Abstract
Optical multiplexing impacts widely in photonics, life science, biomedicine and engineering. Despite intensive efforts, current technology is limited by a longstanding “multiplexing ceiling” from existing optical materials. Here we engineered a novel class of polyyne-based materials for optical super-multiplexing. 20 distinct Raman frequencies are achieved as “Carbon rainbow” through rational engineering of conjugation length, bond-selective isotope doping and end-capping substitution of polyynes. With further probe functionalization, we demonstrated unprecedented 10-color organelle imaging in single living cell with high specificity, sensitivity, and photo-stability. Moreover, optical data storage and identification are realized by combinatorial barcoding, yielding the largest number of distinct spectral barcodes to date. Therefore, these versatile polyynes hold great promises in live-cell imaging and sorting, high-throughput diagnostics and screening, and information technology.
Introduction
Modern science and technology have exploding demands for multiplexing techniques that allow simultaneous measurement of a large number of distinctive species. Notable examples include imaging multiple proteins or organelles with probes of different colors1-3, cell phenotyping, sorting and tracing4, high-throughput detection of antigens for medical diagnostics or small molecules for drug discovery5-7, and high-density information storage and encryption for identity security and anti-counterfeiting8,9. As optical methods readily offer spectral features that can be identified in a non-invasive and non-destructive manner, optical multiplexing is making increasingly broad impact in photonics, biology, medicine and engineering.
Materials are at the core of multiplexing technology. Indeed, many multiplexable optical materials have been developed. Organic dyes and fluorescent proteins are widely applied for multicolor imaging in biology1, and inorganic materials including quantum dots, noble metal nanostructures, rare-earth nanoparticles and upconversion nanocrystals are used in spectral barcoding for high-throughput screening and data security applications7,9. However, because of the intrinsically broad linewidth and/or significant spectral overlap, the number of resolvable features is significantly limited. No more than 6 colors can be imaged in live cells even by sophisticated fluorescence microscopy2. And fewer than 2000 spectral barcodes can be practically reached10, due to unavoidable cross-talk in organic dyes, energy transfer between quantum dots11,12, and limited number of suitable features for straightforward decoding in rare-earth nanocrystals and metal nanoparticles9,13,14. Yet, many more colors and barcodes are essential for next-generation multiplexing. Therefore, new optical materials are urgently needed to overcome the spectral limitation and break the longstanding “multiplexing ceiling”.
Herein we report the development of new polyyne-based materials and exploit their outstanding optical properties for super-multiplexed encoding and detection. Polyyne is a linear chain of sp-hybridized carbon atoms with alternating single and tripe bonds, which is also known as carbon-atom wire15. Compared to other well-known carbon materials such as sp3-hybridized diamonds and sp2-hybridized graphene, carbon nanotube and fullerene, sp-hybridized polyyne is one of the least studied carbon allotropes with true 1-D structure16. Although theory predicts them with many appealing physical properties15,17, difficulty in accessing stable polyynes had largely hindered their exploration. Recently, significant progress has been made toward chemical synthesis of polyynes with well-defined composition. Indeed, isolable polyyne with 44 contiguous carbon atoms was successfully synthesized18.
Results
Raman spectroscopy of phenyl-capped polyynes
We first synthesized a series of polyynes with phenyl end-capping groups (Supplementary Notes). Using the classic Glaser-Hay and Cadiot-Chodkiewicz coupling19, we developed an efficient and robust route to prepare both odd- and even-numbered polyynes with 2 to 6 triple bonds (C≡C) (Figure 1). A polar hydroxymethyl group is introduced on the phenyl ring for facile purification and serves as an active site for subsequent functionalization. UV-Vis spectra show the longest wavelength of absorption in polyynes shift from UV (337 nm) to visible (476 nm) region with clear vibronic progression (Supplementary Figure 1), as the length increases from 2-yne to 6-yne with lowered HOMO-LUMO bandgap. All polyyne compounds in our study show good chemical stability under ambient conditions, as characterized by NMR and mass spectrometry.
Figure 1. Raman spectroscopy of parental phenyl-capped polyynes.
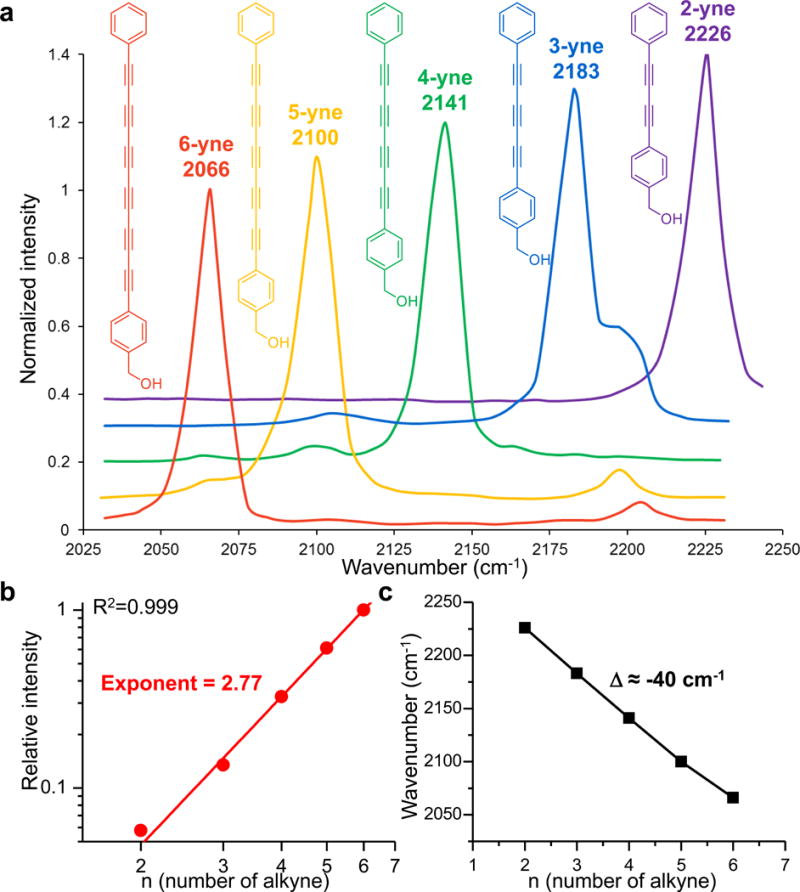
(a) Normalized Raman spectra of polyynes from 2-yne to 6-yne and their structures. The spectra are vertically offset for clarity. (b) Raman peak intensity of polyynes increases superlinearly with the conjugation length, following a power-law dependence. Solid line shows a power function fitting in the double logarithmic plot (R2=0.999, n=5 compounds). (c) Raman frequency of polyynes decreases almost linearly with increasing length, with an interval of ~40 cm−1.
