Abstract
Single-cell microscopy is a powerful tool for studying gene functions using strain libraries, but suffers from throughput limitations. Here we describe the Strain Library Imaging Protocol (SLIP), a high-throughput, automated microscopy workflow for large strain collections with minimal user involvement. SLIP involves transferring arrayed bacterial cultures from multi-well plates onto large agar pads using inexpensive replicator pins and automatically imaging the resulting single cells. The acquired images are subsequently reviewed and analyzed by custom MATLAB scripts that segment single-cell contours and extract quantitative metrics. SLIP yields rich datasets on cell morphology and gene expression, which illustrate the function of certain genes and the connections among strains in a library. For a library arrayed on 96-well plates, image acquisition can be completed within 4 min per plate.
Keywords: single-cell microscopy, high-throughput, multi-well format, automated microscopy
INTRODUCTION
Cell morphogenesis is directly coupled to environmental perturbations and is highly and quantitatively dependent on gene functions1,2. For example, a single mutation in the Escherichia coli actin homolog MreB can cause a ~50% increase in cell width, which contributes to a growth advantage when cells exit stationary phase in certain media2. Cell morphology across a population can be highly heterogeneous, especially under stressed conditions such as antibiotic treatment3 or long-term nutrient depletion4. Such shape heterogeneity, which can contribute to behaviors such as antibiotic persistence5, is masked in population-level measurements and thus motivates single-cell observations. Taken together, quantitative single-cell analyses of bacterial populations across chemical, environmental, and genetic perturbations hold the potential to reveal core aspects of bacterial physiology.
Advances in biotechnology have enabled the creation of large genetic libraries of bacteria, such as single nonessential gene-deletion libraries in E. coli6 and Salmonella enterica7, deep mutational scanning of genes of interest8 in a variety of organisms, and a CRISPR interference (CRISPRi)-based essential gene knockdown library in Bacillus subtilis9. Population-level screening of these libraries has generated useful insights into gene functions related to environmental changes7,9,10. These libraries contain hundreds to thousands of strains, and thus screens are typically performed in 96-, 384-, or 1536-well format in multi-well liquid media or on rectangular agar plates. There are many established instruments available for handling arrayed strain libraries, such as the ROTOR robot (Singer Instrument Company), which performs high-throughput sample transfer, picking, and arraying in multi-well formats for both liquid cultures and colonies on solid agar surfaces10.
Despite the extensive potential for knowledge generation via morphological investigations of strain libraries, single-cell microscopy currently still largely relies on manual, strain-by-strain investigations, which constitutes a strong bottleneck for the acquisition of datasets across large strain libraries. This reliance on manual labor limits not only the number of strains that can be characterized at the single-cell level but also the number of environmental conditions that are amenable to screening at the single-cell level. The development of automated control over microscope stage movements, autofocusing, and digital cameras have increased the speed of and reduced the human effort for image acquisition11,12. μManager, an open-source software package that integrates the control of microscopes, stage controllers, cameras, and light sources, is widely used for automated image acquisition12, providing the opportunity to further increase throughput. A recent study screened the E. coli ASKA collection of fluorescent-protein fusions13 on large-format agarose pads, acquiring images for 48 strains in 6–8 min14. However, manual selection of imaging positions was still required before acquisition began, which becomes a rate-limiting step with larger libraries.
Here we present the Strain Library Imaging Protocol (SLIP), a pipeline that empowers single-cell imaging of large strain libraries under a variety of conditions. This protocol involves transferring liquid cultures from multi-well plates onto agar pads in batch. User specification of the plate layout (96- or 384-well plate) and the location of two strains for calibration then permit automatic acquisition of multiple images for each strain. Output images can be subsequently analyzed using custom MATLAB scripts. This protocol allows for the rapid imaging of large strain libraries (4 min per 96-well plate, 9 min per 384-well plate for one image per well) with minimal human involvement, which reduces human error and eliminates biases.
SLIP makes use of large agar pads (Fig. 1a, b) as substrates for sample preparation (although it can also be used with glass slides of any size). The pads are the same size as multi-well plates, allowing liquid cultures to be easily transferred onto agar pads via inexpensive 96- or 384-pin replicators in batch (Fig. 1c, d). The pads are then covered with large-format coverslips for imaging (Fig. 1e, f). With this setup, we have successfully performed high-throughput phase-contrast microscopy with species such as B. subtilis9, E. coli, and S. enterica. We have also grown CRISPRi-induced B. subtilis cells overnight on these agar pads and imaged terminal morphology phenotypes after depletion of essential proteins9. Wild-type B. subtilis cells grew into homogeneous lawns on large agar pads, reflecting the pads’ suitability as a growth substrate9. Strains depleted of essential proteins exhibited rich and diverse terminal phenotypes, allowing us to characterize the phenotypes of cell growth, morphology, and lysis at the single-cell level9; such information is difficult to extract from batch cultures. Since agar is a common reagent for growing microbes on solid surfaces, SLIP should be adaptable to the vast majority of other culturable microbial species.
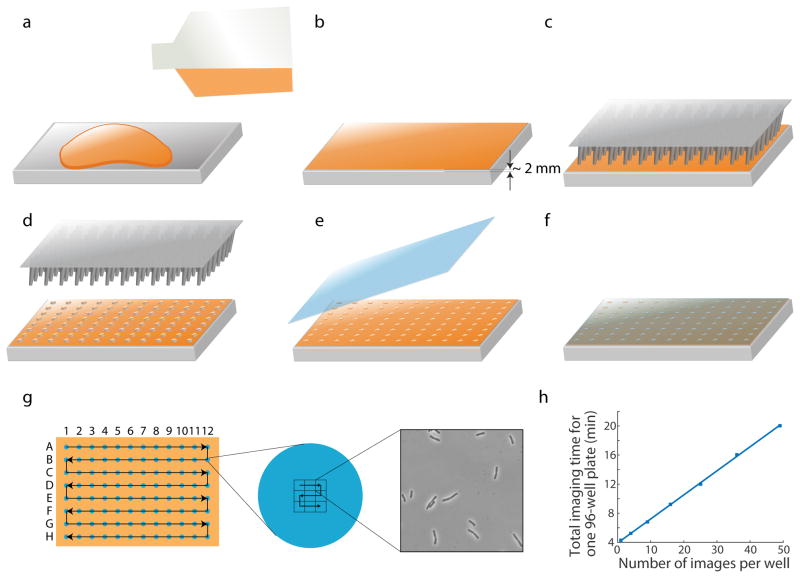
Figure 1. Schematic of pad preparation and imaging.
