Abstract
RNA sequencing (RNA-seq) offers a snapshot of cellular RNA populations, but not temporal information about the sequenced RNA. Here we report TimeLapse-seq, a chemical method that uses oxidative-nucleophilic-aromatic-substitution to convert 4-thiouridine into cytidine analogues, yielding apparent U-to-C mutations that mark new transcripts upon sequencing. TimeLapse-seq is a single molecule approach that is adaptable to many applications, and reveals RNA dynamics and induced differential expression concealed in traditional RNA-seq.
Rapid global changes in regulated transcription on the timescale of minutes to hours have been observed for numerous mammalian systems, including circadian rhythms and the immune response, by identifying new transcripts that co-fractionate with chromatin1, 2 or unspliced transcripts3, 4. New RNA populations can also be identified by examining sites of active RNA polymerase II through biochemical enrichment of transcripts in the process of being synthesized (e.g., PRO-seq5, NET-seq6) or metabolic labeling and enrichment of new transcripts (e.g., TT-seq7, s4U-seq8, 9). These techniques require large amounts of input sample, extensive handling, and present challenges when normalizing enrichment and estimating contamination.
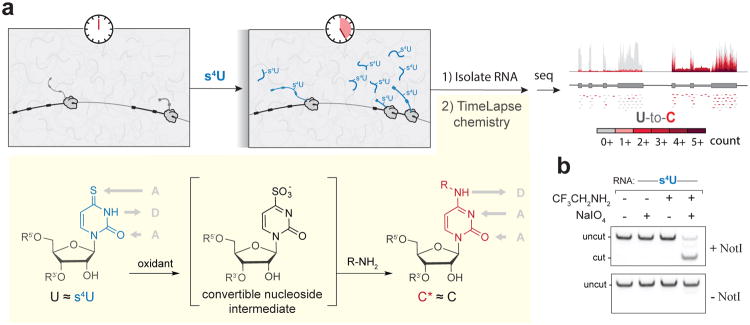
To capture temporal information about RNA directly in a sequencing experiment without biochemical enrichment, we developed TimeLapse-seq (Fig.1a), a method in which cells are exposed to a non-canonical nucleoside that becomes incorporated only into new transcripts. Rather than enriching the metabolically labeled RNAs, we developed chemistry that recodes the hydrogen bonding pattern of the uridine analogue 4-thiouridine (s4U) to match the hydrogen bonding pattern of cytosine, thereby causing mutations in a sequencing experiment. This strategy is similar to bisulfite sequencing, which uses chemically induced mutations to recode nucleobase hydrogen bonding to provide insight into DNA methylation. In our strategy, the recoded nucleosides distinguish which RNAs were transcribed during the time of the experiment. TimeLapse-seq results are internally normalized, as both pre-existing and new transcripts are present in the same library. These mutations reveal which RNAs were recently synthesized by the cell, thereby capturing the rich dynamics of the transcriptome.
Fig.1.
TimeLapse-seq uses a convertible nucleoside approach to identify new transcripts in a sequencing experiment. (a) Scheme of TimeLapse-seq. Metabolically labeled RNAs are isolated and treated with TimeLapse chemistry, converting s4U into a modified cytosine (C*) that is identified through increased numbers of T-to-C mutations upon sequencing (increasingly dark colors of red). s4U is transformed into a convertible nucleoside intermediate through oxidation, which is then converted to C* through aminolysis. (b)Results from a restriction enzyme digestion assay indicating efficient (∼80%) T-to-C* conversion with optimized TimeLapse chemistry
To develop TimeLapse-seq we focused on s4U because of its utility in RNA metabolic labeling experiments10, 11 and the orthogonal reactivity of its thione relative to other functional groups found in RNA. The s4U base itself leads to low levels of U-to-C transitions upon reverse transcription,12 but at levels too low to robustly identify new transcripts. While recent applications of s4U have focused on the thione as a nucleophile8, or for UV crosslinking11, 13, we were inspired by less explored reactivity---transforming s4U using oxidative-nucleophilic-aromatic substitution14. We reasoned that oxidation of s4U would transform it into a convertible nucleoside, providing an intermediate that could be converted into an analogue of cytosine by aminolysis (Fig.1a). The s4U base retains uridine's Watson-Crick hydrogen bonding pattern, and while other chemical conditions used to modify s4U (e.g., alkylation) change the base's hydrogen bonding pattern, they do not recode the base to match C's native hydrogen bonding pattern. While not widely explored, the oxidative reactivity of s4U has precedent in UV crosslinking studies, where sites of s4U-protein crosslinks are enriched for T-to-C mutations, or in mapping the locations of s4U bases in E. coli tRNA11, 15. If conducted before an RNA-seq analysis, this reaction could reveal sites of s4U incorporation through T-to-C mutations stably introduced in the cDNA.
We explored chemistry to convert the free nucleoside (s4U) to cytidine derivatives (Fig.1a and Supplementary Fig.S1) while minimizing oxidation of guanosine (Supplementary Fig.S2) and using amines with low pKa values that remain deprotonated under neutral reaction conditions. We found that treating s4U with 2,2,2-trifluoroethylamine (TFEA) and meta-chloroperoxybenzoic acid (mCPBA) results in near-complete consumption of s4U, producing only small amounts of the hydrolysis product uridine, and mostly the desired trifluoroethylated cytidine (C*, Supplementary Fig.S3a). Similar conditions were successful in the context of an oligoribonucleotide. Optimization of the nucleophile, oxidant, temperature, and time through a restriction enzyme digestion assay (see Online Methods, Supplementary Fig.S4a-c, Supplementary Table S1a) led us to the combination of TFEA and sodium periodate (NaIO4; Fig.1b). These reagents cause clean transformation of 4-thiouracil to N4-trifluoroethylcytosine by 1H NMR (Supplementary Fig.S3b). When RNA with a single s4U was subjected to these conditions (45°C, 1h), reverse transcriptase could efficiently transcribe the product and the majority of the resulting DNA (∼80%) had the desired T-to-C mutation (Supplementary Figs.S3c,d, 4d). NaIO4 is an oxidant commonly used in RNA biology to oxidize the 3′-end vicinal diol of RNAs with minimal effects on other functional groups, even through multiple rounds of oxidation16. To test NaIO4 and TFEA with cellular s4U-RNA, we exposed mouse and human cells to a range of concentrations of s4U. After RNA isolation and chemical treatment, we examined the apparent U-to-C conversion rates (inferred from T-to-C mutations in the cDNA, hereafter referred to as T-to-C) by targeted RT-PCR coupled to paired-end sequencing (Supplementary Table S1b). We observed a notable and specific increase in T-to-C transitions in chemically treated samples (Supplementary Fig.S5).