These phenyl-capped polyynes possess intriguing vibrational spectroscopy. They all display an intense Raman peak with narrow linewidth (13 cm−1) (Figure 1a), originated from a collective out-of-phase bond-length alternation oscillation of both single and triple bonds20. Such a single sharp peak in the Raman-silent spectral region promises sensitive and specific detection. In addition, as the number of triple bonds increases from 2 to 6, the Raman intensity grows superlinearly with a power-law exponent of 2.77±0.06 (Figure 1b). This trend is similar to the dependence observed in the second hyperpolarizability of other conjugated oligomers21, suggesting much higher detection sensitivity than a single alkyne22. More importantly, going from 2-yne to 6-yne, the peak frequencies of polyynes shift almost linearly from 2226 cm−1 to 2066 cm−1 (Figure 1c), naturally separating these polyynes in the frequency domain. Therefore, the unique spectral features of single intense peak, narrow linewidth and the natural frequency spacing of different lengths render these polyynes an ideal scaffold for optical multiplexing.
Engineering polyynes for frequency tuning and expansion
To achieve super-multiplexing, we sought to further engineer the polyynes to vastly expand the number of vibrational frequencies (Figure 2a). We first exploited isotope doping which is an effective approach to modify the reduced mass of the vibrational mode23,24. With multiple triple bonds present in polyynes, we optimized the 13C labeling pattern and were capable of modularly doping one or more triple bonds synthetically. The frequency shifting effect of doping multiple triple bonds appears to be additive, as illustrated by singly, doubly and triply 13C-labeled 4-yne series (Figure 2b). Thus such bond-selective 13C doping allows us to tune the frequency in a large range of 20-80 cm−1 (Figure 2b). We noted the appearance of minor peaks and attributed it to the breakdown of centrosymmetry with non-uniform 13C labeling and the violation of mutual Raman-IR exclusion25.
Figure 2. Chemical strategies for Raman frequency expansion of polyynes.
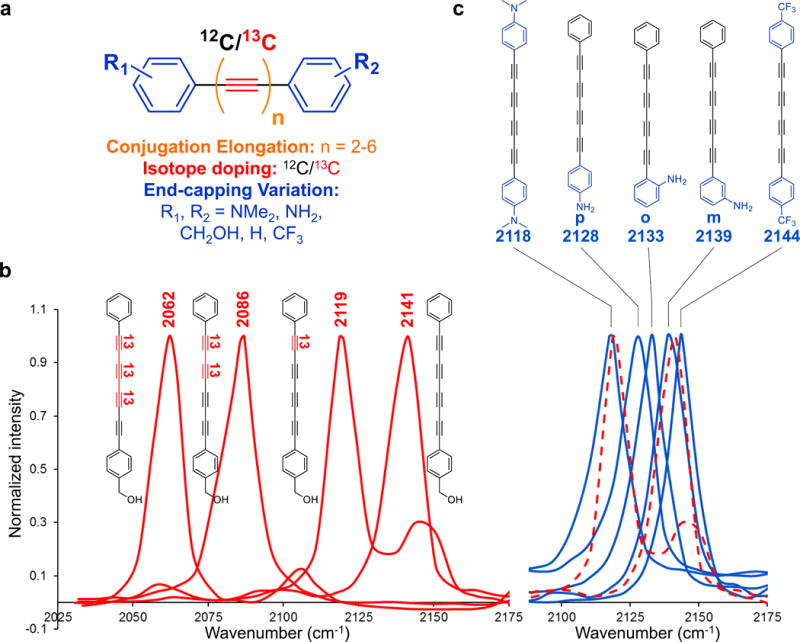
(a) Frequency modulation on the unified polyyne scaffold by conjugation elongation, isotope doping and end-capping variations. (b) Frequency coarse-tuning with bond-selective 13C isotope labeling (in red). (c) Frequency fine-tuning with electron-donating and electron-withdrawing groups (in blue). The red dashed spectra are the 2119 cm−1 and 2141 cm−1 peaks from (b).
Additional frequency modulation strategy is needed, as bond-selective 13C doping only offers coarse tuning with relatively large increments. Theory suggests that enhanced π-electron delocalization can increase electron-phonon coupling, which will weaken the stretching force constant of the vibrational mode and red-shift the peak position20. We hypothesize that substituting the end-capping phenyl ring with electron-donating or -withdrawing groups might be able to tune the vibrational frequency by influencing π-electron delocalization on the polyyne chain (Figure 2c). Indeed, with electron-donating dimethylamine -N(CH3)2, doubly substituted 4-yne shows a significant redshift of 23 cm−1, whereas the electron-withdrawing trifluoromethyl -CF3 blue-shifts by 3 cm−1, compared to 4-yne at 2141 cm−1. The exact substitution position also matters. With the same -NH2, para-position shows the largest redshift (2128 cm−1) due to the strong mesomeric effect. Ortho-position (2133 cm−1) is next, and meta-position (2139 cm−1) is the weakest. Thus, end-capping substitution provides the frequency fine-tuning (2-20 cm−1), which, to our delight, is perfectly complementary to the coarse-tuning (20-80 cm−1) from 13C doping.
Polyynes with 20 resolvable frequencies: Carbow
Guided by above principles, we synthesized a library of polyynes for frequency super-multiplexing through rational combination of length modulation, bond-selective 13C labeling and end-capping substitutions (Supplementary Figure 2). 20 structures with distinct Raman frequencies are selected and termed as “Carbon rainbow” (Carbow, Figure 3), representing the largest number of resolvable frequency in the Raman-silent window. Note that using alkyl instead of phenyl as additional end-capping group can blue-shift further to 2262 cm−1. Combined with 6 reported Raman frequencies in the fingerprint region from commercial dyes26 and 4 commonly available fluorescent channels, 30 colors are resolvable, which is the highest reported for parallel optical detection. Compared to 14 MARS dyes recently reported in this range26, Carbow peaks are more evenly spaced and well resolved with significantly less cross-talk. For instance, the spectral separation of the closest Carbow peaks nearly doubles that in the MARS dyes.
Figure 3. Super-multiplexed polyynes.
