Abstract
Three dimensional (3D) printing is highly amenable to the fabrication of tissue-engineered organs of a repetitive microstructure such as the liver. The creation of uniform and geometrically repetitive tissue scaffolds can also allow for the control over cellular aggregation and nutrient diffusion. However, the effect of differing geometries, while controlling for pore size, has yet to be investigated in the context of hepatocyte function. In this study, we show the ability to precisely control pore geometry of 3D-printed gelatin scaffolds. An undifferentiated hepatocyte cell line (HUH7) demonstrated high viability and proliferation when seeded on 3D-printed scaffolds of two different geometries. However, hepatocyte specific functions (albumin secretion, CYP activity, and bile transport) increases in more interconnected 3D-printed gelatin cultures compared to a less interconnected geometry and to 2D controls. Additionally, we also illustrate the disparity between gene expression and protein function in simple 2D culture modes, and that recreation of a physiologically mimetic 3D environment is necessary to induce both expression and function of cultured hepatocytes.
Keywords: Liver, Tissue Engineering, 3D Printing, Scaffold Geometry
Graphical abstract
1. Introduction
Currently the only viable treatment for end stage liver disease is orthotropic liver transplantation. Transplantation is unfortunately severely limited by the deficit of available donor organs [1]. Temporary transplantation alternatives include extracorporeal liver assist devices, whereas tissue engineered livers offer a permanent solution. An exceptional variety of approaches to liver tissue engineering have been developed over the past several decades with a minority of these approaches leading to clinical results [2]. Nevertheless, development of new tissue engineering strategies continually provides useful insights into in vitro tissue generation, liver biology, and cell-material interactions. For example, the in vitro proliferative capacity and phenotypic stability (and therefore translatable potential) of primary isolated hepatocytes is severely limited, leading researchers to utilize model systems such as cell lines, hepatoblasts, or more translatable sources such as iPS-derived hepatocytes [3, 4]. Regardless of the hepatocyte source in question, an aspect of liver tissue engineering determined early on to be critical for increasing the long-term viability, function, and phenotypic stability of hepatocytes is aggregation [5]. The importance of aggregation has led to the development of a variety of methods to induce the formation and maintenance of aggregates, with the predominant strategy being to form the aggregates in some external system and then embed the aggregates (also termed hepatospheres, spheroids, etc.) within a tissue engineering scaffold or implant them directly in vivo [6, 7]. Approaches to encourage aggregation based on traditional tissue engineering approaches include encapsulation or top-seeding single cell suspensions onto a porous scaffold [8, 9]. Scaffold design must be tailored in a number of ways to, for example limit aggregate size to prevent necrotic core formation or to encourage interconnectivity between adjacent aggregates to facilitate nutrient diffusion and vascularization upon in vivo implantation [6, 10]. Modulation of aggregation within large three dimensional (3D) scaffolds is tied to the limitations inherent in scaffold fabrication strategies such as electrospinning, freeze casting, salt leaching, or gas foaming [11–15]. In these methods, only a loose degree of control over scaffold pore size, geometry, and interconnectivity is possible.
Recent advances in 3D-printing and additive manufacturing technology have expanded into the field of tissue engineering [16]. 3D printing holds several advantages over traditional scaffold fabrication techniques including uniform and reproducible manufacture, reduction of user error, and precise control over scaffold pore size, interconnectivity, and geometry [17]. Hexagonal lobule-like geometries have been shown to have beneficial effects on cultured hepatocytes [18, 19]. While the lobule organization in vivo is known to have biological significances in terms of blood and bile flow as well hepatocyte phenotype and zonality, this effect is largely attributed to gradients in nutrient concentration [20]. Utilization of an engineered system to recreate precise lobule structure would therefore need to demonstrate the significance of said organization and its advantages over other organizations that may have other practical or biological advantages.
The effects of 3D-printed scaffold pore geometry have therefore not been rigorously investigated for tissues outside the differentiation capacity of mesenchymal stem cells or for applications in soft tissue engineering. This may in part be due to the difficulties in 3D printing of soft materials, mainly hydrogels. We have previously reported the development of an extrusion-based gelatin 3D-printing platform that has led to the restoration of fertility and increased survivability and function of seeded mouse ovarian follicles [21]. We sought to extend this investigation by studying the influence of scaffold pore geometry on a seeded suspension of cells while keeping pore size constant and comparing to a 2D surface of an identical material. We employ HUH7 human hepatocellular carcinoma cell line as a model hepatocyte system. Primary isolated hepatocytes rapidly de-differentiate and lose proliferation capacity in-vitro, limiting their applications in tissue engineering [22]. HUH7 cells serve as a model for other proliferative hepatocyte-like cells, such as hepatoblasts, hepatic stem cells, or iPS-derived hepatocytes [4, 23]. HUH7 cells also show a relatively un-differentiated phenotype when cultured on traditional 2D tissue culture plastic, but begin to differentiate when in aggregates in more biomimetic environments [24]. We demonstrate that 3D manipulation of a relatively simple material, such a gelatin, can have significant effects on downstream tissue engineering applications.
2. Materials and Methods
2.1. Scaffold Fabrication
3D-printed gelatin scaffolds were fabricated as previously described [21]. Briefly, a 10% (w/v) gelatin type A (Sigma) solution was 3D-printed using the pneumatic extrusion piston-driven EnvisionTEC (GMBH) 3D-Bioplotter. The gelatin solution was maintained at a constant temperature of 30°C and extruded on to glass slides cooled to 10°C. The nozzle diameter, strut spacing, and layer slicing were all held constant at 200 μm, 700 μm, and 156 μm respectively, leading to a constant scaffold pore size. The strut angle of orientation between subsequent layers was either 90° or 60° leading to strut angles alternating between 0° and 90° or in increasing increments of 60° (60°, 120°, 180°, 240°, 300°, 360°) (Figure 1A). Scaffolds were printed 6 layers high with footprints of 15mm × 15mm boxes. To stabilize for long-term culture, printed scaffolds were then cross-linked in a solution of 15mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and 6mM N-hydroxysuccinimide (NHS) (both Sigma) for 1 hour followed by several washes in PBS to remove residual cross-linker. Scaffolds were sterilized in the presence of 70% ethanol and UV irradiation. Prior to cell seeding, scaffolds were rinsed several times in PBS to remove residual ethanol. Scaffolds were removed from glass slides using sterile blades and biopsy punched using 6mm biopsy punches (Miltex) to create circular scaffold disks (Figure 1B, C).
