Summary
Seasonal flu as well as potential pandemic flu outbreaks continuously underscores the importance of the preventive and therapeutic measures against influenza viruses. During screening of natural and synthetic small molecules against influenza A and B virus, we identified juniferdin as a highly effective inhibitor against both viruses in cells. Since juniferdin is known to inhibit protein disulfide isomerases (PDIs), multiple PDI inhibitors were tested against these viruses. Among PDI inhibitors, 16F16, PACMA31, isoquercetin, epigallocatechin-3-gallate or nitazoxanide significantly reduced the replication of influenza A and B viruses in MDCK and A549 cells. Furthermore, siRNAs specific to three PDI family members (PDI1, PDIA3 or PDIA4) also significantly reduced the replication of influenza A and B viruses in cells. These results suggest that PDIs may serve as excellent targets for the development of new anti-influenza drugs.
1. Introduction
Influenza viruses belong to the family Orthomyxoviridae, which contains seven genera, influenza virus A, B, C, D, Isavirus, Quaranjavirus and Thogotovirus (Adams et al., 2017). Influenza A viruses are further divided into subtypes on the basis of two viral envelope proteins, hemagglutinin (HA) and neuraminidase (NA). Influenza A viruses infect humans and animals including pigs, horses and birds but influenza B virus is found only in humans. Two subtypes of Influenza A viruses, H1N1 and H3N2, and influenza B viruses cause seasonal flu in humans. In any given year seasonal flu causes 3-5 million cases of severe illness and up to 500,000 deaths worldwide (WHO, 2015). In the U.S alone, up to 48,000 deaths (1.4 to 16.7 deaths per 100,000 people during 1976–2007) occur due to influenza-related complications each year (CDC, 2010). Furthermore, recent emergence of new highly pathogenic influenza viruses in humans such as influenza A H7N9 virus in China in early 2013 and highly pathogenic avian influenza H5 strains (including H5N2, H5N8 and H5N1) in the domestic and wild birds in the US indicate that influenza viruses continue to be a public health threat (USDA-APHIS, 2015).
Currently, four FDA-approved drugs, amantadine, rimantadine, oseltamivir, and zanamivir, are available for influenza infection (Palese and Shaw, 2007; Yen, 2016). However, the M2 channel blockers, amantadine and rimantadine, are no longer recommended for use due to widespread resistance among seasonal influenza viruses. The majority of seasonal flu viruses, including 2009 H1N1 as well as H3N2, are still susceptible to neuraminidase (NA) inhibitors, oseltamivir and zanamivir. However, there is a growing concern that resistance to NA inhibitors may rise in the future, rendering the current influenza drugs unusable (Poland et al., 2009). Therefore, identification of new drug target for the development of new influenza drug broadly effective against influenza A and B viruses and less likely be associated with drug resistance is urgently needed.
PDIs are oxidoreductases of the thioredoxin superfamily and more than 20 members of PDIs are found in eukaryotic cells (Appenzeller-Herzog and Ellgaard, 2008). PDIs participate in diverse physiological and pathophysiolocal roles by functioning as redox-dependent chaperones and catalyzing the formation and rearrangement of disulfide bonds (Ali Khan and Mutus, 2014; Appenzeller-Herzog and Ellgaard, 2008). PDIs have also been implicated in the entry of HIV (Barbouche et al., 2003; Fenouillet et al., 2001; Gallina et al., 2002; Khan et al., 2011; Markovic et al., 2004; Ryser et al., 1994), Sindbis virus (Abell and Brown, 1993), mouse polyomavirus virus (Gilbert et al., 2006) and Newcastle disease virus (Jain et al., 2007). In the case of influenza virus infection, knockdown of the expression of PDIA3 (also known as ERp57) by siRNA in cells was reported to inhibit maturation of HA protein (Solda et al., 2006) and influenza virus replication (Roberson et al., 2012b), which necessitates further investigation on the effects of PDIs in influenza virus infection.
In this study we found that juniferdin, a natural compound with a potent PDI inhibitory activity, significantly inhibited the replication of influenza A and B virus in cells. We also identified synthetic PDI inhibitors that suppress influenza A and B viruses in cells. In addition, siRNAs specific to PDI1, PDIA3 or PDIA4 significantly suppressed the replication of influenza A and B viruses in cell culture. These results suggest that PDIs play important roles in influenza virus replication and may serve as the potential therapeutic targets for the antiviral drug development against influenza viruses.
2. Materials and methods
2.1. Cells and viruses
Madin-Darby canine kidney (MDCK), A549 (human lung cell line), or MA104 cells (monkey kidney cell line) were maintained in Minimum Essential Medium (MEM) containing 10% fetal bovine serum and antibiotics (penicillin and streptomycin). Human influenza A virus (A/WS/33 [H1N1]) and B virus (B/Lee/40) were obtained from ATCC (Manassas, VA). Swine influenza A virus (A/swine/OH/511445/2007 [H1N1], Oh7), a triple human/avian/swine reassortant strain, was isolated from pigs at Ohio county fair that infected both pigs and humans in 2007 (Vincent et al., 2009). Those influenza viruses were propagated in MDCK cells. Group A rotavirus SA11 was also obtained from ATCC and propagated in MA104 cells (Dutta et al., 2011).
2.2. Influenza virus titration. Tissue Culture Infectious Dose 50 (TCID50) method
A standard TCID50 method with the 10-fold dilutions of each sample was used for virus titration (Reed and Muench, 1938). Real-Time qRT-PCR. Viral RNAs were extracted from cells using the RNeasy kit according to the manufacturer’s directions (Qiagen, Valencia, CA). Virus specific primers and probe were synthesized based on literature (Schweiger et al., 2000). The primer sequences for influenza A virus M gene were: Forward 5′-CATGGAATGGCTAAAGACAAGACC-3′ and reverse 5′-AAGTGCACCAGCAGAATAACTGAG-3′. The probe sequence used was: FAM-5′-CTGCAGCGTAGACGCTTTGTCCAAAATG-3′-Iowa Black. Using the One-step Platinum qRT-PCR kit (Invitrogen, Carlsbad, CA), the qRT-PCR amplification was performed in a SmartCycler (Cepheid, Sunnyvale, CA) with the following parameters: 45°C for 30 min, and 95°C 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 50°C for 1 min and elongation at 72°C for 30 s. Western blot analysis. Western blot analysis was performed with cell lysates at 24 hr post infection (PI) before extensive cytopathic effects (CPE) appeared. Cell lysates from virus infected MDCK cells with or without various treatments were prepared by adding sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer containing 2% β-mercaptoethanol and sonication for 20 s. Then the proteins were resolved in a 10% Novex Bis-Glycin gel (Invitrogen, Carlsbad, CA) and transferred to a nitrocellulose membrane. The transferred nitrocellulose membranes were incubated with primary goat antibodies to influenza viruses (whole virions) (Santa Cruz Biotech, Santa Cruz, CA) overnight, and then with the secondary antibodies conjugated with peroxidase for 2 hr. Following incubation with a chemiluminescent substrate (SuperSignal West Pico Chemiluminescent Substrate, Pierce biotechnology, Rockford, IL), the signals were visualized by FOTO/Analyst Luminary/FX Systems (Fotodyne Inc, Harland, WI).