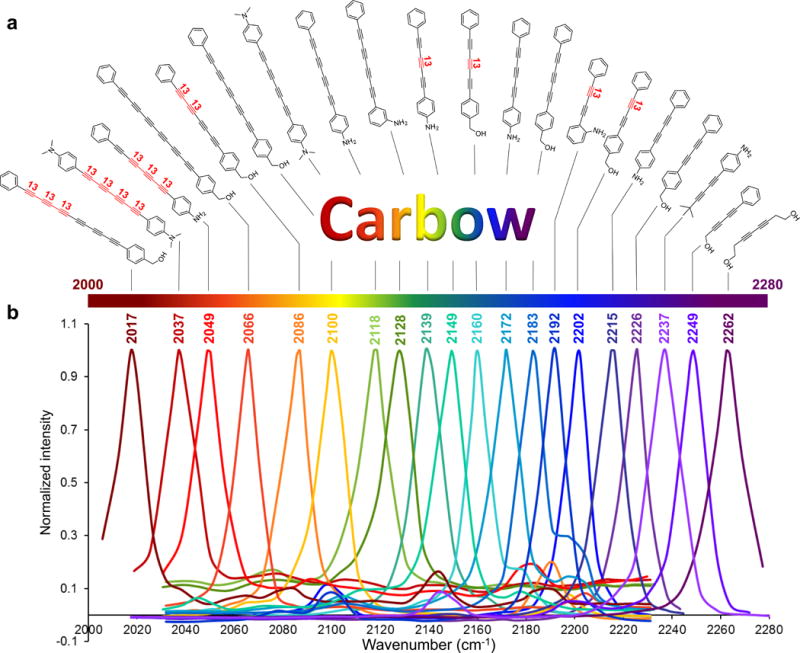
(a) Chemical structures of 20 polyynes with distinct Raman frequencies, which are termed as Carbon rainbow (i.e., Carbow). (b) Highly resolved Raman peaks of Carbow in the silent spectral window.
Optical imaging in cells with Carbow
The multiplexing capability of Carbow is highly demanded in biological imaging for simultaneous visualization of multiple species. Detection sensitivity of Carbow is first characterized using stimulated Raman scattering (SRS) microscopy with ultrahigh sensitivity and specificity27. Down to 630 nM (S/N=1) can be detected for 4-yne with 1 ms time constant (< 5 μM for most polyynes) under our SRS microscope (Figure 4a). This is nearly 500 times more sensitive than previous SRS detection of a single alkyne tag28. Given the sub-μM sensitivity, immuno-staining of specific proteins is demonstrated by conjugating polyynes to secondary antibodies for SRS imaging (Figure 4b and Supplementary Figure 3). Pattern of α-tubulin filament structures is visualized with good contrast. Tuning the wavelength away by 3 nm shows negligible background signal in the off-resonance channel, demonstrating the sharp vibrational feature. Thus, Carbow is fully compatible with standard immuno-staining procedures for protein-specific imaging.
Figure 4. Super-multiplexed optical imaging with polyynes.
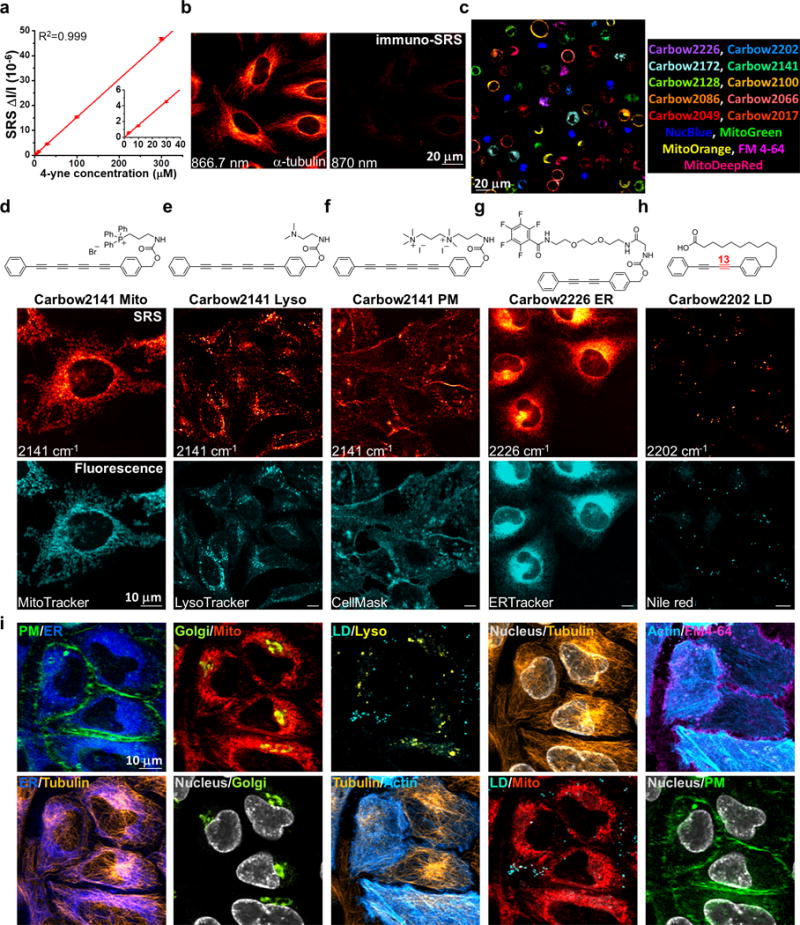
(a) Linear concentration dependence of 4-yne with sub-μM SRS detection sensitivity. Error bars: mean ± s.d., n=3 measurements. Solid line shows a linear fitting (R2=0.999, n=5 concentrations). (b) Immuno-staining and imaging of α-tubulin in fixed HeLa cells with 4-yne conjugated antibody. (c) 15-color tandem fluorescence-SRS imaging of live HeLa cells with corresponding Carbow and fluorescent molecules. (d-h) Chemical structures of five organelle-targeted probes based on polyynes for live-cell imaging, including mitochondria Mito (d), lysosome Lyso (e), plasma membrane PM (f), endoplasmic reticulum ER (g) and lipid droplet LD (h). (i) 10-color optical imaging of PM (Carbow2141), ER (Carbow2226), Golgi (BODIPY TR), Mito (Carbow2062), LD (Carbow2202), Lyso (Carbow2086), nucleus (NucBlue), tubulin (SiR650), actin (GFP) and FM 4-64 in living HeLa cells. Overlay of two species are shown in each image.