Figure 1.
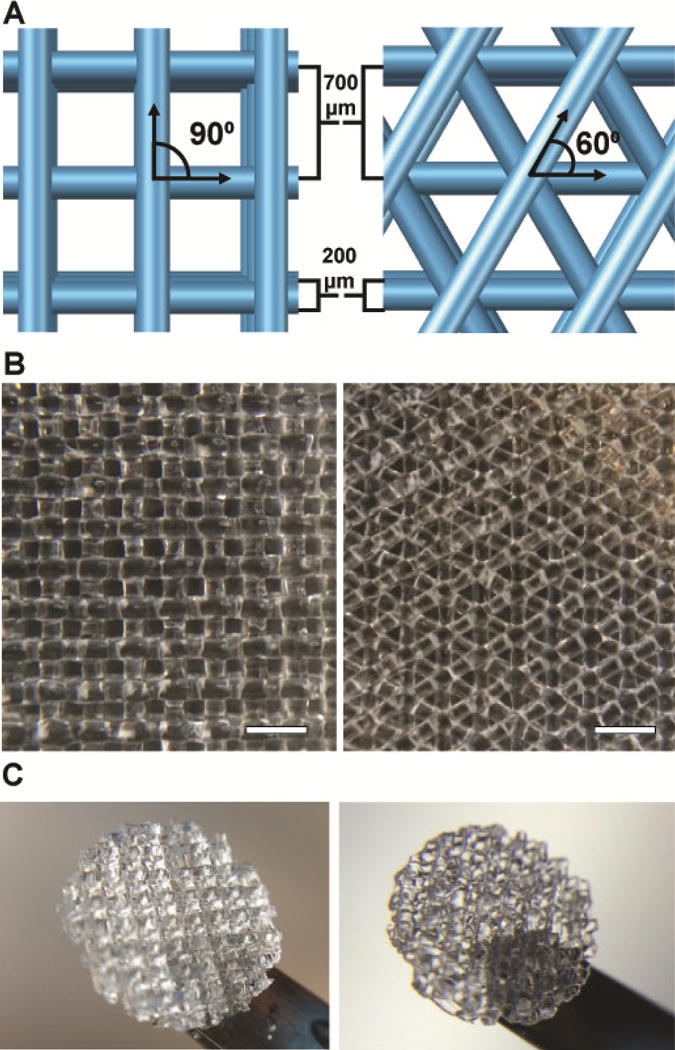
A. Schematic of 3D printed structure with strut spacing (700 μm), nozzle diameter (200 μm) and angle between adjacent layers (90 or 60 degrees). B. Large 3D printed gelatin structure. Scale bars=1mm. C. Cross-linked and biopsy punched scaffolds. Biopsy punches are 6mm in diameter.
2.2. Cell Culture and Seeding
HUH7 human hepatocellular carcinoma cell line was expanded in DMEM (Gibco) supplemented with 10% fetal bovine serum (Gibco) and penicillin/streptomycin antibiotic antimitotic solution (Gibco). Cells were trypsinized using 0.05% Trypsin/EDTA (Gibco) and concentrated to a suspension of 3.3×106 cells/mL. Scaffolds were placed into non-adherent environments and HUH7 cells (passage 8) were seeded in 30 μL volumes, allotting approximately 0.1×106 cells/scaffold. In order to encourage cell adhesion, seeded scaffolds were allowed to incubate for 1 hour prior to the addition of additional culture medium. An equivalent amount of cells were plated into 6-well plates gelatin-coated and EDC/NHS-cross linked as 2D control. Cells were cultured at 37°C at 90% humidity and 5% CO2 atmosphere, with media changed every 2 days.
2.3. DNA Quantification
Scaffolds were collected 1, 3, 7, 10, and 14 (n=3) days after seeding, 2D controls were trypsinized and frozen (−80°C) until analysis. Scaffolds (cell pellets for 2D) were digested using a solution of 1 μg/mL proteinase K (Sigma) in a 60°C for 24 hours. Digests were assayed using PicoGreen dsDNA Quantification Kit (Molecular Probes) according to manufacturer specifications. Fluorescence was read on a Cytation3 Automated Microscope Plate Reader (BioTek). Percent seeding efficiency was determined by dividing total DNA content of day 1 (24 hr) samples by the DNA content of an aliquot of 0.1×106 cells. Cells per scaffold were determined by normalizing DNA content per scaffold to DNA content per cell.
2.4. Viability Staining and Imaging
Scaffolds and 2D controls were analyzed for viability on days 1, 3, 7, 10, and 14. Samples were briefly washed using warmed PBS and incubated for 30 minutes in Live/Dead (calcein AM/ethidium homodimer) solution (Molecular Probes). Live samples were then immediately imaged using a Nikon C2+ confocal laser scanning microscope. To facilitate thick construct imaging, laser power was increased linearly with the depth of focal plane.
2.5. Gene Expression Analysis
Collected scaffolds (n=3) were homogenized using TRIzol (Life Technologies) reagent and subjected to RNA isolation as described in [25]. Total RNA was converted to cDNA using qScript cDNA synthesis kit (Quanta Bioscience). Quantitative PCR (qPCR) was performed using the QuantiTect SYBR Green PCR kit (Qiagen) with an Applied Biosystems Prism 7300 Sequence Detection System (Applied Biosystems). Relative expression for the particular genes of interest were calculated using the ddCt method with cyclophilin as a reference gene. Primer sequences (IDT) for target genes are listed in Table 1.
Table 1.