2.3. Small molecule inhibitors
Because juniferdine is known a PDI inhibitor (Khan et al., 2011), other known inhibitors of PDI including bacitracin A (Jain et al., 2007), Di-tert-butyl nitroxide (DTBN) (Jain et al., 2007), 16F16 (Hoffstrom et al., 2010), PACMA31(Xu et al., 2012), isoquercetin (IQ) (Furie and Flaumenhaft, 2014) and NTZ (Muller et al., 2008) were obtained to test against influenza viruses. EGCG was also included because it was reported to have anti-influenza virus activity (Nakayama et al., 1993; Song et al., 2005; Steinmann et al., 2013; Thapa et al., 2012). The structures of some of these compounds are shown in Figure 1. As controls, amantadine and oseltamivir phosphate (oseltamivir) were included as controls for anti-influenza virus activity. Hsp90 inhibitors, geldanamycin (GDA) and 17-N-Allylamino-17-demethoxygeldanamycin (17-AAG), are reported to inhibit the replication of rotavirus and are used as controls for rotavirus inhibition. All compounds were purchased from Sigma-Aldrich (St. Louis, MO), except for juniferdine which was obtained from Namiki Shonki Co, Tokyo, Japan. Each compound was dissolved in dimethyl sulfoxide (DMSO), and 0.1% DMSO was used as a negative control. Confluent monolayers of MDCK or A549 cells were prepared in 12 well plates. Influenza virus (A/WS/33, Oh7 or B/Lee/40) was inoculated to the cells at a multiplicity of infection (MOI) of 0.1 (for TCID50) or 5 (for Western blot and qRT-PCR) in the presence of each compound at various concentrations (0.1 μM – 1 mM) for 1 hr. Then, the cells were washed with phosphate buffered saline (PBS) and fresh media containing the compound and trypsin (2 μg/ml) was added to the cells for further incubation at 37°C for up to 2 days. Amantadine or oseltamivir served as positive controls. Virus replication was assessed with various methods including Western blot analysis, virus titration with TCID50 method, and/or real time qRT-PCR. The effective concentration that reduces virus replication by 50% (EC50) was determined for each compound by conducting nonlinear regression analyses of dose-response curves of virus titers against log inhibitor concentrations (variable slope) using GraphPad Prism software (GraphPad Software, San Diego, CA). Each compound was also tested against the replication of rotavirus (SA11) in MA104 cells following the same procedure, including the use of trypsin (2 μg/ml) in the media.
Figure 1.
Structures of small molecule compounds with anti-PDI activity that is previously reported (juniferdin, PACMA31, 16F16, IQ and NTZ), or identified in this study (EGCG). These compounds showed antiviral effects against influenza A and B viruses in cell culture.
2.4. Nonspecific cytotoxic effects of the compounds
We determined the cytotoxic concentration for 50% cell death (CC50) for each compound in MDCK and A549 cells. About 60% confluent MDCK or A549 cells grown in 96-well plates were treated with various concentrations (1 µM – 1 mM) of each compound for 24 hr. Cell cytotoxicity was measured by a CytoTox 96® non-radioactive cytotoxicity assay kit (Promega, Madison, WI) and CC50 values were calculated using GraphPad Prism software.
2.5. PDI1 assay
The inhibitory activity of each compound against PDI1 enzyme was determined by using the ProteoStat™ PDI assay kit (Enzo Life Sciences, NY) (Huang et al., 2005; Lundstrom and Holmgren, 1990). This assay is based on the detection of insulin aggregates that are formed by the PDI1-catalyzed reduction of insulin in the presence of dithiothreitol (DTT). Each compound including 16F6, PACMA31, juniferdin, IQ, NTZ, bacitracin A, DTNB, EGCG, GDA, amantadine and oseltamivir was tested for anti-PDI1 activity at 10 or 50 µM following the manufacture’s procedures. The percent reduction to control (mock-treatment) was calculated for each compound.
2.6. siRNA study
To study the role of PDIs (PDI1, PDIA3 and PDIA4), siRNA knockdown assay and mRNA quantitation with real time qRT-PCR were established. These three PDI members are chosen because they are known to be expressed at high levels in the ER, and involved in the replication of various viruses. Grp78 was reported to be essential for influenza virus replication and siRNAs for Grp78 were included as a control (Booth et al., 2015; Hogue and Nayak, 1992). The sources for the design of siRNA, and primers and probes of canine specific genes for qRT-PCR in MDCK cells are; PDI1 or P4HB: XM_540488; PDIA3: XM_535453; PDIA4: XM_843145; and Grp78: XM_858292.4. The siRNAs, primers and probes for each gene were synthesized by Integrated DNA Technology (Coralville, IA). At least three siRNAs for each gene were synthesized and the siRNAs that are effective in reducing target gene in MDCK and A549 cells were identified and used in this study. The sequences for effective siRNA, primers and probes used in this study are listed in Table 1. One-day old MDCK cells in 12-well plates were transfected with mock-medium (Mock), irrelevant siRNA (irre, 100 nM), or target siRNA at 100 or 10 nM. At 48 hr post-transfection. Cell lysates were collected and total RNA was extracted for real time qRT-PCR for each target gene or β-actin. Real time qRT-PCR was stopped after 25 cycles for PDI1, PDIA3 or PDIA4, or after 15 cycles for Grp78 due to its abundant presence in the cells. The RT-PCR products were visualized on the agarose gel.