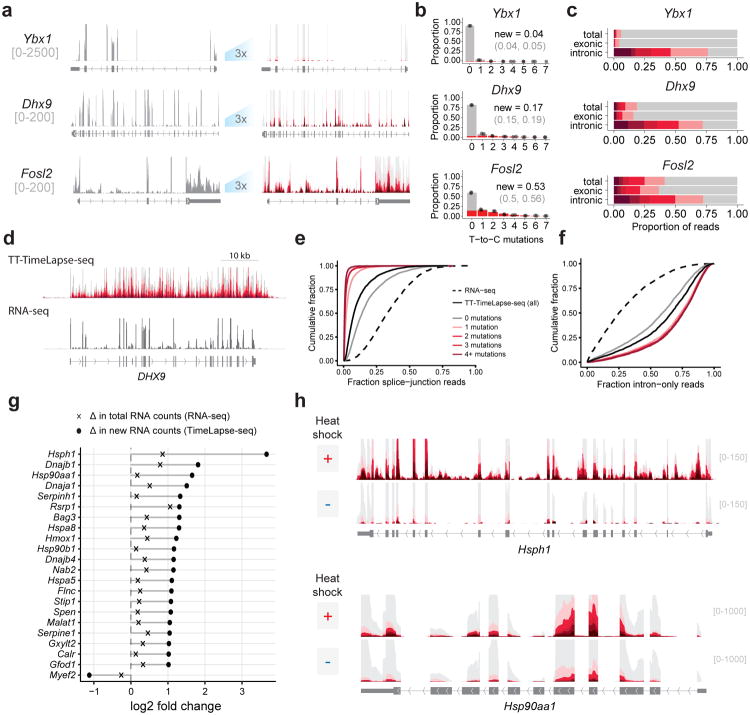
To examine the dynamics of cellular RNAs, we treated MEF cells with s4U for 1h (where no s4U toxicity was observed, Supplementary Fig.S6a-c) and performed TimeLapse chemistry prior to sequencing. The total transcript counts from each sample were highly correlated irrespective of s4U exposure or chemical treatment (Pearson's r≥0.97, Supplementary Fig.S6d), demonstrating that TimeLapse-seq retains information from a traditional RNA-seq experiment. By counting the mutations in each aligned read pair, we found a specific and reproducible increase in T-to-C mutations dependent on both metabolic labeling with s4U and chemical treatment (Supplementary Fig.S7,8). Other mutation rates remained below background levels of T-to-C mutations in untreated samples (e.g. the small increase in G-to-T mutations, Supplementary Fig.S2c,d). Additionally, the reaction was efficient even in regions of RNA secondary structure (Supplementary Fig.S9). The T-to-C mutation counts were dramatically higher in fast-turnover transcripts (e.g., Myc and Fosl2), compared to more stable transcripts (e.g., Dhx9 and Ybx1) (Fig.2a-b, Supplementary Fig.S10). We observed an enrichment of T-to-C mutations in intronic reads (Fig.2c), consistent with the fast turnover of intronic RNA. To quantify these results, we modeled reads as arising from two populations: pre-existing RNAs (background mutation rate) and new RNAs (high T-to-C mutation rate; Fig.2b, Methods). Reads from newly synthesized RNAs had an average of 2.2 mutations per read, corresponding to a ∼3% mutation rate per uridine (compared to ∼0.1% T-to-C mutation rates in controls and for pre-existing RNAs). From each gene, we determined the fraction of newly made transcripts (r≥0.94, 2992 genes, Supplementary Table 2), and estimated transcript half-lives which correlated with those reported previously17 (Supplementary Fig.S11). As expected, the fast turnover RNAs (top 10%, n=360) were enriched for transcripts such as transcription factors (DNA-templated transcription, p<10-20), while the slow turnover RNAs (top 10%, n=361) were enriched for those that are involved in translation (ribosomal biogenesis, p<10-6; translation, p<10-27). Estimates of the fraction of newly synthesized RNA were particularly robust when the new transcripts represented ∼200 reads in the experiment (Supplementary Fig.S12, Supplementary Note).
Fig.2.
Global analysis of steady state and transient RNA dynamics using TimeLapse-Seq. (a) (left) Tracks depicting coverage from all reads (gray) for transcripts with slow (Ybx1), moderate (Dhx9) or fast (Fosl2) rates of turnover. (right) Tracks from reads with increasing numbers of T-to-C mutations (see scale) displaying mutational content provided by TimeLapse chemistry (right, y-axis zoom 3x). (b) Distribution of reads with each number of T-to-C mutations (points) overlaid on a model of the estimated distribution of reads from new transcripts (red) and pre-existing transcripts (gray) for Ybx1, Dhx9, and Fosl2. The estimated fraction of new reads is indicated for each plot. Light gray: 95%CI. (c) Distribution of T-to-C mutations found in reads mapping to Ybx1, Dhx9, and Fosl2, separated by total, exonic, or intronic reads. (d) TT-TimeLapse-seq and RNA-Seq tracks of DHX9. (e) Cumulative distribution plot of reads containing splice-junctions in RNA-seq, and TT-TimeLapse-seq. (f) Cumulative distribution plot of intron-only reads in RNA-seq and TT-TimeLapse-seq with the same scale as in e. (g) Using TimeLapse-seq to distinguish new RNAs after heat shock. Log2 fold changes after heat shock in total RNA-seq counts and new RNA counts for the top RNAs identified in b as significantly changed upon heat shock (padj < 0.01). (h) RNA-seq and TimeLapse-seq tracks of Hsph1 (top) and Hsp90aa1 (bottom) upon heat shock.
Very transient RNA species, such as reads beyond the poly-A termination signal in a gene body, provide insight into transcriptome dynamics but are generally too rare to be observed at high levels by RNA-seq. While these dynamics can be studied through biochemical enrichment of very recently made RNAs after short (5 min) s4U treatments (TT-seq7), biochemically enriched s4U-RNA always contains contaminating reads from unlabeled RNAs (estimated to be up to 30% in some experiments12). This contaminating background can limit analyses; for example, abundant spliced transcripts observed in RNA enriched after short s4U pulses has been interpreted as fast splicing18, but these results could also be explained by contaminating background (e.g., from fully spliced mature RNAs). To test if TimeLapse chemistry could be used in conjunction with transient transcriptome sequencing (TT-seq) to distinguish bona fide new RNAs from contaminating background, K562 cells were labeled for 5 min with s4U, and biochemical enrichment was performed as in TT-seq7, except with more efficient MTS chemistry to biotinylate the s4U-RNA9 (Supplementary Fig.S13a). After enrichment and prior to sequencing, we performed TimeLapse chemistry. As expected, transient RNA species were enriched for introns (two-sample Kolmogorov-Smirnov test, p<10-15, Figs. 2d-f, Supplementary Fig.S13) but depleted for splice junctions (p<10-15). Both enrichment of introns and depletion of splice junctions were slightly greater than previously observed7 (Supplementary Fig.S13c,d), likely due to the efficiency of MTS-chemistry. Even with only 5 min of s4U treatment, the majority of the biochemically enriched reads contained TimeLapse-induced mutations (Fig. 2d). Mutation-containing reads represented a subpopulation that was further enriched for introns, and depleted for splice junctions (Figs. 2e,f, Supplementary Fig.S13c,d). This suggests that mutated reads effectively capture the profile of new RNAs, while the reads without mutations represent a subpopulation that is contaminated by unlabeled reads. We estimated that 15-20% of total TT-seq reads arise from contaminating RNA (estimate from splice-junction content: 17-20%; from intronic content: 18-20%, see methods), similar to estimates from previous s4U experiments12. Reads without mutations were enriched for contaminating reads (estimate from splice junctions, 33-39%; estimate from introns, 35-40%), while reads containing mutations are depleted in contamination. For reads with a single mutation, contaminating reads make up <5% of the signal; for reads with two mutations, the contamination is <1%. Taken together, RNA contamination contributes to the signal at the level of RNA-seq, but TimeLapse chemistry-induced mutations can be used to discriminate between signal from new RNAs and contaminating reads. These results demonstrate transcripts including ACTB (Supplementary Fig.S13b) are not highly spliced on this timescale (5 min), and highlight how TimeLapse chemistry can provide an extra specificity filter when analyzing rare, transient RNAs.