Primers for qPCR gene expression analysis. Cyclophilin was used as an endogenous control.
| Gene | Primer Sequence (5′-3′) |
|---|---|
| Cyclophilin-F | CACAGGAGGAAAGAGCATCTAC |
| Cyclophilin-R | CACAGACGGTCACTCAAAGAA |
| Albumin-F | AGCATGGGCAGTAGCTCGCCT |
| Albumin-R | AGGTCCGCCCTGTCATCAGCA |
| CYP2C9-F | GGATTTGCCTCTGTGCCGCC |
| CYP2C9-R | GCAGCCAGGCCATCTGCTCTT |
| CYP3A4-F | CTGCTTCTCACGGGACTATTT |
| CYP3A4-R | CCTCCCAAACTGCTAGGATTAC |
| SLCO1B3-F | TGGAGCAACAGTACGGTCAG |
| SLCO1B3-R | TGCTTTCGCAGATTAGAGGGAA |
| ABCC2-F | CCCTGCTGTTCGATATACCAATC |
2.6. Albumin Secretion and Quantification
Culture media was collected after every change (n=3), diluted 1:10 in DPBS, and quantified according to manufacturer specifications using the human Albumin ELISA kit (Abcam). Absorbance was read on a Cytation3 Automated Microscope Plate Reader (BioTek). The Human Albumin ELISA has cross-reactivity with bovine albumin, and the albumin content of DMEM + 10% FBS was subtracted from all quantities. Cumulative albumin secretion was determined by summing all quantities and normalizing to cell number which was determined via dsDNA quantification described above.
2.7. Cytochrome P450 Oxidase Activity
Activities of cytochrome P450 oxidase (CYP) subtypes 3A4 and 2C9 were quantified using Luciferin-IPA or Luciferin-H substrates, respectively (Promega). Scaffolds (n=3 per enzyme test) were incubated in enzyme substrate for 60 minutes (Luciferin-IPA) or 4 hours (Luciferin-H). Samples were subjected to the lytic protocol, wherein samples (as opposed to supernatant) are incubated with the Luciferin detection reagent. Lysates were assayed for luminescence on Cytation3 Automated Microscope Plate Reader (BioTek). All samples were first normalized to cell number, then to Day 0 activity of tissue culture plastic cultured cells.
2.8. Bile Canaliculus Imaging
Structural organization and formation of bile canaliculi between hepatocytes was assayed via cholyl-lysyl-fluorescein (CLF) (Corning) dye localization. Scaffolds (or 2D controls) were first rinsed in warm HBSS (HyClone). Samples were then incubated in a solution of 5 μM CLF in HBSS for 30 minutes to allow for cellular uptake of the dye. This was followed by a brief wash and subsequent incubation in HBSS and Hoechst 33342 (Life Technologies) for 30 minutes to allow for canalicular localization of the CLF. Samples were then imaged using a Nikon C2+ confocal laser scanning microscope. Individual canaliculi were quantified using the 3D visualization and analysis software Imaris (Bitplane) using representative images of scaffold pores (n=5). Canaliculi within random scan areas of 2D controls were quantified as a comparison.
2.9. Statistical Analysis
Paired, 2-tailed Student’s T-tests were performed to determine statistical significance.
3. Results
3.1. Seeding Efficiency, Proliferation, and Viability
Initial seeding efficiency was not determined to be significantly different between two scaffolds (Figure 2A), despite the more intertwined struts seen in the 60° scaffolds, although pore tortuosity has been known to play a role in seeding efficiency [26]. The proliferative nature of HUH7 cells caused seeding efficiency values for 2D controls to appear greater than 100% because samples taken 24 hours post-seeding were used in this calculation. Proliferation across time points up to 14 days was also determined to be relatively uniform, with the exception of 2D controls (Figure 2B). 3D samples demonstrated similar viability to 2D samples throughout the culture period (Figure 3A). Initial seeded scaffolds showed a larger concentration of cells at the scaffold top surface. After 7 days, 2D controls began to demonstrate hyperconfluency, small aggregates began protruding from the 2D surface whereas 3D scaffolds showed an even distribution of cells throughout (Figure 3B). After 14 days, proliferation on seeded scaffolds began to hinder adequate viability imaging while proliferation in 2D samples was effectively halted by this time.
Figure 2.
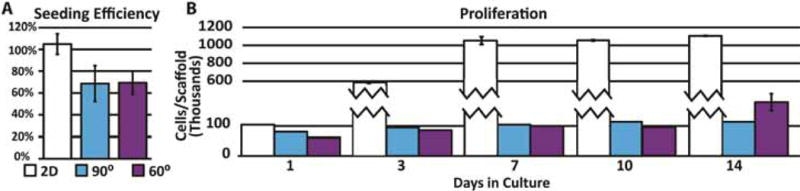
A. Percent seeding efficiency of cell suspensions. Greater than 100% values indicate growth in 2D well plate controls after seeding and before 24 hour sample harvest. B. Proliferation of hepatocytes assessed via DNA quantification. Error bars ± SD.
Figure 3.
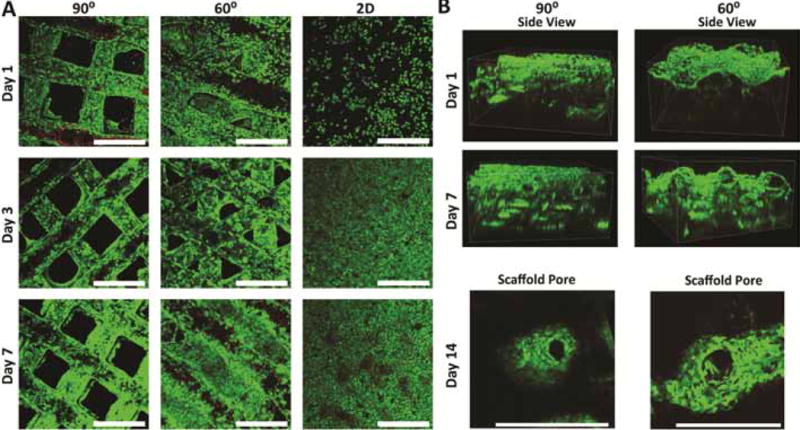
A. Viability and morphology of seeded structures over 7 days. Samples visualized with Live/Dead (green/red) assay and confocal microscopy (10X). B. Side views of scaffolds after 1 and 7 days of culture and higher magnification (20X) of pore aggregation after 14 days. Note the relatively uniform cellular distribution on day 7. Scale bars=500μm.