Table 1.
Sequences of siRNA and real time qRT-PCR primers and probe for PDI1, PDIA3, PDIA4 and Grp78 used in this study.
| siRNA | Target | Sequences (duplex of synthetic RNA) |
| PDI1 | rGrArU rCrUrC rArGrA rArCrC rUrUrC rUrGrC rCrUrU rCrArG rCrUrU* | |
| rArGrC rUrUrC rUrCrU rGrCrC rArGrC rUrGrU rGrUrA rCrUrC rCrCrU | ||
| rCrCrA rUrCrC rUrUrG rCrUrC rArGrC rUrGrG rUrArU rUrUrG rGrArG | ||
| PDIA3 | rArUrC rCrUrC rArArU rGrGrC rArUrC rUrUrC rUrUrU rArUrU rGrGrA * | |
| rUrGrA rGrArG rGrArA rCrUrG rArGrG rCrUrG rGrUrC rCrUrG rCrCrU | ||
| rUrGrU rGrArA rGrGrA rCrGrA rArArC rArArG rGrUrG rArUrA rCrCrC | ||
| PDIA4 | rCrUrC rCrUrC rUrUrC rCrUrC rGrUrC rGrUrC rGrUrC rCrUrC rCrUrC * | |
| rCrArA rUrArA rArGrA rArGrA rUrGrC rCrArU rUrGrA rGrGrArU | ||
| rGrGrA rGrGrA rCrGrA rCrGrA rCrGrA rGrGrA rArGrA rGrGrArG | ||
| Grp78 | rUrArG rUrGrA rGrArA rCrCrA rUrGrG rCrArG rArArA rUrUrU rCrUrU * | |
| rArCrU rCrArA rUrUrU rCrArA rUrUrC rUrUrG rCrUrU rGrArU rGrCrU | ||
| rCrArA rGrUrG rUrUrC rCrArG rArUrC rUrCrG rGrUrU rUrCrC rGrUrC | ||
| Real time qRT-PCR | Target | Sequences (5′ to 3′ DNA) |
| PDI1 | FWD-TCA AGG GCA AGA TCC TGT TTA T | |
| REV-CGG GCA CTC CTC TTT CTT TAG | ||
| Probe-/56-FAM/TT CAT CGA C/Zen/A GCG ACC ACA CTG AC/3IABkFQ/ | ||
| PDIA3 | FWD-GCC AGC AAC TTG AGG GAT AA | |
| REV-GGA AGG ACG AAA CAA GGT GAT A | ||
| Probe-/56-FAM/CC GGT TTG C/Zen/A CAC ACC AAT GTT GA/3IABkFQ | ||
| PDIA4 | FWD-CTA ACA GCC TGA GAG AGG ATT AC | |
| REV-CGT AGC TCT TGG GCT CAT ATT | ||
| Probe-/56-FAM/TG CAG CCT G/Zen/A GAA ATT CCA GTC CA/3IABkFQ/ | ||
| Grp78 | FWD-GTT CTT GTT GGT GGC TCT ACT | |
| REV-ACA GCC TCA TCT GGG TTT ATG | ||
| Probe-/56-FAM/TT TAA TGG C/Zen/A AGG AGC CAT CCC GT/3IABkFQ | ||
| β-actin | FWD-GGCATCCACGAAACTACCTT | |
| REV-AGCACTGTGTTGGCGTACAG | ||
| Probe-/HEX-ATCATGAAGTGTGACGTGGACATCCG-TAMRA |
The listed siRNAs were confirmed to reduce both virus replication and target mRNA.
For siRNA study, one-day old semi-confluent MDCK or A546 cells were transfected with siRNAs of PDI1, PDIA3, PDIA4 or Grp78 at 100 nM and cells were further grown for 48 hr. The confluent monolayers were infected with influenza virus (A/WS/33, Oh7 or B/Lee/40) at an MOI of 0.1 (for TCID50) or 5 (for Western blot and qRT-PCR) and incubated in the presence of trypsin (2 µg/ml) for up to 48 hr. Virus replication was monitored at various time points including 2, 12 and 48 hr post inoculation (PI) with real time qRT-PCR (2 or 12 hr PI), TCID50 (48 hr PI) assay or Western blot analysis (24 hr PI). For the detection of viral gene at 2 or 12 hr PI, total RNA was extracted from cell lysates before apparent CPE appeared and real time qRT-PCR was performed. Each RNA sample was also monitored for target gene (PDI1, PDIA3, PDIA4 or Grp78) by real time qRT-PCR. Irrelevant siRNA (OriGen Tech) was also included in the study as a control. β-actin levels were also assessed to determine cytotoxicity resulting from siRNA transfection and for normalization of the mRNA levels of genes of interest.
2.7. Statistics
The student t-test was used to compare the significance of the unpaired sample means. P values <0.05 were considered significant. All experiments were repeated two or three times independently.
3. Results
3.1. PDI inhibitors suppress the replication of influenza A and B viruses
All tested PDI inhibitors and EGCG (its anti-PDI activity was not previously reported) markedly reduced the replication of A/WS/33 and B/Lee/40, except for bacitracin A and DTBN (Table 2, Figure 2A). The EC50 values of 16F16, PACMA31 and juniferdine were determined to be 5.1, 6.1 and 1.5 µM against A/WS/33 and 7.6, 8.8 or 3.1 µM against B/Lee/40, respectively (Table 2). The EC50 values of IQ, EGCG or NTZ were determined to be 0.5, 4.5 or 0.8 µM against A/WS/33 and 1.2, 5.6 or 1.5 µM against B/Lee/40, respectively (Table 2). Interestingly, DTBN showed weak antiviral activity with the EC50 values at 210 and 351 µM against A/WS/33 or B/Lee/40, respectively, and bacitracin A did not show anti-influenza activity at up to 500 µM (Table 2). The replication of A/WS/33 and B/Lee/40 were potently inhibited by oseltamivir, and A/WS/33 by amantadine, as expected (Table 2). Hsp90 inhibitors, GDA and its derivative, 14-AAG, inhibited the replication of group A rotavirus SA11 strain in MA104 cells (EC50 < 1 µM), but they did not show any inhibitory activity against influenza virus A/WS/33 and B/Lee/40. For rotavirus, other than Hsp90 inhibitors, none of tested compound inhibited virus replication at up to 10 µM. The CC50 for each compound in MDCK cells are summarized in Table 2. Selectivity index (CC50/EC50) for A/WS/33 was also listed in Table Western blot analysis with various concentrations of PACMA31 or juniferdine confirmed their inhibitory activities against A/WS/33 (Figure 2B). The reduction of viral proteins (NP or M1) was dose-dependent for PACMA31 or juniferdine, but the β-actin levels remained unchanged from mock-treated cells (Figure 2B). Antiviral effects of the compounds in A549 cells were comparable to those in MDCK cells.