We next demonstrated 15-color imaging of spatially-resolved living cells (Figure 4c). Each cell is singularly stained with either 5 fluorescent or 10 Carbow molecules. All cells in 15-channels can be identified unequivocally in one setting, without the need of complicated unmixing (Supplementary Figure 4). This is especially challenging for fluorescence imaging with broad and overlapping spectra, and also excels MARS dyes which require predetermined matrix owing to substantial cross-talk between channels26. Therefore, Carbow has afforded multiplexed optical imaging at a number that is rarely reported for live cells with straightforward detection and analysis.
Super-multiplexed imaging of organelles in living cells
Furthermore, owing to the appealing neutral scaffold and high membrane permeability, Carbow can be functionalized into live-cell organelle-specific imaging probes. Through a carbamate linker on the phenyl ring, different targeting groups are introduced (Figure 4d-h). Triphenylphosphonium (TPP+) is a well-known motif with high affinity to mitochondrial matrix due to the positive charge29. TPP+ attached 4-yne (Carbow2141 Mito) shows highly specific localization to mitochondria (Figure 4d). Lysosome lumen is acidic, where basic unit is easily protonated and trapped inside, and thus we used a dimethylamine group to target into lysosomes (Carbow2141 Lyso, Figure 4e). Similarly, a cationic diammonium group (Carbow2141 PM) is chosen to stain plasma membrane through interaction with anionic phosphate headgroups, and a pentafluorobenzamide group with ethylene glycol chain (Carbow2226 ER) is used to target endoplasmic reticulum (Figure 4f, g). Lastly, a carboxylate terminated C12 alkyl chain (Carbow2202 LD) is attached to mimic fatty acids, which is incorporated efficiently into lipid droplets (Figure 4h). All five imaging probes show co-localized patterns with the corresponding fluorescent markers in live cells (Supplementary Figure 5 and 6). Carbow probes also exhibit superb photo-stability (>98%) after 100 continuous frames (Supplementary Figure 7) and little cytotoxicity (Supplementary Figure 8).
With the good specificity, sensitivity, photo-stability and live-cell compatibility, we combined five organelle-targeted Carbow probes and five fluorescent reporters to achieve tandem 10-color optical imaging of subcellular structures in live cells (Figure 4i and Supplementary Table 1; including plasma membrane, ER, Golgi, mitochondria, lysosome, lipid droplets, nucleus, tubulin and actin). Recognizing the essential roles of organelle interactome in cellular activities, up to 6-color organelle imaging has recently been achieved by fluorescence microscopy with spectral unmixing and color compensation2. Here the unprecedented 10-color organelle imaging in live cells doesn’t require any unmixing or color compensation in image processing, which is extremely difficult to achieve by other means. To our knowledge, this is the highest number demonstrated for multi-target imaging inside living cells, and relies critically on Carbow’s well-resolved frequencies and outstanding live-cell compatibility.
Optical barcoding with combinatorial Carbow
The need to multiplex goes beyond imaging. Systems biology and personalized medicine demand high-throughput analysis of cells and biomolecules such as antigens and drugs, the core technology of which often requires distinguishable barcoding on micro-objects such as beads7,30. We hence applied Carbow to optical data storage (i.e., encoding) and identification (i.e., decoding). Harnessing polymer beads as the information carrier, we demonstrated that spectral barcoding with 10 resolvable frequencies at 3 distinct intensity levels (i.e., ternary digit) is readily achievable using Carbow (Figure 5). 3-μm sized polystyrene beads are loaded with specified polyyne mixtures via a swelling-diffusion technique31, and each bead is encoded with desired spectral information in a single preparation step (Figure 5a). As illustrated by four representative codes decoded by SRS readout, the intensity at each of the 10 specified frequencies can be unambiguously digitalized as either 0, or 1 or 2 (Figure 5b and Supplementary Table 2). Spectral barcodes can also be read out by conventional spontaneous Raman spectroscopy, and 10 spectral patterns are decoded at single particle level (Supplementary Figure 9 and Table 3). All 9 combinations from two adjacent frequencies are well identified (Supplementary Figure 10), demonstrating robustness against cross-talk. Thus, 310–1=59048 distinct barcodes can be generated as the largest set to date using spectral coding, whereas the recently reported record10 is around 1000. Compared to previous optical barcoding materials, Carbow not only affords many more resolvable frequencies, which greatly benefit combinatorial coding with exponential growth, but is also free from photo-bleaching or complication of energy transfer as in fluorescence.
Figure 5. Super-multiplexed optical barcoding with polyynes.
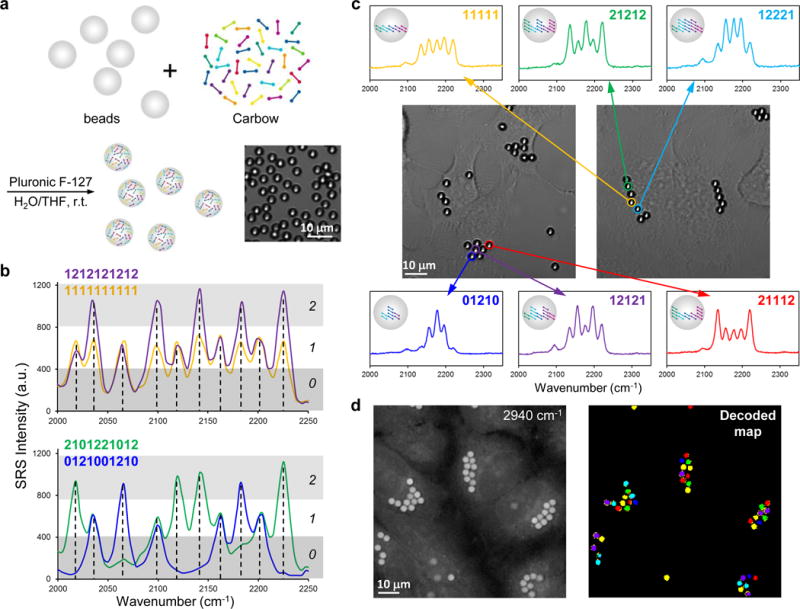
(a) Polymer beads are readily encoded by combinatorial loading of polyynes. Inset shows the bright-field image of barcoded polystyrene beads. (b) Spectral barcoding of polyynes at 10 frequencies and 3 intensities (0, or 1 or 2) with SRS readout, which can obtain 310-1=59048 distinct barcodes. (c) Cells are labeled with multiple encoded beads, as shown by the bright-field images. The barcode information is well preserved in live HeLa cells with clear readout by spontaneous Raman measurement. (d) Rapid decoding and spatial visualization of bead identities with SRS microscopy. Left: 2940 cm−1 image of unidentified beads inside living HeLa cells; Right: decoded beads in the field of view by hyperspectral SRS imaging at characteristic frequencies. The color of each bead (six different types) in the decoded map corresponds to the spectral barcode in (c).