3.2. Albumin Secretion
Expression of albumin in both 3D printed scaffolds was determined to be significantly less than that of 2D controls on day 1 (Figure 4A). However, by day 7, 60° scaffolds began to show increased amounts of albumin expression, while 90° scaffolds remained inferior to both 60° and 2D controls. Measurements of cumulative albumin secretion indicate 60° scaffolds lead to a higher albumin secretion on a per-cell basis, having secreted approximately 40 picograms per cell more than 90° scaffolds (Figure 4B).
Figure 4.
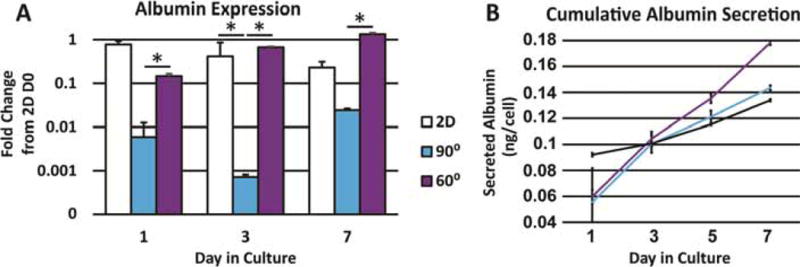
A. Albumin expression at different time points in culture (n=3). B. Cumulative albumin secretion over 7 days normalized to number of cells. Error bars ± SD. * p≤0.05
3.3. Cytochrome P450 Oxidase Activity
Higher expression levels of CYP3A4 were measured in 2D controls (Figure 5) compared to the 3D groups, however, this was not reflected in activity assays, where CYP3A4 activity was significantly less in 2D controls compared to both 3D-printed geometries. Activity and expression of CYP2C9, however, were directly correlated with one another, with 60° scaffolds showing the highest activity and expression and 90° scaffolds and 2D controls showing less activity and expression.
Figure 5.
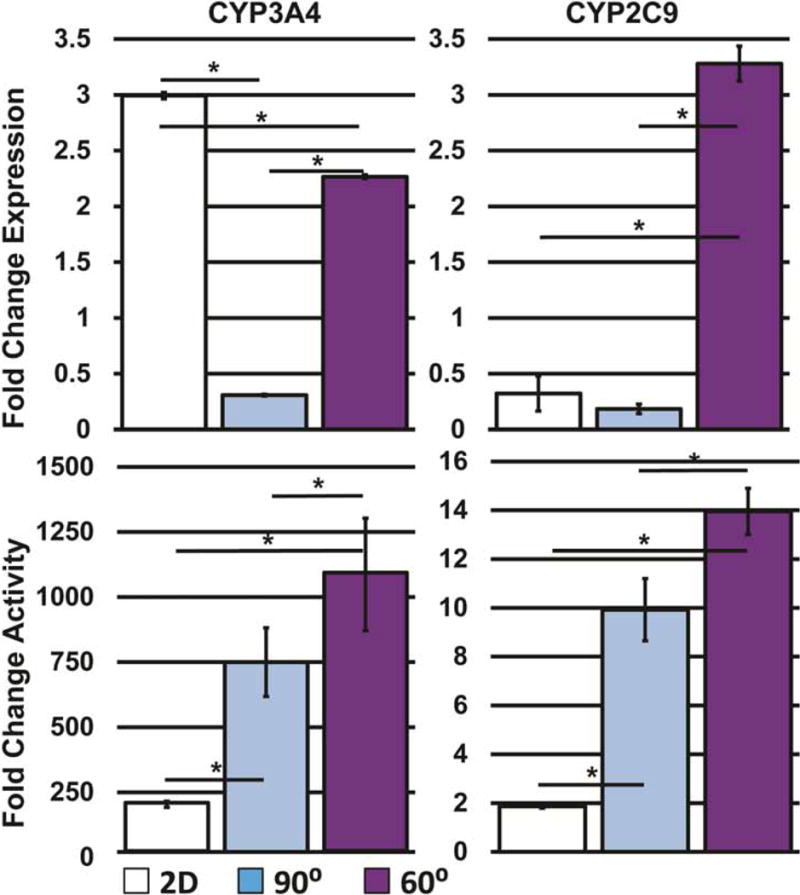
Comparison of expression of cytochrome P450 oxidase enzymes and their respective activities (n=3). Error bars ± SD. * p≤0.05
3.4. Bile Salt Transporter Expression and Function
Expression of uptake channel SCLO1B3 (OATP1B3), normally positioned basolaterally in polarized, differentiated hepatocytes, was significantly higher in the 2D control group compared to the 3D groups, but showed no significant difference between the 60° and 90° scaffold conditions (Figure 6C). Export channel ABCC2 (MRP2), positioned apically in the canalicular space of polarized hepatocytes, showed significantly higher expression in 2D controls compared to either 3D scaffold. However, imaging of samples incubated in CLF indicates an increased amount of canaliculus formation in 3D scaffolds, as is evident by the higher number of green puncta, compared to random scan areas of 2D controls (Figure 6A, B).
Figure 6.