(a) Melted agar is poured onto the bottom surface of a Singer PlusPlate. (b) Melted agar is spread evenly by gently tilting and shaking the plate. A flat agar surface is generally achieved when the plate is left on the benchtop and solidifies without disturbance. The agar layer is ~2 mm in thickness. (c, d) Bacteria cultures are transferred onto an agar pad using a 96-well replicator pin. (e, f) After the agar pad absorbs all the liquid (~5–10 min depending on temperature), a large glass cover slip is used to cover the agar surface. (g) Schematic of stage movement during image acquisition for a 96-well plate. Left: to minimize travel distance, the stage moves across the wells in a zigzag manner, first from A1 to A12, then backward from B12 to B1, etc. Middle: for each strain, the stage moves across an imaging grid in a similar zigzag pattern. An example of a 3x3 grid is shown. The grid size is enlarged relative to the droplet size for illustration purposes. Right: a typical image acquired by SLIP. (h) For a 96-well plate, total image acquisition time scales linearly with number of images per strain, with a slope of 200 ms/strain/image or 0.33 min for each additional image across 96 strains.
As a pad-based protocol, SLIP has some intrinsic limitations for single-cell imaging. The large pad size may lead to oxygen depletion, especially for cells in the middle of the pad. Although we did not detect a position-dependent terminal phenotype in B. subtilis cells grown overnight on the pads9, we recommend testing the oxygen depletion effect when working with other species. Moreover, the pad-based approach makes it challenging to dynamically alter the chemical environment at will when the cells are on the pad. Several microfluidic devices allow image acquisition in concert with media switching15,16, but these devices may be difficult to scale up for large strain libraries.
Since strains on the pad are arrayed in 96- or 384-well format, after calibration SLIP can automatically locate strain positions based on standard metrics of 96- and 384-well plates, avoiding the manual definition of positions for every strain. SLIP calibrates plate position by taking as input the positions of two strains on a pad and calculating the relative translation and rotation for that pad. After calibration, SLIP automatically generates a list of coordinates for the centers of each strain. For each strain, SLIP assigns a grid of neighboring, non-overlapping imaging sites around the center. After the camera acquires an image at one imaging site, the stage moves to the next site for the next exposure. To minimize the stage travel distance, SLIP acquires images in a zigzag manner, moving from A1 to A12, then backward from B12 to B1, etc. (Fig. 1g). If multiple images are acquired within each strain, SLIP moves in the imaging grid in a similar zigzag pattern (Fig. 1g) to reduce acquisition time. Note that the image acquisition software does not necessarily require a large pad, it is compatible with a smaller number of strains arrayed in a grid with the same spacing as a multi-well plate. SLIP is also compatible with other types of samples that are arrayed in 96- or 384-well formats. Thus, with appropriate adaptations to sample preparation, SLIP can be applied to experiments involving intracellular bacteria, host-cell invasion, or tissue colonization and infection. Since the SLIP software communicates with the microscope hardware through μManager12, the extensive collection of hardware supported by μManager can be controlled by SLIP with little to no adaptations.
To rapidly acquire many images, it is desirable to minimize delays between camera exposure and stage movement. However, computers have limited ability to quickly and precisely control all connected hardware; many computer-controlled protocols involve delays. To overcome this challenge, SLIP exploits direct triggering between hardware using Transistor-Transistor Logic (TTL) signals, which eliminates delays and improves efficiency. Many microscope hardware components (e.g. cameras, light sources, and stage controllers) have inputs and outputs for TTL signals via BNC connectors. TTL signals generally have two states, inactive (low voltage) and active (high voltage), and the hardware can communicate through the changes of states. For instance, when the camera finishes an exposure, its TTL signal switches from active to inactive; this change signals the stage controller to start movement. Once stage movement is complete, the TTL state of the stage controller also switches, thus signaling back to the camera for another exposure at the new imaging site. Thusly, TTL triggering in SLIP reduces stage movement time between imaging sites by more than 80%; a 96-well plate can be imaged within 4 min, with one image acquired for each strain. Total acquisition time scaled linearly with the number of images per strain, with a total of 0.33 min required for each additional image across 96 strains (Fig. 1h). Acquisition of z-stacks can also be implemented with TTL signaling if the stage controller supports z-direction movement, allowing for similar speedups for 3D imaging.
Our laboratory previously developed a MATLAB package, Morphometrics, for analyzing phase images with sub-pixel precision17. This package is ideal for large-scale data analysis because it can run without a graphical user interface (GUI) and is amenable to parallelization. There are also other free programs that serve similar purposes. For example, Oufti18 and MicrobeJ19 are open-source software that segments and defines sub-pixel-resolved cell contours. Thus, it may be useful to test the effectiveness of various software packages for particular imaging datasets.
Experimental design
Strain library preliminary analysis and preparation
Before imaging, it is important to measure growth curves for each strain, especially to implement imaging of exponentially growing cells. Since lag times and growth rates can differ by strain, it is critical to identify a suitable back-dilution protocol to ensure that imaging captures most strains during exponential phase, that the cell densities are optimal (enough cells for the analysis of interest in one field of view without over-crowding that would disrupt cell segmentation), and that the cell densities are roughly consistent across strains. In our experiments with E. coli and B. subtilis, a 1:200 dilution of overnight cultures followed by 2 h of outgrowth at 37 °C was effective.
