
Effects of stearoyl coA desaturase-1 (SCD) knockdown on stem cell phenotypes
Genes involved in lipid metabolism were identified as differentially expressed with curcumin treatment in both cellular fractions (Figure 3 G&H). One key gene involved in metabolism of saturated fatty acids, Steroyl-CoA desaturase-1 (SCD), was downregulated with curcumin treatment and curcumin and piperine treatment in both stem cell fractions (Figure 3E, Supplemental Tables 4 and 5), and was one of the most differentially expressed genes in the curcumin treated ALDH+ cells (Figure 3A). As SCD was one of only a few genes to be downregulated in both cell fractions with curcumin treatment, and was associated with the most differentially expressed pathway in ALDH+ cells, we hypothesized that SCD may play a role in the regulation of breast stemness. To test this hypothesis, we first knocked down SCD expression in MCF10A non-tumorigenic breast cells. SCD siRNA concentrations of 25nM and 50nM were found to significantly decrease SCD gene expression after 24 hours treatment (Figure 4A), and SCD protein expression at 48 and 96 hours, with a return to homeostasis by 7 days (Figure 4B; Supplemental Figure 3). SCD knockdown had no effect on cell proliferation at 24 hours, but significantly decreased cell proliferation at 48 hours (Supplemental Figure 2). SCD knockdown for 24 hours followed by plating in anchorage independent conditions for 10 days lead to a decrease in primary mammospheres, a qualitative decrease in overall mammosphere size, and a decrease in secondary sphere formation (Figure 4C). SCD knockdown also caused a decrease in the proportion of CD44+/CD24− MCF10A cells following 72 hours treatment (Figure 4D).
Figure 4.
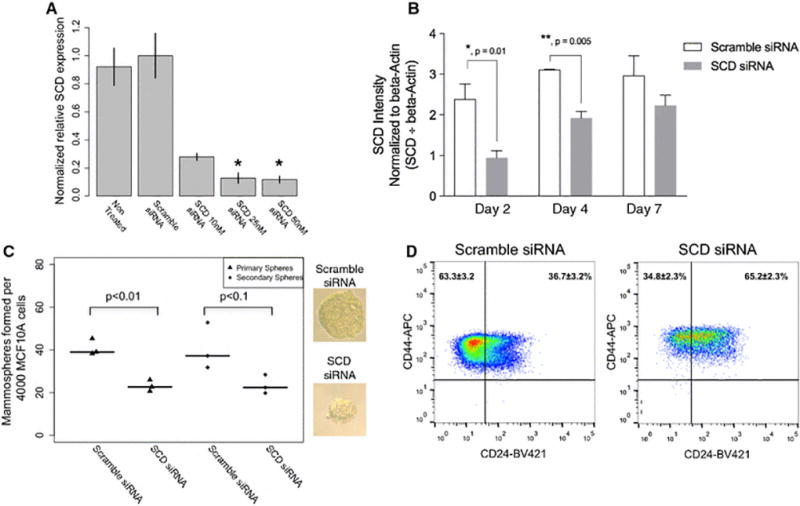
Effects of SCD knockdown on stem cell phenotypes. (A) Effects of SCD siRNA on SCD RNA expression (normalized to GAPD and ACTB) in MCF10A cells. (B) SCD protein expression at 2, 4, and 7 days, including 24 hours of treatment with SCD or Scramble siRNA. (C) Effects of SCD siRNA treatment on primary and secondary MCF10A mammosphere number and primary sphere size. (D) Effects of treatment of MCF10A cells with SCD siRNA on proportions of CD44+/CD24− cells, compared to scrambled siRNA control.
Effects of palmitoleic acid and curcumin treatment on mammosphere formation
To better understand what proportion of the effects of curcumin on mammosphere formation are due to downregulation of SCD, we co-treated normal breast epithelial cells from three individuals with both curcumin and palmitoleic acid, the major monosaturated fatty acid product of SCD.. Cotreatment with palmitoleic acid significantly reduced the inhibitory effects of 5 µM curcumin treatment (mean proportion of sphere formation relative to vehicle control: 61% vs 84%) (Figure 5A). A similar trend was observed for the 10 µM curcumin and palmitoleic acid cotreatment (mean proportion of sphere formation relative to vehicle control: 33% vs 49%), although the results did not reach statistical significance. There was no significant difference observed in secondary mammosphere formation (Figure 5B). Treatment with palmitoleic acid alone increased the number of primary (mean increase: 75%, p<0.05) and secondary (mean increase: 45%, p=0.1) mammospheres formed compared to vehicle control.