Table 2.
The effects of various compounds on the replication of influenza A (A/WS/33 and A/swine/OH/2007) and B (B/Lee/40) and cytotoxicity of the compounds in MDCK cells.
| EC50 (µM) | CC50 (µM) | SI* for A/WS/33 | Target | |||
|---|---|---|---|---|---|---|
| A/WS/33 | A/swine/OH/2007 | B/Lee/40 | ||||
| Juniferdin | 1.5 (1.4) | 1.8 (1.1) | 3.1 (3.5) | 13.2 (4.7) | 8.8 | PDI |
| Isoquercetin (IQ) | 0.5 (0.2) | 1.2 (0.8) | 1.2 (0.3) | 45.5 (5.1) | 90 | PDI |
| Epigallocatechin 3-gallate (EGCG) | 4.5 (0.3) | 8.1 (2.1) | 5.6 (1.1) | 35.4 (2.3) | 7.8 | PDI# |
| Bacitracin A | > 500 | >500 | > 500 | NA+ | NA | PDI |
| Di-tert-butyl nitroxide (DTBN) | 210 (25) | NA | 351 (54) | > 1000 | >4.8 | PDI |
| 16F16 | 5.1 (0.9) | 5.8 (2.3) | 7.6 (1.3) | 15.0 (3.4) | 2.9 | PDI |
| PACMA31 | 6.1 (1.2) | 7.5 (2.8) | 8.8 (2.3) | 12.5 (3.6) | 1.9 | PDI |
| NTZ | 0.8 (0.4) | 1.3 (0.7) | 1.5 (0.5) | 12.8 (2.1) | 16 | PDI |
| GDA | >10 | >10 | >10 | NA | NA | Hsp90 |
| 17-AAG | >10 | >10 | >10 | NA | NA | Hsp90 |
| Amantidine | 1.2 (0.6) | 1.4 (0.7) | >100 | > 100 | >83.3 | M2 |
| Oseltamivir | 0.15 (0.3) | 0.5 (0.6) | 1.1 (0.5) | > 100 | >660 | NA |
SI: selectivity index.
The target for EGCG was newly determined in this study. The targets of other listed compounds were determined elsewhere.
NA: not available. The numbers in parentheses are standard deviations of the means. Listed compounds may have additional targets for anti-influenza effects.
Figure 2.
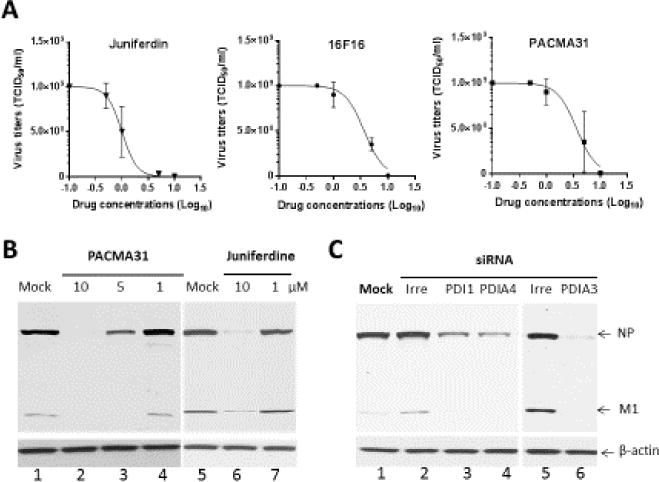
Effects of PDI inhibitors or knockdown of PDI1, PDIA3 or PDIA4 expression by siRNAs on the replication of influenza virus in MDCK cells. A) Dose-response curves of the antiviral activities of Juniferdin, 16F16 or PACMA31. Confluent MDCK cells were infected with influenza A virus, A/WS/33, at an MOI of 0.1 in the presence of Juniferdin, 16F16 or PACMA31 at various concentrations or mock-medium for 1hr. Then, the cells were washed with phosphate buffered saline (PBS), and fresh media containing trypsin (2 μg/ml) and the compound or mock-medium was added to the cells. Virus titers were determined at 2 days post infection using the TCID50 method. B) Expression of influenza virus proteins in the presence of various concentrations of PACMA31 or Juniferdine. Cells were infected with A/WS/33 at an MOI of 5 MOI and incubated in the presence of mock-medium or various concentrations of PACMA31 or juniferdine for 24 hrs. C) Effects of knockdown of PDI1, PDIA3 or PDIA4 expression by siRNAs on the replication of influenza virus in MDCK cells. Cells were mock-transfected or transfected with irreverent RNA or siRNAs targeting each gene. At 24 hr of transfection, cells were infected with A/WS/33 at an MOI of 5. Western blot analysis of cell lysates in B and C was performed with antibodies against whole influenza virus or β-actin. Viral NP and M1 proteins and β-actin are shown.
3.2. EGCG inhibits PDI1 activity
EGCG was previously reported to inhibit influenza virus replication by our group and others (Kesic et al., 2011; Kim et al., 2013; Nakayama et al., 1993; Song et al., 2005), but its effect on PDI has not been reported. In our study, EGCG reduced PDI1 activity by 32% at 10 µM and 72% at 50 µM (Table 3), indicating that EGCG is a PDI1 inhibitor. Other known PDI inhibitors, bacitracin A, DTNB, 16F6, PACMA31, juniferdine, NTZ or IQ also reduced PDI1 activity, compared to the control (Table 3). However, amantadine, oseltamivir or GDA had little effect on PDI1 activity at 10 or 50 µM.
Table 3.