To test if TimeLapse-seq could reveal induced changes in RNA populations, we subjected MEF cells to a mild heat shock (42°C, 1h), where only modest changes in total RNA levels are apparent19-21. We observed induction of a few transcripts such as Hspa1b by RNA-seq (Supplementary Fig.S14a), but TimeLapse-seq revealed the induction of many transcripts encoding heat shock proteins in the new transcript pool that are not apparent by RNA-seq alone (Fig.2g, Supplementary Fig.S14). For example, whereas RNA-seq is less sensitive to the small absolute changes in Hsph1 and Hsp90aa1 (as they are already abundant prior to heat shock; RNA-seq fold-change: Hsph1 = 1.8 fold, Hsp90aa1 = 1.1 fold, DEseq2), TimeLapse-seq reveals substantial induction of both transcripts in the new transcript pool (TimeLapse-seq fold change: Hsph1 = 12.7 fold; Hsp90aa1 = 3.1 fold, DEseq2) (Fig.2h). Unlike PRO-seq and NET-seq, however, which are not sensitive to changes in RNA populations after transcription has completed, TimeLapse-seq captures changes in RNA processing: we observed the induction of a new terminal exon in Rsrp1 upon heat shock (Supplementary Fig.S14c,d), as well as post-transcriptional down regulation of histone mRNAs upon heat shock (Supplementary Fig.S14f,g), neither of which would be apparent from analysis of nascent RNA.
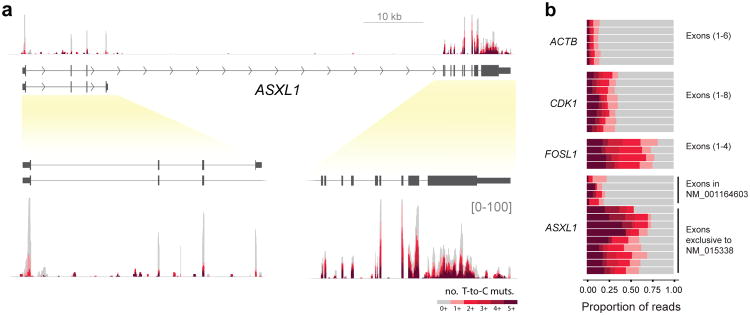
We applied TimeLapse-seq using treatment conditions optimized for studying mRNA turnover (4h s4U)22 in a chronic myelogenous leukemia model cell line (K562). We obtained highly reproducible half-life estimates that correlated with previous observations23 (Supplementary Fig.S11). Inspection of individual transcripts revealed reads mapping to both a shorter isoform of ASXL1 (NM_001164603), as well as a longer isoform (NM_015338) of ASXL1. The ASXL1 protein is involved in epigenetic regulation of chromatin, and mutations in the longer isoform of this gene are implicated in myelodysplastic syndromes (MDS).24 Analysis of the mutational content of the individual exons from ASXL1 demonstrated that reads mapping to the longer isoform had substantially greater turnover than those mapping to the first four exons (Figs. 3a,b), a conclusion supported by transcriptional inhibition (Supplementary Fig. S15b,c). The different stability of ASXL1 isoforms is particularly intriguing given the importance of RNA-processing to many pathologies, including MDS25.
Fig. 3.
TimeLapse-seq reveals differential transcript isoform stability of the ASXL1 transcript. (a) ASXL1 tracks from TimeLapse-seq (4h s4U treatment) with exon-containing regions expanded (lower panel). (b) Exonic T-to-C mutation distributions for ASXL1 in comparison with three transcripts with different stabilities, ACTB, CDK1, FOSL1.
TimeLapse chemistry provides a chemical means of recoding metabolically labeled nucleotides from the hydrogen bonding pattern of one base (s4U) to another (C*). TimeLapse-seq is a single-molecule approach to monitor transcriptome dynamics. The method reveals different rates of RNA turnover, changes in RNA processing, and acute changes in the transcriptome that are not apparent using standard RNA-seq. TimeLapse-seq is broadly applicable to applications that use metabolic labeling (e.g., TT-seq), providing a flexible platform to investigate dynamic biological systems.
Online Methods
Materials
All commercially available materials were purchased from the indicated suppliers and used without further purification. 4-thiouridine (s4U), and meta-chloroperoxybenzoic acid (mCPBA) were purchased from Alfa Aesar (Haverhill, MA). 4-thiouridine-5′-triphosphate (s4UTP) was purchased from TriLink BioTechnologies (San Diego, CA). 2,2,2-trifluoroethylamine (TFEA), sodium acetate, EDTA, Tris hydrochloride, acrylamide/bis-acrylamide 30% solution, phenol:chloroform:isoamyl alcohol (25:24:1), and actinomycin D were purchased from Sigma Aldrich (St. Louis, MO). Sodium periodate (NaIO4) and ammonium bicarbonate were purchased from Acros Organics (Geel, Belgium). Methane thiosulfonate biotin-XX (MTSEA-biotin-XX) was purchased from Biotium. Dynabeads MyOne Streptavidin C1 beads were purchased from Thermo Fisher Scientific. Agencourt RNAClean XP beads were purchased from Beckman Coulter (Brea, CA). Phusion HF PCR master mix and Dithiothreitol (DTT) were purchased from Thermo Fisher Scientific (Waltham, MA). Phosphate buffered saline (PBS) was purchased from AmericanBio (Natick, MA). Dulbecco's Modified Eagle Medium (DMEM), fetal bovine serum (FBS), Trizol reagent, TURBO DNase and SuperScript III Reverse Transcriptase were purchased from Life Technologies (Carlsbad, CA). KAPA Taq Ready Mix was purchased from Kapa Biosystems Inc (Wilmington, MA). DMSO-d6, penicillin-streptomycin (P/S) and 33 mm 0.45 μm PDVF syringe filters were purchased from EMD Millipore (Billerica, MA). ATCC MTT Cell Proliferation Assay kit was purchased from American Type Culture Collection (Manassas, VA). NotI HF restriction enzyme was purchased from New England Biolabs (Ipswich, MA). SMARTer Stranded Total RNA Kit (Pico Input) was purchased from Takara Bio USA (Mountain View, CA). Hypersil Gold 3 μm, 160 × 2.1 mm column was purchased from Thermo Fisher Scientific (Waltham, MA). K562 cells were a gift from the Slavoff Lab, Yale Department of Chemistry. T7 RNA polymerase was a gift from the Strobel Lab, Yale Department of Molecular Biophysics and Biochemistry.