Figure 5.
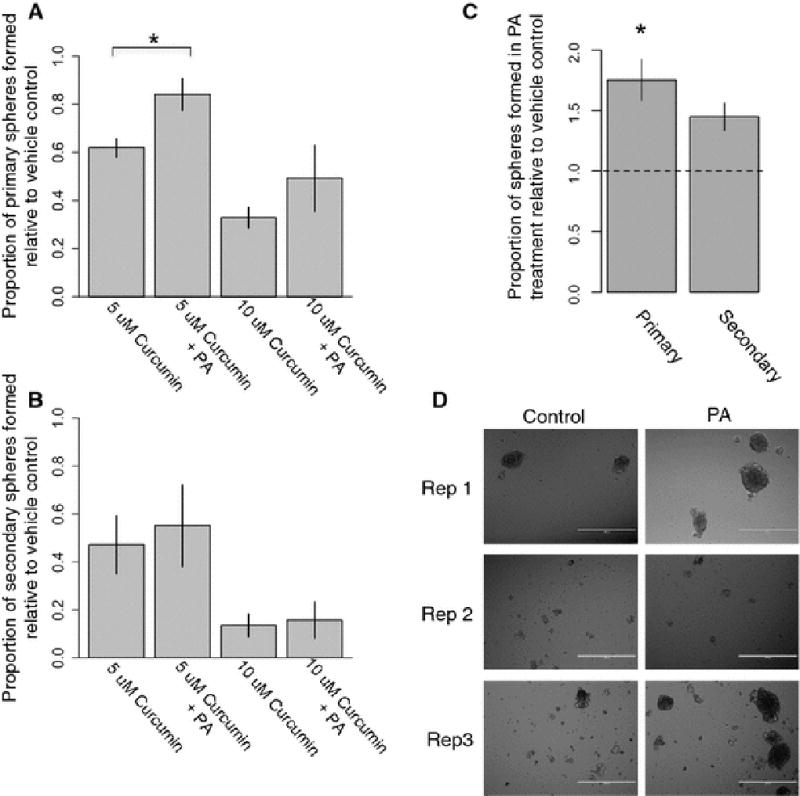
Effects of palmitoleic acid supplementation on normal breast cells (A) Effects of curcumin and palmitoleic acid (PA) co-treatment on primary mammosphere formation of primary breast epithelial cells (n=3 individuals). (B) Effects of curcumin and PA treatment on secondary mammosphere formation. (C) Primary and secondary sphere formation rates following treatment with PA alone, relative to control. (D) Representative images of secondary sphere size, comparing PA treatment to vehicle control.
DISCUSSION
Curcumin has shown promise as a cancer preventive compound in preclinical models, particularly when paired with piperine as an adjuvant to increase bioavailability. Here, we confirm and extend previous findings of curcumin targeting self-renewal of both normal and cancer stem cell populations. We report, for the first time, genome-wide expression profiles of FACS-isolated ALDH+ and ALDH-CD44+CD24- normal breast cells treated with curcumin and piperine. These results highlight the efficacy of pairing flow cytometry sorting of normal stem and progenitor cell populations with RNA-sequencing methods to precisely define mechanisms of action of cancer preventive compounds in relevant cell populations.
Mounting evidence from the past decade shows that breast tumors likely arise from, and are sustained by, a population of stem-like cells that harbor dysregulated self-renewal capacity [11]. Cancer stem cells exist in two distinct phenotypic states, an epithelial-like, proliferative state, and a mesenchymal-like state that is quiescent and invasive, with cancer stem cells being able to interconvert between the two states [13]. These results are consistent with findings from lineage tracing studies in the mouse mammary gland, where distinct stem cell populations that are typically lineage restricted to generating either a luminal or basal progeny [12]can recapitulate a full mammary gland when transplanted into a cleared fat pad [22]. Here, we show that the widely used breast stem cell markers ALDH+ and CD44+/CD24− identify distinct epithelial-like and mesenchymal-like cell populations, respectively, in the normal breast.