The effects of each compound on PDI activity at 50 or 10 µM.
| Mock | IQ | EGCG | NTZ | 16F16 | PACMA31 | ||
|---|---|---|---|---|---|---|---|
| PDI1 activity relative to mock | 50 µM | 100 | 13 (8)* | 28 (5)* | 65 (4)* | 59 (6)* | 71 (9)* |
| 10 µM | 100 | 32 (5)* | 68 (7)* | 81 (2)* | 89 (6) | 91 (3) | |
| Juniferdin | DTBN | Bacitracin A | GDA | Amantadine | Oseltamivir | ||
| PDI1 activity relative to mock | 50 µM | 6 (4)* | 25 (9)* | 53 (12)* | 96 (2) | 103 (4) | 95 (5) |
| 10 µM | 41 (5)* | 67 (6)* | 81 (3)* | 102 (5) | 102 (5) | 103 (6) |
Statistically significant differences from the mock (p < 0.05). Results are the mean percent inhibition values. The numbers in parentheses are standard deviations of the means.
3.3. PDI1, PDIA3, PDIA4 or Grp78 siRNAs significantly reduced influenza viruses in MDCK or A549 cells
The siRNAs yielded over 80% silencing of mRNA of PDI1, PDIA3, PDIA4 or Grp78 at 48 hr post transfection without showing significant changes in β-actin levels (Table 4, Figure 3A) and those siRNAs were used for virus infection study. The siRNA for PDIA1, PDIA3, PDIA4 or Grp78 significantly reduced the replication of A/WS/33, Oh7 and B/Lee/40 in MDCK or A549 cells, determined by the TCID50 assay at 48 hr PI (Table 4). Reduction in viral titers (TCID50) by PDIA4 siRNA was consistently greater than those by PDI1, PDIA3 or Grp78 siRNAs in both MDCK and A549 cells (Table 4). Of note, virus reduction by each siRNA was higher in MDCK cells than that in A549 cells (Table 4), which may be due to lower levels of virus replication in A549 cells, compared to MDCK cells (Table 4). The irrelevant siRNA did not lead to significant changes in viral titers, compared to mock (distilled water)-transfected control, in MDCK or A549 cells. Similar results were observed with swine influenza A virus (A/swine/OH/2007) in both cell lines (Table 4). Reduction of replication of A/WS/33 after transfection with each siRNA of PDI1 or PDIA4 was also demonstrated by Western blot analysis (Figure 3B). The real time qRT-PCR assay showed that the RNA levels of A/WS/33 at 12 hr PI were significantly reduced in the cells transfected with siRNAs of PDI1, PDIA3, PDIA4 or Grp78 (Table 5). However, the viral RNA levels at 2 h PI were not significantly different among mock and the siRNA treatment groups (Table 5).
Table 4.
Effects of the siRNA treatment for PDI1, PDIA3, PDIA4 or Grp78 on the replication of group A and B influenza viruses in MDCK and A549 cells.
| siRNA | Virus titer (Log10 TCID50/ml) | |||||
|---|---|---|---|---|---|---|
| MDCK | A549 | |||||
| A/WS/33 | A/swine/OH/2007 | B/Lee/40 | A/WS/33 | A/swine/OH/2007 | B/Lee/40 | |
| Mock (water) | 8.3 (0.2) | 7.3 (0.5) | 8.7 (0.5) | 5.3 (0.2) | 4.6 (0.3) | 5.9 (0.5) |
| PDI1 (P4HB, PDIA1) | 4.1 (0.6)* | 4.7 (0.5)* | 5.0 (0.3)* | 3.2 (0.4)* | 3.5 (0.3)* | 4.4 (0.3)* |
| PDIA3 (Erp57) | 4.8 (0.6)* | 5.1 (0.6)* | 5.1 (0.7)* | 3.6 (0.5)* | 3.8 (0.3)* | 4.9 (0.2)* |
| PDIA4 (Erp72) | 3.5 (0.4)* | 3.7 (0.4)* | 4.5 (0.4)* | 3.2 (0.3)* | 2.9 (0.3)* | 4.1 (0.5)* |
| Grp78 (BiP) | 5.2 (0.4)* | 5.1 (0.4)* | 5.5 (0.5)* | 4.2 (0.5)* | 3.8 (0.5)* | 4.8 (0.3)* |
| Irrelevant siRNA | 8.6 (0.5) | 7.5 (0.6) | 8.5 (0.4) | 5.3 (0.5) | 4.7 (0.4) | 6.1 (0.5) |
Statistically significant differences from the mock (p < 0.05). The numbers in parentheses are standard deviations of the means.
Figure 3.
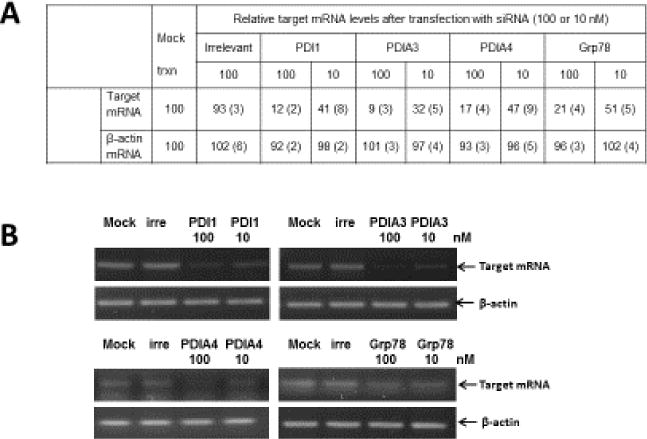
The effects of siRNA treatment targeting PDI1, PDIA3, PDIA4 or Grp78 on the replication influenza A and B viruses. A) Relative mRNA levels of target or β-actin gene following siRNA treatment was calculated using the relative standard curve quantitation method by plotting ct values with standard curves generated using serial dilutions of each mRNA in MDCK cells. The numbers indicate the means and the standard deviations of the means of the relative target mRNA levels. B) Effects of siRNA treatment targeting irrelevant, PDI1, PDIA3, PDIA4 or Grp78 on the replication influenza A and B viruses. One-day old MDCK cells in 12-well plates were transfected with mock-medium (Mock), irrelevant siRNA (irre, 100 nM), or target siRNA at 100 or 10 nM. At 48 hr post-transfection. Cell lysates were collected and total RNA was extracted for real time qRT-PCR for each target gene or β-actin and then the RT-PCR products were visualized on the agarose gel.