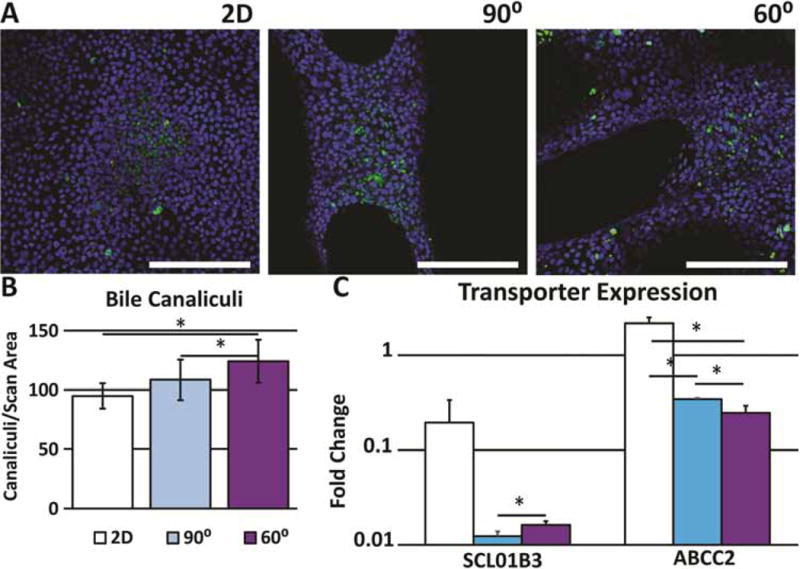
A. Localization of CLF in bile canalicular interior 2D scaffold and representative pores. Green=CLF (canaliculi), Blue=Hoechst 33342 (nuclei) (20X). B. Quantification of the number of canaliculi per scaffold pore and random scan area of 2D controls (n=5). C. Expression of CLF importer (SLCO1B3) and exporter (ABCC2). Error bars ± SD, Scale bars=250μm. * p≤0.05
4. Discussion
Within the past several years, strides in tissue engineering have made tremendous progress, particularly in the realm of 3D printing. The custom, computer-aided design of scaffolds allows for the 3D patterning of cells and materials in progressively more precise and biologically relevant organizations. However, precise placement of each facet of a tissue would take a massive amount of time to complete a single organ, and the necessity to incorporate such details is still unclear. The prevailing strategy is rather to pattern cells and materials in a manner that will guide rapid and functional tissue morphogenesis, such as seeding endothelial cells within cast vascular networks [27, 28]. Effects of 3D-printed scaffold geometry have been investigated in the context of metastasis [29], adult mesenchymal stem cell differentiation [30, 31], bone tissue engineering [32, 33], and cardiac muscle tissue engineering [34, 35]. While there have been numerous examples of liver tissue engineering using 3D printing or additive manufacturing technologies, the vast majority of approaches only seek to replicate a hexagonal (lobule like) motif [18, 19, 36]. However, there have been few investigations into the significance scaffold geometry plays in a liver tissue engineering setting.
In this study, we demonstrated differential gene expression and function of a proliferative human hepatocyte model cell line seeded within 3D-printed gelatin scaffolds of differing geometry. While 3D printing systems have the capacity to vary a near-infinite amount of scaffold architectural features, we chose to focus on a single variable’s effects on multiple biological outcomes. Varying the angle of orientation between adjacent layers was tested because this varies the pore geometry of the scaffold while keeping pore size constant. 3D-printing gelatin into well-defined structures demonstrated here and by Laronda & Rutz et al is only possible due to precise temperature maintenance during printing [21]. Other materials systems such as alginate [37] are able to achieve similar or greater shape fidelity, however most reports of gelatin 3D-printing show low shape fidelity with rough strut surfaces [38]. Without improved shape fidelity, studies into the biological effects of pore geometry would not be possible. Such effects are explored in this study by seeding a suspension of a relatively undifferentiated hepatocyte cell line, which has been shown in the past to respond positively to 3D culture and aggregation [24]. We first demonstrate that our approach is able to reliably seed an equal amount of cells per scaffold, despite changes in interconnectivity as a result of differing pore geometry. The consistent quantity of cells within the 3D scaffolds allows us to conclude that any subsequent changes in gene expression or function are in fact due to changes in geometry and not due to simply an increase in cell number. The lack of proliferation in 3D scaffolds, compared to 2D controls, is hypothesized to be an effect of cellular crowding. While a 3D printed scaffold may be porous and have a large surface area, the proximity between separate surfaces (struts) is small (~400 μm) enough to prevent massive hyperconfluence.
Viability staining visualizes cell survival, but also provides insights on how cells aggregate on the different geometries. This crowding and aggregation leads to a stable cell number, but also leads to increased functional output, such as albumin secretion. Albumin is a critical component of human blood and a ubiquitous marker for hepatocyte maturity. We speculate the initial higher albumin secretion and expression in 2D samples is an effect of the higher seeding efficiency, as samples seeded on 3D scaffolds have not proliferated enough for cells to come into close contact with one another. As HUH7 cells proliferate, there is an upper limit of cell-cell contact formation in 2D scaffolds, while 3D scaffolds encourage aggregation and cell-cell contact in 3D, which has led to an increase in functional response. The increase in albumin expression and secretion in 60° scaffolds is an indication that an increase in pore interconnectivity may lead to an increase in translation of liver-specific genes [5]. This supports previous researchers’ data associating smaller foam pore size with increased albumin secretion [15]. Xenobiotic metabolism gene CYP 3A4 is responsible for detoxification of the vast majority of chemicals to which humans are exposed, including many clinically relevant drugs [39]. CYP 2C9 in contrast is important in drug-drug interactions and the metabolism of drugs with low therapeutic indices, such as warfarin [40]. Function and expression of most cytochrome P450 enzymes is evidence for hepatocyte maturation. Activity of CYP3A4 and CYP2C9 is increased in 60° geometry scaffolds, evidence that scaffold architecture can significantly affect hepatocyte function [41]. When we assay for expression of CYP enzymes, we see a large discrepancy between expression and activity of CYP3A4 in 2D controls. This may be an indication that cell aggregation and presenting a 3D structure are important for adequate activity of CYP3A4. CYP enzymes are localized to the endoplasmic reticulum whose 3D organization is intimately tied to the cell’s polarization in vivo [39]. The reasoning as to why an identical disparity is not observed in the expression/activity relationship of CYP2C9 is unclear, but may indicate CYP2C9 expression is not as sensitive to HUH7 confluence as CYP3A4 expression is [42]. Biologists have known for some time that transcription does not necessarily imply translation, and this experiment may be a manifestation of that phenomenon. This may also be a manifestation of the HUH7 cell line’s sensitivity to 2D versus 3D culture where 3D culture