Fluorescence imaging
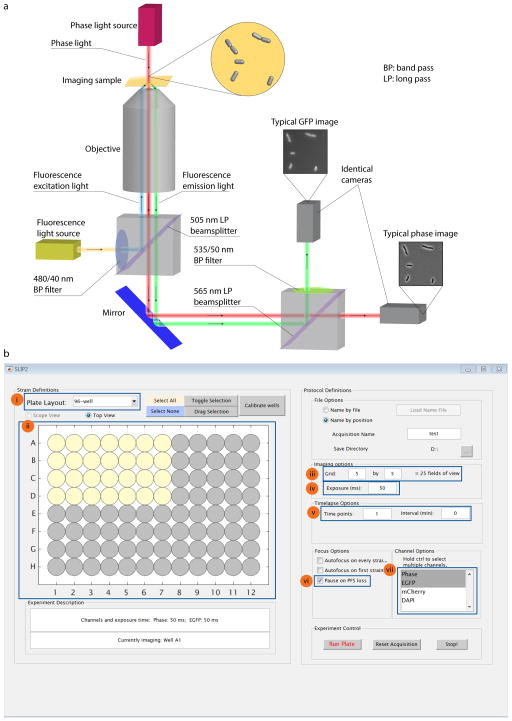
Due to SLIP’s utilization of the TTL triggering system, only one exposure can be achieved at each imaging site. However, dual-color camera setups allow the simultaneous capture of two images at different wavelengths using two identical cameras with only one exposure20. Briefly, the sample is simultaneously exposed to long-wavelength monochromatic phase light and fluorescence excitation light; the resulting light is passed through a long-pass beamsplitter to split the phase light and the fluorescence-emission light in orthogonal directions based on wavelengths. The phase light is detected by one camera, while the fluorescence-emission light passes a band-bass filter and is detected by another identical camera. Thus, with only one exposure, both phase and fluorescence images for a field of view are acquired (Fig. 2a). SLIP can be adapted in a relatively straightforward manner to include such a system.
Figure 2. Experimental setup.
(a) Configuration for dual-color cameras. (b) User interface of SLIP software. The key setup parameters are highlighted. (i) Plate layout selection: the user selects 96-, 384-, or 1536-well format. (ii) Illustration of the whole plate and the selected wells (yellow) for image acquisition. (iii) Imaging grid setting: for each well, SLIP generates a grid of non-overlapping neighboring imaging sites and acquires images at these sites. (iv) Exposure time setting. (v) Time-lapse settings. (vi) ‘Pause on PFS’ setting, which allows the user to manually correct any instance of loss of PFS. (vii) Channel setting: the highlighted channels are selected for image acquisition.
SLIP is also adaptable for fluorescence imaging in which phase and fluorescence signals are sequentially obtained without TTL triggering. Such acquisition is slower but provides more flexibility, and further allows multi-color fluorescence imaging.
Other bacterial species
A major component of SLIP is a user-friendly GUI (Fig. 2b) that facilitates a broad user base. Our laboratory has used SLIP for strain libraries of E. coli (Fig. 3a–c), B. subtilis9 (Fig. 3d), and S. enterica. We expect that with minimal adaptation, SLIP will be suitable for diverse microbial cells and environments. For instance, the agar pad can be prepared in an anaerobic chamber with proper sealing, allowing the screening of strictly anaerobic species. The analysis software Morphometrics17 has also been applied to a wide range of species with diverse morphologies21. The filtering algorithm to remove false-positive segmentations may require adaptation for different cell shapes; it is currently optimized for rod-shaped cells.
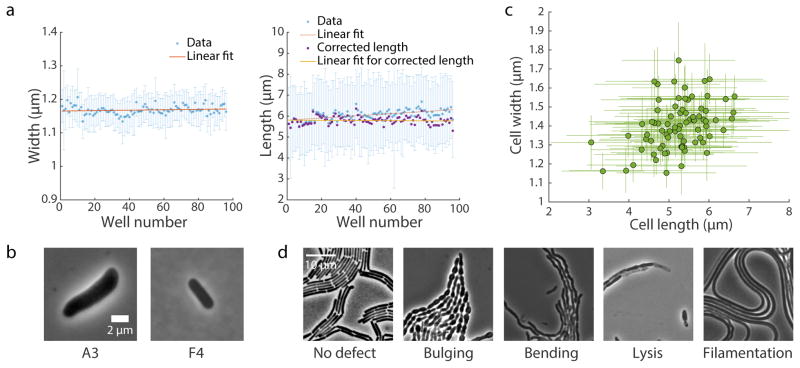
Figure 3. Typical results from SLIP.
(a) Changes in mean cell width (left) and length (right) during imaging of an E. coli MG1655 culture in exponential growth transferred to all 96 wells of a plate. Twenty-five images (5x5 grid) were acquired at each well to visualize enough cells for accurate quantification of cellular dimensions. Image acquisition required ~12 min. Due to growth on the pad, cell length increased by ~10% from the first well to the 96th, while cell width remained roughly constant. Blue data points are mean ± standard deviation. Red lines are the best linear fit of mean values. Purple data points are cell length corrected for the linear growth effect represented by the red line, and the linear fit to the corrected data is shown in yellow. For clarity, the error bars for purple data points are not shown. (b) Typical images from a SLIP screen of E. coli MreB-msfGFP mutants. Mutants with altered cell dimensions (larger or smaller) are easily identified. (c) Cellular dimensions of strains in an MreB-msfGFP mutant library quantified by the MATLAB software package Morphometrics17. Data points are mean ± standard deviation (n > 200 cells per strain). (d) Typical terminal phenotypes of strains in a B. subtilis CRISPRi essential gene knockdown library, where cells were grown on an agar pad with the CRISPRi inducer xylose overnight to deplete one essential protein in each strain. Wild-type cells showed no shape defect and grew into homogeneous lawns (left), whereas we visually identified a variety of morphological and/or growth defects by depleting essential proteins.