The findings that normal breast stem and breast cancer stem cells exist in multiple, interconvertable states has important implications for breast cancer chemoprevention. In breast cancer, ALDH+ cells typically localize in the tumor interior, while CD44+/CD24− cells localize at the invasive edge of the tumor [13]. Thus, ALDH+ cells may be responsible for maintaining the growth of the bulk of the tumor, while CD44+/CD24− cells are responsible for invasion and metastasis. With respect to cancer prevention efforts, recent pathologic evidence suggests that population expansion of ALDH+ cells may represent an important early step in carcinogenesis. In histologically normal tissue isolated from BRCA1/BRCA2 mutation carriers, or women with a family history of breast cancer, increased numbers of ALDH positive cells were observed in the breast ductules compared to control patients [23]. A comparison of benign breast biopsy tissues isolated from women who went on to develop breast cancer, or not, found increased ALDH1 staining in both epithelial and stromal breast cells isolated from women who later went on to develop breast cancer [24]. Additionally, ALDH1A1 tumor staining was strongly associated with early recurrence and metastasis of breast cancer, regardless of ER, PR, or HER2 status [25]. CD44+/CD24− breast cancer cells were originally identified as the cells that contained the tumor initiating fraction in breast cancers [11]. Further work identified that these cells have a basal phenotype and increased invasive capacity in tumors [26], and that these cells can be generated through an EMT in immortalized human mammary epithelial cells [27]. The most efficacious cancer preventive compounds, therefore, should target both of these stem cell phenotypes, particularly if cells with each phenotype can interconvert. Here, we showed that curcumin downregulated genes associated with breast “stemness”, including ALDH1A3 and PROM1 (CD133) in ALDH+ cells, and TP63 and ITGA6 (CD49f) in ALDH−/CD44+/CD24− cells. These results, paired with the functional results here and previously [9] show that curcumin downregulates genes and pathways relevant to stem cells.
In addition to downregulating genes involved in stem cell self-renewal, 24-hour curcumin treatment induced significant expression changes in a large number of genes and pathways in both normal ALDH+ and ALDH−/CD44+/CD24− breast cells that have also been previously reported in other cell types. Induction of HMOX1 by curcumin treatment has been previously identified in a number of model systems [28–30]. We observed HMOX1 upregulation in both stem cell fractions following curcumin treatment. Pathways involved in unfolded protein response, ligase activity, and response to heat were upregulated following curcumin treatment, corroborating previous findings of curcumin activating a heat shock and proteasome response [31, 32]. Interestingly, cancer stem cells have been shown to be more sensitive to inhibition of heat shock proteins than bulk cancer cells. Targeting HSP90 with low concentrations of the inhibitory drug 17-AAG significantly inhibited lymphoma stem cells [33]. Treatment of breast cancer cells with both HSP90 and HSP27 inhibitors lead to a significant reduction in the stem- like cell population [34]. Here, we observe an increase in expression of heat shock genes in concert with a decrease in expression of genes involved in breast stemness. Further research is required to characterize the role of heat shock proteins and maintenance of stem cell state in the normal breast.
Curcumin treatment also modified Notch signaling in ALDH−/CD44+/CD24− cells, potentially through downregulation of JAG1, which has previously been reported in curcumin treated esophageal cancer cell lines [35]. Curcumin decreased the CD44+/CD24− fraction of BT- 549 cells, in addition to decreasing the number of tubulin microtentacles, which are involved in the reattachment of suspended cells [36]. Microtentacle formation has been shown to be dependent on vimentin and tubulin [37], both of which we identified as downregulated in ALDH+ curcumin treated cells, suggesting these processes may also be important in inhibition of mammosphere formation in normal breast cells.
Surprisingly, despite finding that piperine treatment significantly inhibited mammosphere formation in both breast cancer cells and normal breast cells, corroborating previous reports [9], we did not identify any differentially expressed genes with piperine treatment at 24 hours. Since the mammosphere formation assay measures the treatment effects over the course of days, it is possible that the 24-hour window was not appropriate to observe the effects of piperine. A future time course experiment would help to shed light on piperine’s effects on breast stem cells. The effects of piperine on stem cell self-renewal, and its potentiating effects on curcumin treatment, are thought to be mediatedhrough inhibition of drug transporters and metabolizing enzymes [38]. Intriguingly, piperine decreased cholesterol uptake in Caco-2 cells by internalizing cholesterol transport proteins without changing overall cellular expression of these proteins [39]. Thus the effects of piperine on stemness may lie in the inhibition and modification of the localization of drug and lipid transporters.