Instrumentation
LC-MS measurements were carried out on an Agilent 6550A Q-TOF (Yale West Campus Analytical Core). NMR spectroscopy was performed on an Agilent DD2 400 MHz spectrometer with an Agilent OneNMR probe. Analysis of fluorescent RNAs was carried out on a GE Healthcare Typhoon FLA 9500. Sequencing was performed on Illumina HiSeq 2500 and Illumina HiSeq 4000 instruments at the Yale Center for Genome Analysis (YCGA).
LC-MS analysis of nucleosides
To a solution of s4U (50 μM) and ammonium bicarbonate (10 mM) was added TFEA (600 mM). mCPBA (10 mM) was dissolved in ethanol and added dropwise to the reaction mixture. After 1h at 25°C the reaction was analyzed by reverse-phase LC-MS with a Hypersil GOLD column (Thermo, 3 μm, 160 × 2.1 mm) using chromatography conditions described previously (Duffy et al. 2015). Masses were collected using positive ion mode and extracted ions were identified and integrated using Agilent MassHunter software.
NMR analysis of nucleobase chemistry
4-thiouracil (4.3 mg, 1 equiv) was dissolved in DMSO-d6, and TFEA (3.4 μl, 1.3 equiv) was added to the solution. After mixing, a solution of NaIO4 in DMSO-d6 (12.3 mg, 1.7 eq) was added to the nucleobase and amine solution, and the reaction was allowed to proceed at 45°C for 4h. 1H NMR spectra were processed using the MestReNova software.
NotI restriction endonuclease assay
An RNA containing a single s4U nucleotide was in vitro-transcribed (IVT) from a synthetic DNA template (see Supplementary table S1a) strand using T7 RNA polymerase and s4UTP in place of UTP for 16h at 37°C. The reaction mixture was treated with TURBO DNase for 1h at 37°C. The RNA was purified using denaturing PAGE, and the resulting band was extracted by crushing the gel slice and soaking it in extraction buffer (1 mM EDTA, 1 mM DTT, 20 mM Tris, 300 mM NaOAc pH 5.2) at 4°C for 4h. The supernatant was passed through a 0.45 μM syringe filter, and the RNA was ethanol precipitated and washed with 75% ethanol prior to resuspension in nuclease-free water.
IVT RNAs were screened for optimal TimeLapse chemistry as follows: RNA (120 ng) was added to a mixture of amine and water. A solution of oxidant was then added drop wise and the reaction mixture was incubated at the temperature and time indicated (see Supplementary Fig.S4). The RNA was then ethanol precipitated and washed three times with 75% ethanol prior to resuspension in nuclease-free water.
After chemical treatment, IVT RNA (50 ng) was reverse transcribed with SuperScript III according to the manufacturer's directions. The cDNA was PCR amplified for 30 cycles with a fluorescent forward primer, then amplified an additional 2 cycles using 1/5 of the previous PCR reaction material with non-labeled primers. The amplified PCR product was then incubated with NotI HF for 1h at 37°C. The fluorescent products were visualized using native PAGE followed by scanning with a Typhoon FLA imager and the proportion of cut product was determined relative to a positive control (with C in the RNA instead of s4U) using ImageJ.
Primer extension assay
IVT RNA containing a single s4U nucleotide (200 ng RNA) was treated with TimeLapse chemistry and purified as described above. Chemically treated IVT RNA (34 ng) was then annealed to a Cy5 5′ end-labeled primer, and reverse transcription was performed according to manufacturer's instructions using the SmartScribe First Stand cDNA Synthesis kit (15 min). The reaction was then treated with RNase H, and the fluorescent products were visualized using urea PAGE followed by scanning with a Typhoon FLA imager. Full length and truncated RT products were quantified by densitometry using ImageJ.
Targeted TimeLapse sequencing
MEF cells were grown at 37°C in DMEM containing 10% FBS and 1% P/S At approximately 60% confluence, the media was replaced with media supplemented with s4U (700 μM). After 2h, the cells were rinsed with PBS, resuspended in TRIzol reagent, and stored overnight at -80°C. Following chloroform extraction, total RNA was ethanol precipitated including 1 mM DTT to prevent oxidation of the s4U RNA, and washed with 75% ethanol. Total RNA was resuspended and treated with TURBO DNase, then extracted with acidic phenol:chloroform:isoamyl alcohol and ethanol precipitated and washed as described above. Isolated total RNA was added to a mixture of TFEA (600 mM), EDTA (1 mM) and sodium acetate (pH 5.2, 100 mM) in water. A solution of NaIO4 (10 mM) was then added drop wise and the reaction mixture was incubated for 1h at 45°C. Potassium chloride (300 mM) and sodium acetate (pH 5.2, 300 mM) were added and the reaction mixture was allowed to stand on ice for 10 min. prior to centrifugation (>10000 rpm, 30 min, 4°C) to precipitate remaining periodate. The RNA in the supernatant was then ethanol precipitated and washed three times with 75% ethanol prior to resuspension in nuclease-free water. The chemically treated RNAs were then reverse transcribed using a mixture of mouse Actb and Gapdh-specific mRNA RT primers (see Supplementary table S1b). The resulting cDNA was then amplified with Phusion polymerase using corresponding forward PCR primers to produce PCR amplicons approximately 150 nt in length. An Illumina sequencing library was constructed using the Illumina TruSeq Index adapters. Paired-end 75 bp sequencing was performed on an Illumina HiSeq 2500 instrument. Sequencing reads were trimmed to remove adapter sequences and aligned to the mouse genome using Bowtie226. Aligned reads were parsed to identify mutations at each nucleotide position in the Actb and Gapdh mRNAs using a published software package.27 Raw mutation probabilities were determined by dividing the number of recorded mutation events by the number of reads at that position. Mutation probabilities were normalized to appropriate control samples and filtered by read depth (only positions with depth > 3000 were included in analyses). Analyses and figure plot generation were performed in R using the tidyverse, corrplot, and multiplot packages28, 29. The enrichment in mutation rates was tested for significance using a two-sided Wilcoxon test. Targeted sequencing was performed in duplicate using biologically distinct samples.