Besides in vitro detection, barcoded beads can potentially be used as a unique ID to tag individual cell and map out the interaction of cells with time, as in the human cell atlas project (studying ~ 1013 cells). Owing to the small micron-size, multiple beads can be combined in labeling single cell. We show that HeLa cells can take up several beads containing different barcodes. The encoded information is well retained inside live cells after 24 hours, which can be read out by spontaneous Raman spectroscopy (Figure 5c). Further, besides measuring the spectrum of one bead at a time, hyperspectral SRS imaging at discretized frequencies can decode all beads in the field of view (Figure 5d and Supplementary Figure 11), allowing rapid visualization of cell identity. Hence, if using 3 encoded beads to tag each cell, we would be able to generate 59048C3=3×1013 IDs, sufficient to identify all cells in the human body. This is many orders of magnitudes greater than the limit of current state-of-the-art techniques based on organic dyes, quantum dots and upconversion nanocrystals or whispering-gallery mode detection of bead diameters32.
Discussion
We have engineered polyynes into an extraordinary material for optical super-multiplexing, surpassing the existing “multiplexing ceiling”. Compared to a recent report26, the strong signals of polyynes arise from superlinear second hyperpolarizability enhancement of the linearly conjugated triple bonds under electronic non-resonant condition, whereas MARS dyes derive their signals from electronic pre-resonant enhancement of planarly conjugated chromophores. Application-wise, the current work is much more general, as it doesn’t require stringent laser pre-resonance condition, has further expanded and better separated frequencies with less cross-talk and no need of unmixing, and is more suitable for live cell application and spectral barcoding due to the neutral scaffolds of polyynes. The current technology can be further improved. From material perspective, more colors can be accessible with longer polyynes. The frequencies of polyynes can red-shift towards 1850 cm−1 with increasing length33, providing an expanded window of 150 cm−1. From microscopy perspective, SRS imaging of single organelle in live cells can be achieved in as short as 2 seconds per frame (Supplementary Figure 12), and high-speed hyperspectral SRS imaging34,35 can be applied to capture fast dynamics of live cells. From barcoding perspective, beads functionalized with antibodies or enzymes can be applied for medical diagnostics and drug discovery. SRS flow cytometry36 can also be employed to detect tens of thousands of beads per second. Lastly, as a proof of principle for identity security and anti-counterfeiting, frequency encryption in microscopic pattern is also demonstrated (Supplementary Figure 13). Therefore, polyynes represent a new class of optical material that could find wide applications in super-multiplexed cell imaging and sorting, barcoding, high-throughput screening and diagnosis, and identity security, providing exciting opportunities in photonics, life science, medicine, and information technology.
Online Methods
Chemical synthesis
Methods for chemical synthesis and characterization of all new compounds can be found in the Supplementary Notes.
Stimulated Raman scattering (SRS) microscopy
An integrated laser system (picoEMERALD, Applied Physics & Electronics, Inc.) was used to produce two synchronized laser beams at 80 MHz repetition rate. A fundamental Stokes beam (1064 nm, 6 ps pulse width) was intensity modulated at 8 MHz by an electro-optic-modulator with >90% modulation depth, and a tunable pump beam (720-990 nm, 5-6 ps pulse width) was produced by a build-in optical parametric oscillator. The pump and Stokes beams were spatially and temporally overlapped using two dichroic mirrors and a delay stage inside the laser system, and coupled into an inverted laser-scanning microscope (FV1200, Olympus) with optimized near-IR throughput.
The lasers were focused on the sample through a 25 × water objective (XLPlan N, 1.05 N.A. MP, Olympus) or a 60 × water objective (UPlanAPO/IR, 1.2 N.A., Olympus) with high near-IR transmission. The beam sizes of pump and Stokes laser were adjusted to match the back-aperture of the objective. After the sample, both beams were effectively collected by a high N.A. oil condenser lens (1.4 N.A., Olympus) in Kohler illumination and the laser-scanning motion was descanned with a telescope. By blocking the Stokes beam with a high O.D. bandpass filter (890/220 CARS, Chroma Technology), only the pump beam was detected by a large-area (10 mm × 10 mm) silicon photodiode (FDS1010, Thorlabs) reverse-biased at 64 DC voltage to maximize saturation threshold and response bandwidth. The output current of photodiode was filtered with an 8 MHz electronic bandpass filter (KR 2724, KR electronics), and terminated with 50 Ω before entering a RF lock-in amplifier (SR844, Stanford Research Systems or HF2LI, Zurich instrument).
The stimulated Raman loss signal was extracted from the pump beam by demodulation at the 8 MHz frequency with near short-noise-limited sensitivity. The in-phase signal from the lock-in amplifier at each pixel was sent to the analog interface box (FV10-ANALOG, Olympus) of the microscope to generate the SRS image by scanning across the whole field of view. SRS images were acquired using 25 × objective with 30 μs time constant from the lock-in amplifier and 100 μs pixel dwell time (10.2 s per 320 × 320 frame), unless otherwise specified. Measured after the objective, the pump power was from 12-48 mW and the Stokes power was from 40-120 mW for all cell images (Figure 4). For bead (Figure 5d and Supplementary Figure 11) and pattern (Supplementary Figure 13) imaging, the pump power was from 12-24 mW and the Stokes power was 20 mW. For immuno-imaging (Figure 4b and Supplementary Figure 3), 100 μs time constant and 200 μs pixel dwell time were used. 10-color live-cell organelle imaging (Figure 4i) was performed using 60 × objective and 80 μs time constant. Photo-stability experiments (Supplementary Figure 7) were performed with 3 μs time constant and 4 μs pixel dwell time (0.4 s per 320 × 320 frame). Stimulated Raman scattering spectra were acquired by scanning the pump beam across the selected wavelength range point by point.
All bright-field and fluorescence images were collected using the Olympus FV1200 confocal microscope with CW laser excitation (488, 543 and 635 nm) and standard bandpass filter sets. Two-photon fluorescence images of NucBlue were collected with 780 nm pump laser excitation and detected by non-descanned photomultiplier tubes. All images were analyzed and assigned color by ImageJ.