We identified a number of genes and pathways involved in lipid metabolism differentially expressed in normal breast stem cells following curcumin treatment. Our finding of curcumin modulating arachidonic acid metabolism in ALDH−/CD44+/CD24− cells supports a growing body of literature identifying the effects of curcumin on PGE2 regulation [40, 41]. Additionally, in both stem cell fractions, curcumin, and curcumin plus piperine, treatment downregulated expression of SCD, an enzyme involved in the synthesis of monounsaturated fatty acids from saturated fatty acids. To the best of our knowledge, only two other studies have reported effects of curcumin on SCD expression in mammalian cells or animals. Human multiple myeloma cells treated with curcumin and two different curcumin analogs had significantly reduced expression of SCD [42]. In apo E−/− mice, curcumin treatment significantly attenuated increased expression of scd-1 due to western diet [43]. SCD is implicated in carcinogenesis, as fatty acid synthesis is essential for plasma membrane formation and glycolysis regulation [44]. Relevant to cancer stem cell function, SCD is required for lung cancer initiating cell spheroid production [45]. Downregulation of SCD inhibits of beta-catenin and Wnt signaling in MCF7 and MDA-MB231 cells [46]. SCD is required for Wnt protein biogenesis by synthesizing palmitoleic acid, the monosaturated fatty acid substrate that is incorporated onto Wnt3a and Wnt5a protein and mediates their proper trafficking in the cell [47]. Through our unbiased approach, we identified SCD as a novel target of curcumin in breast stem cells and confirmed a role for SCD in breast stem cell regulation by siRNA knockdown experiments in MCF10A cells. Palmitoleic acid treatment increased primary and secondary mammosphere formation in normal breast cells, providing further evidence for SCD activity regulating breast stemness. We further confirmed a role for SCD downregulation in curcumin’s effects on mammosphere formation by co-treating primary breast cells grown in mammosphere conditions with both curcumin and palmitoleic acid. Palmitoleic acid supplementation significantly reduced the inhibition of mammosphere formation caused by 5µM curcumin treatment. Interestingly, however, palmitoleic acid did not completely rescue the effects of curcumin, likely reflecting curcumin’s pleiotropic effects and its modulation of multiple pathways regulating stem cells. Curcumin treatment was recently shown to decrease the palmitoylation of integrin β4 in breast cancer cells, providing additional indirect evidence for curcumin’s influence on this pathway [48]. Our findings support a growing body of literature pointing to a role for SCD in the maintenance of normal and cancer stem cell populations, further highlighting this enzyme as an attractive target for breast cancer chemoprevention and treatment efforts using compounds such as curcumin.
4.6 Conclusion
The results from this study extend our previous findings of the effects of curcumin and piperine on normal and cancer stem cells using an experimental paradigm that combines FACS sorting of normal human breast cells into stem-enriched fractions, treatment in vitro, and RNA-seq. The use of primary tissues provides novel information about stem cell regulation in the normal human breast that may not be available from studies utilizing cell lines or model animals. This experimental methodology can thus be applied to understand the effects of carcinogens or cancer preventive compounds in specific cell populations. Here, we utilized these methods to identify novel mechanisms by which curcumin inhibits stem cells, providing biomarkers of efficacy of curcumin treatment for future breast cancer prevention clinical trials and molecular targets for cancer prevention and treatment efforts.
Supplementary Material
Acknowledgments
Support for this study was provided by a grant from the National Cancer Institute (R03 CA167700). Support for JAC was provided by a Rackham Predoctoral Fellowship from the University of Michigan and Institutional Training Grants from the National Institute of Environmental Health Sciences (NIEHS) (T32 ES007062) and the National Human Genome Research Institute (NHGRI) (T32 HG00040).
Footnotes
Conflict of Interest
The authors have no conflicts of interest to declare.