Controls
Our experiments revealed some potential sources of error. Here we list the main factors, which can be avoided by the means provided below (Table 1), or can be corrected through analysis of control experiments.
Defects in agar pads
Some agar pads have defects due to environmental disturbances during solidification or unmelted agar particles. These local defects may disrupt an infrared laser-based perfect focus system (PFS) and thus images taken at these positions can end up blurred and introduce errors in subsequent analyses. Agar defects can be minimized by careful preparation of agar pads and by inspecting each pad before use.
Growth phase and cellular physiology
Bacterial physiology is highly sensitive to the population density and growth phase. For instance, in an E. coli culture growing in LB, cell size decreases above OD600~0.3, which is only ~1/20 of the final OD reached in stationary phase22. Therefore, strains may display differences in phenotypes of interest due to being in different growth phases. Thus, it is recommended that users specifically test their strains for growth phase-dependent phenotypes when implementing SLIP.
Differences in inoculation cell density
As noted above, bacterial cells constantly modify their shape during growth, especially in rich media. We have shown that differences in the initial numbers of cells in a liquid culture can affect cellular dimensions at certain time points9. Therefore, to reduce variation due to inoculation, we recommend using large volumes of cell cultures for back dilution or performing serial dilutions.
Cell growth on agar pads
Although SLIP has been optimized to achieve much faster imaging speeds than manual operation, the time interval between imaging the first strain and the last strain for a 96-well plate is comparable to the doubling time of E. coli during rapid growth (~20 min). This duration permits some bacterial cells to grow a significant amount on an agar pad and to undergo changes in morphology; our experiments estimate this imaging lag results in a <10% difference in length between the 1st and 96th strain, while width remains essentially constant (Fig. 3a). Since the extent of growth scales linearly with the time interval between when a strain is placed on the pad and when it is imaged (Fig. 3a), the growth effect can be corrected based on a control experiment. Therefore, if precise quantification beyond the resolution prescribed by growth during the imaging interval is critical to subsequent analyses, it is recommended to perform a control experiment using SLIP with one or two strains and to characterize how the time that a strain spends on the pad affects its morphology. The results of the control experiment can be used to correct high-throughput measurements (Fig. 3a, c).
MATERIALS
REAGENTS
Granulated agar (BD Diagnostic Systems, BD cat. no. 214530)
#1.5 large glass cover slips (NEXTERION® Coverslip custom, #1.5; Glass: D263; Size (mm): 110 x 74)
Immersion oil (for Nikon Ti-E: Nikon immersion oil type NF, cat. no. MXA22024)
Growth medium (specific to species and condition of interest)
Singer PlusPlates (Singer Instrument Company, cat. no. PLU-001)
Singer RePads replicate pins (Singer Instrument Company, cat. no. REP-001 for 96-well plates, cat. no. REP-003 for 384-well plates)
500 mL Corning glass bottle (Corning Inc., cat. no. 1395-500)
Bacteria for imaging (e.g. E. coli MG1655 cells, CGSC #6300)
EQUIPMENT
Computer (for controlling microscope acquisition, installed with software necessary for controlling microscope stage, focus, shutters, and cameras)
Inverted fluorescence microscope and objective (e.g. Nikon Eclipse Ti-E with 100X/NA 1.40 oil-immersion objective; Nikon Instruments)
Motorized xy stage controller (Applied Scientific Instrumentation, cat. no. MS-2000-WK, firmware 9.2h, with ARRAY MODULE included)
Camera (e.g., Neo sCMOS camera, cat. no. NEO-5.5 or Zyla sCMOS camera, cat. no. ZYLA-5.5, Andor Technology) with BNC cables
μManager 1.4 (UCSF, www.micro-manager.org)
Antivibration table (Thorlabs, cat. no. B3636F)
Compressed air pump
Dry heat block (Fisher Scientific, cat. No. 11-718)
MATLAB R2014b (MathWorks)
(Optional) TwinCam dual-emission imaging system (Cairn Research, TwinCam LS)
(Optional) Fluorescence light source (e.g., Nikon Intensilight mercury lamp, cat. no. MBF72665, or X-Cite 120LED, Lumen Dynamics, cat. no. P010-00157)
(Optional) BNC T-shape adaptor (Thorlabs, cat. no. T3285)
REAGENT SETUP
Preparation of agar pads
To 200 mL of liquid growth medium in a 500 mL Corning glass bottle, add 1.5% (w/v) granulated agar (3 g) and dissolve by microwaving at 50% power. While microwaving, check the solution frequently to avoid overheating or boiling. If the experiment requires the addition of heat-sensitive chemicals, keep the melted agar on a 65 °C heat block until thermal equilibrium is achieved and then add the chemicals. CRITICAL: Make the agar medium fresh each time and keep bottles capped to avoid evaporation and contamination.
Remove the lid of a Singer PlusPlate and place the bottom (which is usually used for holding agar) upside-down onto a flat surface such as a benchtop. Pour melted agar onto the flat bottom surface of the plate (Fig. 1a, Supplementary Figure 1a), gently tilt and shake the plate to spread the agar evenly over the entire plate bottom, and use a flame to remove any bubbles if necessary. The surface tension of the hot agar will create a flat surface. One plate requires ~25 mL of melted agar. Leave the plates at room temperature (~22 °C) for 20–30 min to solidify the agar (Fig. 1b, Supplementary Figure 1b). CRITICAL: The plate must remain on a flat surface until the agar solidifies in an undisturbed environment to yield a flat and smooth agar surface for imaging.