Targeted TimeLapse-seq of K562 RNA was performed similarly with the following exceptions. Cells were grown at 37°C in RPMI containing 10% FBS and 1% P/S. At approximately 50% confluence, the media was supplemented with a range of s4U concentrations (10 μM-40 μM) for 1h. Total RNA was isolated and chemically treated as described previously. The chemically treated RNAs were then reverse transcribed using a mixture of human MYC-specific mRNA RT primers (see Supplementary table S1b). A targeted sequencing library was prepared and analyzed as described above.
Further information concerning experimental design using biological materials can be found in the Life Sciences Reporting Summary.
Cell viability
MEF cells were grown at 37°C in DMEM containing 10% FBS and 1% P/S. Cells were plated at 106 cells/mL in a 96-well microtiter plate and allowed to recover overnight. Cells were then treated in triplicate with increasing concentrations of s4U (0-1 mM) for 1h, and the ATCC MTT Cell Proliferation Assay kit was used according to manufacturer's instructions to assess cell viability.
Transcriptome-wide TimeLapse-seq
MEF cells were grown at 37°C in DMEM containing 10% FBS and 1% P/S. At approximately 60% confluence, the media was replaced and supplemented with s4U (1 mM). The cells were incubated at 37°C for 1h, at which point total RNA was isolated and chemically treated as described in the targeted sequencing section. For heat shock analyses, at approximately 60% confluence, the media was replaced and supplemented with s4U (1 mM), and heat shocked cells were incubated at 42°C for 1h. RNA was prepared as described for the Targeted TimeLapse-seq libraries. For each sample, 10 ng of total RNA was used to construct a sequencing library using the Clontech SMARTer Stranded Total RNA-Seq kit (Pico Input) with ribosomal cDNA depletion. Paired-end 100 bp sequencing was performed on an Illumina HiSeq 4000 instrument. TimeLapse-seq was performed in duplicate using biologically distinct samples for experimental samples both with and without heat shock. Raw and processed sequencing data have been submitted to the GEO database.
TT-TimeLapse-seq
K562 cells were grown at 37°C in RPMI containing 10% FBS and 1% P/S. At approximately 50% confluence, the media was supplemented with s4U (1 mM). The cells were incubated at 37°C for 5 min, at which point total RNA isolation and genomic DNA depletion were performed as described above. 50 μg of total RNA was subjected to MTS chemistry, followed by biotinylation and streptavidin enrichment essentially as previously described (Duffy et al., 2015)9 with the following modification: after SAV beads were washed three times with high salt wash buffer (1 M NaCl, 100 mM Tris pH 7.4, 10 mM EDTA, 0.05% Tween), beads were incubated in TE buffer (10 mM Tris pH 7.4, 1 mM EDTA) at 55°C for 15 min, followed by two washes with pre-warmed 55°C TE buffer. After elution from SAV beads, enriched RNA was purified using one equivalent volume of Agencourt RNAclean XP beads according to manufacturer's instructions instead of purification by ethanol precipitation. Enriched RNA and input RNA were chemically treated as described previously. Chemically treated RNA was purified using 1 equivalent volume of Agencourt RNAclean XP beads according to manufacturer's instructions. Purified material was then incubated in a reducing buffer (10 mM DTT, 100 mM NaCl, 10 mM Tris pH 7.4, 1 mM EDTA) at 37°C for 30 min, followed by a second RNAclean bead purification. For each sample, all enriched material or 10 ng of total RNA input was used to construct a sequencing library using the Clontech SMARTer Stranded Total RNA-Seq kit (Pico Input) with ribosomal cDNA depletion. Paired-end 150bp sequencing was performed on an Illumina HiSeq 4000 instrument. TimeLapse-seq was performed in duplicate using biologically distinct samples for experimental samples. Raw and processed sequencing data have been submitted to the GEO database.
Samples for TimeLapse-seq analysis of K562 mRNA
K562 cells were grown as described previously. At approximately 50% confluence, the media was supplemented with s4U (100 μM). The cells were incubated at 37°C for 4h, at which point total RNA was isolated using the RNeasy mini kit with the following modifications: buffers RLT and RPE were supplemented with 1% final 2-mercaptoethanol (BME); an additional 80% EtOH wash was performed after the RPE step; and the column was spun at maximum speed for 5 min to dry prior to elution with water. The isolated RNA was then chemically treated and purified as described previously. For each sample, 10 ng of total RNA was used to construct a sequencing library using the Clontech SMARTer Stranded Total RNA-Seq kit (Pico Input) with ribosomal cDNA depletion. Paired-end 150bp sequencing was performed on an Illumina HiSeq 4000 instrument. TimeLapse-seq was performed in duplicate using biologically distinct samples for experimental samples. Raw and processed sequencing data have been submitted to the GEO database.
Transcriptional inhibition
K562 cells were grown as described above. At approximately 50% confluence, cells were treated in duplicate with actinomycin D (2 μg/mL final) for 30 min, 1h, 3h, 5h, and 9h, or left untreated. Total RNA isolation and genomic DNA depletion was then performed as described previously. RT was performed using the SuperScript VILO cDNA synthesis kit and qPCR was performed using primers specific to ACTB, DHX9, and ASXL1. qPCR ct values for DHX9 and ASXL1 were then averaged and normalized to those of ACTB for each time point. The normalized fraction remaining was estimated for each primer pair by dividing the relative abundance of each time point by the relative abundance at t = 0.
Sequencing alignment and mutational analysis
Reads were filtered for unique sequences using FastUniq30, trimmed using cutadapt31 to remove Illumina adapter sequences filtering for reads greater than 20 nt (--minimum-length=20) and aligned to the mouse GRCm38 or human GRCh38 genome and transcriptome annotations using HISAT232, using default parameters and --mp 4,2. Files were further processed with Picard tools (http://broadinstitute.github.io/picard/) including FixMateInformation, SortSam and BuildBamIndex. The samtools33 software was used to retain only reads that aligned uniquely (flag: 83/163, 99/147), with MAPQ ≥ 2, and without insertions (because of ambiguity in mutational analysis) for further analysis.