Table 5.
Effects of the siRNA treatment for PDI1, PDIA3, PDIA4 or Grp78 on the replication of A/WS/33 in MDCK cells determined by real time qRT-PCR at 2 and 12 hr after virus inoculation.
| siRNA | Relative viral RNA levels compared to mock transfection* | |
|---|---|---|
| A/WS/33 | ||
| 2 hr | 12 hr | |
| Mock (water) | 100 | 100 |
| PDI1 (P4HB, PDIA1) | 92 (3.3) | 9 (1.4)# |
| PDIA3 (Erp57) | 96 (1.9) | 12 (3.1) # |
| PDIA4 (Erp72) | 103 (2.7) | 7 (2.9)# |
| Grp78 (BiP) | 94 (3.8) | 18 (4.3)# |
| Irrelevant siRNA | 102 (2.7) | 93 (2.7) |
The relative RNA levels of the virus M gene following the siRNA treatment were determined as percentage relative to the mock transfection following normalization with β actin.
Statistically significant differences from the mock (p < 0.05). The numbers in parentheses are standard deviations of the means.
4. Discussion
Discovery of host factors as potential drug targets requires understanding of the interplay between the host cells and virus. Recent research on host-virus interaction has led to the findings that several host proteins in chaperone system such as PDIs and heat shock proteins play an important role in viral infection (Stertz and Shaw, 2011; Watanabe et al., 2010). In this study, we found that juniferdine, a PDI inhibitor, is highly effective against the replication of influenza A and B viruses with EC50 values in low micromolar range. Subsequent evaluation of other small molecule PDI inhibitors showed that 16F16 and PACMA31, IQ and NTZ also have anti-influenza activities with varying potency in cells. Their anti-influenza activity has not been previously reported, except for IQ (Kim et al., 2010) and NTZ (Rossignol et al., 2009; Tilmanis et al., 2017). Among the tested PDI inhibitors, IQ and NTZ have most potent anti-influenza activity in cells and varying degree of inhibitory activity against PDI1. In our study, the anti-PDI1 activities of these compounds were similar to those reported previously (Furie and Flaumenhaft, 2014; Hoffstrom et al., 2010; Jain et al., 2007; Khan et al., 2011; Muller et al., 2008; Xu et al., 2012).
Previously, our group reported that IQ, a plant flavonoid, has a potent anti-influenza activities in cell culture and in a mouse model of influenza virus infection (Kim et al., 2010). The mechanism of action of IQ was undetermined but later it was reported that IQ possess anti-PDI activity (Furie and Flaumenhaft, 2014). EGCG is another plant flavonoid with anti-influenza activity (Nakayama et al., 1993; Song et al., 2005; Steinmann et al., 2013; Thapa et al., 2012). The anti-influenza activity of EGCG was suggested to be associated with its antioxidant effect with the induction of the transcription factor NF-E2-related factor 2 (Nrf2) (Kesic et al., 2011) or disruption during entry process (Kim et al., 2013). In this study, we found that EGCG markedly inhibits PDI1. These findings suggest that their anti-PDI activity may contribute to anti-influenza effects of these flavonoids.
The mode of actin of NTZ against the replication of influenza A virus was mediated by selectively blocking the maturation of HA protein (Rossignol et al., 2009). Glycosylation of HA0 in the ER leads to the assembly of HA homotrimers, which are rapidly transported to the Golgi complex where the sugar component is further processed to the final mature glycoprotein. Rossignol et al (Rossignol et al., 2009) showed that NTZ inhibited the transport of HA0 to the trans-Golgi compartment and significantly reduced plasma membrane levels of HA0. However, the anti-influenza virus activity of NTZ has not been clearly linked to PDI inhibition (Di Santo and Ehrisman, 2013). In our study, antiviral activity of the tested compounds, including NTZ, did not correlate well with their anti-PDI1 activity. One of the possible explanations for this discrepancy is that the compounds may also have activity against other PDI enzymes. One of the PDI enzymes abundantly present in the ER is PDIA3 (ERp57). PDIA3 was shown to be important for proper cysteine paring of HA and subsequent formation and forward transport of HA homotrimer (Solda et al., 2006) and the replication of influenza virus in cells (Roberson et al., 2012a). Therefore NTZ may target PDI enzymes other than PDI1, although it is also possible that they have additional target for suppression of influenza virus. Interestingly, the PDI inhibitors with limited or no cell permeability, bacitracin A and DTNB, (Di Santo and Ehrisman, 2013; Ryser et al., 1994) did not reduce the replication of influenza virus in cell culture. Bacitracin A and DTNB block the entry of HIV-1 (Ryser et al., 1994) and Newcastle disease virus (Jain et al., 2007) by preventing the reduction of virus surface protein by PDIs that are required for fusion between virus and cell membrane. The PDIs involved in this process is located on cell membrane and accessible to non-permeable inhibitors or antibodies. Since PDIs are presumed to be involved in the maturation of influenza HA proteins in the ER, impermeability of those compounds may explain the lack of their anti-influenza virus activity.
Since only PDIA3 has been shown to be directly involved in the replication of influenza virus (Roberson et al., 2012a; Solda et al., 2006), we conducted knockdown studies using siRNAs for PDI1, PDIA3 or PDIA4, which are ER-residing PDIs, to further elucidate the effects of these individual PDI enzyme on the replication of influenza viruses. We also investigated the effect of knockdown of Grp78, a chaperone protein found in the ER. The downregulation of PDI1, PDIA3 or PDIA4 expression led to significant reduction of influenza A and B virus replication in cells with PDIA4 suppression being most effective. This finding indicates that each of these PDI enzymes are playing important roles in influenza virus replication. Of note, virus RNA levels at 2 hr after virus infection in cells transfected with siRNA for PDI1, PDIA3, PDIA4 were comparable to those transfected with irrelevant siRNA. This finding suggests that the PDI-mediated antiviral effect is directed at post-virus entry event, which is consistent with the previous findings by others that PDI function is associated with the maturation of HA (Roberson et al., 2012a; Solda et al., 2006). We also observed suppression of Grp78 expression led to reduction of virus replication, although milder than those with suppression of PDIs. Grp78 is the most abundant chaperone protein in the ER and facilitates folding and assembly of nascent proteins (Hendershot, 2004), and it was reported that Grp78 is transiently associated with newly synthesized influenza NA protein (Hogue and Nayak, 1992), implying a role of Grp78 in influenza virus replication. While we examined only influenza A H1 subtype in our study, PDI inhibitors or siRNA against PDIs are likely to be effective against other subtypes such as H3, H5, and H7 because the disulfide bond formation during influenza HA trimerization is essential for influenza virus replication.