EQUIPMENT SETUP
Software installation and setup
SLIP runs on MATLAB and controls a μManager12 GUI for image acquisition. Install μManager 1.4 (or the current version; UCSF, www.micro-manager.org) and MATLAB (MathWorks), and then open the SLIP source code (Supplementary Source Code, or visit https://bitbucket.org/kchuanglab/slip2 for the current version) in MATLAB to perform the initial setup as detailed in Box 1.
Box 1. Initial software setup after installation.
On our computer, μManager is installed in the default folder “C:/Program Files/Micro-Manager-1.4”. The path is hard-coded in some scripts and may require modification. The steps below are for a computer running Windows.
Find all.jar files located in the μManager folder: run MMsetup_javaclasspath.m in MATLAB. This script creates MMjavaclasspath.txt under the MATLAB default directory.
Run edit([prefdir ‘/MMjavaclasspath.txt’]) in MATLAB to open the MMjavaclasspath.txt file in the MATLAB editor. Then run edit([prefdir ‘/javaclasspath.txt’]). Copy all contents of MMjavaclasspath.txt to javaclasspath.txt.
Save javaclasspath.txt.
In MATLAB, run edit(‘librarypath.txt’). At the end of the file, add a line with the μManager installation directory, for example, C:/Program Files/Micro-Manager-1.4. Save librarypath.txt.
Restart MATLAB.
Right-click ‘Computer’ on the desktop or start menu.
Click ‘Properties → Advanced system settings’, highlight the ‘Advance’ tab, and click ‘Environment Variables…’.
On the ‘System variables’ list, highlight ‘Path’ and click ‘Edit…’. Do not delete anything. At the end, add ;C:/Program Files/Micro-Manager-1.4 or the appropriate μManager installation directory (do not omit the leading semicolon).
Save the changes and restart the computer to make the change of path effective.
TTL setup
During image acquisition, SLIP uses TTL signals to trigger between the camera and stage controller, circumventing the external controls between each image acquisition and thus speeding up the experiment. TTL signals have inactive (low voltage) and active (high voltage) states, and hardware can communicate through the switch of states. In this setup, when the camera finishes one exposure, its TTL signal switches from the active to the inactive state. This change at the camera’s “arm” port sends a TTL signal to the stage controller “input” port, which directs the stage controller to move to the next imaging site. Once the stage movement is done, its TTL state switches and sends a signal through its “output” port to the camera’s “external triggering” port, directing the camera to start another exposure. To set up TTL, first use BNC or other compatible cables to connect the camera and the stage controller to each other for TTL signals: connect the camera “arm” port to the stage controller “input” port, and then connect the camera “external triggering” port to the stage controller “output” port. An optional step is to connect the camera “fire” port to the “input” port of the light shutter, such that the shutter opens only during camera exposure. This step can be important for long-term time-lapse imaging to minimize photo-toxicity.
Since TTL is hardware-dependent, we recommend that users optimize their hardware settings to achieve the fastest possible acquisition. The main factors that determine total imaging time are camera exposure and stage movement. Acquisition time scales linearly with camera exposure time (Supplementary Figure 2a), thus reducing exposure time can speed up acquisition, but potentially at the cost of reducing image quality due to lower signal intensity. For stage movement, the time-consuming part is stage acceleration and deceleration between two imaging sites. We have found that the acceleration speed can impact total imaging time by ~3-fold, with an optimal time for acceleration for our system of 40–50 ms that minimizes total imaging time (Supplementary Figure 2b). We recommend that users test their stage controller to achieve the best results.
Dual camera setup (optional)
The dual camera setup enables the simultaneous capture of two images at different wavelengths using two identical cameras (Fig. 2a). The cameras should be aligned before imaging. For TTL setting, one camera (the master camera) communicates with the stage controller to coordinate exposure and stage movement; at the same time, it triggers the other camera (the slave camera) for a synchronized exposure in the other channel. To set up the cameras, first connect both cameras to the computer. Open μManager. Click ‘Tool → Device Property Browser’, and find the fields ‘Multi Camera-Physical Camera 1’ and ‘Multi Camera-Physical Camera 2’. Set their values to be the two cameras installed. Assign the slave camera to ‘Multi Camera-Physical Camera 1’, and the master camera to ‘Multi Camera-Physical Camera 2’. In addition to the TTL connections for the master camera, also connect the “fire” port of the master camera to the “external triggering” port of the slave camera. An optional step is to connect the master camera “fire” port to the “input” ports of the light shutters for both phase and fluorescence light sources to control exposure. A BNC T-shape adaptor may be needed for this optional step.
PROCEDURE
Growth of bacteria TIMING 0.5 d
-
1
Grow overnight liquid cultures of bacteria from stocks in deep 96-well or 384-well plates. It is recommended to include appropriate internal controls (e.g. wild-type cells) on the plate.
-
2
The next day, dilute overnight cultures 1:200 (or as desired) into fresh medium. Incubate until cells reach a desired density.
CRITICAL STEP Cell density directly affects the number of cells acquired for each strain during imaging, as well as the cellular physiological state. For E. coli and B. subtilis, we recommend incubating 2–3 h after the 1:200 dilution to reach a cell density of OD600~0.1, which typically yields enough cells (usually tens to a hundred) in each field of view. With this density, most cells are well separated from others, permitting easier and more accurate segmentation. We recommend that users perform preliminary tests to determine an optimal cell density for their particular applications. Such tests are facilitated by SLIP, whereby the user can serially dilute an overnight culture in a 96-well plate and rapidly acquire images of populations from different dilutions.
Preparation of imaging samples TIMING 10 min
-
3
Use a replicator pin to transfer the liquid culture from Step 2 onto a solidified agar pad (see REAGENT SETUP). Each transfer carries ~0.1 μL of liquid from each well. If desired, repeat the transfer several times to increase cell densities (Fig. 1c, d, Supplementary Figure 1c, d).