Reads that uniquely map to the human GRCh38 version 26 (Ensembl 88) or mouse GRCm38 (p6) were identified using HTSeq-count using union mode34. Reads mapping to only mature isoforms or to anywhere in the gene body were determined separately and compared to identify intron-only reads. To determine the number of uridine residues inferred from each read, and the sites of T-to-C mutations, the aligned bam files were processed in R using Rsamtools (http://bioconductor.org/packages/release/bioc/html/Rsamtools.html) and the sites and numbers of mutations were determined using a custom R function (available upon request). Only mutations at positions with a base quality score of greater than 45, that were at least three nt from the end of the read were counted. Reads were excluded where there were greater than five T-to-C mutations and these mutations did not account for at least one third of the observed mutations (NM tag). Without adequate filtering, SNPs could interfere with TimeLapse analysis. To identify sites of SNPs (or RNA modifications that could be mis-identified as TimeLapse mutations), we used the following two strategies. First, we identified T-to-C SNP sites in control samples using bcftools35 with default options and excluded these sites from our analysis. Second, we compiled locations where T-to-C mutations were high in non-s4U treated controls and excluded these sites from analysis. Once the putative SNPs were filtered, the total number of unique mutations in each read pair was counted. To examine the distribution of reads with each minimum number of T-to-C mutations, the bam files were filtered using Picard tools. To make genome-coverage tracks, STAR aligner (inputAlignmentsFromBam mode, outWigType bedGraph) was used and the tracks were normalized using factors derived from RNA-seq analyses using values from DESeq2 (estimateSizeFactors)35. Tracks were converted to binary format (toTDF, IGVtools) and visualized in IGV36.
Secondary structure analysis
Aligned reads from the 4h K562 TimeLapse-seq experiment overlapping the 5′ stem loop of 7SK were extracted using samtools. A Python script developed for analyses of chemical probing data (RTEventsCounter28), was used to calculate the U-to-C mutation frequency for each uridine nucleotide. These frequencies were normalized by subtracting mutation frequencies of control samples that were not subjected to TimeLapse chemistry. The frequencies of mutations at each position were binned and mapped onto a conformational model of this region of human 7SK37. Each nucleotide was classified as either single stranded or basepaired. A two-sided Wilcoxon test was used to determine the significance of differences between mutation rates of the basepaired and single stranded nucleotides.
Estimation of the fraction of new transcripts and transcript half-lives
Two different models were used to examine the mutation distribution in TimeLapse-seq data set: a simpler Poisson model (which does not take into account the uridine content of different reads) and a binomial model that does take the number of uridines into account. We obtained consistent results from both models. For the simpler Poisson model, for each sample (sj), the distribution of T-to-C mutations (Yi) was determined in each read, and the reads were grouped based on the transcripts to which they map. A negative control sample (no s4U treatment) was used to estimate the background rate of read-pairs containing T-to-C mutations that map to each transcript. These frequencies depended on the cell line used (MEF samples required higher s4U treatment to obtain similar levels of mutations compared to K562 cells) as well as the sequencing experiment (different samples led to different background rates independent of chemistry or s4U treatment). See Supplementary Note. The mutation rate and fraction of new transcripts was modeled as a two-component mixture of Poisson distributions with probability mass function:
where θn− is the fraction of new transcripts, λO is the rate of background mutations (determined from –s4U controls), λn is the rate of mutations found in new transcripts, and yi is the number of passing T to C mutations found in read i. Reasonable estimates of these values could be approximated by examining the mutation rates in fast turnover RNAs such as introns. To obtain more objective estimates of the global parameters λO and λn while allowing for low levels of transcript-to-transcript variability, we used a Bayesian hierarchical modeling approach using RStan software (Version 2.16.238) that uses no-U-turn Markov Chain Monte Carlo (MCMC) sampling. To estimate a global mean and standard deviation for λO and λn, we used weakly informative priors (see below). We estimated gene specific rates by drawing from the global mean and standard deviation, with a mixing rate with an uninformative prior (θn ∼ Uniform(0,1)) where the mixing rate (θn) estimates the fraction of each transcript that was new:
Global Parameters:
Priors:
for read i ∈ {1, 2, …, ng}:
Attempts to model entire TimeLapse-seq data sets using this approach were computationally challenging, but we found that consistent results were obtained using 20 representative transcripts from each sample. The majority of these transcripts were chosen randomly from all reasonably expressed transcript (> 200 reads), but we included few transcripts that were hand chosen to ensure the modeling included both fast and slow turnover RNAs such as Myc and Actb. The results using 20 transcripts were consistent with results from 200 transcripts. In the case of the MEF samples shown if Fig. 2, the λO was estimated as 0.07 mutations/read (50% credible interval 0.062-0.074), and λn was estimated as 2.3 mutations/read (2.298 mutation/read, 50% CI 2.10-2.30 for heat shock; 2.288 mutation/read, 50% CI 1.90-2.29 for untreated).
Once these global parameters were determined, they were used to estimate the fraction of new transcripts (θnew), using expectation maximization by minimizing the log likelihood using the nlm function in the MASS package in R:
The 95% Wald confidence interval was calculated using the Hessian (nlm option hessian = TRUE), to calculate:
To ensure the mutations were both s4U-treatment and TimeLapse-chemistry dependent, we only included transcripts where there was sufficient data (reads > 100 counts in at least two samples), and where the fit converged (-0.05 < θn < 1.05; hessian > 1000). The inferred new read counts were determined by multiplying the estimated fraction of new transcripts by the total RNA-seq transcript count. Correlations between replicates were determined using the log10 transformed counts (Supplementary Fig.S8). While the reproducibility of the data was generally high when all converged transcripts were included (Pearson's r > 0.91), filtering for transcripts with at least 75 inferred new reads provided slightly more reproducible results (n = 3603, r = 0.934) and this filter was used for further analysis.
To account for differences in the number of uridine residues in each read pair, an alternative model was used based on the binomial distribution. Specifically, the data was modeled as mixture of two binomial distributions:
where po, pn are the probabilities of mutation at each uridine nucleotide for old and new transcripts, and nu is the number of uridines observed for read i. To determine the global mutation rate, we used Bayesian hierarchical modeling as described above for the Poisson model but using a mixture of binomial distributions. From this analysis, we estimate the background mutation rate (po) to be 0.0012 mutations/uridine (50% CI 0.00121, 0.00123) and the mutation rate for new reads (pn) to be 0.0332 mutations/uridine (50% CI 0.0329, 0.0335). In other words, ∼0.1% of Us are mutated to C in pre-existing reads, and in new reads ∼3% of Us are mutated to C. Using these global parameters, the distributions of individual genes were fit with nlm similarly to what is described above, except by minimizing the log likelihood of the binomial model instead:
In addition to computing the confidence interval using the hessian, we also examined the quality of the fit by plotting the observed frequency of mutations in each replicate in the TimeLapse data (gray points in distribution plots), to a simulated distribution of the expected new and old reads based on the binomial model (Figs.2b and S16). Estimates of the fraction new were highly similar between those determined using the binomial model and the Poisson model.