may also influence protein and intracellular membrane trafficking in addition to gene expression [24]. We further explored whether changes in 3D-printed scaffold geometry can influence cellular structure by assessing polarization. Cholyl-lysyl-fluorescein (CLF) is a fluorescent bile salt used to determine in vivo liver function as well as to assess function of certain transporters [43]. In hepatocytes, CLF is taken up by the organic anion transporter protein 1B3 (OATP1B3, also known as SLCO1B3) and expelled into the canalicular space by the multidrug resistance-associated protein 2 (MRP2, also known as ABCC2) [44]. In physiologic conditions, OATP1B3 is positioned basolaterally in contact with blood while MRP2 is positioned apically in the canalicular membrane. Previous research indicates a complete lack of canalicular localization of CLF in 2D HepG2 cultures, with dramatically increased localization in HepG2 aggregates [41]. Our experiments indicate a significant difference in expression between different scaffold geometries in addition to higher expression in 2D controls. However, image quantification of canalicular localization of CLF within scaffold pores indicates an increase in formation of canalicular spaces in 3D scaffolds compared to 2D cultures, with 60° scaffolds again demonstrating the highest amount of function. This is additional evidence for the discrepancy between expression and function, and is likely another manifestation of the sensitivity of HUH7 cells and hepatocytes in general to 3D culture. The increased CLF localization quantified in multiple pores combined with the homogeneity of scaffold seeding and cell aggregation, especially after 7 days (Figure 3), is indication that pore-to-pore heterogeneity in terms of function and/or gene expression is unlikely. While some researchers have conducted similar experiments using chitosan-coated alginate as opposed to gelatin, and HepaRG cells as opposed to HUH7, changes in expression and function were not observed with scaffolds of differing geometries [37]. We hypothesize this difference in observation is more likely due to the difference in cell type since HUH7 cells are far more proliferative than HepaRG and have a greater tendency to aggregate in scaffold pores in addition to spreading along scaffold struts. This is evident in Figure 2, where HUH7 cells even in 2D culture will begin to aggregate at 7 days, while after 14 days cell aggregation in 3D begins to fill in scaffold pores. Because of this, geometry may play a more important role for cells with a higher tendency for aggregation. However, we cannot completely rule out the role the difference in biomaterial may play in these observations.
5. Conclusion
We have demonstrated that scaffold geometry, using well-defined 3D-printed gelatin constructs, significantly influences the function of seeded hepatocytes. Functional tests demonstrated here indicate the increased function of the 60° scaffolds with regard to albumin secretion, cytochrome p450 oxidase activity, and bile salt transport activity. HUH7 cells in this study can serve as a model for proliferative and undifferentiated forms of hepatocytes, such as hepatoblasts or iPS-derived hepatocytes, which have proven difficult to mature in vitro [45]. Future investigations are necessary to optimize scaffold design especially when incorporating multiple cell types, which is a requirement for organ tissue engineering. Furthermore, organizing multiple cell types within three dimensions to encourage complex tissue formation (including vascular structures) will require much more sophisticated 3D designs that incorporate several different biomaterials and signaling factors.
Statement of Significance.
Three dimensional (3D) printing provides tissue engineers the ability spatially pattern cells and materials in precise geometries, however the biological effects of scaffold geometry on soft tissues such as the liver have not been rigorously investigated. In this manuscript, we describe a method to 3D print gelatin into well-defined repetitive geometries that show clear differences in biological effects on seeded hepatocytes. We show that a relatively simple and widely used biomaterial, such as gelatin, can significantly modulate biological processes when fabricated into specific 3D geometries. Furthermore, this study expands upon past research into hepatocyte aggregation by demonstrating how it can be manipulated to enhance protein function, and how function and expression may not precisely correlate in 2D models.
Acknowledgments
The authors would like to thank Dr. Alexandra Rutz for the development and optimization of gelatin printing protocols. Dr. Jason Wertheim and his laboratory provided human liver qPCR primer sequences, Green lab members Dr. Xiaoying Liu and Brian LeCuyer provided additional guidance with PCR. Imaging work was performed at the Northwestern University Center for Advanced Microscopy generously supported by NCI CCSG P30 CA060553 awarded to the Robert H Lurie Comprehensive Cancer Center. Cytation 3 Plate reader usage was performed in the Analytical BioNanoTechnology Core Facility of the Simpson Querrey Institute (SQI) at Northwestern University. The U.S. Army Research Office, the U.S. Army Medical Research and Materiel Command, and Northwestern University provided funding to develop SQI and ongoing support is being received from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF NNCI-1542205).
Funding Sources
This work was supported by the National Institutes of Health grant number 1K01DK099454 and Northwestern University’s Biotechnology Training Program Cluster Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
P.L.L.: Designed experiments, optimized 3D printing methods, collected and analyzed data, wrote the manuscript, and designed figures. R.M.G.: Provided HUH7 cells, assisted in experimental design, interpretation of data, manuscript writing and editing. R.N.S.: Principle investigator, assisted in experimental design, methods optimization, interpretation of data, manuscript writing and editing.
Disclosure Statement
R.N.S. has financial interests in Dimension Inx, LLC which could potentially benefit from the outcomes of this research.