In summary, our findings provide evidence that multiple PDI enzymes are involved in the replication of influenza A and B viruses. The small molecule PDI inhibitors such as IQ, EGCG, juniferdine, 16F16 and PACMA31 may provide the starting platform for the development of new therapeutics for both influenza A and B infections.
Highlights.
Small molecule inhibitors for PDI showed antiviral effects on influenza A and B viruses
EGCG has anti-PDI1 activity
siRNA for PDI1, PDIA3 or PDIA4 significantly reduces the replication of influenza A and B virus
The results suggest PDI as potential targets for influenza virus infection
Acknowledgments
This work was generously supported by an NIH grant (R01AI109039). We thank David George and Vinay Shivanna for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abell BA, Brown DT. Sindbis virus membrane fusion is mediated by reduction of glycoprotein disulfide bridges at the cell surface. J Virol. 1993;67:5496–5501. doi: 10.1128/jvi.67.9.5496-5501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert M, Sabanadzovic S, Sanfacon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Gorbalenya AE, Davison AJ. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017) Arch Virol. 2017;162:2505–2538. doi: 10.1007/s00705-017-3358-5. [DOI] [PubMed] [Google Scholar]
- Ali Khan H, Mutus B. Protein disulfide isomerase a multifunctional protein with multiple physiological roles. Front Chem. 2014;2:70. doi: 10.3389/fchem.2014.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appenzeller-Herzog C, Ellgaard L. The human PDI family: versatility packed into a single fold. Biochimica et biophysica acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- Barbouche R, Miquelis R, Jones IM, Fenouillet E. Protein-disulfide isomerase-mediated reduction of two disulfide bonds of HIV envelope glycoprotein 120 occurs post-CXCR4 binding and is required for fusion. J Biol Chem. 2003;278:3131–3136. doi: 10.1074/jbc.M205467200. [DOI] [PubMed] [Google Scholar]
- Booth L, Roberts JL, Cash DR, Tavallai S, Jean S, Fidanza A, Cruz-Luna T, Siembiba P, Cycon KA, Cornelissen CN, Dent P. GRP78/BiP/HSPA5/Dna K is a universal therapeutic target for human disease. J Cell Physiol. 2015;230:1661–1676. doi: 10.1002/jcp.24919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Estimates of deaths associated with seasonal influenza — United States, 1976–2007. MMWR Morbidity and mortality weekly report. 2010;59:1057–1062. [PubMed] [Google Scholar]
- Di Santo N, Ehrisman J. Research perspective: potential role of nitazoxanide in ovarian cancer treatment. Old drug, new purpose? Cancers. 2013;5:1163–1176. doi: 10.3390/cancers5031163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Chattopadhyay S, Bagchi P, Halder UC, Nandi S, Mukherjee A, Kobayashi N, Taniguchi K, Chawla-Sarkar M. Active participation of cellular chaperone Hsp90 in regulating the function of rotavirus nonstructural protein 3 (NSP3) J Biol Chem. 2011;286:20065–20077. doi: 10.1074/jbc.M111.231878. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fenouillet E, Barbouche R, Courageot J, Miquelis R. The catalytic activity of protein disulfide isomerase is involved in human immunodeficiency virus envelope-mediated membrane fusion after CD4 cell binding. J Infect Dis. 2001;183:744–752. doi: 10.1086/318823. [DOI] [PubMed] [Google Scholar]
- Furie B, Flaumenhaft R. Thiol isomerases in thrombus formation. Circulation research. 2014;114:1162–1173. doi: 10.1161/CIRCRESAHA.114.301808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina A, Hanley TM, Mandel R, Trahey M, Broder CC, Viglianti GA, Ryser HJ. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry. J Biol Chem. 2002;277:50579–50588. doi: 10.1074/jbc.M204547200. [DOI] [PubMed] [Google Scholar]
- Gilbert J, Ou W, Silver J, Benjamin T. Downregulation of protein disulfide isomerase inhibits infection by the mouse polyomavirus. J Virol. 2006;80:10868–10870. doi: 10.1128/JVI.01117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM. The ER function BiP is a master regulator of ER function. Mt Sinai J Med. 2004;71:289–297. [PubMed] [Google Scholar]
- Hoffstrom BG, Kaplan A, Letso R, Schmid RS, Turmel GJ, Lo DC, Stockwell BR. Inhibitors of protein disulfide isomerase suppress apoptosis induced by misfolded proteins. Nature chemical biology. 2010;6:900–906. doi: 10.1038/nchembio.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue BG, Nayak DP. Synthesis and processing of the influenza virus neuraminidase, a type II transmembrane glycoprotein. Virology. 1992;188:510–517. doi: 10.1016/0042-6822(92)90505-j. [DOI] [PubMed] [Google Scholar]
- Huang SG, Oksenberg D, Urfer R. High-throughput turbidometric assay for screening inhibitors of protein disulfide isomerase activity, US. 2005 doi: 10.1177/1087057104265292. [DOI] [PubMed] [Google Scholar]
- Jain S, McGinnes LW, Morrison TG. Thiol/disulfide exchange is required for membrane fusion directed by the Newcastle disease virus fusion protein. J Virol. 2007;81:2328–2339. doi: 10.1128/JVI.01940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesic MJ, Simmons SO, Bauer R, Jaspers I. Nrf2 expression modifies influenza A entry and replication in nasal epithelial cells. Free radical biology & medicine. 2011;51:444–453. doi: 10.1016/j.freeradbiomed.2011.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MM, Simizu S, Lai NS, Kawatani M, Shimizu T, Osada H. Discovery of a small molecule PDI inhibitor that inhibits reduction of HIV-1 envelope glycoprotein gp120. ACS Chem Biol. 2011;6:245–251. doi: 10.1021/cb100387r. [DOI] [PubMed] [Google Scholar]