CRITICAL STEP It is important that the replicator pin does not directly contact the agar surface; the liquid should drop onto the surface. Direct contact of the pin with the agar may damage the flat agar surface and cause cell clustering and focus drifting, which lead to poor-quality images.
-
4
Wait for the agar pad to absorb liquid droplets. Keep the plate on a flat surface to avoid liquid flow and cross-contamination. The agar pad takes ~10 min to absorb all droplets at room temperature, or ~5 min in a 37 °C warm room. Check progress visually by inspecting light reflected from the surface at a tilted angle.
CRITICAL STEP All droplets should be completely absorbed by the agar pad before the next step. Unabsorbed liquid allows the cells to move above the surface of the pad, causing blurred images and potentially leading to cross-contamination.
-
5
(Optional) Mark the locations of the droplets at A1 and H12 (or other strain positions that will be used for calibration) on the back of the plate after the droplets have been absorbed. These marks will make it easy to find the strains during calibration.
-
6
Clean a large glass cover slip (110 x 74 mm) with compressed air and place the cover slip on top of the agar pad (Fig. 1e, f, Supplementary Figure 1e). Touch the pad with one side of the cover slip and slowly put down the other side, avoiding large air bubbles. If large bubbles occur, gently press the cover slip with a gloved finger to push bubbles to the edge of the cover slip, where they will dissipate.
(Optional) If the experiment requires cell growth before imaging, the pad can be incubated at the desired conditions before imaging. We have performed overnight incubation at 37 °C and have not observed significant drying of the pad.
-
7
(Optional, only for oil objectives) Add immersion oil to the cover glass. Spread the oil evenly across the entire imaging surface (Supplementary Figure 1f).
CRITICAL STEP Due to the large imaging area, it is important to spread the oil over the entire imaging surface such that the objective is always in contact with the oil during imaging, which reduces focus loss. Be careful not to add too much oil, especially on an inverted microscope, as excess oil may drip onto the objective turret.
-
8
Mount the agar plate onto the microscope stage, with the sample side facing the objective. Manually move the stage to the A1 strain and focus on the cells.
Imaging setup TIMING 5–10 min
-
9
Start SLIP by running SLIP.m in MATLAB. This script will open the μManager GUI. Choose the appropriate μManager configuration file, and the program will initialize (Fig. 2b).
-
10
Click ‘Calibrate’ in the SLIP window, and SLIP will display the calibration menu. After specifying the strain locations for calibration, use the “Live” imaging feature in μManager to locate the center of these strain positions. The positions of all strains on the plate will be calculated based on the calibration.
CRITICAL STEP Note that on an inverted microscope, the plate has to be inverted on the stage. Therefore, well A1 will be on the top right, if viewed from the top of the plate holder. Once calibrated, the SLIP GUI will also recognize this flipping and save images acquired at the top right corner as position A1.
TROUBLESHOOTING
-
11
Set the desired imaging parameters (plate layout, strains to image, number of images for each strain, exposure time, etc.; Fig. 2b(i–iv)). Typically, imaging a 5x5 grid (25 images in total) for each strain yields data from enough cells for the statistical analysis of interest. Variation in mean E. coli cell width and length across fields from the same well is ~3% (Supplementary Figure 3). The number of images should be adjusted based on cell densities. Since image acquisition is only a small fraction of the total time required for SLIP, the imaging grid size can be selected based on the strain(s) with the lowest density without substantially affecting the SLIP time.
-
12
For time-lapse imaging, set the desired imaging interval and total time points (Fig. 2b(v)). Typically, a full 96-well plate requires at least ~4 min to image one time point, and acquisition time scales linearly with the number of images acquired for each well (Fig. 1h); the defined interval should be longer than the total acquisition time for one time point.
-
13
In the SLIP window, check that ‘Pause on PFS loss’ (Fig. 2b(vi)) is enabled.
CRITICAL STEP With such a large imaging area, PFS loss is likely. Enabling ‘Pause on PFS loss’ allows SLIP to pause image acquisition for the user to manually fix the focus before the next strain. Thus, although SLIP does not require PFS-loss checking, this strategy improves data quality.
-
14
Select channels for imaging in the selection box (Fig. 2b(vii)). Note that for two or more channels without a dual camera setup (Fig. 2a), image acquisition will take significantly longer.
Image acquisition TIMING 4–60 min
-
15
Once all parameters are set, start image acquisition. Fix the focus when necessary.
CRITICAL STEP If ‘Pause on PFS loss’ is enabled, imaging progress should be monitored so that acquisition can continue after PFS loss.
TROUBLESHOOTING
Data analysis TIMING ~tens of minutes to 1 d
-
16
Segment the cells. Our laboratory has developed a custom MATLAB software, Morphometrics, for segmenting and analyzing phase images of bacteria cells17. General steps to use the software are included below; other software packages, e.g. Oufti18 and MicrobeJ19, can be applied similarly. The segmentation algorithm masks dark objects in a phase-contrast image and thereby identifies the cells. Next, it implements a custom filter to exclude non-cell objects based on their contour shapes. In principle, parameters for cell segmentation depend on the contrast and brightness of images and need to be optimized for each image. However, in our experience, since lighting and exposure conditions are very similar for all images in one experiment, parameters can be set based on a small subset of images (e.g. three images from three strains across the plate) and then used to segment the entire dataset.
-
17
Compile the data. After segmentation, properties such as cell area, contour curvature, and contour length are readily calculated and stored. We then apply filtering algorithms to remove micro-colonies that were not accurately segmented based on contour curvatures and a pill meshing algorithm23 to calculate an internal coordinate system and neutral axis for each cell. For rod-shaped cells, cell length and width are extracted from these measurements. The analyzed results for each cell are stored in a MATLAB structure with multiple fields, and the structures for all cells are saved as .mat files. The user can choose fields for further analyses or incorporate custom metrics as new fields into the data structure.