References
- 1.Vernon G, Baranova A, Younossi ZM. Systematic review: The epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011;34(3):274–285. doi: 10.1111/j.1365-2036.2011.04724.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia SN, Underhill GH, Zaret KS, Fox IJ. Cell and tissue engineering for liver disease. Sci Transl Med. 2014;6(245):245sr2. doi: 10.1126/scitranslmed.3005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng N, Wauthier E, Reid LM. Mature Human Hepatocytes from Ex Vivo Differentiation of Alginate-Encapsulated Hepatoblasts. Tissue Eng Part A. 2008;14(1):1–7. doi: 10.1089/ten.a.2007.0131. [DOI] [PubMed] [Google Scholar]
- 4.Wang B, Jakus AE, Baptista PM, Soker S, Soto-Gutierrez A, Abecassis MM, Shah RN, Wertheim JA. Functional maturation of induced pluripotent stem cell hepatocytes in extracellular matrix - a comparative analysis of bioartificial liver microenvironments. Stem Cells Transl Med. 2016;5:1257–1267. doi: 10.5966/sctm.2015-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Zijl F, Mikulits W. Hepatospheres: Three dimensional cell cultures resemble physiological conditions of the liver. World J Hepatol. 2010;2(1):1–7. doi: 10.4254/wjh.v2.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J. 2008;3(9–10):1172–1184. doi: 10.1002/biot.200700228. [DOI] [PubMed] [Google Scholar]
- 7.Li CY, Stevens KR, Schwartz RE, Alejandro BS, Huang JH, Bhatia SN. Micropatterned Cell-Cell Interactions Enable Functional Encapsulation of Primary Hepatocytes in Hydrogel Microtissues. Tissue Eng Part A. 2014 Apr;(617) doi: 10.1089/ten.tea.2013.0667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Underhill GH, Chen Aa, Albrecht DR, Bhatia SN. Assessment of hepatocellular function within PEG hydrogels. Biomaterials. 2007;28:256–270. doi: 10.1016/j.biomaterials.2006.08.043. [DOI] [PubMed] [Google Scholar]
- 9.Glicklis R, Shapiro L, Agbaria R, Merchuk JC, Cohen S. Hepatocyte Behavior Within Three-Dimensional Porous Alginate Scaffolds. 2000 doi: 10.1002/(sici)1097-0290(20000205)67:3<344::aid-bit11>3.0.co;2-2. no. M. [DOI] [PubMed] [Google Scholar]
- 10.Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, Solorzano RD, Yang MT, Miller JS, Bhatia SN, Chen CS. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc Natl Acad Sci U S A. 2013;110(19):7586–91. doi: 10.1073/pnas.1217796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lien SM, Ko LY, Huang TJ. Effect of pore size on ECM secretion and cell growth in gelatin scaffold for articular cartilage tissue engineering. Acta Biomater. 2009;5(2):670–679. doi: 10.1016/j.actbio.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Yang S-T. Effects of three-dimensional scaffolds on cell organization and tissue development. Biotechnol Bioprocess Eng. 2001;6(5):311–325. [Google Scholar]
- 13.Lowery JL, Datta N, Rutledge GC. Effect of fiber diameter, pore size and seeding method on growth of human dermal fibroblasts in electrospun poly(ε-caprolactone) fibrous mats. Biomaterials. 2010;31(3):491–504. doi: 10.1016/j.biomaterials.2009.09.072. [DOI] [PubMed] [Google Scholar]
- 14.Lee SB, Kim YH, Chong MS, Hong SH, Lee YM. Study of gelatin-containing artificial skin V: Fabrication of gelatin scaffolds using a salt-leaching method. Biomaterials. 2005;26(14):1961–1968. doi: 10.1016/j.biomaterials.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 15.Ranucci CS, Kumar A, Batra SP, Moghe PV. Control of hepatocyte function on collagen foams: Sizing matrix pores toward selective induction of 2-D and 3-D cellular morphogenesis. Biomaterials. 2000;21(8):783–793. doi: 10.1016/s0142-9612(99)00238-0. [DOI] [PubMed] [Google Scholar]
- 16.Murphy SV, Atala A. 3D bioprinting of tissues and organs. Nat Biotechnol. 2014 Aug;32(8):773–785. doi: 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 17.Gariboldi MI, Best SM. Effect of Ceramic Scaffold Architectural Parameters on Biological Response. Front Bioeng Biotechnol. 2015;3:1–11. doi: 10.3389/fbioe.2015.00151. no. October. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Tsang V, Chen Aa, Cho LM, Jadin KD, Sah RL, DeLong S, West JL, Bhatia SN. Fabrication of 3D hepatic tissues by additive photopatterning of cellular hydrogels. FASEB J. 2007 Mar;21(3):790–801. doi: 10.1096/fj.06-7117com. [DOI] [PubMed] [Google Scholar]
- 19.Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, Liu J, Wang P, Lai CSE, Zanella F, Feng G-S, Sheikh F, Chien S, Chen S. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci. 2016;113(8):2206–2211. doi: 10.1073/pnas.1524510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hailfinger S, Jaworski M, Braeuning A, Buchmann A, Schwarz M. Zonal gene expression in murine liver: Lessons from tumors. Hepatology. 2006;43(3):407–414. doi: 10.1002/hep.21082. [DOI] [PubMed] [Google Scholar]