Literature Cited
- Jemal A, et al. Cancer statistics 2010. CA Cancer J Clin. 2010;60(5):277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- Brandberg Y, et al. Psychological reactions, quality of life, and body image after bilateral prophylactic mastectomy in women at high risk for breast cancer: a prospective 1-year follow-up study. J Clin Oncol. 2008;26(24):3943–9. doi: 10.1200/JCO.2007.13.9568. [DOI] [PubMed] [Google Scholar]
- Fisher B, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90(18):1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- Vogel VG, et al. Update of the National Surgical Adjuvant Breast and Bowel Project Study of Tamoxifen and Raloxifene (STAR) P-2 Trial: Preventing breast cancer. Cancer Prev Res (Phila) 2010;3(6):696–706. doi: 10.1158/1940-6207.CAPR-10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A. The endocrine prevention of breast cancer. Best Pract Res Clin Endocrinol Metab. 2008;22(4):615–23. doi: 10.1016/j.beem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Noorafshan A, Ashkani-Esfahani S. A review of therapeutic effects of curcumin. Curr Pharm Des. 2013;19(11):2032–46. [PubMed] [Google Scholar]
- Shoba G, et al. Influence of piperine on the pharmacokinetics of curcumin in animals and human volunteers. Planta Med. 1998;64(4):353–6. doi: 10.1055/s-2006-957450. [DOI] [PubMed] [Google Scholar]
- Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarala M, et al. Targeting breast stem cells with the cancer preventive compounds curcumin and piperine. Breast Cancer Res Treat. 2010;122(3):777–85. doi: 10.1007/s10549-009-0612-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux G, et al. BRCA1 Basal-like Breast Cancers Originate from Luminal Epithelial Progenitors and Not from Basal Stem Cells. Cell Stem Cell. 2010;7(3):403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479(7372):189–93. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Liu S, et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of their Normal Counterparts. Stem Cell Reports. 2013;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995:289–300. [Google Scholar]
- Lim E, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- Svedberg J, et al. Free-fatty acid inhibition of insulin binding, degradation, and action in isolated rat hepatocytes. Diabetes. 1990;39(5):570–4. doi: 10.2337/diab.39.5.570. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- R Core Team, R. A language and environment for statistical computing. R Foundation for Statistical Computing: Vienna, Austria: 2013. [Google Scholar]
- Eirew P, et al. Aldehyde Dehydrogenase Activity Is a Biomarker of Primitive Normal Human Mammary Luminal Cells. STEM CELLS. 2012;30(2):344–348. doi: 10.1002/stem.1001. [DOI] [PubMed] [Google Scholar]
- Keller PJ, et al. Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci U S A. 2012;109(8):2772–7. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isfoss BL, et al. Women with familial risk for breast cancer have an increased frequency of aldehyde dehydrogenase expressing cells in breast ductules. BMC Clin Pathol. 2013;13(1):28. doi: 10.1186/1472-6890-13-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunju LP, et al. EZH2 and ALDH-1 mark breast epithelium at risk for breast cancer development. Mod Pathol. 2011;24(6):786–93. doi: 10.1038/modpathol.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, et al. Expression of ALDH1 in breast invasive ductal carcinoma: an independent predictor of early tumor relapse. Cancer Cell Int. 2013;13(1):60. doi: 10.1186/1475-2867-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan C, et al. CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8(5):R59. doi: 10.1186/bcr1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, et al. Curcumin upregulates transcription factor Nrf2, HO-1 expression and protects rat brains against focal ischemia. Brain Res. 2009;1282:133–41. doi: 10.1016/j.brainres.2009.05.009. [DOI] [PubMed] [Google Scholar]
- McNally SJ, et al. Curcumin induces heme oxygenase 1 through generation of reactive oxygen species, p38 activation and phosphatase inhibition. Int J Mol Med. 2007;19(1):165–72. [PubMed] [Google Scholar]