Spontaneous Raman spectroscopy
Raman spectra were collected with the LabSpec 6 software on a confocal Raman microscope (Xplora, Horiba Jobin Yvon) at room temperature. The samples were excited through a 50 × air objective (MPlan N, 0.75 N.A., Olympus) by a 532-nm diode laser (27 mW after the objective). The acquisition time for bead solution samples (Supplementary Figure 9 and 10) was 5 s and for beads in live cells (Figure 5c) was 10 s.
Secondary antibody conjugation with 4-yne Sulfo-NHS ester
Goat-anti-Rabbit secondary antibody solution (2 mg/mL, Millipore, AP132) was adjusted to pH~8.3 with sodium bicarbonate solution. 50 μL 4 mg/mL 4-yne Sulfo-NHS ester in DMSO solution was added dropwise to 250 μL protein solution while stirring. Reaction was kept under gentle stirring at RT for 1 h. The labeled antibodies were purified using gel permeation chromatography with Sephadex® G-25 (Sigma, G25150). Sephadex® G-25 gel was first swelled in PBS buffer at 90°C for 1 h and settled down at room temperature. The gel was then exchanged with fresh PBS buffer and stored at 4 °C. For gel chromatography, the column (diameter ~1 cm and length >12 cm) was loaded with swelled gels and equilibrated with PBS buffer. The antibody solution was applied and eluted with PBS. The first band with light color for 4-yne conjugated antibodies was collected. The solution was then centrifuged briefly and the supernatant was concentrated using Amicon® Ultra Centrifugal Filters (Millipore, UFC501096) to a final concentration of ~2 mg/mL in PBS with 5 mM sodium azide and stored at -20 °C.
Sample preparations for SRS and fluorescence imaging
HeLa and COS-7 cells were cultured with DMEM medium (Invitrogen, 11965) supplemented with 10% fetal bovine serum (Invitrogen, 16000) and 1% penicillin-streptomycin (Invitrogen, 15140). All cell cultures were maintained in a humidified environment at 37 °C and 5% CO2.
All samples were assembled into a chamber using an imaging spacer (Sigma, GBL654008) filled with PBS solution for imaging.
Immuno-staining for SRS imaging in fixed HeLa cells (Figure 4b and Supplementary Figure 3)
HeLa cells were seeded on a glass coverslip in a 4-well plate with ~1 mL of culture media for 24 h. Cells were fixed with 4% PFA for 20 min or methanol for 25 min, washed with 10% goat serum supplemented with 1% bovine serum albumin (BSA) and 0.3 M glycine twice, and permeabilized with 0.1% Tween PBS solution for 40 min. Primary antibody in rabbit (Anti-α-tubulin, Abcam ab18251; Anti-Histone H2B, Abcam ab1790; Anti-Tom20, Santa Cruz Biotechnology sc-11415) was added to the cells at 1:50 dilution in 3% BSA solution and incubated overnight at 4 °C. The cells were then blocked with 10% goat serum for 30 min and incubated with 4-yne conjugated goat-anti-rabbit secondary antibody by 1:25 dilution in 10% goat serum at 4 °C overnight. The cells were washed with PBS twice before imaging.
15-color imaging of live Hela cells with super-multiplexed polyynes (Figure 4c and Supplementary Figure 4)
HeLa cells were separately seeded in 15 wells of a 24-well plate for 24 h. Each well was labeled with a single color of fluorescent dyes or polyynes in culture media at 37 °C, including Carbow2226 Mito (5 μM), Carbow2172 (3 μM), Carbow2141 (10 μM), Carbow2128 (2 μM), Carbow2100 (4 μM), Carbow2086 (10 μM), Carbow2066 (10 μM), Carbow2049 (5 μM), and Carbow2017 (10 μM) for 4 h, Carbow2202 LD (4 μM) for 24 h, MitoTracker Green (100 nM, Invitrogen, M7514), MitoTracker Orange CMTMRos (50 nM, Invitrogen, M7510), and MitoTracker Deep Red (50 nM, Invitrogen, M22426) for 1 h, NucBlue® Live ReadyProbes Reagent (1 drop, Invitrogen, R37605) and FM 4-64 (5 μg/mL, Invitrogen, T3166) for 30 min. All cells were then washed with PBS once, detached from each well with trypsin for 2 min, and mixed together in fresh culture media. The cell mixture was centrifuged at 1000 rpm for 1.5 min and the cell pellet was mixed with PBS. Cells were added to the imaging chamber and settled for 1 h to reduce the movement before imaging, and each cell was maintained with a single color during the imaging period.
Mitochondria imaging in live HeLa or COS-7 cells with Carbow2141 Mito (Figure 4d, Supplementary Figure 5 and 6)
Cells were cultured on a glass coverslip in a 4-well plate for 24 h, and 2 μM Carbow2141 Mito was added in the media for 1 h at 37 °C. Cells were washed with PBS for three times before imaging. For co-localization, 100 nM MitoTracker Deep Red was used to stain mitochondria for 1 h.
Lysosome imaging in live HeLa or COS-7 cells with Carbow2141 Lyso (Figure 4e, Supplementary Figure 5 and 6)
Cells were cultured on a glass coverslip in a 4-well plate for 24 h, and 4 μM Carbow2141 Lyso was added to cells for 30 min at 37 °C. Cells were washed with PBS for three times before imaging. For co-localization, 100 nM LysoTracker Red (Invitrogen, L7528) was used to stain lysosome for 30 min.
Plasma membrane imaging in live HeLa or COS-7 cells with Carbow2141 PM (Figure 4f, Supplementary Figure 5 and 6)
Cells were cultured on a glass coverslip in a 4-well plate for 24 h, and were incubated with 3-5 μM Carbow2141 PM in the culture media for 20-30 min at 37 °C and were washed with PBS for three times before imaging. For co-localization, 0.5 μg/mL CellMask Deep Red (Invitrogen, C10046) was used to stain cell membrane for 5 min.
Endoplasmic reticulum imaging in live HeLa or COS-7 cells with Carbow2226 ER (Figure 4g, Supplementary Figure 5 and 6)
Cells were cultured on a glass coverslip in a 4-well plate for 24 h, and were incubated with 10 μM Carbow2226 ER for 2 h at 37 °C. Cells were washed with PBS for three times before imaging. For co-localization, 1 μM ERTracker Green (Invitrogen, E34251) was used to stain endoplasmic reticulum for 2 h.