- Kim M, Kim SY, Lee HW, Shin JS, Kim P, Jung YS, Jeong HS, Hyun JK, Lee CK. Inhibition of influenza virus internalization by (-)-epigallocatechin-3-gallate. Antiviral research. 2013;100:460–472. doi: 10.1016/j.antiviral.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Kim Y, Narayanan S, Chang KO. Inhibition of influenza virus replication by plant-derived isoquercetin. Antiviral Res. 2010;88:227–235. doi: 10.1016/j.antiviral.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Lundstrom J, Holmgren A. Protein disulfide-isomerase is a substrate for thioredoxin reductase and has thioredoxin-like activity. J Biol Chem. 1990;265:9114–9120. [PubMed] [Google Scholar]
- Markovic I, Stantchev TS, Fields KH, Tiffany LJ, Tomic M, Weiss CD, Broder CC, Strebel K, Clouse KA. Thiol/disulfide exchange is a prerequisite for CXCR4-tropic HIV-1 envelope-mediated T-cell fusion during viral entry. Blood. 2004;103:1586–1594. doi: 10.1182/blood-2003-05-1390. [DOI] [PubMed] [Google Scholar]
- Muller J, Naguleswaran A, Muller N, Hemphill A. Neospora caninum: functional inhibition of protein disulfide isomerase by the broad-spectrum anti-parasitic drug nitazoxanide and other thiazolides. Experimental parasitology. 2008;118:80–88. doi: 10.1016/j.exppara.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Suzuki K, Toda M, Okubo S, Hara Y, Shimamura T. Inhibition of the infectivity of influenza virus by tea polyphenols. Antiviral research. 1993;21:289–299. doi: 10.1016/0166-3542(93)90008-7. [DOI] [PubMed] [Google Scholar]
- Palese P, Shaw ML. Orthomyxoviridae: The viruses and Their Replication. In: Knipe DM, et al., editors. Fields Virology Wolters. Kluwer/Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 1647–1690. [Google Scholar]
- Poland GA, Jacobson RM, Ovsyannikova IG. Influenza virus resistance to antiviral agents: a plea for rational use. Clin Infect Dis. 2009;48:1254–1256. doi: 10.1086/598989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty percent endpoints. Amercan Journal of Hygiene. 1938;27:493–497. [Google Scholar]
- Roberson EC, Tully JE, Guala AS, Reiss JN, Godburn KE, Pociask DA, Alcorn JF, Riches DW, Dienz O, Janssen-Heininger YM, Anathy V. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-beta release in lung epithelial cells. American journal of respiratory cell and molecular biology. 2012a;46:573–581. doi: 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson EC, Tully JE, Guala AS, Reiss JN, Godburn KE, Pociask DA, Alcorn JF, Riches DWH, Dienz O, Janssen-Heininger YMW, Anathy V. Influenza Induces Endoplasmic Reticulum Stress, Caspase-12-Dependent Apoptosis, and c-Jun N-Terminal Kinase-Mediated Transforming Growth Factor-beta Release in Lung Epithelial Cells. Am J Resp Cell Mol. 2012b;46:573–581. doi: 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol JF, La Frazia S, Chiappa L, Ciucci A, Santoro MG. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J Biol Chem. 2009;284:29798–29808. doi: 10.1074/jbc.M109.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser HJ, Levy EM, Mandel R, DiSciullo GJ. Inhibition of human immunodeficiency virus infection by agents that interfere with thiol-disulfide interchange upon virus-receptor interaction. Proc Natl Acad Sci U S A. 1994;91:4559–4563. doi: 10.1073/pnas.91.10.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger B, Zadow I, Heckler R, Timm H, Pauli G. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J Clin Microbiol. 2000;38:1552–1558. doi: 10.1128/jcm.38.4.1552-1558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solda T, Garbi N, Hammerling GJ, Molinari M. Consequences of ERp57 deletion on oxidative folding of obligate and facultative clients of the calnexin cycle. J Biol Chem. 2006;281:6219–6226. doi: 10.1074/jbc.M513595200. [DOI] [PubMed] [Google Scholar]
- Song JM, Lee KH, Seong BL. Antiviral effect of catechins in green tea on influenza virus. Antiviral research. 2005;68:66–74. doi: 10.1016/j.antiviral.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Steinmann J, Buer J, Pietschmann T, Steinmann E. Anti-infective properties of epigallocatechin-3-gallate (EGCG), a component of green tea. British journal of pharmacology. 2013;168:1059–1073. doi: 10.1111/bph.12009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S, Shaw ML. Uncovering the global host cell requirements for influenza virus replication via RNAi screening. Microbes and infection/Institut Pasteur. 2011;13:516–525. doi: 10.1016/j.micinf.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thapa M, Kim Y, Desper J, Chang KO, Hua DH. Synthesis and antiviral activity of substituted quercetins. Bioorg Med Chem Lett. 2012;22:353–356. doi: 10.1016/j.bmcl.2011.10.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilmanis D, van Baalen C, Oh DY, Rossignol JF, Hurt AC. The susceptibility of circulating human influenza viruses to tizoxanide, the active metabolite of nitazoxanide. Antiviral Res. 2017;147:142–148. doi: 10.1016/j.antiviral.2017.10.002. [DOI] [PubMed] [Google Scholar]
- USDA-APHIS. Avian Influenza Disease 2015 [Google Scholar]
- Vincent AL, Swenson SL, Lager KM, Gauger PC, Loiacono C, Zhang Y. Characterization of an influenza A virus isolated from pigs during an outbreak of respiratory disease in swine and people during a county fair in the United States. Vet Microbiol. 2009;137:51–59. doi: 10.1016/j.vetmic.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell host & microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Influenza Seasonal. Geneva: Media Center; 2015. Available from: http://www.who.int/mediacentre/factsheets/fs211/en/ [Google Scholar]
- Xu S, Butkevich AN, Yamada R, Zhou Y, Debnath B, Duncan R, Zandi E, Petasis NA, Neamati N. Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc Natl Acad Sci U S A. 2012;109:16348–16353. doi: 10.1073/pnas.1205226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HL. Current and novel antiviral strategies for influenza infection. Curr Opin Virol. 2016;18:126–134. doi: 10.1016/j.coviro.2016.05.004. [DOI] [PubMed] [Google Scholar]