To account for any specific loss of transcripts that might arise from biased loss of s4U-RNA transcripts independent of TimeLapse chemistry, or TimeLapse-depended loss due to reverse transcription termination, we developed a means of estimating the loss of fast turnover transcripts in the data. This correction was only used when estimating transcript half-lives after observing a modest, but statistically significant loss of reads from high turnover RNAs (see Supplementary Fig.S3d). To estimate the fraction of new reads missing, we used the R package nlm to fit the equation:
where sy and so are scale factors that adjust for library sizes determined using DESeq2 with the total (RNA-seq) transcript counts for the experimental sample and control, respectively; Ny and No are the counts for each transcript, and θn is the unadjusted fraction new of each transcript. This equation was fit using transcripts where 0.8 < θn for K562 RNA, but 0.5 < θn in the case of MEF RNA (the shorter s4U treatment lead to fewer transcripts with high θn so the threshold was lowered to increase the number of transcripts). In the case of the comparison shown in Fig.S3d, the adjustment factor determined for chemistry-induced dropout was ∼5% (i.e., x = 0.05 in the equation above, which leads a transcript with 75% new reads to be adjusted to 79% and a transcript with 25% new reads would be adjusted to 26% new reads).
The transcript half-lives were determined using the adjusted fraction of new RNA assuming a simple exponential model of their kinetics. The half-life values were compared to similar reports and the r2 determined using the lm function in R.
GO analysis
GO analysis from the PANTHER database (version 12.0)39 was performed using a statistical overrepresentation test (default parameters) on the complete biological process annotation set using the top 10% slow or top 10% fast turnovers RNAs in our 1h MEF TimeLapse-seq data as determined by the half-life analyses described above.
Differential expression analysis
Differential expression analysis was performed using DESeq2. To examine the inferred differences in the new transcript pool based on TimeLapse mutations, we used the unadjusted estimates of the fraction of new RNA to infer the number of counts resulting from new transcripts as described above. As TimeLaspse-seq data is internally controlled, we used the size factors determined from total counts to scale each dataset (i.e., we ran DESeq2 on the total RNA-seq data, and used the sizeFactors function to scale the inferred new RNA counts to the RNA-seq determined values) with default conditions including the Benjamini-Hochberg40 adjusted p-value (padj in text). RNA-Seq analysis was performed on all reads (i.e., reads that had zero or more T-to-C mutations) using DESeq2 with default parameters.
Estimation of contaminating reads in TT-TimeLapse-seq
Reads from TT-TimeLapse-seq were processed and analyzed as for TimeLapse-seq. Junction-containing reads were determined from the presence of “N” characters in the CIGAR string in the aligned bam file using bamtools (version 2.3, https://hcc-docs.unl.edu/display/HCCDOC/BamTools). The levels of contaminating reads were estimated by assuming the contaminating reads have the same ratios as RNA-seq data, and that reads with three or more mutations constitute the true ratio of reads. We use of reads with three or more mutations as true positives because the probability of a read containing three or more mutations without s4U is <10-5. We used the fraction of intron or junction containing reads for the RNA-seq data (ro), the total in the true positive population (rtp), and the total for each population (rx). In each analysis, we only considered reads that had non-zero ratios and ratios that were less than one. The fraction of reads from contamination (cx) was then estimated:
For comparisons with the TT-TimeLapse-seq data presented here, the data from Schwalb et al.7, (SRR4000390, SRR4000391 and SRR4000397) were aligned and processed using the same pipeline described for TimeLapse-seq. For this comparison, we reprocessed our TT-TimeLapse-seq data using only 75 nt of each read, and this was performed on fastq files prior to alignment. This trimming was performed because the probability of a sequencing read containing a splice junction or being an intron-only read is dependent on the read length. Otherwise, all processing was handled equally between data sets.
Supplementary Note
Important parameters in TimeLapse-seq
TimeLapse-seq builds upon previous work using s4U to metabolically label RNA, and many of the considerations when designing experiments are shared with previous work and have been discussed in depth elsewhere22, including the time of s4U treatment required to accurately estimate transcript half-lives. Considerations that are specific to TimeLapse-seq are discussed below.
Each read-pair in TimeLapse-seq data reports mutations that are present in a single molecule of RNA that was either made prior to the s4U treatment, or was made after s4U was added to the cells. For new RNA, there is an s4U- and chemistry-dependent increase in the probability of a T-to-C mutation at each nucleotide. For any given region of an RNA molecule that is copied into a sequencing read of a given length (lr), our ability to accurately identify whether the read pair is from a new RNA or not is dependent on the following: nu, the number of uridine residues that could be substituted with s4U; pnew, the probability a s4U residue substitutes for U at each position; ychem, the efficiency of the conversion from s4U to C*; and pold, the background mutation rate in untreated samples. At the population level, the accuracy of the estimates for the newly made faction of any feature (e.g., transcript, exon, etc.) depends on the read depth (nreads).
The background mutation rates (po) are constrained by the methods and technology used for RNA-seq and estimated using negative controls. The number of uridines (nu) in the read is dependent on the U-content of the RNA feature and on the read length (lr) in the sequencing experiment (e.g., single-end 75 nt reads vs paired end 150 nt reads). The probability of s4U incorporation (pn) depends on ratio of s4UTP/UTP in the nucleotide pool, which is dependent on the s4U concentration used in the feed, the cell line used and the time of the experiment. The rate of incorporation of s4U into the UTP pool is quite fast. This is clear from the observation that many reads in the TT-TimeLapse-seq experiment have multiple mutations, suggesting that even within 5 min at 1 mM s4U treatment, the nucleotide pool builds up substantial concentration of s4UTP. There is also cell-type variability in the influence of s4U treatment (e.g., we found TimeLapse-seq in MEF cells worked best with 1 mM s4U, whereas labeling of K562 cells was successful with 100 μM s4U used in the 4h treatment). In practice 10 μM – 1 mM treatments have been successful. The chemical efficiency (ychem) determines the number of s4U residues that are converted to C, which we have estimated to be 80% (Supplementary Fig.S4).
To explore how deeply any RNA feature must be sequenced in order to detect changes in the new transcript pool by TimeLapse-seq, we simulated data according to the following model:
where the ith read (out of nreads total) with nu uridine residues is determined to arise from either a new or old RNA according to a Bernoulli distribution with the fraction of new RNA (θn). If the transcript is new, it is modeled to have a number of mutations (Yi) defined by a binomial distribution with nu trials and probability of mutation the probability of s4U incorporation (pn) attenuated by the yield of the chemistry (ychem). If the RNA is old, the number of mutations is modeled by a binomial distribution nu trials and a background probability of mutation (po). The data from these simulated trials were treated as the output from a TimeLapse-seq experiment in which the fraction new was modeled as described in the methods (using likelihood maximization to estimate θn), and the number of new reads inferred using this estimate. Different fold changes in the new transcript pool (x) were modeled in duplicate, with duplicate controls to match the design we used in this manuscript. To provide a conservative estimate of the sensitivity of the approach, these counts were added to a real RNA-seq data set (from the differential expression of heat shock expression) and the significance determined using DESeq2 with default parameters. We favored this approach because the dispersion estimates used to determine the significance in the simulation are influenced by the distribution of real TimeLapse-seq data. This simulation was repeated 250 times for each set of parameters, and the average number of times the simulation provided a significant difference was plotted (Supplementary Fig.S12). For each simulation, conditions were held constant that were similar to (or more conservative than) the actual parameters for the MEF experiment presented in Fig.2.