- Motterlini R, et al. Curcumin, an antioxidant and anti-inflammatory agent, induces heme oxygenase-1 and protects endothelial cells against oxidative stress. Free Radic Biol Med. 2000;28(8):1303–12. doi: 10.1016/s0891-5849(00)00294-x. [DOI] [PubMed] [Google Scholar]
- Shen SQ, et al. Protective effect of curcumin against liver warm ischemia/reperfusion injury in rat model is associated with regulation of heat shock protein and antioxidant enzymes. World J Gastroenterol. 2007;13(13):1953–61. doi: 10.3748/wjg.v13.i13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsmore KE, Chen PG, Wong HR. Curcumin, a medicinal herbal compound capable of inducing the heat shock response. Crit Care Med. 2001;29(11):2199–204. doi: 10.1097/00003246-200111000-00024. [DOI] [PubMed] [Google Scholar]
- Newman B, et al. HSP90 inhibitor 17-AAG selectively eradicates lymphoma stem cells. Cancer Res. 2012;72(17):4551–61. doi: 10.1158/0008-5472.CAN-11-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, et al. Inhibition of heat shock protein (Hsp) 27 potentiates the suppressive effect of Hsp90 inhibitors in targeting breast cancer stem-like cells. Biochimie. 2012;94(6):1382–9. doi: 10.1016/j.biochi.2012.02.034. [DOI] [PubMed] [Google Scholar]
- Subramaniam D, et al. Curcumin induces cell death in esophageal cancer cells through modulating Notch signaling. PLoS One. 2012;7(2):e30590. doi: 10.1371/journal.pone.0030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier MS, et al. Curcumin targets breast cancer stem-like cells with microtentacles that persist in mammospheres and promote reattachment. Cancer Res. 2014;74(4):1250–60. doi: 10.1158/0008-5472.CAN-13-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple RA, et al. Vimentin filaments support extension of tubulin-based microtentacles in detached breast tumor cells. Cancer Res. 2008;68(14):5678–88. doi: 10.1158/0008-5472.CAN-07-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj RK, et al. Piperine, a Major Constituent of Black Pepper, Inhibits Human P-glycoprotein and CYP3A4. Journal of Pharmacology and Experimental Therapeutics. 2002;302(2):645–650. doi: 10.1124/jpet.102.034728. [DOI] [PubMed] [Google Scholar]
- Duangjai A, et al. Black pepper and piperine reduce cholesterol uptake and enhance translocation of cholesterol transporter proteins. Journal of Natural Medicines. 2013;67(2):303–310. doi: 10.1007/s11418-012-0682-7. [DOI] [PubMed] [Google Scholar]
- Hong J, et al. Modulation of arachidonic acid metabolism by curcumin and related β-diketone derivatives: effects on cytosolic phospholipase A2, cyclooxygenases and 5-lipoxygenase. Carcinogenesis. 2004;25(9):1671–1679. doi: 10.1093/carcin/bgh165. [DOI] [PubMed] [Google Scholar]
- Lev-Ari S, et al. Down-regulation of prostaglandin E2 by curcumin is correlated with inhibition of cell growth and induction of apoptosis in human colon carcinoma cell lines. J Soc Integr Oncol. 2006;4(1):21–6. [PubMed] [Google Scholar]
- Kudo C, et al. Novel curcumin analogs, GO-Y030 and GO-Y078, are multi-targeted agents with enhanced abilities for multiple myeloma. Anticancer Res. 2011;31(11):3719–26. [PubMed] [Google Scholar]
- Shin HS, et al. Anti-atherosclerosis and hyperlipidemia effects of herbal mixture, Artemisia iwayomogi Kitamura and Curcuma longa Linne, in apolipoprotein E-deficient mice. J Ethnopharmacol. 2014;153(1):142–50. doi: 10.1016/j.jep.2014.01.039. [DOI] [PubMed] [Google Scholar]
- Igal RA. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31(9):1509–15. doi: 10.1093/carcin/bgq131. [DOI] [PubMed] [Google Scholar]
- Noto A, et al. Stearoyl-CoA desaturase-1 is a key factor for lung cancer-initiating cells. Cell Death Dis. 2013;4:e947. doi: 10.1038/cddis.2013.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvoisin D, et al. Decreasing stearoyl-CoA desaturase-1 expression inhibits beta-catenin signaling in breast cancer cells. Cancer Sci. 2013;104(1):36–42. doi: 10.1111/cas.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rios-Esteves J, Resh Marilyn D. Stearoyl CoA Desaturase Is Required to Produce Active, Lipid-Modified Wnt Proteins. Cell Reports. 4(6):1072–1081. doi: 10.1016/j.celrep.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman DT, et al. Curcumin Prevents Palmitoylation of Integrin beta4 in Breast Cancer Cells. PLoS One. 2015;10(5):e0125399. doi: 10.1371/journal.pone.0125399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