Lipid droplets imaging in live HeLa or COS-7 cells with Carbow2202 LD (Figure 4h, Supplementary Figure 5 and 6)
Cells were cultured on a glass coverslip in a 4-well plate for 24 h, and 10 μM Carbow2202 LD was added to cell media for overnight at 37 °C. Cells were washed with PBS for three times before imaging. For co-localization, 1 μM Nile red (Invitrogen, N1142) was used to stain lipid droplets for 10 min.
10-color organelle-specific imaging in live Hela cells with polyyne probes (Figure 4i and Supplementary Table 1)
HeLa cells were seeded on a glass coverslip in a 4-well plate with culture media for 24 h, and actin-GFP plasmids (Invitrogen, C10582) were transfected into cells for 48 h before imaging according to Invitrogen protocol. 10 μM Carbow2202 LD was added in the culture media overnight at 37 °C before imaging. On the day of imaging, cells were incubated with 10 μM Carbow2226 ER and 0.02% Pluronic F-127 (Invitrogen, P3000MP) for 2 h, 2 μM Carbow2086 Lyso, 4 μM Carbow2062 Mito and 1 μM SiR-tubulin (Cytoskeleton, CY-SC002) for 1 h, 3 μM Carbow2141 PM and one drop of NucBlue for 20 min at 37 °C. 1 hour before imaging, cells were labeled with 5 μM BODIPY TR Ceramide (Invitrogen D7540) and 0.1% Pluronic F-127 in Hanks’ buffered salt solution (HBSS, Invitrogen, 14025) for 10 min at 37 °C. After the incubation, cells were washed with PBS solution twice, quickly immersed in 5 μg/mL FM 4-64 HBSS solution (without magnesium or calcium) on ice for 1 minute before imaging.
Cell viability assays (Supplementary Figure 8)
HeLa cells were incubated with organelle-targeted polyynes at specified conditions or illuminated by SRS lasers. Cell viability assays were then performed using Live/Dead viability/cytotoxicity kit for mammalian cells (Invitrogen, L3224) by incubating with 2 μM calcein AM and 4 μM EthD-1 working solution for 20 min at 37 °C.
Spectral barcoding in polystyrene beads with super-multiplexed polyynes
10 μL 3.0 μm polystyrene beads (10% in aqueous solution, Sigma, LB30) were mixed with 5 μL Pluronic F-127 (20% in DMSO) and 85 μL deionized water. Selected polyynes (in DMSO) were diluted to designated concentrations (Supplementary Table 2 and 3) in a solution of 100 μL deionized water and 100 μL THF (Sigma, 401757). The polyyne THF/water solution was then mixed with 100 μL of bead and F-127 solution. After vortexing for 10 minutes, the mixture was further agitated on a shaker for 2 h at room temperature and then washed with deionized water for three times. For live-cell labeling, barcoded beads were incubated in 1 wt% poly-L-lysine aqueous solution (Mw=30000-70000, Sigma, P2636) for 30 min at room temperature and washed with water for three times before use.
Live-cell tagging with spectral barcoded beads
HeLa cells were seeded on a glass coverslip in a 4-well plate with culture media for 24 h. Cell were incubated with barcoded beads for 24 h before spontaneous Raman measurement (Figure 5c) or SRS imaging (Figure 5d and Supplementary Figure 11). A custom MATLAB program was used to decode the spectral barcodes of beads in the whole field of view based on the hyperspectral SRS images.
Code availability
The MATLAB code is available from the corresponding author on request.
Fabrication of microscopic pattern and frequency encryption with super-multiplexed polyynes
Poly(methyl methacrylate) (PMMA, 495K A4 + 950K A9, MicroChem) was spin coated onto Si/SiO2 substrate and baked at 170 °C for 10 min. The substrate was exposed to e-beam lithography and a solution of developer to generate the microscopic pattern of Columbia logo. The polydimethylsiloxane (PDMS) precursor was prepared using Sylgard 184 Silicone Elastomer Kit (Fisher, NC9644388). Elastomer and curing agent (10:1 ratio) were mixed and poured onto the patterned Si/SiO2 substrate. After heating at 80 °C for 1 h, the PDMS pattern was peeled off from the substrate.
Frequency encryption was carried out by immersing the PDMS with Columbia logo in ethanol solution (Fisher, BP2818) with specified polyynes (0.2-1.2 mM) as security codes at room temperature overnight. After ethanol evaporation, the pattern was rinsed with deionized water before imaging (Supplementary Figure 13).
Statistics
For correlation analysis in spectroscopy measurements (Figure 1b and Figure 4a), least-squares regression is applied with Origin software. Reported n values represents the number of compounds (Figure 1b) and concentrations (Figure 4a) measured independently in the experiments. For the immuno-staining and imaging experiments (Figure 4b), experiments were repeated four times independently with similar results. For the 15-color imaging experiments (Figure 4c), experiments were repeated three times independently with similar results. For the live-cell organelle-targeted experiments (Figure 4d-h), experiments were repeated five times independently with similar results. For the 10-color organelle-targeted imaging (Figure 4i), experiments were repeated three times independently with similar results. For the bead labeling in live cells by spontaneous Raman readout (Figure 5c) or SRS imaging (Figure 5d), experiments were repeated three times independently with similar results.
Data availability
The data that support the findings of this study are provided in the Supplementary Information and are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We are grateful for the discussion with L. E. Brus, Y. Shen and Z. Chen. W. M. acknowledges support from NIH Director’s New Innovator Award (1DP2EB016573), R01 (EB020892), the US Army Research Office (W911NF-12-1-0594), and the Camille and Henry Dreyfus Foundation.
Footnotes
Author Contributions
F. H. performed the spectroscopy, microscopy and biological studies and analyzed the data with the help of Y. M., L. W., and Q. X.; C. Z. performed the chemical synthesis together with R. L.; F. H., and W. M. conceived the concept; F. H., C. Z., and W. M. designed the experiments and wrote the manuscript with input from all authors.
Competing Financial Interests
Columbia University has filed a patent application based on this study.
A Life Sciences Reporting Summary is available online.