-
18
Analyze the data. Depending on the biological system and questions of interest, the compiled dataset can be used to extract information about the dimensions and growth (in the case of time-lapse experiments) across strains and/or chemical environments. For instance, the mean and standard deviation of morphological parameters such as width and length can be calculated for each strain in the 96- or 384-well plate (Fig. 3a, b).
TROUBLESHOOTING
Table 1.
Troubleshooting advice for common problems, with the associated step and possible explanation.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 10 | Cells are motile (and hence difficult to image in a single plane) | Liquid droplets not fully absorbed by pad before placing the cover slip | Wait longer until all droplets are absorbed before Step 6 |
| Too many/too few cells | Cell density of the liquid culture not optimal | Adjust back-dilution volume and/or incubation time before spotting cells onto agar pad; note that incubation time can affect cell size and/or shape24 | |
| 15 | PFS loss for many positions | Agar particles not fully melted | Fully melt agar before pouring pads. Melted agar should be homogenous without visible particles |
| Agar surface not flat during solidification | When preparing agar pads, keep the plate on a flat surface until the agar solidifies in an undisturbed environment | ||
| Replicator pin touched agar surface in Step 3 and damaged the agar surface | Avoid direct contact of replicator pin with agar surface; surface tension should be sufficient to transfer droplets to the agar surface | ||
| Insufficient immersion oil on the cover slip | Spread more immersion oil evenly across the imaging region | ||
| PFS loss for some positions on the edge | Agar pad not flat at the edge | Some extent of unevenness at the edge is expected due to surface tension, and can be minimized by using the right volume of melted agar so that the agar surface is at the same height as the plate edge | |
| No cells are found in certain strain positions | Little or no cell growth in the liquid culture | Check the OD of the liquid culture before transferring onto the pad to make sure cultures are sufficiently dense | |
| Calculated strain positions differ from the actual positions | Repeat calibration and make sure to set the centers of the calibration droplets |
TIMING
Steps 1 and 2, Growth of bacteria: ~0.5 d for E. coli in LB at 37 °C (varies by bacterial species)
Steps 3–8, Preparation of imaging samples: 10 min
Steps 9–14, Imaging setup: 5–10 min
Step 15, Image acquisition: 4 min per 96-well plate per time point, or 9 min per 384-well plate, with one image acquired for each strain. Total acquisition time for one plate scales linearly with the number of images per strain (Fig. 1h), with a total of 0.33 min required for each additional image across 96 strains, or 1.32 min for each additional image across 384 strains.
Steps 16–18, Data analysis: ~tens of minutes to 1 d per 96-well plate per time point (depending on the details of analysis, the time frame can be reduced substantially by parallelization)
ANTICIPATED RESULTS
Controls
To account for the imaging time required for SLIP, which can be a nontrivial fraction of the cell cycle of fast-growing bacteria, we performed control experiments to correct for growth effects. By performing a SLIP experiment on identical E. coli MG1655 cells in exponential phase in 96-well format, we found that mean cell width remained roughly constant across the populations throughout image acquisition (Pearson’s R = 0.13, p = 0.21), whereas mean cell length increased linearly with the temporal ordering of well position imaging: in the last well position imaged, the median cell length was 10% longer than that at the first well position (Fig. 3a), and the length increased linearly during imaging (Pearson’s R = 0.61, p < 1010); the pad represents an environment with fresh medium (unlike the supernatant of the exponential-phase culture from which the cells originated), and the increase in length is thus reflective of how long the cells have experienced the new environment of the pad24. Since E. coli grown at 37 °C on LB has one of the shortest doubling times of all bacterial species, typical increases during a SLIP experiment are likely to be <10%. The growth effect can be corrected based on strain positions in subsequent experiments (Fig. 3a, right panel, purple data points). The variation in mean width across wells is ~1% of the mean value (< 20 nm) and the variation in corrected length is ~3% of the mean value, indicating that errors introduced by SLIP are expected to be small.
Strains in exponential growth
Using the above protocol, we acquired images for an E. coli library of mutants in MreB, an actin homolog that dictates cell shape25. Each strain has a single mutation in a sandwich fusion of MreB to monomeric superfolder GFP (MreBsw-msfGFP). The mutations were selected via fluorescence-activated cell sorting based on forward scatter. We grew the library in a 96-well plate and acquired phase images for exponentially growing cells using SLIP (Fig. 3b), then used Morphometrics17 to extract quantitative morphological information from these images for direct comparison across strains. After correcting for the cell growth effect, mean cell length and width were highly correlated (Fig. 3c, Pearson’s R = 0.38, p < 0.001). In addition, other geometric properties such as population heterogeneity in cellular dimensions can be quantified from the imaging data through relatively straightforward additional programming.
Strains grown on agar pads
To demonstrate the ability to perform imaging after extended periods of growth on agar pads, we examined a CRISPRi-based library of knockdowns of essential genes in B. subtilis9. We first grew the strains in 96-well format to exponential phase without inducer in liquid culture, and then transferred cells onto agar pads with xylose, the inducer of the CRISPRi system. The agar pads were incubated overnight at 37 °C, during which time cells were depleted of an essential gene. Using SLIP, we imaged the terminal phenotypes of cells from each strain and observed a variety of morphological and/or growth defects (Fig. 3d).
Supplementary Material
Figure S1: Photographs of agar pad and sample preparation. (a) Melted agar is poured onto the bottom surface of a Singer PlusPlate and spread evenly by gently tilting and shaking the plate. (b) A flat agar surface is generally achieved when the plate is left on the benchtop to solidify without disturbance. The agar layer is ~2 mm in thickness. (c, d) Bacterial cultures are transferred onto an agar pad using a 96-well replicator pin. (e) After the agar pad absorbs all the liquid from the cultures, a large glass cover slip is used to cover the agar surface. (g) Immersion oil is applied evenly onto the cover slip for oil-immersion objectives.