In general, many conditions lead to reliable detection of differential expression when there are hundreds-to-thousands of reads. Under the conditions of these simulations, neither the chemical efficiency nor the read length have dramatic impact unless they are greatly reduced. One practical consequence of this observation is that improving the efficiency of the reaction from 80% would have very little impact, and even a drop to 50% yields would only have a small impact on the sensitivity of the experiment. On the other hand, to be able to sensitively detect changes, the fraction of new RNA must be large enough to detect (> 5%), but less than half the RNA. Large fold differences (>2) are straight forward to detect even at very low coverage, but much higher coverage is necessary to confidently detect transcripts that have a 1.5-fold induction in the new RNA pool. Both depletion and enrichment were detected, and the specificity was very high (the false positive rate by this metric was too low to detect). The background mutational rates (∼0.1% in the samples presented in this manuscript) are predicted to have minimal impact unless they are increased five-to-ten-fold. While decreasing the amount of s4U can decrease sensitivity, increasing it is predicted to only lead to a modest increase in sensitivity. In summary, experimental design regarding the timing of the s4U treatment is critical.
Code availability
All software and parameters used is described above, and custom scripts and functions are available upon request.
Accession codes
Data are available in the Gene Expression Omnibus (GEO) under accession number GSE95854.
Supplementary Material
Acknowledgments
We thank J. Steitz, A. Schepartz, D. Söll, D. Canzio and the Simon Lab for insightful comments, Y. Wang and A. Sexton for assistance and scripts used in mutational analysis of targeted sequencing data. This work was supported by the NIH NIGMS T32GM007223 (J.A.S. and E.E.D); NSF Graduate Research Fellowship (E.E.D); NIH New Innovator Award DP2 HD083992-01 (M.D.S.), and a Searle scholarship (M.D.S.).
Footnotes
Author contributions: J.A.S. and M.D.S. designed experiments. J.A.S., E.E.D., and L.K. carried out experiments. J.A.S., M.C.S., and M.D.S. performed computational analyses of data. J.A.S. and M.D.S. wrote the manuscript with assistance from all authors.
Competing financial interests: We declare no competing financial interests.
Note added in revision: During the revision, a manuscript was published reporting the use of alkylation chemistry to produce mutations from s4U-metabolically labeled RNAs (Herzog et al. 2017).
References
- 1.Bhatt DM, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menet JS, Rodriguez J, Abruzzi KC, Rosbash M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. Elife. 2012;1:e00011. doi: 10.7554/eLife.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wada Y, et al. A wave of nascent transcription on activated human genes. PNAS. 2009;106:18357–18361. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaidatzis D, Burger L, Florescu M, Stadler MB. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotechnol. 2015;33:722–729. doi: 10.1038/nbt.3269. [DOI] [PubMed] [Google Scholar]
- 5.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwalb B, et al. TT-seq maps the human transient transcriptome. Science. 2016;352:1225–1228. doi: 10.1126/science.aad9841. [DOI] [PubMed] [Google Scholar]
- 8.Rabani M, et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy EE, et al. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol Cell. 2015;59:858–866. doi: 10.1016/j.molcel.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melvin WT, Milne HB, Slater AA, Allen HJ, Keir HM. Incorporation of 6-thioguanosine and 4-thiouridine into RNA. Application to isolation of newly synthesised RNA by affinity chromatography. Eur J Biochem. 1978;92:373–379. doi: 10.1111/j.1432-1033.1978.tb12756.x. [DOI] [PubMed] [Google Scholar]
- 11.Rabani M, et al. High-resolution sequencing and modeling identifies distinct dynamic RNA regulatory strategies. Cell. 2014;159:1698–1710. doi: 10.1016/j.cell.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishima Y, Steitz JA. Site-specific cross linking of 4-thiouridine-modified Human tRNA(3Lys) to reverse transcriptase from human immunodeficiency virus type I. Embo J. 1995;14:2679–2687. doi: 10.1002/j.1460-2075.1995.tb07266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano M, Hayatsu H. Permanganate oxidation of 4-thiouracil derivatives. Isolation and properties of I-substituted 2-pyrimidone 4-sulfonates. Biochim Biophys Acta. 1970;199:303–315. [PubMed] [Google Scholar]
- 15.Ziff EB, Fresco JR. A method for locating 4-thiouridylate in the primary structure of transfer ribonucleic acids. Biochemistry. 1969;8:3242–3248. doi: 10.1021/bi00836a016. [DOI] [PubMed] [Google Scholar]
- 16.Dai Q, et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat Methods. 2017;14:695–698. doi: 10.1038/nmeth.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 18.Mukherjee N, et al. Integrative classification of human coding and noncoding genes through RNA metabolism profiles. Nat Struct Mol Biol. 2017;24:86–96. doi: 10.1038/nsmb.3325. [DOI] [PubMed] [Google Scholar]
- 19.Trinklein ND, Murray JI, Hartman SJ, Botstein D, Myers RM. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol Biol Cell. 2004;15:1254–1261. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-wide Transcriptional Regulation. Mol Cell. 2016;62:63–78. doi: 10.1016/j.molcel.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalgi R, Hurt JA, Lindquist S, Burge CB. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 2014;7:1362–1370. doi: 10.1016/j.celrep.2014.04.044. [DOI] [PubMed] [Google Scholar]
- 22.Russo J, Heck AM, Wilusz J, Wilusz CJ. Metabolic labeling and recovery of nascent RNA to accurately quantify mRNA stability. Methods. 2017;120:39–48. doi: 10.1016/j.ymeth.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedel CC, Dolken L, Ruzsics Z, Koszinowski UH, Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res. 2009;37:e115. doi: 10.1093/nar/gkp542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelsi-Boyer V, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145:788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- 25.Scotti MM, Swanson MS. RNA mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sexton AN, Wang PY, Rutenberg-Schoenberg M, Simon MD. Interpreting Reverse Transcriptase Termination and Mutation Events for Greater Insight into the Chemical Probing of RNA. Biochemistry. 2017;56:4713–4721. doi: 10.1021/acs.biochem.7b00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP) Nat Methods. 2014;11:959–965. doi: 10.1038/nmeth.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei T. Package ‘corrplot’. Statistician. 2015;56:316–324. [Google Scholar]
- 30.Xu H, et al. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS One. 2012;7:e52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M. Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet journal. 2011;17:10–12. [Google Scholar]
- 32.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Herreweghe E, et al. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. Embo J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpenter B, et al. STAN: A Probabilistic Programming Language. J Statistical Software. 2017;76 doi: 10.18637/jss.v076.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.