- 21.Laronda MM, Rutz AL, Xiao S, Whelan KA, Duncan FE, Roth EW, Woodruff TK, Shah RN. A bioprosthetic ovary created using 3D printed microporous scaffolds restores ovarian function in sterilized mice. Nat Commun. 2017;8:1–10. doi: 10.1038/ncomms15261. no. May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffith LG, Wells A, Stolz DB. Engineering Liver. Hepatology. 2014;3 doi: 10.1002/hep.27150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SY, Kim HJ, Choi D. Cell sources, liver support systems and liver tissue engineering: Alternatives to liver transplantation. Int J Stem Cells. 2015;8(1):36–47. doi: 10.15283/ijsc.2015.8.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sainz B, TenCate V, Uprichard SL. Three-dimensional Huh7 cell culture system for the study of Hepatitis C virus infection. Virol J. 2009;6(1):103. doi: 10.1186/1743-422X-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Henkel AS, LeCuyer BE, Schipma MJ, Anderson KA, Green RM. Hepatocyte X-box binding protein 1 deficiency increases liver injury in mice fed a high-fat/sugar diet. Am J Physiol Gastrointest Liver Physiol. 2015;309(12):G965–74. doi: 10.1152/ajpgi.00132.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobral JM, Caridade SG, Sousa RA, Mano JF, Reis RL. Three-dimensional plotted scaffolds with controlled pore size gradients: Effect of scaffold geometry on mechanical performance and cell seeding efficiency. Acta Biomater. 2011;7(3):1009–1018. doi: 10.1016/j.actbio.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat Mater. 2012 Sep;11(9):768–74. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA. Three-dimensional bioprinting of thick vascularized tissues. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1521342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu W, Holmes B, Glazer RI, Zhang LG. 3D printed nanocomposite matrix for the study of breast cancer bone metastasis. Nanomedicine. 2015;12(1):69–79. doi: 10.1016/j.nano.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 30.Kolan KCR, Thomas A, Leu MC, Hilmas G. In vitro assessment of laser sintered bioactive glass scaffolds with different pore geometries. Rapid Prototyp J. 2015;21(2):152–158. [Google Scholar]
- 31.Ferlin KM, Prendergast ME, Miller ML, Kaplan DS, Fisher JP. Influence of 3D printed porous architecture on mesenchymal stem cell enrichment and differentiation q. Acta Biomater. 2016;32:1–9. doi: 10.1016/j.actbio.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Rumpler M, Woesz A, Dunlop JW, van Dongen JT, Fratzl P. The effect of geometry on three-dimensional tissue growth. J R Soc Interface. 2008;5(27):1173–1180. doi: 10.1098/rsif.2008.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Bael S, Chai YC, Truscello S, Moesen M, Kerckhofs G, Van Oosterwyck H, Kruth JP, Schrooten J. The effect of pore geometry on the in vitro biological behavior of human periosteum-derived cells seeded on selective laser-melted Ti6Al4V bone scaffolds. Acta Biomater. 2012;8(7):2824–2834. doi: 10.1016/j.actbio.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 34.Engelmayr GC, Cheng M, Bettinger CJ, Borenstein JT, Langer R, Freed LE. Accordion-like honeycombs for tissue engineering of cardiac anisotropy. Nat Mater. 2008;7(12):1003–1010. doi: 10.1038/nmat2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park H, Larson BL, Guillemette MD, Jain SR, Hua C, Engelmayr GC, Freed LE. The significance of pore microarchitecture in a multi-layered elastomeric scaffold for contractile cardiac muscle constructs. Biomaterials. 2011;32(7):1856–1864. doi: 10.1016/j.biomaterials.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis PL, Shah RN. 3D Printing for Liver Tissue Engineering: Current Approaches and Future Challenges. Curr Transplant Reports. 2016:100–108. [Google Scholar]
- 37.Colosi C, Costantini M, Latini R, Ciccarelli S, Stampella A, Barbetta A, Massimi M, Devirgiliis C. Rapid prototyping of chitosan-coated alginate scaffolds through the use of a 3D fiber deposition technique. J Mater Chem B Mater Biol Med. 2014;2:6779–6791. doi: 10.1039/c4tb00732h. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Yan Y, Pan Y, Xiong Z, Liu H, Cheng J, Liu F, Lin F, Wu R, Zhang R, Lu Q. Generation of three-dimensional hepatocyte/gelatin structures with rapid prototyping system. Tissue Eng. 2006;12(1):83–90. doi: 10.1089/ten.2006.12.83. [DOI] [PubMed] [Google Scholar]
- 39.Monga SPS, Cagle PT. Molecular Pathology of Liver Diseases. 5th. Springer Science & Business Media; 2010. [Google Scholar]
- 40.Rettie AE, Jones JP. Clinical and Toxicological Relevance of CYP2C9: Drug-Drug Interactions and Pharmacogenetics. Annu Rev Pharmacol Toxicol no 45. 2005:477–494. doi: 10.1146/annurev.pharmtox.45.120403.095821. [DOI] [PubMed] [Google Scholar]
- 41.Ramaiahgari SC, Den Braver MW, Herpers B, Terpstra V, Commandeur JNM, Van De Water B, Price LS. A 3D in vitro model of differentiated HepG2 cell spheroids with improved liver-like properties for repeated dose high-throughput toxicity studies. Arch Toxicol. 2014;88(5):1083–1095. doi: 10.1007/s00204-014-1215-9. [DOI] [PubMed] [Google Scholar]
- 42.Sivertsson L, Edebert I, Palmertz MP, Ingelman-sundberg M, Neve EPA. Induced CYP3A4 Expression in Confluent Huh7 Hepatoma Cells as a Result of Decreased Cell Proliferation and Subsequent Pregnane X Receptor Activation s. Mol Pharmacol. 2013;(83):659–670. doi: 10.1124/mol.112.082305. [DOI] [PubMed] [Google Scholar]
- 43.Matsui H, Takeuchi S, Osada T, Fujii T, Sakai Y. Enhanced bile canaliculi formation enabling direct recovery of biliary metabolites of hepatocytes in 3D collagen gel microcavities. Lab Chip. 2012;12(10):1857. doi: 10.1039/c2lc40046d. [DOI] [PubMed] [Google Scholar]
- 44.De Waart DRDR, Ha S, Vlaming MLHLH, Kunne C, Ha E, Gruss H, Elferink RPJOPJO, Stieger B. Hepatic Transport Mechanisms of Cholyl- L -Lysyl-Fluorescein. Jpet. 2010;334:78–86. doi: 10.1124/jpet.110.166991. [DOI] [PubMed] [Google Scholar]
- 45.Berger DR, Ware BR, Davidson MD, Allsup SR, Khetani SR. Enhancing the Functional Maturity of Induced Pluripotent Stem Cell–Derived Human Hepatocytes by Controlled Presentation of Cell–Cell Interactions In Vitro. Hepatology. 2014;61(4):1370–1381. doi: 10.1002/hep.27621. [DOI] [PubMed] [Google Scholar]