References
- 1.Dean KM, Palmer AE. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat Chem Biol. 2014;10:512–523. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Valm AM, et al. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature. 2017;546:162–167. doi: 10.1038/nature22369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niehorster T, et al. Multi-target spectrally resolved fluorescence lifetime imaging microscopy. Nat Methods. 2016;13:257–262. doi: 10.1038/nmeth.3740. [DOI] [PubMed] [Google Scholar]
- 4.Krutzik PO, Nolan GP. Fluorescent cell barcoding in flow cytometry allows high-throughput drug screening and signaling profiling. Nat Methods. 2006;3:361–368. doi: 10.1038/nmeth872. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Li YG, Cu YTH, Luo D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat Biotechnol. 2005;23:885–889. doi: 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leng Y, Sun K, Chen X, Li W. Suspension arrays based on nanoparticle-encoded microspheres for high-throughput multiplexed detection. Chem Soc Rev. 2015;44:5552–5595. doi: 10.1039/c4cs00382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zijlstra P, Chon JW, Gu M. Five-dimensional optical recording mediated by surface plasmons in gold nanorods. Nature. 2009;459:410–413. doi: 10.1038/nature08053. [DOI] [PubMed] [Google Scholar]
- 9.Lu YQ, et al. Tunable lifetime multiplexing using luminescent nanocrystals. Nat Photonics. 2014;8:33–37. [Google Scholar]
- 10.Nguyen HQ, et al. Programmable Microfluidic Synthesis of Over One Thousand Uniquely Identifiable Spectral Codes. Adv Opt Mater. 2017;5:1600548. doi: 10.1002/adom.201600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fournier-Bidoz S, et al. Facile and rapid one-step mass preparation of quantum-dot barcodes. Angew Chem Int Ed. 2008;47:5577–5581. doi: 10.1002/anie.200800409. [DOI] [PubMed] [Google Scholar]
- 12.Han M, Gao X, Su JZ, Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 13.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 14.Jin RC, Cao YC, Thaxton CS, Mirkin CA. Glass-bead-based parallel detection of DNA using composite Raman labels. Small. 2006;2:375–380. doi: 10.1002/smll.200500322. [DOI] [PubMed] [Google Scholar]
- 15.Casari CS, Tommasini M, Tykwinski RR, Milani A. Carbon-atom wires: 1-D systems with tunable properties. Nanoscale. 2016;8:4414–4435. doi: 10.1039/c5nr06175j. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch A. The era of carbon allotropes. Nat Mater. 2010;9:868–871. doi: 10.1038/nmat2885. [DOI] [PubMed] [Google Scholar]
- 17.Liu MJ, Artyukhov VI, Lee H, Xu FB, Yakobson BI. Carbyne from First Principles: Chain of C Atoms, a Nanorod or a Nanorope. ACS Nano. 2013;7:10075–10082. doi: 10.1021/nn404177r. [DOI] [PubMed] [Google Scholar]
- 18.Chalifoux WA, Tykwinski RR. Synthesis of polyynes to model the sp-carbon allotrope carbyne. Nat Chem. 2010;2:967–971. doi: 10.1038/nchem.828. [DOI] [PubMed] [Google Scholar]
- 19.Luu T, et al. Synthesis, structure, and nonlinear optical properties of diarylpolyynes. Org Lett. 2005;7:51–54. doi: 10.1021/ol047931q. [DOI] [PubMed] [Google Scholar]
- 20.Milani A, Tommasini M, Del Zoppo M, Castiglioni C, Zerbi G. Carbon nanowires: Phonon and pi-electron confinement. Phys Rev B. 2006;74:153418. [Google Scholar]
- 21.Lucotti A, et al. Absolute Raman intensity measurements and determination of the vibrational second hyperpolarizability of adamantyl endcapped polyynes. J Raman Spectrosc. 2012;43:1293–1298. [Google Scholar]
- 22.Yamakoshi H, et al. Alkyne-Tag Raman Imaging for Visualization of Mobile Small Molecules in Live Cells. J Am Chem Soc. 2012;134:20681–20689. doi: 10.1021/ja308529n. [DOI] [PubMed] [Google Scholar]
- 23.Chen ZX, et al. Multicolor Live-Cell Chemical Imaging by Isotopically Edited Alkyne Vibrational Palette. J Am Chem Soc. 2014;136:8027–8033. doi: 10.1021/ja502706q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, et al. Multiplexed multicolor Raman imaging of live cells with isotopically modified single walled carbon nanotubes. J Am Chem Soc. 2008;130:13540–13541. doi: 10.1021/ja806242t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lucotti A, et al. Evidence for Solution-State Nonlinearity of sp-Carbon Chains Based on IR and Raman Spectroscopy: Violation of Mutual Exclusion. J Am Chem Soc. 2009;131:4239–4244. doi: 10.1021/ja078198b. [DOI] [PubMed] [Google Scholar]
- 26.Wei L, et al. Super-multiplex vibrational imaging. Nature. 2017;544:465–470. doi: 10.1038/nature22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freudiger CW, et al. Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science. 2008;322:1857–1861. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei L, et al. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat Methods. 2014;11:410–412. doi: 10.1038/nmeth.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamakoshi H, et al. A sensitive and specific Raman probe based on bisarylbutadiyne for live cell imaging of mitochondria. Bioorg Med Chem Lett. 2015;25:664–667. doi: 10.1016/j.bmcl.2014.11.080. [DOI] [PubMed] [Google Scholar]
- 30.Wilson R, Cossins AR, Spiller DG. Encoded microcarriers for high-throughput multiplexed detection. Angew Chem Int Ed. 2006;45:6104–6117. doi: 10.1002/anie.200600288. [DOI] [PubMed] [Google Scholar]
- 31.Lee JH, Gomez IJ, Sitterle VB, Meredith JC. Dye-labeled polystyrene latex microspheres prepared via a combined swelling-diffusion technique. J Colloid Interface Sci. 2011;363:137–144. doi: 10.1016/j.jcis.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 32.Humar M, Yun SH. Intracellular microlasers. Nat Photonics. 2015;9:572–576. doi: 10.1038/nphoton.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agarwal NR, et al. Structure and chain polarization of long polyynes investigated with infrared and Raman spectroscopy. J Raman Spectrosc. 2013;44:1398–1410. [Google Scholar]
- 34.Ozeki Y, et al. High-speed molecular spectral imaging of tissue with stimulated Raman scattering. Nat Photonics. 2012;6:844–850. [Google Scholar]
- 35.Liao CS, et al. Microsecond scale vibrational spectroscopic imaging by multiplex stimulated Raman scattering microscopy. Light Sci Appl. 2015;4:e265. doi: 10.1038/lsa.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C, et al. Stimulated Raman scattering flow cytometry for label-free single-particle analysis. Optica. 2017;4:103–109. doi: 10.1364/optica.4.000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are provided in the Supplementary Information and are available from the corresponding author upon request.