Figure S2: Optimization of TTL setup. (a) Imaging time for a 5x5 grid of images for one strain scales linearly with camera exposure time. Reducing exposure time can expedite image acquisition. (b) Total imaging time for a 5x5 grid is dependent on stage acceleration time. In our configuration, an acceleration time of 40–50 ms is optimal.
Figure S3: Variation in mean cellular dimensions calculated from individual fields of view is small. The coefficient of variation of mean cell width (a) and length (b) across the fields of view from one of the wells in the experiment shown in Fig. 3a was ~3%. Data points are mean ± standard deviation, horizontal solid lines are the mean value for cells across all fields of view, and dashed lines are ±5% of the mean. Position #7 happened to have no cells and thus is not included in the plot.
Footnotes
Author contributions
H.S., A.C., T.K.L., and K.C.H. designed the research; H.S., A.C., and T.K.L. performed research; H.S. and K.C.H. analyzed the data; H.S. and K.C.H. wrote the manuscript.
Competing financial interests
The authors declare that they have no competing financial interests.
Source code for SLIP software. For the most up-to-date version of SLIP, visit https://bitbucket.org/kchuanglab/slip2.
References
- 1.Dörr T, et al. Differential requirement for PBP1a and PBP1b in in vivo and in vitro fitness of Vibrio cholerae. Infection and immunity. 2014;82:2115–2124. doi: 10.1128/IAI.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Monds RD, et al. Systematic perturbation of cytoskeletal function reveals a linear scaling relationship between cell geometry and fitness. Cell reports. 2014;9:1528–1537. doi: 10.1016/j.celrep.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao Z, Kahne D, Kishony R. Distinct single-cell morphological dynamics under beta-lactam antibiotics. Molecular cell. 2012;48:705–712. doi: 10.1016/j.molcel.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zambrano MM, Kolter R. GASPing for life in stationary phase. Cell. 1996;86:181–184. doi: 10.1016/s0092-8674(00)80089-6. [DOI] [PubMed] [Google Scholar]
- 5.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular systems biology. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bogomolnaya LM, et al. Identification of novel factors involved in modulating motility of Salmonella enterica serotype typhimurium. PloS One. 2014;9:e111513. doi: 10.1371/journal.pone.0111513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peters JM, et al. A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. Cell. 2016;165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nichols RJ, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young JW, et al. Measuring single-cell gene expression dynamics in bacteria using fluorescence time-lapse microscopy. nature protocols. 2012;7:80–88. doi: 10.1038/nprot.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelstein AD, et al. Advanced methods of microscope control using μManager software. Journal of biological methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA research. 2006;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 14.Kuwada NJ, Traxler B, Wiggins PA. Genome-scale quantitative characterization of bacterial protein localization dynamics throughout the cell cycle. Molecular microbiology. 2015;95:64–79. doi: 10.1111/mmi.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang P, et al. Robust growth of Escherichia coli. Current biology. 2010;20:1099–1103. doi: 10.1016/j.cub.2010.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gefen O, Fridman O, Ronin I, Balaban NQ. Direct observation of single stationary-phase bacteria reveals a surprisingly long period of constant protein production activity. Proceedings of the National Academy of Sciences. 2014;111:556–561. doi: 10.1073/pnas.1314114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ursell Tristan, et al. Rapid, precise quantification of bacterial cellular dimensions across a genomic-scale knockout library. 2016 doi: 10.1186/s12915-017-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paintdakhi A, et al. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Molecular microbiology. 2016;99:767–777. doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ducret A, Quardokus EM, Brun YV. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature Microbiology. 2016;1:16077. doi: 10.1038/nmicrobiol.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bacus JW, Hernicz RS. Dual color camera microscope and methodology for cell staining and analysis. US patent. 1991
- 21.Chang F, Huang KC. How and why cells grow as rods. BMC biology. 2014;12:54. doi: 10.1186/s12915-014-0054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sezonov G, Joseleau-Petit D, D’Ari R. Escherichia coli physiology in Luria-Bertani broth. Journal of bacteriology. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Molecular microbiology. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjeldgaard N, Maaløe O, Schaechter M. The transition between different physiological states during balanced growth of Salmonella typhimurium. Microbiology. 1958;19:607–616. doi: 10.1099/00221287-19-3-607. [DOI] [PubMed] [Google Scholar]
- 25.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Photographs of agar pad and sample preparation. (a) Melted agar is poured onto the bottom surface of a Singer PlusPlate and spread evenly by gently tilting and shaking the plate. (b) A flat agar surface is generally achieved when the plate is left on the benchtop to solidify without disturbance. The agar layer is ~2 mm in thickness. (c, d) Bacterial cultures are transferred onto an agar pad using a 96-well replicator pin. (e) After the agar pad absorbs all the liquid from the cultures, a large glass cover slip is used to cover the agar surface. (g) Immersion oil is applied evenly onto the cover slip for oil-immersion objectives.
Figure S2: Optimization of TTL setup. (a) Imaging time for a 5x5 grid of images for one strain scales linearly with camera exposure time. Reducing exposure time can expedite image acquisition. (b) Total imaging time for a 5x5 grid is dependent on stage acceleration time. In our configuration, an acceleration time of 40–50 ms is optimal.
Figure S3: Variation in mean cellular dimensions calculated from individual fields of view is small. The coefficient of variation of mean cell width (a) and length (b) across the fields of view from one of the wells in the experiment shown in Fig. 3a was ~3%. Data points are mean ± standard deviation, horizontal solid lines are the mean value for cells across all fields of view, and dashed lines are ±5% of the mean. Position #7 happened to have no cells and thus is not included in the plot.